Abstract
Minute virus of mice (MVM) replicates via a linearized form of rolling-circle replication in which the viral nickase, NS1, initiates DNA synthesis by introducing a site-specific nick into either of two distinct origin sequences. In vitro nicking and replication assays with substrates that had deletions or mutations were used to explore the sequences and structural elements essential for activity of one of these origins, located in the right-end (5′) viral telomere. This structure contains 248 nucleotides, most-favorably arranged as a simple hairpin with six unpaired bases. However, a pair of opposing NS1 binding sites, located near its outboard end, create a 33-bp palindrome that could potentially assume an alternate cruciform configuration and hence directly bind HMG1, the essential cofactor for this origin. The palindromic nature of this sequence, and thus its ability to fold into a cruciform, was dispensable for origin function, as was the NS1 binding site occupying the inboard arm of the palindrome. In contrast, the NS1 site in the outboard arm was essential for initiation, even though positioned 120 bp from the nick site. The specific sequence of the nick site and an additional NS1 binding site which directly orients NS1 over the initiation site were also essential and delimited the inboard border of the minimal right-end origin. DNase I and hydroxyl radical footprints defined sequences protected by NS1 and suggest that HMG1 allows the NS1 molecules positioned at each end of the origin to interact, creating a distortion characteristic of a double helical loop.
Nickase molecules that initiate rolling-circle replication (RCR) generally bind to specific duplex recognition sequences in the replicon origin before introducing a site-specific, single-strand nick at an adjacent initiation site. The trans-esterification reaction which creates this nick leaves the initiator protein covalently attached to the new 5′ end of the DNA while liberating a base-paired 3′ nucleotide to serve as a primer for a host DNA polymerase. NS1, the nickase encoded by the parvovirus minute virus of mice (MVM), carries consensus catalytic site motifs which suggest a close evolutionary link with archetypal, prokaryotic RCR initiators, and it fulfills the same spectrum of functions during replication (24, 34), but in its eukaryotic host MVM has evolved additional roles for this highly expressed site-specific DNA binding protein. MVM has a linear, negative-sense, single-stranded DNA genome, but it replicates through a series of multimeric duplex intermediates in which forms of the NS1 recognition sequence, (ACCA)2-3, are repeated at least once every hundred base pairs (4, 12; S. F. Cotmore and P. Tattersall, unpublished data). NS1 exhibits both ATPase and helicase activity (33, 43) and has an acidic carboxy-terminal transcriptional activation domain (27). NS1 binding sites in the transcriptional promoter P38 position it to modulate viral transcription (6, 28), while others distributed throughout the genome are believed to have roles in viral chromatin structure and in progeny genome encapsidation (16, 21). Replicative-form (RF) duplex MVM DNA also contains frequent, albeit somewhat degenerate, repetitions of the consensus initiation site (CTWWTCA), so the nickase function of NS1 must be tightly suppressed to confine this otherwise disruptive phosphodiesterase activity to the viral origins. This suppression is achieved by the requirement for activation of the nickase by a host-encoded cofactor which is specific for origin sequence or structure and is different for the origins present at each end of the genome.
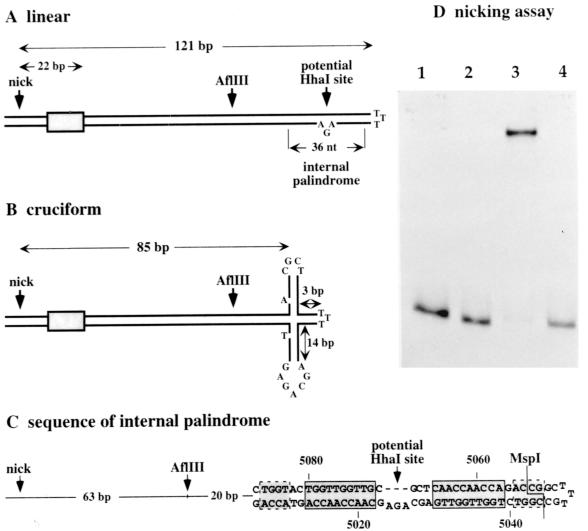
The MVM right-end telomere can serve as a competent replication origin in its covalently closed hairpin configuration (3, 17). Two distinguishable forms of the terminus, dubbed “flip” and “flop,” are generated in equimolar amounts in vivo, and both give rise to viral origins (2, 17). These two forms, which are the inverted complements of each other, are most simply depicted as duplex stem structures of 121 bp interrupted by a triplet of unpaired nucleotides that forms a small asymmetric “bubble” near the distal end of one strand, illustrated in Fig. 1A, plus three unpaired bases which form the cross-link at the tip of hairpin. However, these stems harbor a small internal palindrome surrounding the asymmetric bubble, so an alternative structure, in which the stem assumes an asymmetric cruciform configuration, is also thermodynamically probable, as shown in Fig. 1B. This structure has been implicated in the control of replication (1, 38), although it is not clear whether it influences initiation or the subsequent unrolling and reformation of the hairpin during strand displacement synthesis.
FIG. 1.
(A) The 5′ (right-end) hairpin of MVM displayed as a simple duplex stem structure in the flip configuration. A small internal palindrome positioned near the top of the stem potentially allows this region to reconfigure into the asymmetric cruciform structure shown in panel B. At its most extended position, the four-way junction at the core of this cruciform is 85 bp from the nick site. As detailed in panel C, the internal palindrome contains two 10-mer [(ACCA)2.5] sequence repeats, boxed with solid lines, which constitute NS1 binding sites positioned in opposite orientations around the twofold symmetry axis. Additional tetranucleotide motifs which likely contribute to NS1 binding are boxed with dashed lines. In panels A and B, the shaded boxes represent binding sites which position NS1 over the nick site. (D) Autoradiograph of a nondenaturing polyacrylamide gel showing a 32P-, 3′-end-labeled hairpin origin before (lane 1) and after (lane 2) cleavage with MspI. Following incubation with NS1 and HMG1 in the presence of ATP, the hairpin substrate is nicked and covalently associated with NS1 (lane 3), while the substrate precut with MspI remains unmodified (lane 4).
Right-end hairpin substrates can be nicked site specifically in vitro by recombinant NS1, but only in the presence of cellular proteins from the high-mobility group, HMG1/2, family (17). HMG1/2 molecules bind with low affinity to both single-stranded and duplex DNA in a non-sequence-specific manner, but they bind bent DNA and four-way junctions with significantly higher affinity (reviewed by Pöhler and colleagues [36]). Since a four-way junction would be created at the base of the putative cruciform depicted in Fig. 1B, this structural element could serve to recruit the necessary accessory protein, in which case efficient initiation would depend upon the ability of the origin to adopt a cruciform configuration. Alternatively, HMG1/2 proteins can be delivered to particular sites in the DNA as a result of direct, but relatively low-affinity, protein-protein interactions with other site-specific DNA binding proteins, resulting in the formation of local tertiary complexes (11, 45, 46). Theoretically, then, HMG1/2 could be localized specifically to the NS1 binding sites in the origin as a result of direct interactions with NS1. Alternatively, after it is brought to the origin by such direct interactions, HMG1/2 could adopt a unique position in the nucleoprotein complex, as is seen for the bacterial sequence-independent DNA binding protein HU in the context of the Mu intasome (26). In this study we set out to define the minimal sequences and secondary-structure elements which control the interaction between NS1, HMG1/2, and target DNA in this unusual origin.
MATERIALS AND METHODS
Plasmids.
The pUC19-based plasmid pREB1412 has been described previously (14). The insert from this construct, diagrammed in Fig. 2A, was excised and then cut twice near its axis of symmetry with MspI, and the two arms of the palindrome were recloned separately into pUC19 to yield plasmids pHha and pAGA. PCR products generated using these two constructs as templates and oligonucleotide primers R1 and R2, etc., as described below were cloned into the T/A vector pCRII (Invitrogen, San Diego, Calif.).
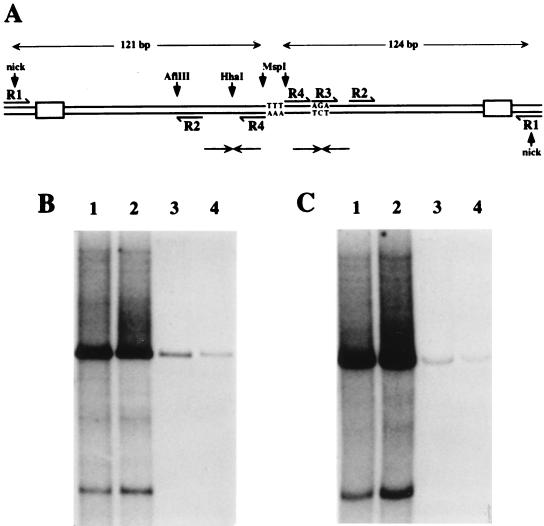
FIG. 2.
(A) Schematic representation of the tetramer bridge region of RF DNA, created by unfolding and copying the right-end hairpin sequence shown in Fig. 1A. The unpaired nucleotides at the top of the stem (TTT) now form the dyad symmetry axis of a palindromic duplex junction. Pairs of small opposing arrows indicate the positions of the internal palindromes detailed in Fig. 1C. Sequences used as primers for PCR-mediated subcloning of this region are labeled R1-R4. Shaded boxes near the nick sites represent the inboard NS1 binding sites. (B and C) Autoradiographs of native agarose gels showing ScaI-linearized products of NS1-driven in vitro replication assays programmed with plasmids containing inserts derived from single arms of the bridge before (B) and after (C) immunoprecipitation with anti-NS1 serum. Replication substrates were pR1-4AGA (lane 1), pR1-4 Hha (lane 2), pR1-2 (lane 3), and pR1-3 (lane 4). Autoradiographs were overexposed to show residual levels of replication seen with defective substrates. In all cases, digestion with ScaI reduced replication products to a single major species which comigrated with unit-length plasmid substrate DNA. In addition, such long exposures revealed a minor, smaller product and multiple rarer species, none of whose structures are currently understood.
Expression vectors and recombinant proteins.
Recombinant histidine-tagged MVM NS1 was in expressed in HeLa cells by coinfection with vaccinia virus vTF7-3, which expresses T7 polymerase, and vv-his-NS1wt, which expresses full-length copies of NS1 from the T7 promoter. NS1 was purified by nickel-chelate chromatography as previously described (32). Recombinant baculoviruses expressing histidine-tagged HMG1 sequences were prepared as follows. HMG1 cDNA was amplified from HeLa cell mRNA by reverse-transcription-PCR using oligonucleotide primers positioned just outside the coding sequence (GenBank sequence X12597) and cloned into pCR2.1-Topo (Invitrogen). Following PCR reamplification using primers which added cloning sites to each end of the product, the HMG1 cDNA was recloned between SalI and XbaI sites in baculovirus transfer vector pBac-PAK-His3 (Clontech Laboratories, Inc., Palo Alto, Calif.), and recombinant viruses were generated as described by the supplier. HMG1, His tagged at its N terminus, was harvested from whole extracts of Sf9 cells 42 h after infection and partially purified by chromatography through phosphocellulose as previously described (17). The high-salt eluate from this column (fraction P-cell 3) was then purified to apparent homogeneity on a nickel-chelate column, eluted in 20 mM HEPES-KOH (pH 7.8)–10% glycerol–0.1 mM dithiothreitol (DTT)–50 mM NaCl–0.01% NP-40–100 mM imidazole, and stored at −80°C.
Nicking substrates and assays.
A 3′-end-labeled hairpin substrate in the flip orientation containing 418 bp (and six unpaired nucleotides) of viral sequence extending from an SspI site at MVM nucleotide 4626 to the tip of the right-end hairpin was prepared as previously described (17), and the gel-purified product was cut with MspI or left intact as indicated below. Linear nicking substrates (380 to 386 nucleotides) were excised from wild-type and mutant plasmids by using Sau3A, which leaves 211 nucleotides of vector sequence flanking the nick site end of the insert and ∼40 nucleotides of vector sequence abutting the internal palindrome. Fragments were quantitated against the hairpin substrates by ethidium staining, cut or not with MspI, and 3′ end labeled with a mixture of [32P]dATP and [32P]dCTP. Nicking assays were performed in a final volume of 15 μl as described previously (17).
Replication assays.
Replication extracts were prepared from uninfected HeLa cells essentially as described by Wobbe and colleagues (44), except that they were dialyzed against 20 mM Tris-HCl (pH 7.5)–0.2 mM EDTA–25 mM NaCl–0.5 mM DTT–10% glycerol–20% sucrose, which concentrated the extracts approximately fourfold. Assay mixtures (20 μl) contained recombinant vaccinia virus NS1 (10 μg/ml), deoxynucleotides, MgCl2, ATP and an ATP-regenerating system, template DNA (5 μg/ml), and a 32P-labeled deoxynucleoside triphosphate. Assays were terminated and results were analyzed as previously described (13).
DNase I and hydroxyl radical footprinting.
For footprinting reactions, plasmid pR1-4AGA was digested with either HindIII or XbaI and 3′ end labeled on one strand with [32P]dATP. The substrates were then redigested with XbaI or HindIII, and DNA fragments were purified from agarose gels. Reaction mixtures (25 μl) contained NS1 (50 to 200 ng) and/or HMG1 (50 to 200 ng), as indicated below, and DNA fragments (approximately 4 × 104 cpm) in a solution containing 25 mM HEPES-KOH (pH 7.8), 75 mM sodium acetate, 0.1 mM EDTA, 2 mM MgCl2, 2.5 mM DTT, 1 mM γS-ATP, 250 μg of bovine serum albumin per ml, 0.005% NP-40, 0.125 μg of an unrelated double-stranded oligonucleotide (“SCRAM” [detailed below]), and 0.125 μg of blunt-ended, nonspecific duplex DNA fragments (100 to 800 bp in length) derived from vector plasmid pCRII by digestion with RsaI and NciI. SCRAM (“scramble” oligonucleotide; 5′-GAT CTA GAG AGT CGA TGT ATC TGC AGA TC-3′) is a blunt-ended, nonspecific duplex 29-mer oligonucleotide competitor. Binding was allowed to proceed for 15 min at room temperature (RT) and for 45 min on ice before samples were reequilibrated to RT (3 min) prior to addition of the footprinting reagent.
For DNase I protection assays, 1.0 U of DNase I (Boehringer Mannheim GmbH, Mannheim, Germany) was added and incubation was continued at RT for 1.75 min. Reactions were terminated with 0.15 ml of a solution containing 10 mM Tris-HCl (pH 8.0), 10 mM EDTA, 0.5% sodium dodecyl sulfate (SDS), and 50 μg of proteinase K per ml; termination was followed by incubation at 50°C for 1 h. For hydroxyl radical cleavage, methidium propyl-EDTA (MPE) (originally from Peter Dervan; kindly provided by Nigel Grindley and Cathy Joyce) was used to generate MPE-Fe(II) complexes, essentially as described by Hatfull and colleagues (19). After incubation for 1.75 min at RT, reactions were terminated by addition of 1 μl of 50 mM bathophenanthroline disulfonate. All samples were then extracted with phenol-chloroform, precipitated with ethanol, using oyster glycogen as a carrier, and analyzed by electrophoresis through denaturing 7% acrylamide gels. DNA probes were also chemically cleaved at G residues by the procedure of Maxam and Gilbert (29) and electrophoresed as markers to allow sequence alignment.
RESULTS
Removal of nine nucleotides from the extreme tip of the MVM right-end hairpin prevents NS1-mediated initiation.
NS1 initiates replication by means of a trans-esterification reaction in which the hydroxyl group of tyrosine210 attacks the phosphodiester bond of adenine 5170 in the covalently closed right-end hairpin sequence of duplex RF DNA, leaving the NS1 molecule covalently attached to the 5′ nucleotide (16). This reaction is ATP dependent and can be reproduced in vitro using 32P-, 3′-end-labeled hairpin substrates that mimic the right end of covalently closed monomer duplex RF molecules (17). In the example shown in Fig. 1D, reaction products were first incubated in SDS and then analyzed by electrophoresis through a nondenaturing polyacrylamide gel in the presence of SDS, which destroys noncovalent protein-DNA or protein-protein interactions so that only successfully formed covalent initiation complexes are retarded. Initiation was compared before and after digestion of the hairpin substrate with MspI, which removes just nine nucleotides from the extreme tip of the hairpin, as shown in Fig. 1C, creating a modified probe that still effectively comigrated with the wild-type substrate (Fig. 1D, cf. lanes 1 and 2). However, when incubated with recombinant NS1 and HMG1, only the hairpin probe was nicked and covalently linked to NS1 (Fig. 1D, cf. lanes 3 and 4). Thus, even though the deleted sequences are 120 bp (and three unpaired bases) distant from the nick site, their deletion inactivates the origin.
There appear to be two distinct ways in which removal of these nine nucleotides might disrupt origin function. Firstly, the MspI cut may destabilize an essential structural element in the origin, such as a four-way junction. As shown in Fig. 1B, extrusion of the cruciform is dependent upon the presence of a cross-link at the tip of the hairpin. If this connection is removed, the separate strands of the palindrome might still melt out and refold back on themselves, but the resulting four-way junction is unlikely to be stable.
Alternatively, the MspI cut might remove or destabilize essential recognition sequences located near the tip of the hairpin. This alternative is particularly plausible because the palindrome which creates the cruciform contains two opposing consensus 10-mer NS1 binding sites, (ACCA)2.5, and there are additional ACCA tetranucleotides, including a degenerate one which forms part of the MspI site (Fig. 1C). If nicking occurs only when NS1 is bound to this distal region of the hairpin, removal of just a few nucleotides might be enough to destabilize its interaction with the site.
To discriminate between these possibilities, we wished to generate mutant templates. However, the asymmetric, hairpinned substrates used in the nicking assay were themselves generated from the cloned palindromic tetramer bridge sequence, illustrated in Fig. 2A, which is relatively difficult to mutate using standard PCR-based procedures. Instead, we used nonhairpinned, fully duplex forms of the origin sequences obtained by cloning each arm of the tetramer bridge separately into plasmid vectors. These single-origin circular constructs were then used as substrates for NS1-mediated RCR in a HeLa S100 in vitro DNA replication extract, labeling the newly synthesized DNA with 32P-labeled deoxynucleoside triphosphates. This provided a sensitive and rapid way to screen mutants for NS1-dependent origin activity.
Linear, nonhairpinned sequences from the right-end origin can support NS1-dependent replication in vitro.
The two arms of the palindromic tetramer bridge fragment in plasmid pREB1412 (14) were recloned separately as described in Materials and Methods. The two inserts were almost identical, except that one of them, in pAGA, contained a 3-bp insertion which corresponds to the unpaired three-nucleotide bubble (AGA) present in the lower strand of the hairpin structure illustrated in Fig. 1A. The absence of these three residues in the duplex form of the upper strand (pHha) creates an HhaI site at this position. Both of these plasmids supported NS1-dependent DNA synthesis in vitro (data not shown). However, since the cloned inserts extended to an XbaI site in the MVM coding sequence (at MVM nucleotide 4342), they still contained several hundreds of base pairs of viral sequence derived from regions outside the terminal hairpin. To delineate the minimal right-end origin sequence, and to minimize vector influences, we used PCR to subclone sequences from the hairpin regions of these two clones and selected constructs with inserts in the same orientation. PCR products were generated using oligonucleotide primer R1, which spans the nick site and extends 12 nucleotides into non-hairpin-derived sequence, and R4, which lies over the MspI site (shown in Fig. 2A), and the products were cloned into pCRII. These constructs, pR1-4Hha and pR1-4AGA, both served as efficient NS1-dependent replication substrates (Fig. 2B, lanes 1 and 2). Since such assays routinely allow low levels of nonspecific repair synthesis, this result alone does not prove that the inserts function as NS1-dependent origins. However, because initiation results in the formation of a covalent NS1-DNA complex, correctly initiated reaction products remain attached to NS1 after incubation in 1% SDS at 60°C and can be immunoprecipitated with anti-NS1 sera (Fig. 2C).
Thus, an effective, NS1-dependent minimal replication origin need only extend 12 nucleotides (or less) inboard of the nick site and can be expressed in a fully duplex sequence of 133 bp (pHha) or 136 bp (pAGA) which extends to a position near the tip of the telomeric hairpin. Although lacking the bubble asymmetry, these sequences still contain an internal palindrome and so can theoretically still reconfigure into cruciform structures, although such transitions would be thermodynamically less favored in the absence of the mismatched triplet.
Minimal linear origins must extend to a position near the tip of the hairpin.
In contrast to pR1-4AGA and pR1-4Hha, plasmids containing PCR-derived viral inserts of 95 bp (pR1-2), which contain all of the hairpin stem but not the potential cruciform structure, failed to support replication, as did constructs with inserts of 116 bp (pR1-3AGA), which contained sequences up to the midpoint of the palindrome, including the asymmetric AGA sequence (Fig. 2B and C, lanes 3 and 4, respectively). These data therefore support the conclusions drawn from the hairpin nicking assay presented in Fig. 1D, suggesting that sequences near the tip of the hairpin are required for successful initiation.
To check that linear origin sequences exhibited the same cofactor requirements as the hairpin origin, we used a 386-bp Sau3A fragment from pR1-4AGA, which contained the entire viral insert plus flanking plasmid sequences at each end, as a substrate in the direct nicking assay described in Materials and Methods. As shown in Fig. 3, this sequence could be nicked in vitro with NS1 in an HMG1-dependent manner, although it appeared to be a less efficient substrate than its hairpin counterpart (compare lanes 1 and 2 with lanes 5 and 6). However, like the hairpin form, the linear origin was inactivated by cleavage with MspI (Fig. 3, lanes 7 and 8).
FIG. 3.
Autoradiograph of a nondenaturing polyacrylamide gel showing the products of nicking reactions carried out in the presence of NS1 and HMG1 or NS1 alone. Substrates were 32P-, 3′-end-labeled hairpin sequences in the flip orientation (lanes 1 and 2) or linear Sau3A fragments of plasmid DNA (lanes 3 to 8). Substrates contained viral sequences from R1 to nucleotide 5042 (the minimal origin derived from pAGA [lanes 3 and 4]) or from R1 to nucleotide 5046 (pR1-4AGA [lanes 5 and 6]), with flanking vector sequences at each end of the insert. Lanes 7 and 8 show products obtained with pR1-4AGA precut with MspI.
Minimal origins must extend to MVM position 5042.
To determine which viral sequences from the hairpin tip are required for origin function, we generated a series of PCR-derived constructs with inserts extending from primer R1 to various positions between primer sites R3 and R4 in the AGA arm, yielding constructs 1 through 6 as indicated in Fig. 4A. We were not able to recover all inserts in exactly the constructs we originally intended. Most importantly, inserts ending at MVM nucleotide 5041 were obtained only in plasmids which had sustained small (1-, 5-, or 6-bp) deletions in the immediate vector flanking sequence and were obtained only with a G or T as the first base in the vector. Since replication assays suggested that this was a critical position, we generated additional plasmids using a longer, somewhat degenerate primer which placed four additional nucleotides (5′-CTTN-3′) outboard of residue 5041, in an attempt to obtain clones with all four nucleotides in position 5042 by spacing them and, we presumed, protecting them from the influence of the adjacent vector sequences. Even so, of the 12 clones sequenced, 3 had deletions in the inserts, 1 had a G in position 5042 (construct 5 in Fig. 4A), and 8 had a T in that position (e.g., construct 3 in Fig. 4A). Ligation products were transformed into INV αF, the normal host strain for pCRII, and into the recombination defective SURE strain (Stratagene, La Jolla, Calif.), with approximately the same results. At present, we do not know why plasmids with an A or C residue in this position were not obtained. Replication products and anti-NS1 immunoprecipitates obtained using these cloned substrates are shown in Fig. 4B and C. Additional plasmids which had MVM inserts that extended to nucleotide 5037 or 5038 were tested, and both were inactive (data not shown). Parallel assays (not shown) were conducted with ScaI-linearized substrates. Since these assays gave the same results as those shown here for circular plasmid substrates, we conclude that the torsional stress induced by supercoiling does not play a role in the specificity of the nicking reaction. When excised as a linear fragment with Sau3A and end labeled with [32P]dATP, the insert from construct 4 (i.e., the minimal origin) could be nicked by NS1 in an HMG1-dependent manner, albeit rather inefficiently (Fig. 3, lanes 3 and 4), while inserts from constructs 1 and 2 were inactive (data not shown). Thus, when assayed as either supercoiled or linearized substrates, active origins must extend to MVM residue 5042 or beyond.
FIG. 4.
(A) Details of deletion mutations introduced at the outboard junction of the MVM insert in pR1-4AGA. In constructs 1, 2, 6, and 7, the final nucleotide of viral sequence (as indicated) is linked directly to the first nucleotide of vector sequence, indicated in lightface. In construct 4, MVM nucleotide 5042 is linked directly to the T residue seven nucleotides into the vector sequence in the bottom strand. Constructs 3 and 5 have three nucleotides, 5′-CTT-3′ in the bottom strand, inserted between the viral and vector sequences. (B and C) Autoradiographs of total replication products (B) and anti-NS1 immunoprecipitates of replication products (C) generated using substrates 1 through 7, with each lane numbered according to the substrate used.
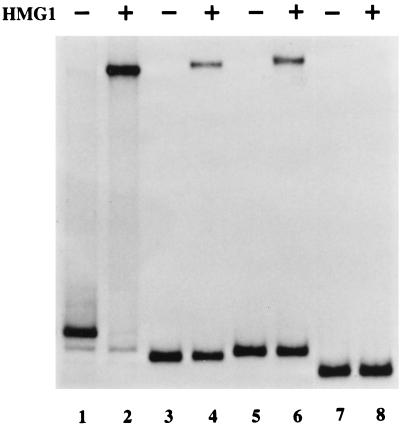
Does residue 5042 form part of an essential, extended cruciform structure?
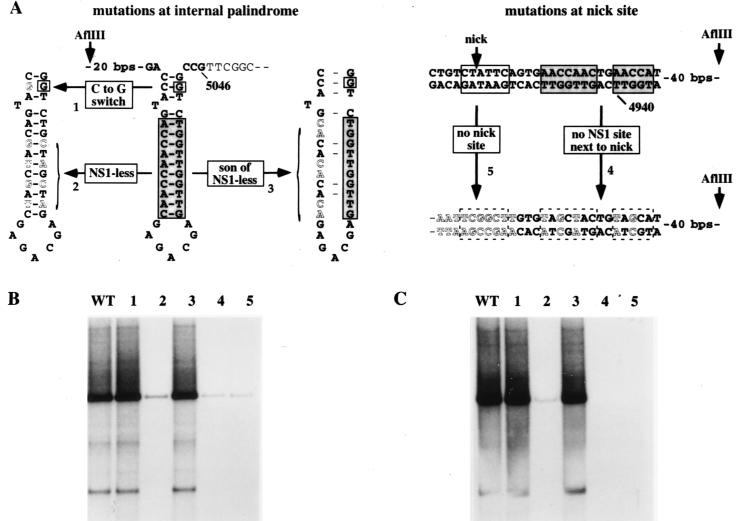
Since the potential cruciform structure at the outboard end of the minimal origin is created from a palindrome, the length of its cruciform arms must fluctuate as the sequences transition, in a concertina-like fashion, between the linear and cruciform configurations. At its maximum extension, depicted for the lower strand of the AGA arm in Fig. 5A, the stem of each arm is 14 bp long but would include a single extrahelical T residue between 3 and 4 bp from the four-way junction. This structure is less favored thermodynamically than a shorter cruciform, with arms of 11 bp, which extends only to the mismatched base. However, if the critical CG base pair at nucleotide 5042 is important because it forms part of an essential cruciform structure, that structure must be one which is in the maximally extended position. To explore the need for such a cruciform we made the C-to-G-switch mutant shown in Fig. 5A, in which G5042 is wild type but its opposing base in the cruciform arm is mutated from a C to a G, so that it cannot base pair in the cruciform. Since this mutated template (mutant 1) was an efficient NS1-dependent origin (Fig. 5B and C), we conclude that G5042 is essential for reasons other than forming part of an extended cruciform. This does not indicate that the cruciform itself is dispensable but merely indicates that G5042 does not have to be included within such a structure. By default, it suggests that G5042 more likely forms part of an NS1 binding site required for origin function.
FIG. 5.
(A) Details of substitution mutations within the internal palindrome and around the nick site of the MVM insert in pR1-4AGA. Letters indicating wild-type nucleotides are black, and those indicating mutations are white. Mutant 1 has a C-to-G switch at the position in the inboard arm of the internal palindrome which would base pair with G5042 in the cruciform structure. In mutant 2, NS1-less, the core of all possible NS1 binding sites is destroyed by switching every second nucleotide between strands of the cruciform. In mutant 3, son-of-NS1-less, only the binding site core in the inner arm of the palindrome is mutated, by switching three pairs of alternating bases along the strand. Mutant 4 has alternating nucleotides through the NS1 binding site switched between strands. Mutant 5 has the entire consensus nick site deleted; the juxtaposed vector sequences are shown in white letters. (B and C) Autoradiographs of total replication products (B) and anti-NS1 immunoprecipitates of replication products (C) generated using substrates 1 through 5, with each lane numbered according to the substrate used. WT, wild type.
NS1 binding sites in the internal palindrome are essential for origin function.
NS1 binding sites were originally identified, by coimmunoprecipitation, as sequences containing the motif (ACCA)2-3 (12). Thus, the two opposing 10-mer motifs B and B′ (Fig. 4A) in the internal palindrome of the right-end origin would appear to constitute credible NS1 binding sites both in the linear, extended form of this sequence, shown in Fig. 4, and in its cruciformed configuration, as shown in Fig. 5A. However, SELEX analysis (R. Gottlieb, S. F. Cotmore, and P. Tattersall, unpublished observations) suggests that favored binding sites should contain three ACCA tetramers, of which one can be 50% or less degenerate, while small interruptions of one to three nucleotides between tetramers are well tolerated. When bound to DNA, NS1 complexes protect at least 43 nucleotides from digestion with DNase I, giving asymmetric footprints that terminate abruptly five nucleotides to the 3′ side of the most 3′ ACCA motif but project up to 31 nucleotides on the 5′ side of the most 5′ motif (6, 12). Since the length of the 5′ extension varies somewhat with the size of the ACCA repeat, it seems likely that it is the position of the most 3′ ACCA tetramer that determines the precise position that NS1 occupies on the DNA. Viewed from this perspective, the C residue which is the complement of G5042 could form part of a degenerate third 3′ tetramer, A′ (Fig. 4A). Use of this degenerate tetranucleotide motif would be expected to position the NS1 complex 5 bp further away from the nick site than would a site which simply involved the B′ 10-mer sequence.
To explore whether any of the potential NS1 binding sites near the tip of the hairpin are required for origin function, we mutated both B and B′ 10-mer ACCA motifs, without touching G5042 itself. Since this removes the core of the binding site, the residual degenerate tetramer could not bind NS1 effectively. To make this mutant, dubbed “NS1-less,” we switched every second base between strands along the arm of the potential cruciform (mutant 2) (Fig. 5A), creating a sequence in which the NS1 binding sites in both the inner and outer arms of the linear palindrome were destroyed but a palindrome of the normal size and base composition, which could theoretically adopt an equivalent cruciform structure, was preserved. This mutant origin did not support NS1-mediated replication in vitro (Fig. 5B and C), indicating that one or both of the NS1 binding sites in the linear sequence of this region, or the NS1 binding site in the arms of the cruciform structure, were essential for initiation.
Neither the NS1 binding site in the inner arm of the palindrome nor the cruciform structure is required for initiation.
We constructed a further mutant, “son-of-NS1-less” (Fig. 5A), in which the NS1 site in the outer arm of the palindrome was intact, while the site in the inner arm was mutated by switching three alternating pairs of bases within motif B. This mutant (mutant 3) is not palindromic and so could not assume a cruciform configuration, but it proved to be as good an NS1- and HMG1-dependent origin in vitro as the wild-type sequence (Fig. 5B and C). Thus, the cruciform structure created by the internal palindrome is not required for initiation, and of the two juxtaposed NS1 binding sites present in the linear sequence, only the outer site is required, despite being positioned some 109 to 120 bp away from the nick site.
The minimal origin requires a specific nick site with an adjacent NS1 binding site.
We also created mutants in which the normal nick site was replaced by vector sequences (mutant 5) (Fig. 5A) or in which the nearby consensus NS1 binding site was destroyed while maintaining the normal base composition of the region (mutant 4) (Fig. 5A). As shown in Fig. 5B and C, both mutants 4 and 5 were inactive as substrates in replication assays, indicating that both of these sequences are critical for initiation. Thus, the minimal element required to create an NS1-dependent origin is a fully duplex, noncruciform stretch of ∼132 bp which contains a specific nick site and two critical NS1 binding sites, one oriented next to the nick site in such a way that its footprint should extend over the nick site and the second positioned in the same orientation but some 120 bp away.
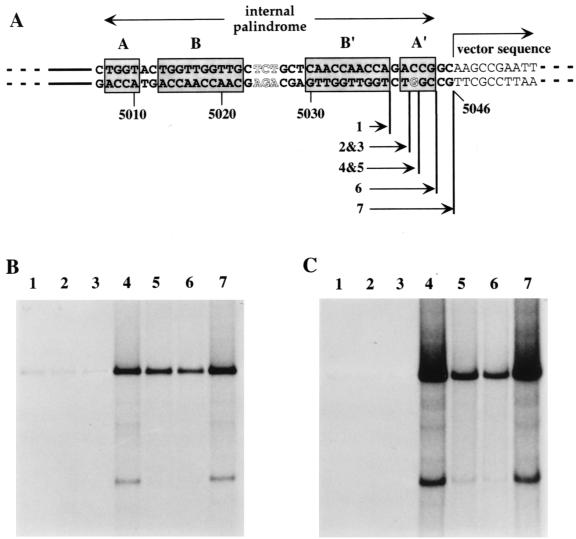
MPE-Fe(II) and DNase I footprints confirm that NS1 complexes are positioned over both the nick site and G5042 in the internal palindrome.
The genetic data suggested that the minimal MVM right-end origin requires two NS1 binding sites spaced approximately 100 nucleotides apart and that the spacing between these sites is important. To confirm that NS1 makes contact with the DNA at the expected positions, and to look for evidence of other interactions within the complex, we carried out MPE-Fe(II) and DNase I protection assays in the presence or absence of the activating cofactor HMG1 (Fig. 6). As seen previously (6, 12), NS1 footprints were obtained only when ATP, in the form of its nonhydrolyzable analog γS-ATP, was present in the reaction, and this dependence was not altered by the addition of HMG1 (not shown). In the presence of ATP, NS1 alone was seen to protect regions of DNA in the vicinity of the three sets of ACCA sequences already identified in this report.
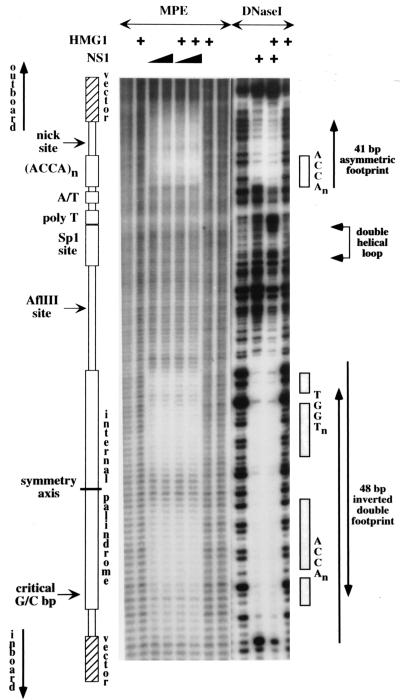
FIG. 6.
A schematic representation of the origin is aligned down the left-hand side of autoradiographs from denaturing polyacrylamide gels displaying hydroxyl radical (MPE) and DNase I cleavage profiles obtained with naked 32P-, 3′-end-labeled DNA or following its incubation with NS1 and/or HMG1 as indicated. Each lane displays products from the upper strand, containing the nick site. The positions of the ACCA tetranucleotide motifs that make up the NS1 binding sites are indicated down the right-hand side, together with a region of overcutting and undercutting, obtained with DNase I in the presence of both NS1 and HMG1, that reflects the formation of a double helical DNA loop.
At the nick site, NS1 protects an asymmetric 41-bp sequence from digestion with DNase I, extending from residue 4949, 5 bp 3′ of the ACCA motif boxed in Fig. 6, to a position 22 bp 5′ of this motif. This end of the footprint is 14 bp beyond the nick site, or 2 bp beyond the end of the MVM insert, in vector DNA. Since DNase I is a relatively large molecule and is easily excluded from the DNA by overlying protein complexes, we used hydroxyl radical footprinting in order to fine map points of close contact between NS1 and its DNA binding site. This technique involves oxidative cleavage of the deoxyribose backbone via a diffusable hydroxyl radical generated from a small molecule, MPE-Fe(II), that intercalates via the minor groove with little regard for DNA sequence specificity. Due to the short half-life of the free radical, cleavage only occurs, with progressively diminishing frequency, at positions up to four residues 3′ to the site of intercalation (20). Surprisingly, MPE-Fe(II) footprints show that NS1 makes extremely close contact with the DNA throughout much of the region protected from DNase I. Two zones of protection are apparent in Fig. 6: one is centered over the ACCA motifs (covering MVM residues 4929 to 4946) and is separated by four to five residues of unprotected DNA from the second one, which is more diffuse, extending over the nick site itself.
In the vicinity of the internal palindrome, NS1 excluded DNase I from a region extending for 48 bp from residue 5002, located five nucleotides 3′ of the first ACCA motif in the inner arm of the palindrome (motif A) (Fig. 4A), to a position 5 bp beyond the 3′ end of the degenerate motif A′ in the outer arm, which contains G5042. In the construct used here, the latter border falls in vector DNA, 3 bp beyond the end of the MVM insert. By analogy with all other mapped NS1 sites, this positioning indicates that NS1 does indeed recognize and bind to the degenerate A′ motif, as discussed above.
We had expected that an NS1 complex bound to one of the two opposing sites in this palindrome might interfere with binding at the other site simply because the 5′ end of one bound complex would project over and protect most of the ACCA sequences in the opposing site. Footprints derived from such a competitive interaction would show only partial protection at each site, indicating that NS1 was bound to each side only half of the time. However, the two opposing sites on the internal palindrome showed no sign of competition with each other. On the contrary, they created a higher affinity site than the one positioned over the nick site. This enhanced protection is apparent in the footprint shown in Fig. 6 and has been confirmed by quantitative footprinting of numerous active and inactive origin sequences (data not shown). This suggests that multiple NS1 complexes organized on the DNA in an antiparallel configuration interact with each other in such a way as to enhance binding. When one or other of the two sites in the palindrome was mutated, binding to this entire region dropped dramatically (relative to binding over the nick site), even though one such mutant, son-of-NS1-less, still functioned as an active origin (data not shown).
MPE-Fe(II) footprints over the internal palindrome were, for the most part, restricted to the ACCA motifs themselves. This was especially clear over the inner arm, where two nucleotides separating the two protected ACCA motifs (motifs A and B) (Fig. 4A) were cleaved normally, creating a bipartite footprint which highlights the modular nature of the NS1 binding site. However, replication assays with the mutant substrate son of NS1-less, described above, indicated that this inner binding site is not essential for origin function. MPE-Fe(II) footprints over the essential binding site in the outer palindromic arm were more complex. In this case, the 10-mer ACCA motif (motif B) (Fig. 4A) was highly protected, while both the degenerate A′ tetranucleotide, which contains G5042, and the next five nucleotides were protected to a somewhat lesser extent. At this essential site, therefore, NS1 makes tight contact with the DNA over an extended area that projects beyond its actual recognition element to the end of the DNase I footprint and hence includes several vector nucleotides whose actual sequence is not critical.
NS1 complexes positioned over the nick site and internal palindrome interact with HMG1 to create a double helical loop in the intervening DNA.
Compared to naked DNA, NS1 binding resulted in enhanced DNase I cleavage at most positions between the two NS1 footprints (Fig. 6). There was also hypercutting of specific residues inboard of the nick site and outboard of the internal palindrome, suggesting that the minor groove of the DNA is particularly exposed at these positions.
When recombinant HMG1 was included in the complex, the margins of the protected regions changed slightly. The footprint at the nick site was extended an additional four to five nucleotides 3′ of the ACCA motif, while hypercutting at its inboard end became more pronounced. At the internal palindrome, hypercutting at the outboard border of the palindrome was diminished and the footprint extended a few nucleotides further inboard of the inner binding site. Moreover, HMG1 tended to enhance the affinity with which NS1 bound to both sites. Although this is not as apparent in the example presented in Fig. 6, modest increases in the binding affinity of NS1 at both sites have been observed in many footprints of active right-end origin sequences upon addition of HMG1 but have not been seen with inactive, mutant origins or at NS1 binding sites located outside the right-end origin.
HMG1 alone had no effect on the cutting pattern of otherwise naked DNA, even when added at five times the concentration required for maximal nicking. However, in the presence of NS1, HMG1 induced a marked change in the pattern of DNase I cutting between the two protected sites. Instead of overcutting at every base, DNase I cleavage now showed a 10- to 11-base periodicity on both strands (Fig. 6), which is characteristic of looped, double helical DNA (18, 39). DNase I is positioned over the minor groove of the DNA and cuts phosphate groups symmetrically to either side, according to the local architecture and twofold symmetry of the double helix. When the DNA is looped, all grooves on the inside of the circle narrow somewhat due to compression, while those on the outside become correspondingly wider, resulting in alternating bands of overcutting and undercutting. Studies are in progress to determine whether all active, but mutant, origin sequences reconfigure in this way upon addition of HMG1, but all three active mutant sequences tested to date do so, whereas six of the seven inactive substrates tested failed to show any evidence of HMG1-induced rearrangement. The sole exception was the nick site deletion mutant, mutant 5 (Fig. 4D). Thus, in the presence of both NS1 and HMG1, a higher-order nucleoprotein complex assembles on the origin, creating a specific loop in the intersite DNA. A more detailed analysis of this interaction is currently in progress.
DISCUSSION
The MVM initiator nickase NS1 is an ATP-dependent, site-specific DNA binding protein recognizing the sequence (ACCA)2-3, present both in the viral origins and at multiple sites throughout the genome. However, when bound at most sites the nickase function of this initiator remains tightly sequestered, and even at the origins it is unmasked only through the cooperation of specific cellular accessory proteins. Surprisingly, and for reasons which we do not yet understand, the two viral origins are very different from each other in size, sequence, and potential secondary structure, and the NS1 nickase function is activated by different cellular cofactors and through the establishment of very different structural complexes.
The minimal left (3′) origin is a fully duplex 50-bp sequence which extends from the binding site for its cellular cofactor, parvovirus initiation factor (p96-p79PIF), at one end, through a single NS1 binding site, a poly-T track, the nick site, and seven nucleotides of flanking sequence (15). p96-p79PIF is a heterodimeric complex of two closely related polypeptides which functions as a transcriptional modulator in the host cell (8). It binds coordinately to two ACGT sequences spaced five nucleotides apart, positioned in the origin so that the DNase I footprints of PIF and NS1 abut perfectly over a critical spacer element called the bubble sequence (7). In contrast, we show here that the minimal right-end origin is around 130 bp in length and can operate both in its mismatched hairpin configuration and as a fully duplex linear sequence derived from either arm of a palindromic tetramer bridge replication intermediate. This minimal sequence extends from guanidine residue 5042, located near the tip of the hairpin, to a position some 12 nucleotides upstream of the nick site, so all viral sequences located outside this region, which may serve as additional binding sites for NS1 (42) or for cellular factors, such as those described previously (5, 9, 37, 41), are not required for initiation. Nicking assays do suggest that hairpinned sequences are nicked more efficiently than their fully duplex counterparts, but we have not attempted to quantitate this effect or to explore the physical basis for the discrepancy.
Possibly, the presence of an unpaired nucleotide triplet in the stem of the right-end hairpin contributes to their efficiency difference since others (10) have shown that MVM genomes which lack such a mismatch remain infectious but replicate less effectively than wild-type genomes in vivo, giving rise to duplex replicative intermediate molecules with an unusually high proportion of uncleaved “turnaround” forms of the 5′ hairpin. However, when analyzed in a semiquantitative in vitro resolution assay, this same mutant origin appeared to be nicked and extended in the presence of recombinant NS1 with approximately the same efficiency as the wild-type sequence (42), suggesting that removal of the bubble asymmetry may have little influence on initiation efficiency per se or that its effect is slight when measured in vitro in a single round of terminal resolution. Presumably, the cumulative effects of even a slight reduction in efficiency would put the genome at a selective disadvantage in vivo. In the studies reported here, we have not attempted to quantitate or evaluate intriguing, but relatively minor, template differences of this type. Instead, we have focused on defining the minimal elements required to activate the nickase.
The essential sequences contain two widely spaced NS1 binding sites, one oriented so that its footprint extends over the nick site and a second positioned in the same orientation but some 100 bp further away. Footprints show that a third candidate NS1 binding site, in the inner arm of the palindrome, also binds NS1 efficiently, but this site is not required for origin activity. In the absence of HMG1, there is no evidence to suggest that the two essential NS1 complexes interact, but if HMG1 is added, a higher-order nucleoprotein complex is formed in which intersite DNA, in the vicinity of a GC-rich Sp1 site, is distorted in a way that strongly suggests that a double helical loop has been formed. As would be expected, by itself HMG1 can be shown to bend the origin sequence dramatically in a circularization/ligation assay, but it does so in a non-sequence-specific manner that cannot be detected by DNase I footprinting. In contrast, NS1 fails to induce circularization of this sequence or to enhance HMG1-induced circularization (Cotmore and Tattersall, unpublished data). Instead, the two spaced NS1 complexes appear to interact in the presence of HMG1, stabilizing the bend at a particular site in the complex.
Surprisingly, the NS1 binding site near the tip of the hairpin must include a GC base pair at nucleotide 5042 as part of a degenerate form of its recognition module (in the tetranucleotide ACNN), even though this arm of the palindrome already contains a contiguous ACCA 10-mer, which would appear to satisfy the known criteria for NS1 recognition (12). We do not know if this additional motif enhances the affinity of the site, but if nucleotide 5042 is mutated, the position of the NS1 footprint shifts (Cotmore and Tattersall, unpublished data), so recognition of the G residue at position 5042 influences the spacing, and hence orientation, of the two essential NS1 complexes. Preliminary SELEX analysis indicates that NS1 favors sequences which contain three copies of the ACCA tetranucleotide, of which one can be substantially degenerate, and that at least one of these motifs can be separated from the others by up to three nucleotides (Gottlieb et al., unpublished observations). Accordingly, all three binding sites in the right-end origin contain ACCA tetranucleotides which are not in strict register with each other, a fact which strongly suggests that they are recognized as separate modular “half-sites,” possibly by individual NS1 molecules which bind DNA as dimers or higher-order concatemers. Even so, the ability to tolerate variable spacing between recognition half-sites is rare, although it has been described for γδ resolvase (30), matα2 (40), and, more recently, for p96-p79PIF heterodimers (8).
We also show that NS1 must encounter a specific sequence at the nick site if it is to carry out the trans-esterification reaction. This sequence must interact with residues in the catalytic site of the nickase, but mutating active-site tyrosine210 both prevents nicking and severely impairs the ability of NS1 to bind ACCA reiterations (34). This suggests that Y210 is located within, or very close to, amino acids which are responsible for site-specific binding, a dual role that could readily be accommodated if NS1 bound DNA as an antiparallel complex in which one NS1 molecule (or oligomer) made contact with ACCA motifs while the opposing molecule made contact with the nick site across a twofold symmetry axis. That such an interaction would be possible in three dimensions is suggested by the finding, described here, that NS1 complexes, interacting with the internal palindrome of the right-end origin, bind with enhanced affinity when assembled into what appears to be an interdigitating, antiparallel supercomplex. Thus, although individual NS1 complexes partially cover the DNA recognition sequences normally used by the opposing complex, they do not compete for the substrate but rather interact to create a more stable complex. In this regard, it is perhaps significant that these two overlapping NS1 binding sites would be rotated through approximately a third of a turn of the helix with respect to one another.
In the origins of most prokaryotic RCR replicons, and in the MVM left-end origin, the initiator protein binds at a single site prior to nicking, but in the right-end origin two NS1 sites are required, positioned in such a way that they can interact with each other in the presence of HMG1 to create a higher-order nucleoprotein complex. In more complex systems, where nickases function as part of recombination mechanisms, binding site reiterations of this type, often associated with the use of DNA-bending proteins, are common (25, 30, 31, 35). Like NS1, such nickases remain catalytically silent until the entire nucleoprotein complex is assembled correctly, in part because when bound site specifically to otherwise naked DNA, their catalytic sites fail to make direct contact with the DNA at the nick site. In such cases complex formation promotes nicking by bending the duplex DNA through a particular path in which both the binding site and catalytic site of the nickase contact DNA simultaneously (22, 23). In MVM the situation is slightly different because it seems likely that the duplex DNA of the nick site must be melted, via an NS1-mediated, ATP-dependent unwinding reaction, before it can be cleaved. Recent studies indicate that when the nick site of the MVM left-end origin is presented as a single strand, nicking becomes both ATP and cofactor (PIF) independent (J. P. F. Nuesch, R. Corbau, V. Duverger, and J. Rommelaere, Program Abstr. 1st Parvovirus Euroconf., abstr. 2.1, 1999). By analogy, this would suggest that the NS1-NS1 interactions and three-dimensional structure of the right-end nucleoprotein complex may be required both to allow NS1 to unwind its duplex substrate and to facilitate direct contact between its catalytic residues and a single-stranded form of the initiation site.
ACKNOWLEDGMENTS
We thank Cathy Joyce and Nigel Grindley for providing MPE and advice on its use.
This work was supported by Public Health Service grants AI26109 and CA29303 from the National Institutes of Health.
REFERENCES
- 1.Astell C R. Terminal hairpins of parvovirus genomes and their role in DNA replication. In: Tijssen P, editor. Handbook of parvoviruses. Boca Raton, Fla: CRC Press; 1990. pp. 59–80. [Google Scholar]
- 2.Astell C R, Chow M B, Ward D C. Sequence analysis of the termini of virion and replicative forms of minute virus of mice DNA suggests a modified rolling hairpin model for autonomous parvovirus DNA replication. J Virol. 1985;54:171–177. doi: 10.1128/jvi.54.1.171-177.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldauf A Q, Willwand K, Mumtsidu E, Nüesch J P F, Rommelaere J. Specific initiation of replication at the right-end telomere of the closed species of minute virus of mice replicative-form DNA. J Virol. 1997;71:971–980. doi: 10.1128/jvi.71.2.971-980.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodnar J W. Sequence organization in regulatory regions of DNA of minute virus of mice. Virus Genes. 1989;2:167–182. doi: 10.1007/BF00315260. [DOI] [PubMed] [Google Scholar]
- 5.Brunstein J, Astell C R. Analysis of the internal replication sequence indicates that there are three elements required for efficient replication of minute virus of mice minigenomes. J Virol. 1997;71:9087–9095. doi: 10.1128/jvi.71.12.9087-9095.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen J, Cotmore S F, Tattersall P. Minute virus of mice transcriptional activator protein NS1 binds directly to the transactivation region of the viral P38 promoter in a strictly ATP-dependent manner. J Virol. 1995;69:5422–5430. doi: 10.1128/jvi.69.9.5422-5430.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen J, Cotmore S F, Tattersall P. Parvovirus initiation factor PIF: a novel human DNA-binding factor which coordinately recognizes two ACGT motifs. J Virol. 1997;71:5733–5741. doi: 10.1128/jvi.71.8.5733-5741.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen J, Cotmore S F, Tattersall P. Two new members of the emerging KDWK family of combinatorial transcription modulators bind as a heterodimer to flexibly spaced PuCGPy half-sites. Mol Cell Biol. 1999;19:7741–7750. doi: 10.1128/mcb.19.11.7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cossons N, Zannis-Hadjopoulos M, Tam P, Astell C R, Faust E A. The effect of regulatory sequence elements upon the initiation of DNA replication of the minute virus of mice. Virology. 1996;224:320–325. doi: 10.1006/viro.1996.0535. [DOI] [PubMed] [Google Scholar]
- 10.Costello E, Sahli R, Hirt B, Beard P. The mismatched nucleotides in the 5′-terminal hairpin of minute virus of mice are required for efficient viral DNA replication. J Virol. 1995;69:7489–7496. doi: 10.1128/jvi.69.12.7489-7496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costello E, Saudan P, Winocour E, Pizer L, Beard P. High mobility group chromosomal protein 1 binds to the adeno-associated virus replication protein (Rep) and promotes Rep-mediated site-specific cleavage of DNA, ATPase activity and transcriptional repression. EMBO J. 1997;16:5943–5954. doi: 10.1093/emboj/16.19.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotmore S F, Christensen J, Nüesch J P F, Tattersall P. The NS1 polypeptide of the murine parvovirus minute virus of mice binds to DNA sequences containing the motif [ACCA]2–3. J Virol. 1995;69:1652–1660. doi: 10.1128/jvi.69.3.1652-1660.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cotmore S F, Nüesch J P, Tattersall P. In vitro excision and replication of 5′ telomeres of minute virus of mice DNA from cloned palindromic concatemer junctions. Virology. 1992;190:365–377. doi: 10.1016/0042-6822(92)91223-h. [DOI] [PubMed] [Google Scholar]
- 14.Cotmore S F, Tattersall P. In vivo resolution of circular plasmids containing concatemer junction fragments from minute virus of mice DNA and their subsequent replication as linear molecules. J Virol. 1992;66:420–431. doi: 10.1128/jvi.66.1.420-431.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cotmore S F, Tattersall P. An asymmetric nucleotide in the parvoviral 3′ hairpin directs segregation of a single active origin of DNA replication. EMBO J. 1994;13:4145–4152. doi: 10.1002/j.1460-2075.1994.tb06732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cotmore S F, Tattersall P. DNA replication in the autonomous parvoviruses. Semin Virol. 1995;6:271–281. [Google Scholar]
- 17.Cotmore S F, Tattersall P. High-mobility group 1/2 proteins are essential for initiating rolling-circle-type DNA replication at a parvovirus hairpin origin. J Virol. 1998;72:8477–8484. doi: 10.1128/jvi.72.11.8477-8484.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drew H R, Travers A A. DNA bending and its relation to nucleosome positioning. J Mol Biol. 1985;186:773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- 19.Hatfull G F, Noble S M, Grindley N D. The gamma delta resolvase induces an unusual DNA structure at the recombinational crossover point. Cell. 1987;49:103–110. doi: 10.1016/0092-8674(87)90760-4. [DOI] [PubMed] [Google Scholar]
- 20.Hertzberg R P, Dervan P B. Cleavage of DNA with methidiumpropyl-EDTA-iron(II): reaction conditions and product analyses. Biochemistry. 1984;23:3934–3945. doi: 10.1021/bi00312a022. [DOI] [PubMed] [Google Scholar]
- 21.Kestler J, Neeb B, Struyf S, Van Damme J, Cotmore S F, D'Abramo A, Tattersall P, Rommelaere J, Dinsart C, Cornelis J J. Cis-requirements for the efficient production of recombinant DNA vectors based on autonomous parvoviruses. Hum Gene Ther. 1999;10:1618–1632. doi: 10.1089/10430349950017626. [DOI] [PubMed] [Google Scholar]
- 22.Kim S, Landy A. Lambda Int protein bridges between higher order complexes at two distant chromosomal loci attL and attR. Science. 1992;256:198–203. doi: 10.1126/science.1533056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim S, Moitoso de Vargas L, Nunes-Duby S E, Landy A. Mapping of a higher order protein-DNA complex: two kinds of long-range interactions in lambda attL. Cell. 1990;63:773–781. doi: 10.1016/0092-8674(90)90143-3. [DOI] [PubMed] [Google Scholar]
- 24.Koonin E V, Ilyina T V. Computer-assisted dissection of rolling circle DNA replication. Biosystems. 1993;30:241–268. doi: 10.1016/0303-2647(93)90074-m. [DOI] [PubMed] [Google Scholar]
- 25.Lavoie B D, Chaconas G. Transposition of phage Mu DNA. Curr Top Microbiol Immunol. 1996;204:83–102. doi: 10.1007/978-3-642-79795-8_4. [DOI] [PubMed] [Google Scholar]
- 26.Lavoie B D, Shaw G S, Millner A, Chaconas G. Anatomy of a flexer-DNA complex inside a higher-order transposition intermediate. Cell. 1996;85:761–771. doi: 10.1016/s0092-8674(00)81241-6. [DOI] [PubMed] [Google Scholar]
- 27.Legendre D, Rommelaere J. Targeting of promoters for trans activation by a carboxy-terminal domain of the NS1 protein of the parvovirus minute virus of mice. J Virol. 1994;68:7974–7985. doi: 10.1128/jvi.68.12.7974-7985.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lorson C, Burger L R, Mouw M, Pintel D J. Efficient transactivation of the minute virus of mice P38 promoter requires upstream binding of NS1. J Virol. 1996;70:834–842. doi: 10.1128/jvi.70.2.834-842.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maxam A M, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 30.Murley L L, Grindley N D F. Architecture of the gamma-delta resolvase synaptosome—oriented heterodimers identify interactions essential for synapsis and recombination. Cell. 1998;95:553–562. doi: 10.1016/s0092-8674(00)81622-0. [DOI] [PubMed] [Google Scholar]
- 31.Nash H A. The HU and IHF proteins: accessory factors for complex protein-DNA assemblies. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. R. G. Austin, Tex: Landes Company; 1996. pp. 149–179. [Google Scholar]
- 32.Nüesch J P, Corbau R, Tattersall P, Rommelaere J. Biochemical activities of minute virus of mice nonstructural protein NS1 are modulated in vitro by the phosphorylation state of the polypeptide. J Virol. 1998;72:8002–8012. doi: 10.1128/jvi.72.10.8002-8012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nüesch J P, Cotmore S F, Tattersall P. Expression of functional parvoviral NS1 from recombinant vaccinia virus: effects of mutations in the nucleotide-binding motif. Virology. 1992;191:406–416. doi: 10.1016/0042-6822(92)90202-z. [DOI] [PubMed] [Google Scholar]
- 34.Nüesch J P, Cotmore S F, Tattersall P. Sequence motifs in the replicator protein of parvovirus MVM essential for nicking and covalent attachment to the viral origin: identification of the linking tyrosine. Virology. 1995;209:122–135. doi: 10.1006/viro.1995.1236. [DOI] [PubMed] [Google Scholar]
- 35.Pedulla M L, Lee M H, Lever D C, Hatfull G F. A novel host factor for integration of mycobacteriophage L5. Proc Natl Acad Sci USA. 1996;93:15411–15416. doi: 10.1073/pnas.93.26.15411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pöhler J R, Norman D G, Bramham J, Bianchi M E, Lilley D M. HMG box proteins bind to four-way DNA junctions in their open conformation. EMBO J. 1998;17:817–826. doi: 10.1093/emboj/17.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Russnak R H, Candido E P, Astell C R. Interaction of the mouse chromosomal protein HMG-I with the 3′ ends of genes in vitro. J Biol Chem. 1988;263:6392–6399. [PubMed] [Google Scholar]
- 38.Salvino R, Skiadopoulos M, Faust E A, Tam P, Shade R O, Astell C R. Two spatially distinct genetic elements constitute a bipartite DNA replication origin in the minute virus of mice genome. J Virol. 1991;65:1352–1363. doi: 10.1128/jvi.65.3.1352-1363.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salvo J J, Grindley N D. The gamma delta resolvase bends the res site into a recombinogenic complex. EMBO J. 1988;7:3609–3616. doi: 10.1002/j.1460-2075.1988.tb03239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith D L, Johnson A D. A molecular mechanism for combinatorial control in yeast: MCM1 protein sets the spacing and orientation of the homeodomains of an alpha 2 dimer. Cell. 1992;68:133–142. doi: 10.1016/0092-8674(92)90212-u. [DOI] [PubMed] [Google Scholar]
- 41.Tam P, Astell C R. Multiple cellular factors bind to cis-regulatory elements found inboard of the 5′ palindrome of minute virus of mice. J Virol. 1994;68:2840–2848. doi: 10.1128/jvi.68.5.2840-2848.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willwand K, Baldauf A Q, Deleu L, Mumtsidu E, Costello E, Beard P, Rommelaere J. The minute virus of mice (MVM) nonstructural protein NS1 induces nicking of MVM DNA at a unique site of the right-end telomere in both hairpin and duplex conformations in vitro. J Gen Virol. 1997;78:2647–2655. doi: 10.1099/0022-1317-78-10-2647. [DOI] [PubMed] [Google Scholar]
- 43.Wilson G M, Jindal H K, Yeung D E, Chen W, Astell C R. Expression of minute virus of mice major nonstructural protein insect cells: purification and identification of ATPase and helicase activities. Virology. 1991;185:90–98. doi: 10.1016/0042-6822(91)90757-3. [DOI] [PubMed] [Google Scholar]
- 44.Wobbe C R, Dean F, Weissbach L, Hurwitz J. In vitro replication of duplex circular DNA containing the simian virus 40 DNA origin site. Proc Natl Acad Sci USA. 1985;82:5710–5714. doi: 10.1073/pnas.82.17.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zappavigna V, Falciola L, Citterich M H, Mavilio F, Bianchi M E. HMG1 interacts with HOX proteins and enhances their DNA binding and transcriptional activation. EMBO J. 1996;15:4981–4991. [PMC free article] [PubMed] [Google Scholar]
- 46.Zwilling S, Konig H, Wirth T. High mobility group protein 2 functionally interacts with the POU domains of octamer transcription factors. EMBO J. 1995;14:1198–1208. doi: 10.1002/j.1460-2075.1995.tb07103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]