Abstract
The PA subunit of the influenza virus polymerase complex is a phosphorylated protein that induces a proteolytic process that decreases its own accumulation levels and those of coexpressed proteins. The amino-terminal third of the protein is responsible for the induction of proteolysis. We mutated five potential casein kinase II phosphorylation sites located in the amino-terminal third of the protein. Mutations affecting position 157 almost completely abrogated proteolysis induction, whereas a mutation at position 162 produced a moderate decrease and mutations at positions 151, 200, and 224 did not affect proteolysis induction. Reconstitution of the influenza virus polymerase in vivo with viral model RNA containing the chloramphenicol acetyltransferase (CAT) gene indicated that the CAT activity obtained correlated with the capacity of each PA mutant to induce proteolysis. RNA protection assays of the products obtained with viral polymerase, reconstituted in vivo with model RNAs, indicated that mutations at position 157 led to a selective loss of the ability to synthesize cRNA from the viral RNA template but not to transcribe viral RNA, while a mutation affecting position 162 showed an intermediate phenotype. Collectively, these data provide a link between PA-mediated induction of proteolysis and the replication activity of the polymerase.
The influenza virus RNA polymerase is a heterotrimer formed by the PB1, PB2, and PA subunits. It associates with nucleoprotein (NP)-complexed viral RNA (vRNA) to form virion ribonucleoproteins (vRNPs). In influenza virus-infected cells, the vRNPs direct two types of RNA synthesis: mRNA synthesis (transcription) and vRNA amplification (replication). For mRNA synthesis, 5′-capped oligonucleotides derived from cellular mRNAs by cap-snatching are used as primers (21). These primers are elongated until polyadenylation occurs at a signal of five to seven U residues close to the 5′ end of the template (24, 32–34). Replication, in contrast, occurs without primer. The vRNA template is copied to form full-length positive-stranded RNA (cRNA), which serves as a template for vRNA synthesis (18, 21). Free NP is required as an antitermination factor to ignore the polyadenylation signal during the synthesis of cRNA (39). However, a detailed picture of the mechanism of the transcription-replication switch is still lacking.
The PB1 subunit contains several sequence motifs characteristic of the vRNA-dependent RNA polymerases (31). These motifs have been shown to be essential for vRNA synthesis (6), suggesting that PB1 is the polymerase itself. PB2 protein binds CAP1 structures (7, 41) and might contain the endonucleolytic activity responsible of the cleavage of host mRNA precursors (8, 23). The phenotype of viral temperature-sensitive (ts) mutants indicates that the PA subunit is involved in vRNA replication (reviewed in reference 25), but its precise role in this process is unknown. The PA subunit induces a generalized proteolytic process when expressed individually from cloned cDNA (36), and the amino-terminal third of the molecule (positions 1 to 247) is sufficient to activate this proteolysis (38). We recently showed that the PA protein is phosphorylated in vivo and that it is a substrate of casein kinase II in vitro (37). PA protein contains 11 potential phosphorylation sites for casein kinase II in its molecule, 8 of them located in a cluster inside the first 247 N-terminal amino acids. Therefore we produced point mutations of several putative casein kinase II phosphorylation sites located at the amino-terminal third of the protein and studied the consequences of these genetic changes in the activity of the mutated PA proteins. Some of these PA mutants presented decreased ability to induce proteolysis. Interestingly, the capacity of these mutants to support replication of model vRNA in a polymerase reconstituted in vivo from cloned cDNAs strongly correlated with their proteolysis induction, but all mutants were as active as wild-type (wt) PA in their transcription activity.
MATERIALS AND METHODS
Biological materials.
The COS-1 cell line (14) was obtained from Y. Gluzman. Cell cultures were grown in Dulbecco's modified Eagle medium (DMEM) containing 5% fetal bovine serum. Vaccinia virus vTF7-3 is a recombinant virus that expresses the phage T7 RNA polymerase (12) and was provided by B. Moss.
Recombinant plasmids encoding the influenza virus polymerase and NP proteins (pGPB1, pGPB2, pGPA, pGNP, and pGNPpA) have been described (26, 29). A plasmid-expressing ribozyme construct that originates in vivo a vNSCAT model RNA (pT7vNSCAT-RT) was generated as follows: first, the intermediate cloning vector pUC19RT was generated by inserting the SmaI-XbaI fragment of plasmid 2.0, which contains the cDNA copy of the hepatitis δ virus ribozyme and the T7 RNA terminator (1), into pUC19. Next, a PCR fragment was amplified using as template plasmid pIVACAT1/S (30). The primers used were 5′-AGCAAAAGCAGG-3′, which is complementary to the 3′ end of the NS RNA segment, and 5′-GCCTGGTACCTAATACGCCTCACTATAAGTAGAAACAAGG-3′, which contains an Asp718 restriction site, the T7 RNA polymerase promoter (underlined) and the 5′-terminal sequence of NS segment. Finally, the PCR fragment was digested with Asp718 restriction nuclease and ligated with the SmaI/Asp718-digested pUC19RT. A plasmid-expressing ribozyme construct that originates in vivo a cNSCAT model RNA (pT7cNSCAT-RT) was kindly provided by P. Palese. The deleted versions of these plasmids, pT7vNSΔCAT-RT and pT7cNSΔCAT-RT, were constructed from the original undeleted plasmids by making an internal deletion inside the chloramphenicol acetyltransferase (CAT) gene by digestion with BsmI endonuclease and autoligation.
Construction of mutants.
Mutant plasmids pGPAT151A, pGPAT157A, and pGPAT162A were produced with the Transformer site-directed mutagenesis kit (Clontech), using as template pGPA plasmid and a degenerated oligonucleotide with the sequence 5′-TTC(A/G)CTGGGGAGGAAATGGCC(A/G)CAAAGGCCGACTAC(A/G)CTCTT-3′ as mutagenic primer. Plasmids pGPAT157E, pGPAT200A, and pGPAS224A were constructed using the same protocol, with pGPA as template and an oligonucleotides with the sequence 5′-GGAAATGGCCGAAAAGGCCG-3′, 5′-CTTCAATCGCTTCTTCGCC-3′, or 5′-CAAGGCACGCGAAGTTCGGCGG-3′ as mutagenic primer.
To construct a vaccinia virus able to express the PAT157A mutant gene, the mutant cDNA was cloned into expression vector pTM1 (12) so that the ATG codon of the gene became part of the unique NcoI site of the plasmid. Plasmid pTMPAT157A was then used to transfer the influenza virus gene into the thymidine kinase gene locus of vaccinia virus by in vivo recombination. Recombinant VPA-T157A thus generated was capable of expressing high levels of PAT157A protein by dual infection with vTF7-3 virus.
Infection and transfection.
For vaccinia virus infection, COS-1 cells were inoculated with vTF7-3 plus either VPA or VPA-T157A vaccinia virus recombinants at 5 PFU per cell of each virus in DMEM plus 2% fetal bovine serum. After adsorption for 1 h, the inoculum was removed and the cultures were incubated for 24 h at 37°C in the same medium. For transfection experiments, subconfluent monolayers of COS-1 cells were infected with vTF7-3 virus at 5 to 10 PFU per cell. After 1 h at 37°C, cells were transfected with the indicated plasmids by the liposome-mediated method using cationic liposomes (35). The total amount of transfected DNA per dish was kept constant by adjustment, if necessary, with pGEM3 plasmid. Cells were incubated at 37°C in serum-free DMEM for 16 to 20 h.
Western blotting.
Western blotting was done as described previously (36). The following primary antibodies were used: for PB2 protein, PARB2 8N, a rabbit antiserum prepared by immunizing animals with a carboxyl-truncated form of PB2 (1/100 dilution); for PA protein, a mixture of monoclonal antibodies (MAbs) (MAbs 9, 11, 12, and 14; 1/40 dilution from culture supernatant [2]); and for PB1 protein, a rabbit antiserum prepared by immunization with a fusion protein containing the N-terminal 250 amino acids (17) (1/100 dilution).
Two-dimensional analysis.
Cultures of COS-1 cells in 16-mm-diameter dishes were mock infected or infected with vTF7-3 virus plus either VPA or VPA-T157A at 5 PFU/cell for each virus. For radioactive phosphate incorporation, cells were starved for 90 min in phosphate-free DMEM and labeled for 2 h with 1 mCi of 32Pi (Amersham) per ml in the same medium. Cells were washed twice with phosphate-buffered saline and resuspended in lysis buffer. Two-dimensional gel electrophoresis was performed as described previously (37). Some gels were transferred to nitrocellulose membranes as reported previously (37) and processed for Western blotting as described above.
CAT assays.
Influenza virus RNA polymerase activity was reconstituted by the infection-transfection protocol, as described previously (26, 29). For CAT assays, total cell extracts were prepared in 0.25 M Tris-HCl, pH 7.5, by three cycles of freezing-thawing and CAT activity was analyzed by the phase extraction method (9, 28).
RNA analysis.
vTF7-3-infected COS-1 cells (60-mm dishes) were transfected with 1.5 μg each of pGPB1 and pGPB2 plasmids plus 300 ng of pGPA or variable amounts of mutant plasmids, 6 μg of pGNPpA, and 2 μg of ribozyme construct pT7vNSΔCAT-RT or pT7cNSΔCAT-RT, as described above. Total RNAs were isolated 14 to 16 h postinfection using the Ultraspect RNA isolation reagent from Biotex. The poly(A)+ RNA was isolated as described previously (42) except that 1% Sarkosyl was substituted for sodium dodecyl sulfate in all buffers and the binding and washing steps were done at 4°C. Oligo(dT) retained and unretained RNAs were ethanol precipitated and used for protection assays as previously described (29).
RESULTS
Characterization of PA point mutants affected in the induction of proteolysis.
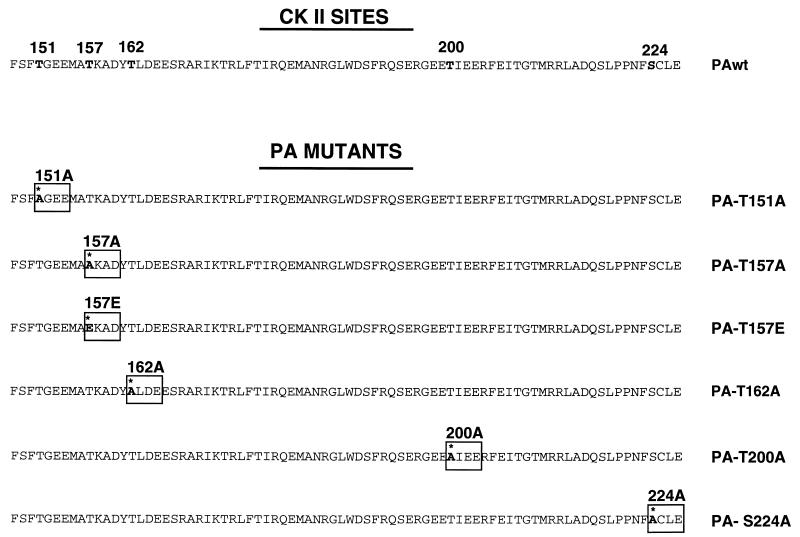
We have previously shown that the expression of PA protein leads to a generalized proteolysis that reduces its own steady-state level and that of coexpressed proteins. Deletion analysis revealed that the first 247 amino-terminal amino acids are sufficient to bring about the induction of proteolysis. Recently, it has been described that PA is phosphorylated in vivo by a cellular kinase and in vitro by casein kinase II (37). Among the 11 potential phosphorylation sites for casein kinase II present in the PA sequence, we observed a cluster of 8 sites within the 247 amino-terminal residues of the protein. Then we tested whether alteration of several potential casein kinase II phosphorylation sites, located around the two regions of the protein where the nuclear translocation signal has been identified in this amino-terminal part (amino acids 124 to 139 and 186 to 247) (27), could affect the activity of proteolysis induction caused by PA. We carried out site-directed mutagenesis to obtain T-to-A or S-to-A single mutants for these sites and analyzed their activity as inducers of proteolysis. A scheme of the point mutants generated is presented in Fig. 1.
FIG. 1.
Scheme of mutant PA proteins. The sequences of wt PA protein in the region comprising amino acids 148 to 227 and the five potential sites for casein kinase II (CK II) phosphorylation in this region are shown. Positions 151, 157, 162, 200, and 224 were mutated from threonine or serine to alanine to generate PA-T151A, PA-T157A, PA-T162A, PA-T200A, and PA-S224A, respectively. In addition, position 157 was mutated to glutamic acid to generate mutant PA-T157E.
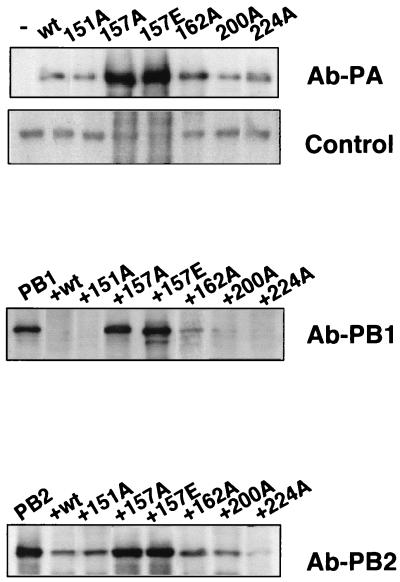
As previously described, PA protein itself is a substrate for the proteolytic activity induced by its own expression, and the steady-state level of a PA mutant is inversely correlated to its capacity to induce proteolysis (38). To check the possible loss of this activity in the point mutants, wt PA or the different mutants were expressed by transfection of the corresponding plasmids into vTF7-3 virus-infected COS-1 cells. In addition, each PA-expressing plasmid was cotransfected with plasmids expressing PB1 or PB2 proteins as reporters. Total cell extracts from the infected-transfected cells were used to analyze the levels of accumulation of PA or the reporter proteins by Western blotting. The results are presented in Fig. 2. The accumulation of mutant PA-T157A was much higher than that of wt PA (Fig. 2, Ab-PA). Mutant protein PA-T162A presented an intermediate accumulation level (Fig. 2, Ab-PA) (38), while mutants PA-T151A, PA-T200A, and PA-S224A showed levels indistinguishable from the wild type. Conversely, the stationary levels of PB1 (Fig. 2, Ab-PB1) and PB2 (Fig. 2, Ab-PB2) proteins were strongly reduced by coexpression of wt PA, PA-T151A, PA-T200A, or PA-S224A; slightly reduced by coexpression of PA-T162A (Fig. 2) (38); and unaffected by coexpression of PA-T157A protein. As loading control, we included the signal obtained by cross-reaction with a protein of the COS-1-infected cells by using the Ab-PA antibody (Fig. 2, Control). These results suggested that elimination of the potential phosphorylation site at position 157, and to a lesser extent at position 162, leads to a reduction in the ability of the mutant proteins to induce proteolysis.
FIG. 2.
Effect of PA point mutations on proteolysis induction. Cells were infected with vTF7-3 and transfected with 0.5 μg of each reporter plasmid (pGPB1 or pGPB2) alone or in combination with 1 μg of either pGPA or pGPA mutant plasmids. Total cell extracts were prepared and analyzed by Western blotting for the accumulation of PA (Ab-PA) or each reporter protein (Ab-PB1 or Ab-PB2) using specific antibodies. As a loading control, the signal obtained with a protein of the COS-1-infected cells that cross-reacts with the Ab-PA antibody was included (Control).
PA-T157A mutant protein is defective in phosphorylation.
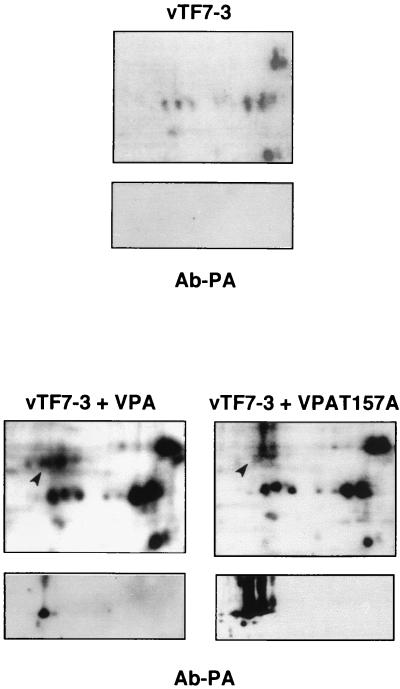
Since the influenza virus PA polymerase subunit is a phosphoprotein (37), we tested whether a Thr-to-Ala substitution affected the level of phosphorylation of the protein. As mutant PA-T157A showed a severe phenotype in the loss of induction of proteolysis, this mutant was chosen to follow phosphate incorporation studies. COS-1 cells were infected with vTF7-3 virus either alone or together with VPA or VPA-T157A recombinant viruses and labeled with 32Pi for 2 h at 20 h postinfection. The cells were collected and the extracts were processed for two-dimensional analysis followed by transfer to nitrocellulose. The filters were autoradiographed and then analyzed by Western blotting to identify specific PA spots (Fig. 3). The top panels show the 32P-labeled spots, while the bottom panels (Ab-PA) show the immunodetection. Using the same amounts of cell extracts, VPA-T157A-infected cells showed a much higher accumulation of PA protein than VPA-infected cells, consistent with the decreased induction of proteolysis of the mutant protein. The comparison of the 32P-labeled spots present in wt or mutant infected cells (lower part, top panels) revealed the presence of at least four PA-specific phosphorylated spots in the cell extract from VPA-infected cells which were not detectable in VPA-T157A-infected cells, in spite of the much higher PA accumulation levels in the latter. It should be emphasized that the main PA-specific spot is a phosphoisoform in VPA-infected cells (Fig. 3, arrow), but it is not labeled in VPA-T157A cells (Fig. 3, arrow).
FIG. 3.
PA157 protein is underphosphorylated. COS-1 cells were infected with vTF7-3 alone or in combination with VPA wt (vTF7-3+VPA) or VPA-T157A mutant (vTF7-3+VPA-T157A) and pulse labeled with 32Pi for 2 h. Total cell extracts were analyzed by two-dimensional gel electrophoresis. The gels were transferred to nitrocellulose membranes and autoradiographed (top panels). The pH gradient from right to left is acidic to basic. The same membranes whose autoradiographs are shown were developed with MAbs specific for PA protein (bottom panels [Ab-PA]). The arrow shows the phosphorylation of the main PA isoform (marked by a solid lane) in wt PA, while the arrow indicates the absence of 32P label in the most prominent PA isoform present in PA-T157A-infected cells.
As it has been described that substitution of phosphorylated threonine or serine residues by aspartic or glutamic acids can mimick a constitutive phosphorylated state of the protein (13, 19, 20), we constructed mutant PA-T157E, in which the original threonine has been changed to a glutamic acid residue (Fig. 1), and checked its ability to induce proteolysis. The results are presented in Fig. 2. Mutant PA-T157E did not revert to the wt phenotype but showed a proteolysis defect more intense that mutant PA-T157A. Thus, mutant PA-T157E showed higher accumulation levels than PA-T157A when expressed individually (Fig. 2, Ab-PA) and was unable to reduce the steady-state levels of PB1 and PB2 when these proteins were used as reporters (Fig. 2, Ab-PB1 and Ab-PB2). These results indicated that although the T157A mutation leads to a dephosphorylated PA protein, a clear correlation between phosphorylation and induction of proteolysis could not be established.
PA point mutants defective in proteolysis are able to reconstitute active influenza virus polymerase complexes.
The phenotypic changes shown by some of the PA mutants could be a reflection of gross alterations in the folding of the protein that would abolish in toto their biological activity. To test this possibility, we studied the capacity of PA mutants to form active RNA polymerase complexes. Reconstitution experiments were followed by transfection of viral cDNAs and a CAT model vRNA (26). Thus, plasmids expressing PB1, PB2, NP, and either wt or mutant PA proteins were transfected into vTF7-3-infected COS-1 cells. The cells were further transfected with an NS-CAT model vRNA, and the CAT activity generated was used as a measure of the RNA polymerase activity reconstituted. In order to have a meaningful comparison of the biological activities of wt PA and mutant proteins, the reconstitution has to fulfill two conditions: (i) the levels of expression of PA proteins have to be similar, and (ii) the rest of the elements of the system have to be present at a saturating level. To comply with the first condition, dose-response experiments were done to determine the relative amounts of wt PA and mutant-expressing plasmids that produced similar levels of protein in the context of the reconstitution of the polymerase. In this way we could have an estimation of the polymerase specific activity of each PA protein. Western blotting of PB1 was carried out in parallel and used as a control that the other elements of the system accumulated as expected regarding the presence of a proteolytic or nonproteolytic PA in the polymerase complex. The CAT activities obtained under these conditions are shown in Table 1. The data indicated that, whereas the polymerases reconstituted with proteolytic PA proteins (PA-T151A, PA-T200A, or PA-S224A) were similar to the wild type, the enzymatic activities of the polymerases reconstituted with the nonproteolytic PA-T157A and PA-T157E were lower. The PA-T162A mutant was intermediate, both in proteolytic induction and CAT activity. These results indicate that the mutant PA proteins are able to interact with the other polymerase components to form an active enzyme. To check that the other elements in the system were present in excess over PA protein, CAT activity was assayed in extracts from COS-1 cells transfected with PA-expressing plasmids and decreasing amounts of plasmids expressing PB1, PB2, and NP influenza virus proteins. Although the accumulation of the other polymerase components, as measured by PB1 detection, was diminished when PA proteins able to induce proteolysis were used, the enzymatic activity did not change even when 10 times less PB1, PB2, and NP plasmids were used for transfection.
TABLE 1.
CAT activity of polymerases reconstituted with PA point mutantsa
| PA protein | CAT activity (%) |
|---|---|
| wt | 100 |
| T151A | 88 ± 26 |
| T157A | 30 ± 10 |
| T157E | 13 ± 7 |
| T162A | 51 ± 10 |
| T200A | 91 ± 8 |
| S224A | 124 ± 41 |
COS-1 cells (35-mm dishes) were infected with vTF7-3 (10 PFU) and transfected with 250 ng of wt PA or variable amounts of mutated PA proteins plus 1 μg of pGPB1 and pGPB2, 2 μg of pGNPpA plasmid, and 2 μg of a ribozyme construct expressing an influenza virus-like NSCAT RNA. After Western blot analysis, extracts containing equal amounts of PA proteins were processed to determine CAT activity, as described in Materials and Methods. Enzymatic activities are shown as percentages of the value obtained with wt PA-expressing extracts.
vRNA polymerases reconstituted with nonproteolytic PA proteins are able to transcribe but unable to replicate.
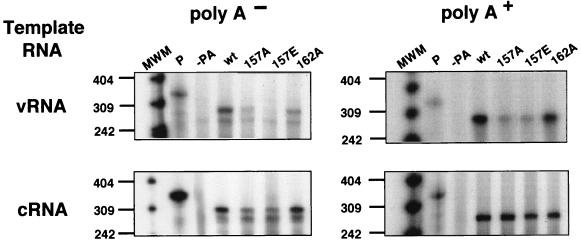
The decrease in the CAT activity obtained by reconstitution of viral polymerase with nonproteolytic PA mutants could be the consequence of either reduced replication, reduced transcription, or both. To characterize the phenotype of the mutant polymerases at the RNA level, we determined the cRNA, vRNA, and mRNA levels accumulated after polymerase reconstitution in vivo, as described previously (29). To analyze this, cultures of vTF7-3-infected COS-1 cells were transfected with plasmids expressing NP, PB1, PB2, and either wild-type or mutated PA proteins. In addition, the cells were transfected with ribozyme constructs that produce either vNSΔCAT or cNSΔCAT model RNAs in vivo. Each experiment included a dose effect of mutant PA-expressing plasmids, and the relative accumulation of wt and mutant PA proteins was first ascertained by Western blotting. Cultures parallel to these showing similar accumulation levels of wt and mutated PAs were processed for RNA analysis. Total cell RNA was isolated and separated into poly(A)+ and poly(A)− fractions. These RNAs were then analyzed by RNase protection assays using probes specific to detect cRNA, vRNA, and mRNA. The results are presented in Fig. 4. The PA-T157A- and PA-T157E-containing polymerases were consistently unable to synthesize cRNA from the vRNA model (Fig. 4, vRNA [A−]); as a consequence of the lack of genome amplification, the accumulation of mRNA was severely affected (Fig. 4, vRNA [A+]). The synthesis of vRNA from the cRNA model was reduced but not abolished (Fig. 4, cRNA [A−]), and mRNA production was not affected. Quantitation of several experiments indicated that the relative amount of cRNA synthesized from the system reconstituted with PA-T157A was around 10 and 5% for PA-T157E-reconstituted polymerase compared with that for wt PA-containing polymerase. PA-T162A-reconstituted polymerase showed an intermediate phenotype. These results indicate that alterations of the capacity of PA proteins to induce proteolysis are parallel to a defect in vRNA replication, particularly at the vRNA-to-cRNA step, but do not affect vRNA transcription.
FIG. 4.
Nonproteolytic PA point mutants are defective in replication. Accumulation of RNAs in a transcription-replication system reconstituted with negative-sense (top panels) or positive-sense (bottom panels) RNA templates is shown. Cultures of COS-1 cells were infected with vTF7-3 and transfected with a mixture of pGPB1, pGPB2, pGNPpA, and pT7vNSΔCAT-RT (vRNA) or pT7cNSΔCAT-RT (cRNA) plasmids as well as the appropriate amounts of wt pGPA or mutated pGPA plasmids to express equal amounts of wtPA or mutated PA proteins. Alternatively, the transfection mixtures were deficient in one of the plasmids, as indicated. After transfection, total cell RNA was isolated and fractionated on oligo(dT) columns. Aliquots of each RNA sample were assayed by RNase protection, as indicated in Materials and Methods. The protected RNAs were analyzed in a 4% polyacrylamide-urea gel. P, undigested probe; poly A− and poly A+, results obtained with poly(A)− or poly(A)+ RNA, respectively. The numbers to the left indicate the lengths of the molecular weight markers (MWM), in nucleotides.
DISCUSSION
Pleiotropic effects of a single amino acid change in PA protein.
The changes at position 157 in the PA protein sequence led to profound alterations in its phenotype. The T157A mutant protein was underphosphorylated, suggesting that Thr at position 157 is critical for this modification of the protein, and, interestingly, it was defective in the induction of proteolysis (Fig. 2). These results, together with the loss of proteolytic activity produced by the T-to-E change at the same position, suggest that mutations introduced at position 157 of PA affect an especially sensitive regulatory region of the protein. Alternatively, the mutations at position 157 might induce a complete misfolding of the protein. Such a possibility was ruled out because active complexes were obtained when the polymerase was reconstituted in vivo by transfection of all subunits, the NP and a vNSCAT model RNA, although the specific activity of mutant PA-containing polymerases was lower compared to the wt polymerase (Table 1). Altogether, these results are consistent with a regulatory role for the amino-terminal region of the PA protein.
The phenotype of PA157 mutants indicates a link between proteolytic activity and vRNA replication.
In addition to the phenotypic changes discussed above, polymerases containing PA mutations at position 157 were found to be defective in vRNA replication. Thus, the level of protected RNA signal corresponding to cRNA was very reduced when the polymerase was reconstituted with model vRNA and was slightly lowered when a model cRNA was used in the reconstitution assay (Fig. 4). In contrast to their inability to replicate, these mutant polymerases were capable of transcribing model vRNAs (Fig. 4). Interestingly, polymerase reconstituted with PA-T162A, which presented an intermediate phenotype as an inducer of proteolysis, was only partially capable to synthesize cRNA from a model vRNA (Fig. 4), indicating that a correlation exists between the capacity of a particular PA mutant to induce proteolysis and its ability to replicate. In this context it should be emphasized that the other PA point mutants used in this work, which preserved the ability to induce proteolysis, were similarly active in the reconstitution of functional polymerase complexes, further indicating a correlation between proteolysis induction and enzymatic activity.
The replication-defective phenotype of polymerases with mutant PA proteins is in line with the phenotype of viral ts mutants affected in the PA gene (reviewed in reference 25). The fact that these mutant PA-containing polymerases were able to sustain transcription, but not replication, suggests that PA protein is involved in the transcription-replication shift. This mechanistic change implies alterations in three steps: (i) initiation of RNA synthesis should become primer independent, (ii) RNA chain elongation has to be coupled to RNA encapsidation with NP molecules, and (iii) polyadenylation (i.e., premature termination) should be inhibited to allow full-length synthesis of cRNA template. For antitermination and polyadenylation, a complex cis signal is required that includes an oligo(U) sequence (34) and RNA secondary structures involving the ends of the genomic segments (10, 11, 32, 33) that may mediate interaction with the polymerase (15, 16). The coupling of replicative RNA synthesis and encapsidation is reflected by the requirement of newly synthesized NP for replication to occur (4) and may be mediated by specific polymerase-NP interactions (5). However, the change in the mechanism of RNA synthesis initiation is not understood. In the transcription-replication shift, the use of a capped oligonucleotide as a primer has to be inhibited. In this regard, the results described in this report, which show a concomitant defect in RNA replication and proteolysis induction by the T157A and T157E mutations in PA protein, suggest possible regulation of the viral polymerase by proteolytic modification. Although such a possibility would be a novel mechanism in the Orthomyxoviridae, it is not without precedent in other virus families. Thus, synthesis of negative-polarity versus positive-polarity RNA in Sindbis virus is regulated by proteolytic processing of P123 precursor protein. The nsP4 RNA polymerase, together with the P123 precursor, is responsible for the synthesis of negative-polarity RNA, while elimination of the latter by the activity of the nsP2 proteinase is required to switch to positive-polarity RNA synthesis (22, 40). In the case of poliovirus, the proteolytic processing of a cellular factor has been reported as necessary for the synthesis in vitro of complete virion sense RNA (3). It is conceivable that a proteolytic alteration of the polymerase or the NP is important for influenza virus replication to occur. Such an alteration might block its capacity for the generation or usage of a capped primer and prompt de novo RNA synthesis initiation. Alternatively, the modification of a cellular cofactor might be required for vRNA replication. In this regard, identification of the specific protein target will be an important goal for future research.
ACKNOWLEDGMENTS
We are indebted to Agustín Portela and José A. Melero for critical comments on the manuscript. We thank B. Moss, P. Palese, and T. Zürcher for providing biological materials. The technical assistance of Y. Fernández and J. Fernández is gratefully acknowledged.
J. Ortega was a fellow of Instituto de Estudios Turolenses, and P. Gastaminza was a fellow of Gobierno Vasco. This work was supported by Programa Sectorial de Promoción General del Conocimiento (grants PM-0015, PB94-1542, and PB97-1160).
REFERENCES
- 1.Ball L A. Cellular expression of a functional nodavirus RNA replicon from vaccinia virus vectors. J Virol. 1992;66:2335–2345. doi: 10.1128/jvi.66.4.2335-2345.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bárcena J, de la Luna S, Ochoa M, Melero J A, Nieto A, Ortín J, Portela A. Monoclonal antibodies against the influenza virus PB2 and NP polypeptides interfere with the initiation step of viral mRNA synthesis in vitro. J Virol. 1994;68:6900–6909. doi: 10.1128/jvi.68.11.6900-6909.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barton D J, Black E P, Flanegan J B. Complete replication of poliovirus in vitro: preinitiation RNA replication complexes require soluble cellular factors for the synthesis of VPg-linked RNA. J Virol. 1995;69:5516–5527. doi: 10.1128/jvi.69.9.5516-5527.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaton A R, Krug R M. Transcription antitermination during influenza viral template RNA synthesis requires the nucleocapsid protein and the absence of a 5′ capped end. Proc Natl Acad Sci USA. 1986;83:6282–6286. doi: 10.1073/pnas.83.17.6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biswas S K, Boutz P L, Nayak D P. Influenza virus nucleoprotein interacts with influenza virus polymerase proteins. J Virol. 1998;72:5493–5501. doi: 10.1128/jvi.72.7.5493-5501.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biswas S K, Nayak D P. Mutational analysis of the conserved motifs of influenza A virus polymerase basic protein 1. J Virol. 1994;68:1819–1826. doi: 10.1128/jvi.68.3.1819-1826.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blaas D, Patzelt E, Keuchler E. Identification of the cap binding protein of influenza virus. Nucleic Acids Res. 1982;10:4803–4812. doi: 10.1093/nar/10.15.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blok V, Cianci C, Tibbles K W, Inglis S C, Krystal M, Digard P. Inhibition of the influenza virus RNA-dependent RNA polymerase by antisera directed against the carboxy-terminal region of the PB2 subunit. J Gen Virol. 1996;77:1025–1033. doi: 10.1099/0022-1317-77-5-1025. [DOI] [PubMed] [Google Scholar]
- 9.de la Luna S, Martín J, Portela A, Ortín J. Influenza virus naked RNA can be expressed upon transfection into cells co-expressing the three subunits of the polymerase and the nucleoprotein from SV40 recombinant viruses. J Gen Virol. 1993;74:535–539. doi: 10.1099/0022-1317-74-3-535. [DOI] [PubMed] [Google Scholar]
- 10.Flick R, Neumann G, Hoffmann E, Neumeier E, Hobom G. Promoter elements in the influenza vRNA terminal structure. RNA. 1996;2:1046–1057. [PMC free article] [PubMed] [Google Scholar]
- 11.Fodor E, Palese P, Brownlee G G, García-Sastre A. Attenuation of influenza A virus mRNA levels by promoter mutations. J Virol. 1998;72:6283–6290. doi: 10.1128/jvi.72.8.6283-6290.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuerst T R, Earl P L, Moss B. Use of a hybrid vaccinia virus-T7 RNA polymerase system for expression of target genes. Mol Cell Biol. 1987;7:2538–2544. doi: 10.1128/mcb.7.7.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao Y, Lenard J. Multimerization and transcriptional activation of the phosphoprotein (P) of vesicular stomatitis virus by casein linase-II. EMBO J. 1995;14:1240–1247. doi: 10.1002/j.1460-2075.1995.tb07107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gluzman Y. SV40 transformed simian cells support the replication of early SV40 mutants. Cell. 1981;23:175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- 15.González S, Ortín J. Characterization of the influenza virus PB1 protein binding to viral RNA: two separate regions of the protein contribute to the interaction domain. J Virol. 1999;73:631–637. doi: 10.1128/jvi.73.1.631-637.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez S, Ortín J. Distinct regions of influenza virus PB1 subunit recognize vRNA and cRNA templates. EMBO J. 1999;18:3767–3775. doi: 10.1093/emboj/18.13.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.González S, Zürcher T, Ortín J. Identification of two separate domains in the influenza virus PB1 protein involved in the interaction with the PB2 and PA subunits: a model for the viral RNA polymerase structure. Nucleic Acids Res. 1996;24:4456–4463. doi: 10.1093/nar/24.22.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hay A J. Characterization of influenza virus RNA complete transcripts. Virology. 1982;116:517–522. doi: 10.1016/0042-6822(82)90144-1. [DOI] [PubMed] [Google Scholar]
- 19.Huang W, Erikson R L. Constitutive activation of Mek1 by mutation of serine phosphorylation sites. Proc Natl Acad Sci USA. 1994;91:8960–8963. doi: 10.1073/pnas.91.19.8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang L N, Englund N, Das T, Banerjee A K, Pattnaik A K. Optimal replication activity of vesicular stomatitis virus RNA polymerase requires phosphorylation of a residue(s) at carboxy-terminal domain II of its accessory subunit, phosphoprotein P. J Virol. 1999;73:5613–5620. doi: 10.1128/jvi.73.7.5613-5620.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krug R M, Alonso-Kaplen F V, Julkunen I, Katze M G. Expression and replication of the influenza virus genome. In: Krug R M, editor. The influenza viruses. New York, N.Y: Plenum Press; 1989. pp. 89–152. [Google Scholar]
- 22.Lemm J A, Rumenapf T, Strauss E G, Strauss J H, Rice C M. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 1994;13:2925–2934. doi: 10.1002/j.1460-2075.1994.tb06587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Licheng S, Summers D F, Peng Q, Galarza J M. Influenza A virus polymerase subunit PB2 is the endonuclease which cleaves host cell mRNA and functions only as the trimeric enzyme. Virology. 1995;208:38–47. doi: 10.1006/viro.1995.1127. [DOI] [PubMed] [Google Scholar]
- 24.Luo G X, Luytjes W, Enami M, Palese P. The polyadenylation signal of influenza virus RNA involves a stretch of uridines followed by the RNA duplex of the panhandle structure. J Virol. 1991;65:2861–2867. doi: 10.1128/jvi.65.6.2861-2867.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahy B W J. Mutants of influenza virus. In: Palese P, Kingsbury D W, editors. Genetics of influenza viruses. Vienna, Austria: Springer-Verlag; 1983. pp. 192–253. [Google Scholar]
- 26.Mena I, de la Luna S, Albo C, Martín J, Nieto A, Ortín J, Portela A. Synthesis of biologically active influenza virus core proteins using a vaccinia-T7 RNA polymerase expression system. J Gen Virol. 1994;75:2109–2114. doi: 10.1099/0022-1317-75-8-2109. [DOI] [PubMed] [Google Scholar]
- 27.Nieto A, de la Luna S, Bárcena J, Portela A, Ortín J. Complex structure of the nuclear translocation signal of the influenza virus polymerase PA subunit. J Gen Virol. 1994;75:29–36. doi: 10.1099/0022-1317-75-1-29. [DOI] [PubMed] [Google Scholar]
- 28.Perales B, de la Luna S, Palacios I, Ortín J. Mutational analysis identifies functional domains in the influenza A virus PB2 polymerase subunit. J Virol. 1996;70:1678–1686. doi: 10.1128/jvi.70.3.1678-1686.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perales B, Ortín J. The influenza A virus PB2 polymerase subunit is required for the replication of viral RNA. J Virol. 1997;71:1381–1385. doi: 10.1128/jvi.71.2.1381-1385.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piccone M E, Fernandez S A, Palese P. Mutational analysis of the influenza virus vRNA promoter. Virus Res. 1993;28:99–112. doi: 10.1016/0168-1702(93)90129-b. [DOI] [PubMed] [Google Scholar]
- 31.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pritlove D C, Poon L L, Devenish L J, Leahy M B, Brownlee G. A hairpin loop at the 5′ end of influenza A virus virion RNA is required for synthesis of poly(A)+ mRNA in vitro. J Virol. 1999;73:109–114. doi: 10.1128/jvi.73.3.2109-2114.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pritlove D C, Poon L L, Fodor E, Sharps J, Brownlee G G. Polyadenylation of influenza virus mRNA transcribed in vitro from model virion RNA templates: requirements for 5′ conserved sequences. J Virol. 1998;72:1280–1286. doi: 10.1128/jvi.72.2.1280-1286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robertson J S, Schubert M, Lazzarini R A. Polyadenylation sites for influenza mRNA. J Virol. 1981;38:157–163. doi: 10.1128/jvi.38.1.157-163.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rose J K, Buonocore L, Whitt M A. A new cationic liposome reagent mediating nearly quantitative transfection of animal cells. BioTechniques. 1991;10:520–525. [PubMed] [Google Scholar]
- 36.Sanz-Ezquerro J J, de la Luna S, Ortín J, Nieto A. Individual expression of influenza virus PA protein induces degradation of coexpressed proteins. J Virol. 1995;69:2420–2426. doi: 10.1128/jvi.69.4.2420-2426.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanz-Ezquerro J J, Férnandez-Santarén J, Sierra T, Aragón T, Ortega J, Ortín J, Smith G L, Nieto A. The PA influenza virus polymerase subunit is a phosphorylated protein. J Gen Virol. 1998;79:471–478. doi: 10.1099/0022-1317-79-3-471. [DOI] [PubMed] [Google Scholar]
- 38.Sanz-Ezquerro J J, Zürcher T, de la Luna S, Ortín J, Nieto A. The amino-terminal one-third of the influenza virus PA protein is responsible for the induction of proteolysis. J Virol. 1996;70:1905–1911. doi: 10.1128/jvi.70.3.1905-1911.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shapiro G I, Krug R M. Influenza virus RNA replication in vitro: synthesis of viral template RNAs and virion RNAs in the absence of an added primer. J Virol. 1988;62:2285–2290. doi: 10.1128/jvi.62.7.2285-2290.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shirako Y, Strauss J H. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J Virol. 1994;68:1874–1885. doi: 10.1128/jvi.68.3.1874-1885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ulmanen I, Broni B A, Krug R M. Role of two of the influenza virus core P proteins in recognizing cap 1 structures (m7GpppNm) on RNAs and in initiating viral RNA transcription. Proc Natl Acad Sci USA. 1981;78:7355–7359. doi: 10.1073/pnas.78.12.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valcárcel J, Portela A, Ortín J. Regulated M1 mRNA splicing in influenza virus-infected cells. J Gen Virol. 1991;72:1301–1308. doi: 10.1099/0022-1317-72-6-1301. [DOI] [PubMed] [Google Scholar]