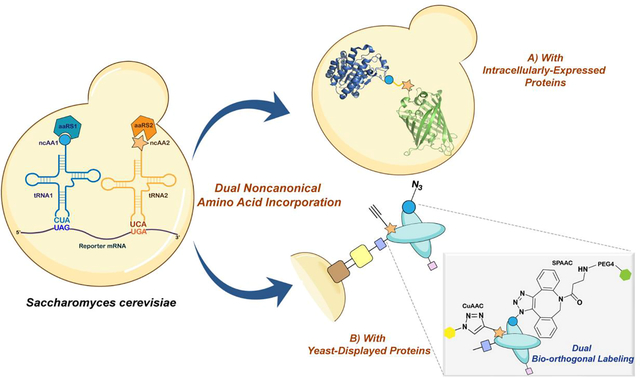
Abstract
Incorporation of more than one non-canonical amino acid (ncAA) within a single protein endows the resulting construct with multiple useful features such as augmented molecular recognition or covalent crosslinking capabilities. Herein, for the first time, we demonstrate the incorporation of two chemically distinct ncAAs into proteins biosynthesized in Saccharomyces cerevisiae. To complement ncAA incorporation in response to the amber (TAG) stop codon in yeast, we evaluated opal (TGA) stop codon suppression using three distinct orthogonal translation systems. We observed selective TGA readthrough without detectable cross-reactivity from host translation components. Readthrough efficiency at TGA was modulated by factors including the local nucleotide environment, gene deletions related to the translation process, and the identity of the suppressor tRNA. These observations facilitated systematic investigation of dual ncAA incorporation in both intracellular and yeast-displayed protein constructs, where we observed efficiencies up to 6% of wildtype protein controls. The successful display of doubly-substituted proteins enabled the exploration of two critical applications on the yeast surface - A) antigen-binding functionality; and B) chemoselective modification with two distinct chemical probes through sequential application of two bioorthogonal click chemistry reactions. Lastly, by utilizing a soluble form of a doubly-substituted construct, we validated the dual incorporation system using mass spectrometry and demonstrated the feasibility conducting selective labeling of the two ncAAs sequentially using a ”single-pot” approach. Overall, our work facilitates the addition of a 22nd amino acid to the genetic code of yeast and expands the scope of applications of ncAAs for basic biological research and drug discovery.
Keywords: Noncanonical amino acids, yeast display, genetic code expansion, orthogonal translation system, Strain-promoted azide-alkyne cycloaddition, Copper(I)-catalyzed azide-alkyne cycloaddition
Graphical Abstract
Introduction:
The genetic encoding of noncanonical amino acids (ncAAs) in proteins allows for precise manipulation of their native structures and the introduction of new properties and chemical entities1–7. The number of ncAAs that can be encoded during protein biosynthesis and the organisms capable of encoding them is constantly expanding8,9. The most successful dual ncAAs incorporation systems have been demonstrated in bacterial and mammalian expression systems; these approaches have enabled a wide range of applications such as exploring protein dynamics, protein-protein crosslinking, and mimicking epigenetic modifications10–18. However, to the best of our knowledge, such dual ncAA incorporation systems have not yet been implemented in S. cerevisiae or any other yeast. With reasonably fast growth kinetics and biological machinery analogous to mammalian cells, S. cerevisiae is a critical model organism for understanding fundamentals of eukaryotic biology, functions of disease-implicated genes as well as serving as workhorse for generating complex recombinant proteins19–21. Additionally, powerful genetic, genomic, and engineering tools are now available in yeast, including single-gene knockout collections, strains with synthetic chromosomes, and yeast display systems22–26. Therefore, engineering a dual ncAA incorporation system in S. cerevisiae would enable applications in areas of protein engineering, chemical biology, synthetic biology, and other related disciplines27.
Incorporating two distinct ncAAs in cells requires the use of two orthogonal codons and two corresponding aminoacyl-tRNA-synthetase/tRNA pairs (aaRS/tRNA pairs); also referred to as orthogonal translation systems (OTSs). These pairs must be orthogonal to one another as well as to the host cell’s native translation machinery. Such orthogonal codons can be repurposed sense codons, nonsense (“stop”) codons, or engineered frameshift codons. For both bacteria and mammalian cells, these orthogonal codons have been discovered and utilized in various combinations, but in yeast, only the amber (TAG) codon has been repurposed for ncAA incorporation11–13,16,17,28–31. The lack of additional validated orthogonal codons and mutually-orthogonal translation systems in yeast has hindered the development of dual ncAA incorporation systems, in contrast to systems in bacteria and mammalian cells that support the decoding of multiple orthogonal codons with structurally and functionally diverse ncAAs11,16,30,32–35.
Efforts are underway in yeast to expand the range of available OTSs for genetic code manipulation. Recent studies by He et al and Tan et al demonstrated the isolation of E. coli aaRS variants that support translation with new ncAAs including sulfotyrosine36,37. Parallel efforts from our lab have also led to the discovery of a broad range of E. coli tyrosyl-tRNA synthetase (EcTyrRS) and E. coli Leucyl-tRNA synthetase (EcLeuRS) variants with a variety of ncAA incorporation properties, including new-to-yeast ncAAs and improved selectivity for known ncAAs38. Similar efforts by Chatterjee and coworkers in engineered E. coli strains has led to the identification of additional EcaaRS variants derived from EcTyRS and EcTrpRS with unique substrate preferences for use in mammalian cells33,39. While not yet evaluated in yeast, such EcaaRS variants have the potential to further broaden the range of OTSs available for use in this organism. Beyond E. coli-derived OTSs, translation machinery derived from archaeal Pyrrolysyl-tRNA synthetases (PylRS)-tRNA pairs continues to mature10, 40–45. PylRS-based OTSs from several organisms and their engineered derivatives support diverse ncAA incorporation and efficient decoding of a range of orthogonal codons. We and others have advanced the use of such PylRS-tRNA pairs in yeast, which presents opportunities for the identification of mutually orthogonal translation systems46–49. The growing versatility of OTSs available in yeast sets the stage for pursuing dual ncAA incorporation in this important model organism.
Herein, for the first time, we report the systematic establishment of dual ncAA incorporation systems in S. cerevisiae suitable for applications involving both intracellular and yeast-displayed proteins. We repurpose the opal (TGA) nonsense codon to function alongside the amber (TAG) nonsense codon in yeast and demonstrate the successful decoding of these two orthogonal codons within the same mRNA transcript by different combinations of mutually orthogonal OTSs. Our detailed investigations identify potential non-cognate interactions that can interfere with the fidelity of the system and favorable genetic conditions augmenting the decoding efficiencies of both TAG and TGA codons. This engineered dual ncAA incorporation system enables site-specific incorporation of distinct ncAAs pairs, allowing for the generation of doubly-labeled proteins through sequential strain-promoted azide-alkyne cycloaddition (SPAAC) and copper-catalyzed azide-alkyne cycloaddition (CuAAC) reactions on the yeast surface. These reactions and corroborating investigations on soluble proteins demonstrate the incorporation and selective addressability of two distinct chemical functionalities within proteins biosynthesized in yeast.
Results:
A second orthogonal codon identified in yeast
For complementing the TAG codon as a location for ncAA incorporation in yeast, another nonsense codon—opal (TGA) codon—was selected due to its low usage within yeast genome and prior use for ncAA incorporation in mammalian systems17,50–53. However, for TGA to be repurposed as an orthogonal codon in yeast, it must fulfill two important criteria: i) Orthogonality; indicating that TGA should not be decoded at high levels by the endogenous translational machinery or non-cognate OTSs; and ii) Decoding efficiency; meaning that TGA should be efficiently decoded only by OTSs that utilize tRNAUCAs and not by OTSs utilizing tRNACUAs or other exogenous machinery. As there are examples of natural TGA readthrough in yeast by endogenous translation machineries under selected conditions (which also holds true for TAG), it is critical that the second criteria is met to minimize aberrant readthrough54. Thus, to systematically evaluate both criteria at TGA codons, a dual-plasmid approach was used, where the OTS was encoded on one plasmid and the reporter protein bearing the orthogonal codon was encoded on another. This dual-plasmid approach allows for the parallel assessment of different cognate and non-cognate interactions: aaRS:tRNA, aaRS:ncAA, aaRS:cAA (canonical amino acid), and codon:anticodon (Figure 1A).
Figure 1.
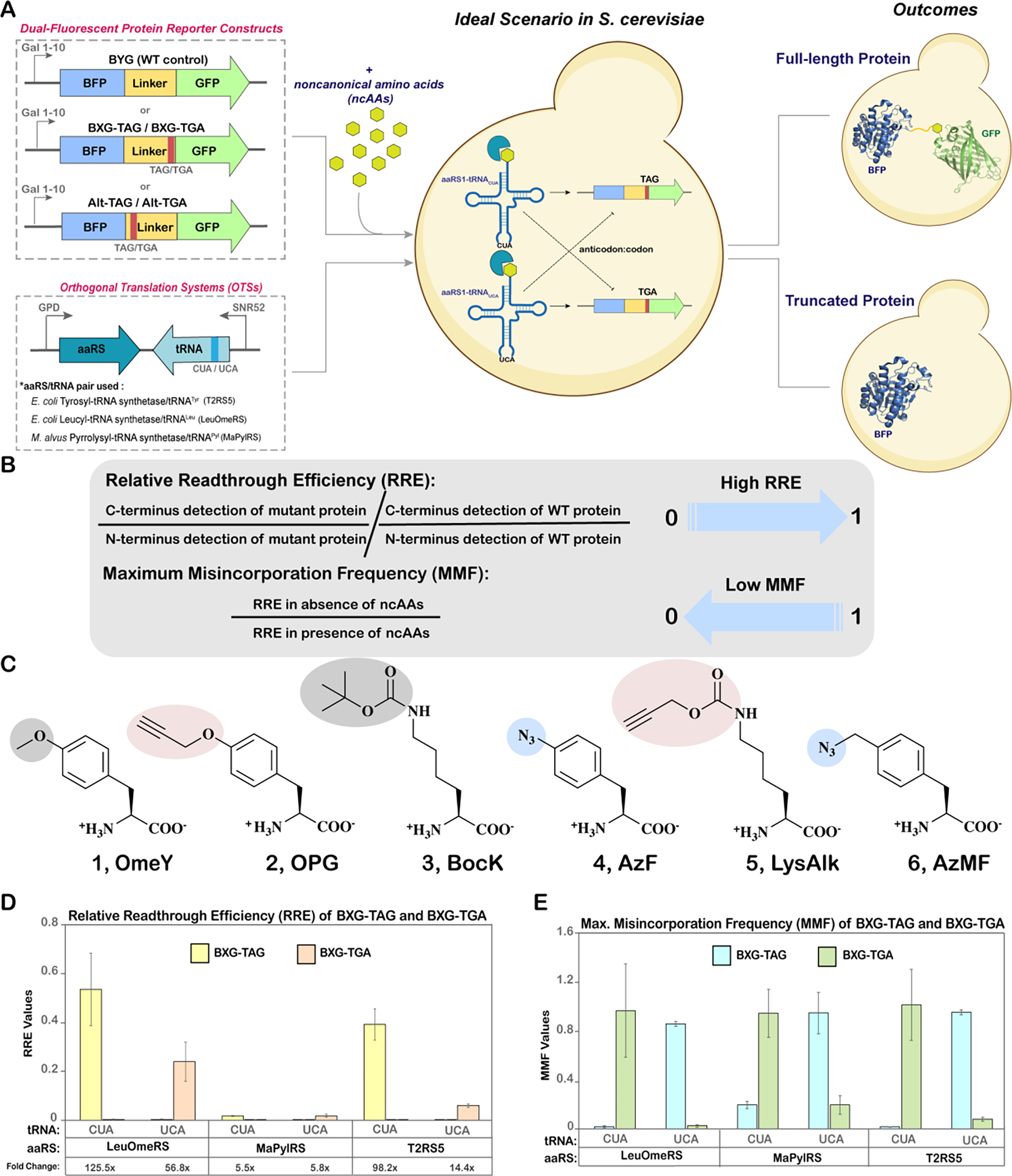
Evaluation of TAG versus TGA stop codon suppression. A) Schematic overview of dual plasmid approach outlining the architecture of dual-fluorescent reporter constructs and orthogonal translation system (OTS) constructs. The possible cognate and non-cognate codon:anticodon interactions of amber and opal suppressor tRNAs are shown as bold arrows and dotted lines, respectively. B) Equations used for determining relative readthrough efficiency (RRE) and maximum misincorporation frequency (MMF), quantitative parameters for evaluating ncAA incorporation efficiency and fidelity, respectively. C) Structures of ncAAs used in this study. Shaded regions highlight functional groups, some of which can undergo specific bioorthogonal reactions. 1, O-methyl-L-tyrosine (OmeY); 2, Nε-Boc-L-Lysine (BocK); 3, p-propargyloxyl-L-phenylalanine (OPG); 4, p-azido-L-phenylalanine (AzF); 5, Nε-propargyloxycarbonyl-L-lysine (LysAlk); 6, 4-azidomethyl-L-phenylalanine (AzMF). D) Relative Readthrough Efficiency (RRE) of BXG-TAG and BXG-TGA reporters with indicated tRNA and aaRS constructs calculated from flow cytometry experiments. The fold changes reported below the plots (e.g. 125.5x) indicate the fold change in readthrough between BXG-TAG and BXG-TGA by a particular OTS; these values were calculated from the ratios of median fluorescence intensities (MFIs) of readthrough events of these reporter constructs (the larger value was used as the numerator). E) Maximum Misincorporation Frequency (MMF) calculated for BXG-TAG and BXG-TGA reporter constructs. Flow cytometry dot plots and and bar graphs of MFIs for all data in this figure are depicted in Figure S1 and Figure S2, respectively. The error bars in D and E were derived from measurements of biological triplicates, starting with standard deviations of MFI values and propagated using error propagation equations (during calculations of RRE and MMF).
Employing our established, galactose-inducible dual fluorescent protein reporter system, we evaluated the incorporation of ncAAs at TGA and TAG codons55,56. The reporter system is a fusion protein containing a N-terminal fluorescent protein (BFP) and a C-terminal fluorescent protein (GFP) connected by a linker with desired orthogonal codon at a permissive site. The orthogonal codon was either a TAG (referred to as BXG-TAG) or TGA (referred to as BXG-TGA); a “wild-type” linker containing no stop codon served as a positive control (referred to as BYG; Figure 1A and Table S4)68. This dual-fluorescent reporter system enabled the use of the quantitative metrics Relative Readthrough Efficiency (RRE) and Maximum Misincorporation Frequency (MMF) to evaluate characteristics of protein translation with ncAAs (Figure 1B). RRE quantifies the readthrough of an orthogonal codon with respect to wildtype protein synthesis, with values typically ranging between 0 and 1. An RRE value of 0 indicates that all proteins produced are truncated at the orthogonal codon, while an RRE value of 1 indicates readthrough efficiency equivalent to wildtype protein synthesis. MMF measures the probability of unintentional canonical amino acid incorporation at the orthogonal codon, with values approaching zero during high fidelity ncAA incorporation and one or higher as the misincorporation frequency increases. An ideal scenario representing efficient and site-selective ncAA incorporation should have a high RRE and a low MMF value.
We utilized variants of three yeast-compatible OTSs: E. coli tyrosyl tRNA synthetase/tRNATyr, E. coli leucyl tRNA synthetase/tRNALeu and M. alvus pyrrolysyl tRNA synthetase/tRNAPyl, and cloned each of them separately into the pRS315 plasmid backbone under constitutive promoters (Figure 1A). Recognition of TAG or TGA codons during mRNA translation was facilitated by mutating the anticodon sequences of suppressor tRNAs to CUA or UCA, respectively. This simple approach facilitated evaluation of cognate and non-cognate aminoacylation events to determine whether the aaRSs can tolerate changes in anticodons, as anticodon identity is known to modulate tRNA recognition and aminoacylation in many aaRSs57. For the EcLeuRS/tRNALeu pair, we used the polyspecific aaRS variant LeuOmeRS with a T252A mutation in its editing domain and employed the ncAA O-methyl-L-tyrosine (1; OmeY) as a substrate58. For the EcTyrRS/tRNATyr pair, we employed a polyspecific aaRS variant, T2RS5, identified through our prior high-throughput screening and used it with the ncAA p-propargyloxyl-L-phenylalanine (2; OPG) as a substrate38. Lastly, for the PylRS/tRNAPyl pair, we used the M. alvus PylRS with the ncAA Nε-Boc-L-Lysine (3; BocK) as a substrate (Figure 1C)61.
For our initial investigation of ncAA incorporation at TGA codons, we conducted a series of flow cytometry experiments to determine RRE and MMF at “cognate” and “noncognate” orthogonal codons for each of the OTSs (Figure 1D). The three aaRS/tRNA pairs encoding plasmids with either tRNACUA or tRNAUCA were separately co-transformed into the S. cerevisiae strain RJY100 with a reporter construct encoding a cognate or a non-cognate orthogonal codon, or a WT reporter (absence of orthogonal codons; Figure 1A)59. All transformants were induced either in the absence or in the presence of 1 mM cognate ncAA and subjected to flow cytometry analysis and RRE and MMF calculations (Figure 1A; Figure S1). Notably, all three OTSs supported ncAA incorporation in response to TGA when the OTSs contained tRNAs with complementary UCA anticodon, but not a noncognate anticodon. Additionally, quantitative analyses indicated high RRE and low MMF values for all cognate OTS:reporter combinations tested, and the opposite trend for non-cognate OTS:reporter combinations. These data demonstrated that these OTSs exhibit specificity at the codon:anticodon interaction level, without any detectable cross-reactivity (Figure 1C and 1D). Moreover, both flow cytometry dot plots and median fluorescence intensity (MFI) graphs revealed insignificant background fluorescence levels for both TAG and TGA codons in the absence of any ncAAs, suggesting negligible participation of endogenous translation machinery in codon readthrough events (Figure S1 and S2). On direct comparison with TAG, TGA was decoded 2–6 fold less efficiently by both T2RS5/tRNAUCATyr and LeuOmeRS/tRNAUCALeu pairs (Figure 1C). The observation of lower levels of TGA readthrough by the two E. coli OTSs in comparison to TAG readthrough is consistent with the prior observations of Chatterjee and coworkers with TGA and TAG readthrough in mammalian cells17. In contrast, for the MaPylRS/tRNAPyl pair, the decoding efficiencies of both TAG and TGA codons were at equivalent levels when paired with tRNAs containing the “cognate” anticodon. However, RRE values determined with these systems were observed to be indistinguishable from samples lacking ncAAs. Hence, to further substantiate the translational activities of these samples, we evaluated median fluorescence intensities of C-terminal reporter expression and observed clear differences in fluorescence between samples induced in the absence and presence of ncAAs (Figure S2). Overall, these data validated TGA as an orthogonal codon in yeast for site-specific ncAA incorporation, with no detectable cross-reactivity with TAG at the codon:anticodon level.
Sequence context impacts TGA suppression efficiency
Prior to combining constructs encoding both TAG and TGA codons for dual ncAA incorporation, we sought to better understand the underlying factors that can influence ncAA incorporation in response to TGA. Previous studies have shown that nucleotides upstream and downstream of TAG codons can affect ncAA incorporation efficiency in bacteria, mammalian, and yeast expression systems28,56,60–63. These observations and the fact that natural TGA readthrough events in yeast (with a cAA) are also dictated by their neighboring nucleotides, motivated us to explore whether the same trend holds true for ncAA incorporation at TGA codons50,64. For our evaluation, we selected an alternate location within the linker of our reporter construct, referred to as ‘Alt-TAG’, which in our prior work demonstrated moderate but significantly decreased readthrough across different OTS and ncAA combinations56. We replaced the TAG in this construct with TGA to generate a new construct, referred to as Alt-TGA (Figure 1A, Table S4). For quantifying solely the changes related to orthogonal codon position, other experimental parameters including OTS identities, induction conditions, and ncAA types were all kept identical to the experimental parameters used in combination with BXG reporters.
Flow-cytometric analysis revealed similar ncAA incorporation efficiency and fidelity trends at the Alt-TGA position as at the BXG-TGA position, irrespective of the OTS used (Figure 2A, S3, and S4A). For both E. coli OTSs, ncAA incorporation in response to TAG was more efficient than incorporation in response to TGA, while for the MaPylRS/tRNAPyl OTS, ncAA incorporation in response to either TAG or TGA (with corresponding “cognate” tRNAs) was comparable; this is evident in evaluations of RRE, MMF, and median fluorescence intensities of C-terminal GFP fluorescence levels (Figure 2A and S4). Interestingly, comparing RRE values between the two stop codon positions (BXG- versus Alt- reporters) confirmed that the overall readthrough at the position within the Alt- construct was substantially lower than the corresponding readthrough at the position within the BXG- construct. This corroborated our and other research groups’ findings that the nucleotide environment around the orthogonal codon impacts the readthrough efficiency in yeast, and suggests that this trend holds irrespective of whether the readthrough occurs at a TAG or TGA codon56. To further assess changes in readthrough performance that occur between the two stop codon positions, we determined the ratios of the MFIs of GFP fluorescence for cognate OTS:reporter combinations at the BXG or BXG-TGA position to the Alt-TAG or Alt-TGA position (Figure 2B). Surprisingly, when comparing changes in readthrough at these two positions, the resulting fold changes in TGA readthrough across the three OTSs was distinct in comparison to corresponding fold changes in TAG. Theoretically, the fold change for a particular nonsense codon should not change between OTSs if the nucleotide context of the reporter is the sole contributing factor to readthrough efficiency. We attribute the observed differences in TGA readthrough (in comparison to TAG readthrough) to the anticodon change in the tRNA and to differences in the nucleotide sequences (and presumably identity elements) of the tRNAs in each OTS. Therefore, our data indicates that efficient codon:anticodon interactions are governed by specific features of both the stop codon sequence context in the reporter and the nucleotide sequence of the tRNA component of the OTS.
Figure 2.
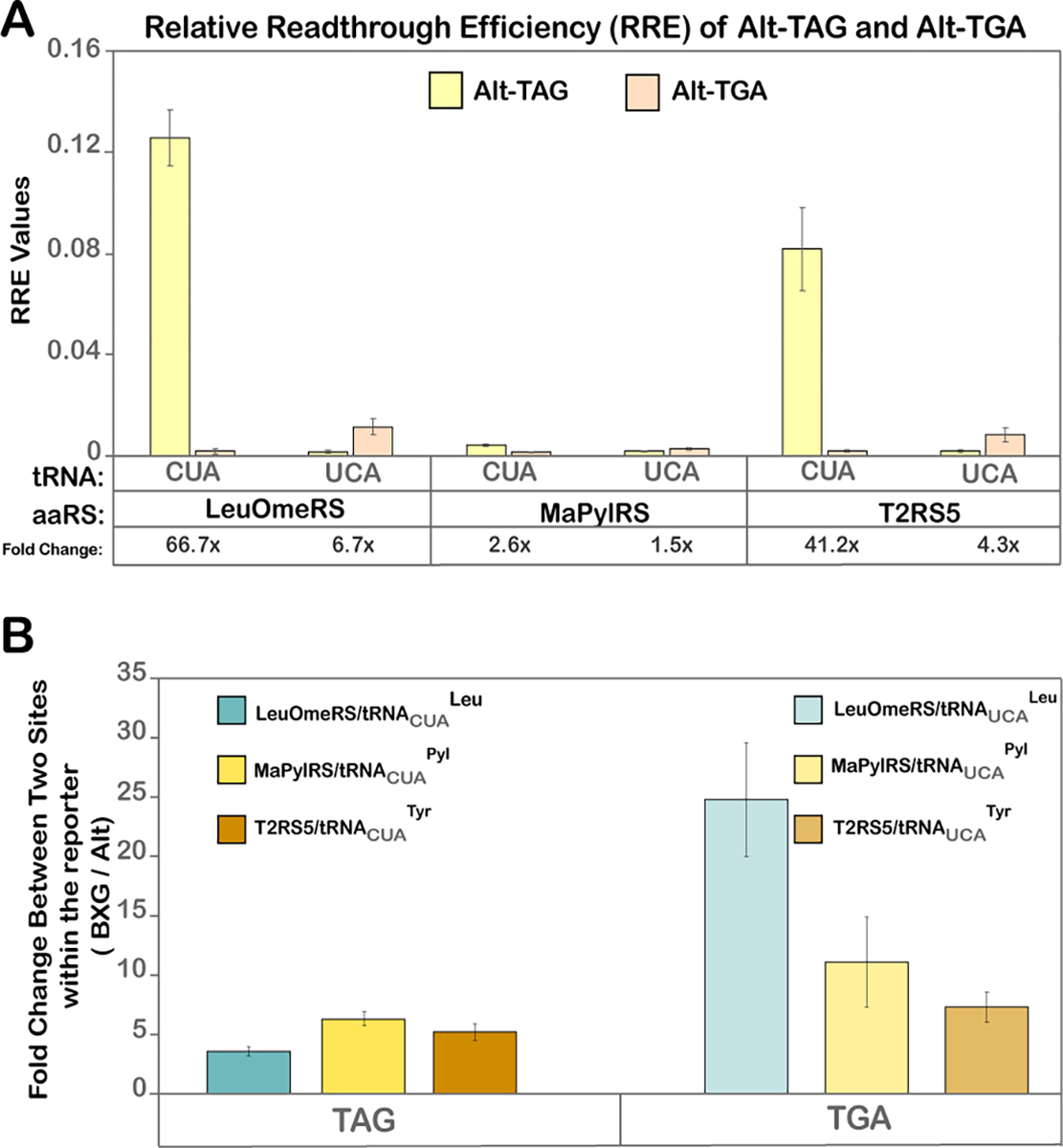
Quantitative analysis of readthrough events for Alt-TAG and Alt-TGA constructs. A) Relative Readthrough Efficiency (RRE) for Alt-TAG and Alt-TGA reporter constructs calculated from MFI values obtained from flow cytometry dot plot data, depicted in Figure S3. The fold-changes in readthrough (eg: 66.7X) in the table below indicate the fold change in readthrough between Alt-TAG and Alt-TGA by a particular OTS; these values were calculated from the ratios of MFIs of readthrough events of these reporter constructs (the larger value was used as the numerator). The corresponding MMF graph and the MFI bar graph are represented in Figure S4. B) Fold change in readthrough performance between the two codon positions in the reporter (BXG and Alt) obtained from the ratio of MFI values of cognate OTS:reporter interaction at the BXG-TAG (or BXG-TGA) site to the Alt-TAG (or Alt-TGA) site. The error bars in both A and B were calculated from the standard deviation of MFI values and then propagated in subsequent calculations using error propagation equations.
Single-gene knockout yeast strains enhance TGA readthrough efficiency
Numerous engineering strategies are known to augment stop codon readthrough, including single-gene deletions, whole-genome synthesis, and engineering various components of the translation machinery1,26,30,38,65,66. Prior work from our group showed that single-gene knockout strains of S. cerevisiae improved ncAA incorporation efficiencies in response to the TAG codon for various OTSs and ncAAs combinations 56,66,67. Based on our past work, we narrowed investigations here to two single-gene knockout yeast strains, ppq1Δ and tpa1Δ; deletions of these genes are also known to enhance nonsense suppression with near-cognate tRNAs68. TPA1 regulates mRNA poly(A) tail length and interacts with eukaryotic release factors eRF1 and eRF3, while PPQ1 is a serine/threonine phosphatase with an undefined role in translation termination69,70. We employed a BY4741 yeast strain background due to the ready availability of single-gene knockouts in this background. This necessitated transferring the BXG- and Alt- reporter constructs from a pCTCON2-plasmid backbone (TRP1 marker) to a pRS416-plasmid backbone (URA3 marker), since trp1 is not an available auxotrophic marker in BY4741. Otherwise, all experimental conditions were kept identical to the conditions used above, and we included TAG-containing reporter constructs for side-by-side comparison of readthrough effects in ppq1Δ and tpa1Δ strains.
NcAA incorporation efficiencies were evaluated via flow cytometry for all strains using each of the 3 OTSs and 2 stop codon positions summarized in Figure 1A. Our analysis (both RRE and MFIs) revealed that the ppq1Δ strain demonstrated statistically significant (2–3 fold) enhancements in TGA readthrough across different OTSs and codon positions compared to the WT BY4741 strain (Figure 3, Figure S9). These improvements were more readily apparent for the Alt-TGA (and Alt-TAG) constructs than for the corresponding BXG constructs (Figure 3; Figure S5–S7, S9). On other hand, for the tpa1Δ strain, slight increases in readthrough efficiency were only observed in some cases where EcTyrRS/tRNATyr or EcLeuRS/tRNALeu OTSs were used. In the case of MaPylRS/tRNAPyl, no enhancements in TGA readthrough efficiencies were observed in tpa1Δ under the conditions tested. Under all conditions tested here, improvements in ncAA incorporation efficiency in knockout strains were not accompanied by detectable changes to the apparent fidelity of ncAA incorporation, as evidenced by the lack of changes in MMF (Figure S8–S9). The overall trends in RRE values determined in BY4741 strains were consistent with the trends observed in RJY100. Most notably, RRE values for ncAA incorporation at TGA codons were lower than RRE values at TAG codons under corresponding conditions (Figure 1D and 3A; Figure 2A and 3B). Interestingly, when changing the plasmid backbone from pCTCON2 to pRS416, there was a modest decrease in RRE values with LeuOmeRS/tRNALeu OTSs for both TAG and TGA, but not for the other two OTSs. These observations are consistent with our previous findings, indicating that changes in plasmid backbone can impact the apparent performance of some OTSs for reasons that have not yet been elucidated56. In summary, the ppq1Δ strain enhanced ncAA incorporation across both orthogonal codons, independent of the codon position or OTS type.
Figure 3.
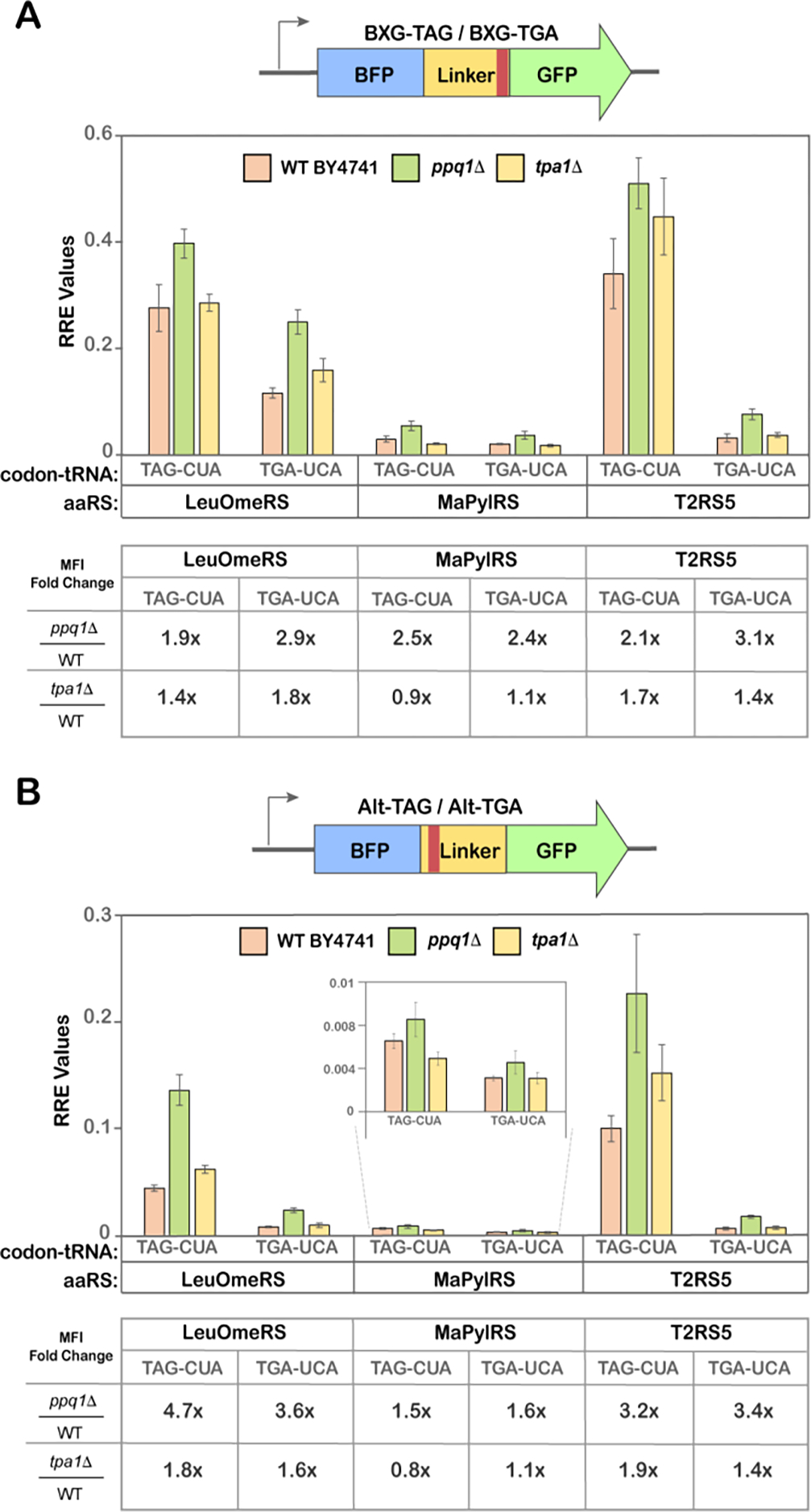
Quantitative readthrough measurements in single-gene knockout yeast strains. A) Relative Readthrough Efficiency (RRE) of BXG-TAG and BXG-TGA reporters with indicated tRNA and aaRS constructs calculated from flow cytometry experiments for the yeast strain BY4741 and single-gene knockouts ppq1Δ and tpa1Δ. B) Relative Readthrough Efficiency (RRE) of Alt-TAG and Alt-TGA reporters with indicated tRNA and aaRS constructs calculated from flow cytometry experiments for the yeast strain BY4741 and single-gene knockouts ppq1Δ and tpa1Δ. O-methyl-L-tyrosine (1; OmeY) was used as substrate for LeuOmeRS/tRNALeu, Nε-Boc-L-Lysine (2; BocK) was used as a substrate for MaPylRS/tRNAPyl; and p-propargyloxyl-L-phenylalanine (3; OPG) was used as a substrate for T2RS5/tRNATyr. The error bars are representative of three independent experiments, calculated from the standard deviations and processed through error propagation equations. The fold change in readthrough values (e.g. 1.9x) are represented in table format (below each RRE plot in A and B) and were obtained from the ratios of MFI values of a particular codon:OTS interaction in a knockout yeast strain to the corresponding MFI value in the WT BY4741 strain. The corresponding MMF graph and the MFI bar graphs for the data in this figure are depicted in supplementary Figure S8 and S9, respectively. One way-ANOVA was performed on MFI plots (Figure S9) to demonstrate the statistical significance between WT and KO strains.
Dual ncAA incorporation system for intracellular proteins
With a broad understanding of factors influencing ncAA incorporation at both TAG and TGA sites in separate constructs, we next sought to enable dual ncAA incorporation within a single protein. For facilitating expression of two OTSs, we replaced the reporter plasmid with our previously described single plasmid system encoding both a TAG-suppressing OTS and the inducible reporter system (Figure 4A)81. The reporter was modified to encode both TAG and TGA within the linker, at the “BXG” and “Alt” positions in both possible orientations, resulting in two mutant reporters: BX2G-AO (A: Amber (TAG); O: Opal (TGA)) and BX2G-OA. These plasmids were co-transformed with a separate plasmid encoding the second, TGA-suppressing OTS. We also used a single-plasmid version of the BYG reporter (orthogonal codons absent) as a positive control (Figure 4A). For initial analyses of dual ncAA incorporation, we selected the EcTyrRS/tRNATyr pair to suppress the TAG codon because it exhibited the highest TAG codon readthrough in the BY4741 background among all the OTSs and across different codon positions. We employed the TyrOmeRS variant in place of the T2RS5 variant because TyrOmeRS exhibits a narrower ncAA substrate preference, reducing potential crosstalk with other OTSs71. For suppressing TGA, we used the same OTS variants as were used in the investigations above, resulting in the following combinations of OTSs for dual ncAA incorporation experiments: EcTyrRS(TyrOmeRS)/tRNACUATyr + EcLeuRS(LeuOmeRS)/tRNAUCALeu (hereafter, referred as ‘TyrRSTAG + LeuRSTGA’) and EcTyrRS(TyrOmeRS)/tRNACUATyr + MaPylRS/tRNAUCAPyl (hereafter, referred as ‘TyrRSTAG + PylRSTGA’) (Figure 4A). For these experiments, we induced BY4741 and ppq1Δ yeast transformed with the dual systems under three sets of conditions: i) absence of ncAAs; ii) presence of one ncAA; and iii) presence of both ncAAs. These allowed systematic evaluation of cognate readthrough events as well as identification of aberrant readthrough events that may result from non-cognate interactions such as aaRSs:tRNA or aaRSs:ncAA (Figure 4A). For minimizing off-target aaRS:ncAA interactions, the ncAAs recognized most efficiently by their “cognate” OTSs were selected: 4-Azidomethyl-L-phenylalanine (6) as a substrate for LeuOmeRS, 4-Azido-L-phenylalanine (4) for TyrOmeRS and, Nε-Boc-L-Lysine (5) for MaPylRS.
Figure 4.
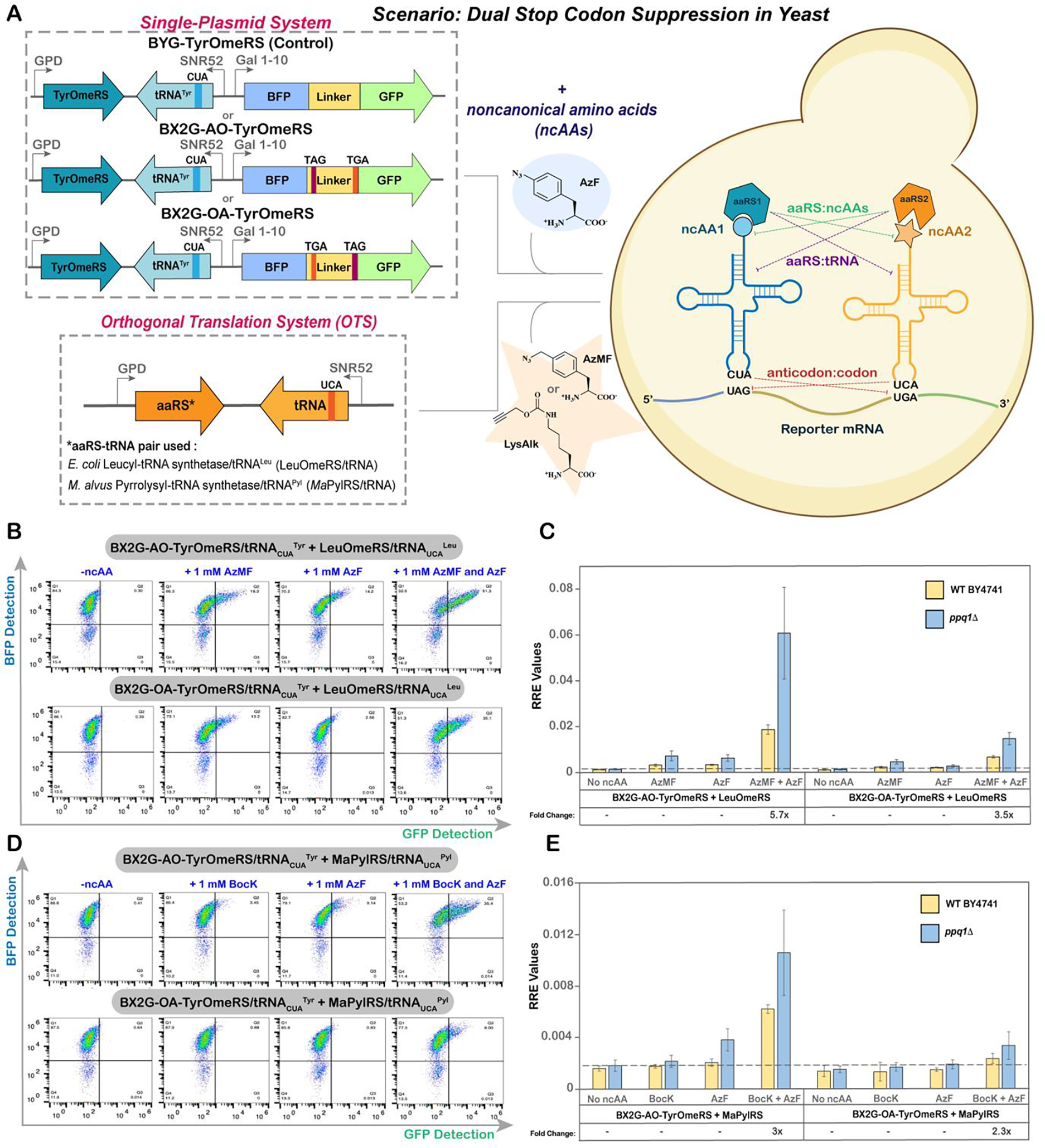
Evaluation of dual ncAA incorporation system in yeast. A) Experimental outline of dual plasmid approach to facilitate expression of two OTSs in the host cell. Dotted lines (---) and color-matched labels indicate potential non-cognate interactions that can occur. B) Flow cytometry dot plots of readthrough events of BX2G-AO and BX2G-OA in ppq1Δ, employing the OTS combination of EcTyrRS/tRNACUATyr + EcLeuRS/tRNAUCALeu and four different induction conditions. C) Corresponding Relative Readthrough Efficiency (RRE) calculated for OTS combination of EcTyrRS/tRNACUATyr + EcLeuRS/tRNAUCALeu for the reporters BX2G-AO and BX2G-OA in BY4741 and ppq1Δ strains. D) Flow cytometry dot plots of readthrough events of BX2G-AO and BX2G-OA in ppq1Δ, employing the OTS combination of EcTyrRS/tRNACUATyr + MaPylRS/tRNAUCAPyl and four different induction conditions. E) Corresponding Relative Readthrough Efficiency (RRE) measurements for the OTS combination of EcTyrRS/tRNACUATyr + MaPylRS/tRNAUCAPyl for the reporters BX2G-AO and BX2G-OA in WT BY4741 and ppq1Δ strains. The error bars are representative of three independent experiments, calculated from the standard deviation values and processed through error propagation equations. The fold change in readthrough represented in the graphs (e.g. 6x; panels C and E) are obtained from the ratios of MFI values of readthrough events in ppq1Δ and BY4741 strains. Corresponding maximum misincorporation frequency (MMF) graphs can be found in Supplementary Figure S12.
Following induction under all conditions, we conducted flow cytometry to explore dual ncAA incorporation. Dot plots and RRE calculations revealed clear evidence of full-length reporter expression in the presence of both ncAAs, for both OTS combinations and both orientations of orthogonal codon positions, (BX2G-AO or BX2G-OA) (Figure 4B–E, Figure S10–S11). Quantitatively, the amount of full-length reporter detected was 1–6% of wild-type reporter expression levels (Figure 4C and 4E). As expected, readthrough efficiency in ppq1Δ was higher than in BY4741, improving RRE for the TyrRSTAG + LeuRSTGA combination by 3–6 fold, while the RRE for the TyrRSTAG + PylRSTGA combination was improved by 2–3 fold (Figure 4C and 4E). Readthrough levels remained at background levels in the absence of ncAAs, but low levels of full-length reporter expression (<1% of the BYG WT control reporter) were observed when induced in the presence of a single ncAA. These aberrant readthrough events were more pronounced with the TyrRSTAG + LeuRSTGA combination of OTSs, likely due to non-cognate aaRSs:tRNA or aaRSs:ncAA interactions (Figure 4B–C). This observation is in line with prior work from Zheng et al in mammalian cells, where it was demonstrated that mutating the E. coli tRNA anticodon sequence resulted in recognition by non-cognate E. coli aaRSs17. In addition, since AzF and AzMF only differ by a methylene unit, there is a possibility of either of the aaRSs recognizing the “non-cognate” but structurally related ncAA. Also, there is some evidence for low-level AzF incorporation by LeuOmeRS in our earlier work62,68. In comparison to TyrRSTAG + LeuRSTGA, the TyrRSTAG + PylRSTGA OTS combination exhibited much lower RRE values for both orientations of stop codons. This is consistent with our above results and with our prior work (Figure 3)61. Readthrough efficiencies were also affected by the orientation of the two orthogonal codons within the reporter. Readthrough experiments with BX2G-AO led to approximately 4-fold higher levels of full-length reporter than experiments with BX2G-OA for both of the OTS combinations evaluated here. These results are in agreement with our observations of individual TAG and TGA codon readthrough events at BXG- and Alt- stop codon positions. As highlighted in Figure 2A and C, the BXG position is more permissive for orthogonal codon readthrough than the Alt position. As TGA is decoded at lower levels than TAG by the OTSs used here, having TGA at a more permissive position (BX2G-AO rather than BX2G-OA) facilitated higher overall readthrough (Figure 4C and E). In conclusion, these experiments demonstrate the feasibility of conducting dual ncAA incorporation in yeast, with the combination of TyrRSTAG + LeuRSTGA yielding up to 6% of wild-type reporter levels.
Dual ncAA incorporation system established in yeast display format
Next, we extended our dual ncAA incorporation system to yeast-displayed proteins, as yeast-display is a powerful platform to engineer proteins and supports myriad assays for evaluating properties like affinity, stability, activity, and specificity3,23,72. We began by identifying two sites in a previously reported synthetic antibody fragment (Donkey1.1 scFv) – L93 (located in complementarity determining region L3; CDR-L3) and H54 (located in CDR-H2) for encoding ncAAs in response to TAG and TGA (Figure 5A)3. Both of these sites have been previously demonstrated to tolerate different ncAAs inserted in response to the TAG codon. The two codons were incorporated interchangeably at both sites, giving rise to two yeast display reporter constructs: Donkey1.1-AO and Donkey1.1-OA (Figure 5A; the nomenclature for AO and OA is the same as stated in the previous section). We employed N- and C-terminal epitope tags (HA and c-myc) for quantitative flow cytometric evaluation of protein display levels. A wild-type version of the reporter protein was used as a control and to facilitate calculation of RRE and MMF. Similar to the set of plasmids used for the intracellular dual ncAA incorporation system, a single-plasmid yeast display + ncAA incorporation system was generated by cloning a TAG-suppressing OTS upstream of the reporter in the pCTCON2 vector backbone, with TGA-suppressing OTS cloned separately into the pRS315 vector backbone. Both of the plasmids were co-transformed into yeast-display strain RJY100, which facilitates the use of induction procedures identical to those used for dual fluorescent reporters59. For preparation of a “mutually orthogonal” EcTyrRS/tRNACUATyr + EcLeuRS/tRNAUCALeu combination, we used the TyrAcFRS variant as part of an EcTyrRS/tRNACUATyr OTS to suppress TAG, and a newly-identified variant termed LysAlkRS3 as part of the EcLeuRS/tRNAUCALeu OTS to suppress TGA38. These OTSs were chosen based on their nonoverlapping substrate specificities, where the ncAA AzF (4) can be encoded with TyrAcFRS/tRNACUATyr, and the ncAA LysAlk (5) can be encoded with LysAlkRS3/tRNAUCALeu. Importantly, choosing ncAAs with bio-orthogonal reactive handles in these experiments facilitated later investigations of double labeling strategies on the yeast surface.
Figure 5.
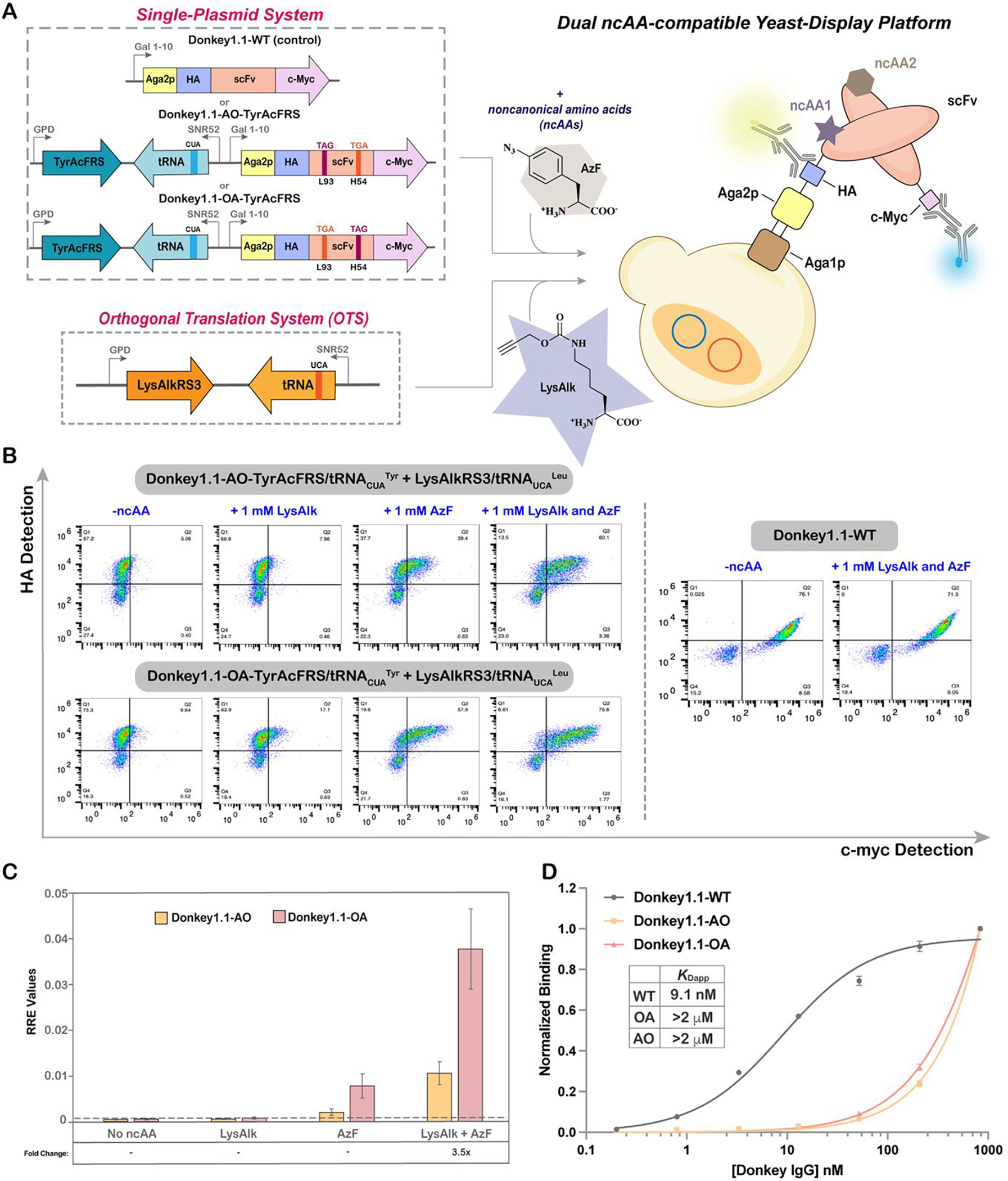
Evaluation of dual ncAA incorporation in yeast display format. A) Experimental outline for setting up a dual ncAA incorporation system with yeast-display platform. HA and c-Myc tags were employed to evaluate full-length readthrough of yeast-displayed scFvs using flow cytometry. B) Flow cytometry dot plots of readthrough experiments conducted in the RJY100 yeast display strain. Display of Donkey1.1-AO, Donkey1.1-OA and Donkey1.1 WT reporters following induction in the presence of 0, 1, or 2 ncAAs. Donkey1.1-WT was used as a positive control. C) RRE measurements comparing readthrough efficiencies between Donkey1.1-AO and Donkey1.1-OA scFv reporters across different induction conditions. Errors bars are representative of experiments conducted with three biological replicates and were determined via error propagation. The fold changes in readthrough represented in the graph (e.g. 3.5x) were obtained from the ratio of MFI values of readthrough events of Donkey1.1-OA to Donkey1.1-AO by the OTS combination of EcTyrRS/tRNACUATyr + EcLeuRS/tRNAUCALeu. The corresponding MMF data can be found in Supplementary Figure S13. D) Yeast display titration experiments with doubly substituted and WT synthetic antibody clones. The graph depicts normalized antigen binding levels of the three yeast-displayed scFvs obtained over a range of antigen concentrations using flow cytometry. Corresponding dot plots can be found in Supplementary Figure S14.
Flow cytometry evaluations revealed the presence of doubly-positive populations (HA+ and c-myc+) for both stop codon orientations following induction in the presence of AzF and LysAlk; this provides tangible evidence of full-length protein display on the yeast surface (Figure 5B). RRE analysis indicated that the full-length antibody display ranged between 1–4% of WT protein display levels. As observed with intracellular reporters, low levels of full-length protein (<1% of the WT protein) were also detected following induction in the presence of a single ncAA. This signal was most apparent when cultures were induced in the presence of AzF, which could be the result of non-cognate interactions between TyrAcFRS and tRNALeu; LysAlkRS3 and AzF; or LysAlkRS3 and canonical amino acids38. A comparison of full-length protein display levels revealed that the orientation of the orthogonal codons within the Donkey1.1-OA construct leads to 3.5-fold higher full-length protein than the orientation of the orthogonal codons within the Donkey1.1-AO construct. This implies that the L93 position is more permissive for TGA readthrough than H54, consistent with our prior observation with TAG readthrough events (Figure 5C)3. Furthermore, we assessed the binding functionalities of the doubly-substituted yeast-displayed clones through yeast surface titration experiments and included a wild-type version of the reporter (absence of any ncAAs) as a positive control. For both doubly substituted clones, we observed clear antigen binding for concentrations as low as 3.3 nM. However, their apparent binding affinities were decreased by roughly 2 orders of magnitude in comparison to the WT protein (KDapp: 9.1 nM; Figure 5D and S14). For both Donkey1.1- OA and Donkey1.1-AO, apparent KDs were estimated to be in range of approximately 3–5 μM. Given that we previously observed some loss of binding affinity upon single ncAA substitution at both L93 and H54, it seems reasonable to observe further loss of binding affinity upon the introduction of a second ncAA within the same construct. Another possibility is that the low display levels of doubly substituted constructs, which result in reduced avidity on the yeast surface, decrease the apparent affinities of the constructs for the dimeric IgG antigen. Nevertheless, these data clearly demonstrate the feasibility of performing dual ncAA incorporation on the yeast surface, and further provide evidence for the retention of antigen binding functionality in doubly substituted synthetic antibody variants.
To further corroborate dual ncAA incorporation, mass spectrometry was performed with soluble forms of Donkey1.1-OA and Donkey1.1-WT. Using plasmids encoding secreted scFv-Fc constructs, we prepared and isolated pure forms of each protein via protein A affinity chromatography as determined by SDS-PAGE (Figure S15)3. Like yeast-displayed constructs, these secreted constructs must traverse the secretory pathway prior to secretion. Following tryptic digests, it was possible to identify the L93 and H54 peptide fragments in both WT and OA samples (Figure 6A, B). For the WT sample, the L93 fragment was observed at m/z of 1108.5016, z=4 with a neutral mass of 4429.97 (expected 4429.9849; −3.36 ppm) and the H54 fragment was observed at m/z of 1117.5346 z=2 with a neutral mass of 2233.0549 (expected 2233.0539; 0.43 ppm) (Figure 6A). For the OA sample, the L93 fragment was observed at m/z of 1139.2668 z=4 with a neutral mass of 4553.0371 (expected 4553.0533; −3.56 ppm), suggesting LysAlk incorporation, and the H54 fragment was observed at m/z = 1155.057 z=2 with a neutral mass of 2308.1015 (expected 2308.1012; 0.15 ppm) (Figure 6B). Thus, the observed mass change associated with ncAA incorporation in the OA sample is [M+123] for the L93 site, as expected for LysAlk incorporation. However, it is only [M+75] instead of the anticipated [M+101] for pAzF incorporation in the H54 peptide. As we and others have previously observed, pAzF is readily converted to p-aminophenylalanine (pAmF) during sample preparation and mass spectrometry analysis38,73. The H54 fragment we identified is consistent (within 0.15 ppm) with the presence of pAmF, which we can use a surrogate for pAzF incorporation prior to analysis.
Figure 6.
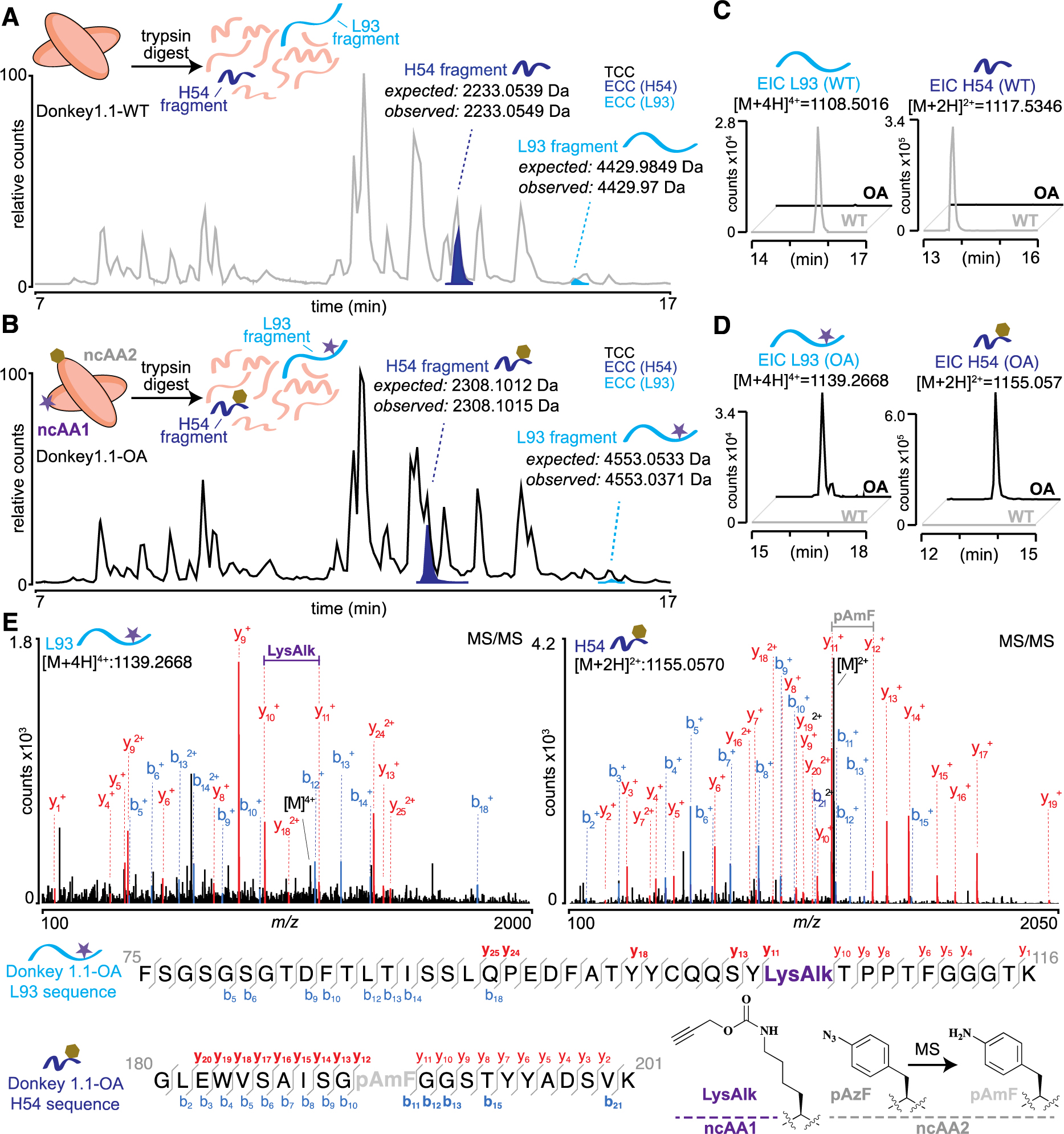
Confirmation of dual ncAA incorporation by mass spectrometry. After tryptic digestion of A) Donkey1.1-WT or B) Donkey1.1-OA, the resulting peptide fragments were analyzed by liquid-chromatography mass spectrometry (LC-MS). Total compound chromatograms (TCC) show the resulting digest mixture. Extracted compound chromatograms (ECC) show the peaks corresponding to L93 (cyan) and H54 (blue) tryptic fragments. A shift in retention time can be observed for ncAA-containing peptides in the OA sample, as compared to WT. Extracted ion chromatograms for C) WT L93 and H54 peptides or D) ncAA-containing L93 and H54 peptides show that the identified peptides are specific to either WT or OA samples. E) MS/MS analysis of ncAA-containing L93 and H54 peptides from the OA sample. Observed ions that are specific only to ncAA-containing (but not WT) peptides are shown in bold. blue, b ions; red, y ions.
Further confirmation of these assignments was accomplished by using extracted ion chromatograms (EIC) to search for specific tryptic fragments in both WT and OA samples. Using this approach, we confirmed that the unmodified (WT) H54 and L93 fragments could not be found in the OA digest (Figure 6C). Likewise, we also found that the LysAlk-containing L93 fragment and pAmF-containing H54 fragment could not be detected in the WT digest (Figure 6D). Finally, using MS/MS analysis, it was possible to identify diagnostic b+ and y+ ions confirming that both AzF (pAmF) and LysAlk were incorporated at the expected sites within the observed tryptic fragments (Figure 6E, Figures S16 and S17). Taken together, these results allow us to unambiguously confirm that Donkey1.1-OA contains both azide and alkyne functionalities imparted by dual ncAA incorporation.
Chemoselective double labeling of yeast-displayed and soluble proteins
Covalent attachment of two distinct chemical probes to dual ncAA-substituted proteins has enabled diverse biological applications, including FRET for studying protein conformation and dynamics12,13,16. Here, we investigate, for what we believe is the first time, the combination of SPAAC and CuAAC reactions to install two different chemical probes within yeast-displayed proteins. Generally, SPAAC and CuAAC reactions are not considered mutually compatible, as they both require azide reactants. However, prior studies have demonstrated that sequential use of SPAAC and CuAAC reactions can support selective biomolecule labeling, both in vivo and in vitro74–76. For facilitating exploration of such sequential reactions with our system, we used the yeast-displayed dual ncAA-containing antibody reporters Donkey1.1-AO and Donkey1.1-OA (Figure 5A; see previous section) along with the corresponding wild-type control. Figure 7A outlines the reaction conditions and probes used for labeling. Consistent with previous studies, it was essential to perform SPAAC on the azide present in the displayed protein of interest before conducting CuAAC to ensure that all free azides were consumed prior to exogenous addition of an azide probe for CuAAC. Flow cytometry analysis was used to evaluate all SPAAC and CuAAC reactions performed on doubly substituted proteins displayed on the yeast surface.
Figure 7.
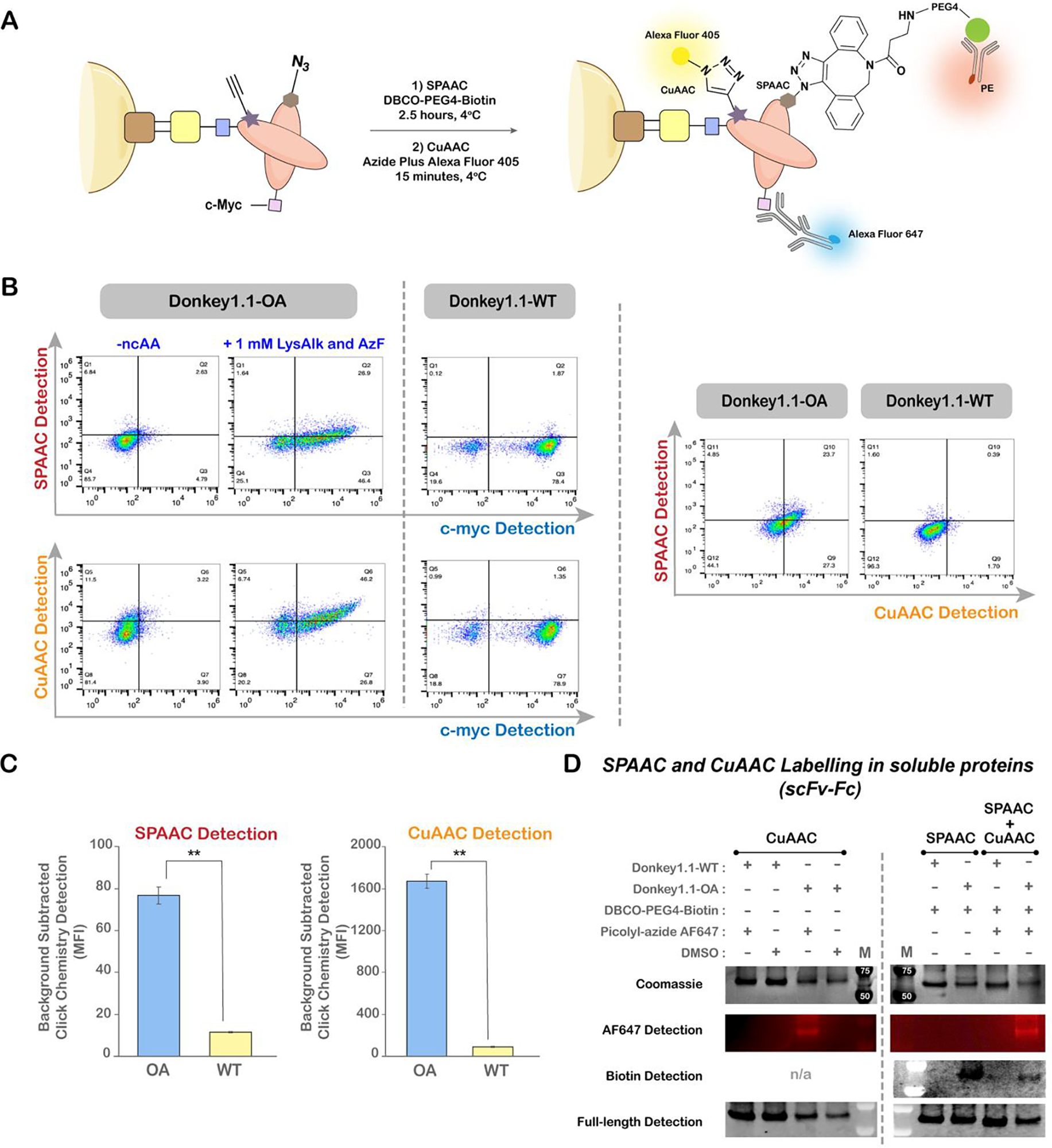
Site-specific double labeling of yeast-displayed and soluble proteins. A) Reaction and detection schemes for double labeling of yeast displayed, doubly substituted antibody constructs. TAG was employed to encode AzF (4; SPAAC reactions), while TGA was employed to encode LysAlk (5; CuAAC reactions). SPAAC products were detected using a PE-labeled anti-biotin antibody, CuAAC reaction products were detected with an Alexa Fluor 405-labeled azide probe, and c-myc (full length proteins) was detected using an anti-c-myc primary antibody followed by an Alexa Fluor 647-labeled secondary antibody. B) Flow cytometry dot plots of SPAAC and CuAAC reactions along with full-length (c-myc) display detection for Donkey1.1-OA under different induction conditions. Donkey1.1-WT (no ncAA substitutions) served as a negative control. C) Median fluorescence intensities of SPAAC and CuAAC reaction products in c-myc+ populations for Donkey1.1-OA and Donkey1.1-WT constructs. Error bars denote standard deviations obtained from three independent experiments. Unpaired t-tests were performed to evaluate statistical significance (**: p<0.005). D) Single and double labeling of soluble Donkey1.1-OA and Donkey1.1-WT scFv-Fcs. SPAAC was performed using a DBCO-PEG4-biotin probe and detected by western blot using Streptavidin 488. CuAAC was performed using Picolyl-azide Alexa Fluor 647 and detected by fluorescent gel imaging. Full-length protein expression was detected by western blot using goat anti-human IgG Fc Dy488. Coomassie staining was performed to verify sample purity. “M” indicates gel lane with molecular weight marker. Corresponding dot plots for all control flow cytometry experiments can be found in Supplementary Figure S18.
We first examined each reaction individually with both Donkey1.1-OA and Donkey1.1-AO constructs to identify efficient reaction conditions. Flow cytometry data from single-labeling studies with SPAAC and CuAAC reactions revealed double-positive populations for Donkey1.1-OA signifying that the full-length OA construct (c-myc+) had successfully undergone either SPAAC (biotin+) or CuAAC (Alexa Fluor 405+) reactions (Figure S18). On other hand, for the Donkey1.1-AO reporter protein, the flow cytometry data provided clear evidence for SPAAC, but not for CuAAC reactions, where high background fluorescence was observed. This made it difficult to delineate specific CuAAC labeling (Alexa Fluor 405+) of full-length proteins (c-myc+) from non-specific labeling of yeast cells (Figure S18). We attribute the lack of specific signal to low levels of LysAlk (5) at the H54 site, which is observed to be a less permissive site for TGA suppression. Thus, we proceeded to perform sequential double labeling with the Donkey1.1-OA construct as it unambiguously supported selective bioorthogonal reactions (SPAAC and CuAAC) (Figure 7A). Dot plots indicated elevated detection levels of both biotin (SPAAC) and Alexa Fluor 405 (CuAAC) probes in c-myc+ populations of Donkey1.1-OA samples. Such signals were undetected in the case of c-myc– cells of Donkey1.1-OA or in any cells of Donkey1.1-WT samples (Figure 7B, left panel). Additionally, dot plots of biotin detection versus Alexa Fluor 405 detection confirmed that both the SPAAC and CuAAC probes were present in the same population of Donkey1.1-OA cells (Figure 7B, right panels). Quantitative evaluation of the c-myc+ populations for the SPAAC (biotin) and CuAAC (Alexa Fluor 405) probes further substantiated statistically significant increases in the fluorescent signal of Donkey1.1-OA samples in comparison to Donkey1.1-WT samples (Figure 7C). All these data provide direct evidence of double-labeling of yeast-displayed proteins and confirm that both azide and alkyne functionalities encoded within the Donkey1.1-OA antibody construct can be selectively addressed using sequential SPAAC and CuAAC reactions.
To validate our observations of double labeling in yeast-display format, we used the soluble form of Donkey1.1-OA to conduct evaluations of SPAAC and CuAAC reactivities in solution via SDS-PAGE and Western blotting; donkey1.1-WT protein was utilized as a negative control. With individual SPAAC (probe: DBCO-PEG4-biotin) and CuAAC reactions (probe: picolyl azide Alexa Fluor 647) in solution, we detected protein labeling only with Donkey1.1-OA and not with the Donkey1.1-WT control (Figure 7D), revealing the presence of two noncanonical functionalities in Donkey1.1-OA and confirming the selectivity of individual SPAAC and CuAAC reactions in solution. Finally, we explored double labeling of Donkey1.1-OA in single pot reactions (Figure 7D). Clear signals for both probes were evident in the Donkey1.1-OA sample subjected to sequential bioorthogonal reactions. To achieve these signals, we treated samples with low concentrations of the SPAAC probe for extended reaction times to attempt to minimize interference of unreacted DBCO with the CuAAC probe (picolyl azide Alexa Fluor 647). Overall, for the first time, we have successfully demonstrated the feasibility of performing two distinct conjugations on yeast-prepared, doubly substituted proteins in both yeast display format and on secreted, purified proteins.
Discussion
In this study, we have identified and systematically characterized OTSs, gene deletions, and stop codon contexts that support (and enhance) the incorporation of diverse ncAAs in response to opal (TGA) codons in yeast. Combining these systems with mutually orthogonal amber suppression machineries, we established dual ncAA incorporation in yeast for the first time with both intracellular and yeast-displayed proteins. The successful incorporation of two distinct ncAAs at pre-defined sites within a protein was further confirmed using ESI-MS and tandem mass spectrometry analysis. Furthermore, in both formats, we could exploit the encoded ncAAs for biorthogonal conjugation reactions to achieve double labeling, demonstrating the utility of our engineered system for presenting multiple, distinct functionalities within a single protein.
In our initial investigations of ncAA incorporation in response to TGA, we determined that variants from three distinct classes of OTSs in yeast – EcTyrRS/tRNATyr, EcLeuRS/tRNALeu and MaPylRS/tRNAPyl – supported significant ncAA incorporation without any detectable readthrough from either orthogonal tRNACUAs or from endogenous aaRS-tRNA pairs. Such versatile decoding of TGA allowed us to identify two different OTS combinations that support dual ncAA incorporation in yeast: TyrRS/tRNACUATyr + LeuRS/tRNAUCALeu and TyrRS/tRNACUATyr + PylRS/tRNAUCAPyl. Both combinations facilitated incorporation of two distinct ncAAs in proteins expressed intracellularly, and the TyrRS/tRNACUATyr + LeuRS/tRNAUCALeu combination further facilitated dual ncAA incorporation in yeast display format. However, we did observe some evidence for aberrant production of full-length protein when the dual incorporation systems were induced in the presence of only a single ncAA (most notably with TyrRS/tRNACUATyr + LeuRS/tRNAUCALeu). This may be attributable to one or more of the following: 1) mischarging of tRNA by non-cognate aaRSs (for example, tRNACUATyr by LeuRS); 2) recognition of a non-cognate amino acid substrate by an aaRS (for example, TyrRS recognizing AzMF, instead of its “cognate” AzF substrate); or 3) mischarging of a tRNA with a cAA (for example, tRNACUATyr is charged with tryptophan rather than the ncAA of interest). The first possibility is a known phenomenon observed by Chatterjee and coworkers when using a combination of E. coli aaRSs in mammalian cells17. Mutating the tRNA anticodon sequence can result in the tRNA becoming a substrate of a non-cognate E. coli aaRS. In the future, such crosstalk could be mitigated by engineering or designing tRNAs to reduce such phenomena. The second possibility (noncognate ncAA charging) arises due to the polyspecific nature of many engineered aaRSs. This challenge could be avoided by utilizing aaRSs with non-overlapping sets of substrates. High-throughput engineering efforts from our lab and others have demonstrated the possibility of engineering highly specific aaRSs that discriminate against closely related ncAA substrates13,38,78. Lastly, the third possibility (canonical amino acid mischarging) is a known challenge associated with engineering aaRSs for genetic code expansion38,78–79. While a portion of this challenge can be mitigated by using high ncAA concentrations during induction of protein synthesis, the discovery of additional high-performing aaRS variants that discriminate against the canonical amino acids will be important going forward. Dual ncAA incorporation in yeast will certainly benefit from ongoing efforts to expand the availability and performance of OTSs to access a broader palette of genetically encodable ncAAs with useful chemistries10,33,34,36–39.
Identification of mutually orthogonal pairs of OTSs in yeast enabled us to prepare doubly substituted synthetic antibodies with unique reactive functionalities. Albeit with a decrease in affinity, these synthetic antibodies retained their binding function and facilitated efficient labelling with two bioorthogonal chemistries. Sequential SPAAC and CuAAC reactions led to doubly functionalized antibodies in both yeast display and soluble formats, which has not previously been achieved in yeast-based ncAA incorporation systems. Other promising biorthogonal conjugation chemistries for double labelling explorations on the yeast surface and in yeast-produced proteins include strain-promoted inverse electron-demand Diels-Alder cycloaddition (SPIEDAC) and chemoselective rapid azo-coupling reaction (CRACR)11–13,17, 80,81. Doubly labeled proteins on the yeast surface are expected to enable applications such as FRET to study protein conformation and dynamics, as evidenced by prior work in mammalian cells16,82–84. Beyond bioconjugation strategies, proteins with dual ncAAs can support applications including intramolecular protein stapling, tethering strategies for small molecules or other drug modalities, and simultaneous presentation of two distinct chemistries such as crosslinking chemistries or posttranslational modifications3,12,13,18,72,85. Thus, the emerging dual ncAA incorporation strategies in this work along with the expanding collection of OTSs supporting diverse ncAA incorporation in yeast will streamline the realization of these intricate protein manipulations in the near future.
Despite the successful implementation of dual ncAA incorporation in yeast, the systems described here exhibit relatively low efficiencies due to the lower readthrough efficiency of TGA compared to TAG by the E. coli OTSs. One likely explanation is that the anticodon change from CUA to UCA is not well-tolerated within the tRNAs, indicating that the tRNA identity elements play a role in determining TGA decoding efficiency86. Such inefficiencies from poor tRNA recognition can be alleviated either by enhancing the expression levels of these tRNAs to increase tRNA abundance, or by engineering suppressor tRNAs to improve tRNA recognition35,65,87,88. In addition to tRNA properties, the nucleotide context surrounding opal codons also dictates ncAA incorporation efficiency. The effects of changing the context around an opal codon appear to be similar to the effects of changing the context around an amber codon. This is consistent with prior work from our own group and work from other groups investigating the role of nucleotide context in influencing ncAA incorporation efficiency (bacteria, mammalian cells and yeast)56,60–62,67. Besides engineering individual translation components, genetic and genomic engineering approaches offer a complementary set of tools for further enhancing dual ncAA incorporation efficiency in yeast. Single-gene deletion yeast strains ppq1Δ and tpa1Δ demonstrated significant improvements in ncAA incorporation in response to TGA across different OTSs, codon positions, and a range of ncAAs. The enhancements observed are consistent with the enhancements measured for ncAA incorporation in response to TAG in our previous work56. Our findings suggest that improving our fundamental understanding of eukaryotic protein biosynthesis may lead to further enhancements in genetic code manipulation in yeast (and other eukaryotes). Recently, our group reported a genome-wide screen to identify single-gene deletions that enhance ncAA incorporation efficiency in response to the amber (TAG) codon66. Out of the several dozen hits we identified, many of the genes had functions that are apparently unrelated to protein biosynthesis, indicating that there are numerous routes available for engineering enhanced dual ncAA incorporation systems. Finally, bold genome engineering strategies such as the construction of a synthetic yeast genome (Sc2.0 project), free up codons for the addition of ncAAs to the genetic code26,89,90. “Blank” codons in such synthetic genomes are expected to greatly enhance the efficiency of ncAA incorporation and should be compatible with the dual ncAA incorporation machinery described in this work.
Conclusions
The yeast S. cerevisiae is one of the most important model organisms for elucidating fundamental biology and is a valuable chassis for protein engineering and synthetic biology. Empowering yeast with the ability to encode multiple ncAAs in individual proteins will lead to exciting biotechnological advances including new classes of protein-based therapeutics, biomaterials, artificial enzymes, and biosensors. More fundamentally, elucidating the processes that support and enhance dual ncAA incorporation systems is expected to lead to a deeper understanding of eukaryotic translation processes and the identification of unknown factors that can enhance genetic code expansion in yeast. In our current work, the tools and conditions identified are indispensable elements for realizing such applications and provide general insights that can be applied to the implementation of multiple ncAA incorporation systems in additional eukaryotic cells and organisms.
Supplementary Material
Acknowledgements
This research was supported by grants from the National Institute of General Medical Sciences of the National Institutes of Health (1R35GM133471 to J.A.V. and R01GM132422 to R.A.S.). The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors would also like to thank Heather Amoroso and Alla Leshinsky at the Koch Institute Biopolymers and Proteomics Core for their assistance with MALDI mass spectrometry data collection. Additionally, the authors would like to thank Rebecca Hershman for her valuable feedback on the manuscript.
Footnotes
Conflicts of Interest
P.L. and J.A.V declare competing financial interests: A provisional patent application (Application No.- 63/319,729) has been filed by Tufts University for the dual ncAA incorporation systems described in this work. T2RS5, a variant of EcTyrRS, and LysAlkRS3, a variant of EcLeuRS used in this study, are sequence variants disclosed in a patent application filed by Tufts University- U.S. Patent Application No. PCT/US22/29775.
Supporting Information
Experimental details, supplemental flow cytometry plots, fluorescence intensity graphs, RRE and MMF graphs, SDS-PAGE gel, MALDI mass spectrometry data, list of primers utilized for different plasmid construction, list of vector backbones for all the reporters and OTSs, list of yeast strains and plasmid combinations, DNA sequences of ‘Linker’ region present in dual fluorescent reporter construct and DNA sequence of scFv reporters with orthogonal codon incorporation sites.
References:
- (1).Chin JW Expanding and Reprogramming the Genetic Code of Cells and Animals. Annu. Rev. Biochem. 2014, 83 (1), 379–408. 10.1146/annurev-biochem-060713-035737. [DOI] [PubMed] [Google Scholar]
- (2).Chung CZ; Amikura K; Söll D Using Genetic Code Expansion for Protein Biochemical Studies. Front Bioeng Biotechnol 2020, 8. 10.3389/fbioe.2020.598577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Islam M; Kehoe HP; Lissoos JB; Huang M; Ghadban CE; Berumen Sánchez G; Lane HZ; Van Deventer JA Chemical Diversification of Simple Synthetic Antibodies. ACS Chem. Biol. 2021, 16 (2), 344–359. 10.1021/acschembio.0c00865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Lewis AK; Harthorn A; Johnson SM; Lobb RR; Hackel BJ Engineered Protein-Small Molecule Conjugates Empower Selective Enzyme Inhibition. Cell Chem Biol 2021. 10.1016/j.chembiol.2021.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Shandell MA; Tan Z; Cornish VW Genetic Code Expansion: A Brief History and Perspective. Biochemistry 2021, 60 (46), 3455–3469. 10.1021/acs.biochem.1c00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Vanderschuren K; Arranz-Gibert P; Khang M; Hadar D; Gaudin A; Yang F; Folta-Stogniew E; Saltzman WM; Amiram M; Isaacs FJ Tuning Protein Half-Life in Mouse Using Sequence-Defined Biopolymers Functionalized with Lipids. PNAS 2022, 119 (4). 10.1073/pnas.2103099119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Zubi YS; Seki K; Li Y; Hunt AC; Liu B; Roux B; Jewett MC; Lewis JC Metal-Responsive Regulation of Enzyme Catalysis Using Genetically Encoded Chemical Switches. Nat Commun 2022, 13 (1), 1864. 10.1038/s41467-022-29239-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Dumas A; Lercher L; Spicer CD; Davis BG Designing Logical Codon Reassignment – Expanding the Chemistry in Biology. Chem. Sci. 2015, 6 (1), 50–69. 10.1039/C4SC01534G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Vargas-Rodriguez O; Sevostyanova A; Söll D; Crnković A Upgrading Aminoacyl-tRNA Synthetases for Genetic Code Expansion. Curr Opin Chem Biol 2018, 46, 115–122. 10.1016/j.cbpa.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Dunkelmann DL; Willis JCW; Beattie AT; Chin JW Engineered Triply Orthogonal Pyrrolysyl–tRNA Synthetase/tRNA Pairs Enable the Genetic Encoding of Three Distinct Non-Canonical Amino Acids. Nat. Chem. 2020, 12 (6), 535–544. 10.1038/s41557-020-0472-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Zheng Y; Mukherjee R; Chin MA; Igo P; Gilgenast MJ; Chatterjee A Expanding the Scope of Single- and Double-Noncanonical Amino Acid Mutagenesis in Mammalian Cells Using Orthogonal Polyspecific Leucyl-tRNA Synthetases. Biochemistry 2018, 57 (4), 441–445. 10.1021/acs.biochem.7b00952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Bednar RM; Jana S; Kuppa S; Franklin R; Beckman J; Antony E; Cooley RB; Mehl RA Genetic Incorporation of Two Mutually Orthogonal Bioorthogonal Amino Acids That Enable Efficient Protein Dual-Labeling in Cells. ACS Chem. Biol. 2021, 16 (11), 2612–2622. 10.1021/acschembio.1c00649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Meineke B; Heimgärtner J; Eirich J; Landreh M; Elsässer SJ Site-Specific Incorporation of Two NcAAs for Two-Color Bioorthogonal Labeling and Crosslinking of Proteins on Live Mammalian Cells. Cell Rep 2020, 31 (12), 107811. 10.1016/j.celrep.2020.107811. [DOI] [PubMed] [Google Scholar]
- (14).Sachdeva A; Wang K; Elliott T; Chin JW Concerted, Rapid, Quantitative, and Site-Specific Dual Labeling of Proteins. J. Am. Chem. Soc. 2014, 136 (22), 7785–7788. 10.1021/ja4129789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Venkat S; Sturges J; Stahman A; Gregory C; Gan Q; Fan C Genetically Incorporating Two Distinct Post-Translational Modifications into One Protein Simultaneously. ACS Synth. Biol. 2018, 7 (2), 689–695. 10.1021/acssynbio.7b00408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Wang K; Sachdeva A; Cox DJ; Wilf NM; Lang K; Wallace S; Mehl RA; Chin JW Optimized Orthogonal Translation of Unnatural Amino Acids Enables Spontaneous Protein Double-Labelling and FRET. Nat Chem 2014, 6 (5), 393–403. 10.1038/nchem.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Zheng Y; Addy PS; Mukherjee R; Chatterjee A Defining the Current Scope and Limitations of Dual Noncanonical Amino Acid Mutagenesis in Mammalian Cells. Chem. Sci. 2017, 8 (10), 7211–7217. 10.1039/C7SC02560B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Zheng Y; Gilgenast MJ; Hauc SC; Chatterjee A Capturing Post-Translational Modification-Triggered Protein-Protein Interactions Using Dual Noncanonical Amino Acid Mutagenesis. ACS Chem Biol 2018, 13 (5), 1137–1141. 10.1021/acschembio.8b00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Botstein D; Fink GR Yeast: An Experimental Organism for 21st Century Biology. Genetics 2011, 189 (3), 695–704. 10.1534/genetics.111.130765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Smith MG; Snyder M Yeast as a Model for Human Disease. Curr Protoc Hum Genet 2006, Chapter 15, Unit 15.6. 10.1002/0471142905.hg1506s48. [DOI] [PubMed] [Google Scholar]
- (21).Mattanovich D; Branduardi P; Dato L; Gasser B; Sauer M; Porro D Recombinant Protein Production in Yeasts. Methods Mol Biol 2012, 824, 329–358. 10.1007/978-1-61779-433-9_17. [DOI] [PubMed] [Google Scholar]
- (22).Chao G; Lau WL; Hackel BJ; Sazinsky SL; Lippow SM; Wittrup KD Isolating and Engineering Human Antibodies Using Yeast Surface Display. Nat Protoc 2006, 1 (2), 755–768. 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- (23).Cherf GM; Cochran JR Applications of Yeast Surface Display for Protein Engineering. Methods Mol Biol 2015, 1319, 155–175. 10.1007/978-1-4939-2748-7_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Giaever G; Nislow C The Yeast Deletion Collection: A Decade of Functional Genomics. Genetics 2014, 197 (2), 451–465. 10.1534/genetics.114.161620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).McMahon C; Baier AS; Pascolutti R; Wegrecki M; Zheng S; Ong JX; Erlandson SC; Hilger D; Rasmussen SGF; Ring AM; Manglik A; Kruse AC Yeast Surface Display Platform for Rapid Discovery of Conformationally Selective Nanobodies. Nat Struct Mol Biol 2018, 25 (3), 289–296. 10.1038/s41594-018-0028-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Sanders J; Hoffmann SA; Green AP; Cai Y New Opportunities for Genetic Code Expansion in Synthetic Yeast. Curr Opin Biotechnol 2022, 75, 102691. 10.1016/j.copbio.2022.102691. [DOI] [PubMed] [Google Scholar]
- (27).Li X; Liu CC Biological Applications of Expanded Genetic Codes. ChemBioChem 2014, 15 (16), 2335–2341. 10.1002/cbic.201402159. [DOI] [PubMed] [Google Scholar]
- (28).Chen Y; He X; Ma B; Liu K; Gao T; Niu W; Guo J Noncanonical Amino Acid Mutagenesis in Response to Recoding Signal-Enhanced Quadruplet Codons. Nucleic Acids Research 2022, 50 (16), 1–12. 10.1093/nar/gkac474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Niu W; Schultz PG; Guo J An Expanded Genetic Code in Mammalian Cells with a Functional Quadruplet Codon. ACS Chem. Biol. 2013, 8 (7), 1640–1645. 10.1021/cb4001662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).de la Torre D; Chin JW Reprogramming the Genetic Code. Nat Rev Genet 2021, 22 (3), 169–184. 10.1038/s41576-020-00307-7. [DOI] [PubMed] [Google Scholar]
- (31).Wiltschi B Incorporation of Non-Canonical Amino Acids into Proteins in Yeast. Fungal Genetics and Biology 2016, 89, 137–156. 10.1016/j.fgb.2016.02.002. [DOI] [PubMed] [Google Scholar]
- (32).Biddle W; Schwark DG; Schmitt MA; Fisk JD Directed Evolution Pipeline for the Improvement of Orthogonal Translation Machinery for Genetic Code Expansion at Sense Codons. Front. Chem. 2022, 10. 10.3389/fchem.2022.815788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Grasso KT; Singha Roy SJ; Osgood AO; Yeo MJR; Soni C; Hillenbrand CM; Ficaretta ED; Chatterjee A A Facile Platform to Engineer Escherichia Coli Tyrosyl-tRNA Synthetase Adds New Chemistries to the Eukaryotic Genetic Code, Including a Phosphotyrosine Mimic. ACS Cent. Sci. 2022. 10.1021/acscentsci.1c01465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Italia JS; Addy PS; Wrobel CJJ; Crawford LA; Lajoie MJ; Zheng Y; Chatterjee A An Orthogonalized Platform for Genetic Code Expansion in Both Bacteria and Eukaryotes. Nat Chem Biol 2017, 13 (4), 446–450. 10.1038/nchembio.2312. [DOI] [PubMed] [Google Scholar]
- (35).Kelemen R; Jewel D; Huang R; Zhu Z; Cao X; Pasha M; Anthony J; Opijnen T. van; Chatterjee A. Virus-Assisted Directed Evolution of Enhanced Suppressor tRNAs in Mammalian Cells. bioRxiv 2022. 10.1101/2022.01.21.477302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).He X; Chen Y; Beltran DG; Kelly M; Ma B; Lawrie J; Wang F; Dodds E; Zhang L; Guo J; Niu W Functional Genetic Encoding of Sulfotyrosine in Mammalian Cells. Nat Commun 2020, 11 (1), 4820. 10.1038/s41467-020-18629-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Tan L; Zheng Z; Xu Y; Kong W; Dai Z; Qin X; Liu T; Tang H Efficient Selection Scheme for Incorporating Noncanonical Amino Acids Into Proteins in Saccharomyces Cerevisiae. Front. Bioeng. Biotechnol. 2020, 8, 569191. 10.3389/fbioe.2020.569191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Stieglitz JT; Van Deventer JA High-Throughput Aminoacyl-tRNA Synthetase Engineering for Genetic Code Expansion in Yeast. ACS Synth. Biol. 2022, 11 (7), 2284–2299. 10.1021/acssynbio.1c00626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Osgood AO; Zheng Y; Roy SJS; Loynd C; Jewel D; Chatterjee A An Efficient Opal-Suppressor Tryptophanyl Pair Creates New Routes for Simultaneously Incorporating up to Three Distinct Noncanonical Amino Acids into Proteins in Mammalian Cells. bioRxiv 2022, 19. 10.1101/2022.08.02.502539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Ding W; Zhao H; Chen Y; Zhang B; Yang Y; Zang J; Wu J; Lin S Chimeric Design of Pyrrolysyl-tRNA Synthetase/tRNA Pairs and Canonical Synthetase/tRNA Pairs for Genetic Code Expansion. Nat Commun 2020, 11 (1), 3154. 10.1038/s41467-020-16898-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Fischer JT; Söll D; Tharp JM Directed Evolution of Methanomethylophilus Alvus Pyrrolysyl-tRNA Synthetase Generates a Hyperactive and Highly Selective Variant. Front. Mol. Biosci. 2022, 9. 10.3389/fmolb.2022.850613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Hohl A; Karan R; Akal A; Renn D; Liu X; Ghorpade S; Groll M; Rueping M; Eppinger J Engineering a Polyspecific Pyrrolysyl-tRNA Synthetase by a High Throughput FACS Screen. Sci Rep 2019, 9 (1), 11971. 10.1038/s41598-019-48357-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Meineke B; Heimgärtner J; Lafranchi L; Elsässer SJ Methanomethylophilus Alvus Mx1201 Provides Basis for Mutual Orthogonal Pyrrolysyl tRNA/Aminoacyl-tRNA Synthetase Pairs in Mammalian Cells. ACS Chem. Biol. 2018, 13 (11), 3087–3096. 10.1021/acschembio.8b00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Schmied WH; Elsässer SJ; Uttamapinant C; Chin JW Efficient Multisite Unnatural Amino Acid Incorporation in Mammalian Cells via Optimized Pyrrolysyl tRNA Synthetase/tRNA Expression and Engineered ERF1. J. Am. Chem. Soc. 2014, 136 (44), 15577–15583. 10.1021/ja5069728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Sharma V; Zeng Y; Wang WW; Qiao Y; Kurra Y; Liu WR Evolving the N-Terminal Domain of Pyrrolysyl-tRNA Synthetase for Improved Incorporation of Noncanonical Amino Acids. ChemBioChem 2018, 19 (1), 26–30. 10.1002/cbic.201700268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Hancock SM; Uprety R; Deiters A; Chin JW Expanding the Genetic Code of Yeast for Incorporation of Diverse Unnatural Amino Acids via a Pyrrolysyl-tRNA Synthetase/tRNA Pair. J. Am. Chem. Soc. 2010, 132 (42), 14819–14824. 10.1021/ja104609m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Kleiner RE; Ti S-C; Kapoor TM Site-Specific Chemistry on the Microtubule Polymer. J. Am. Chem. Soc. 2013, 135 (34), 12520–12523. 10.1021/ja405199h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Mukai T; Kobayashi T; Hino N; Yanagisawa T; Sakamoto K; Yokoyama S Adding L-Lysine Derivatives to the Genetic Code of Mammalian Cells with Engineered Pyrrolysyl-TRNA Synthetases. Biochem Biophys Res Commun 2008, 371 (4), 818–822. 10.1016/j.bbrc.2008.04.164. [DOI] [PubMed] [Google Scholar]
- (49).Stieglitz JT; Lahiri P; Stout MI; Van Deventer JA Exploration of Methanomethylophilus Alvus Pyrrolysyl-tRNA Synthetase Activity in Yeast. ACS Synth. Biol. 2022, 11 (5), 1824–1834. 10.1021/acssynbio.2c00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Beznosková P; Pavlíková Z; Zeman J; Echeverría Aitken C; Valášek LS. Yeast Applied Readthrough Inducing System (YARIS): An Invivo Assay for the Comprehensive Study of Translational Readthrough. Nucleic Acids Res 2019, 47 (12), 6339–6350. 10.1093/nar/gkz346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Blanchet S; Cornu D; Argentini M; Namy O New Insights into the Incorporation of Natural Suppressor tRNAs at Stop Codons in Saccharomyces Cerevisiae. Nucleic Acids Res 2014, 42 (15), 10061–10072. 10.1093/nar/gku663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Kohli J; Kwong T; Altruda F; Söll D; Wahl G Characterization of a UGA-Suppressing Serine TRNA from Schizosaccharomyces Pombe with the Help of a New in Vitro Assay System for Eukaryotic Suppressor TRNAs. J Biol Chem 1979, 254 (5), 1546–1551. 10.1016/S0021-9258(17)37806-7. [DOI] [PubMed] [Google Scholar]
- (53).Macino G; Coruzzi G; Nobrega FG; Li M; Tzagoloff A Use of the UGA Terminator as a Tryptophan Codon in Yeast Mitochondria. Proc. Natl. Acad. Sci. 1979, 76 (8), 3784–3785. 10.1073/pnas.76.8.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Dabrowski M; Bukowy-Bieryllo Z; Zietkiewicz E Translational Readthrough Potential of Natural Termination Codons in Eucaryotes – The Impact of RNA Sequence. RNA Biol 2015, 12 (9), 950–958. 10.1080/15476286.2015.1068497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Stieglitz JT; Kehoe HP; Lei M; Van Deventer JA A Robust and Quantitative Reporter System To Evaluate Noncanonical Amino Acid Incorporation in Yeast. ACS Synth. Biol. 2018, 7 (9), 2256–2269. 10.1021/acssynbio.8b00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Potts KA; Stieglitz JT; Lei M; Van Deventer JAV. Reporter System Architecture Affects Measurements of Noncanonical Amino Acid Incorporation Efficiency and Fidelity. Mol. Syst. Des. Eng. 2020, 5 (2), 573–588. 10.1039/C9ME00107G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Reynolds NM; Vargas-Rodriguez O; Söll D; Crnković A The Central Role of tRNA in Genetic Code Expansion. Biochim Biophys Acta 2017, 1861 (11 Pt B), 3001–3008. 10.1016/j.bbagen.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Wu N; Deiters A; Cropp TA; King D; Schultz PG A Genetically Encoded Photocaged Amino Acid. J. Am. Chem. Soc. 2004, 126 (44), 14306–14307. 10.1021/ja040175z. [DOI] [PubMed] [Google Scholar]
- (59).Van Deventer JA; Le DN; Zhao J; Kehoe HP; Kelly RL A Platform for Constructing, Evaluating, and Screening Bioconjugates on the Yeast Surface. Protein Eng Des Sel 2016, 29 (11), 485–494. 10.1093/protein/gzw029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Bartoschek MD; Ugur E; Nguyen T-A; Rodschinka G; Wierer M; Lang K; Bultmann S Identification of Permissive Amber Suppression Sites for Efficient Non-Canonical Amino Acid Incorporation in Mammalian Cells. Nucleic Acids Res 2021, 49 (11), e62. 10.1093/nar/gkab132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Pott M; Schmidt MJ; Summerer D Evolved Sequence Contexts for Highly Efficient Amber Suppression with Noncanonical Amino Acids. ACS Chem. Biol. 2014, 9 (12), 2815–2822. 10.1021/cb5006273. [DOI] [PubMed] [Google Scholar]
- (62).Xu H; Wang Y; Lu J; Zhang B; Zhang Z; Si L; Wu L; Yao T; Zhang C; Xiao S; Zhang L; Xia Q; Zhou D Re-Exploration of the Codon Context Effect on Amber Codon-Guided Incorporation of Noncanonical Amino Acids in Escherichia Coli by the Blue–White Screening Assay. ChemBioChem 2016, 17 (13), 1250–1256. 10.1002/cbic.201600117. [DOI] [PubMed] [Google Scholar]
- (63).Anzalone AV; Zairis S; Lin AJ; Rabadan R; Cornish VW Interrogation of Eukaryotic Stop Codon Readthrough Signals by in Vitro RNA Selection. Biochemistry 2019, 58 (8), 1167–1178. 10.1021/acs.biochem.8b01280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Beznosková P; Gunišová S; Valášek LS Rules of UGA-N Decoding by near-Cognate TRNAs and Analysis of Readthrough on Short UORFs in Yeast. RNA 2016, 22 (3), 456–466. 10.1261/rna.054452.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).DeBenedictis EA; Carver GD; Chung CZ; Söll D; Badran AH Multiplex Suppression of Four Quadruplet Codons via TRNA Directed Evolution. Nat Commun 2021, 12 (1), 5706. 10.1038/s41467-021-25948-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Zackin MT; Stieglitz JT; Van Deventer JA Genome-Wide Screen for Enhanced Noncanonical Amino Acid Incorporation in Yeast. ACS Synth. Biol. 2022, 11 (11), 3669–3680. 10.1021/acssynbio.2c00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Stieglitz JT; Potts KA; Van Deventer JA Broadening the Toolkit for Quantitatively Evaluating Noncanonical Amino Acid Incorporation in Yeast. ACS Synth. Biol. 2021, 10 (11), 3094–3104. 10.1021/acssynbio.1c00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).von der Haar T; Tuite MF Regulated Translational Bypass of Stop Codons in Yeast. Trends in Microbiology 2007, 15 (2), 78–86. 10.1016/j.tim.2006.12.002. [DOI] [PubMed] [Google Scholar]
- (69).Keeling KM; Salas-Marco J; Osherovich LZ; Bedwell DM Tpa1p Is Part of an mRNP Complex That Influences Translation Termination, mRNA Deadenylation, and mRNA Turnover in Saccharomyces Cerevisiae. Mol Cell Biol 2006, 26 (14), 5237–5248. 10.1128/MCB.02448-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Song JM; Liebman SW Allosuppressors That Enhance the Efficiency of Omnipotent Suppressors in Saccharomyces Cerevisiae. Genetics 1987, 115 (3), 451–460. 10.1093/genetics/115.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Chin JW An Expanded Eukaryotic Genetic Code. Science 2003, 301 (5635), 964–967. 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- (72).Alcala-Torano R; Islam M; Cika J; Lam KH; Jin R; Ichtchenko K; Shoemaker CB; Van Deventer JA Yeast Display Enables Identification of Covalent Single-Domain Antibodies against Botulinum Neurotoxin Light Chain A. ACS Chem. Biol. 2022. 10.1021/acschembio.2c00574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Li Y; Hoskins JN; Sreerama SG; Grayson SM MALDI-TOF Mass Spectral Characterization of Polymers Containing an Azide Group: Evidence of Metastable Ions. Macromolecules 2010, 43 (14), 6225–6228. 10.1021/ma100599n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Holstein JM; Muttach F; Schiefelbein SHH; Rentmeister A Dual 5′ Cap Labeling Based on Regioselective RNA Methyltransferases and Bioorthogonal Reactions. Eur. J. Chem. 2017, 23 (25), 6165–6173. 10.1002/chem.201604816. [DOI] [PubMed] [Google Scholar]
- (75).Simon C; Lion C; Spriet C; Baldacci-Cresp F; Hawkins S; Biot C One, Two, Three: A Bioorthogonal Triple Labelling Strategy for Studying the Dynamics of Plant Cell Wall Formation In Vivo. Angew Chem 2018, 130 (51), 16907–16913. 10.1002/ange.201808493. [DOI] [PubMed] [Google Scholar]
- (76).Truong F; Yoo TH; Lampo TJ; Tirrell DA Two-Strain, Cell-Selective Protein Labeling in Mixed Bacterial Cultures. J Am Chem Soc 2012, 134 (20), 8551–8556. 10.1021/ja3004667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Winz M-L; Linder EC; Becker J; Jäschke A Site-Specific One-Pot Triple Click Labeling for DNA and RNA. Chem. Commun. 2018, 54 (83), 11781–11784. 10.1039/C8CC04520H. [DOI] [PubMed] [Google Scholar]
- (78).Amiram M; Haimovich AD; Fan C; Wang Y-S; Aerni H-R; Ntai I; Moonan DW; Ma NJ; Rovner AJ; Hong SH; Kelleher NL; Goodman AL; Jewett MC; Söll D; Rinehart J; Isaacs FJ Evolution of Translation Machinery in Recoded Bacteria Enables Multi-Site Incorporation of Nonstandard Amino Acids. Nat Biotechnol 2015, 33 (12), 1272–1279. 10.1038/nbt.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Jones CM; Robkis DM; Blizzard RJ; Munari M; Venkatesh Y; Mihaila TS; Eddins AJ; Mehl RA; Zagotta WN; Gordon SE; Petersson EJ Genetic Encoding of a Highly Photostable, Long Lifetime Fluorescent Amino Acid for Imaging in Mammalian Cells. Chem. Sci. 2021, 12 (36), 11955–11964. 10.1039/D1SC01914G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Italia JS; Addy PS; Erickson SB; Peeler JC; Weerapana E; Chatterjee A Mutually Orthogonal Nonsense-Suppression Systems and Conjugation Chemistries for Precise Protein Labeling at up to Three Distinct Sites. J. Am. Chem. Soc. 2019, 141 (15), 6204–6212. 10.1021/jacs.8b12954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Row RD; Prescher JA Constructing New Bioorthogonal Reagents and Reactions. Acc. Chem. Res. 2018, 51 (5), 1073–1081. 10.1021/acs.accounts.7b00606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Cui Z; Mureev S; Polinkovsky ME; Tnimov Z; Guo Z; Durek T; Jones A; Alexandrov K Combining Sense and Nonsense Codon Reassignment for Site-Selective Protein Modification with Unnatural Amino Acids. ACS Synth. Biol. 2017, 6 (3), 535–544. 10.1021/acssynbio.6b00245. [DOI] [PubMed] [Google Scholar]
- (83).Maurel D; Comps-Agrar L; Brock C; Rives M-L; Bourrier E; Ayoub MA; Bazin H; Tinel N; Durroux T; Prézeau L; Trinquet E; Pin J-P Cell-Surface Protein-Protein Interaction Analysis with Time-Resolved FRET and Snap-Tag Technologies: Application to GPCR Oligomerization. Nat Methods 2008, 5 (6), 561–567. 10.1038/nmeth.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Wu B; Wang Z; Huang Y; Liu WR Catalyst-Free and Site-Specific One-Pot Dual-Labeling of a Protein Directed by Two Genetically Incorporated Noncanonical Amino Acids. ChemBioChem 2012, 13 (10), 1405–1408. 10.1002/cbic.201200281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Ren C; Wu Q; Xiao R; Ji Y; Yang X; Zhang Z; Qin H; Ma J-A; Xuan W Expanding the Scope of Genetically Encoded Lysine Post-Translational Modifications with Lactylation, β-Hydroxybutyrylation and Lipoylation. ChemBioChem 2022, 23 (18), e202200302. 10.1002/cbic.202200302. [DOI] [PubMed] [Google Scholar]
- (86).Uhlenbeck OC; Schrader JM Evolutionary Tuning Impacts the Design of Bacterial TRNAs for the Incorporation of Unnatural Amino Acids by Ribosomes. Curr Opin Chem Biol 2018, 46, 138–145. 10.1016/j.cbpa.2018.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Fan C; Xiong H; Reynolds NM; Söll D Rationally Evolving tRNAPyl for Efficient Incorporation of Noncanonical Amino Acids. Nucleic Acids Res 2015, 43 (22), e156. 10.1093/nar/gkv800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Guo J; Melançon III CE; Lee HS; Groff D; Schultz PG Evolution of Amber Suppressor TRNAs for Efficient Bacterial Production of Proteins Containing Nonnatural Amino Acids. Angew Chem Int Ed Engl 2009, 48 (48), 9148–9151. 10.1002/anie.200904035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Annaluru N; Muller H; Mitchell LA; Ramalingam S; Stracquadanio G; Richardson SM; Dymond JS; Kuang Z; Scheifele LZ; Cooper EM; Cai Y; Zeller K; Agmon N; Han JS; Hadjithomas M; Tullman J; Caravelli K; Cirelli K; Guo Z; London V; Yeluru A; Murugan S; Kandavelou K; Agier N; Fischer G; Yang K; Martin JA; Bilgel M; Bohutskyi P; Boulier KM; Capaldo BJ; Chang J; Charoen K; Choi WJ; Deng P; DiCarlo JE; Doong J; Dunn J; Feinberg JI; Fernandez C; Floria CE; Gladowski D; Hadidi P; Ishizuka I; Jabbari J; Lau CYL; Lee PA; Li S; Lin D; Linder ME; Ling J; Liu J; Liu J; London M; Ma H; Mao J; McDade JE; McMillan A; Moore AM; Oh WC; Ouyang Y; Patel R; Paul M; Paulsen LC; Qiu J; Rhee A; Rubashkin MG; Soh IY; Sotuyo NE; Srinivas V; Suarez A; Wong A; Wong R; Xie WR; Xu Y; Yu AT; Koszul R; Bader JS; Boeke JD; Chandrasegaran S Total Synthesis of a Functional Designer Eukaryotic Chromosome. Science 2014, 344 (6179), 55–58. 10.1126/science.1249252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Shen Y; Gao F; Wang Y; Wang Y; Zheng J; Gong J; Zhang J; Luo Z; Schindler D; Deng Y; Ding W; Lin T; Swidah R; Zhao H; Jiang S; Zeng C; Chen S; Chen T; Wang Y; Luo Y; Mitchell L; Bader JS; Zhang G; Shen X; Wang J; Fu X; Dai J; Boeke JD; Yang H; Xu X; Cai Y Dissecting Aneuploidy Phenotypes by Constructing Sc2.0 Chromosome VII and SCRaMbLEing Synthetic Disomic Yeast. bioRxiv 2022, 2022.09.01.506252. 10.1101/2022.09.01.506252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


