Abstract
Biallelic pathogenic variants in the PNPLA6 gene cause a broad spectrum of disorders leading to gait disturbance, visual impairment, anterior hypopituitarism and hair anomalies. PNPLA6 encodes neuropathy target esterase (NTE), yet the role of NTE dysfunction on affected tissues in the large spectrum of associated disease remains unclear.
We present a systematic evidence-based review of a novel cohort of 23 new patients along with 95 reported individuals with PNPLA6 variants that implicate missense variants as a driver of disease pathogenesis. Measuring esterase activity of 46 disease-associated and 20 common variants observed across PNPLA6-associated clinical diagnoses unambiguously reclassified 36 variants as pathogenic and 10 variants as likely pathogenic, establishing a robust functional assay for classifying PNPLA6 variants of unknown significance. Estimating the overall NTE activity of affected individuals revealed a striking inverse relationship between NTE activity and the presence of retinopathy and endocrinopathy. This phenomenon was recaptured in vivo in an allelic mouse series, where a similar NTE threshold for retinopathy exists.
Thus, PNPLA6 disorders, previously considered allelic, are a continuous spectrum of pleiotropic phenotypes defined by an NTE genotype:activity:phenotype relationship. This relationship, and the generation of a preclinical animal model, pave the way for therapeutic trials, using NTE as a biomarker.
Keywords: spastic paraplegia type 39, Gordon-Holmes syndrome, Boucher-Neuhäuser syndrome, Laurence-Moon syndrome, Oliver-McFarlane syndrome, neurodegeneration
Biallelic pathogenic variants in PNPLA6, which encodes neuropathy target esterase (NTE), cause a broad spectrum of neurological disorders. Liu et al. show that NTE activity predicts neurological phenotypes among affected individuals, suggesting that PNPLA6 disorders are a continuous spectrum of pleiotropic phenotypes defined by the activity of NTE.
Introduction
Biallelic pathogenic variants in the Patatin-like phospholipase domain containing 6 (PNPLA6, MIM #603197) gene cause a broad spectrum of neurological disorders, including spastic paraplegia type 39 (SPG39, MIM #612020), Gordon-Holmes syndrome (GDHS, MIM #212840), Boucher-Neuhäuser syndrome (BNHS, MIM #215470), Laurence-Moon syndrome (LNMS, MIM #245800) and Oliver-McFarlane syndrome (OMCS, MIM #275400).1-5 These disorders exhibit significant pleiotropy involving the CNS and peripheral nervous system (PNS), with endocrine, ophthalmic and hair anomalies. Patients diagnosed with SPG39 typically present with cerebellar ataxia, upper motor neuron deficits and peripheral neuropathy. Patients with GDHS and BNHS present with hypogonadotropic hypogonadism, with BNHS additionally accompanied by chorioretinal dystrophy. Childhood-onset disorders, OMCS and LNMS, also include presenting features such as chorioretinal dystrophy and anterior pituitary hormone deficiency, with OMCS patients having trichomegaly and alopecia.
PNPLA6 encodes neuropathy target esterase (NTE), an endoplasmic reticulum-associated enzyme that is highly expressed in the developing human brain and eye.1 The phospholipase B activity of NTE is critical for phospholipid homeostasis and membrane trafficking.6-8 NTE is 1375 amino acids long with five protein domains: an N-terminal transmembrane domain, three cyclic nucleotide binding (CNB) domains, and a NEST domain (NTE-esterase domain) at the C-terminal end. Although the function of the CNB domains remains unknown, the NEST domain contains the catalytic residues required for phospholipid remodelling.7,9
Previous research has shown circumstantial evidence that NTE levels may be a key driver in disease pathogenesis. Work done by our team and others utilizing patient fibroblasts with missense and predicted loss-of-function (pLOF; nonsense, frameshift and truncating) variants in PNPLA6 lead to a reduction in NTE activity.1,10 Furthermore, skin fibroblasts from OMCS patients with biallelic PNPLA6 missense variants had significantly less NTE activity compared to SPG39-associated genotypes, providing evidence that different clinical subtypes might be influenced by NTE activity.1
In affected individuals, biallelic pLOF variants are exceptionally rare. While it has been proposed that single pLOF variants are associated with retinopathy,11 truncating variants are seen in all clinical diagnoses seen in PNPLA6 patients. Therefore, the putative relationship between the clinical subtype, genotype and NTE levels in PNPLA6 disorders remains unclear. Such relationships, especially if recapitulated in a preclinical animal model, would open the door to therapeutic options with NTE levels as a potential biomarker.
In this study, we examine the fundamental role of NTE activity on disease onset among PNPLA6-associated disorders. To investigate the genotype-phenotype correlations connected with PNPLA6 variants, a novel cohort of 23 cases with varying clinical diagnoses contains 17 novel variants in the PNPLA6 gene. A systematic evidence-based review of this cohort and 95 reported individuals reveals missense variants in the enzymatic domain as a driver of disease pathogenesis due to their recurrence in specific clinical subtypes. Using a modified NTE enzymatic assay to incorporate full-length protein analytics,12,13 we characterize the activities of 66 missense, truncating and common PNPLA6 variants in vitro that were able to resolve uncertainty in all variants tested. Estimating an affected individual’s overall NTE activity by taking the average in vitro activity of their two variants demonstrates a relationship between residual NTE activity and presence of clinical subtypes. This phenomenon was recapitulated in a preclinical mouse model of the PNPLA6 disorder spectrum, where retinopathy and mouse viability are predicated on NTE activity. Overall, these experiments uncover a novel genotype:NTE activity:phenotype relationship in PNPLA6-associated disorders that lay the foundation for preclinical trials using NTE activity as a novel biomarker.
Materials and methods
Systematic evidence-based review of previously published patient data
Published clinical data of patients with PNPLA6 variants were found by the following search terms in PubMed: ‘PNPLA6’, ‘Neuropathy target esterase’, ‘Oliver McFarlane Syndrome’, ‘Boucher Neuhäuser Syndrome’, ‘Gordon Holmes Syndrome’, ‘Spastic Paraplegia type 39’, ‘Ataxia’. Available patient data were recorded, including genotype, phenotype (with tissue age of onsets if available), age, sex and diagnosis. The last search was done in May 2023.
All systematic evidence-based reviews were done by patient pedigree (except for tissue age of onset analysis). Analysis of the age of onsets (AOO) in affected tissues was done with patients with available numerical AOO information. Clinical definitions for each tissue system containing key words are found in Supplementary Table 3. PNPLA6 variant location were defined by RefSeq transcript NM_001166111.2. PNPLA6 amino acid and domain locations were defined by RefSeq protein NP_001159583.1.
In vitro NTE-specific activity assay
NTE enzymatic activity assay was performed using previously described colorimetric assays.12,13 Briefly, cell lysate was incubated in 40 μM paraoxon and 50 μM mipafox for 20 min. Phenyl valerate (0.5 mM, 0.03% w/v Tx-100) was added and incubated for 20 min. Thereafter, 1.23 mM 4-aminoantipyrine (33.25 mg/ml SDS) was added to stop the reaction. Potassium ferricyanide (12.1 mM) was used for colorimetric determination. NTE activity is defined as the difference in phenyl valerate hydrolysis activity inhibited by paraoxon (inhibits background esterase activity) and paraoxon + mipafox (inhibits NTE in addition to background esterase activity). End point absorbance was measured at 486 nm using a Synergy 2 microplate reader (Biotek). Specific NTE protein concentration was determined by SDS PAGE and Coomassie using previously defined methods.14 Li-Cor Odyssey DLx and Image Studio Lite (Li-Cor) were used to image and quantify the intensity of the bands. Band intensity was converted to a protein concentration using a BSA standard curve ranging from 0.25–1 mg/ml on the same gel. The final protein concentration was determined by taking the average concentration of at least three technical replicates. End point absorbance was normalized to specific NTE concentration. This assay defines one unit of NTE-specific activity as 1 μmol phenol produced per minute per milligram protein.
Molecular modelling and simulation
The human PNPLA6 amino acid sequence (acc# Q8IY17) was obtained from the UniProt database (https://www.uniprot.org). Residues 968 to 1286 were extracted from the PNPLA6 sequence and were used to model the human patatin domain. The following files were downloaded from the RCSB website (https://www.rcsb.org/): (i) Pseudomonas aeruginosa patatin-like protein structure, PLPD (5fya); (ii) human cytosolic phospholipase A2 dimer (1cjy); and (iii) Solanum cardiophillum SeMet patatin (1oxw). Both structures, 1cjy and 1oxw, function as lipid acyl hydrolases with Ser-Asp catalytic dyad in an active site. The homology model of the human patatin dimer was generated using these proteins as structural templates in the molecular graphics, modelling and simulation program YASARA (http://www.yasara.org/). The homology model of the human patatin dimer was equilibrated by 10 ns molecular dynamics in water using YASARA’s ‘run. Mcr’ macro. Ion concentration was added as a mass fraction with 0.9% NaCl. The simulation temperature was set to 298 K with a water density of 0.997 g/ml. The cell size extended to 10 Å beyond each side of the protein in the shape of a cube with dimensions 119.7 Å × 119.7 Å × 119.7 Å. Each simulation was run in YASARA using an AMBER14 forcefield, with a timestep of 1.0 fs. Simulation snapshots were outputted for every 0.1 ns.
Global computational mutagenesis
Global mutagenesis was conducted on a model of the human patatin dimer and each mutant was characterized by a thermodynamic change in Gibbs free energy (DDG) and by the fraction of protein in the unfolded state for each subunit (A and B) of the dimer.15,16 Gibbs’s free energy changes were calculated using the semi-empirical method FoldX.17 The fraction of protein in the unfolded state was standardized on a 0–1 scale as the unfolding fraction u.15 This parameter is a sum of severity-weighted unfolding propensities for the 19 variants generated at a specific residue. To correlate the result with the enzymatic activity change caused by the mutation the unfolding parameters of subunits A and B were averaged, and standard deviations were calculated.
Generation of allelic series mice
Pnpla6 missense variants were generated using CRISPR/Cas9-mediated homologous recombination, as previously described.18 The truncating variant p.Met1030Valfs*2 (c.3088_3089delAT) used in this study was a by-product of indel mutagenesis from the production of c.3088A>G missense mutation. C57BL/6 zygotes were microinjected with a mixture of SpCas9 protein, a gRNA specific for each target and a mutagenic oligo that carries the intended knock-in mutation and short homology arms. Injected zygotes were transferred into the oviducts of female mice and newborn F0 mice were screened for the intended knock-in mutations by PCR and Sanger sequencing spanning the knock-in or deletion area. F0 founders carrying the desired mutations are genetic mosaics and were backcrossed to C57BL/6J mice for germline transmission of each mutant allele. Mice backcrossed greater than F4 generation were used for retinal function and structure analysis. Animals used for function and structure assays had evenly distributed males and females. sgRNA and mutagenic oligos used for this study are described in Supplementary Table 4.
Mouse brain preparation and activity assay
Allelic series mice at 3 and 12 months were sacrificed and the brain was resected for activity assay analysis. The protocol was adapted from Quistad et al.19 Brain (stored at −80°C) was homogenized at 20% w/v in 50 mM Tris HCl containing 0.2 mM EDTA (pH 8.0). Activity assay used the supernatant fraction after centrifugation at 700g for 10 min. Brain lysate was frozen down and then subsequently used for activity analysis. Activity assay of mouse brain homogenate (20 μl) was incubated in 40 μM paraoxon and 500 μM mipafox for 20 min. Phenyl valerate (1.5 mM, 0.03% w/v Tx-100) was added and incubated for 20 min. End point absorbance was measured at 486 nm using a Synergy 2 microplate reader (Biotek). Bradford assay (Cat# 5000201, Bio-Rad) was used to determine the total protein concentration of each sample. End point absorbance was normalized to total protein concentration. This assay defines one unit of NTE-specific activity as 1 nmol phenol produced per minute per milligram protein.
Electroretinogram recording
Electroretinogram (ERG) recordings used the Espione E3 system (Diagnosys LLC.). Mice were dark-adapted overnight (>12 h) and anaesthetized with an intraperitoneal injection of ketamine (100 mg/kg) and xylazine (6 mg/kg). All procedures were done under dim red-light conditions. Pupils were dilated with 0.5% tropicamide and 2.5% phenylephrine, and anaesthetized with 0.5% proparacaine topical anaesthesia. Animals were placed on a heating plate to keep their body temperature at 37°C. Responses were recorded using a gold loop wire electrode placed at the centre of the cornea, a reference electrode in the mouth and a ground electrode at the tail. A 2.5% hypromellose ophthalmic demulcent solution was used for corneal hydration. Dark-adapted ERG was performed using flashes with intensities ranging from 0.0001 to 10 sc cd.s/m2. Light adaptation was performed with white light at 20 sc cd/m2 for 2 min. Photopic ERG response was recorded using flashes with intensities ranging from 0.3 to 100 sc cd.s/m2.
Optomotor response
Optomotor response (OMR) was measured using the Optodrum (Striatech). Optodrum recommended parameters were used for the experiment. Square wave gratings were used at a fixed 99.72% contrast and rotation speed at 12°/s. The number of cycles/degree varied by a staircase algorithm and the rotation of the drum was both clockwise and counterclockwise. The final determination of mouse visual acuity was determined by two successful and three unsuccessful trials.
Optical coherence tomography imaging
Cross-sections of mouse retinas were imaged and acquired using the Heidelberg Spectralis HRA plus optical coherence tomography (OCT) system (Heidelberg Engineering). Mice were anaesthetized with an intraperitoneal injection of ketamine (100 mg/kg) and xylazine (6 mg/kg). Pupils were dilated with 0.5% tropicamide and 2.5% phenylephrine and anaesthetized with 0.5% proparacaine topical anaesthesia. Hypromellose ophthalmic demulcent solution and Systane Ultra (2.5%) were used for corneal hydration until eyes were imaged. OCT imaging for each eye was centred around the optic nerve head. Infrared reflectance (IR) plus OCT volume scans were obtained using Spectralis automatic real-time mode (ART), acquiring 55 cross-sectional images that were 200 μm apart. The individual layer thickness was measured using an artificial intelligence (AI) OCT segmentation tool developed by the NEI IT Bioteam. Briefly, B-scan images that cross the centre of the optic nerve were exported as a .tiff image. The AI segmentation tool outlines six lines on the OCT, totalling five retinal layers. Values for each layer were calculated from a region 350 to 650 μm away from the centre of the optic nerve head on each side. This region was chosen to have a relatively flat retinal layer thickness. Total retinal thickness is the summation of each retinal layer value.
Study approval
All animal experiments were conducted in accordance with recommendations of the Guide for the Care and Use of Laboratory animals of the National Institutes of Health (Protocol #NEI-680).
Patient consent
This study was conducted under institutional review board-approved protocols in accordance with the Declaration of Helsinki for the release of clinical information and family history. Informed consent was obtained after the explanation of the study’s risks and benefits.
Ethics approval
This study followed the tenets of the Declaration of Helsinki and was approved by all local ethics committees involved.
Statistical analysis
Statistical analyses performed on GraphPad Prism 9.4.1 (GraphPad Software) included Welch’s t-test and one-way ANOVA (Brown-Forsythe and Welch ANOVA) with post hoc Tukey test. Chi-squared and Fisher’s exact tests with an α-value threshold of 0.05 was used to determine statistical significance.
Results
Missense variants define the PNPLA6 genotype-phenotype spectrum
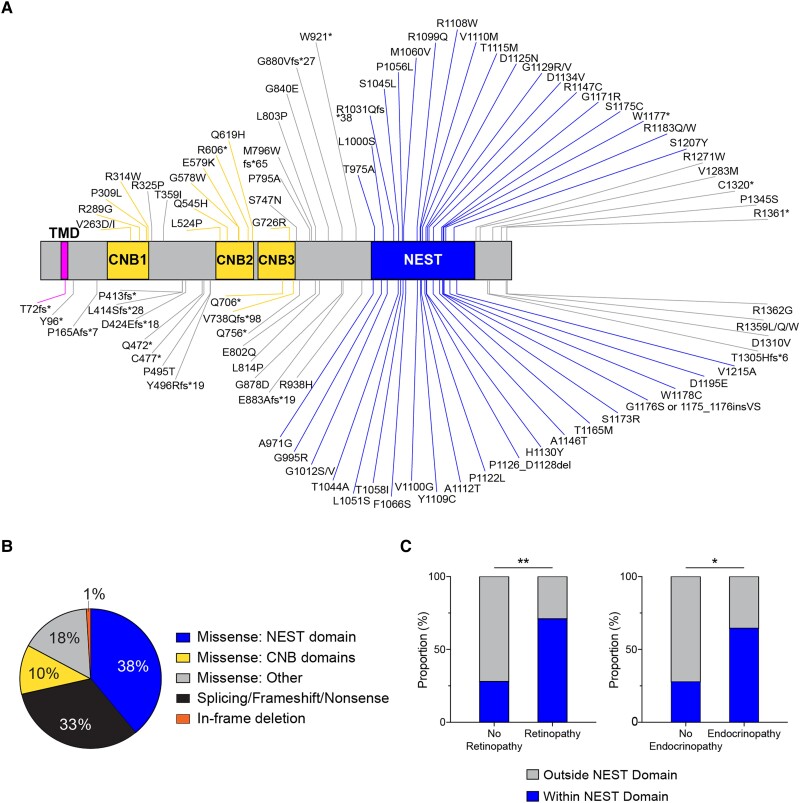
To investigate the genotype-phenotype associations among PNPLA6 disorders, we performed a systematic evidence-based review of reports of individuals with variants in the PNPLA6 gene1-5,11,20-47 along with a novel cohort of 23 patients carrying 12 reported and 17 novel variants in the PNPLA6 gene (Supplementary Table 1). To May 2023, 118 individuals have been reported (including this study) with PNPLA6 disorders and biallelic genotypes containing 106 unique PNPLA6 variants (Fig. 1A). In total, 70/106 of variants were missense substitutions, 35/106 were pLOF, including splice-altering, nonsense or frameshift variants, and 1/106 was in-frame deletion (Fig. 1B). Categorizing missense and in-frame deletion variants based on protein functional domain revealed that a majority (41/71) are located within the NTE catalytic domain of the protein. Interestingly, several missense variants, such as p.Gly1129Arg, were observed to recur in association with specific clinical diagnoses. Given the prevalence of missense variants and the near absence of reported individuals with homozygous null alleles, we hypothesized that PNPLA6 missense alleles are a key driver in NTE disease pathogenesis and determining patient phenotype.
Figure 1.
Summary of systematic evidence-based review of patients with PNPLA6 variants. (A) Schematic of the protein domain architecture and the location of 93 PNPLA6 missense, truncating, nonsense and in-frame variants (excluding 11 splice variants, one duplication and one deletion) known since May 2023 (NCBI reference NP_001159583.1). Neuropathy target esterase (NTE) contains a single pass transmembrane domain (aa60-80), three cyclic nucleotide binding (CNB) domains, and a NTE-esterase (NEST) domain. Figure generated using IBS Cuckoo illustration software.48 (B) Frequency distribution of variants based on their locations on the protein and variant type. Missense: NEST domain, n = 40; splicing/frameshift/nonsense, n = 35; Missense: CNB domains, n = 11; Missense: Other, n = 19; in-frame deletion: n = 1. (C) Missense variants categorized within the NEST domain (aa964–1269) and outside the nest domain (aa1–964, aa1270–1375) were grouped by their association with/without retinopathy and endocrinopathy. (C) Used a Fisher’s exact test with α = 0.05. Additional genotypic information can be found in the Supplementary material, ‘Supporting data values’ file.
To understand the role of missense alleles in disease severity and organ-specific phenotypes, we categorized missense variants located outside the NEST domain [amino acids (aa) 1–963, 1270–1375] and within the NEST domain (aa 964–1269) and compared their frequencies between individuals with and without retinopathy and endocrinopathy. Significantly more missense alleles located within the NEST domain are associated with disorders causing syndromic retinopathy (27/40, P = 0.002) and endocrinopathy (31/40, P = 0.01) compared to those not associated with retinopathy (7/40) or endocrinopathy (5/40) (Fig. 1C). In contrast, this pattern was not observed in association with missense variants located outside the NEST domain. Hence, missense variants within the NEST domain are observed more frequently in individuals with retinopathy and endocrinopathy, suggesting NTE function in retinal and endocrine-related tissues is more severely affected by variants in the NEST domain than outside the NEST domain.
PNPLA6 missense variants modulate NTE activity
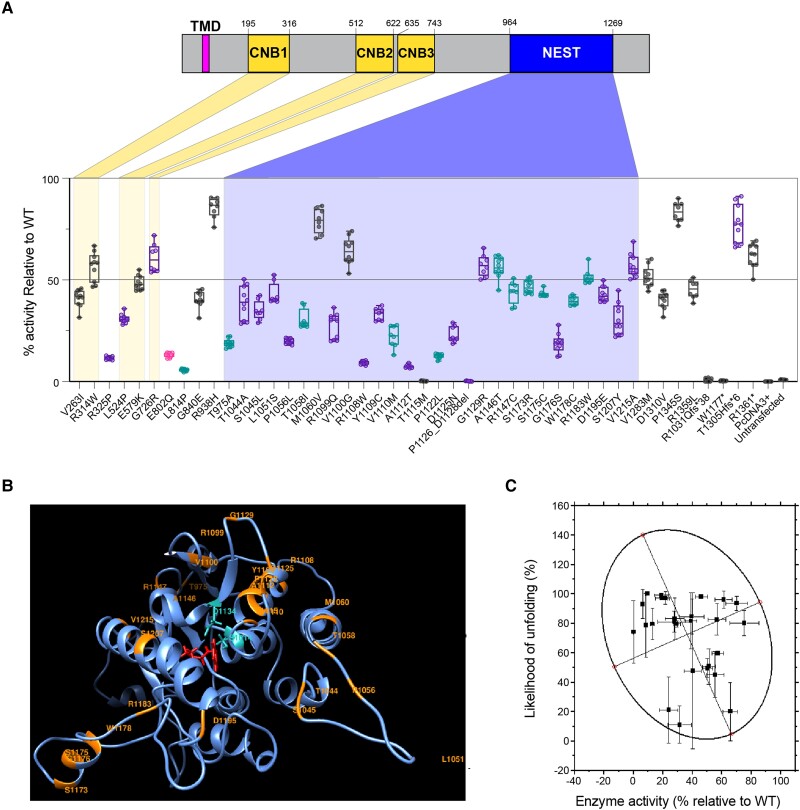
We hypothesized that the reduction in enzymatic activity correlates with disease severity, including the risk of retinopathy and endocrinopathy. To evaluate the effect of missense variants on NTE enzymatic activity, we developed an enzymatic assay for testing variants in full-length PNPLA6 protein, using previous NTE assays.1,10,49 HEK293 suspension cells were transfected with full-length PNPLA6 harbouring individual missense mutations located throughout the gene, and enzymatic activity was measured using a colorimetric assay that directly measures NTE hydrolase activity by cleavage of phenol valerate (Fig. 2A). We tested 41 missense variants, 20 common variants, four truncating variants and one in-frame deletion variant. The NTE activity of all disease-associated missense/in-frame and truncating variants was significantly reduced compared to the wild-type control, whereas 20 common variants were not significantly different, establishing our assay’s robustness to measure NTE activity accurately.
Figure 2.
PNPLA6 variants reduce in vitro NTE activity and increase likelihood of in silico misfolding. (A) Variants colour-coded by the syndrome they associate with (black = SPG39; pink = GDHS; teal = BNHS; purple = OMCS/LNMS). Highlighted areas indicate the location of variants within the protein’s domains (RefSeq protein NP_001159583.1). ‘% activity’ is the activity of variants relative to wild-type (WT). The activity of variants was normalized to the wild-type samples run in parallel. Each run had a wild-type, a previously tested missense variant, Arg1031Glnfs*38 and untransfected samples. Box and whisker plots extend from the 25th to 75th percentiles, with whiskers extending to the minimum and maximum values in the dataset. The median is the line plotted between boxes. See Supplementary material, ‘Supporting data values’ file for exact mean, n and standard deviation values. (B) The homology model of the human patatin domain. The ribbon structure of the patatin domain is shown in blue. Positions of 27 missense variants in the domain are labelled orange. Two residues forming a catalytic dyad, Ser1014-Asp1134, are shown in cyan. A patatin substrate, phenyl pentanoate, was docked to the active site and shown in red. (C) Global computational mutagenesis analysis comparing predicted unfolding fraction versus residual NTE activity. A 95% confidence ellipse is fitted in a scatter plot. R2 = 0.7. BNHS = Boucher-Neuhäuser syndrome; CNB = cyclic nucleotide binding; GDHS = Gordon-Holmes syndrome; LNMS = Laurence-Moon syndrome; NEST = NTE-esterase domain; NTE = neuropathy target esterase; OMCS = Oliver-McFarlane syndrome; SPG39 = spastic paraplegia type 39.
Truncating variants that disrupt the NEST domain (p.Arg1031Glnfs*38, p.Trp1177*) produced zero NTE hydrolase activity, as expected. Interestingly, truncating variants located outside the NEST domain and near the C-terminal end of the protein (p.Thr1305Hisfs*6, p.Arg1361*) exhibited residual esterase activity, suggesting that truncation of the protein near the C-terminal end results in preserved, yet decreased residual esterase activity. Three novel splice variants were clarified using a minigene splicing assay and digital droplet PCR (ddPCR). Splice variant c.1635+3G>T produced two alternative splice products, whereas c.1635+10_1635+15del produced a mixture of canonical and alternatively spliced products (Supplementary Fig. 1A). This mixture of products was also seen in the splice variant c.1636-3C>A, with ddPCR quantifying the total levels of each transcript (Supplementary Fig. 1B and C).
Because phenol valerate is a synthetic substrate, we validated this observation using a biological substrate. The individual phospholipase A1 activity on a fluorescently labelled lysophosphatidylethanolamine and A2 activity on a fluorescently labelled phosphatidylcholine of select missense and truncating variants was confirmed (refer to the ‘Materials and methods’ section), producing comparable activities to phenol valerate cleavage and to reported patient fibroblast activity assays1,10 (Supplementary material, ‘Supporting data values’ file). Individual variant activity and genotype (biallelic variant) activities correlated well between phenol valerate and phospholipase assays to measure NTE activity.
Next, we assessed the value of the NTE hydrolase activity assay in variant classification using the ClinGen standards for functional evidence. We applied the OddsPath equation developed by Brnich and colleagues,50 which produced an OddsPath of 20 for pathogenic variants and 0.022 for benign variants, indicating the level of evidence as PS3 and BS3_Strong (Supplementary material, ‘Supporting data values’ file). Using the 2015 ACMG criteria51 and the point-based system developed by Tavtigian et al.,52 100% of disease-reported variants were able to be classified as likely pathogenic or pathogenic, including reclassification of 36 variants as pathogenic and 10 variants as likely pathogenic (Supplementary material, ‘Supporting data values’ file).
To further evaluate the relationship of missense variants in protein function, protein stability was assessed in 27 missense variants in the NEST domain using a homology model (Fig. 2B) from available crystal structures of patatin-like proteins (refer to the ‘Materials and methods’ section). Applying global computational mutagenesis to determine the unfolding propensities of missense variants in the NEST domain revealed an inverse relationship between protein unfolding and residual NTE activity (Fig. 2C, R2 = 0.7). This result indicates that reduced catalytic function could be associated with a decrease in protein stability in the NEST domain.
PNPLA6 disorder spectrum is determined by residual NTE activity levels
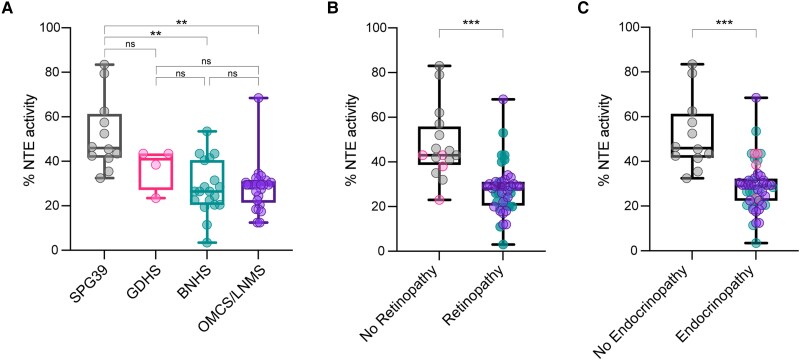
To test the hypothesis that total residual NTE activity is predictive of PNPLA6 disorder severity, we calculated the estimated (‘synthetic’) residual activity of biallelic variants and compared them to phenotypic expression. Individual genotype activity levels were determined by the average activity of the two individual variants. Next, categorization of individuals by clinical diagnosis showed a significant difference in average NTE activity levels in SPG39 patients (51%, n = 12) compared to BNHS patients (28%, n = 19, P = 0.002 against SPG39) or OMCS/LNMS patients (28%, n = 26, P = 0.002 against SPG39) (Fig. 3A). These results suggest that patient genotype dictates NTE activity levels in affected individuals and that disease severity and synthetic NTE activity levels are inversely correlated.
Figure 3.
Inverse relationship between residual NTE activity and disease severity and tissue manifestation. (A) Comparing synthetically determined patient neuropathy target esterase (NTE) activity categorized by PNPLA6-associated syndromes. (B) Comparing synthetically determined patient NTE activity between individuals with and without retinopathy. (C) Comparing synthetically determined patient NTE activity between individuals with and without endocrinopathy. ‘% activity’ is the activity of variants relative to wild-type (WT). Data-points for B and C correspond to the following: black = SPG39; pink = GDHS; teal = BNHS; purple = OMCS/LNMS. Box and whisker plots extend from the 25th to 75th percentiles, with whiskers extending to the minimum and maximum values in the dataset. The median is the line plotted between boxes. (A) Used a Brown-Forsythe ANOVA with post hoc Tukey test with α = 0.05. (B and C) Used a Welch’s t-test with α = 0.05. P > 0.05 (not significant, ns), *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. See Supplementary material, ‘Supporting data values’ file for exact mean, n and standard deviation values. BNHS = Boucher-Neuhäuser syndrome; GDHS = Gordon-Holmes syndrome; LNMS = Laurence-Moon syndrome; OMCS = Oliver-McFarlane syndrome; SPG39 = spastic paraplegia type 39.
As clinical diagnoses reflect the age of onset and type of affected tissues, we compared NTE activity levels in those with and without retinopathy and in those with and without endocrinopathy. NTE activity levels in individuals with retinopathy (28%, n = 45, P = 0.0001) and endocrinopathy (28%, n = 49, P = 0.0004) were significantly lower than those in individuals without retinopathy (48%, n = 16) or endocrinopathy (51%, n = 12), respectively (Fig. 3B and C). Altogether, these results propose a novel genotype:NTE activity:phenotype relationship of the PNPLA6 disorder spectrum, where lower residual activity results in a higher likelihood of more severe disease outcomes, as well as retinopathy or endocrinopathy. Therefore, these conditions can be considered a single pleiotropic spectrum of PNPLA6-related disorders.
Identifying tissue-specific patterns between residual NTE activity and onset of disease
To demonstrate the relationship between NTE activity and the onset of disease, the age of tissue manifestation for unreported and reported individuals1,4,34 with biallelic PNPLA6 variants was correlated to an individual’s overall NTE activity (Supplementary Fig. 3). There was no apparent relationship between the activity of NTE and the onset of retinopathy or endocrinopathy. A more detailed analysis looking at the onset of CNS and PNS clinical disease, with resulting ataxia, spasticity/pyramidal tract signs and peripheral neuropathy, did not reveal noticeable patterns either (unpublished observations).
NTE activity drives retinal degeneration in mice
To demonstrate that the genotype:NTE activity:phenotype relationship observed can be replicated in vivo, we developed a murine allelic series. Mice were generated with a missense variant seen exclusively in SPG39 patients (c.3088A>G, p.Met1030Val) in the homozygous state (‘MV’ allele) and a truncating variant ‘delAT’ (c.3088_3089delAT, p.Met1030Valfs*2) that exhibits 0% residual activity due to its truncation prior to the enzymatic domain. We hypothesized that mice with the MV/MV genotype would have 80% NTE activity and lack retinopathy, consistent with the human phenotype in SPG39, while mice with the genotype MV/delAT (not seen in patients) and a much lower residual NTE activity, would develop retinopathy.
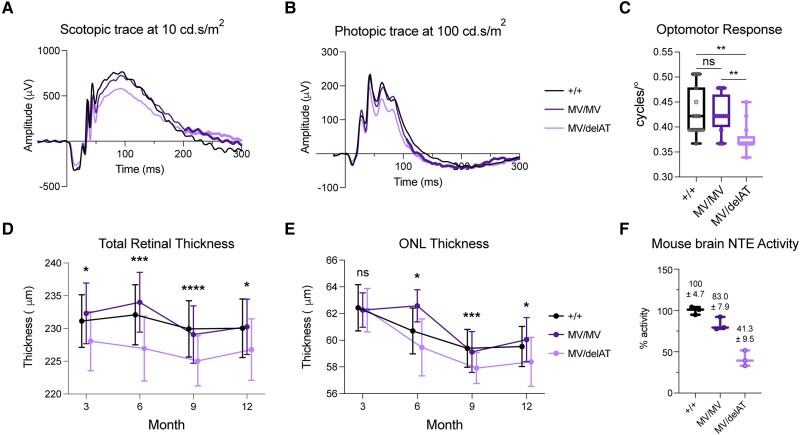
Mice homozygous for MV and compound heterozygous for MV/delAT were viable with typical Mendelian ratios, whereas delAT/delAT mice were not viable as predicted (Supplementary Table 2). Testing the visual function of control, MV/MV and MV/delAT via (ERG) over the course of 12 months revealed a significant reduction in photopic and scotopic a- and b-wave amplitudes between control and MV/delAT mice (Supplementary Fig. 4). An example of this reduction in visual function can be seen in the representative scotopic and photopic traces at the maximum stimulus intensity (10 cd.s/m2 for scotopic, 100 cd.s/m2 for photopic) of our assay protocol at 3 months (Fig. 4A and B). Traces show a discernible reduction in both b-wave amplitudes under scotopic and photopic conditions and smaller, yet still significant, differences in a-wave amplitudes. Additionally, there was no significant difference in amplitudes between control and MV/MV mice over several time points, reiterating the phenotypic observations seen in SPG39 patients who do not exhibit a retinopathy phenotype. Testing the visual acuity of mice in our allelic series at 3 months using the OMR showed similar results, where control and MV/MV mice did not exhibit significant differences in visual acuity [+/+ = 0.43 cycles/degree (cpd), MV/MV = 0.425 cpd, P = 0.80], whereas MV/delAT mice visual acuity was significantly reduced compared to controls (MV/delAT = 0.378 cpd, P = 0.008, Fig. 4C). Overall, the reduction in ERG amplitudes and OMR provide evidence that reduction in NTE activity in a dose-dependent manner causes reduced retinal function in mice.
Figure 4.
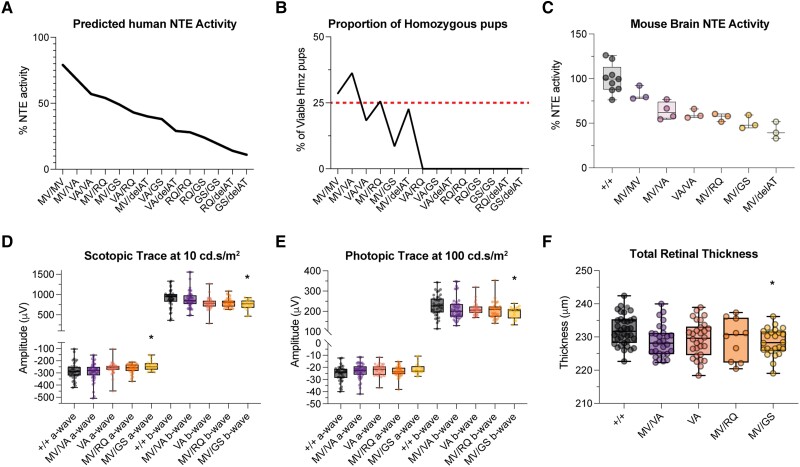
Residual NTE activity determines the loss of visual function and structure in PNPLA6 allelic series mice in a dose-dependent manner. (A) Scotopic electroretinogram (ERG) trace at maximum stimulus intensity (10 cd.s/m2) for our photopic imaging protocol. (B) Photopic ERG trace at maximum stimulus intensity (100 cd.s/m2) for our scotopic imaging protocol. (C) Optomotor response (OMR) (+/+ n = 15, MV/MV n = 12, MV/delAT n = 13) of allelic series mice at 3 months. (D) Time course of total retinal thickness of optical coherence tomography (OCT) images. Significant values are between Control and MV/delAT values. Control and MV/MV line were not significantly different, and MV/MV and MV/delAT were significantly different. (E) Time course of outer nuclear layer (ONL) thickness of OCT images. Significant values are between Control and MV/delAT values. Control and MV/MV line were not significantly different, and MV/MV and MV/delAT were significantly different. (F) Neuropathy target esterase (NTE) activity of allelic series mice brain homogenate. Each time point took measurements from littermates, and bars in E and F are offset for cleaner presentation. All error bars in standard deviations. Box and whisker plots extend from the 25th to 75th percentiles, with whiskers extending to the minimum and maximum values in the dataset. The median is the line plotted between boxes. Statistical tests from these figures used one-way ANOVA with post hoc Tukey test with α = 0.05. P > 0.05 (not significant, ns), *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. See Supplementary material, ‘Supporting data values’ file for exact mean, n and standard deviation values.
To examine the morphological features of the retinas in our allelic series, we obtained cross-sectional mouse retinal images by spectral domain (SD)-OCT from 3 to 12 months. Using an AI-driven OCT segmentation tool, the thickness of individual retinal layers was measured (Supplementary Fig. 5). The total retinal thickness measured from the retinal nerve fibre layer to the retinal pigment epithelium (RPE) showed a significant reduction in retinal thickness between control and MV/delAT mice from 3 to 12 months (Fig. 4D). Similar to the visual function data, there was a significant difference in total retinal thickness at all time points between control and MV/delAT mice, but not between control and MV/MV mice. To identify the specific retinal layers affected by the reduction of NTE activity, individual layers (photoreceptor and RPE layer, outer nuclear layer, inner nuclear layer, inner plexiform layer, nerve fibre layer) were measured and showed intermittent differences in thickness across all time points between control and MV/delAT mice. Noticeably, the outer nuclear layer (ONL) was consistently thinner across all time points between control and MV/delAT mice (except at 3 months), and not significantly thinner between control and MV/MV mice (Fig. 4E). These results reinforce the idea that NTE activity level predicts the onset of retinopathy in a dose-dependent manner.
We then measured the NTE activity of mouse brain in our murine allelic series to further validate our relative enzymatic activities measured from overexpression experiments. Using a modified NTE protocol19 for mouse tissue, mouse brain homogenate showed similar results, as observed in our in vitro results, highlighting the remarkable predictability between genotype and enzymatic activity in a dose-dependent manner (Figs 4F, 5A and C). Testing the viability of our Pnpla6 murine allelic series revealed that <40% NTE activity was embryonic lethal in mice, demonstrating that a threshold of residual NTE activity is needed for proper embryonic development (Fig. 5B). Evaluating the visual function and structure of viable homozygous mice in our allelic series indicated that retinal degeneration only occurs in the MV/GS (p.Met1030Val in trans with p.Gly1146Ser, a missense variant found in BNHS and OMCS patients) mouse line, which has an overall activity level of 50% (Fig. 5D–F). This indicates that the onset of retinal degeneration occurs between 40–50% residual NTE activity, which is similar to overall NTE activity levels in patients with retinopathy. Overall, these results underpin our clinical and in vitro observations, where residual NTE activity is critical for the onset of retinopathy in individuals with PNPLA6 biallelic variants.
Figure 5.
Residual NTE activity dictates the onset of retinopathy and embryonic lethality in PNPLA6 missense allelic series mice. (A) Predicted overall neuropathy target esterase (NTE) activity of allelic series mice using human in vitro variant activities (Fig. 2). (B) The proportion of viable homozygous pups in the PNPLA6 allelic series. The red line denotes the normal Mendelian ratio expected (25%). See Supplementary Table 2 for exact numbers. (C) NTE activity of viable homozygous allelic series mice brain homogenate. (D) Six-month scotopic electroretinogram (ERG) wave amplitudes at a stimulus intensity of 10 cd.s/m2. (E) Six-month photopic ERG wave amplitudes at a stimulus intensity of 100 cd.s/m2. (F) Total retinal thickness measured from spectral domain-optical coherence tomography (SD-OCT) images at 6 months. Box and whisker plots extend from the 25th to 75th percentiles, with whiskers extending to the minimum and maximum values in the dataset. The median is the line plotted between boxes. D–F used one-way ANOVA with post hoc Tukey test with α=0.05. P > 0.05 (not significant, ns), *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Significant values in D–F indicate significance between Control and MV/GS values. See Supplementary material, ‘Supporting data values’ file for exact mean, n and standard deviation values.
Discussion
In this study, we describe a novel relationship in PNPLA6 disorders, correlating genotype, NTE activity and phenotype to predict the presence of retinopathy and endocrinopathy. Through a systematic evidence-based review of a novel cohort and reported patients, missense alleles were found to comprise a majority of the genotypic landscape, in particular those located within the NEST domain. This implies an important relationship between missense variants and NTE activity. Subsequent measurement of NTE activity in 66 missense, common and truncating variants demonstrated that variants and genotypes recurrent among PNPLA6 clinical diagnoses exhibit a broad range of residual activity. Synthetically reconstructing expected individual NTE activities based on their genotypic information produced a striking relationship between genotype, phenotype and NTE activity, where residual NTE activity is inversely correlated with the presence of signs and symptoms of this group of disorders. We validated this relationship in a preclinical mouse model, where allelic series mice with Pnpla6 genotypes associated with SPG39 do not develop retinopathy, while Pnpla6 genotypes associated with OMCS develop retinopathy. Collectively, our research has established that PNPLA6 disorders comprise a clinical spectrum in which the presence of affected tissues is determined by residual protein activity.
A systematic evidence-based review of previously studied individuals revealed that missense alleles directly correlate with tissue pathology. Sixty-six per cent of known variants in the affected population to date are missense alleles, with 41/71 missense variants localizing within the enzymatic domain of the protein and biasing towards the more severe forms of the disease (BNHS, OMCS, LNMS). Testing the hydrolase activity of missense variants exhibited a wide range of activities, where missense variants located within the NEST domain exhibited a lower average activity compared to variants located outside the NEST domain (43% versus 35%). Interestingly, several missense variants located near amino acids 1112–1122 had extremely low activity levels. This is possibly due to the proximity and interaction with the asparagine at position 1134, which is a crucial component of the catalytic reaction by acting as a general base and acid.9 Intriguingly, several missense variants located outside the enzymatic domain exhibited low residual activities (p.Arg325Pro, p.Leu814Pro). This could be due to disruption of the CNB domains that have unknown function thus far. Structure-function analyses of the missense variants that produce a drastic decrease in NTE activity located within and outside the NEST domain may reveal details about the abnormal catalytic mechanism and the function of the CNB domains, respectively.
Synthetically reconstructing an affected individual’s overall NTE activity revealed a striking relationship between the activity of the protein and the severity of disease that was dictated by patient genotype (Fig. 3). One hundred and fourteen of 116 genotyped individuals have at least one missense allele, implicating missense alleles as a key driver of residual NTE activity and thus, disease severity. To date, there are two patients with two frameshift variants that predict to have 0% activity.21,47 Based on the results of this study, truncations prior to amino acid position 1177 produce 0% NTE activity, while truncations past this position produce relatively high residual esterase activity (e.g. c.4081C>T or p.Arg1361* homozygous patient having mild age of onset symptoms and diagnosis of SPG39). These findings uncover 16 additional truncating variants prior to amino acid position 1177 that are predicted to have 0% activity in the patient population. Interestingly, both patients with two truncating variants with 0% predicted NTE activity were diagnosed with GDHS. In mouse, truncation of the protein produces similar viability results, where mice with two truncating alleles (p.Met1030Valfs*2) near the start of the enzymatic domain produce non-viable pups.
Previously, PNPLA6-associated disorders were defined by the presence, absence or AOO of manifestations in specific organs or tissues.53 For example, OMCS is defined by the presence of ‘trichomegaly, chorioretinal dystrophy and congenital or childhood hypopituitarism’.53 Although these disorders were previously defined, several individuals with biallelic PNPLA6 variants do not fit the definition of a specific syndrome, such as OMCS, due to the asynchronous onset of affected tissues (e.g. hypopituitarism manifesting later) or the presence/absence of effects on other tissues (e.g. CNS symptoms in OMCS patients). Additionally, many missense variants are seen recurrently throughout the affected population and are present in multiple syndromes, lending support to the theory that variants in PNPLA6 are involved in a set of pleiotropic disorders. Our study demonstrates that phenotypic onset, especially in the pituitary and the eye, is determined by residual NTE activity that is predicted by their genotype. A striking example of this can be seen with the missense variant p.Arg1183Trp (52% activity), which has been observed in individuals with SPG39, BNHS and OMCS. One individual homozygous for the variant is diagnosed with SPG39 and predicts to have an overall activity of 52%, three individuals with BNS have the missense variant in trans with a splicing or frameshift allele and predicts to have an overall activity of 26%, and two individuals with OMCS have the missense variant in trans with another missense variant (p.Ala1112Thr) and predicts to have an overall activity of 30%. This remarkable observation validates the conclusion that PNPLA6-associated disorders are not distinct, but a continuous spectrum of disease1,4 that is influenced by the activity of NTE. Therefore, a strong case can be made for syndrome nomenclature associated with variants in the PNPLA6 gene (SPG39, GDHS, BNHS, OMCS, LNMS) to be referred to as syndromes associated with the PNPLA6 disorder spectrum.
Studies in our Pnpla6 murine allelic series are the first to model PNPLA6-associated retinopathy by measuring the visual function and structure of affected mice. Measuring the NTE activity of viable homozygous mice in our preclinical mouse model produced a remarkable activity gradient that is dictated by genotype. This is highlighted in Fig. 4, where the MV/MV genotype, seen exclusively in SPG39 patients, does not exhibit signs of retinopathy in either mice or humans. In contrast, the MV/delAT genotype, although not seen in the affected population to date, halves the overall NTE activity compared to the MV/MV genotype and has a NTE activity level similar to OMCS individuals. Evaluation of other homozygous viable mice in our allelic series indicated that <50% NTE activity is required for the onset of retinopathy and >40% NTE activity is required for embryonic viability. Taken together, these results correlate a decrease in NTE activity with a decrease in retinal function and structure in a dose-dependent manner, signifying the importance of NTE activity in proper retinal function in mice. Comparing the average difference in magnitude of the scotopic b-wave ERG (∼16%), scotopic a-wave (∼12%), photopic a- and b-wave (∼10%), and total retinal thickness (∼2%) measurements between control and MV/delAT mouse lines across all time points implies that visual function may not be affected by the total loss of cells in the retina/RPE, but disruptions to cellular connections or function. Further work characterizing the molecular mechanisms and lipid profiles that lead to retinopathy will help provide clarity on the role Pnpla6 plays in retinal degeneration.
Testing the BMI of mice in our allelic series at 3 months yielded no significant differences between control and mutant mice, implying that hormone levels are not affected when NTE activity is reduced in homozygous viable mice (unpublished observations). In contrast, previous papers showing that conditionally knocking out Pnpla6 using nestin-cre in the CNS and PNS caused mice to become noticeably smaller compared to littermate controls.6,54 These findings demonstrate the limitations in using Pnpla6 allelic mouse models to look at pituitary and growth defects, as much have a low survival rate in the setting of decreased NTE activity.
Although NTE activity can predict the occurrence of retinopathy or endocrinopathy, aggregating reported and unreported patients did not show striking patterns between the onset of clinical manifestations and activity of NTE (Supplementary Fig. 3). This could potentially be due to the nature of reporting the exact onset of tissue disease phenotypes, which can be difficult. Collecting the numerical age of onsets of affected tissues in PNPLA6-affected individuals showed that CNS and PNS phenotypes did not show significant differences in onset across clinical categories (Supplementary Fig. 6D and E). In contrast, ophthalmic and endocrine phenotypes occurred significantly earlier in patients with OMCS/LNMS (Supplementary Fig. 6A and B). This can also be seen when categorizing patients with either a ‘0’ or ‘1’ truncating allele, where individuals with ‘1’ truncating allele are more likely to experience retinopathy and endocrinopathy than individuals with two missense variants (Supplementary Fig. 6C and F). To gain a better understanding of the relationship between the activity of NTE and AOO, accurate reporting via a prospective natural history study of individuals with PNPLA6 variants may be imperative for further analysis of the disease gene.
With the advancement of next-generation sequencing, it is vital to properly interpret variants of unknown significance (VOUS) to validate variants identified in genes that cause Mendelian disorders. Our study has provided a new and robust method of disambiguating VOUS’s in the PNPLA6 gene that can be adapted in research institutions and CLIA laboratories. Additionally, we were able to verify three novel splice variants using a minigene splicing assay and ddPCR to detect the canonical and alternative spliced products (Supplementary Fig. 1).
In conclusion, in this novel cohort of 23 patients with biallelic variants, we describe 12 reported and 17 novel variants in the PNPLA6 gene, together with their clinical phenotypes. A systematic evidence-based review of 118 individuals, in vitro activity measurements of pathogenic and common variants, genotype-phenotype correlation of pathogenic and common variants, and in vivo characterization of retinal activity and structure in Pnpla6 allelic series mice support a genotype:NTE activity:phenotype relationship that dictates the disease state with NTE activity as the common mechanism. This work outlines the importance for clinicians and geneticists to be aware of the predicted effects of NTE activity on the onset of progression of affected tissues, such as the pituitary and eye. Furthermore, NTE activity can be used as a biomarker to predict the presence or absence of affected tissues, which will be invaluable for mitigation and surveillance. Future studies will look at therapeutic interventions such as gene therapy or enzyme replacement therapy to restore relative levels of NTE activity in affected tissues to ameliorate the symptoms experienced by individuals affected by PNPLA6 variants.
Supplementary Material
Acknowledgements
We would like to thank the participating patients and their families, as well as the healthcare professionals involved in their care. We would like to thank Megan Kopera and her team for helping maintain and oversee the mouse colony. We would also like to thank Rudy Richardson for providing the protocol for the NTE esterase activity assay, as well as recommending vendors for chemicals related to the experiment. We would like to thank Jee Min Kim for editing the figures for the manuscript. We would like to thank Zubair Ahmed, Sally Camper, Ellen Sidransky and Sua Myong for their advice on the manuscript.
Contributor Information
James Liu, Ophthalmic Genetics and Visual Function Branch, National Eye Institute, National Institutes of Health, Bethesda, MD 20892, USA.
Yi He, Fermentation Facility, Biochemistry and Biophysics Center, National Heart, Lung and Blood Institute, Bethesda, MD 20892, USA.
Cara Lwin, Ophthalmic Genetics and Visual Function Branch, National Eye Institute, National Institutes of Health, Bethesda, MD 20892, USA.
Marina Han, Ophthalmic Genetics and Visual Function Branch, National Eye Institute, National Institutes of Health, Bethesda, MD 20892, USA.
Bin Guan, Ophthalmic Genetics and Visual Function Branch, National Eye Institute, National Institutes of Health, Bethesda, MD 20892, USA.
Amelia Naik, Ophthalmic Genetics and Visual Function Branch, National Eye Institute, National Institutes of Health, Bethesda, MD 20892, USA.
Chelsea Bender, Ophthalmic Genetics and Visual Function Branch, National Eye Institute, National Institutes of Health, Bethesda, MD 20892, USA.
Nia Moore, Ophthalmic Genetics and Visual Function Branch, National Eye Institute, National Institutes of Health, Bethesda, MD 20892, USA.
Laryssa A Huryn, Ophthalmic Genetics and Visual Function Branch, National Eye Institute, National Institutes of Health, Bethesda, MD 20892, USA.
Yuri V Sergeev, Ophthalmic Genetics and Visual Function Branch, National Eye Institute, National Institutes of Health, Bethesda, MD 20892, USA.
Haohua Qian, Visual Function Core, National Eye Institute, National Institutes of Health, Bethesda, MD 20892, USA.
Yong Zeng, Visual Function Core, National Eye Institute, National Institutes of Health, Bethesda, MD 20892, USA.
Lijin Dong, Genetic Engineering Core, National Eye Institute, National Institutes of Health, Bethesda, MD 20892, USA.
Pinghu Liu, Genetic Engineering Core, National Eye Institute, National Institutes of Health, Bethesda, MD 20892, USA.
Jingqi Lei, Genetic Engineering Core, National Eye Institute, National Institutes of Health, Bethesda, MD 20892, USA.
Carl J Haugen, Genetic Engineering Core, National Eye Institute, National Institutes of Health, Bethesda, MD 20892, USA.
Lev Prasov, Department of Ophthalmology and Visual Sciences, Kellogg Eye Center, University of Michigan, Ann Arbor, MI 48105, USA; Department of Human Genetics, University of Michigan, Ann Arbor, MI 48105, USA.
Ruifang Shi, Department of Ophthalmology, Peking Union Medical College Hospital, Peking Union Medical College, Chinese Academy of Medical Sciences, 100730 Beijing, China.
Hélène Dollfus, Centre de référence pour les Affections Rares Ophtalmologiques CARGO, Hôpitaux Universitaires de Strasbourg, Université de Strasbourg, UMRS_1112, Strasbourg 67091, France.
Petros Aristodemou, Cyprus Institute of Neurology and Genetics, Nicosia 1683, Cyprus; VRMCy Centre, Limassol 3025, Cyprus.
Yannik Laich, UCL Institute of Ophthalmology, University College London, London EC1V 9EL, UK; Department of Genetics, Moorfields Eye Hospital NHS Trust, London EC1V 2PD, UK.
Andrea H Németh, Oxford Centre for Genomic Medicine, Oxford University Hospitals NHS Foundation Trust, ACE Building, Nuffield Orthopaedic Centre, Oxford OX3 7HE, UK; Nuffield Department of Clinical Neurosciences, University of Oxford, John Radcliffe Hospital, Oxford OX3 9DU, UK.
John Taylor, Oxford Regional Genetics Laboratory, Oxford University Hospitals NHS Foundation Trust, Oxford OX3 9DU, UK.
Susan Downes, Nuffield Department of Ophthalmology, Nuffield Department of Clinical Neuroscience, University of Oxford, Oxford OX3 9DU, UK; Oxford Eye Hospital, Oxford University Hospitals NHS Foundation Trust, Oxford OX3 9DU, UK.
Maciej R Krawczynski, Department of Medical Genetics, Poznan University of Medical Sciences, Poznan 60-512, Poland.
Isabelle Meunier, National Referent Centre for Rare Sensory Diseases, Montpellier University Hospital, Montpellier University, Montpellier 34295, France.
Melissa Strassberg, Invitae Corporation, San Francisco, CA 94103, USA.
Jessica Tenney, Division of Medical Genetics, Department of Pediatrics, UCSF School of Medicine, San Francisco, CA 94143, USA.
Josephine Gao, Division of Medical Genetics, Department of Pediatrics, UCSF School of Medicine, San Francisco, CA 94143, USA.
Matthew A Shear, Division of Medical Genetics, Department of Pediatrics, UCSF School of Medicine, San Francisco, CA 94143, USA.
Anthony T Moore, UCL Institute of Ophthalmology, University College London, London EC1V 9EL, UK; Department of Ophthalmology, UCSF School of Medicine, San Francisco, CA 94143, USA.
Jacque L Duncan, Department of Ophthalmology, UCSF School of Medicine, San Francisco, CA 94143, USA.
Beatriz Menendez, Department of Pediatrics, University of Illinois School of Medicine, Chicago, IL 60612, USA.
Sarah Hull, Department of Ophthalmology, University of Auckland, Auckland 1023, New Zealand.
Andrea L Vincent, Department of Ophthalmology, University of Auckland, Auckland 1023, New Zealand.
Carly E Siskind, Neurology and Neurological Sciences, Stanford School of Medicine, Stanford, CA 94305, USA.
Elias I Traboulsi, The Center for Genetic Eye Diseases, The Cleveland Clinic Eye Institute, Cleveland, OH 44106, USA.
Craig Blackstone, Movement Disorders Division, Department of Neurology, Massachusetts General Hospital, Boston, MA 02114, USA.
Robert A Sisk, Department of Ophthalmology, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH 45229, USA.
Virginia Miraldi Utz, Department of Ophthalmology, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH 45229, USA; Abrahamson Pediatric Eye Institute, Cincinnati Children's Hospital Medical Center, Cincinnati, OH 45229, USA.
Andrew R Webster, UCL Institute of Ophthalmology, University College London, London EC1V 9EL, UK; Department of Genetics, Moorfields Eye Hospital NHS Trust, London EC1V 2PD, UK.
Michel Michaelides, UCL Institute of Ophthalmology, University College London, London EC1V 9EL, UK; Department of Genetics, Moorfields Eye Hospital NHS Trust, London EC1V 2PD, UK.
Gavin Arno, UCL Institute of Ophthalmology, University College London, London EC1V 9EL, UK; Department of Genetics, Moorfields Eye Hospital NHS Trust, London EC1V 2PD, UK.
Matthis Synofzik, Division Translational Genomics of Neurodegenerative Diseases Hertie Institute for Clinical Brain Research, University of Tübingen, Tübingen 72076, Germany; German Center of Neurodegenerative Diseases (DZNE), Tübingen 72076, Germany.
Robert B Hufnagel, Ophthalmic Genetics and Visual Function Branch, National Eye Institute, National Institutes of Health, Bethesda, MD 20892, USA; Department of Genetics and Center for Integrated Healthcare Research, Kaiser Permanente Hawaii Region, Honolulu, HI 98619, USA.
Data availability
The data presented in this study are available from the corresponding author upon request.
Funding
This work was supported by National Eye Institute intramural funds. S.H. and A.V. are supported by Save sight society NZ, Retina NZ, and the Ombler trust. G.A. is supported by the National Institute of Health Research Biomedical Research Centre (NIHR-BRC) at Moorfields Eye Hospital and UCL Institute of Ophthalmology, a Fight For Sight UK Early Career Investigator Award (5045/46) and NIHR-BRC at Great Ormond Street Hospital Institute for Child Health. L.P. is supported by NEI K08-EY032098 and the Research to Prevent Blindness Career Development Award.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Hufnagel RB, Arno G, Hein ND, et al. Neuropathy target esterase impairments cause Oliver-McFarlane and Laurence-Moon syndromes. J Med Genet. 2015;52:85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kmoch S, Majewski J, Ramamurthy V, et al. Mutations in PNPLA6 are linked to photoreceptor degeneration and various forms of childhood blindness. Nat Commun. 2015;6:5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rainier S, Bui M, Mark E, et al. Neuropathy target esterase gene mutations cause motor neuron disease. Am J Hum Genet. 2008;82:780–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Synofzik M, Gonzalez MA, Lourenco CM, et al. PNPLA6 mutations cause Boucher-Neuhauser and Gordon Holmes syndromes as part of a broad neurodegenerative spectrum. Brain. 2014;137(Pt 1):69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Topaloglu AK, Lomniczi A, Kretzschmar D, et al. Loss-of-function mutations in PNPLA6 encoding neuropathy target esterase underlie pubertal failure and neurological deficits in Gordon Holmes syndrome. J Clin Endocrinol Metab. 2014;99:E2067–E2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Read DJ, Li Y, Chao MV, Cavanagh JB, Glynn P. Neuropathy target esterase is required for adult vertebrate axon maintenance. J Neurosci. 2009;29:11594–11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van Tienhoven M, Atkins J, Li Y, Glynn P. Human neuropathy target esterase catalyzes hydrolysis of membrane lipids. J Biol Chem. 2002;277:20942–20948. [DOI] [PubMed] [Google Scholar]
- 8. Zhu L, Wang P, Sun YJ, Xu MY, Wu YJ. Disturbed phospholipid homeostasis in endoplasmic reticulum initiates tri-o-cresyl phosphate-induced delayed neurotoxicity. Sci Rep. 2016;6:37574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Atkins J, Glynn P. Membrane association of and critical residues in the catalytic domain of human neuropathy target esterase. J Biol Chem. 2000;275:24477–24483. [DOI] [PubMed] [Google Scholar]
- 10. Hein ND, Rainier SR, Richardson RJ, Fink JK. Motor neuron disease due to neuropathy target esterase mutation: Enzyme analysis of fibroblasts from human subjects yields insights into pathogenesis. Toxicol Lett. 2010;199:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu S, Sun Z, Zhu T, et al. Novel variants in PNPLA6 causing syndromic retinal dystrophy. Exp Eye Res. 2021;202:108327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kayyali US, Moore TB, Randall JC, Richardson RJ. Neurotoxic esterase (NTE) assay: Optimized conditions based on detergent-induced shifts in the phenol/4-aminoantipyrine chromophore spectrum. J Anal Toxicol. 1991;15:86–89. [DOI] [PubMed] [Google Scholar]
- 13. Johnson MK. Improved assay of neurotoxic esterase for screening organophosphates for delayed neurotoxicity potential. Arch Toxicol. 1977;37:113–115. [DOI] [PubMed] [Google Scholar]
- 14. Luo S, Wehr NB, Levine RL. Quantitation of protein on gels and blots by infrared fluorescence of Coomassie blue and Fast Green. Anal Biochem. 2006;350:233–238. [DOI] [PubMed] [Google Scholar]
- 15. McCafferty CL, Sergeev YV. In silico mapping of protein unfolding mutations for inherited disease. Sci Rep. 2016;6:37298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McCafferty CL, Sergeev YV. Dataset of eye disease-related proteins analyzed using the unfolding mutation screen. Sci Data. 2016;3:160112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: An online force field. Nucleic Acids Res. 2005;33:W382–W388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang H, Yang H, Shivalila CS, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Quistad GB, Barlow C, Winrow CJ, Sparks SE, Casida JE. Evidence that mouse brain neuropathy target esterase is a lysophospholipase. Proc Natl Acad Sci U S A. 2003;100:7983–7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tarnutzer AA, Gerth-Kahlert C, Timmann D, et al. Boucher-Neuhäuser syndrome: Cerebellar degeneration, chorioretinal dystrophy and hypogonadotropic hypogonadism: Two novel cases and a review of 40 cases from the literature. J Neurol. 2015;262:194–202. [DOI] [PubMed] [Google Scholar]
- 21. Yoon G, Baskin B, Tarnopolsky M, et al. Autosomal recessive hereditary spastic paraplegia-clinical and genetic characteristics of a well-defined cohort. Neurogenetics. 2013;14:181–188. [DOI] [PubMed] [Google Scholar]
- 22. Rump P, Hamel BC, Pinckers AJ, van Dop PA. Two sibs with chorioretinal dystrophy, hypogonadotrophic hypogonadism, and cerebellar ataxia: Boucher-Neuhäuser syndrome. J Med Genet. 1997;34:767–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Deik A, Johannes B, Rucker JC, et al. Compound heterozygous PNPLA6 mutations cause Boucher-Neuhäuser syndrome with late-onset ataxia. J Neurol. 2014;261:2411–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Koh K, Kobayashi F, Miwa M, et al. Novel mutations in the PNPLA6 gene in Boucher-Neuhäuser syndrome. J Hum Genet. 2015;60:217–220. [DOI] [PubMed] [Google Scholar]
- 25. Wiethoff S, Bettencourt C, Paudel R, et al. Pure cerebellar ataxia with homozygous mutations in the PNPLA6 gene. Cerebellum. 2017;16:262–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Langdahl JH, Frederiksen AL, Nguyen N, Brusgaard K, Juhl CB. Boucher Neuhäuser Syndrome—A rare cause of inherited hypogonadotropic hypogonadism. A case of two adult siblings with two novel mutations in PNPLA6. Eur J Med Genet. 2017;60:105–109. [DOI] [PubMed] [Google Scholar]
- 27. Teive HAG, Camargo CHF, Sato MT, et al. Different cerebellar ataxia phenotypes associated with mutations of the PNPLA6 gene in Brazilian patients with recessive ataxias. Cerebellum. 2018;17:380–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zheng R, Zhao Y, Wu J, et al. A novel PNPLA6 compound heterozygous mutation identified in a Chinese patient with Boucher-Neuhäuser syndrome. Mol Med Rep. 2018;18:261–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. DeNaro BB, Dhrami-Gavazi E, Rubaltelli DM, et al. Chorioretinal changes in a genetically confirmed case of Boucher-Neuhäuser syndrome. Retin Cases Brief Rep. 2021;15:179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Patsi O, De Beaufort C, Kerschen P, et al. A new PNPLA6 mutation presenting as Oliver McFarlane syndrome. J Neurol Sci. 2018;392:1–2. [DOI] [PubMed] [Google Scholar]
- 31. Coutelier M, Hammer MB, Stevanin G, et al. Efficacy of exome-targeted capture sequencing to detect mutations in known cerebellar ataxia genes. JAMA Neurol. 2018;75:591–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Salgado P, Carvalho R, Brandão AF, et al. Gordon Holmes syndrome due to compound heterozygosity of two new PNPLA6 variants—A diagnostic challenge. eNeurologicalSci. 2018;14:9–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. O'Neil E, Serrano L, Scoles D, et al. Detailed retinal phenotype of Boucher-Neuhäuser syndrome associated with mutations in PNPLA6 mimicking choroideremia. Ophthalmic Genet. 2019;40:267–275. [DOI] [PubMed] [Google Scholar]
- 34. Makuloluwa AK, Dodeja R, Georgiou M, et al. Oliver McFarlane syndrome and choroidal neovascularisation: A case report. Ophthalmic Genet. 2020;41:451–456. [DOI] [PubMed] [Google Scholar]
- 35. Donaldson L, Tarnopolsky MA, Martin JA, Rodriguez AR. Severe chorioretinal atrophy in Boucher-Neuhauser syndrome. Can J Ophthalmol. 2020;55:e26–e28. [DOI] [PubMed] [Google Scholar]
- 36. Sen K, Finau M, Ghosh P. Bi-allelic variants in PNPLA6 possibly associated with Parkinsonian features in addition to spastic paraplegia phenotype. J Neurol. 2020;267:2749–2753. [DOI] [PubMed] [Google Scholar]
- 37. D'Amore A, Tessa A, Casali C, et al. Next generation molecular diagnosis of hereditary spastic paraplegias: An Italian cross-sectional study. Front Neurol. 2018;9:981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu F, Ji Y, Li G, Xu C, Sun Y. Identification of Oliver-McFarlane syndrome caused by novel compound heterozygous variants of PNPLA6. Gene. 2020;761:145027. [DOI] [PubMed] [Google Scholar]
- 39. Emekli AS, Samanci B, Şimşir G, et al. A novel PNPLA6 mutation in a Turkish family with intractable Holmes tremor and spastic ataxia. Neurol Sci. 2021;42:1535–1539. [DOI] [PubMed] [Google Scholar]
- 40. Lisbjerg K, Andersen MKG, Bertelsen M, et al. Oliver McFarlane syndrome: Two new cases and a review of the literature. Ophthalmic Genet. 2021;42:464–473. [DOI] [PubMed] [Google Scholar]
- 41. Doğan M, Eröz R, Öztürk E. Chorioretinal dystrophy, hypogonadotropic hypogonadism, and cerebellar ataxia: Boucher-Neuhauser syndrome due to a homozygous (c.3524C>G (p.Ser1175Cys)) variant in PNPLA6 gene. Ophthalmic Genet. 2021;42:276–282. [DOI] [PubMed] [Google Scholar]
- 42. Locci S, Bianchi S, Tessa A, Santorelli FM, Mignarri A. Gordon Holmes syndrome caused by two novel mutations in the PNPLA6 gene. Clin Neurol Neurosurg. 2021;207:106763. [DOI] [PubMed] [Google Scholar]
- 43. Dong Y, Li XY, Wang XL, et al. Genetic, clinical and neuroimaging profiles of sporadic and autosomal recessive hereditary spastic paraplegia cases in Chinese. Neurosci Lett. 2021;761:136108. [DOI] [PubMed] [Google Scholar]
- 44. Suchowersky O, Ashtiani S, Au PB, et al. Hereditary spastic paraplegia initially diagnosed as cerebral palsy. Clin Park Relat Disord. 2021;5:100114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chung EJ, You E, Oh SH, et al. The first Korean family with Boucher-Neuhäuser syndrome carrying a novel mutation in PNPLA6. J Clin Neurol. 2022;18:233–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. He J, Liu X, Liu L, Zeng S, Shan S, Liao Z. Identification of novel compound heterozygous variants of the PNPLA6 gene in Boucher-Neuhäuser syndrome. Front Genet. 2022;13:810537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nanetti L, Di Bella D, Magri S, et al. Multifaceted and age-dependent phenotypes associated with biallelic PNPLA6 gene variants: Eight novel cases and review of the literature. Front Neurol. 2021;12:793547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu W, Xie Y, Ma J, Luo X, Nie P, Zuo Z, et al. IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics (Oxford, England). 2015;31:3359–3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hein ND, Stuckey JA, Rainier SR, Fink JK, Richardson RJ. Constructs of human neuropathy target esterase catalytic domain containing mutations related to motor neuron disease have altered enzymatic properties. Toxicol Lett. 2010;196:67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Brnich SE, Abou Tayoun AN, Couch FJ, et al. Recommendations for application of the functional evidence PS3/BS3 criterion using the ACMG/AMP sequence variant interpretation framework. Genome Med. 2019;12:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tavtigian SV, Harrison SM, Boucher KM, Biesecker LG. Fitting a naturally scaled point system to the ACMG/AMP variant classification guidelines. Hum Mutat. 2020;41:1734–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Synofzik M, Hufnagel RB, Züchner S.. PNPLA6 Disorders. In: Adam MP, Everman DB, Mirzaa GM, et al., eds. GeneReviews®. University of Washington; 1993. [Google Scholar]
- 54. Akassoglou K, Malester B, Xu J, Tessarollo L, Rosenbluth J, Chao MV. Brain-specific deletion of neuropathy target esterase/swisscheese results in neurodegeneration. Proc Natl Acad Sci U S A. 2004;101:5075–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available from the corresponding author upon request.