In this study, Ares et al. provide insight into the biogenesis of a novel class of spliced circular intron RNAs that are distinct from the RNA lariats that form during RNA splicing. Here, the spliceosome catalyzes a previously undefined 3′–5′ linkage, forming full-length and processed RNA circles that may facilitate intron transposition during genome evolution.
Keywords: circular RNA, intron transposition, reverse splicing
Abstract
Rare, full-length circular intron RNAs distinct from lariats have been reported in several species, but their biogenesis is not understood. We envisioned and tested a hypothesis for their formation using Saccharomyces cerevisiae, documenting full-length and novel processed circular RNAs from multiple introns. Evidence implicates a previously undescribed catalytic activity of the intron lariat spliceosome (ILS) in which the 3′-OH of the lariat tail (with optional trimming and adenylation by the nuclear 3′ processing machinery) attacks the branch, joining the intron 3′ end to the 5′ splice site in a 3′–5′ linked circle. Human U2 and U12 spliceosomes produce analogous full-length and processed circles. Postsplicing catalytic activity of the spliceosome may promote intron transposition during eukaryotic genome evolution.
Circular RNAs are widespread in nature. Among the first discovered were viroids and virusoids, whose replication requires a ribozyme to cleave and ligate monomeric circles from the long linear product of rolling circle replication (Lasda and Parker 2014). Other ribozymes, including the group I and group II self-splicing introns, produce circular RNAs by performing splicing-related side reactions during or after splicing (Grabowski et al. 1981; Murray et al. 2001; Lasda and Parker 2014). During the normal course of splicing, group II introns and the spliceosome produce tailed circles called lariats closed by a 2′–5′ linkage between the intron 5′ end and a 2′-OH of an internal branchpoint (BP) residue (Ruskin et al. 1984; Konarska et al. 1985; Reed and Maniatis 1985; Peebles et al. 1986; van der Veen et al. 1986). Nucleolytic trimming of lariat 3′ tails (Ruskin and Green 1985) can produce tailless lariats—circular intron RNAs closed by a 2′–5′ linkage (Gardner et al. 2012; Zhang et al. 2013; Lasda and Parker 2014; Neil and Fairbrother 2019). Using its standard first and second step splicing chemistry, the spliceosome can also create circular exons through backsplicing, an alternative splicing event that joins a downstream 5′ splice site (ss) to an upstream 3′ss (Nigro et al. 1991; Cocquerelle et al. 1992; Zaphiropoulos 1996; Salzman et al. 2012). If the natural order of the splice sites of group I, group II, or spliceosomal introns is permuted, they can also backsplice to create circular exons (Puttaraju and Been 1992; Ford and Ares 1994; Pasman et al. 1996). Circular introns are also produced by protein-based splicing of tRNA introns (Schmidt et al. 2019), archaeal rRNA and tRNA genes (Lykke-Andersen et al. 1997), and nonspliceosomal introns in protein-coding mRNAs such as for Saccharomyces cerevisiae HAC1 (Cherry et al. 2019; Schmidt et al. 2019) or numerous genes in Euglena (Gumińska et al. 2021). In addition to providing important insight into splicing mechanisms, some circular RNAs are functional (Lasda and Parker 2014; Neil and Fairbrother 2019), although the biological roles for most remain mysterious.
During their analysis of RNA-seq reads to identify intron lariat branchpoints, Fairbrother and colleagues (Taggart et al. 2012, 2017) noted that ∼3% of their intron-derived split reads appeared to show “branch formation” at the 3′ss. They validated the existence of intron-derived RNAs in which the 3′ss is directly joined to the 5′ss but noted that the reads lacked characteristic mutations created by reverse transcriptase (RT) reading through the lariat branch, suggesting that they were not 2′–5′-linked and thus not branch points. They speculated that the circles were formed after splicing by an unknown ligation mechanism. Similar circles have since been documented in the distantly related eukaryotic microbe Entamoeba histolytica (Mendoza-Figueroa et al. 2018). To distinguish them from tail-trimmed lariats that have also been called intron circles (Gardner et al. 2012; Zhang et al. 2013; Lasda and Parker 2014; Neil and Fairbrother 2019), we refer to this distinct class putatively lacking 2′–5′ linkages as “true circles.” Group II introns can also form circles that join their 5′ss to their 3′ss through a 2′–5′ linkage (Murray et al. 2001). We wondered whether the spliceosome creates intron circles and, if so, how. We searched in S. cerevisiae and found full-length intron circles for multiple introns—along with a new class of intron circles with extra nontemplated A residues or lacking 3′ nucleotides or both—that we refer to as “processed” circles. We provide evidence supporting a model for their formation that implicates persistent catalytic activity of the spliceosome after spliced exon release. By analogy with the transposable group II introns, we suggest that the modern spliceosome may still be able to transpose introns to new locations.
Results
How could the spliceosome create true intron circles that lack 2′–5′ linkages?
In addition to its two forward splicing reactions, branch formation (step 1) and exon ligation (step 2), the spliceosome catalyzes both reverse reactions (Tseng and Cheng 2008). In the reverse of the first step, exon 1 (E1) attacks the branch phosphate to regenerate the pre-mRNA. If the lariat tail could attack the branch phosphate in a similar way, a circular intron would be formed (Fig. 1A, top). We hypothesize that after the spliced exons are removed from the spliceosome, the resulting intron lariat spliceosome (ILS) would have an empty E1 binding site that could accommodate the lariat tail. Remodeling of the ILS into a C-complex-like conformation with the branch phosphate in the catalytic center could allow the lariat tail 3′-OH to attack and create a circular intron (Fig. 1A, bottom). To explore this hypothesis in a tractable system, we turned to yeast.
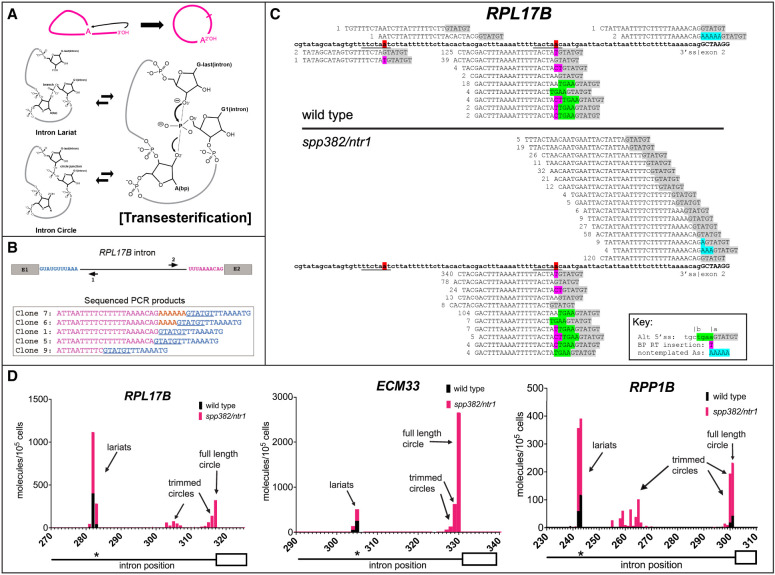
Figure 1.
A hypothesis and detection of intron circles in S. cerevisiae. (A) A possible mechanism for the spliceosome to create intron circles by nucleophilic attack of the lariat tail 3′-OH. (Top) Conversion of the lariat to a circle, releasing the 2′-OH of the A residue from the branch (arrow). (Bottom) Proposed chemical mechanism for attack of the lariat tail on the phosphate at the branch (bold P), starting with the lariat (top left) and then the transition state for the transesterification reaction (right), followed by circle formation (bottom left). The reactions are reversible. (B) RPL17B intron with “inverse” primers to capture circular intron junctions. Sequences of selected cloned PCR products are shown, with the 5′ss underlined and nongenomic As shown in gold. (C) High-throughput sequencing reveals a complex set of processed circular RPL17B introns whose abundance is dramatically increased by the spp382-1 mutation. The intron is in bold. Reads are aligned above (circles) or below (lariats), the 5′ss sequence is highlighted in gray, and nongenomic As are in blue. Deduplicated (unique) read counts from each library are shown in front of each read. Lariats derived from incorrect 5′ splice sites are in green. (D) Different introns produce distinct distributions of intron circles. Graphs show the junction locations (x-axis) and number of unique reads with junctions at that location (y-axis; calibrated to a spiked-in circular RNA) per 105 yeast cells from wild-type (black bars) or spp382-1 (pink bars) cells. The 3′ part of each intron (line) and its second exon (white box) are shown below the x-axis, with the asterisk indicating the position of the BP. Additional introns and data are shown in Supplemental Figure S1 and Supplemental Table S1.
Full-length and novel “processed” circular intron RNAs from multiple S. cerevisiae genes
To search for intron circles, we designed outward-pointing (“inverse”) PCR primers for a highly expressed intron-containing gene, RPL17B, and used them to amplify, clone, and sequence reverse transcripts extending from the 5′ end of the intron past predicted circle junctions (Fig. 1B; Supplemental Table S1). In addition to RNAs containing junctions between the 5′ss and 3′ss of the RPL17B intron analogous to those from mammals and Entamoeba (clone 1), we found unexpected reads that included nontemplated adenosine residues (As) between their 5′ splice sites and 3′ splice sites (clones 6 and 7), as well as those missing nucleotides from the 3′ end of the intron (clones 5 and 9). These unexpected products may also be circles, and we incorporate them into our analysis below, referring to them as “processed” circles. To test other introns, as well as to sample physiological, biochemical, and genetic conditions that might reveal insights into intron circle formation, we devised a multiplexed circular intron amplicon library preparation protocol using inverse primers carrying unique molecular indexes (UMIs) and sample barcoded adaptors for Illumina sequencing of a dozen introns (see the Supplemental Material). We sequenced libraries from wild type and a mutant strain (spp382-1) defective in spliceosome disassembly (Pandit et al. 2006) and readily detected circles for all 12 tested introns. We examined the reads from the RPL17B (Fig. 1C) and ACT1 (Supplemental Fig. S1) introns in detail. For RPL17B in wild-type cells (Fig. 1C, top), lariats (5′ss joined to the BP) are readily observed at both the annotated (TACTAAC) and an upstream cryptic BP (TTCTAAT). The RT mutational signature at the BP is evident (Fig. 1C, in purple) as is the use of an incorrect 5′ss upstream (Fig. 1C, green). Reads derived from an unprocessed full-length circular intron (like clone 1 in Fig. 1B), as well as from a circular intron with five extra nongenomic A residues (like clone 7), were observed. Two circles mapping upstream of the annotated RPL17B BP from the wild-type library would seem to rule out the hypothesis that circles form when the 3′ end of the lariat tail attacks the branch, since they appear to have been trimmed well past the annotated BP. Note, however, that they map downstream from the cryptic BP and could be derived from lariats branched at the cryptic BP via the proposed mechanism. In the spp382-1 mutant (Fig. 1C, bottom), a more complex group of circles missing nested sets of “tail” nucleotides was observed. We conclude that like humans, rodents, and Entamoeba (Taggart et al. 2012, 2017; Mendoza-Figueroa et al. 2018), S. cerevisiae makes full-length intron circles and, in addition, a novel class of processed intron circles. This processed class of circles can also be formed after processing of the lariat by trimming or polyadenylation by the proposed spliceosomal mechanism if the lariat tail retains a 3′-OH after processing.
To estimate the numbers of circles per cell, we prepared libraries from RNA samples spiked with known numbers of a synthetic circular RNA molecule for normalization (see the Supplemental Material). We found that intron circles are rare, occurring at approximately one to 10 per intron per 10,000 wild-type cells (Fig. 1D; Supplemental Fig. S1). The same classes of intron RNA circles observed for the RPL17B (Fig. 1C) and ACT1 (Supplemental Fig. S1) introns are also found (in different proportions) for other introns. In the spp382-1 mutant, circles increase to different extents for different introns—not at all for RPL24B, and ∼10-fold to 100-fold for others (Fig. 1D; Supplemental Fig. S1; also see below). In addition, some introns produce abundant processed circles (e.g., RPL17B, ACT1, and RPP1B), whereas others (e.g., RPL19A and ARF2) produce only untrimmed circles (Fig. 1D; Supplemental Fig. S1). Note that because RT readthrough at the lariat branch is inefficient, the number of lariats reads per circular read should not be directly compared. The number of lariats per cell is likely underestimated by this library method, and thus the y-axes of the plots in Figure 1D refer only to numbers of circles. The relative changes in lariats in the mutant versus wild-type comparisons are valid, however. Taken together, these initial observations show that circular intron RNAs arise by a rarely occurring but general process that operates on most or all introns, and that idiosyncratic differences in transcription rate and intron structure likely contribute to the different numbers and types of circles produced from each intron. Increased circular introns from the spp382-1 mutant provide the first hint that circles are formed by a reaction that is in competition with catalytic inactivation of the spliceosome. Furthermore, the processed intron circles fit the profile expected for RNAs that have visited the nuclear exosome and the TRAMP complex (Chlebowski et al. 2013; Zinder and Lima 2017) before they become circular, suggesting that if they are formed by the spliceosome, the lariat tail can exit and re-enter before circularization.
Intron circles are associated with the spliceosome and are not formed by tRNA ligase
Our working hypothesis invokes the spliceosome as the enzyme that closes the intron circles. The only other known RNA ligase in yeast is Trl1/Rlg1 (Phizicky et al. 1992), which ligates tRNA and HAC1 exons together during nonspliceosomal (tRNA) splicing in yeast (Phizicky et al. 1992; Cherry et al. 2018, 2019; Peschek and Walter 2019). We asked whether circular introns are associated with the spliceosome in vivo and asked separately whether their formation depends on Trl1 (Fig. 2). We tagged the spliceosomal protein Cef1 at its C terminus with a protein A (TAPS) tag in the spp382-1 mutant and performed IgG-sepharose chromatography to capture spliceosomes, detecting the presence of intron circles from ECM33 in the fractions (Fig. 2A,B). To follow the spliceosome through the purification, we used 32P-labeled oligonucleotides complementary to U2 snRNA (as a spliceosome marker) and SCR1 (a cytoplasmic SRP RNA) for detection by primer extension with RT (Supplemental Fig. S2). The ECM33 intron circle is clearly enriched in the bound (Fig. 2B, lane P) spliceosomal fraction of the Cef1-tagged extract but not in the untagged extract (Fig. 2B, cf. lanes 3 and 6), supporting the idea that the spliceosome forms the circular intron RNAs. To address the role of the tRNA ligase, we compared counts of circular introns (normalized to the spike-in control circle) in wild type and a strain lacking the gene (Cherry et al. 2018) and found that circle formation remains robust in the absence of the tRNA ligase Trl1 (Fig. 2C). Based on these observations, we conclude that circular introns are most likely created by an RNA ligation activity of the spliceosome.
Figure 2.
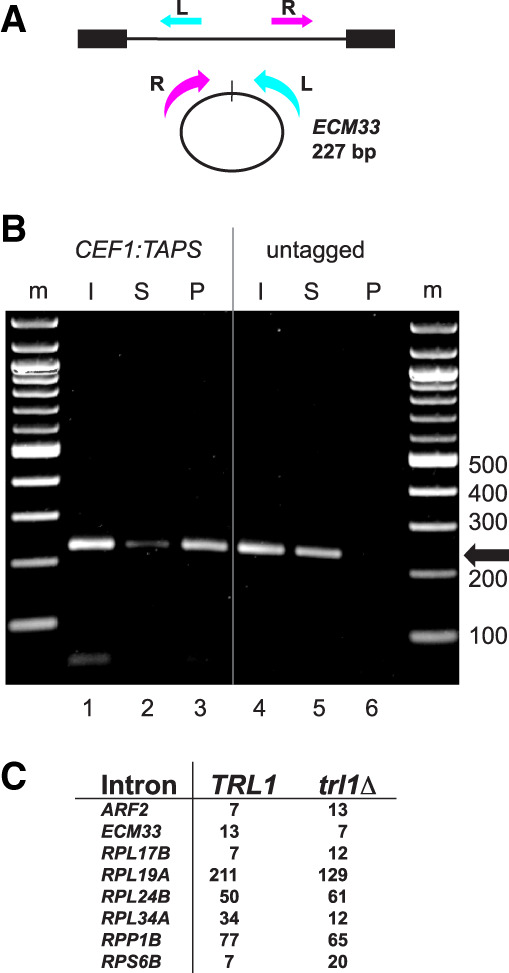
Intron circles are associated with the spliceosome and are not formed by the tRNA ligase Trl1. (A) Detecting circular ECM33 introns. Inverse PCR primers that create a 227 bp product from full-length ECM33 intron circles are shown; the R primer is 3′ of the branchpoint and cannot detect lariats. (B) Association of ECM33 intron circles with the spliceosome in spp382-1 cells. The spp382-1 mutant was fitted with a TAPS tag at the C terminus of its CEF1-coding region for affinity purification of spliceosomes. Extracts from this strain and an untagged control were prepared (input [I]) and bound in batches to IgG-sepharose. Unbound (supernatant [S]) and bound (pellet [P]) fractions were prepared. RNA from each fraction was used for RT-PCR to identify ECM33 circles (227 bp product). The pellet fraction from the tagged (but not the untagged) extract was enriched by primer extension in spliceosomal U2 snRNA and free of cytoplasmic SCR1 (Supplemental Fig. S2). (C) Robust detection of intron circles in a strain lacking tRNA ligase. The table shows spike-in-normalized circular intron counts for eight introns from libraries prepared from strains that differ by deletion of TRL1. Detailed data are shown in Supplemental Table S2.
Intron circles are not closed by a 2′–5′ linkage
Intron circles could be formed by one of the known activities of the spliceosome (either 2′–5′ or 3′–5′ linkage forming) or by some other activity. The absence of a mutational signature at the circle junction (Fig. 1C; Taggart et al. 2017) might not permit the conclusion that intron circles are closed by a 3′–5′ linkage if an internal (i.e., unbranched) 2′–5′ linkage is being formed and does not similarly impede RT; for example, as for a group II introns (Murray et al. 2001). To resolve this directly, we sought to distinguish lariats (with 2′–5′ branched junctions) and circles with at least one internal nonbranched 2′–5′ linkage from true circles (containing only 3′–5′ linkages). The lariat-debranching enzyme Dbr1 hydrolyzes 2′–5′ linkages if they are part of a branched structure (Ruskin and Green 1985; Montemayor et al. 2014; Clark et al. 2022), but we were unable to find published evidence for the ability of purified enzyme to cleave internal, unbranched 2′–5′ linkages. We designed several synthetic RNA oligonucleotides that differ by replacement of a single 3′–5′ linkage with an unbranched 2′–5′ linkage and treated them with purified recombinant yeast Dbr1 protein (Khalid et al. 2005) as described by Qin et al. (2016) (a kind gift of Aiswarya Krishnamohan and Jon Staley) (Fig. 3A). Dbr1 cleaves the oligos containing internal 2′–5′ linkages (Fig. 3A, lanes 2,6) without substantially cleaving 3′–5′ linkages (Fig. 3A, lanes 8,10) and therefore can be used to distinguish the classes of molecules described above. We prepared libraries from equivalent amounts of spp382-1 RNA either treated with Dbr1 or not and sequenced them, tabulating the numbers of lariats and circles for several introns (Fig. 3B). As expected, Dbr1 treatment greatly reduces the recovery of reads corresponding to lariat RNAs; however, intron circles in the same digest are resistant. We conclude that full-length and processed intron circles are not closed by either a branched or internal 2′–5′ linkage and therefore are likely to be composed entirely of 3′–5′ linkages. The bond formed by a reaction similar to the reverse of step 1 would be 3′–5′-linked, consistent with the hypothesis that circles are formed as proposed (Fig. 1A).
Figure 3.
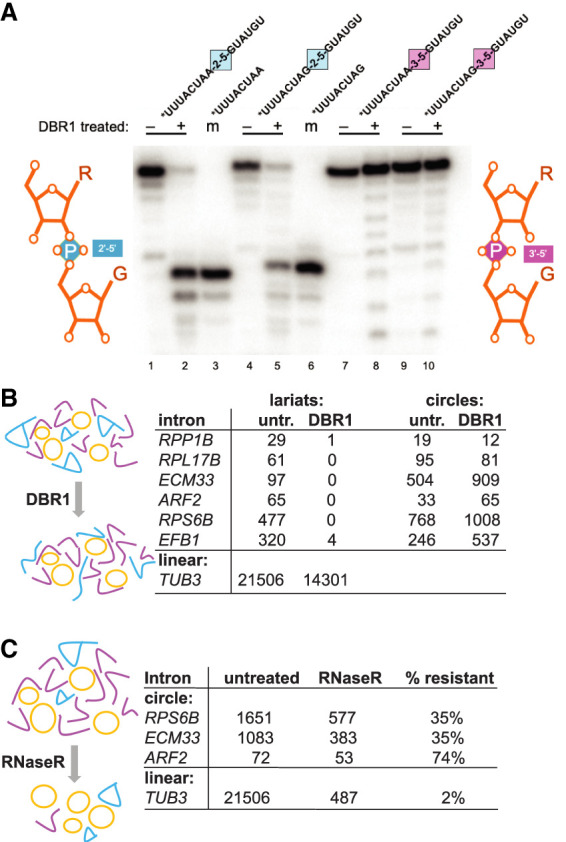
Enzymatic tests of circular intron structure. (A) Dbr1 does not require a 3′ nucleotide at the branch and can cleave an internal 2′–5′ linkage. 32P-5′ end-labeled RNA oligonucleotides carrying either a single 2′–5′ internal linkage in the middle of an otherwise 3′–5′-linked RNA or carrying only 3′–5′-linked nucleotides were incubated (or not) with recombinant Dbr1 and separated by electrophoresis on an 8 M urea/20% acrylamide gel. Two different substrate sequences (bpA-G5ss and 3ssG-G5ss) were made with either a 2′–5′ (blue) or a 3′–5′ (pink) linkage at the junction (marked by the colored squares) and are shown above each reaction. Minus and plus signs indicate enzyme addition. (Lanes 3,6) Two 8-mers identical to the expected Dbr1 cleavage products are labeled as markers (m). Comigration with the marker indicates cleavage at the 2′–5′ linkage. (B) Circle junctions from the spp382-1 mutant are resistant to cleavage with Dbr1, whereas lariat junctions are not. Read counts of lariats and circles for six introns from libraries made with or without prior treatment of the input RNA with Dbr1 are shown. (C) Circle junction-containing RNAs from the spp382-1 mutant are much more resistant to RNase R than a linear pre-mRNA. Read counts of circles for three introns from libraries made with or without prior treatment of the input RNA with RNase R are shown. Detailed data are shown in Supplemental Table S3.
Intron RNAs containing putative circle junctions are resistant to RNase R
RNA sequencing captured cDNA sequences from junctions consistent with circles but did not prove that the original RNA templates were circular. We tested the resistance of the putatively circular RNAs to the strict 3′ exonuclease RNase R (Fig. 3C; Vincent and Deutscher 2006). Equivalent amounts of spp382-1 RNA were either treated with RNase R or not and sequenced. More than 30% of reads with circle junctions are resistant to RNase R, as shown for the three most abundant intron circles (Fig. 3C). In contrast, the recovery of reads for the linear TUB3 control pre-mRNA was only 2%. Some circles may have broken in the cell or during isolation and thus were lost to RNase R; however, intron RNAs carrying circle-closing junctions are at least ∼15–30 times more resistant to RNase R than the linear control intron and therefore are very likely circular.
Mutations in splicing termination factors promote accumulation of intron circles
The general increase in intron circles per cell in the spp382-1 mutant is consistent with the idea that delaying spliceosome disassembly (Pandit et al. 2006) promotes circle formation (Fig. 1C,D; Supplemental Fig. S1). To test the idea that rapid disassembly is in competition with circle formation, we quantified changes in circle numbers in mutants for three factors required for catalytic inactivation: Spp382/Ntr1, the 3′ tail of U6 snRNA, and the DEAH helicase Prp43. Libraries were prepared from strains carrying mutations shown to slow catalytic inactivation and disassembly of the spliceosome—prp43-Q423N (Arenas and Abelson 1997; Martin et al. 2002; Tsai et al. 2005; Boon et al. 2006; Fourmann et al. 2017; Su et al. 2018; Toroney et al. 2019) and short deletions in the tail of U6 snRNA (Toroney et al. 2019)—and changes were evaluated using Fisher's exact test (Fig. 4). The log odds (fold change) and P-values for each intron in each of the three mutants are shown in the inset tables adjacent to each factor's location in a model of the ILS poised for catalytic inactivation (Fig. 4, 5Y88; Wan et al. 2017). For most introns, inhibiting disassembly promotes circle formation (Fig. 4), consistent with the conclusion that circle formation is promoted by delaying spliceosome disassembly. Interestingly, the impact on accumulation of individual introns is different for the three mutations. For example, RPL24B intron circle formation is unaffected by partial loss of function of any of the termination factors, whereas ECM33 circle formation is increased ∼250-fold by spp382-1, 14-fold by shortening of the U6 3′ end by 5 nt, and 2.5-fold by the prp43 cold-sensitive mutation Q423N (Fig. 4; Toroney et al. 2019). The differences between mutants acting on the same intron could be due to qualitative effects of loss of function, quantitative differences in the severity of the tested allele, or both. For example, the greater magnitude of effect of spp382-1 could be explained if Spp382/Ntr1 binding inhibits spliceosome catalytic activity even before Prp43 functions. Differences between introns for the same mutant may be due to differences in the intron-specific intrinsic rate(s) of disassembly steps for spliceosomes carrying different introns. We conclude that the rate of circle formation or the stabilization of intron circles is enhanced when binding of Spp382/Ntr1 and catalytic inactivation of the ILS are delayed, consistent with the idea that circles are formed by the ILS.
Figure 4.
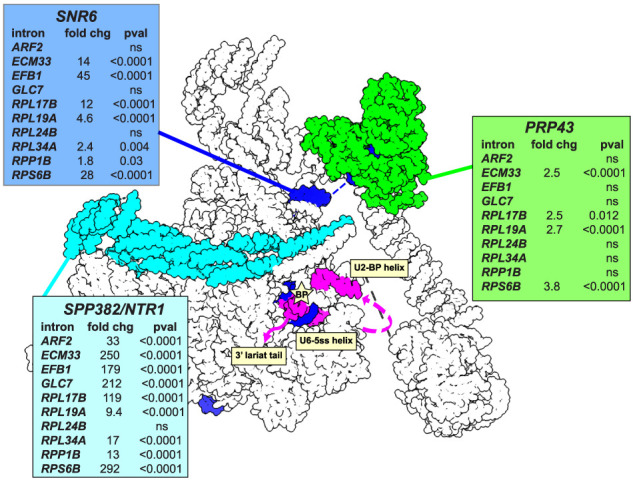
Effects of spliceosome disassembly mutants on the abundance of different intron circles. Catalytic inactivation of the spliceosome is mediated by the Prp43 helicase (green) acting in concert with its G-patch protein, Spp382/Ntr1 (cyan), on the 3′ end of U6 snRNA (SNR6; blue) to pull U6 from the spliceosome (Toroney et al. 2019). Intron circles were counted in pairs of strains carrying wild-type or mutant alleles of each factor, normalized using a spiked-in circle, and tested for significant change using Fisher's exact test (detailed data are shown in Supplemental Table S4). Each table shows the log odds calculation as fold change and the two-sided P-value for each of 10 intron comparisons. (ns) Not significant. The position of the intron is shown in magenta. The image was made using ChimeraX (Meng et al. 2023) and the S. cerevisiae ILS structure model (5Y88; Wan et al. 2017).
Effect of nuclear 3′ decay factors on circle formation
We observed a novel class of intron circles whose 3′ tails have been processed prior to the circularization reaction (Fig. 1). Nuclear 3′ RNA processing machinery includes the nuclear exosome with its exonuclease subunits (Rrp6 and Dis3) and the TRAMP complex, with either of its two paralogous poly(A) polymerases (Trf4 or Trf5) (Chlebowski et al. 2013; Zinder and Lima 2017). RNA 3′ ends are trimmed and polyadenylated using combinations of these activities in a quasidistributive fashion coordinated by the Mtr4 RNA helicase (Chlebowski et al. 2013; Zinder and Lima 2017). To test whether circle formation might be influenced by loss of the nuclear 3′ processing machinery, we made libraries from deletion strains lacking Rrp6 or Trf4 and compared the yield of different kinds of circles. Three introns (RPL19A, RPP1B, and RPL24B) produced enough circles for statistical analysis (Supplemental Table S5). Although we could not detect an effect of loss of Trf4 (possibly because its paralog, Trf5, remains active in these strains), loss of Rrp6 significantly reduces the number of intron circles from RPL24B (approximately twofold, P < 0.001 Fisher's exact test) (Supplemental Table S5) and RPP1B (∼3.7-fold, P < 0.0001, Fisher's exact test) (Supplemental Table S5). In contrast, the number of intron circles from RPL19A is unaffected (Supplemental Table S5). Whereas most of the circles from RPL24B and RPP1B are processed by trimming, polyadenylation, or both, RPL19A produces almost entirely unprocessed circles. A feature that distinguishes these introns is their BP–3′ss distance: RPL24B and RPP1B have long tails (58 and 65 nt, respectively), and RPL19A has a short tail (14 nt). Taken together, this suggests that introns differ in their interactions with the exosome based on the length of their tails, and that Rrp6 function may enhance the efficiency of circle formation for introns with long tails.
To evaluate this idea in more detail, we analyzed the more numerous circles from an spp382-1 mutant that is wild type for nuclear exosome components, comparing circle abundance with the length of tail remaining after processing for 10 introns with a range of tail lengths (Fig. 5; see also Fig. 1C; Supplemental Fig. S1). Two things are evident: One is that introns with tail lengths >22 nt can produce trimmed circles that contain as few as 15–20 nt downstream from the branch but not fewer. The other is that two introns with tails ≤17 nt (RPL19A and ARF2) form robust numbers of circles, virtually all of which are untrimmed (Fig. 5), although a small number have nongenomic As. This suggests that full-length circle formation can occur for introns with at least 14 BP–3′ss nucleotides (and possibly fewer), but that trimming requires introns to have longer BP–3′ss tails. The 15–20 nt tail limit, below which trimmed circles were not observed, is likely enforced by the distance between the BPA and the closest approach of the exosome active site on the outside of the spliceosome. The observation of nongenomic As but not trimming in the ARF2 and RPL19A intron circles indicates that the TRAMP complex can react with shorter tails than the exosome, possibly because its active site is nearer to its surface and the surface of the spliceosome. We conclude that the nuclear 3′ processing machinery interacts with the set of introns with BP–3′ss distances (“tails”) longer than ∼17 nt and enhances circle formation for some of these by shortening the lariat tail, possibly increasing the ease with which it can return to the spliceosome and access the E1 binding site for circularization.
Figure 5.
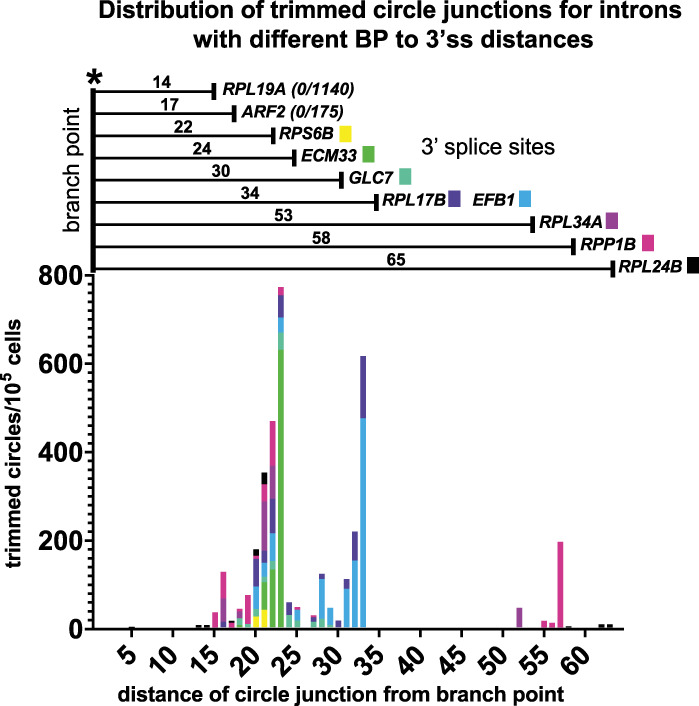
Effects of nuclear 3′ processing on abundance and structure of different intron circles. The distribution of trimmed intron circles for all tested introns has a limit of 15–20 nt of lariat tail remaining, independent of the initial tail length. Two introns with natural BP–3′ss tails of 14 nt (RPL19A) or 17 nt (ARF2) are not trimmed. Detailed data are shown in Supplemental Table S5.
Human U2 and U12 spliceosomes produce analogous full-length and processed intron circles
In their analysis of human and rodent intron circles, Fairbrother and colleagues (Taggart et al. 2012, 2017) described introns joined by the last intron nucleotide for the U2 major spliceosome but did not note the presence of circles among the minor (U12) class of introns. They also did not report processed circles, possibly because these can easily be missed when mapping split reads to the genome, especially when nontemplated nucleotides are added. To ask whether human intron circles are processed, we analyzed human branchpoint mapping data from a different group (Mercer et al. 2015) by creating a 100 nt permuted intron target file in which the 50 3′ nucleotides of each annotated intron are joined to its 50 5′ nucleotides for each annotated intron in the human genome (see the Materials and Methods) and mapping the reads to this target (Fig. 6; Supplemental Table S6). Examples of unprocessed full-length intron circles and circles missing 3′ nucleotides with or without nongenomic As were readily detected for introns spliced by both the major and minor spliceosomes (Fig. 6). In addition, using a similar mapping strategy for yeast, we found circular intron reads for 32 of the ∼270 expressed introns (including seven of the 12 amplicon targets) in standard RNA-seq libraries made from rRNA-depleted total RNA (Supplemental Table S6). Considering previous observations, we suggest that intron circularization can occur for many different introns in many eukaryotes for both major and minor class introns.
Figure 6.
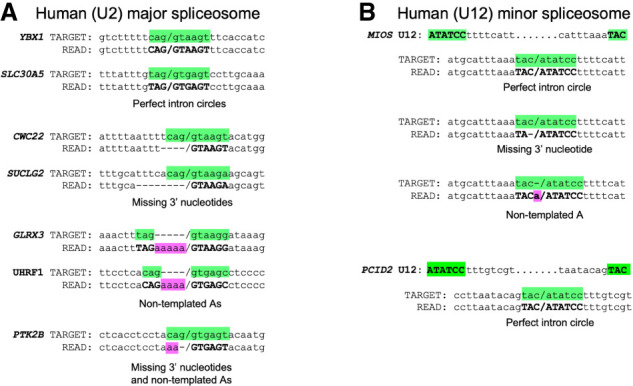
Evidence for full-length and processed circles from human cells. (A) Selected reads from major (U2) spliceosomal introns found by remapping the branchpoint enrichment libraries from Mercer et al. (2015) to permuted intron target sequences from the human genome. Junctions between the 5′ and 3′ intron ends are denoted by a forward slash. Splice site nucleotides from the permuted target intron are highlighted in green. Dashes were added to the target sequence to maintain alignment when nongenomic As are present in the read. Nucleotides in the read that match the splice sites are in bold, and nongenomic As are highlighted in pink. Dashes were added to the read sequence to maintain alignment where nucleotides from the intron 3′ end are missing. (B) Selected reads from minor (U12) spliceosomal introns found by remapping the branchpoint enrichment libraries (Mercer et al. 2015) to permuted intron target sequences from the human genome. Highlights and dash placement are the same as for A. Additional examples with read identifiers are shown in Supplemental Table S6.
Discussion
A model for formation of circular intron RNAs by a catalytically active ILS
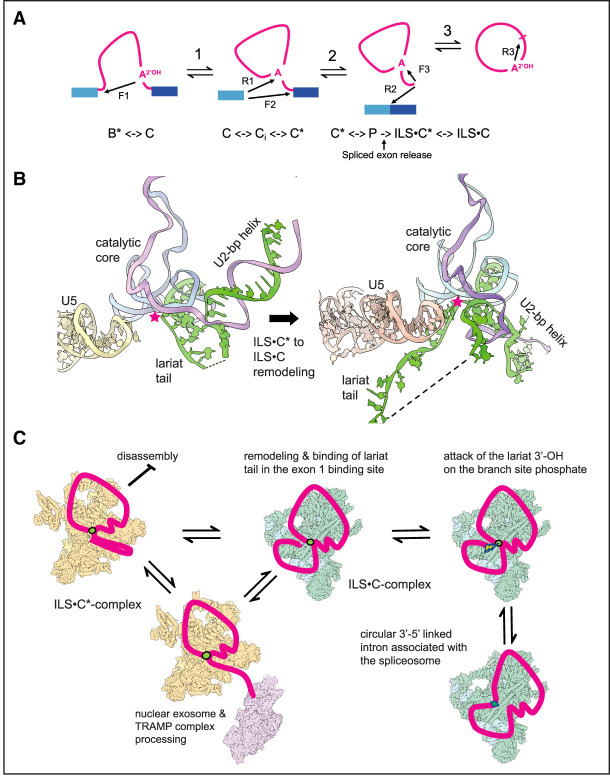
Our observations support a model that explains the formation of intron circles (Fig. 7). Circles possess an intact 5′ss at the closing junction whether processed or not (Fig. 1) and are resistant to Dbr1 (Fig. 3), as would be expected if the 3′-OH of the lariat tail attacks the branch phosphate to form a 3′–5′ linkage (Fig. 7A,B, reaction F3; see also Fig. 1A). The origin of the processed circles is explained by lariat tails longer than ∼17 nt visiting the nuclear 3′ processing machinery outside the spliceosome before returning to the E1 binding site (Figs. 5, 7C). The E1 binding site is known to accommodate a wide variety of exon sequences during forward splicing, consistent with the many different circle junction sequences observed (Fig. 1C; Supplemental Fig. S1). Circles are associated with the spliceosome, and Trl1 (the only other RNA ligase in yeast) is not required (Fig. 2), indicating that this reaction is catalyzed by the spliceosome. The cartoon in Figure 7C shows an overall pathway for formation of the observed classes of intron circles. Since the same classes of circles arise from introns of the major and minor spliceosome (Fig. 6), this mechanism appears general for all spliceosomes and most introns across eukaryotes.
Figure 7.
Model for the formation of intron circles. (A) Transesterification reactions catalyzed by the spliceosome. (F) Forward reactions, (R) reverse reactions (numbered 1–3). The arrows indicate the direction of nucleophilic attack. (F1) Attack of the BP 2′-OH on the phosphate (P) at the 5′ss. (F2) Attack of the 3′-OH at the end of E1 on the P at the 3′ss. (F3) Attack of the lariat tail 3′-OH on the BP P to form the intron circle. Below the pathway are spliceosome complexes within which these reactions occur. The active site of the C complex configuration has the 5′ss P and the BP 2′-OH in the active site for F1, whereas the C* complex configuration has the U2 BP helix pulled out of the active site to create the 3′ss binding site for F2. (ILS) Intron lariat spliceosome. The ILS is formed by spliced exon release (arrow) in the C* configuration (ILS•C*). For F3, it must remodel to the C configuration (ILS•C). (B) Cartoon showing remodeling of the RNA in the core of the ILS•C* complex to the proposed ILS•C complex and docking of the lariat tail for circularization. (C) Cartoon showing events leading to mixtures of processed and unprocessed full-length intron circles. Yellow spliceosomes represent ILS•C* complexes, and green spliceosomes represent ILS•C complexes. The nuclear exosome (purple) represents nuclear 3′ processing machinery interacting with ILS•C*, but it is not known which ILS forms permit processing. (Red) Intron, (green circles) 2′–5′ linkages, (green diamonds) 3′–5′ linkages. The lightning bolt indicates nucleophilic attack. The cartoons were made using ChimeraX as above.
The spliceosome is already known to catalyze a reaction similar to the circularization reaction, as revealed by the discovery of spliceosomal reversibility (Tseng and Cheng 2008, 2013). The first forward step of splicing (F1, attack of the BPA 2′-OH on the 5′ss P) (Fig. 7A) is readily reversible in vitro (R1, attack of the E1 3′-OH on the branch P) to recreate the pre-mRNA (Tseng and Cheng 2008, 2013). Our proposed forward circularization reaction, F3, is formally similar to R1, replacing the lariat tail for E1 (Fig. 7B, left, asterisk; see also Fig. 1A). After exons are released, the lariat tail would leave its position in the intron lariat spliceosome (ILS) and find its way to the E1 binding site. During forward splicing, the active site is remodeled from a C complex conformation (Galej et al. 2016; Wan et al. 2016; Bertram et al. 2020) to C* (Bertram et al. 2017; Fica et al. 2017; Yan et al. 2017) on its way to completing exon ligation in the P complex (Liu et al. 2017; Wilkinson et al. 2017; Zhan et al. 2022). Our model necessitates that the ILS, which retains the C* configuration (Yan et al. 2015; Wan et al. 2017; Zhang et al. 2019), must return from the C* configuration (ILS•C*) to a C configuration with the branch P in the catalytic center (ILS•C). Once the lariat tail docks in the E1 binding site of ILS•C (Fig. 7B, right, asterisk), the reaction can proceed (Figs. 1A, 7B). The requirement for this remodeling seems a fair expectation given that P complex spliceosomes (still carrying spliced exons) must also remodel backward from C* to C while retaining catalytic activity (Fig. 7A, R2 and R1) during reverse splicing (Tseng and Cheng 2008). The ILS•C* complex would need to perform a parallel set of rearrangements (Fig. 7A) but in the absence of bound exons. Recent refinements of C-complex-like spliceosomes identified the Ci complex, a low free-energy-state complex on the pathway between C and C* (Wilkinson et al. 2021; Aupič et al. 2023), further demonstrating the flexibility of spliceosome transitions between the C and C* complexes. This study also identified a long-suspected monovalent metal binding site (Tseng and Cheng 2008, 2013; Marcia and Pyle 2012) that, when occupied by K+, favors F1 over R1 during the first step (Wilkinson et al. 2021; Aupič et al. 2023). This predicts that the proposed circularization reaction (F3), like R1, may be favored when K+ is absent or if Li+ is bound in the monovalent metal site of the catalytic core.
Biological implications of a persistent and active ILS
There are two consequences of extending the lifetime of the ILS: (1) slowing of overall splicing rate and (2) persistence of a catalytically active spliceosome with empty exon binding sites. Slowing spliceosome disassembly quickly inhibits the early steps of splicing (Han et al. 2017; Mendoza-Ochoa et al. 2019; Buerer et al. 2023), suggesting that rapid disassembly of the ILS helps satisfy the demand for continued splicing. The consequences of continued catalytic activity of the ILS are less clear. The circular RNAs described here are rare (Fig. 1) and in most cases likely harmless, but an active ILS might also catalyze events thatwe have yet to detect in vivo. Unusual full-length linear forms of introns have been described (Morgan et al. 2019; Yao et al. 2022) and in yeast have been implicated in growth control (Morgan et al. 2019; Parenteau et al. 2019), but whether these are formed within the ILS is not yet clear. In addition, circle formation does not conform to the established tail length rule for the much more abundant linear introns described previously (Morgan et al. 2019). Among the most interesting possible activities of the ILS are reverse splicing reactions that might lead to intron transposition, analogous to those of the group II introns.
Extending the parallels between group II introns and the spliceosome
Ideas that explain the birth and persistence of introns and the spliceosome in eukaryotic genomes commonly invoke an ancient self-splicing transposable group II intron that spreads itself as (or before) it fragments during its evolution into the modern spliceosome (Lynch and Richardson 2002; Lambowitz and Belfort 2015). This hypothesis has become ever more cemented by the near superimposability of high-resolution structures for the spliceosome and group II introns at various steps in forward splicing (Xu et al. 2023; Haack et al. 2024). If group II introns and the spliceosome shared a common ancestor, they each must have retained, acquired, or lost different abilities as their evolution proceeded along divergent paths. The modern group II introns are highly selected for transposition after splicing, but only to new DNA sites that satisfy the sequence requirements for forward splicing, enforced by the same (in reverse) interactions during reverse splicing into DNA (Lambowitz and Belfort 2015). In contrast, the modern spliceosome is not known to reverse splice into DNA. As introns became fixed in eukaryotic genomes and as the spliceosome became essential for gene expression, it may have lost the luxury of lingering with its intron on the chance that transposition might occur, possibly through the evolution of the rapid disassembly processes needed to maintain high rates of forward splicing. Although the specific circularization reaction described here may not be on the direct path to transposition, the existence of ILS catalytic activity opens the possibility that the ILS could reverse splice its lariat intron into a new genomic location, provided it could access a substrate for reverse splicing.
A pathway by which the ILS could acquire a reverse splicing substrate for transposition is revealed by cryoEM structures of the ILS (Zhang et al. 2019). After the spliced exons and several proteins are released from the P complex to form the ILS, conserved conformational changes in several Prp8 domains create an exon release tunnel between the catalytic center and the outside of the spliceosome (Zhang et al. 2019). This tunnel might also provide a path for a single-stranded DNA or RNA to access the exon binding sites and the catalytic core of a still active ILS for reverse splicing. Structures of group II introns poised or engaged with DNA substrates are revealing how reverse splicing into DNA leads to transposition (Haack et al. 2019; Chung et al. 2022). In particular, comparisons of structures at different steps in reverse splicing document conformational changes in the positions of the branch helix and the catalytic core (Haack et al. 2019) analogous to the transition of the ILS•C* to ILS•C (Fig. 7).
How the genome got its introns: an unfinished story
The modern spliceosome might still be creating new introns. Diverse microbial genomes show clear evidence of recent repeated insertion of sequence families of spliceosomal introns called “introners” (Worden et al. 2009; Simmons et al. 2015; Gozashti et al. 2022). A major class has signatures of protein transposase-mediated spread (terminal inverted repeats [TIRs], target site duplications [TSDs], and LTRs); however, a distinct class of introners lack retroposon or DNA transposon features (van der Burgt et al. 2012; Collemare et al. 2015; Simmons et al. 2015). Members of this class are more often found in algal and fungal genomes, are <200 nt long, do not code for proteins, and might be transposed by the spliceosome (Simmons et al. 2015; Gozashti et al. 2022). Using a special reporter to capture new intron insertions in the laboratory, two instances of intron transposition have been observed in S. cerevisiae (Lee and Stevens 2016), supporting the idea that the spliceosome could be the catalyst. We favor the idea that the spliceosome might reverse splice introns directly into the displaced single-stranded DNA of an R-loop (Simmons et al. 2015), which would also place the new intron in the correct orientation on the transcribed strand for splicing. Introns tend to suppress R-loop formation (Bonnet et al. 2017), offering the possibility that new intron insertion at certain locations might be adaptive. The idiosyncratic ability of some introns to form circles more efficiently than others (Fig. 1) suggests that certain introns may evade disassembly more efficiently or have enhanced rates of reverse splicing. These special features may explain how certain introns might access an ancient pathway back to the genome that may yet live in the modern spliceosome.
Materials and methods
Yeast strains, plasmids, and oligonucleotides
S. cerevisiae strains deleted for various single genes came from the yeast deletion collection (Giaever et al. 2002). The spp382-1 mutant strain was provided by Brian Rymond (Pandit et al. 2006). The TRL1 strains were provided by Jay Hesselberth (Cherry et al. 2018). Jon Staley provided PRP43 strains and plasmids carrying U6 mutants (Toroney et al. 2019). Details on these resources are provided in Supplemental Table S7.
Traditional cloning of circle PCR products
Outward-pointing PCR primers were nested with an intron-specific RT primer to capture circular intron sequence junctions. RNA was converted to cDNA with SuperScript III using the intron-specific primer in combination with anchored oligo(dT) and random hexamers, and cDNA was purified on Zymogen Clean and Concentrate columns. PCR was done using intron-specific outward-pointing primers (Supplemental Table S7) and Phusion DNA polymerase (NEB). Purified PCR products were incubated with Taq to add a 3′ A residue and cloned using a Topo cloning kit (Invitrogen). Plasmid inserts were sequenced by Sanger methods, and the sequence trace and raw read data are shown in Supplemental Table S1.
RNA extraction and synthesis of the circular GFP spike-in control RNA
Yeast cells were grown in YEPD medium at 30°C according to standard protocols (Guthrie and Fink 1991) and harvested in log phase (OD600 = 0.3–0.5). About five OD600 units of cells were extracted by a hot phenol method (Ares 2012) and evaluated using an Agilent Bioanalyzer to assure intactness. Total RNA was used directly for some libraries; for others, RNA was depleted of rRNA by the Illumina RiboZero Gold kit for yeast (no longer available) as described in each case below. Additional treatments before library preparation included digestion of the RNA with the enzymes Dbr1 or RNase R (see below). Plasmid pFiniteGFP (see Supplemental Table S7 for the sequence; Perriman and Ares 1998) was cut with Hind III and used as a template for T7 transcription overnight according to the instructions in the Megaprime T7 transcription kit (Invitrogen). RNA from the reaction was digested with RNase R, purified, and run on 8 M urea/5% acrylamide gels, and the very slow migrating circular RNA band was excised and purified. After purification, an aliquot was rerun on the same type of gel, and inevitably a small amount of 812 nt linear RNA appeared due to breakage during the preparation of the circular RNA. The estimated fraction of broken circles was subtracted from the absorbance measurement of RNA to estimate the true number of intact circles in the spike-in sample.
Dbr1 digestion
Debranching enzyme Dbr1 digestion was performed according to Qin et al. (2016) with minor modifications. Recombinant Dbr1 (Khalid et al. 2005) at 6 mg/mL in 50 mM Tris-Cl (pH 7.0) was a gift from Aiswaria Krishnamohan in Jon Staley's group. For the gel in Figure 3, RNA oligonucleotides were purchased from IDT. Oligos (20 pmol) were labeled at their 5′ ends using 3000 Ci/mmol γ-32P-rATP and polynucleotide kinase (New England Biolabs) and purified on 7 M urea/8% polyacrylamide gels by elution from excised gel pieces. Digestions were performed in 20 μL with ∼105 cpm of labeled substrate, 50 ng of unlabeled total yeast BY4741 RNA, and 0.25–1 ng of enzyme (0.5–2 μL of a 1:10 dilution of a 50 ng/μL [∼1 μM] working stock) in Dbr1 digestion buffer (50 mM Tris–HCl at pH 7.0, 4 mM MnCl2, 2.5 mM DTT, 25 mM NaCl, 0.01% Triton X-100, 0.1 mM EDTA, 0.15% glycerol) for 10 min at 30°C. Digestion products were separated on an 8 M urea/20% acrylamide gel in TBE. For library preparation of Dbr1-treated RNA, ∼1 μg of rRNA-depleted RNA (RiboZero, Illumina; no longer available) from wild-type BY4741 or spp382-1 yeast or ∼3 μg of total RNA from BY4742 dbr1Δ::KanMX4 was digested in a 10 μL reaction with 50 ng of Dbr1 for 10 min at 30°C, diluted with 200 μL of 0.3 M sodium acetate (pH 5.2), extracted with phenol:chloroform:isoamyl alcohol (25:24:1), ethanol-precipitated, and resuspended in 20 μL of water. Recovery was >90%, and the RNA was used directly for library preparation.
RNase R digestion
For library preparation of RNase R-treated RNA, ∼1 μg of rRNA-depleted RNA (RiboZero) from wild-type BY4741 or spp382-1 yeast or ∼3 μg of total RNA from BY4742 dbr1Δ::KanMX4 was digested in a 10 μL reaction in the reaction buffer provided by the manufacturer (Epicenter Biotechnologies) containing 20 U of RNase R for 10 min at 37°C, diluted with 200 μL of 0.3 M sodium acetate (pH 5.2), extracted with phenol:chloroform:isoamyl alcohol (25:24:1), ethanol-precipitated, and resuspended in 20 μL of water. Recovery was ∼30%–40% for the rRNA-depleted samples and ∼20% for the total RNA samples. The RNA was used directly for library preparation.
Library preparation
Five-hundred nanograms of RNA processed in various ways (see below) was used as input for library preparation. Reverse transcription was performed using SuperScript IV (Thermo) and first strand primers complementary to a region just downstream from the 5′ss of each target intron, preceded by four random nucleotides and then by partial Illumina adapters (Supplemental Table S7). After purification of first strand product on Zymo Clean and Concentrator-5 columns (Zymogen Research), second strand synthesis was done using Phusion polymerase (NEB) and a set of primers complementary to the cDNA (i.e., same sense as the RNA) of sequences upstream of the intron branchpoint, preceded by four random nucleotides (UMI) and partial Illumina adapter sequences for the same set of introns. Double-stranded cDNA >150 bp was purified using Zymo Select-A-Size columns. Primer pairs for the control GFP spike-in RNA circle were designed similarly using a first strand primer downstream from the group I 3′ss and a second strand primer upstream of the group I 5′ss in the exonic GFP sequences that became circular (Perriman and Ares 1998). The linear control TUB3 amplicon was conventionally arranged to amplify a similarly sized internal linear segment of the TUB3 intron (Supplemental Table S7). Double-stranded cDNA was barcoded and amplified with Platinum Taq Hifi polymerase (Invitrogen) with two primers that completed Illumina adapter sequences (2 min at 95°C; 27 cycles of 30 sec at 95°C, 30 sec at 65°C, and 2 min at 68°C for; and 7 min at 68°C). Final sequencing libraries were purified by size-selecting the PCR product using Ampure XP beads to obtain fragments >250 bp. Details are available in the Supplemental Material.
Mapping and quantification of intron circles from amplicon sequencing libraries
Libraries were analyzed as follows (for details, see the Supplemental Material): All reads were filtered to have UMI bases with quality scores >33, and paired-end reads were trimmed to 35 bases × 24 bases. Trimmed reads were mapped using Bowtie2 against fasta files for each intron, the linear control, and the cGFP spike-in to capture reads derived from each target element. Reads mapping to each target element were filtered to ensure that they had the correct orientation and the correct distance between read1 and read2 and contained the correct string from the 5′ss into the intron (the filter range, “RangeE”). Filtered reads for each were deduplicated and counted by their UMIs, and the top 30 unique reads were used to query the target with BLAT to determine the extent of the 3′ tail match. The mapping and filtering details are described in the Supplemental Material, analysis parameters are reported in Supplemental Table S8, and the mapping data are reported in Supplemental Tables S9–S11.
Detection of intron circles in spliceosomes
To test for association of intron circles with spliceosomes, we used the methods described by Shi et al. (2021) with modifications. Briefly, we tagged CEF1 in the spp382-1 mutant strain using a PCR product with homology arms targeting the C terminus of the CEF1-coding region to insert the TAPS tag in which the calmodulin binding domain was replaced with a strep tag (a kind gift of Dr. Kelly Nyugen) (Schreieck et al. 2014). We modified the plasmid by swapping the 1446 bp BglII–EcoRI fragment carrying the KanMX6 cassette for the 1333 bp BglII–EcoRI HIS3MX6 cassette carrying the Lachancea kluyveri HIS3 gene from a plasmid designed for C-terminal GFP tagging (Longtine et al. 1998) to make pFA6a_TAPS_kHISMX6. Integration was confirmed by PCR across both junctions (see Supplemental Table S7 for oligonucleotide and plasmid sequences). Cultures were grown at 23°C (spp382-1 is temperature-sensitive), washed, flash-frozen in liquid nitrogen as described (Shi et al. 2021), and processed in a ball mill at liquid nitrogen temperature. Extract was bound to IgG-sepharose 6 Fast Flow resin and washed according to Shi et al. (2021), except that instead of TEV cleavage, the beads were extracted with phenol:chloroform:isoamyl alcohol (25:24:1), and the aqueous phase was ethanol-precipitated. Aliquots of RNA from the input extract, the supernatant after bead binding, and the beads were adjusted to volume equivalence with respect to the input extract and subjected to primer extension with 32P-labeled oligonucleotides complementary to U2 and SCR1 (Supplemental Table S7) according to Perriman and Ares (2007), as shown in Supplemental Figure S2, or were subjected to RT-PCR as described above. The ECM33 primer pair produced a 227 bp product on the full-length circle from the ECM33 intron, as shown in Figure 2.
Identification of reads derived from circular intron RNAs in published RNA-seq data sets
A target fasta database was constructed using all annotated introns in the human genome (hg19), obtained from the University of California, Santa Cruz (UCSC), genome browser using UCSC “known genes” and the RefSeq annotation tracks (Raney et al. 2024). Individual target files were made by joining the 50 bases from the 3′ end of the intron upstream of the 50 bases from the 5′ end of the intron—creating a 100 bp target with a junction similar to the full intron circle in the middle—for each intron. Each target in this initial database was used to query the genome by BLAT, and any targets with more than three hits were removed. Sequencing reads from Mercer et al. (2015)—samples SRR1049823 and SRR1049825—were converted to fasta and used to query the refined target database by BLAT. The resulting alignments were further filtered, leaving only the longest target alignment and longest read alignment. Hits to the same target were grouped together in the output file for review. To identify yeast reads from intron circles, we followed a similar process using annotated splice junctions from the track “talkish standard introns” on our yeast genome browser (http://intron.ucsc.edu) (Talkish et al. 2019) and the sequencing data in GEO at GSE90105 from Talkish et al. (2019) and in the Sequence Read Archive (SRA) at PRJNA972189 from Hunter et al. (2024). A github deposit with scripts and more technical details is available at https://github.com/donoyoyo/intron_circle_hunt.
Data deposition
Sequence data from amplicon pools for all libraries are available under BioProject accession number PRJNA739208 at the SRA (https://www.ncbi.nlm.nih.gov/sra).
Supplementary Material
Acknowledgments
Funding was provided by National Institute of General Medical Sciences grants R01 GM040478 and R35 GM145266 to M.A. We thank Chris Vollmers for advice on library design; Aiswarya Krishnamohan (Staley laboratory) for Dbr1; Brian Rymond, Rebecca Toroney, Klaus Nielsen, Jon Staley, and Jay Hesselberth for strains; Kelly Nguyen for the TAPS plasmid; and Anna Marie Pyle, Anita Hopper, Jon Staley, Jay Hesselberth, Nav Toor, Alex Worden, Jen Quick-Cleveland, Sebastian Fica, Soo-Chen Cheng, and Russ Corbett-Detig for enthusiastic discussions and comments. Special thanks to Doug Black, Tracy Johnson, and Peter and Olivia Narins for providing amazing sabbatical support of different kinds.
Author contributions: M.A. conceived the study. M.A., H.I., S.K., and J.P.D. developed the methods of the study. M.A. and H.I. acquired the data. M.A., H.I., S.K., and J.P.D. analyzed the data. M.A. created the figures and tables and wrote the first draft of the manuscript. M.A., H.I., S.K., and J.P.D. reviewed and edited the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.351764.124.
Competing interest statement
The authors declare no competing interests.
References
- Arenas JE, Abelson JN. 1997. Prp43: an RNA helicase-like factor involved in spliceosome disassembly. Proc Natl Acad Sci 94: 11798–11802. 10.1073/pnas.94.22.11798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ares M. 2012. Isolation of total RNA from yeast cell cultures. Cold Spring Harb Protoc 2012: 1082–1086. [DOI] [PubMed] [Google Scholar]
- Aupič J, Borišek J, Fica SM, Galej WP, Magistrato A. 2023. Monovalent metal ion binding promotes the first transesterification reaction in the spliceosome. Nat Commun 14: 8482. 10.1038/s41467-023-44174-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram K, Agafonov DE, Liu W-T, Dybkov O, Will CL, Hartmuth K, Urlaub H, Kastner B, Stark H, Lührmann R. 2017. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature 542: 318–323. 10.1038/nature21079 [DOI] [PubMed] [Google Scholar]
- Bertram K, El Ayoubi L, Dybkov O, Agafonov DE, Will CL, Hartmuth K, Urlaub H, Kastner B, Stark H, Lührmann R. 2020. Structural insights into the roles of metazoan-specific splicing factors in the human step 1 spliceosome. Mol Cell 80: 127–139.e6. 10.1016/j.molcel.2020.09.012 [DOI] [PubMed] [Google Scholar]
- Bonnet A, Grosso AR, Elkaoutari A, Coleno E, Presle A, Sridhara SC, Janbon G, Géli V, de Almeida SF, Palancade B. 2017. Introns protect eukaryotic genomes from transcription-associated genetic instability. Mol Cell 67: 608–621.e6. 10.1016/j.molcel.2017.07.002 [DOI] [PubMed] [Google Scholar]
- Boon K-L, Auchynnikava T, Edwalds-Gilbert G, Barrass JD, Droop AP, Dez C, Beggs JD. 2006. Yeast ntr1/spp382 mediates prp43 function in postspliceosomes. Mol Cell Biol 26: 6016–6023. 10.1128/MCB.02347-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerer L, Clark NE, Welch A, Duan C, Taggart AJ, Townley BA, Wang J, Soemedi R, Rong S, Lin C-L, et al. 2023. The debranching enzyme Dbr1 regulates lariat turnover and intron splicing. Res Sq 10.21203/rs.3.rs-2931976/v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry PD, White LK, York K, Hesselberth JR. 2018. Genetic bypass of essential RNA repair enzymes in budding yeast. RNA 24: 313–323. 10.1261/rna.061788.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry PD, Peach SE, Hesselberth JR. 2019. Multiple decay events target HAC1 mRNA during splicing to regulate the unfolded protein response. Elife 8: 42262. 10.7554/eLife.42262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlebowski A, Lubas M, Jensen TH, Dziembowski A. 2013. RNA decay machines: the exosome. Biochim Biophys Acta 1829: 552–560. 10.1016/j.bbagrm.2013.01.006 [DOI] [PubMed] [Google Scholar]
- Chung K, Xu L, Chai P, Peng J, Devarkar SC, Pyle AM. 2022. Structures of a mobile intron retroelement poised to attack its structured DNA target. Science 378: 627–634. 10.1126/science.abq2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark NE, Katolik A, Taggart AJ, Buerer L, Holloway SP, Miller N, Phillips JD, Farrell CP, Damha MJ, Fairbrother WG. 2022. Metal content and kinetic properties of yeast RNA lariat debranching enzyme Dbr1. RNA 28: 927–936. 10.1261/rna.079159.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquerelle C, Daubersies P, Majérus MA, Kerckaert JP, Bailleul B. 1992. Splicing with inverted order of exons occurs proximal to large introns. EMBO J 11: 1095–1098. 10.1002/j.1460-2075.1992.tb05148.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collemare J, Beenen HG, Crous PW, de Wit PJGM, van der Burgt A. 2015. Novel introner-like elements in fungi are involved in parallel gains of spliceosomal introns. PLoS One 10: e0129302. 10.1371/journal.pone.0129302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fica SM, Oubridge C, Galej WP, Wilkinson ME, Bai X-C, Newman AJ, Nagai K. 2017. Structure of a spliceosome remodelled for exon ligation. Nature 542: 377–380. 10.1038/nature21078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford E, Ares M Jr. 1994. Synthesis of circular RNA in bacteria and yeast using RNA cyclase ribozymes derived from a group I intron of phage T4. Proc Natl Acad Sci 91: 3117–3121. 10.1073/pnas.91.8.3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourmann J-B, Tauchert MJ, Ficner R, Fabrizio P, Lührmann R. 2017. Regulation of Prp43-mediated disassembly of spliceosomes by its cofactors Ntr1 and Ntr2. Nucleic Acids Res 45: 4068–4080. 10.1093/nar/gkw1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galej WP, Wilkinson ME, Fica SM, Oubridge C, Newman AJ, Nagai K. 2016. Cryo-EM structure of the spliceosome immediately after branching. Nature 537: 197–201. 10.1038/nature19316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner EJ, Nizami ZF, Talbot CC Jr, Gall JG. 2012. Stable intronic sequence RNA (sisRNA), a new class of noncoding RNA from the oocyte nucleus of Xenopus tropicalis. Genes Dev 26: 2550–2559. 10.1101/gad.202184.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391. 10.1038/nature00935 [DOI] [PubMed] [Google Scholar]
- Gozashti L, Roy SW, Thornlow B, Kramer A, Ares M Jr, Corbett-Detig R. 2022. Transposable elements drive intron gain in diverse eukaryotes. Proc Natl Acad Sci 119: e2209766119. 10.1073/pnas.2209766119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski PJ, Zaug AJ, Cech TR. 1981. The intervening sequence of the ribosomal RNA precursor is converted to a circular RNA in isolated nuclei of Tetrahymena. Cell 23: 467–476. 10.1016/0092-8674(81)90142-2 [DOI] [PubMed] [Google Scholar]
- Gumińska N, Zakryś B, Milanowski R. 2021. A new type of circular RNA derived from nonconventional introns in nuclear genes of euglenids. J Mol Biol 433: 166758. 10.1016/j.jmb.2020.166758 [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. 1991. Guide to yeast genetics and molecular biology. Methods Enzymol 194: 1–863. [PubMed] [Google Scholar]
- Haack DB, Yan X, Zhang C, Hingey J, Lyumkis D, Baker TS, Toor N. 2019. Cryo-EM structures of a group II intron reverse splicing into DNA. Cell 178: 612–623.e12. 10.1016/j.cell.2019.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haack DB, Rudolfs B, Zhang C, Lyumkis D, Toor N. 2024. Structural basis of branching during RNA splicing. Nat Struct Mol Biol 31: 179–189. 10.1038/s41594-023-01150-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B, Park HK, Ching T, Panneerselvam J, Wang H, Shen Y, Zhang J, Li L, Che R, Garmire L, et al. 2017. Human DBR1 modulates the recycling of snRNPs to affect alternative RNA splicing and contributes to the suppression of cancer development. Oncogene 36: 5382–5391. 10.1038/onc.2017.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter O, Talkish J, Quick-Cleveland J, Igel H, Tan A, Kuersten S, Katzman S, Donohue JP, S Jurica M, Ares M Jr. 2024. Broad variation in response of individual introns to splicing inhibitors in a humanized yeast strain. RNA 30: 149–170. 10.1261/rna.079866.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalid MF, Damha MJ, Shuman S, Schwer B. 2005. Structure–function analysis of yeast RNA debranching enzyme (Dbr1), a manganese-dependent phosphodiesterase. Nucleic Acids Res 33: 6349–6360. 10.1093/nar/gki934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska MM, Grabowski PJ, Padgett RA, Sharp PA. 1985. Characterization of the branch site in lariat RNAs produced by splicing of mRNA precursors. Nature 313: 552–557. 10.1038/313552a0 [DOI] [PubMed] [Google Scholar]
- Lambowitz AM, Belfort M. 2015. Mobile bacterial group II introns at the crux of eukaryotic evolution. Microbiol Spectr 3: MDNA3-0050-2014. 10.1128/microbiolspec.MDNA3-0050-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasda E, Parker R. 2014. Circular RNAs: diversity of form and function. RNA 20: 1829–1842. 10.1261/rna.047126.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Stevens SW. 2016. Spliceosomal intronogenesis. Proc Natl Acad Sci 113: 6514–6519. 10.1073/pnas.1605113113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Li X, Zhang L, Jiang J, Hill RC, Cui Y, Hansen KC, Zhou ZH, Zhao R. 2017. Structure of the yeast spliceosomal postcatalytic P complex. Science 358: 1278–1283. 10.1126/science.aar3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J, Aagaard C, Semionenkov M, Garrett RA. 1997. Archaeal introns: splicing, intercellular mobility and evolution. Trends Biochem Sci 22: 326–331. 10.1016/S0968-0004(97)01113-4 [DOI] [PubMed] [Google Scholar]
- Lynch M, Richardson AO. 2002. The evolution of spliceosomal introns. Curr Opin Genet Dev 12: 701–710. 10.1016/S0959-437X(02)00360-X [DOI] [PubMed] [Google Scholar]
- Marcia M, Pyle AM. 2012. Visualizing group II intron catalysis through the stages of splicing. Cell 151: 497–507. 10.1016/j.cell.2012.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Schneider S, Schwer B. 2002. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J Biol Chem 277: 17743–17750. 10.1074/jbc.M200762200 [DOI] [PubMed] [Google Scholar]
- Mendoza-Figueroa MS, Alfonso-Maqueira EE, Vélez C, Azuara-Liceaga EI, Zárate S, Villegas-Sepúlveda N, Saucedo-Cárdenas O, Valdés J. 2018. Postsplicing-derived full-length intron circles in the protozoan parasite Entamoeba histolytica. Front Cell Infect Microbiol 8: 255. 10.3389/fcimb.2018.00255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Ochoa GI, Barrass JD, Maudlin IE, Beggs JD. 2019. Blocking late stages of splicing quickly limits pre-spliceosome assembly in vivo. RNA Biol 16: 1775–1784. 10.1080/15476286.2019.1657788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng EC, Goddard TD, Pettersen EF, Couch GS, Pearson ZJ, Morris JH, Ferrin TE. 2023. UCSF chimerax: tools for structure building and analysis. Protein Sci 32: e4792. 10.1002/pro.4792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Clark MB, Andersen SB, Brunck ME, Haerty W, Crawford J, Taft RJ, Nielsen LK, Dinger ME, Mattick JS. 2015. Genome-wide discovery of human splicing branchpoints. Genome Res 25: 290–303. 10.1101/gr.182899.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montemayor EJ, Katolik A, Clark NE, Taylor AB, Schuermann JP, Combs DJ, Johnsson R, Holloway SP, Stevens SW, Damha MJ, et al. 2014. Structural basis of lariat RNA recognition by the intron debranching enzyme Dbr1. Nucleic Acids Res 42: 10845–10855. 10.1093/nar/gku725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JT, Fink GR, Bartel DP. 2019. Excised linear introns regulate growth in yeast. Nature 565: 606–611. 10.1038/s41586-018-0828-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray HL, Mikheeva S, Coljee VW, Turczyk BM, Donahue WF, Bar-Shalom A, Jarrell KA. 2001. Excision of group II introns as circles. Mol Cell 8: 201–211. 10.1016/S1097-2765(01)00300-8 [DOI] [PubMed] [Google Scholar]
- Neil CR, Fairbrother WG. 2019. Intronic RNA: Ad‘junk’ mediator of post-transcriptional gene regulation. Biochim Biophys Acta Gene Regul Mech 1862: 194439. 10.1016/j.bbagrm.2019.194439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B. 1991. Scrambled exons. Cell 64: 607–613. 10.1016/0092-8674(91)90244-S [DOI] [PubMed] [Google Scholar]
- Pandit S, Lynn B, Rymond BC. 2006. Inhibition of a spliceosome turnover pathway suppresses splicing defects. Proc Natl Acad Sci 103: 13700–13705. 10.1073/pnas.0603188103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenteau J, Maignon L, Berthoumieux M, Catala M, Gagnon V, Abou Elela S. 2019. Introns are mediators of cell response to starvation. Nature 565: 612–617. 10.1038/s41586-018-0859-7 [DOI] [PubMed] [Google Scholar]
- Pasman Z, Been MD, Garcia-Blanco MA. 1996. Exon circularization in mammalian nuclear extracts. RNA 2: 603–610. [PMC free article] [PubMed] [Google Scholar]
- Peebles CL, Perlman PS, Mecklenburg KL, Petrillo ML, Tabor JH, Jarrell KA, Cheng HL. 1986. A self-splicing RNA excises an intron lariat. Cell 44: 213–223. 10.1016/0092-8674(86)90755-5 [DOI] [PubMed] [Google Scholar]
- Perriman R, Ares M Jr. 1998. Circular mRNA can direct translation of extremely long repeating-sequence proteins in vivo. RNA 4: 1047–1054. 10.1017/S135583829898061X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriman RJ, Ares M Jr. 2007. Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing. Genes Dev 21: 811–820. 10.1101/gad.1524307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschek J, Walter P. 2019. tRNA ligase structure reveals kinetic competition between non-conventional mRNA splicing and mRNA decay. Elife 8: e44199. 10.7554/eLife.44199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky EM, Consaul SA, Nehrke KW, Abelson J. 1992. Yeast tRNA ligase mutants are nonviable and accumulate tRNA splicing intermediates. J Biol Chem 267: 4577–4582. 10.1016/S0021-9258(18)42872-4 [DOI] [PubMed] [Google Scholar]
- Puttaraju M, Been MD. 1992. Group I permuted intron–exon (PIE) sequences self-splice to produce circular exons. Nucleic Acids Res 20: 5357–5364. 10.1093/nar/20.20.5357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin D, Huang L, Wlodaver A, Andrade J, Staley JP. 2016. Sequencing of lariat termini in S. cerevisiae reveals 5′ splice sites, branch points, and novel splicing events. RNA 22: 237–253. 10.1261/rna.052829.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raney BJ, Barber GP, Benet-Pagès A, Casper J, Clawson H, Cline MS, Diekhans M, Fischer C, Navarro Gonzalez J, Hickey G, et al. 2024. The UCSC genome browser database: 2024 update. Nucleic Acids Res 52: D1082–D1088. 10.1093/nar/gkad987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R, Maniatis T. 1985. Intron sequences involved in lariat formation during pre-mRNA splicing. Cell 41: 95–105. 10.1016/0092-8674(85)90064-9 [DOI] [PubMed] [Google Scholar]
- Ruskin B, Green MR. 1985. An RNA processing activity that debranches RNA lariats. Science 229: 135–140. 10.1126/science.2990042 [DOI] [PubMed] [Google Scholar]
- Ruskin B, Krainer AR, Maniatis T, Green MR. 1984. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell 38: 317–331. 10.1016/0092-8674(84)90553-1 [DOI] [PubMed] [Google Scholar]
- Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. 2012. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7: e30733. 10.1371/journal.pone.0030733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt CA, Giusto JD, Bao A, Hopper AK, Matera AG. 2019. Molecular determinants of metazoan tricRNA biogenesis. Nucleic Acids Res 47: 6452–6465. 10.1093/nar/gkz311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreieck A, Easter AD, Etzold S, Wiederhold K, Lidschreiber M, Cramer P, Passmore LA. 2014. RNA polymerase II termination involves C-terminal-domain tyrosine dephosphorylation by CPF subunit Glc7. Nat Struct Mol Biol 21: 175–179. 10.1038/nsmb.2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Li X, Zhao R. 2021. Detecting circRNA in purified spliceosomal P complex. Methods 196: 30–35. 10.1016/j.ymeth.2021.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons MP, Bachy C, Sudek S, van Baren MJ, Sudek L, Ares M Jr, Worden AZ. 2015. Intron invasions trace algal speciation and reveal nearly identical arctic and antarctic Micromonas populations. Mol Biol Evol 32: 2219–2235. 10.1093/molbev/msv122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y-L, Chen H-C, Tsai R-T, Lin P-C, Cheng S-C. 2018. Cwc23 is a component of the NTR complex and functions to stabilize Ntr1 and facilitate disassembly of spliceosome intermediates. Nucleic Acids Res 46: 3764–3773. 10.1093/nar/gky052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taggart AJ, DeSimone AM, Shih JS, Filloux ME, Fairbrother WG. 2012. Large-scale mapping of branchpoints in human pre-mRNA transcripts in vivo. Nat Struct Mol Biol 19: 719–721. 10.1038/nsmb.2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taggart AJ, Lin C-L, Shrestha B, Heintzelman C, Kim S, Fairbrother WG. 2017. Large-scale analysis of branchpoint usage across species and cell lines. Genome Res 27: 639–649. 10.1101/gr.202820.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkish J, Igel H, Perriman RJ, Shiue L, Katzman S, Munding EM, Shelansky R, Donohue JP, Ares M Jr. 2019. Rapidly evolving protointrons in Saccharomyces genomes revealed by a hungry spliceosome. PLoS Genet 15: e1008249. 10.1371/journal.pgen.1008249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toroney R, Nielsen KH, Staley JP. 2019. Termination of pre-mRNA splicing requires that the ATPase and RNA unwindase Prp43p acts on the catalytic snRNA U6. Genes Dev 33: 1555–1574. 10.1101/gad.328294.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai R-T, Fu R-H, Yeh F-L, Tseng C-K, Lin Y-C, Huang Y-H, Cheng S-C. 2005. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev 19: 2991–3003. 10.1101/gad.1377405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng C-K, Cheng S-C. 2008. Both catalytic steps of nuclear pre-mRNA splicing are reversible. Science 320: 1782–1784. 10.1126/science.1158993 [DOI] [PubMed] [Google Scholar]
- Tseng C-K, Cheng S-C. 2013. The spliceosome catalyzes debranching in competition with reverse of the first chemical reaction. RNA 19: 971–981. 10.1261/rna.038638.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Burgt A, Severing E, de Wit PJGM, Collemare J. 2012. Birth of new spliceosomal introns in fungi by multiplication of introner-like elements. Curr Biol 22: 1260–1265. 10.1016/j.cub.2012.05.011 [DOI] [PubMed] [Google Scholar]
- van der Veen R, Arnberg AC, van der Horst G, Bonen L, Tabak HF, Grivell LA. 1986. Excised group II introns in yeast mitochondria are lariats and can be formed by self-splicing in vitro. Cell 44: 225–234. 10.1016/0092-8674(86)90756-7 [DOI] [PubMed] [Google Scholar]
- Vincent HA, Deutscher MP. 2006. Substrate recognition and catalysis by the exoribonuclease RNase R. J Biol Chem 281: 29769–29775. 10.1074/jbc.M606744200 [DOI] [PubMed] [Google Scholar]
- Wan R, Yan C, Bai R, Huang G, Shi Y. 2016. Structure of a yeast catalytic step I spliceosome at 3.4 Å resolution. Science 353: 895–904. 10.1126/science.aag2235 [DOI] [PubMed] [Google Scholar]
- Wan R, Yan C, Bai R, Lei J, Shi Y. 2017. Structure of an intron lariat spliceosome from Saccharomyces cerevisiae. Cell 171: 120–132.e12. 10.1016/j.cell.2017.08.029 [DOI] [PubMed] [Google Scholar]
- Wilkinson ME, Fica SM, Galej WP, Norman CM, Newman AJ, Nagai K. 2017. Postcatalytic spliceosome structure reveals mechanism of 3′-splice site selection. Science 358: 1283–1288. 10.1126/science.aar3729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson ME, Fica SM, Galej WP, Nagai K. 2021. Structural basis for conformational equilibrium of the catalytic spliceosome. Mol Cell 81: 1439–1452.e9. 10.1016/j.molcel.2021.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden AZ, Lee J-H, Mock T, Rouzé P, Simmons MP, Aerts AL, Allen AE, Cuvelier ML, Derelle E, Everett MV, et al. 2009. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 324: 268–272. 10.1126/science.1167222 [DOI] [PubMed] [Google Scholar]
- Xu L, Liu T, Chung K, Pyle AM. 2023. Structural insights into intron catalysis and dynamics during splicing. Nature 624: 682–688. 10.1038/s41586-023-06746-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Hang J, Wan R, Huang M, Wong CCL, Shi Y. 2015. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science 349: 1182–1191. 10.1126/science.aac7629 [DOI] [PubMed] [Google Scholar]
- Yan C, Wan R, Bai R, Huang G, Shi Y. 2017. Structure of a yeast step II catalytically activated spliceosome. Science 355: 149–155. 10.1126/science.aak9979 [DOI] [PubMed] [Google Scholar]
- Yao J, Winans S, Xu H, Wu DC, Ferrick-Kiddie EA, Ares M, Lambowitz AM. 2022. Human cells contain myriad excised linear intron RNAs with links to gene regulation and potential utility as biomarkers. bioRxiv 10.1101/2020.09.07.285114 [DOI] [Google Scholar]
- Zaphiropoulos PG. 1996. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci 93: 6536–6541. 10.1073/pnas.93.13.6536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Lu Y, Zhang X, Yan C, Shi Y. 2022. Mechanism of exon ligation by human spliceosome. Mol Cell 82: 2769–2778.e4. 10.1016/j.molcel.2022.05.021 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang X-O, Chen T, Xiang J-F, Yin Q-F, Xing Y-H, Zhu S, Yang L, Chen L-L. 2013. Circular intronic long noncoding RNAs. Mol Cell 51: 792–806. 10.1016/j.molcel.2013.08.017 [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhan X, Yan C, Zhang W, Liu D, Lei J, Shi Y. 2019. Structures of the human spliceosomes before and after release of the ligated exon. Cell Res 29: 274–285. 10.1038/s41422-019-0143-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder JC, Lima CD. 2017. Targeting RNA for processing or destruction by the eukaryotic RNA exosome and its cofactors. Genes Dev 31: 88–100. 10.1101/gad.294769.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.