Abstract
Glycoprotein B (gB; gpUL55) of human cytomegalovirus (HCMV) plays a critical role in virus entry and cell-to-cell spread of infection. To define the structure-function relationships in gB, a panel of linker-insertion mutations was generated throughout the coding region. This strategy yielded a panel of 22 mutants with four amino acid insertions and 3 large truncation mutants. Assessment of the mutant proteins' biosynthetic properties and folding patterns analyzed in context with predicted secondary features revealed novel insights into gB's structure and trafficking properties. All of the insertion mutants were able to assemble into oligomers, suggesting that oligomerization is tolerant of small insertions and/or that multiple regions of the protein may be involved. Computer algorithm predictions of gB's secondary structure indicate that the furin-recognized cleavage site falls within an exposed loop. This loop may be particularly sensitive to structural alterations, since insertions upstream and downstream of the cleavage site rendered the mutant proteins cleavage defective. In addition, a strong correlation existed between terminal folding and cleavage of gB. Interestingly, terminal folding was not correlated with delivery to the cell surface but may influence the rate of transport to the cell surface. Nine mutants, containing insertions in both the extracellular and intracellular portions of gB, retained wild-type structural properties. This panel of characterized gB mutants, the first of this type for an HCMV protein, will be a useful tool in dissecting the role of gB during HCMV infection.
Human cytomegalovirus (HCMV), a member of the Herpesviridae, is a ubiquitous pathogen that causes significant morbidity and mortality in immunocompromised individuals (1). The frequency and severity of HCMV diseases and the paucity of desirable therapeutic options indicate the need to identify and understand the structures and functions of key potential antiviral targets. An ideal target for either prevention or therapy would be to block entry and/or cell-to-cell spread of HCMV into and between cells. To rationally develop drugs, a detailed understanding of the viral and cellular components necessary for virus entry is needed. While HCMV entry is not completely understood, significant progress has been made in recent years identifying the major viral components involved in entry (reviewed in reference 13). Entry of HCMV into host cells requires a complex series of sequential interactions between multiple viral and cellular molecules (7, 13, 28). Viral entry can be divided into two distinct stages: attachment and penetration. Virus binding to heparan sulfate proteoglycans (HSPGs) is the initial step in the entry pathway (15, 27). Virus bound to cell surface HSPG is rapidly converted to a more stable binding state likely mediated by interaction of the virion with a second cellular receptor (15). Once stably attached, the virion penetrates into the cell by direct fusion of the virus envelope and the cellular plasma membrane (14).
A central player in all facets of HCMV entry is glycoprotein B (gB). gB is a 906-amino-acid (aa) type I transmembrane protein encoded by the UL55 gene of HCMV and is one of only five glycoproteins conserved throughout the herpesvirus family; the others are homologs of herpes simplex virus glycoproteins gH, gL, gM, and gN (7, 12, 16, 28). The gB protein of HCMV is the major structural protein of the viral envelope and is present on the surface of infected cells (5, 19, 29). Independent lines of evidence have suggested that gB functions as a ligand and may facilitate virus attachment to host cells. Heparin affinity chromatography (11, 15, 21) directly demonstrated gB's ability to bind heparin. Binding studies with cell lines containing or lacking HSPGs have shown that gB can directly bind two different cell surface receptors in a dose-dependent, saturable, and specific manner (3). In those studies, cell surface HSPGs were identified as one class of receptors for gB; the second, nonheparin receptor remains to be elucidated (3, 11). Interaction of gB with its nonheparin receptor has immediate consequences to the cell, resulting in the initiation of intracellular signaling and the induction of cellular gene expression (4). In addition to its role as a ligand, gB is also required for fusion activity. gB-expressing cell lines and antibody inhibition studies have directly implicated gB both in the fusion step of entry and in cell-to-cell spread during HCMV infection (26, 42, 43).
Analysis of gB has been hampered by the difficulty in obtaining recombinant HCMV containing either null or modified forms of the protein. Researchers have circumvented this problem by making specific mutations within the coding region, producing the recombinant products in various expression systems, and then analyzing the molecule for specific properties or defects (3, 8, 24, 37, 42). This approach has been useful in defining some of the structural features and functional properties of gB (reviewed in reference 7). For example, deletion analysis of the large hydrophobic segment of gB revealed that aa 751 to 771 are necessary and sufficient to retain the molecule within the cell membrane (33, 47). However, a number of important questions about HCMV gB remain unanswered. What domains of gB confer its ligand binding properties? Which segments of the protein contain elements essential for fusion? What regions are important in cell-to-cell spread? Which portions of the molecule are critical during viral maturation? What functions are contained within the large (135-aa) cytoplasmic tail of gB? Last, what are the structural correlates for these functional domains?
Structure-function relationships of proteins are often deciphered by the analysis of many individual mutations. To this end, a systematic global mutagenesis approach of gB was undertaken. Using an oligonucleotide-directed mutagenesis approach, we generated 22 mutants containing four amino acid insertions at sites throughout the coding region of gB; three large truncation mutations prior to the transmembrane anchor of gB were also generated. A triage system utilizing information of the gB maturation pathway in virally infected cells was established to analyze the structural integrity of the mutants. Processing of gB is a complex step wise pathway that begins with nascent gB quickly assuming a disulfide-linked oligomeric structure in the endoplasmic reticulum (ER) (2, 10). Export from the ER requires a series of prolonged folding steps, many of which can be distinguished on the basis of recognition with various anti-gB monoclonal antibodies (MAbs) (2, 10). Following transport out of the ER, gB enters the Golgi apparatus, where it undergoes glycosylation modifications and is cleaved into two fragments by the cellular protease, furin (44). The monomeric form of gB is composed of two fragments (gp116 and gp55) held together by intramolecular disulfide bonds (2). The mature product is then delivered to the surface of an infected cell, where it is cycled between endosomal vesicles and the plasma membrane and eventually incorporated into virions (32). Our triage screen examined four structural properties of gB. Initially, primary structural characteristics of gB mutants were analyzed by using MAbs specific for linear, conformational, and oligomeric epitopes on gB. Next, oligomer formation and proteolytic processing of the modified proteins were directly examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Finally, the ability of the gB mutants to be transported to the cell surface was assessed by using two independent delivery assays. The rationale for examining the structural characteristics of the mutant proteins was threefold: first, key structural elements required for gB trafficking and ultimate localization to virion maturation sites may be identified; second, if a mutant was aberrant in a number of processing steps, it would likely be uninformative in functional studies due to gross structural defects and should be eliminated from functional studies; third, examining the processing characteristics of the mutants gives us a framework of information that can be applied to understanding results from functional studies.
MATERIALS AND METHODS
Construction of gB insertion mutants.
All enzymatic reactions were performed with manufacturer-supplied buffers and protocols. The entire 2.8-kb gB open reading frame (ORF) of HCMV (strain AD169) was isolated from a cosmid library and placed into plasmid pSP72 (Promega) by using available restriction sites (34). The resulting plasmid, p72-gB, was digested individually with one of a number of restriction enzymes (BsaAI, EheI, HindII, MscI, PvuII, and RsaI) under conditions which produced a singly cut linearized plasmid containing free blunt ends. The restricted DNA was separated by gel electrophoresis, and the linearized full-length plasmid was excised, eluted, and treated with calf intestinal phosphatase. A double-stranded oligonucleotide 12 bp in length containing a NotI restriction site (5′ GAGCGGCCGCTC 3′) was then ligated onto the purified plasmid. The insertion of the oligonucleotide introduced 4 additional in-frame aa into the coding region of gB: primarily Glu-Arg-Pro-Leu, Ser-Gly-Arg-Ser, or Ala-Ala-Ala-Arg, depending on the site of insertion. In a small minority of the insertions (3 of 25), a stop codon was introduced into the coding region of gB. After ligation, the p72-gB oligonucleotide insertion construct was digested with NotI to ensure that only one copy of the NotI oligonucleotide had been introduced into the plasmid. The restricted DNA was again separated by gel electrophoresis, and the mutagenized p72-gB linearized plasmid was excised, eluted, and ligated to itself. The circularized plasmids were propagated in Escherichia coli DH5α cells (Gibco-BRL), and small-batch plasmid DNA purification was performed. The resulting plasmids were screened for the presence of the introduced NotI restriction site. The mutant constructs were mapped by restriction site analysis to determine the site of insertion. A mutagenesis strategy parallel to the one described above but utilizing additional restriction enzymes and complementary 12-bp oligonucleotides (BsrFI [5′ CCGGGCGGCCGC 3′], BssHII [5′ CGCGGCGGCCGC 3′], and NsiI and PstI [5′ GCGGCCGCTGCA 3′]) was performed to complete the mutagenesis of the HCMV gB gene. These oligonucleotides gave rise to insertion mutants I-885 (BsrFI), I-256 (BssHII), I-832 (NsiI), and I-484 (PstI) (Fig. 1). Fidelity of all mutants was confirmed by sequencing of the constructs around the area of the oligonucleotide insertion (UW Biotechnology Center, DNA Synthesis and Sequencing Facility). The mutagenized gB ORFs were then placed into the pRC/CMV vector (Invitrogen) for expression in mammalian cells.
FIG. 1.
Schematic representation of the 25 gB mutants. The site of insertion is designated with an X, and the three mutants below represent the truncation mutants. Some important features of the HCMV gB molecule are highlighted: signal sequence (Seq), important antigenic regions (AD1 and AD2), furin cleavage site, transmembrane anchor, and phosphorylation site.
Cell maintenance.
The generation and characterization of 293T cells (originally referred to as 293tsA1609neo) have been previously described (17). The 293T cells were cultured in Dulbecco's minimal medium (DMEM; Cellgro) supplemented with 10% fetal bovine serum (FBS; HyClone), 1.0% penicillin-streptomycin-amphotericin B (Fungizone) (BioWhittaker), and 0.3% l-glutamine (BioWhittaker).
Transient expression of the gB mutants.
Subconfluent 293T cells in 60-mm-diameter dishes were transfected with 3 μg of gB constructs in serum-free DMEM by using the lipofection agent GenePORTER (Gene Therapy Systems). After 5 h of incubation, the DNA-GenePORTER-containing medium was replaced with DMEM containing 10% FBS. Fresh DMEM containing 10% FBS was added to the cells 24 h posttransfection, and the 293T cells were harvested 40 h posttransfection.
Analysis of extracts from gB mutant-expressing cells by using MAbs.
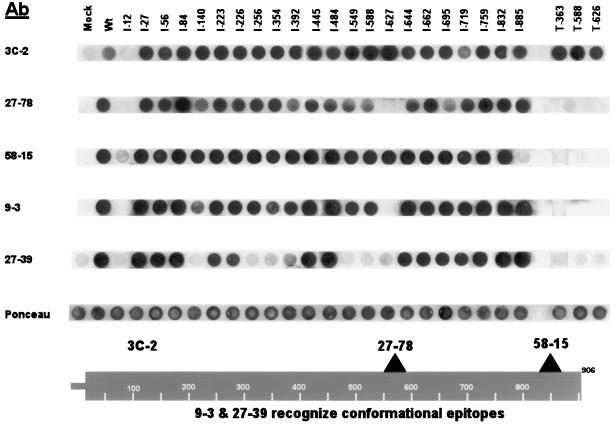
After collection, washing, and counting of transfected 293T cells, detergent extracts were made by lysing the cells in a phosphate-buffered saline (PBS) solution containing 1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS; Sigma). Clarified detergent extracts from 105 cell equivalents were transferred to nitrocellulose membranes via a 96-well dot blot apparatus (Millipore). Analysis of the lysates was conducted with MAbs reacting to various epitopes of gB, utilizing standard procedures detailed previously for immunoblots (11, 31). The mouse MAbs used in this study were 3C-2 (recognizes an epitope specific for the amino terminus of gB [22]), 27-78 (recognizes an epitope in the carboxyl-terminal fragment of gB at an immunodominant region termed AD1 [5]), 58-15 (recognizes an epitope in the extreme carboxyl terminus of gB [2, 10]), 9-3 (recognizes a neutralizing conformational epitope of gB [5, 10]), and 27-39 (recognizes a conformational epitope of gB found specifically on the oligomer form of gB [2, 10]). MAb 3C-2 was a gift from Mark Stinski (University of Iowa); MAbs 27-78, 58-15, 9-3, and 27-39 were donated by William J. Britt (University of Alabama at Birmingham).
SDS-PAGE analysis and immunoblotting.
Transfected cell extracts were prepared as described above. Extracts containing 5 × 105 cell equivalents were subjected to SDS-PAGE as previously described (11, 31). For comparison purposes, human fibroblast cells were infected with the AD169 strain of HCMV at a multiplicity of infection of 5. The cells were harvested 7 days postinfection, and detergent lysates were prepared as described for transfected cells. Cell lysates were resuspended in sample buffer (0.06 M Tris [pH 6.8], 10% glycerol, 2% SDS) either lacking or containing a reducing agent (0.4 M dithiothreitol). Resolved proteins were transferred to a nitrocellulose membrane and probed with the indicated antibodies, using procedures previously described (11, 31).
Cell surface biotinylation.
Mutant constructs were transfected into 293T cells as described above. Forty hours posttransfection, cells were washed twice with PBS and then incubated at 0°C for 45 min with PBS containing 0.75 mg of Sulfo-NHS-LC-biotin (a membrane-impermeable form of biotin; Pierce) per ml. The reaction was quenched with the addition of 0.5 ml of 20 mM glycine in PBS. The cells were harvested and lysed in a 1% CHAPS solutionin in PBS. The cell extracts were incubated overnight with 30 μl of a 50% (vol/vol) slurry of streptavidin-agarose beads that had been preequilibrated with a 1% CHAPS solution in PBS. Following the incubation, the streptavidin-agarose beads were washed three times with a 1% CHAPS solution in PBS. The streptavidin-agarose beads were then resuspended in 2× sample buffer containing 4% SDS and heated to 100°C for 5 min. The resulting supernatants were transferred to nitrocellulose membranes via a dot blot apparatus and probed with MAbs 58-15 or 3C-2, using standard procedures for immunoblots (11, 31). To determine total gB content of transfectants, cells were treated as described above but lysed in 1% CHAPS solution in PBS prior to incubation with Sulfo-NHS-LC-biotin. For control experiments, the Erk1/2 protein products were detected with a commercially available polyclonal rabbit antibody specific for the cytoplasmic (inactive) forms of Erk1/2 (anti-mitogen-activated protein [MAP] kinase 1/2 [Erk1/2-CT; Upstate Biotechnology]). For additional control experiments, the HCMV immediate-early (IE) protein was expressed by using the pRC/CMV vector (Invitrogen) in 293T cells and detected with the commercially available MAb GICR 1203 (Goodwin Institute for Cancer Research).
Cell surface proteinase K sensitivity assay.
Mutant constructs were transfected into 293T cells as described above. Forty hours posttransfection, cells were washed twice with PBS and then incubated at 0°C for 60 min with PBS containing 400 mg of proteinase K (Sigma) per ml. The cells were collected by centrifugation, washed three times with FBS, and then subjected to additional three washes in PBS to remove the proteinase K. The rinsed cells were lysed in a 1% CHAPS solution in PBS. Clarified cell lysates were resuspended in sample buffer containing a reducing agent (0.4 M dithiothreitol) and subjected to SDS-PAGE analysis as described above. Resolved proteins were transferred to a nitrocellulose membrane and probed with MAb 58-15, using procedures described above. The cytoplasmic proteins HSP90 and Erk1/2 were detected in control experiments using commercially available antibodies: anti-HSP90 (Transduction Laboratories) and anti-MAP kinase 1/2 (Erk1/2-CT; Upstate Biotechnology).
RESULTS
Using an insertional mutagenesis approach, we generated 25 independent mutations (22 insertions and 3 truncations) throughout the extracellular and cytoplasmic domains of HCMV gB (Fig. 1). The specific insertion mutagenesis strategy that we used had a number of significant advantages: (i) it had the potential to generate a large number of mutations throughout the gB coding region, (ii) the specific oligonucleotides chosen minimized the probability of producing truncation mutants, and (iii) the insertion of 4 aa reduced the risk of severe disruptions with gB's tertiary structure. Once the panel of gB mutants was generated, we wished to examine their synthesis and processing in mammalian cells. These data provide insights into the structural requirements necessary for gB to be exported to the cell surface. In addition, knowledge of the biosynthetic properties of the gB mutants will provide a useful background of information that will aid in deciphering future functional analysis. The following specific features of the gB mutants were examined in an experimental triage screen: recognition by a panel of MAbs, ability to assemble into an oligomer, ability to be proteolytically processed, and competence to traffic to the cell surface. The rationale for selecting these specific criteria is that all of these properties have been well defined as intermediates in the processing pathway of viral gB in infected cells (2, 10, 19, 44).
Recognition of gB mutants by a panel of anti-gB MAbs.
To analyze the protein products of the mutant constructs, transient expression experiments were performed in human 293T cells. Lysates were recovered from transfected cells and analyzed by a dot blot procedure using MAbs reacting to various epitopes of gB (Fig. 2). The Ponceau S staining profile demonstrated that near-equivalent amounts of protein were applied to the membrane. MAb 3C-2, which reacts to a linear epitope in the amino terminus of gB, recognized all of the expressed mutant proteins except for the mutant with an insertion at aa 12 (I-12). The I-12 insertion mutant disrupts the predicted gB signal sequence (37) and likely causes the mutant to be translated in the cytoplasm, where it may be rapidly degraded. The I-12 mutant was not significantly recognized by any of the antibodies tested (including conformation-specific MAbs) and was therefore eliminated from further structural analysis. MAb 3C-2 also recognized all of the truncation mutants, including T-363. This finding permits further localization of the unmapped 3C-2 epitope to the first 363 aa of gB.
FIG. 2.
Recognition of gB mutants with a panel of MAbs. Transfected mutant lysates were transferred to nitrocellulose and probed with a panel of MAbs. A representative Ponceau S stain is displayed to indicate that approximately equal concentrations of protein were loaded. Insertion mutants are indicated as I-n, and truncation mutants are shown as T-n. Binding sites of the antibodies are indicated at the bottom (see Materials and Methods for MAb references). The analysis was performed in a standard 96-well dot blot apparatus. The data are shown in a linear form for presentation purposes and were rearranged with the graphics program Adobe Photoshop.
MAb 27-78, reactive against a carboxyl-terminal linear epitope within gB (region AD1 [Fig. 1]), recognized all of the insertion mutant proteins except I-12 and I-627. The I-627 mutant protein was likely not recognized by MAb 27-78 because the insertion disrupts the epitope recognized by 27-78. The three truncation mutants were not recognized by 27-78 because they terminate prior to the epitope recognized by the antibody. MAb 58-15, reactive to a linear epitope in the extreme carboxyl terminus of gB (Fig. 2), recognized all of the insertion proteins except I-12 and I-884. MAb 58-15 failed to recognize the I-884 protein because the insertion likely overlaps or is extremely close to its epitope. As predicted, the three truncation mutants were also not recognized by 58-15.
The last two MAbs tested (9-3 and 27-39) were the most stringent antibodies used in this study because they bind only to conformational epitopes found on mature forms of gB (2, 5, 10). Maturation of gB within the ER is a complex process with folding intermediates forming sequentially. The conformational epitope recognized by 9-3 is formed after oligomerization but before the formation of the epitope recognized by 27-39 (2, 10). The 27-39 epitope is found only on a terminally folded form of gB and occurs with half-times of formation within the ER approaching 200 min (2, 10). MAb 9-3 recognized all but two of the insertion mutants (I-12 and I-627). Recognition by 9-3 is a strong indication that nearly all of the insertion mutants were not globally disrupted in their secondary structure. MAb 27-39 recognized 14 of 22 insertion mutants, indicating that over half of the mutants are processed through this characterized folding epitope. Neither MAb 9-3 nor MAb 27-39 recognized the three truncation mutants, the longest of which terminates at aa 626. A previously characterized truncation mutant (aa 1 to 692) retained many of gB's functional properties and was recognized by MAb 27-39 (3). This finding suggests that the region between aa 626 and 692 is critical for the proper folding of gB. Since our three truncation mutants were not recognized by either of our conformation-specific MAbs, we concentrated our efforts on further evaluating the structural properties of only the insertion mutants.
Oligomer formation of the gB mutants.
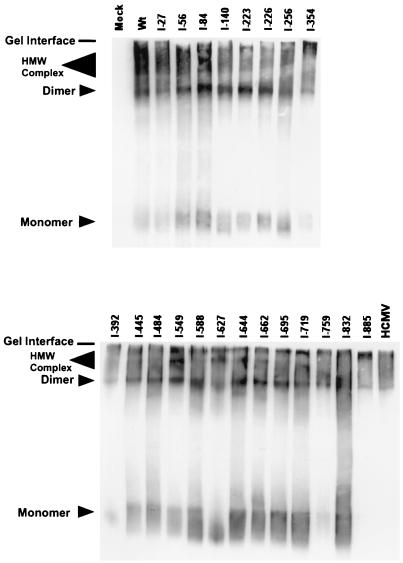
Studies have shown that HCMV gB is a disulfide-linked oligomer, likely a dimer, that assumes its higher-order structure in the ER (2, 10). The oligomeric form of gB is the mature form found in the virion envelope and is likely the active form of the molecule (6). In addition, we have shown that the gB dimer complexes with cellular annexin II (31). The gB-annexin II oligomer and gB dimer migrate as diffuse but distinct high-molecular-weight species analyzed in polyacrylamide gels under nonreducing conditions (10, 31, 47). We determined the ability of the insertion mutants to form oligomers by examining electrophoretic migration patterns of nonreduced protein samples in SDS-PAGE followed by immunoblotting. When wild-type gB is expressed in the context of HCMV-infected cells or in transfected cells, both the dimer and high-molecular-weight oligomer are detected (Fig. 3) (2, 10, 31, 47). In addition, all of the mutants formed similar complexes when expressed in 293T cells (Fig. 3). These results show that none of the insertions affected an early step in gB's biosynthetic processing, that of dimerization (oligomerization).
FIG. 3.
Immunoblot analysis examining oligomer formation of the gB insertion mutants. Transfected cell lysates were resuspended in nonreduced sample buffer, resolved by SDS-PAGE on a 7.5% gel, and electrotransferred to nitrocellulose. The blots were probed with a combination of MAbs 27-78 and 58-15 as described in Materials and Methods. The HCMV lane represents cell lysates from 7-day-infected fibroblast cells; mock lane represents cell lysates from untransfected 293T cells. HMW, high molecular weight.
Proteolytic processing of the gB mutants.
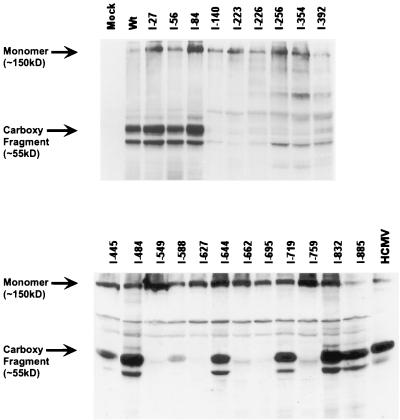
As is the case for many viral fusion proteins, HCMV gB protein is cleaved by the cellular protease, furin. Specifically, gB is cleaved between aa 460 and 461 in the Golgi network and/or endosomal compartment (36, 44), yielding disulfide-linked complexes composed of amino-terminal (115-kDa) and carboxyl-terminal (55-kDa) fragments. It has been suggested that proteolytic cleavage may enhance HCMV infectivity (44). In addition, full activation by furin requires routing through the trans-Golgi network -endosomal system and may be a critical step in virion maturation and egress (25). The ability of the gB mutants to be cleaved was determined by analysis of reduced protein samples in SDS-PAGE and immunoblotting. Cleavage of gB is indicated by the presence of a 55-kDa carboxy-terminal fragment in these experiments. As shown in Fig. 4, the analysis revealed that 10 of 22 mutants were efficiently proteolytically processed. The origin of the reactive species migrating slightly faster than the gB carboxy-terminal fragment is not known but is frequently detected (5, 19, 31, 42, 43, 47). In addition, detectable monomeric gB was evident for all mutants and wild-type (wt) gB and likely represents protein resistant to reduction. Interestingly, mutations immediately flanking the cleavage site (I-445 and I-484) were cleaved normally. However, mutations further from the cleavage site but in the central portion of the molecule generally resulted in a cleavage-defective phenotype.
FIG. 4.
Immunoblot analysis examining proteolytic processing of the gB insertion mutants. Transfected cell lysates were resuspended in reduced sample buffer, resolved by SDS-PAGE on a 10% gel, and electrotransferred to nitrocellulose. The blots were probed with a combination of MAbs 27-78 and 58-15 as described in Materials and Methods. The HCMV lane represents cell lysates from 7-day-infected fibroblast cells; the mock lane represents cell lysates from untransfected 293T cells.
Cell surface expression of the gB mutants.
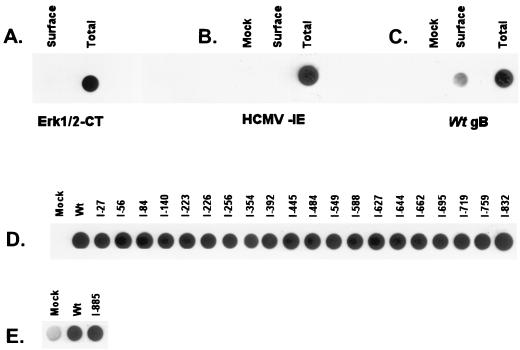
gB traffics to the plasma membrane in infected or transfected cells (5, 19, 29, 33). gB on the surface of infected cells has been shown to play a crucial role in cell-to-cell dissemination of the infection (7). Steady-state levels of gB on the cell surface are greatly influenced by the rate of internalization and recycling (18, 41). Transport competence of the mutants along the exocytotic pathway was determined by a cell surface biotinylation assay. We observed that a small fraction (less than 5%) of the total amount of gB in the transfectants was detectable at steady-state levels on the surface of cells (Fig. 5C). To ensure that the signal seen in the surface samples was not due to contamination by a small fraction of lysed cells, two control experiments examining cytoplasmic and nuclear proteins were conducted. The MAP kinase Erk1/2 serves as a particularly useful control since it resides on or near the cytoplasmic face of the plasma membrane (35, 40). As shown in Fig. 5, Erk was not detected on the cell surface but was readily detectable in the total cell fraction. Additionally, the HCMV IE protein which accumulates in the nucleus was readily detectable in the total cell fraction but not on the cell surface as expected (Fig. 5B). These two controls establish that the signals observed with our gB mutants represent true cell surface expression. Analysis of the gB insertion mutants in the cell surface biotinylation assay (Fig. 5D and E), showed that all 21 of the insertion gB mutants were detectable on the cell surface. We were surprised at the results since some of these proteins have folding defects, and correct folding is often correlated with forward trafficking through the export pathway.
FIG. 5.
Cell surface detection of the gB insertion mutants by using a biotinylation assay. Transfected cells were biotinylated, then lysed in a 1% CHAPS solution, and precipitated with streptavidin-agarose. The biotinylated proteins were boiled in 2× sample buffer and transferred to nitrocellulose. The biotinylated proteins were then detected with antibodies described in Materials and Methods. Total amount of protein in the cells was detected essentially the same, except that the cells were lysed prior to biotinylation. (A) Control examining both cell surface and total expression of the cytoplasmic protein Erk1/2; (B) control examining both cell surface and total expression of the transfected HCMV IE protein; (C) control using wt gB examining both cell surface and total expression (MAb 58-15 was used for detection; data were quantitated with a Bio-Rad phosphorimager); (D) cell surface expression of the insertion mutants, using MAb 58-15 for detection; (E) cell surface expression of I-885; MAb 3C-2 was used for detection because MAb 58-15 reacts poorly with I-885 (Fig. 2).
Thus, we sought to confirm these findings by using a second independent assessment of cell surface delivery. To this end, we developed a protease sensitivity assay to examine cell surface expression of the mutant constructs. Intact cells transfected with wt gB and mutants were treated with proteinase K. Complete proteolytic digestion of a cell surface gB molecule would generate a fragment consisting of the transmembrane and cytoplasmic portions of gB (aa 751 to 906, ca. 17 kDa). This fragment is detectable with the cytoplasmic domain-specific MAb 58-15. The migration patterns of two cytoplasmic proteins, HSP90 and Erk1/2, were examined as controls for the protease sensitivity of the cells. As expected, these proteins were unaltered in protease-treated cells (Fig. 6A). In wt gB-transfected cells treated with protease, a specific fragment of approximately 21 kDa was detected (Fig. 6B). This fragment was not present in either mock-treated or untreated transfected cells (Fig. 6B, lane Wt-ProK). The size of the fragment (21 kDa) indicates that aa 710 to 906 of gB were being protected from proteolysis. A fragment of slightly higher molecular weight is also present in the gB-transfected lanes but is not specific to the protease K treatment. All but two (I-832 and I-885) of the insertion mutants contained the 21-kDa protected fragment indicative of cell surface expression. Mutant I-832 had a diffuse, faster-migrating product ca. 10 kDa in size which was not present in untreated cells (data not shown). The identity of this product remains unknown, but it may be a unique breakdown form of the 21-kDa protected fragment specific for this mutant. No detectable fragment was seen with mutant I-885; MAb 58-15 does not react efficiently with this mutant protein (no other antibody reactive with I-885 and capable of detecting the 21-kDa protected fragment was available). There were differences in the abundance of the protected fragments seen with the different gB proteins. This is likely due to the inherent variability involved in transfection experiments and/or the recycling rates of the various mutants. In summary, two independent measures have indicated that all 21 of our insertion mutant gB proteins are capable of being expressed on the surface of cells.
FIG. 6.
Cell surface detection of the gB insertion mutants, using a protease sensitivity assay. Cells were treated with proteinase K (ProK); following removal of the protease, the cells were lysed in a 1% CHAPS solution. Cell lysates were resuspended in reduced sample buffer, resolved by SDS-PAGE on a 12.5% gel, and electrotransferred to nitrocellulose. The blots were probed with antibodies described in Materials and Methods. (A) Results of the protease treatment on two endogenous cytoplasmic proteins (HSP90 and Erk1/2); (B) Results of the protease treatment on transfected cells as examined by immunoblotting with the cytoplasmic domain-specific MAb 58-15. The arrow points to the presence of a unique protected fragment observed in gB-transfected cells which were treated with protease.
Summary of the triage screen for the mutants.
Using a generalized mutagenesis strategy, we generated 25 mutants (22 insertion and 3 truncation mutants) within the coding region of HCMV gB. The expressed mutant proteins were characterized with respect to four distinct biosynthetic and structural criteria: recognition by MAbs, oligomer formation, proteolytic processing, and cell surface expression (results are summarized in Table 1). Nine insertion mutants (I-27, I-56, I-84, I-445, I-484, I-644, I-719, I-832, and I-885) retained all four characteristics of viral gB.
TABLE 1.
| Construct | MAb
|
Structural property
|
||||||
|---|---|---|---|---|---|---|---|---|
| 3C-2 | 27-78 | 58-15 | 9-3 | 27-39 | Oligomer formation | Proteolytic processing | Surface expression | |
| wt | + | + | + | + | + | + | + | + |
| I-27 (81) | + | + | + | + | + | + | + | + |
| I-56 (168) | + | + | + | + | + | + | + | + |
| I-84 (252) | + | + | + | + | + | + | + | + |
| I-140 (419) | + | + | + | + | N | + | N | + |
| I-223 (669) | + | + | + | + | + | + | N | + |
| I-226 (678) | + | + | + | + | + | + | N | + |
| I-256 (769) | + | + | + | + | N | + | N | + |
| I-354 (1062) | + | + | + | + | N | + | N | + |
| I-392 (1177) | + | + | + | + | N | + | N | + |
| I-445 (1336) | + | + | + | + | + | + | + | + |
| I-484 (1451) | + | + | + | + | + | + | + | + |
| I-549 (1646) | + | + | + | + | N | + | N | + |
| I-588 (1764) | + | + | + | + | N | + | + | + |
| I-627 (1881) | + | N | + | N | N | + | N | + |
| I-644 (1932) | + | + | + | + | + | + | + | + |
| I-662 (1985) | + | + | + | + | + | + | N | + |
| I-695 (2085) | + | + | + | + | + | + | N | + |
| I-719 (2157) | + | + | + | + | + | + | + | + |
| I-759 (2278) | + | + | + | + | + | + | N | + |
| I-832 (2496) | + | + | + | + | + | + | + | + |
| I-885 (2654) | + | + | N | + | + | + | + | + |
Each mutant construct name indicates the approximate area of the amino acid insertion. The exact position (in nucleotides) of the insertion is shown in parentheses. The A of the gB start codon represents base 1, and the number indicated is the last base found in the wt gB sequence (AD169) before the insertion. In two constructs (I-445 and I-759), the fifth amino acid (proline) is an alteration of the wt sequence and not an insertion. All results are qualitative; no quantitative judgments have been made from the experimental results. Relative signal strength close to wt levels were rated as positive (+), and signals near mock controls were rated as negative (N).
DISCUSSION
A comprehensive understanding of the structure-function relationship of any HCMV glycoprotein is lacking. gB plays a vital role in many aspects of the viral life cycle, including entry, cell-to-cell spread, and egress (reviewed in reference 7). The functional dissection of this multifaceted protein has been hampered by the limited number of mutants that exist for this essential gene (K. A. Boyle and T. Compton, unpublished results). The goal of these studies was to perform a systematic global mutagenesis of gB to develop a large panel of mutants that could be used to study the structure-function relationship of gB. Using an oligonucleotide-directed mutagenesis approach, we generated 25 individual mutants within the coding region of gB. As designed, the oligonucleotide insertions minimized the number of truncations, with just three mutants containing stop codons. The insertion mutants span the gB ORF fairly uniformly, with three regions having mutational gaps of greater than 60 aa (aa 140 to 220, 256 to 354, and 759 to 831). All of the insertion (except I-12) and truncation mutants were efficiently expressed and detected by one or more MAbs. Although the dot blot method used was not highly quantitative, the results shown in Fig. 2 (3C-2 panel) indicate that the levels of expression did not vary greatly among the constructs. Thus, a representative panel of gB mutants has been created, the first of this type for a HCMV glycoprotein.
Insights into gB structure.
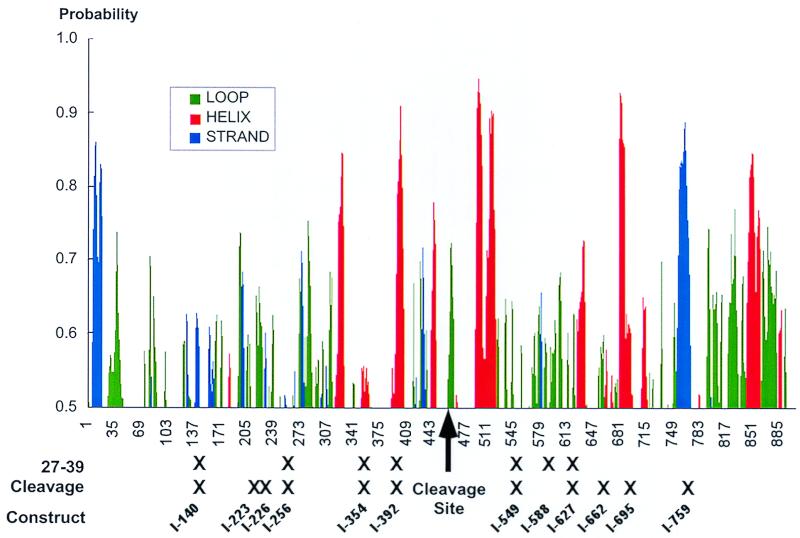
To facilitate interpretation of our results and to put these findings in context with possible structural organization of gB, we subjected the primary sequence of gB to secondary structural predictions and indicated the positions of mutations affecting folding and cleavage (Fig. 7). Our characterization of the synthesis and processing of gB mutants has provided novel insights into the structure of this important protein. (i) The first 22 aa of gB serve as the signal sequence. An insertion within the predicted signal sequence of gB, I-12, resulted in barely detectable levels of protein. We believe that the I-12 insertion was likely translated in the cytoplasm and rapidly degraded by the cell. This result experimentally confirms that the amino terminus of gB is critical for expression of the molecule and presumably functions as a signal sequence. (ii) Oligomerization is tolerant of small insertions. Nascent gB rapidly oligomerizes in the ER, and we found that all of the insertion mutants (except I-12) retained their ability to oligomerize. There are a number of possible reasons for this. It is possible that we lack mutations in the oligomerization domain or that the insertions were not large enough to disrupt the oligomerization domain. Alternatively, there may be multiple and/or redundant oligomerization domains present within gB. Multiple oligomerization domains have been observed with the herpes simplex type 1 gB homologue (23), and it is therefore likely that the HCMV gB molecule also contains multiple oligomerization domains. (iii) Folding and cleavage are coupled. All of our mutants retained the furin consensus cleavage recognition site (R X K/R R) occurring at amino acids 456 to 459, but 10 mutant proteins failed to be cleaved. Alterations both upstream and downstream of the cleavage site resulted in a cleavage-negative phenotype. Since all of the mutants were detected at the cell surface and presumably trafficked through the trans-Golgi network (see below), the failure of individual proteins to be cleaved by furin is not likely due to inaccessibility to the cleavage enzyme. Rather, the data suggest that there is a strong correlation between terminal folding and proteolytic cleavage. Several studies have shown that following dimerization, gB undergoes a prolonged postoligomerization folding associated in part with posttranslational disulfide bond formation (2). This complex folding process can be monitored by using different conformation-specific antibodies. An epitope recognized by 27-39 is the current standard for a fully folded or terminally folded gB molecule. Our analysis revealed that all but one of the 27-39-negative mutants was cleavage defective. As shown in Fig. 7, the cleavage recognition site may reside in an exposed loop. Many of our insertions resulted in the incorporation of a proline residue (Table 1) which would introduce a strong bend into that particular region disrupting adjacent alpha-helical content. It is therefore likely that the ability of furin to either recognize, bind, or cleave at this site was eliminated by a change in the secondary structure of gB introduced by our insertions. In contrast, insertions immediately adjacent to the cleavage site (I-445 and I-484) were in regions devoid of extensive predicated secondary structure and had a silent phenotype.
FIG. 7.
Schematic showing gB secondary structural predictions and insertion mutants. A secondary structural map of gB was generated by using the Internet-based Protein Sequence Analysis System (http://bmerc-www.bu.edu/psa/index.htm). The analysis was done with the mem_span model and based on algorithms previously published (38, 39, 45). Only structures with a probability greater than 50% are shown. The positions of mutants that did not react with the oligomer/conformation-specific MAb 27-39 (Fig. 2) and those that were defective for proteolytic processing (Fig. 4) are indicated.
Insights into gB trafficking.
Quality control of proteins generally occurs within the ER. Oligomerization of gB along with accurate folding with the aid of cellular chaperones is critical for subsequent trafficking steps including cell surface expression (20, 46, 47). Transport along the exocytotic pathway is associated with disulfide bond formation and rearrangement as well as the ordered formation of at least two folding intermediates that can be distinguished with MAbs. Formation of the epitope recognized by 9-3 precedes folding into the form of gB recognized by 27-39 (2). Nearly all of the insertion mutant proteins were recognized by the conformational antibody 9-3, indicating that all of the modified proteins were capable of assuming this folding intermediate described for viral gB (2, 10). However, only 14 of 22 mutant proteins folded into the terminal form recognized by 27-39. We predicted that there would be a correlation between fully folded forms and cell surface delivery and thus expected that 27-39-defective mutant proteins would be impaired or halted during exocytosis. We were therefore surprised that all of the mutants were expressed on the cell surface. Thus, the 27-39 epitope does not appear to be an essential marker for trafficking. A possible hypothesis for this unexpected result is that during the prolonged folding process of gB, the molecule becomes transport competent at or near the time that the 9-3 epitope is formed, but this intermediate may not be efficiently exported to the Golgi complex unless the folded form recognized by MAb 27-39 is present. A partially folded molecule lacking the 27-39 epitope but containing the 9-3 epitope would still be capable of reaching the cell surface but at significantly slower rates. Our current experiments did not measure delivery rates or the efficiency of plasma membrane localization; therefore, it is possible that folding-defective mutant proteins are slowed or poorly efficient in cell surface delivery. We did note overall low steady-state levels (less than 5% of the total intracellular pool) on the surface of cells. Low levels of steady-state surface expression have been observed in other recombinant gB expression systems and may indicate that other HCMV viral products are necessary to achieve high levels of steady-state gB on the surface of infected cells (8, 9). Recent work has shown that the phosphorylation status of aa Ser 900 is a crucial determinant of steady-state cell surface expression of gB (18). Other viral products may influence the phosphorylation state of Ser 900 and may contribute to the high levels of cell surface gB seen in an infected cell. It is interesting that two insertion mutants within the cytoplasmic tail of gB (I-832 and I-885) were expressed on the cell surface. The cytoplasmic portion of gB has also been shown to be important in vectoral targeting in polarized cells (41). It would be interesting to examine the expression of the two cytoplasmic insertion mutants (I-832 and I-885) in polarized cells and see if altered cell surface expression occurs.
gB has at least four distinct functional activities. (i) We have shown that gB has the capacity to bind cell surface HSPGs and to a second, heparin-independent binding site (3, 11). Binding of gB to its receptor is physiologically relevant since virus entry was blocked when gB binding sites were occupied (3, 11). (ii) A cellular consequence of HCMV virion binding is the induction of a intracellular signaling cascade that upregulates genes in the interferon-responsive pathway (4, 8, 49). Recent evidence from our laboratory has shown that the HCMV-mediated signaling is largely attributable to the interaction of gB with its receptor (4). (iii) gB is also necessary for fusion (26, 42, 43). Thus, ligand binding, signal transduction, and fusion may all be regulated at the level of receptor engagement. (iv) HCMV gB also forms a stable interaction with cellular annexin II (31). At present we favor a role of this interaction in virus maturation and egress (30). Clearly a collection of mutants will be extremely useful in creating a functional map for this essential and multifunctional protein.
In summary, we have created a collection of mutants that span the gB gene and characterized the mutant proteins with respect to biosynthetic hallmarks and intracellular trafficking. Our studies illuminate the complexity of gB structural organization. We discovered that there is a tight correlation between folding and proteolytic cleavage of gB, but that these defects do not necessarily impair intracellular sorting and trafficking. Given the prolonged folding pathway and the implications for disulfide bond rearrangement, our current work is aimed at solving the disulfide bond configuration by peptide mapping techniques. In addition, we are endeavoring to solve the structure of gB at high resolution. Such information will be critical to understanding the functional mapping in a structural context and may ultimately be useful in the design of compounds that would interfere with the activity of this essential mediator of CMV entry.
ACKNOWLEDGMENTS
This study was supported by Public Health Service grant RO1 AI-34998 and a Basic Research grant from the March of Dimes Birth Defects Foundation. J.S. was supported in part by National Cancer Institute grant 2-T32-CA09075-21.
We thank Mark Stinski and William J. Britt for gifts of MAbs. We thank Meera Prasanna for help in screening for the mutants, Matthew Lopper for assistance in generating the structural prediction plot, and members of the Compton lab for critical reading of the manuscript.
REFERENCES
- 1.Alford C A, Britt W J. Cytomegalovirus. In: Fields B N, Knipe D M, Chanock R M, Hirsch M S, Melnick J L, Monath T P, Roizman B, editors. Virology. New York, N.Y: Raven Press; 1990. pp. 1981–2010. [Google Scholar]
- 2.Billstrom M A, Britt W J. Postoligomerization folding of human cytomegalovirus glycoprotein B: identification of folding intermediates and importance of disulfide bonding. J Virol. 1995;69:7015–7022. doi: 10.1128/jvi.69.11.7015-7022.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyle K A, Compton T. Receptor-binding properties of a soluble form of human cytomegalovirus glycoprotein B. J Virol. 1998;72:1826–1833. doi: 10.1128/jvi.72.3.1826-1833.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyle K A, Pietropaolo R L, Compton T. Engagement of the cellular receptor for glycoprotein B of human cytomegalovirus activates the interferon-responsive pathway. Mol Cell Biol. 1999;19:3607–3613. doi: 10.1128/mcb.19.5.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Britt W J. Neutralizing antibodies detect a disulfide-linked glycoprotein complex within the envelope of human cytomegalovirus. Virology. 1984;135:369–378. doi: 10.1016/0042-6822(84)90193-4. [DOI] [PubMed] [Google Scholar]
- 6.Britt W J, Auger D. Synthesis and processing of the envelope gp55-116 complex of human cytomegalovirus. J Virol. 1986;58:185–191. doi: 10.1128/jvi.58.1.185-191.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Britt W J, Mach M. Human cytomegalovirus glycoproteins. Intervirology. 1996;39:401–412. doi: 10.1159/000150510. [DOI] [PubMed] [Google Scholar]
- 8.Britt W J, Vugler L, Butfiloski E J, Stephens E B. Cell surface expression of human cytomegalovirus (HCMV) gp55-116 (gB): use of HCMV-recombinant vaccinia virus-infected cells in analysis of the human neutralizing antibody response. J Virol. 1990;64:1079–1085. doi: 10.1128/jvi.64.3.1079-1085.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Britt W J, Vugler L, Stephens E B. Induction of complement-dependent and -independent neutralizing antibodies by recombinant-derived human cytomegalovirus gp55-116 (gB) J Virol. 1988;62:3309–3318. doi: 10.1128/jvi.62.9.3309-3318.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Britt W J, Vugler L G. Oligomerization of the human cytomegalovirus major envelope glycoprotein complex gB (gp55-116) J Virol. 1992;66:6747–6754. doi: 10.1128/jvi.66.11.6747-6754.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlson C, Britt W J, Compton T. Expression, purification, and characterization of a soluble form of human cytomegalovirus glycoprotein B. Virology. 1997;239:198–205. doi: 10.1006/viro.1997.8892. [DOI] [PubMed] [Google Scholar]
- 12.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchinson III C A, Kouzarides T, Martignetti J A, et al. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 13.Compton T. Towards a definition of the HCMV entry pathway. Scand J Infect Dis Suppl. 1995;99:30–32. [PubMed] [Google Scholar]
- 14.Compton T, Nepomuceno R R, Nowlin D M. Human cytomegalovirus penetrates host cells by pH-independent fusion at the cell surface. Virology. 1992;191:387–395. doi: 10.1016/0042-6822(92)90200-9. [DOI] [PubMed] [Google Scholar]
- 15.Compton T, Nowlin D M, Cooper N R. Initiation of human cytomegalovirus infection requires initial interaction with cell surface heparan sulfate. Virology. 1993;193:834–841. doi: 10.1006/viro.1993.1192. [DOI] [PubMed] [Google Scholar]
- 16.Cranage M P, Kouzarides T, Bankier A T, Satchwell S, Weston K, Tomlinson P, Barrell B, Hart H, Bell S E, Minson A C, et al. Identification of the human cytomegalovirus glycoprotein B gene and induction of neutralizing antibodies via its expression in recombinant vaccinia virus. EMBO J. 1986;5:3057–3063. doi: 10.1002/j.1460-2075.1986.tb04606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DuBridge R B, Tang P, Hsia H C, Leong P M, Miller J H, Calos M P. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol Cell Biol. 1987;7:379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fish K N, Soderberg-Naucler C, Nelson J A. Steady-state plasma membrane expression of human cytomegalovirus gB is determined by the phosphorylation state of Ser900. J Virol. 1998;72:6657–6664. doi: 10.1128/jvi.72.8.6657-6664.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gretch D R, Gehrz R C, Stinski M F. Characterization of a human cytomegalovirus glycoprotein complex (gcl) J Gen Virol. 1988;69:1205–1215. doi: 10.1099/0022-1317-69-6-1205. [DOI] [PubMed] [Google Scholar]
- 20.Hurtley S M, Helenius A. Protein oligomerization in the endoplasmic reticulum. Annu Rev Cell Biol. 1989;5:277–307. doi: 10.1146/annurev.cb.05.110189.001425. [DOI] [PubMed] [Google Scholar]
- 21.Kari B, Gehrz R. A human cytomegalovirus glycoprotein complex designated gC-II is a major heparin-binding component of the envelope. J Virol. 1992;66:1761–1764. doi: 10.1128/jvi.66.3.1761-1764.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kari B, Liu Y N, Goertz R, Lussenhop N, Stinski M F, Gehrz R. Structure and composition of a family of human cytomegalovirus glycoprotein complexes designated gC-I (gB) J Gen Virol. 1990;71:2673–2680. doi: 10.1099/0022-1317-71-11-2673. [DOI] [PubMed] [Google Scholar]
- 23.Laquerre S, Person S, Glorioso J C. Glycoprotein B of herpes simplex virus type 1 oligomerizes through the intermolecular interaction of a 28-amino-acid domain. J Virol. 1996;70:1640–1650. doi: 10.1128/jvi.70.3.1640-1650.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y N, Klaus A, Kari B, Stinski M F, Eckhardt J, Gehrz R C. The N-terminal 513 amino acids of the envelope glycoprotein gB of human cytomegalovirus stimulates both B- and T-cell immune responses in humans. J Virol. 1991;65:1644–1648. doi: 10.1128/jvi.65.3.1644-1648.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molloy S S, Thomas L, VanSlyke J K, Stenberg P E, Thomas G. Intracellular trafficking and activation of the furin proprotein convertase: localization to the TGN and recycling from the cell surface. EMBO J. 1994;13:18–33. doi: 10.1002/j.1460-2075.1994.tb06231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Navarro D, Paz P, Tugizov S, Topp K, La Vail J, Pereira L. Glycoprotein B of human cytomegalovirus promotes virion penetration into cells, transmission of infection from cell to cell, and fusion of infected cells. Virology. 1993;197:143–158. doi: 10.1006/viro.1993.1575. [DOI] [PubMed] [Google Scholar]
- 27.Neyts J, Snoeck R, Schols D, Balzarini J, Esko J D, Van Schepdael A, De Clercq E. Sulfated polymers inhibit the interaction of human cytomegalovirus with cell surface heparan sulfate. Virology. 1992;189:48–58. doi: 10.1016/0042-6822(92)90680-n. [DOI] [PubMed] [Google Scholar]
- 28.Pereira L. Function of glycoprotein B homologues of the family herpesviridae. Infect Agents Dis. 1994;3:9–28. [PubMed] [Google Scholar]
- 29.Pereira L, Hoffman M, Gallo D, Cremer N. Monoclonal antibodies to human cytomegalovirus: three surface membrane proteins with unique immunological and electrophoretic properties specify cross-reactive determinants. Infect Immun. 1982;36:924–932. doi: 10.1128/iai.36.3.924-932.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pietropaolo R, Compton T. Interference with annexin II has no effect on entry of human cytomegalovirus into fibroblast cells. J Gen Virol. 1999;80:1807–1816. doi: 10.1099/0022-1317-80-7-1807. [DOI] [PubMed] [Google Scholar]
- 31.Pietropaolo R L, Compton T. Direct interaction between human cytomegalovirus glycoprotein B and cellular annexin II. J Virol. 1997;71:9803–9807. doi: 10.1128/jvi.71.12.9803-9807.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radsak K, Eickmann M, Mockenhaupt T, Bogner E, Kern H, Eis-Hubinger A, Reschke M. Retrieval of human cytomegalovirus glycoprotein B from the infected cell surface for virus envelopment. Arch Virol. 1996;141:557–572. doi: 10.1007/BF01718317. [DOI] [PubMed] [Google Scholar]
- 33.Reschke M, Reis B, Noding K, Rohsiepe D, Richter A, Mockenhaupt T, Garten W, Radsak K. Constitutive expression of human cytomegalovirus glycoprotein B (gpUL55) with mutagenized carboxy-terminal hydrophobic domains. J Gen Virol. 1995;76:113–122. doi: 10.1099/0022-1317-76-1-113. [DOI] [PubMed] [Google Scholar]
- 34.Ruger R, Bornkamm G W, Fleckenstein B. Human cytomegalovirus DNA sequences with homologies to the cellular genome. J Gen Virol. 1984;65:1351–1364. doi: 10.1099/0022-1317-65-8-1351. [DOI] [PubMed] [Google Scholar]
- 35.Seger R, Krebs E G. The MAPK signaling cascade. FASEB J. 1995;9:726–735. [PubMed] [Google Scholar]
- 36.Spaete R R, Saxena A, Scott P I, Song G J, Probert W S, Britt W J, Gibson W, Rasmussen L, Pachl C. Sequence requirements for proteolytic processing of glycoprotein B of human cytomegalovirus strain Towne. J Virol. 1990;64:2922–2931. doi: 10.1128/jvi.64.6.2922-2931.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spaete R R, Thayer R M, Probert W S, Masiarz F R, Chamberlain S H, Rasmussen L, Merigan T C, Pachl C. Human cytomegalovirus strain Towne glycoprotein B is processed by proteolytic cleavage. Virology. 1988;167:207–225. doi: 10.1016/0042-6822(88)90071-2. [DOI] [PubMed] [Google Scholar]
- 38.Stultz C M, Nambudripad R, Lathrop R H, White J V. Predicting protein structure with probabilistic models. In: Bittar E E, editor. Advances in molecular and cell biology. 22B. Protein structural biology in biomedical research. Greenwich, Conn: JAI Press; 1997. pp. 447–506. [Google Scholar]
- 39.Stultz C M, White J V, Smith T F. Structural analysis based on state-space modeling. Protein Sci. 1993;2:305–314. doi: 10.1002/pro.5560020302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sugden P H, Clerk A. Regulation of the ERK subgroup of MAP kinase cascades through G protein-coupled receptors. Cell Signalling. 1997;9:337–351. doi: 10.1016/s0898-6568(96)00191-x. [DOI] [PubMed] [Google Scholar]
- 41.Tugizov S, Maidji E, Xiao J, Zheng Z, Pereira L. Human cytomegalovirus glycoprotein B contains autonomous determinants for vectorial targeting to apical membranes of polarized epithelial cells. J Virol. 1998;72:7374–7386. doi: 10.1128/jvi.72.9.7374-7386.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tugizov S, Navarro D, Paz P, Wang Y, Qadri I, Pereira L. Function of human cytomegalovirus glycoprotein B: syncytium formation in cells constitutively expressing gB is blocked by virus-neutralizing antibodies. Virology. 1994;201:263–276. doi: 10.1006/viro.1994.1291. [DOI] [PubMed] [Google Scholar]
- 43.Tugizov S, Wang Y, Qadri I, Navarro D, Maidji E, Pereira L. Mutated forms of human cytomegalovirus glycoprotein B are impaired in inducing syncytium formation. Virology. 1995;209:580–591. doi: 10.1006/viro.1995.1290. [DOI] [PubMed] [Google Scholar]
- 44.Vey M, Schafer W, Reis B, Ohuchi R, Britt W, Garten W, Klenk H D, Radsak K. Proteolytic processing of human cytomegalovirus glycoprotein B (gpUL55) is mediated by the human endoprotease furin. Virology. 1995;206:746–749. doi: 10.1016/s0042-6822(95)80002-6. [DOI] [PubMed] [Google Scholar]
- 45.White J V, Stultz C M, Smith T F. Protein classification by stochastic modeling and optimal filtering of amino-acid sequences. Math Biosci. 1994;119:35–75. doi: 10.1016/0025-5564(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 46.Yamashita Y, Shimokata K, Mizuno S, Daikoku T, Tsurumi T, Nishiyama Y. Calnexin acts as a molecular chaperone during the folding of glycoprotein B of human cytomegalovirus. J Virol. 1996;70:2237–2246. doi: 10.1128/jvi.70.4.2237-2246.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng Z, Maidji E, Tugizov S, Pereira L. Mutations in the carboxyl-terminal hydrophobic sequence of human cytomegalovirus glycoprotein B alter transport and protein chaperone binding. J Virol. 1996;70:8029–8040. doi: 10.1128/jvi.70.11.8029-8040.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu H, Cong J P, Mamtora G, Gingeras T, Shenk T. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:14470–14475. doi: 10.1073/pnas.95.24.14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu H, Cong J P, Shenk T. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc Natl Acad Sci USA. 1997;94:13985–13990. doi: 10.1073/pnas.94.25.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]