SUMMARY
Alveolar epithelial type I cells (AT1s) line the gas exchange barrier of the distal lung and have been historically challenging to isolate or maintain in cell culture. Here, we engineer a human in vitro AT1 model system via directed differentiation of induced pluripotent stem cells (iPSCs). We use primary adult AT1 global transcriptomes to suggest benchmarks and pathways, such as Hippo-LATS-YAP/TAZ signaling, enriched in these cells. Next, we generate iPSC-derived alveolar epithelial type II cells (AT2s) and find that nuclear YAP signaling is sufficient to promote a broad transcriptomic shift from AT2 to AT1 gene programs. The resulting cells express a molecular, morphologic, and functional phenotype reminiscent of human AT1 cells, including the capacity to form a flat epithelial barrier producing characteristic extracellular matrix molecules and secreted ligands. Our results provide an in vitro model of human alveolar epithelial differentiation and a potential source of human AT1s.
In brief
Kotton and colleagues generate human alveolar epithelial type I cells (AT1s) from induced pluripotent stem cells (iPSCs). The resulting cells can be grown as 3D organoids or in 2D air-liquid interface cultures, displaying many of the molecular, morphologic, and functional phenotypes of primary AT1s.
Graphical abstract
INTRODUCTION
The alveolar epithelium of the lung is vital for gas exchange and consists of two cell types, alveolar epithelial type I cells (AT1s) and type II cells (AT2s). AT2s are cuboidal and produce surfactant, while AT1s cover 95% of the alveolar surface area and are uniquely flattened to allow for the diffusion of oxygen into the capillaries.1 Despite their critical role comprising the majority of the lung’s air-facing surface and recent revelations of the role of AT1 differentiation in fibrotic lung disease,2–4 relatively little is known about the cell biology, origins, and fates of human AT1s.
AT1s are generally assumed to be replenished by adjacent AT2s, based on literature beginning in the 1970s that employed rodent models both in vivo5,6 and after culturing AT2s in vitro.3,4,7,8 More recently, lineage tracing in mice has shown that AT1s can be derived from mature AT2s after injury and during normal homeostasis.7,9,10 The exact signaling mechanisms driving AT1 differentiation are still uncertain, although murine studies have implicated a wide variety of classical pathways, including Wnt, BMP, TGFβ, FGF, and YAP/TAZ signaling.11–18 Additionally, mouse studies have shown that nuclear YAP/TAZ localization is necessary for the maintenance of the AT1 program and that loss of YAP/TAZ in mature AT1s leads to reversion back into an AT2-like cell fate.19,20
The origin and differentiation of human AT1s, however, have been less clear and more difficult to study. Reports involving in vitro 2D culture of primary fetal or adult human AT2s have documented “transdifferentiation” into AT1s expressing markers of unclear specificity, leading to the paradigm that AT1s are a “default state” of AT2s cultured in undefined (e.g., serum containing) conditions21–24; however, 3D models of AT2s have observed little if any evidence of genuine AT1s emerging from either primary human AT2 cells co-cultured with7 or without fibroblasts or from human induced pluripotent stem cell (iPSC)-derived AT2s (iAT2s), even after prolonged time in most culture conditions tested to date,25–27 with three notable exceptions.28–31
Understanding the differentiation of human AT1s is particularly important since damage to AT1s in response to toxic inhalational exposures, radiation, or infection can lead to respiratory failure and severe diseases such as acute respiratory distress syndrome (ARDS).32 Impaired differentiation of AT1s has also been implicated in fibrotic lung diseases.2–4,33 Hence, understanding normal differentiation could provide insights for resolving the aberrant transitional state found in fibrotic lung tissue. Additionally, engineering mice carrying fluorochrome reporters under the regulatory control of AT1 gene promoters has been invaluable in identifying, tracing, and isolating mouse AT1s during development and disease34–36; however, to date, no comparable human reporter has yet been generated to facilitate the study of human AT1s.
Here, we report the in vitro generation of cells expressing the molecular and functional phenotypes of human AT1s via differentiation of iAT2s. We first profile human lung explant tissues at single-cell resolution using RNA sequencing to identify potential AT1-selective marker gene sets and AT1-enriched signaling pathways. We engineer an iPSC line carrying a tdTomato reporter targeted to the endogenous AGER locus, which is specifically upregulated in primary AT1s according to our gene set. The AGERtdTomato reporter enables real-time tracking, quantitation, and purification of the resulting putative iPSC-derived AT1s (iAT1s). We find activated nuclear YAP expression is sufficient to drive the transcriptomic shift from iAT2 to iAT1 programs in a cell-autonomous manner. Further, we have developed a defined, serum-free differentiation medium containing a LATS inhibitor that recapitulates the above process, leading to robust and efficient differentiation of iAT2s into iAT1s.
RESULTS
scRNA-seq defines the transcriptomic program of primary human alveolar epithelial type I cells
To understand the transcriptomic programs of primary human AT1s, we performed profiling by single-cell RNA sequencing (scRNA-seq) of 5 distal human lung explant tissues (partial dataset published previously in Basil et al.), including 1.401 AT1s (Figure S1A).37 Selecting and reclustering epithelial cells (uniform manifold approximation and projection [UMAP]; Figure 1A), we visualized expression of previously reported putative AT1 markers and canonical AT2 marker genes (Figure 1B). We found some human AT1 markers (AGER, CAV1, PDPN, and RTKN2) shared similar qualitative clustered expression patterns to published mouse datasets38,39; however, well-characterized mouse markers, HOPX, AQP5, and IGFBP2,7,34,40 were expressed in other epithelial cells, such as AT2s for HOPX and airway cells for AQP5 and IGFBP2 (Figures 1B–1D and S1B).
Figure 1. Transcriptomic profiling at single-cell resolution of primary adult human AT1s.
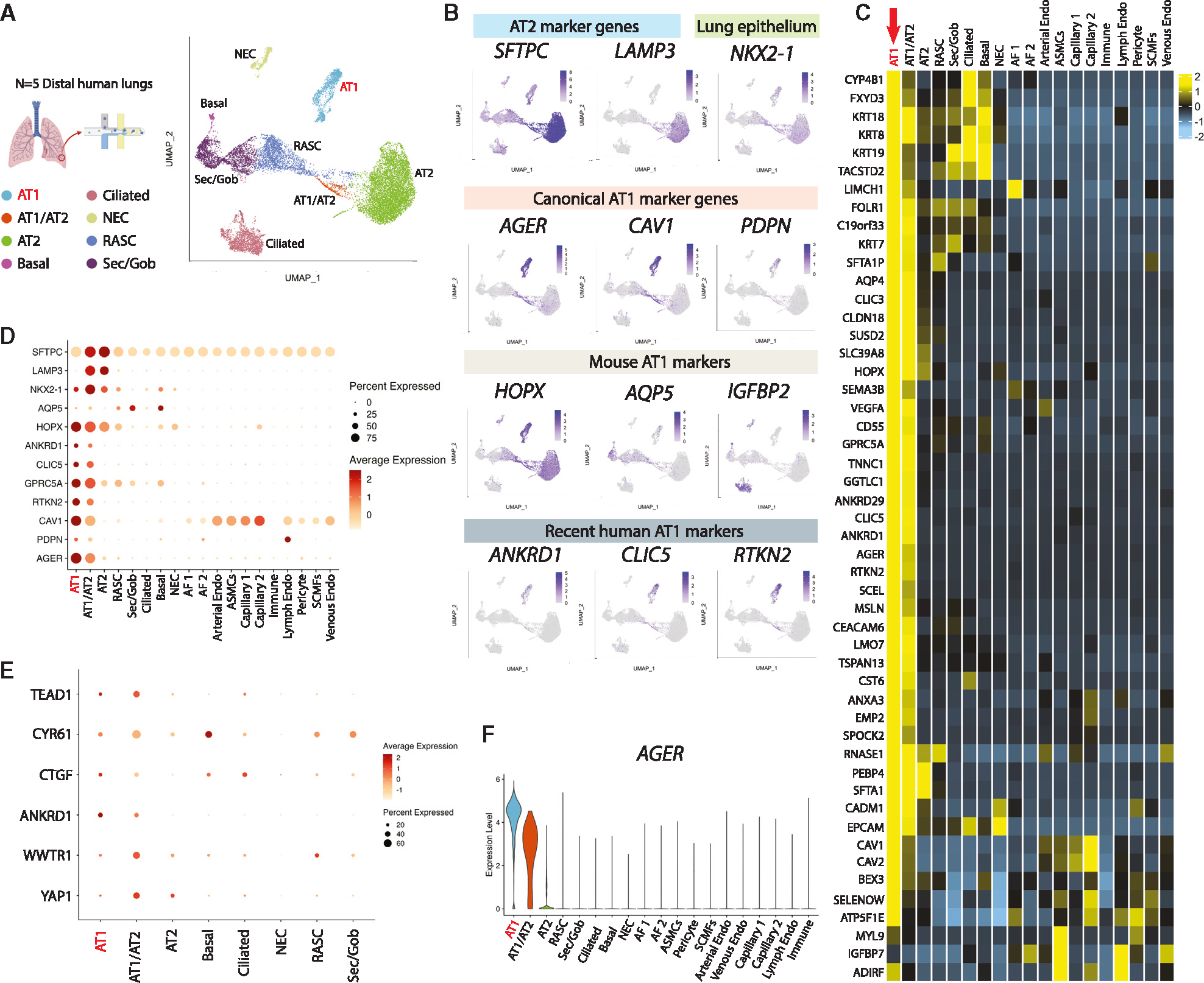
(A) UMAP of an integrated analysis of 15,769 primary distal lung epithelial cells (from Basil et al., N = 5, including 1,401 AT1s and 7,039 AT2s).37
(B) Expression of indicated AT2 marker genes; lung epithelial marker NKX2–1; canonical AT1 marker genes; mouse-specific AT1 marker genes; and more recent human AT1 marker genes.
(C) Heatmap showing average expression for each cell type of top differentially upregulated human AT1 genes compared with all distal lung cells.
(D and E) (D) Expression of selected AT1 and AT2 marker genes and (E) Hippo signaling-related genes across all lung cell types.
(F) AGER expression across indicated cell types. See also Figure S1 and Table S1.
To generate unbiased human AT1 marker 50-gene sets, we identified differentially expressed genes (DEGs) enriched in AT1s using three pairwise comparisons: (1) AT1s vs. all lung cells, (2) AT1s vs. lung epithelial cells, and (3) AT1s vs. AT2s (Figure 1C; Table S1). AGER was the top transcript significantly enriched in each of the 3 comparisons (ranked by logFC), with multiple caveolin (CAV1 and CAV2) and chloride intracellular channel (CLIC3 and CLIC5) gene family members present as well (Figure 1C; Table S1). PDPN, although not in the top 50 DEGs, likely due to its expression in lymphatic endothelium and basal cells, was still selected as an informative AT1 marker as it was significantly upregulated in all comparisons and has been extensively published as a canonical AT1 marker for both mice and humans.38,39,41–43 To confirm the utility of the above markers, we compared their expression levels, frequencies, and relative specificities across all human lung epithelia (Figures 1D–1F and S1B). Taken together, these analyses suggested AGER, CAV1, CLIC5, and PDPN as an informative 4-gene human AT1 marker set, in addition to the more extended 50-gene marker sets (Table S1), each able to reliably distinguish human AT1s.
We noted the canonical YAP/TAZ target gene, ANKRD1, was highly enriched in AT1s in each of our comparisons (Table S1), a finding consistent with recent publications suggesting activated nuclear YAP/TAZ is important for the differentiation and maintenance of the mouse AT1 program.19,44–47 Other downstream targets of YAP/TAZ, including CTGF, CYR61, and TEAD1, were also highly expressed in AT1s and transitioning AT1/AT2s (Figure 1E). However, YAP1 and TAZ (WWTR1) mRNAs were expressed broadly (Figure 1E), consistent with YAP/TAZ activity being primarily defined through post-translational regulation.48 Taken together, these analyses: (1) defined the transcriptomic programs of human AT1s, including multiple extended (50-gene) AT1 marker gene sets (Table S1), (2) validated a quartet of canonical human AT1 transcript markers (AGER, CAV1, CLIC5, and PDPN), and (3) suggested that nuclear YAP/TAZ activation distinguishes AT1s from AT2s in humans.
Nuclear localization of YAP leads to activation of the AT1 program in iAT2s
To examine the roles of nuclear YAP independent of upstream Hippo pathway inputs, we engineered a lentiviral vector encoding a constitutively active nuclear YAP cassette, YAP5SA, which cannot be phosphorylated by LATS kinases48 (Figure 2A). We also generated a control lentivirus encoding constitutively overexpressed wild-type (WT) YAP (Figure S2A), which is predicted to be degraded with minimal nuclear translocation.48 We differentiated human iPSCs carrying a SFTPCtdTomato reporter (clone SPC2-ST-B2)49 into iAT2s using our previously established protocol,25,50 and transduced the resulting iAT2s (>95% SFTPCtdTomato+) with either lentiviral YAP5SA, WT YAP, or no vector control (Mock) (Figure 2B). After 12 days of outgrowth in 3D cultures maintained in our published iAT2 medium,25 we observed significant upregulation of YAP targets, ANKRD1 and CYR61, in the YAP5SA-transduced samples (Figure 2C). Although both WT YAP and YAP5SA overexpression resulted in a significant decrease in the mature AT2 markers SFTPC and NAPSA, only YAP5SA led to a significant increase in the 4-gene AT1 marker set, AGER, CAV1, PDPN, and CLIC5 (Figure 2C). Although lung epithelial marker NKX2–1 was slightly decreased after YAP5SA transduction, we observed no evidence of significant induction of mesenchymal (SNAIL1 and TWIST) or airway (P63, SCGB3A2) markers after YAP5SA lentiviral transduction (Figure S2B). These results suggest that nuclear YAP activity in iAT2s promotes loss of the AT2 program and gain of AT1 markers.
Figure 2. iAT2s upregulate AT1 marker genes in response to activated nuclear YAP.
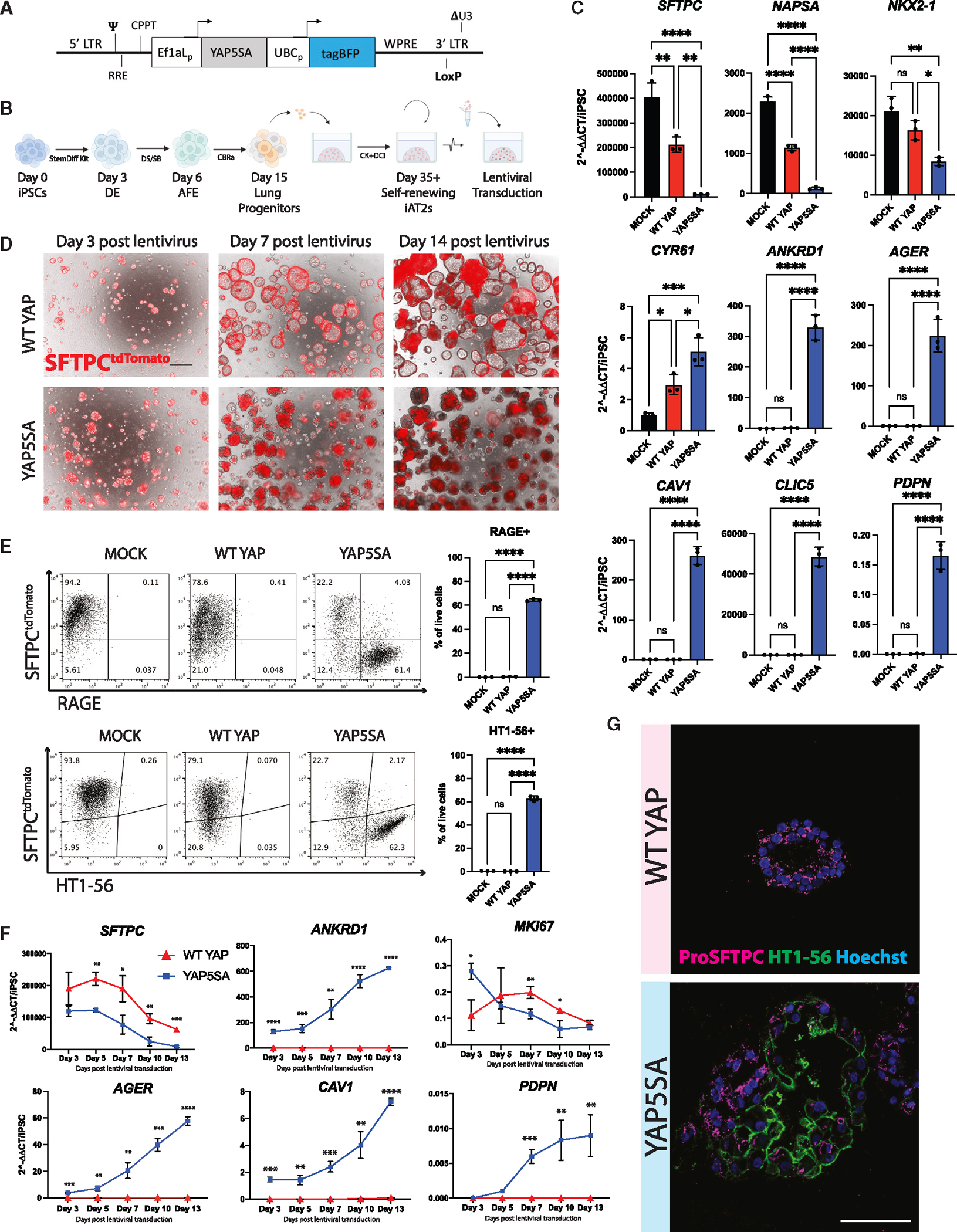
(A) Lentiviral vector encoding dual promoters driving activated nuclear YAP (YAP5SA) and a tagBFP reporter.
(B) Directed differentiation protocol for producing and lentivirally transducing iAT2s (clone SPC2-ST-B2) as previously published.50 (“StemDiff,” definitive endoderm kit; “DE,” definitive endoderm; “AFE,” anterior foregut endoderm; “DS/SB,” dorsomorphin and SB431542; CBRa, Chir, BMP4, retinoic acid; CK+DCI, iAT2 medium as detailed in the STAR Methods).
(C) Expression of indicated genes by RT-qPCR relative to day 0 iPSCs in whole-well RNA extracts taken 14 days post YAP5SA, WT YAP, or Mock lentiviral transduction (N = 3 transductions, one-way ANOVA).
(D) Representative live cell imaging of iAT2s following transduction with either WT YAP or YAP5SA lentivirus (bright field/SFTPCtdTomato overlay, scale bars, 500 μm).
(E) Flow cytometry analysis of SFTPCtdTomato and either RAGE protein or AT1 marker HT1–56 (N = 3 wells per condition, Student’s t test).
(F) Gene expression by RT-qPCR over time following WT YAP vs. YAP5SA transduction of iAT2s, relative to day 0 iPSCs (N = 3 transductions).
(G) Immunofluorescence staining for ProSFTPC (magenta) and HT1–56 (green) (nuclei, blue; scale bars, 100 μm). *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.001, bars represent mean ± SD for all panels. iPSC clone = SPC2-ST-B2. See also Figure S2.
When visualized by live light microscopy, iAT2s transduced with WT YAP continued to grow normally as monolayered epithelial spheres with visible lumens and retained SFTPCtdTomato reporter expression, whereas YAP5SA-transduced cells rapidly lost SFTPCtdTomato (Figures 2D and 2E). By 3 days post-transduction, we observed an altered morphology with aggregated clumps of cells lacking visible lumens, which became more pronounced over time (Figure 2D). By 14 days post-transduction, cells in the YAP5SA well had lost SFTPCtdTomato and gained AT1 membrane proteins, HT1–56 and RAGE (Figure 2E). Further, 73.7% ± 4.5% of cells exposed to YAP5SA were double-positive for both AT1 markers (Figure S2C). Analysis of HT1–56 staining in SFTPCtdTomato+ or lentiviral BFP+ sub-gated cells revealed that HT1–56+ cells were largely lentiviral BFP+ and SFTPCtdTomato−, further suggesting that YAP5SA-transduced cells lose their AT2 program and turn on AT1-specific markers (Figure S2D).
To further study the kinetics of this hypothesized transcriptomic shift, we performed a time series gene expression analysis. By day 3 post-transduction, SFTPC expression was decreased in the YAP5SA-transduced well compared with WT YAP control and continued to decrease over time to a greater degree than the expected decline in WT YAP cells.51 By contrast, YAP5SA transduction resulted in significant upregulation of AT1 markers, AGER, CAV1, and PDPN, as well as YAP downstream target ANKRD1, which continued to increase over time with only PDPN seeming to plateau (Figure 2F).
Consistent with the known non-proliferative state of AT1s in vivo,16 decreased proliferation in the outgrowth of YAP5SA-transduced cells (and resultant drop-out of transduced cells with further passaging) was indicated by: (1) decreasing expression of MKI67 over time (Figure 2F); (2) serial passaging-associated loss of expression of AT1 markers and coincident return of SFTPC (Figure S2E); and (3) loss of YAP5SA-transduced cells after serial passaging using a competition co-culture assay of iAT2s transduced with WT YAP-BFP vs. YAP5SA-GFP lentiviruses (Figures S2A and S2F). Taken together, these results suggest competitive over-growth of residual iAT2s over time, with loss of the less proliferative YAP5SA-transduced AT1-like cells.
Emergence of the AT1 program in YAP5SA-transduced cells was also confirmed by immunofluorescence microscopy (HT1–56 and proSFTPC; Figure 2G). Whereas iAT2s exposed to the WT YAP lentivirus gave rise to epithelial spheres expressing punctate proSFTPC cytoplasmic protein with no detectable HTI-56 staining, iAT2s exposed to YAP5SA lentivirus gave rise to subsets of organoids that exclusively contained either membrane-bound HTI-56-positive cells or proSFTPC-positive cells, with other subsets of organoids containing a mixture of cells positive for either HTI-56 or proSFTPC (Figure 2G).
YAP5SA-induced iAT1 program profiled by scRNA-seq
To further evaluate the effect of nuclear YAP on the global transcriptomic programs of iAT2s, we repeated YAP5SA vs. WT YAP lentiviral transduction and performed profiling by scRNA-seq (SPC2-ST-B2 clone49; Figure 3A). We analyzed the resulting cells at 7 days post transduction, guided by our quantitative reverse-transcription PCR (RT-qPCR) (Figure 2F), and captured all live cells to allow examination of both transduced as well as non-transduced cells within the same well. We observed WT YAP-transduced control iAT2s clustered together and expressed SFTPC, regardless of whether the cells were transduced, as shown by lentiviral BFP expression (UMAP; Figures 3B, 3C, and S2G). The population of cells exposed to YAP5SA lentivirus contained some cells clustered with the WT YAP cells and some that clustered separately (Figure 3B), presumably resulting from heterogeneity in transduction as indicated by BFP reporter expression (Figure S2G). We analyzed expression of AT1 markers AGER, PDPN, and CLIC5 as well as our AT1 50 gene-set signature derived in Figure 1 (Figures 3C–3G and S2H–S2J; Table S1). We observed high AGER and AT1 gene expression exclusively in the new cluster made only from the YAP5SA-transduced cells. This upregulation of the AT1 program coincided with high expression of YAP downstream targets ANKRD1 and CYR61 (Figure 3D) and loss of expression of AT2 markers such as SFTPC, SFTPB, and NAPSA (Figures 3C–3G and S2H).
Figure 3. Nuclear YAP overexpression drives AT1 program in a cell-autonomous manner.
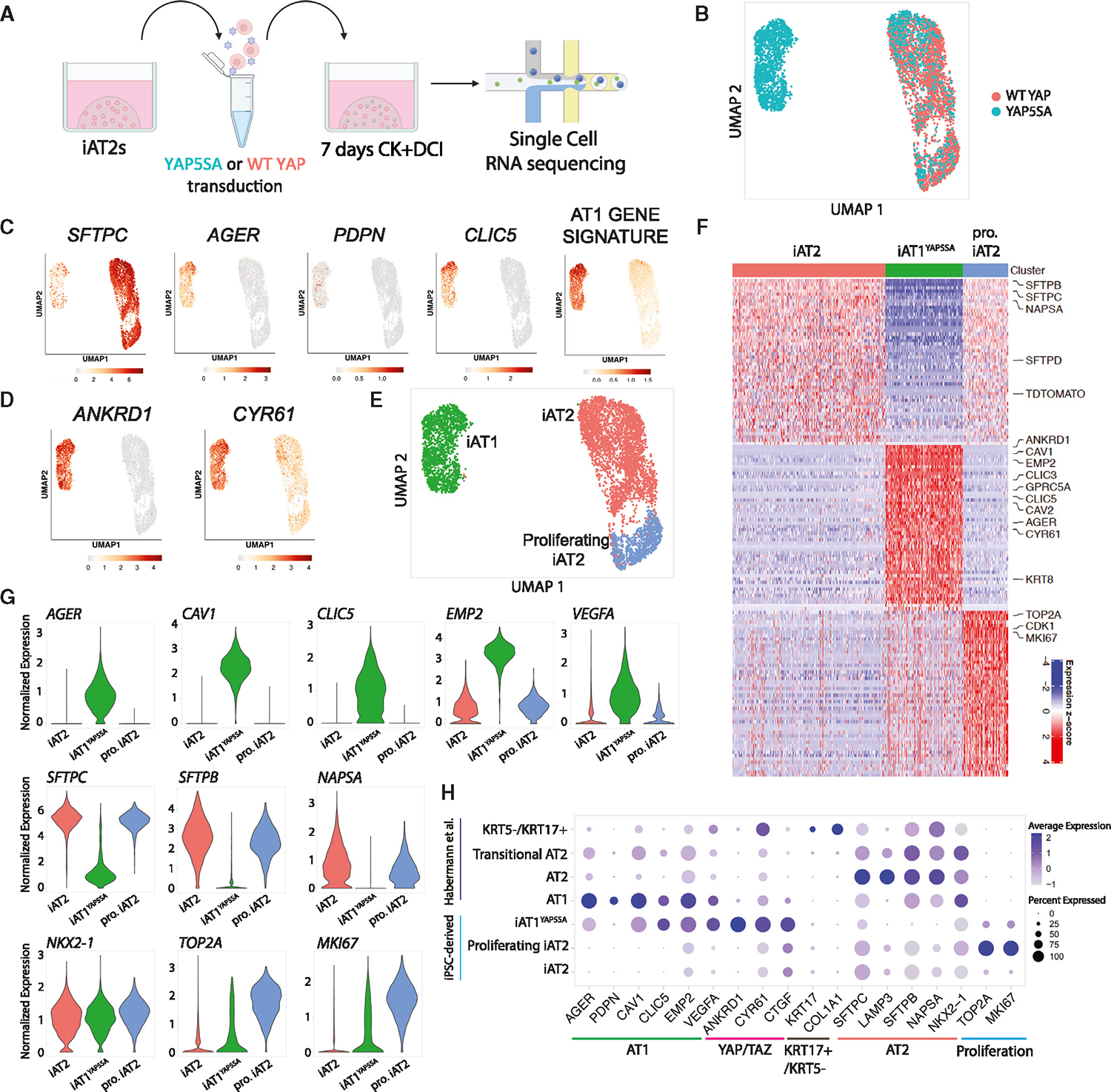
(A) scRNA-seq of iAT2s 7 days post WT YAP or YAP5SA lentiviral transduction (SPC2-ST-B2 clone).
(B) UMAP of WT YAP- and YAP5SA-exposed samples.
(C and D) (C) Gene expression of: select AT2 and AT1 markers and 50-gene AT1 signature (Figure 1; Table S1), and (D) select YAP downstream targets.
(E) Louvain clustering (res 0.05).
(F) Top 50 DEGs for clusters shown in (E).
(G) Gene expression of specific AT2, AT1, and proliferation markers across the clusters shown in (E) (“pro. iAT2,” proliferating iAT2s).
(H) Dot plot showing expression levels and frequencies of the indicated genes in iPSC-derived cells and scRNA-seq profiles of human adult primary populations previously published by Habermann et al.2 See also Figure S2 and Table S2.
We employed Louvain clustering to identify three distinct cell clusters (Figure 3E), annotated based on the top 50 enriched DEGs (Figures 3F and S2H; Table S2). Both iAT2s and proliferating iAT2s expressed high levels of AT2 markers SFTPC and NAPSA, whereas proliferating iAT2s were uniquely enriched in expression of proliferation markers MKI67 and TOP2A (Figures 3F and 3G). The YAP5SA-driven cluster’s (hereafter iAT1YAP5SA) top DEGs included YAP downstream targets, ANKRD1 and CYR61, as well as AT1 markers like AGER, CAV1, CAV2, CLIC3, CLIC5, VEGFA, and EMP2 (Figures 3F and 3G). Known markers for KRT5−/KRT17+ abberant basaloid cells2 were detectable but not enriched in either iAT2 or iAT1YAP5SA populations (Figure S2J). Similarly, we observed no significant differences in the Wnt target, AXIN2, or FGF receptors 1–3 in iAT1YAP5SSA cells and found no further iAT2 subpopulations or heterogeneity based on AXIN2 levels (Figure S2J). Markers of airway epithelia (SCGB3A2, FOXJ1, TP63) were expressed only rarely or at low levels in iAT2s (Figure S2J), with further diminished expression (Figure S2B) in YAP5SA-transduced cells, consistent with our previous studies of iAT2s.25,52
To provide an unbiased assessment of the lineage identity of iAT1YAP5SA cells based on the top DEGs (false discovery rate [FDR] < 0.05, logFC > 1; 92 genes), we employed the Tabula Sapiens gene set library,53 and found these cells ranked highest in similarity to “lung-type I pneumocyte” (Figure S2K). Additional comparisons to primary human lung scRNA-seq datasets published by Habermann et al.2 indicated similar expression frequencies of individual AT1 markers in iAT1YAP5SA cells (Figure 3H). However, iAT1YAP5SA cells had higher frequency of expression of YAP downstream targets, ANKRD1, CYR61, and CTGF, consistent with forced overexpression of activated nuclear YAP (Figure 3H). Taken together, our results suggest that overexpression of nuclear YAP drives a transcriptomic shift from iAT2 to iAT1-like cells in a cell-autonomous manner without evidence of paracrine effects on non-transduced cells.
Cre excision of YAP5SA lentivirus results in reversion to the iAT2 state
We next sought to determine whether iAT1YAP5SA cells stably maintain an AT1 program after removal of lentiviral YAP5SA. Recent publications indicate that YAP/TAZ deletion in murine AT1s leads to reversion to an AT2-like state.19,20 As the lentiviral YAP5SA vector was engineered to be floxed after genomic integration (Figures 2A and S2A), we employed adenoviral Cre-mediated vector excision (AdenoCre) after initial YAP5SA transduction (Figures S3A–S3E). AdenoCre-infected cells significantly augmented SFTPCtdTomato or SFTPC expression compared with YAP5SA cells that were unexposed to AdenoCre, without a statistically significant change in proliferation (EdU incorporation and cell counts). By contrast, YAP5SA-AdenoCre-treated cells decreased expression of YAP downstream targets CTGF and ANKRD1 and AT1 markers, AGER and CAV1, although not to WT YAP levels, suggesting only partial Cre excision. Together, these data suggest that iAT1YAP5SA cells do not exhibit a stable AT1 phenotype after loss of nuclear YAP overexpression when maintained in our medium that has been optimized for iAT2 maintenance.
Development of AGER reporter iPSCs for tracking iAT1s
We next sought to engineer a fluorescent reporter to identify, monitor, and purify human iAT1s. Due to its high expression in and specificity to AT1s in our human primary scRNA-seq dataset (Figure 1F), AGER was selected for reporter targeting. Our lab has previously published an NKX2–1GFP reporter iPSC line (BU3 NG; Hawkins et al.)54 for the visualization and purification of lung epithelial cells. Using CRISPR-Cas9 editing, we inserted a second fluorophore-encoding cassette, tdTomato, at the start codon of the endogenous AGER locus of this line (Figures 4A, S3F, and S3G), generating a karyotypically normal bifluorescent reporter iPSC line carrying NKX2–1GFP and AGERtdTomato, hereafter BU3 “NGAT.”
Figure 4. NKX2–1GFP;AGERtdTomato reporter iPSC line enables tracking and purification of iAT1YAP5SA cells.
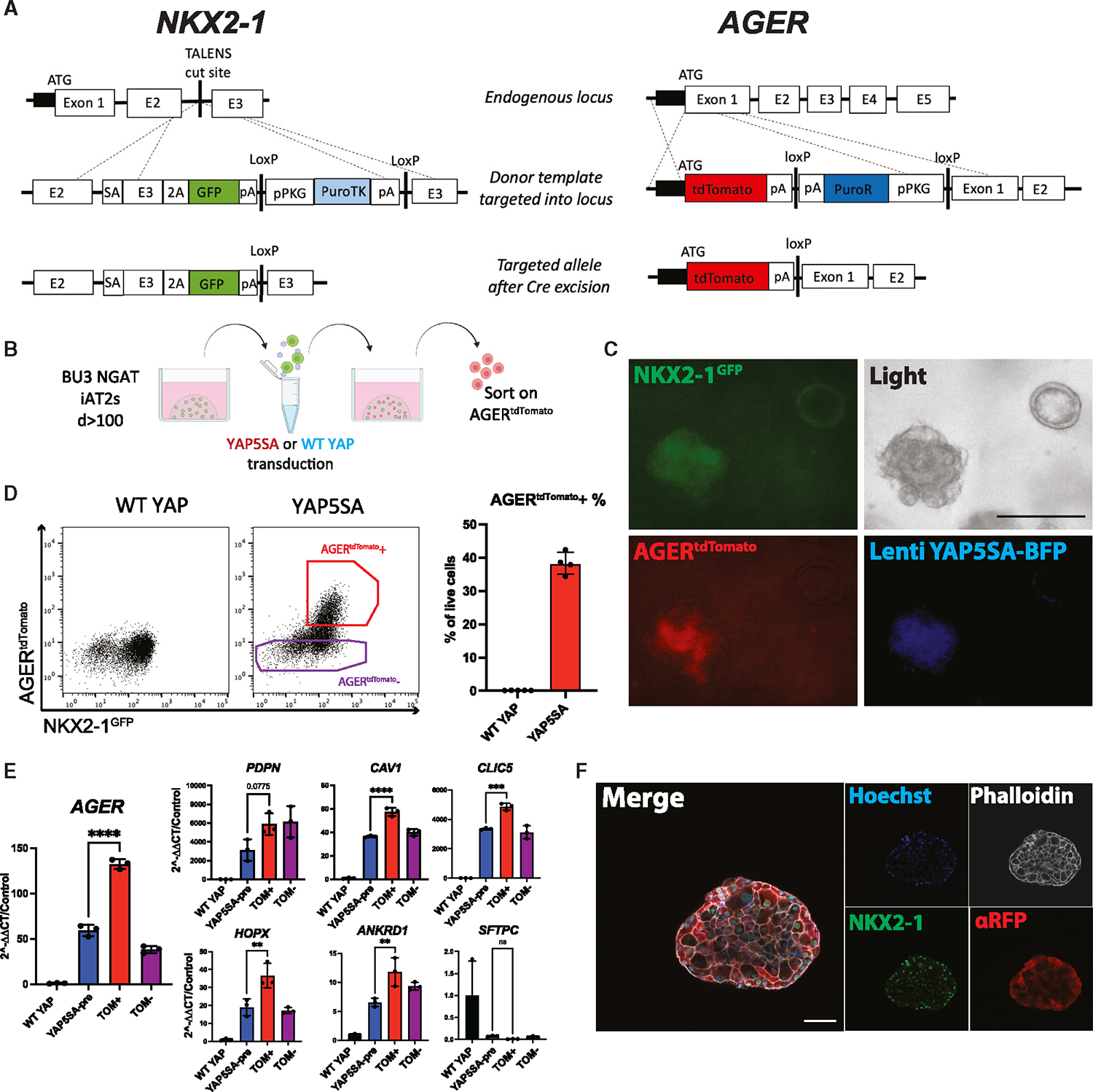
(A) Gene editing strategy to generate BU3 NKX2–1GFP;AGERtdTomato (NGAT) dual reporter iPSC line for tracking lung epithelial lineages and AT1-like cells (BU3 NKX2–1GFP reporter previously published by Hawkins et al.).54
(B) AGERtdTomato+ cells sorted and analyzed 14 days after transduction of iAT2s with lentiviral WT YAP or YAP5SA.
(C) Live cell fluorescence microscopy of YAP5SA-transduced cells growing next to an un-transduced epithelial sphere (GFP, tdTomato, and TagBFP fluorescence; scale bar, 200 μm).
(D) Flow cytometry of iAT2s (showing sorting gate for AGERtdTomato + and − cells with AGERtdTomato+ percentage quantified.
(E) Gene expression in sorted cells from (D). WT YAP, unsorted WT YAP transduced cells; YAP5SA-pre, unsorted YAP5SA transduced cells; TOM+,AGERtdTomato+ sorted cells; TOM−, AGERtdTomato−-sorted cells (N = 3 transductions, one-way ANOVA).
(F) Immunofluorescence staining of NKX2–1 protein and tdTomato (αRFP) (NKX2–1: green, F-actin: phalloidin white, tdTomato: red, nuclei: blue. Scale bars, 50 μm).
*p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.001, bars represent mean ± SD, BU3 NGAT iPSC line for all panels. See also Figure S3.
We characterized BU3 NGAT iPSCs after directed differentiation into distal lung epithelium followed by YAP5SA lentiviral transduction (Figure 4B). Cells transduced with the WT YAP lentivirus did not express tdTomato, while YAP5SA-transduced cells expressed AGERtdTomato specifically in the NKX2–1GFP+ population (Figures 4C and 4D). Additionally, the parental BU3 NG line, when transduced with YAP5SA, showed no tdTomato expression (Figure S3H). BU3 NGAT-derived AGERtdTomato+ cells were sorted and compared with YAP5SA-transduced unsorted (“presort”) cells, tdTomato−-sorted cells, or control unsorted WT YAP transduced samples (Figure 4E). AGERtdTomato+ cells were enriched in expression of AGER and other AT1 markers (Figure 4E), suggesting the utility of the reporter to track and purify iAT1s. Immunofluorescence microscopy further confirmed tdTomato protein expression was specific to cells co-expressing nuclear NKX2–1 protein, and the altered clumped organoid morphology observed in YAP5SA transduced SPC2-ST-B2 iAT2s (Figures 2A–2G) was recapitulated in the BU3 NGAT line (Figures 4C and 4F). Additionally, AGERtdTomato expression was lost over subsequent passaging of YAP5SA-transduced cells (Figure S3I), further suggesting that iAT1YAP5SA cells are outcompeted over passaging in our iAT2 medium (in agreement with Figures 2F, 2G, and S2D).
Serum-free medium-based induction of iAT1s
We next sought to identify factors that could be added to a defined medium to recapitulate iAT2 to iAT1 differentiation without lentivirus. Genetic mouse models of conditional deletion of the Hippo kinases, LATS1/2, in the developing lung epithelium have shown increased expression of mouse AT1 markers, suggesting a role for Hippo-LATS-YAP signaling in AT1 differentiation.45 Recently, the small molecule LATS inhibitor LATS-IN-1 or TRULI (hereafter “L”) has been shown to drive nuclear YAP in vitro.55 Thus, we tested the effect of L supplementation on iAT2s cultured with and without the iAT2 supportive growth factors CHIR99021 (“Chir,” or “C”) and KGF/FGF7 (“K”) in our base medium (“DCI”) (Figures 5A–5E). We found L induced rapid and robust changes including: (1) morphologic changes within 48 h, resembling YAP5SA-transduced cells, and (2) AGERtdTomato induction, detectable within 72 h and increasing over the following 2 weeks to 84.97% ± 1.23% of the NKX2–1GFP+ cells in L+DCI (Figures 5B–5E). AGERtdTomato expression was highest and most frequent in wells without Chir or KGF, suggesting inhibitory effects of these growth factors (Figures 5B, 5D, and 5E). There was little to no AGERtdTomato expression in cells exposed to DCI without L (Figures 5B–5E). To confirm the action of the LATS inhibitor in iAT2s, we performed western blots to examine LATS-mediated phosphorylation of YAP (phospho-Ser127),48 which demonstrated decreased ratios of phosphorylated to total YAP in cells grown in L+DCI media compared with iAT2s grown in CK+DCI (Figure S4A). Notably, L+DCI lysates also showed increased levels of RAGE protein (Figure S4A).
Figure 5. Serum-free medium-based induction of AGERtdTomato.
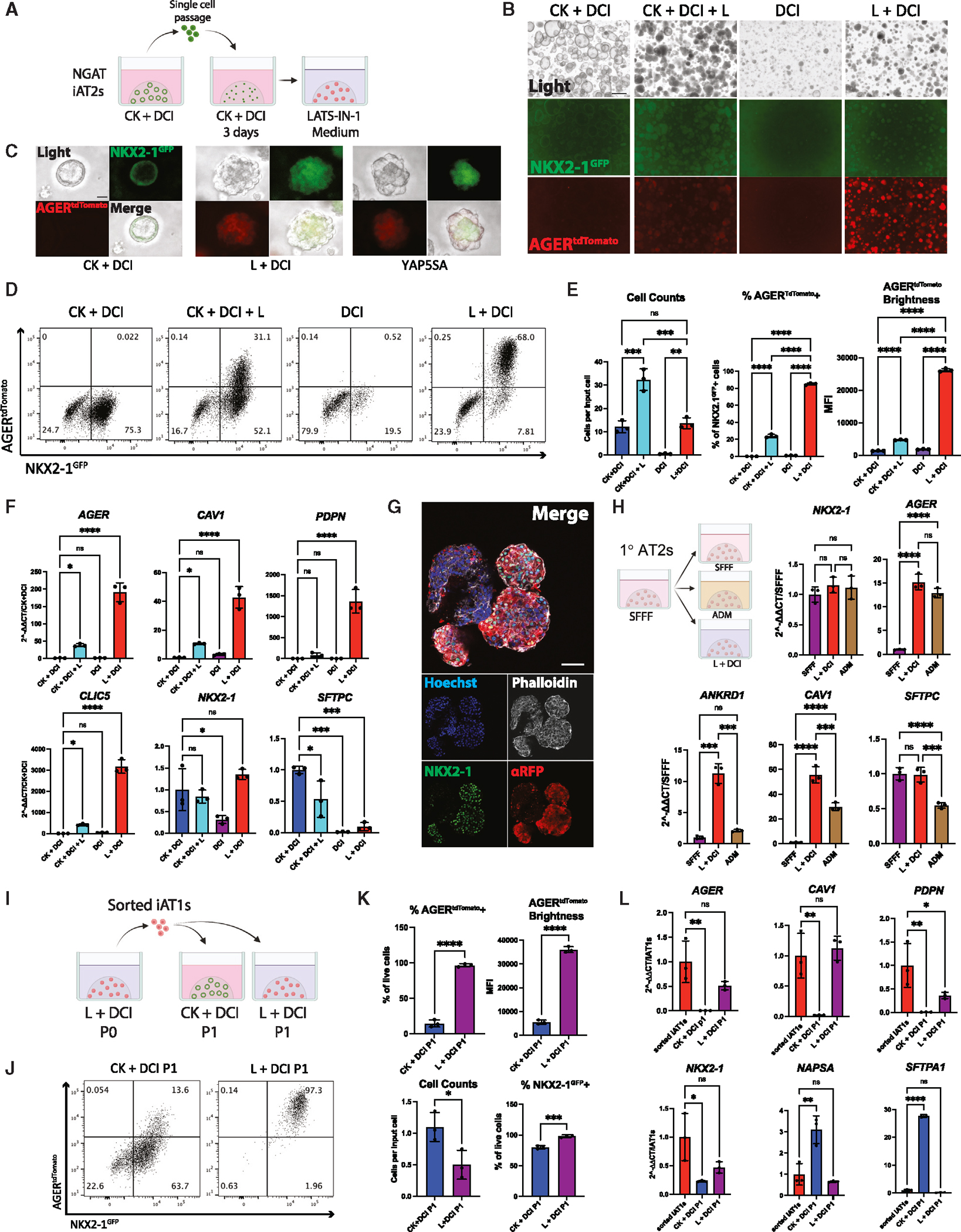
(A) iAT2s were passaged as usual into CK+DCI medium for 3 days. Medium was then kept the same or switched to LATS inhibitor-based media, CK+DCI+L, DCI, or L+DCI (C, Chir; K, rhKGF; L, LATS-IN-1; DCI as defined in Figure 2).
(B and C) (B) Live fluorescence microscopy of iAT2s at 12 days post passage, either without (B) or with (C) YAP5SA transduction comparison (scale bars, 500 μm in B; 50 μm in C).
(D) Representative flow cytometry of NKX2–1GFP and AGERtdTomato 9 days after changing to each indicated medium.
(E) Cell counts and quantification of AGERtdTomato+ percentage and MFI 9 days post medium change (N = 3 wells per condition, one-way ANOVA).
(F) Gene expression of AT1 and AT2 markers by whole-well RT-qPCR (N = 3 wells per condition, one-way ANOVA).
(G) Whole-mount immunofluorescence microscopy of organoids in L+DCI. (NKX2–1: green, F-actin: phalloidin white, tdTomato: red, nuclei: blue. scalebars, 50 μm).
(H) Primary adult human AT2 cells (1AT2s) were cultured as described28 in SFFF or ADM compared with L+DCI (medium changed 7 days post plating). Whole-well RT-qPCR of selected genes 7 days post medium change (N = 3 per condition, one-way ANOVA).
(I) Experimental schematic: iAT1s were grown in L+DCI for 10 days (P0), sorted on AGERtdTomato, and then plated in 3D Matrigel in either L+DCI or CK+DCI. Outgrowths were analyzed after an additional 9 days (P1).
(J) Representative flow cytometry plots P1 from (I).
(K) Quantification of (J): cell counts or transcript expression.
(L) Whole-well RT-qPCR of cells from (I). Control-sorted iAT1s are freshly sorted AGERtdTomato+ from P0 (N = 3 wells per condition; K, Student’s t test; L, one-way ANOVA). *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.001, bars represent mean ± SD. BU3 NGAT iPSC line for all panels. See also Figures S4 and S5.
We examined the individual effects of Chir and KGF in media containing L and saw that both were significantly inhibitory to both AGERtdTomato percentage and mean fluorescence intensity (MFI) (Figures S4B and S4C). Even in the presence of Chir, addition of the LATS inhibitor resulted in diminished expression of Wnt downstream targets AXIN2 and LEF1 (Figure S4D). To test whether the inhibitory effects of Chir and KGF on the AT1 program were specific to the media-based differentiation, we tested withdrawal of each of these factors in our YAP5SA differentiation model. We again saw inhibitory effects of both growth factors on AGERtdTomato as well as AT1 markers by bulk RT-qPCR, suggesting both Chir and KGF need to be removed for the efficient differentiation of iAT1s (Figures S4E–S4I). This is consistent with prior publications suggesting canonical Wnt or KGF signaling is inhibitory to the expression of the AT1 program.17,23,30
After dose-response testing (Figure S5A), we found replacing the CK+DCI medium on day 3 after passaging iAT2s with a medium consisting of 10 μM L plus DCI yielded the greatest frequency and brightness (MFI) of NKX2–1GFP+; AGERtdTomato+ cells 2 weeks post-passage. When NKX2–1GFP+ cells were repurified and exposed to L+DCI beginning 3 days post sort, up to 97% of all cells were AGERtdTomato+ by day 14, suggesting that all iAT2s, rather than a subset, are competent to differentiate into iAT1s (Figure S5B). Additionally, AGERtdTomato+ cells grown in L+DCI were nearly all positive for the human AT1 marker HT1–56 by flow cytometry (92.4% ± 1.8%) (Figure S5C). By whole-well RT-qPCR, AGER, CAV1, PDPN, and CLIC5 were significantly upregulated in wells containing L, with higher levels expressed in conditions without KGF and Chir (Figures 5F and S5D). NKX2–1 remained unchanged between CK+DCI and L+DCI, suggesting maintenance of the lung epithelial program, whereas SFTPC was decreased (though not entirely absent), indicating downregulation of the AT2 program (Figures 5F and S5D). A modified derivative of LATS-IN-1, TDI-011536,56 also generated AGERtdTomato+ cells (Figure S5E), further validating the utility of LATS inhibition in our protocol. Similar responses to L+DCI were seen in SPC2-ST-B2 iAT2s, with loss of SFTPCtdTomato reporter and gain of RAGE protein in 93.8% ± 1.7% of cells (Figures S5F–S5H). Immunostaining showed similar organoid morphologies to that of YAP5SA-transduced organoids, with AGERtdTomato only being expressed in NKX2–1+ cells (Figure 5G). Taken together, these results indicate that treatment with the LATS inhibitor in the absence of Chir and KGF efficiently differentiates iAT2s into iAT1s.
To determine whether L+DCI medium induces the AT1 program in primary adult human AT2s, we culture-expanded primary AT2s in serum-free, feeder-free (SFFF) conditions as reported.28 After 7 days in SFFF, medium was either continued as SFFF or switched to published human serum-containing alveolar differentiation medium (ADM)28 or L+DCI. 7 days later, NKX2–1 expression was similar for all conditions, while AGER was significantly upregulated both differentiation conditions. CAV1 and ANKRD1 expression was higher in L+DCI than ADM, suggesting this differentiation medium is effective in the differentiation of primary human AT2s into AT1s (Figure 5H). By contrast to iPSC-derived cells, SFTPC expression was not decreased in L+DCI conditions in primary cells. In tests using iAT2s instead of primary AT2s, only L+DCI and not ADM promoted iAT1 differentiation evident by no induction of AGER or ANKRD1 (Figure S5I).
To assess the stability of iAT1s induced by LATS inhibition, after differentiation in L+DCI for 11 days (P0), cells were passaged and grown for an additional 14 days in L+DCI (P1) (Figures S5J and S5K). At this point, cells did not appear to have proliferated (yield at end of P1 = 0.97 ± 0.46 cells per input cell, compared with 12.38 ± 3.28 cells per input iAT2 at the end of P0). However, AGERtdTomato expression was slightly increased at the end of P1, indicating that iAT1s are stable when passaged in L+DCI and maintain their AT1 program for at least 25 days, but do not proliferate after passaging (Figures S5J and S5K), as expected for AT1s.
To determine whether iAT1s maintain their phenotype when transitioned back into iAT2-maintenance medium, AGERtdTomato+ cells were sorted after 11 days in L+DCI and replated into either L+DCI or CK+DCI (Figure 5I). After 9 further days in culture, >95% of cells in L+DCI maintained AGERtdTomato expression, while those switched into CK+DCI showed 10%–20% AGERtdTomato+ cells (Figures 5I–5L) and re-expressed AT2 markers, consistent with residual plasticity in iAT1s.
To test whether our iAT1 medium was capable of differentiating iAT1s from developmental lung progenitors, we differentiated BU3 NGAT iPSCs to day 15 NKX2–1GFP+ lung progenitor cells (previously shown to be similar to fetal primordial lung progenitors).25,54,57 When placed into L+DCI medium for 2 weeks, a small percentage of NKX2–1GFP+ cells co-expressed AGERtdTomato (2.02% ± 0.52%; Figure S5L). However, this small population did not proliferate in culture after passage, suggesting that AGERtdTomato+ cells can be derived from lung progenitors using this medium, but not as efficiently as from mature iAT2s.
We tested select other compounds to determine whether they could further mature iAT1s, including addback of KGF, and addition of FGF18 or IL1β, but saw no increase in AGERtdTomato with or without L (Figures S5M–S5O).
Transcriptomic profiling by scRNA-seq of iAT1s
To comprehensively compare the transcriptomic programs of iAT1s generated through our defined iAT1 medium vs. lentiviral overexpression of YAP5SA in CK+DCI, we performed scRNA-seq profiling of iAT1s prepared by either method vs. parallel iAT2 controls, hereafter named iAT1, iAT1YAP5SA, and iAT2, respectively (SPC2-ST-B2 iPSCs; Figure 6A). Clustering analysis revealed 3 predominant cell clusters (Louvain resolution = 0.05; Figures 6A and 6B) visualized by UMAP, which segregated based on the method used to generate the cells. As in Figure 3, cells from the YAP5SA sample segregated into either the iAT1YAP5SA cluster or the iAT2 cluster, depending on whether they were successfully transduced (see inset Figure 6A).
Figure 6. iAT1s generated by defined medium or lentiviral YAP5SA express a broad AT1 transcriptomic program.
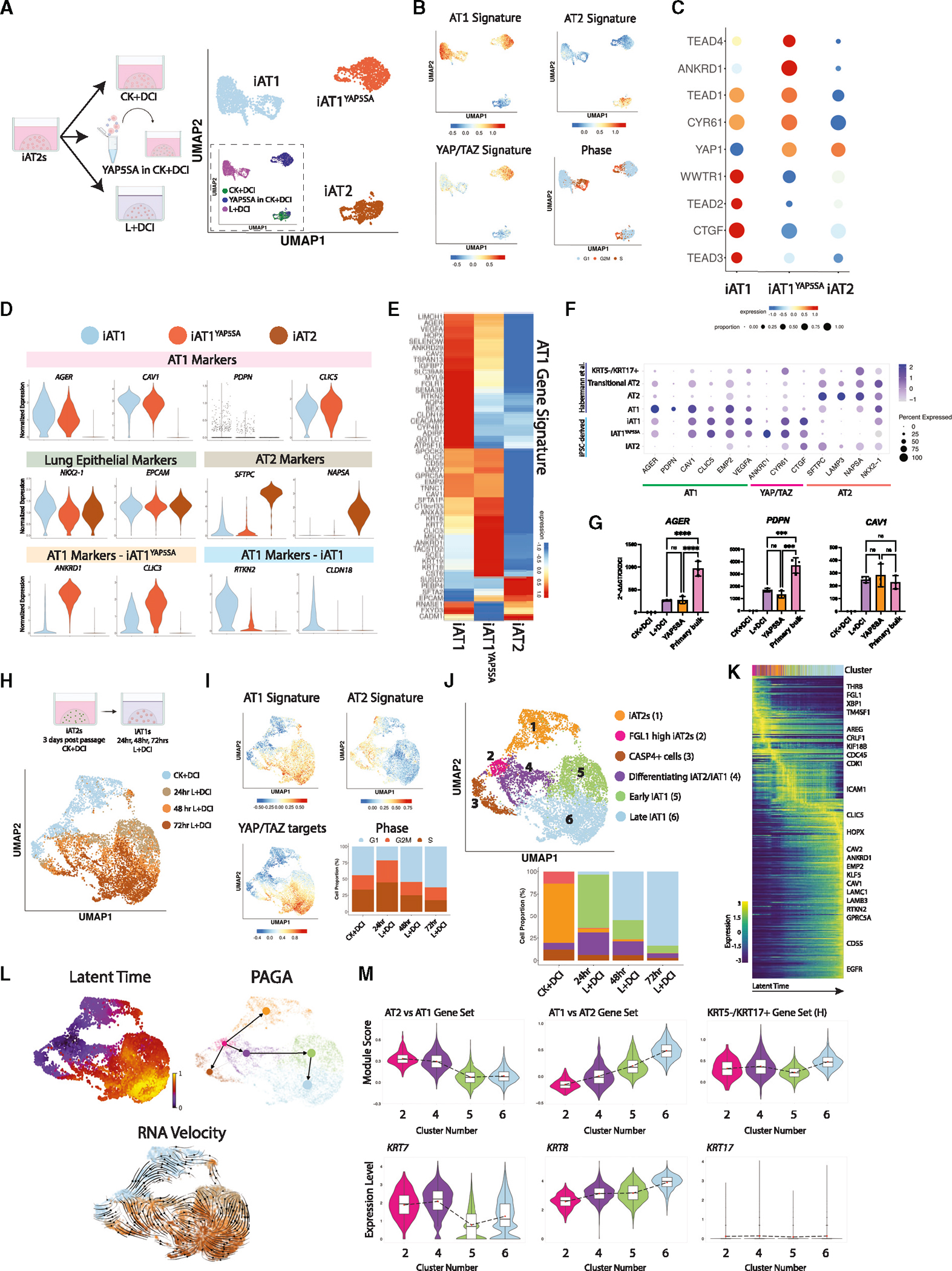
(A) SPC2-ST-B2 iAT2s were grown in CK+DCI either with or without YAP5SA transduction, or grown in CK+DCI for 3 days before switching to L+DCI. 9 days post passage live cells were profiled by scRNA-seq. UMAP visualization of single-cell transcriptomes by original sample name (inset) or after Louvain clustering (res 0.05). Clusters were named iAT2, iAT1YAP5SA, and iAT1.
(B) Gene expression overlays of our AT1 50-gene signature, a 22-gene YAP/TAZ signature,58 human AT2 50-gene signature, or cell cycle phase.
(C) Dot plot of transcript expression levels/frequencies of YAP/TAZ downstream targets and TEADs.
(D) Violin plots quantifying expression of indicated markers.
(E) Heatmap showing average expression (normalized by column) of genes in the AT1 50-gene set.
(F) Dot plot showing expression levels and frequencies of AT1, AT2, and YAP/TAZ targets in this dataset compared with human adult primary populations previously published by Habermann et al.2
(G) Expression of AT1 marker genes by RT-qPCR in whole-well extracts of L+DCI-induced and YAP5SA-transduced iAT1s compared with bulk primary human distal lung tissue; fold change normalized to 18S (2^−DDCT) is calculated relative to iAT2s in CK+DCI (N = 3 wells per condition, one-way ANOVA).
(H) UMAP visualization of single-cell transcriptomes at 24 h intervals over 4 days of iAT2 (CK+DCI) to iAT1 (L+DCI) differentiation in 3D (BU3 NGAT).
(I) Gene expression overlays of human AT1 and AT2 50-gene signatures, 22-gene YAP/TAZ target signature, and cell cycle phase.
(J) UMAP visualization of Louvain clustering.
(K) Heatmap of top 300 driver genes for RNA velocity path.
(L) Pseudotime analysis visualized on UMAP (latent time, partition-based graph abstraction [PAGA], and RNA velocity).
M) Violin plots showing module scores of gene sets or expression of individual genes (H = gene set from Habermann et al.)2 *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.001, bars represent mean ± SD for all panels. See also Figure S6.
We observed that both iAT1 and iAT1YAP5SA populations exhibited downregulation of the AT2 signature with robust upregulation of the AT1 50-gene set signature (Figure 6B; Table S1) and individual AT1 markers (Figures 6C–6G and S6A). Expression of AT1 transcripts was validated by RT-qPCR (Figure 6G), and the frequency of expression of most AT1 marker transcripts was similar to expression profiles of published primary AT1s (Figure 6F). However, absolute expression levels for AGER and PDPN were lower than in primary control distal lung tissue by RT-qPCR (Figures 6G and S6B).
Despite these similarities, iAT1 and iAT1YAP5SA cells clustered separately with 298 transcripts differentially expressed (FDR < 0.05, logFC > 1, Table S2). These differences included: (1) differential upregulation of a YAP/TAZ 22-gene target signature58 (Figure 6B), likely reflecting the higher levels of YAP signaling by lentiviral forced overexpression; and (2) differential expression of other AT1 marker genes, such as ANKRD1, CLIC3, RTKN2, and CLDN18 (Figures 6C, 6D, and S6A). As expected, only iAT1YAP5SA cells expressed the lentiviral tagBFP reporter, and neither iAT1 nor iAT1YAP5SA cells expressed high levels of the iAT2 marker, SFTPCtdTomato (Figure S6C). Additionally, there were notable differences in specific Hippo-LATS-YAP signaling targets. For example, YAP and TEAD4 were upregulated in iAT1YAP5SA cells, whereas TAZ (WWTR1), TEAD2, TEAD3, and CTGF were upregulated in iAT1s (Figure 6C). These differences may be due to the LATS inhibitor affecting both YAP and TAZ, whereas the lentivirus is YAP-specific, or the effect of Chir and KGF in the medium of the lentivirally transduced iAT1YAP5SA cells. Taken together, these results indicate efficient induction of the AT1 program via a serum-free differentiation medium, L+DCI.
iAT1 differentiation time series transcriptomic profile
To further understand the kinetics of differentiation, we performed time series scRNA-seq profiling over 72 h as iAT2s differentiated to iAT1s. iAT2s were passaged, and 3 days later, we began profiling cells before medium change (CK+DCI condition) and 24, 48, and 72 h post medium change to L+DCI (Figure 6H). After 24 h of L+DCI exposure, YAP/TAZ targets were upregulated, and the 50-gene AT1 signature was induced and continuously increased in expression over the next 2 days (Figures 6I–6M). Concurrently, the 50-gene AT2 signature decreased. Although cells at all time points could be found in various phases of cell cycle, the majority of cells in CK+DCI were initially in active cycle (G2/M or S), but by 72 h in L+DCI, the majority of cells were found in G1 cell-cycle phase (Figure 6I).
We performed Louvain clustering and annotated 6 cell clusters based on expression of top DEGs or levels of expression of the AT1 marker gene set, finding 2 clusters of iAT2s (#1–2) in the CK+DCI samples: one of which (#2) expressed higher levels of FGL1. The apparently apoptotic CASP4+ cluster (#3) containing cells from each time point (also high in ribosomal proteins) was not analyzed further. We observed 3 distinct and apparently sequential cell clusters in L+DCI exposed samples (#4–6) and annotated these based on AT1 signature levels (Figures 6I–6M) and a variety of pseudotime analyses, including partition-based graph abstraction (PAGA), latent time, and RNA velocity (Figure 6L), as: “differentiating iAT2/iAT1 (#4),” “early iAT1 (#5),” and “late iAT1 (#6)” (Figures 6J–6L). By 72 h in L+DCI, 80% of cells were in the “late iAT1” cluster.
Since all 3 pseudotime trajectory analyses (Figure 6L) as well as diffusion heatmaps (Figure 6K) suggested a differentiation sequence from CK+DCI cluster 2 through L+DCI treated cell clusters 4 through 6, we next analyzed this putative differentiation trajectory to discern whether cells were passing through a discrete “TC” (transitional cell) or “aberrant basaloid” cell state, as has been described in recent reports.2–4,12,33,59–61 Although some known TC markers were detectable in our dataset, we observed a smooth transition from iAT2s to iAT1s and found no discrete clustering to suggest a distinct TC state. We detected no expression of basal marker KRT5 and little to no TP63 or KRT17 (Figures 6M and S6D), suggesting no evidence of basal cell markers that have been described as part of the human aberrant basaloid cell phenotype. Of known human TC state markers, only KRT7 was differentially upregulated in the differentiating iAT2/iAT1 cluster, whereas KRT17, SFN, FN1, and VIM were not. Other reported TC markers, such as KRT8, KRT18, and SOX4, were most highly expressed in the late iAT1 cluster (Figures 6M and S6D), but these were also differentially enriched in human primary AT1 cells (Figure 1; Table S1; AT1 vs. all lung epithelial cells), suggesting they indicated emergence of the AT1 program rather than a TC state. Additionally, there was little to no expression of “AT0” marker SCGB3A2 in this time series (Figure S6D). Jaccard similarity coefficient indexing indicated little similarity of any cluster to published TC states, such as the KRT5−/KRT17+ cell state of Habermann and colleagues2 (Figure S6E), and the late iAT1 population was most similar to primary AT1s by this method.
iAT1s display functional capacities to form a flattened epithelial barrier, secrete signaling ligands, and express ECM-encoding transcripts
We next evaluated the functional capacity of iAT1s. An emerging literature has established several functions that characterize primary AT1s in vivo, including their potential to: form a flattened epithelial barrier1; produce a characteristic alveolar extracellular matrix (ECM)62; and serve as signaling hubs in the lung through secretion of signaling ligands.63 Since iAT1s in 3D Matrigel cultures do not discernably form flattened epithelial barriers, we transitioned BU3 NGAT iAT1s into 2D transwell cultures in L+DCI and 3 days later aspirated apical medium to establish air-liquid interface (ALI) cultures, hereafter “iAT1 ALI P1.” We observed maintenance of AGERtdTomato reporter expression and positive staining for RAGE protein, suggesting maintenance of the AT1 program (Figures 7A–7D). As in 3D conditions, iAT1 ALI P1 cells were still plastic if transitioned back to CK+DCI medium, losing their AGER, CAV1, and CLIC5 expression (Figure S7A).
Figure 7. iAT1s cultured at air-liquid interface (ALI) express AT1-like molecular and functional phenotypes.
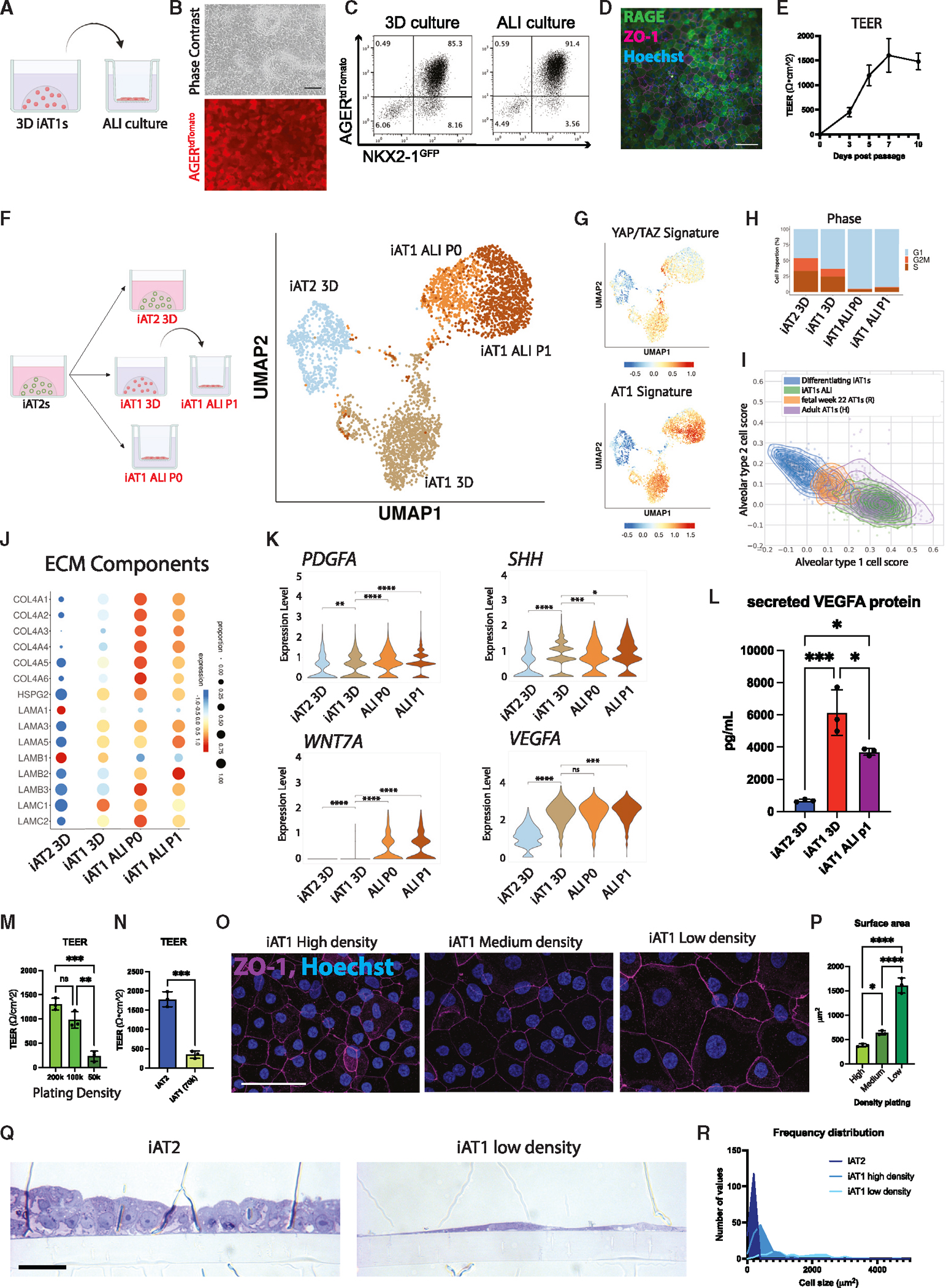
(A) Experimental schematic: iAT1s were cultured in 3D in L+DCI for 8–11 days before passaging and replating onto transwell inserts in L+DCI. Upper chamber media was aspirated after 3 days (airlift) to form ALI.
(B) Live cell imaging showing retention of AGERtdTomato after ALI culture (scale bar, 100 μm).
(C) Representative flow cytometry of NKX2–1GFP and AGERtdTomato expression in 3D or ALI cultures (6 days).
(D) Immunofluorescence microscopy of RAGE and ZO-1 in iAT1s at ALI (scale bar, 100 μM).
(E) Transepithelial electrical resistance (TEER) measurements of BU3 NGAT iAT1s over 10 days of ALI cultures (air lifted at day 3; N = 3).
(F) scRNA-seq profiling of iAT2s in 3D CK+DCI; iAT1s in 3D L+DCI; or iAT1s in ALI cultures, which were plated either from iAT2s into L+DCI (iAT1 ALI P0) or from 3DiAT1s after 9 days pre-culturing in 3D L+DCI (iAT1 ALI P1).
(G) UMAP overlays of YAP/TAZ 22-gene signature58 and primary human AT1 50-gene signature (Table S1).
(H) Cell cycle phase distribution across samples.
(I) Alignment scores to primary human AT1 and AT2s using scTOP64 analysis comparing two iAT1 populations (differentiating iAT1/iAT2—Figure 6 and iAT1s ALI—this figure) with primary human fetal lung at 22 weeks65 and adult primary human AT1s.2
(J and K) Expression of transcripts encoding (J) ECM components and (K) secreted ligands, comparing the samples from (E) (pairwise t test).
(L) Analysis of secreted VEGFA protein in conditioned media at day 10 of culture of each indicated sample (N = 3 wells per condition, one-way ANOVA).
(M) TEER of iAT1s after plating at high, medium, and low densities and outgrowth in ALI cultures in L+DCI (N = 3 wells per condition, one-way ANOVA).
(N) TEER of iAT2s compared with iAT1s plated at low density (SPC2-ST-B2) (N = 3 wells per condition, Student’s t test).
(O) Tight junction protein ZO-1 staining (magenta) at high, medium, and low plating density outgrowths at day 10 (scale bars, 50 μm).
(P) Average surface area of cells calculated using ZO-1 cell outlines at different iAT1 ALI plating densities (N = 3 per condition, averaged from ~150 cells per sample).
(Q) Cross-sectional imaging of SPC2-ST-B2 iAT2s at ALI and iAT1 P1s plated at low densities (Toluidine blue stain, scale bars, 10 μm).
(R) Frequency distribution of cell surface areas of iAT2, iAT1 high density, and iAT1 low density (N = 149, 139, and 79, respectively). *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.001, bars represent mean ± SD for all panels, BU3 NGAT iPSC line unless otherwise noted. See also Figure S7.
We evaluated the functional epithelial barrier-forming capacity of iAT1 ALI P1 cells and found that confluent iAT1s prevented apical medium leakage and increased transepithelial electrical resistance (TEER) over time to 1,479 ± 166 Ω · cm2 by day 10 (Figure 7E). In more extended follow-up experiments, iAT1 ALI P1 cells were able to maintain a non-leaking epithelial barrier for at least 3 months.
We profiled the iAT1s produced in 3D L+DCI (iAT1 3D) vs. iAT1 ALI P1 conditions as well as iAT1s produced in a second ALI condition where iAT2s are only exposed to L+DCI coincident with transwell plating (hereafter “iAT1 ALI P0”; Figures 7F–7K and S7B–S7F). Compared with parallel control iAT2s, we found downregulation of the AT2 program and upregulation of the YAP/TAZ and AT1 programs in iAT1s grown in both ALI cultures, like 3D iAT1s, but with significantly reduced active cell cycling (reduced G2/M phases; Figures 7G, 7H, and S7B). These results indicate that iAT1s transition to a more quiescent state after ALI culture. As we have previously reported for iAT2s,52 a rare subset of cells plated in these conditions upregulated markers of non-lung endoderm (ALB, AFP), suggesting some degree of residual plasticity in rare cells plated in all 4 conditions (Figure S7C).
To understand how these cells compare with primary adult and fetal human AT1s, we employed our recently published algorithm, single-cell type order parameters (scTOP; Figures 7I and S7D), which allows unbiased and quantitative comparison scores of how PSC-derived single-cell transcriptomes align to reference datasets from established scRNA-seq atlases, with less susceptibility to technical batch artifacts than other methods.64,66 We found our iAT1s aligned progressively more closely to primary AT1s over time in L+DCI culture (concurrent with progressive loss of similarity to adult AT2s; Figure S7D). Transcriptomes of iAT1s after ALI culture displayed the closest alignment to adult human primary AT1s, and these cells were more similar to AT1s than any other primary lung cell type in the reference atlas (LungMap: LMEX0000004396; Figure S7D). Additionally, comparisons to human fetal lung epithelial cell atlases at different developmental time points (recently published by Rawlins and colleagues)65 indicated our late iAT1s (in 3D culture) had similar AT1 transcriptomic alignment scores to fetal 22-week primary human AT1s, and after ALI culture, our iAT1s had higher alignment scores to adult AT1s (Figure S7D). We also analyzed each individual iPSC-derived single-cell transcriptome’s similarity score to the adult AT2 and AT1 reference bases and again found individual ALI iAT1 transcriptomes aligned more closely to primary AT1s than AT2s, whereas control primary transcriptomes of 22 weeks fetal AT1s by Rawlins and colleagues (R) and adult AT1s by Habermann and colleagues (H) aligned as expected with intermediate or highly similar scores, respectively, to the AT1 reference basis (Figure 7I).
Since cells cultured at ALI aligned more closely to primary adult AT1s, we sought to better understand the transcriptomic changes following ALI culture. We found genes encoding ECM components, such as COL4A4, featured prominently in those upregulated transcripts (Figure 7J; Table S2). Given the recently reported role of ECM generation by mouse AT1s during development,62 we looked at other components of collagen IV and laminins, including the components of laminin-332 (LAMA3, LAMB3, and LAMC2). As reported for mouse AT1s in vivo,62 human iAT1s at ALI expressed all 6 components of collagen IV and patterns of laminin expression similar to published mouse single-cell data (Figure 7J). Notably, although the expression pattern of these laminins in human lung development is unknown, expression of laminin-332 in mouse data was associated with more mature AT1s.62 Consistent with these observations, the most significantly upregulated pathways in iAT1s plated in 2D ALI compared with 3D culture (Enrichr Reactome pathways based on the top 100 DEGs [FDR < 0.05] ranked by logFC iAT1 ALI vs. iAT1 3D; Figure S7E) were ECM organization, collagen formation and assembly, and integrin cell surface interactions. In addition to collagen and laminin components, iAT1s plated in 2D ALI also increased expression of actin, non-muscle myosin, and cdc42-N-WASP-Arp2/3 complex genes that have been enriched in mouse primary adult AT1s47 (Figure S7F), suggesting a cytoskeletal rearrangement in response to being grown in 2D conditions that mimics the genetic programs recently reported for in vivo post-natal AT1s.
Another recently recognized function of AT1s in vivo is their role as signaling hubs in the distal lung, based on mouse and human scRNA-seq profiles and genetic mouse models that suggest AT1s stimulate local alveolar Shh, Wnt, PDGF, and VEGF signaling during alveologenesis or tissue maintenance.62,63 Consistent with these reports, we found iAT1s after ALI culture upregulated transcripts encoding the signaling ligands, PDGFA, SHH, WNT7A, and VEGFA (Figure 7K) that were also enriched in primary AT1s from 2-month-old human infants as reported by Zepp et al.63 At the protein level, we verified secreted VEGFA was present in conditioned medium from both 3D and ALI iAT1 culture conditions (Figure 7L).
Finally, we evaluated whether iAT1s could form the characteristic flattened epithelial barrier that defines their unique morphology. We tested whether BU3 NGAT iAT1s would flatten at lower plating densities, potentially stretching out to cover the available surface area while still forming tight junctions necessary to form a functional epithelial barrier. The ALI barrier remained intact for all 3 plating densities with measurable TEER and formation of tight junctions, evidenced by ZO-1 staining (Figures 7M–7O). TEER for iAT1s was inversely correlated with plating density, as higher plating density of iAT1s had similar TEER values to published iAT2s,67 (Figures 7M–7O). Average cell surface area was calculated, and cells plated at lower densities had significantly larger cell surface areas on average as well as lower total cell counts (Figures 7O–7R and S7G–S7J). Similar results were observed for SPC2-ST-B2 iAT1s plated at high- and low-density ALI cultures, and iAT1s occupied larger cell surface areas than iAT2s plated at an identical initial cell density (Figures 7O–7R and S7G–S7J). Reported average surface areas of adult human AT1s range from 3,960 to 8,290 μm2, averaging around 5,100 μm2.1,68,69 iAT1s reached the size of cells on the smaller end of this range (Figures 7O–7R and S7G). Additionally, cross-sectional images of SPC2-ST-B2 iAT1s showed thin cells with elongated, flattened nuclei compared with cuboidal iAT2s (Figure 7Q). These results suggest iAT1s readily flatten and stretch to cover available surface area to form and maintain a thin epithelial barrier.
DISCUSSION
In this study, we employed directed differentiation of iPSCs to efficiently generate cells expressing several key features of human AT1s, including a molecular phenotype and the potential to form a flattened epithelial barrier that expresses AT1-associated ligands and matrix components. This flattened morphology, important to the gas exchange function of AT1s, appears to be decoupled from the molecular phenotype in our system, as cells were able to turn on the AT1 program in 3D culture while still maintaining a cuboidal appearance. However, iAT1s were uniquely able to flatten into a thin epithelial barrier when plated in ALI cultures.
Our finding that Hippo-LATS-YAP signaling activates the human AT1 program in lung epithelia is consistent with mouse genetic models that activated this pathway in vivo.19,20,44–46 Although nuclear YAP activation was sufficient to drive the transcriptomic shift from iAT2 to iAT1, withdrawal of canonical Wnt or FGF signaling activators (Chir and KGF) alone was not sufficient to induce this differentiation, and these factors were found to be inhibitory toward the AT1 program. We found that iAT1s grown in our defined medium (L+DCI) resembled YAP5SA-transduced iAT1s in terms of their structure, growth, and expression of most but not all canonical AT1 marker transcripts. Although both populations expressed a broad AT1 transcriptomic program, they differed in expression of several key AT1 transcripts, findings of unclear significance that will require further study. Importantly, L+DCI medium also induced upregulation of several AT1 markers in cultured primary adult human AT2s, providing an identical serum-free defined medium-based approach that can be studied using cells prepared from multiple sources, either primary or engineered.
Notably, we detected no clear evidence of cells passing through a discernible “transitional” or “aberrant basaloid” state found in several mouse and human datasets as a potential disease-relevant state.2–4,12,33,59–61 Although some markers of transitional cells, the most notable being KRT7, were differentially upregulated in our differentiating iAT2/iAT1 cluster, the late iAT1 cluster had the highest similarity coefficient when scored against a 50-gene set panel of transitional markers.70 Many reported transitional markers are expressed at higher levels in primary human AT1s than AT2s, suggesting that an increase in their expression may be indicative of the presence of AT1 features, not a separate transitional cell state. Taken together our time series supports the model introduced by Evans et al. and Adamson and Bowden in the 1970s,5,6,71 where differentiation of rodent AT2s into AT1s proceeds relatively quickly through an “undetermined” or “intermediate” cell type; these intermediate cells exhibit loss of AT2 features with coincident gain of AT1 features. In our time series, most cells proceed smoothly through this continuum, without evoking a discrete transitional cell state. Possibly, disease settings are required to bring out a distinct cell state, as was recently reported.2–4,12,33,59–61 Alternatively, it is possible that our cluster 4 cells or the intermediate cells of Evans may represent cells hypothesized by some to be in a separate state or that we missed a key time point in our study. It is also possible that LATS inhibition acts further downstream in the AT1-promoting pathway than signals required to drive a distinct transitional cell state.
Key to benchmarking our in vitro cells was the identification of gene set markers and pathways that define the in vivo transcriptomic program of human AT1 cells. scRNA-seq of adult primary human AT1 cells provided a comprehensive gene signature as well as a quartet of canonical markers (AGER, PDPN, CAV1, and CLIC5) that can be used for future profiling of these cells. Additionally, these datasets help to distinguish differences between human and mouse marker genes. Although mice are the most common in vivo lung model, several mouse AT1 markers are not identically expressed in humans, including marker genes HOPX, AQP5, and IGFBP2, which are commonly used mouse AT1-specific genetic drivers that do not share this specificity in the human lung.34,36
Limitations of the study
Our results raise several questions that will require further study. First, future work will be required to further mature the iAT1s produced by our methods. Although our iAT1s show some functional properties and transcriptomic similarities to in vivo human AT1 cells, expression levels of most canonical AT1 markers are significantly lower than primary cell controls. A variety of future approaches can now be tested to augment maturation, such as introducing biomechanical cues that more closely recapitulate the distal lung microenvironment.
Overall, our work shows the generation of AT1-like cells from iAT2s cells, providing an in vitro model of human alveolar epithelial differentiation and a potential source of human cells that until now have been challenging to viably obtain from patients. Access to these cells, either in pure form or combined with other lineages, should facilitate a variety of basic developmental studies, disease modeling, and potential engineering of future regenerative therapies.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents may be directed to, and will be fulfilled by, the lead contact, Darrell Kotton (dkotton@bu.edu).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement. Pluripotent stem cell lines used in this study are available from the CReM iPSC Repository at Boston University and Boston Medical Center and can be found at http://www.stemcellbank.bu.edu/Catalog/Item/Home. All plasmids have been made available on Addgene.
Data and code availability
Single-cell RNA-seq datasets have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Interactive visualizations of these datasets will also be available on the Kotton Lab’s Bioinformatics Portal at http://www.kottonlab.com. This paper does not report original code. Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Rabbit anti-NKX2–1 antibody | Abcam | Cat#ab76013; RRID:AB_1310784 |
| Goat anti-RFP antibody | My Biosource | Cat#MBS448122 |
| Rabbit anti-proSFTPC antibody | Seven Hills | Cat#r76694 |
| Goat anti-RAGE antibody | R&D Systems | Cat#AF1145 |
| Mouse anti-HT1–56 antibody | Terrace Biotech | SKU: TB-29AHT1–56 |
| Rabbit anti-ZO-1 antibody | ThermoFisher | Cat#61–7300; RRID:AB_2533938 |
| Rabbit anti-YAP antibody | Cell Signaling Technology | Cat#14074 |
| Rabbit anti-phospho-YAP antibody | Cell Signaling Technology | Cat#13008 |
| Rabbit anti-Hexokinase1 antibody | Proteintech | Cat#19662–1-AP; RRID:AB_10859778 |
| Alexa Fluor 647 donkey anti-rabbit | Invitrogen | Cat#A31573; RRID:AB_2536183 |
| Alexa Fluor 488 donkey anti-mouse | Invitrogen | Cat#A21202; RRID:AB_141607 |
| Alexa Fluor 647 donkey anti-goat | Invitrogen | Cat#A21447; RRID:AB_2535864 |
| Alexa Fluor 546 donkey anti-goat | Invitrogen | Cat#A11056; RRID:AB_2534103 |
| Alexa Fluor 488 donkey anti-rabbit | Invitrogen | Cat#A21208; RRID:AB_2535794 |
| Alexa Fluor 488 donkey anti-goat | Invitrogen | Cat#A11055; RRID:AB_2534102 |
| Anti-rabbit IgG, HRP-linked antibody | Cell Signaling Technology | Cat#7074 |
| Anti-goat IgG, HRP-linked antibody | Jackson ImmunoRsearch | Cat#705–036-147; RRID:AB_2340392 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| Cre Recombinase Adenovirus | Vector Biolabs | Cat#1700 |
|
| ||
| Biological samples | ||
|
| ||
| Primary Human Lung samples | Gift from Dr. Barry Stripp, Cedars Sinai | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Growth Factor Reduced Matrigel | Corning | Cat#356234 |
| mTeSR 1 complete kit | StemCell Technologies | Cat#85850 |
| DMEM | Gibco | Cat#2414671 |
| IMDM | ThermoFisher | Cat#12440053 |
| Glutamax | Life Technologies | Cat#35050–061 |
| Ham's F12 | Cellgro | Cat#10–080-CV |
| B27 supplement | Invitrogen | Cat#17504–44 |
| N2 supplement | Invitrogen | Cat#17502–048 |
| Primocin | Invivogen | Cat#ANTPM1 |
| 7.5% BSA Fraction V | ThermoFisher | Cat#15260–037 |
| 1-thioglycerol (MTG) | Sigma | Cat#M6145 |
| Ascorbic Acid | Sigma | Cat#A4544 |
| SB431542 | Sigma | Cat#S4317 |
| Y-27632 (ROCK inhibitor) | Tocris | Cat#1254 |
| Growth Factor Reduced 3D Matrigel | Corning | Cat#354230 |
| Dispase | Gibco | Cat#17105–041 |
| Retinoic acid | Sigma | Cat#R2625 |
| 0.05% Trypsin-EDTA | Gibco | Cat#25300062 |
| Fetal bovine serum | Gibco | Cat#16141079 |
| Paraformaldehyde | Electron Microscopy Sciences | Cat#19208 |
| Normal donkey serum | Sigma | Cat#D9663 |
| Antigen Unmasking Solution, Citric Acid Based | Vector Laboratories | Cat#H-3300–250 |
| Hoechst 33342 | Thermo Fisher | Cat#H3570 |
| Triton-X | Sigma | Cat#T9284 |
| Calcein Blue | ThermoFisher | Cat#C1429 |
| DRAQ7 | Abcam | Cat#ab109202 |
| Dorsomorphin | Fisher Scientific | Cat#NC0275327 |
| CHIR99021 | Fisher Scientific | Cat#44–235-0 |
| rhKGF (FGF7) | Fisher Scientific | Cat#251KG050 |
| rhBMP4 | R&D systems | Cat#314-BP |
| rh IL-1beta | Gibco | Cat#200–01B |
| rhFGF18 | Gibco | Cat#100–28 |
| TRULI (LATS-IN-1) | MedChem Express | Cat#HY-138489 |
| TDI-011536 | MedChem Express | Cat#HY-150042 |
| Dexamethasone | Sigma | Cat#D4902 |
| 8-bromoadenosine 30,50-cyclic monophosphate sodium salt (cAMP) | Sigma | Cat#B7880 |
| 3-Isobutyl-1-methylxanthine (IBMX) | Sigma | Cat#I5879 |
| Polybrene | Tocris | Cat#7711 |
| Phalloidin (Texas Red) | Invitrogen | Cat#T7471 |
| Puromycin Dihydrochloride | ThermoFisher Scientific | Cat#A1113802 |
| Geneticin Sulfate | Life Technologies | Cat#11811–023 |
| Dimethyl Sulfoxide (DMSO) | Sigma Aldrich | Cat#D2650 |
| ProLong Diamond Antifade Mountant | Invitrogen | Cat#P36965 |
| West Femto Maximum Sensitivity Substrate | Thermo Scientific | Cat#PI34096 |
| Restore PLUS Western Blot Stripping Buffer | Thermo Scientific | Cat#46430 |
| Human Serum | Sigma-Aldrich | Cat#H6914 |
|
| ||
| Critical commercial assays | ||
|
| ||
| RNeasy Mini Kit | QIAGEN | Cat#74104 |
| QIAzol Lysis Reagent | QIAGEN | Cat#79306 |
| TaqMan Fast Universal PCR Master Mix (2X) | Thermo Fisher | Cat#4364103 |
| High-Capacity cDNA Reverse Transcription Kit | Applied Biosytems | Cat#4368814 |
| P3 Primary Cell 4D-Nucleofector X Kit | Lonza | Cat#V4XP-3032 |
| Human VEGFA ELISA Kit | Abcam | Cat#119566 |
| Click-iT EdU Cell Proliferation Kit | ThermoFisher | Cat#C10340 |
|
| ||
| Deposited data | ||
|
| ||
| scRNAseq data: WT YAP, YAP5SA (Figure 3) | This paper | GEO: GSE221342 |
| scRNAseq data: L+DCI, YAP5SA, CK+DCI (Figure 6) | This paper | GEO: GSE221343 |
| scRNAseq data: L+DCI time series (Figure 6) | This paper | GEO: GSE246243 |
| scRNAseq data: 3D vs ALI (Figure 7) | This paper | GEO: GSE221344 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Human: Donor iPSC line targeted with SFTPCtdTomato (SPC2-ST-B2) | Kotton Lab; Hurley et al.52 | http://stemcellbank.bu.edu |
| Human: Normal donor iPSC line targeted with NKX2–1gfp (BU3 NG) | Kotton Lab; Hawkins et al.54 | http://stemcellbank.bu.edu |
| Human: Normal donor iPSC line targeted with NKX2–1gfp and AGERtdTomato (BU3 NGAT) | This paper | http://stemcellbank.bu.edu |
|
| ||
| Oligonucleotides | ||
|
| ||
| Oligonucleotide primers | Integrated DNA Technologies | N/A |
| P1 (AGER left homology arm) | This paper | 5'AGGACTCTTGTCCCAAAGGC 3' |
| P2 (AGER right homology arm) | This paper | 5'CTGGGGTGTGGGGTTAAAGT 3' |
| P3 (Puromycin resistance Fwd) | This paper | 5'ACTTGTGTAGCGCCAAGTGC 3' |
| P4 (Puromycin resistance Rev) | This paper | 5'ACACACACTCGCCTCCTGTT 3' |
| P5 (tdTomato insert Fwd) | This paper | 5'CTGATCCCCTCAGACATTCTCAGGA 3' |
| P6 (tdTomato insert Rev) | This paper | 5'GAGCTGCCGCTGCCGGT 3' |
|
| ||
| Recombinant DNA | ||
|
| ||
| p2701-AGER-tdTomato | This paper | Addgene 216470 |
| p2702-AGER-gRNA1 | This paper | Addgene216471 |
| p2703-AGER-gRNA2 | This paper | Addgene216472 |
| p2704-AGER-gRNA3 | This paper | Addgene 216473 |
| p2706-pHAGE-EF1aL-YAP5SA-UBC-GFP | This paper | Addgene 216475 |
| p2709-pHAGE2-Ef1aL-YAP5SA-UBC-tagBFP-loxP | This paper | Addgene216478 |
| p2710-pHAGE2-Ef1aL-WTYAP-UBC-tagBFP-loxP | This paper | Addgene 216479 |
|
| ||
| Software and algorithms | ||
|
| ||
| ImageJ | National Institutes of Health | https://imagej.nih.gov/ij/ |
| Prism | GraphPad | https://www.graphpad.com |
| FlowJo | Becton Dickinson & Company | https://flowjo.com/solutions/flowjo |
|
| ||
| Other | ||
|
| ||
| StemDiff Definitive Endoderm Kit | StemCell Technologies | Cat#05110 |
| Gentle Cell Dissociation Reagent | StemCell Technologies | Cat#07174 |
| 0.4μm pore Polyester Membrane Transwell Insert | Corning | Cat#3470 |
| Accutase | Sigma | Cat#A6964 |
| TaqMan probe: 18S | Thermo Fisher | 4318839 |
| TaqMan probe: SFTPC | Thermo Fisher | Hs00161628_m1 |
| TaqMan probe: NAPSA | Thermo Fisher | Hs00362192_m1 |
| TaqMan probe: NKX2–1 | Thermo Fisher | Hs00968940_m1 |
| TaqMan probe: CYR61 | Thermo Fisher | Hs00155479_m1 |
| TaqMan probe: ANKRD1 | Thermo Fisher | Hs00173317_m1 |
| TaqMan probe: AGER | Thermo Fisher | Hs00542584_g1 |
| TaqMan probe: CAV1 | Thermo Fisher | Hs00971716_m1 |
| TaqMan probe: CLIC5 | Thermo Fisher | Hs00213494_m1 |
| TaqMan probe: PDPN | Thermo Fisher | Hs00366766_m1 |
| TaqMan probe: MKI67 | Thermo Fisher | Hs04260396_g1 |
| TaqMan probe: HOPX | Thermo Fisher | Hs05028646_s1 |
| TaqMan probe: SFTPA1 | Thermo Fisher | Hs00831305_s1 |
| TaqMan probe: SNAI1 | Thermo Fisher | Hs00195591_m1 |
| TaqMan probe: TWIST1 | Thermo Fisher | Hs04989912_s1 |
| TaqMan probe: ECAD (CDH1) | Thermo Fisher | Hs01023895_m1 |
| TaqMan probe: TP63 | Thermo Fisher | Hs00978340_m1 |
| TaqMan probe: SCGB3A2 | Thermo Fisher | Hs00369678_m1 |
| TaqMan probe: CTGF | Thermo Fisher | Hs00170014_m1 |
| TaqMan probe: AXIN2 | Thermo Fisher | Hs00610344_m1 |
| TaqMan probe: LEF1 | Thermo Fisher | Hs01547250_m1 |
| TaqMan probe: VEGFA | Thermo Fisher | Hs00900055_m1 |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Cell lines
Human iAT2s and iAT1s were generated using previously published iPSC lines BU3 NG54 and SPC2-ST-B249,52 as well BU3 NGAT, newly generated as described in the method details. Culture conditions for different cell types (iPSC, iAT2, and iAT1) are described below. All iPSC differentiations were performed under regulatory approval of the Institutional Review Board of Boston University (protocol H33122). The reprogramming and characterization of the original human iPSC clones employed in this study (BU3 and SPC2-ST-B2; aka “SPC2B2”) were previously published.49,52,54 All iPSC lines had a normal karyotype (Cell Line Genetics G-banding analysis), both before and after gene-editing, and were maintained on hESC qualified Matrigel (Corning, 8774552) in feeder-free conditions in mTeSR1 medium (STEMCELL Technologies, 05850) at 37C. Gentle Cell Dissociation Reagent (STEMCELL Technologies, 07174) was used for passaging. All iPSC differentiations were performed under regulatory approval of the Institutional Review Board of Boston University. Additional details and protocols for iPSC derivation, culture, and characterization can be downloaded at https://crem.bu.edu/cores-protocols/. All iPSC lines are available from the CReM repository upon request, https://stemcellbank.bu.edu.
Primary adult AT2 cells (DD0479), a generous gift of Purushothama Rao Tata (Duke University), were cultured in Serum Free Feeder Free (SFFF) medium in 3D Matrigel as reported at 37C.28
METHOD DETAILS
CRISPR targeting of tdTomato to AGER locus in iPSCs
The iPSC line BU3 NG,54 previously engineered to carry an NKX2–1GFP reporter was used for targeting a tdTomato reporter to the human AGER endogenous locus using CRISPR gene editing as follows. Left and right homology arms of about 700bp in length to the left and right of the AGER endogenous start codon were generated by PCR amplification using gDNA extracts from BU3 NG iPSCs. These arms were cloned into our previously published plasmid backbone p1303-DV-SFTPC-tdTomato25 replacing the SFTPC homology arms, generating the p2701-AGER-tdTomato plasmid. Guide RNAs (gRNA1: CACCGCCAGGCTCCAACTGCTGTTC; gRNA2: CACCGATGGCTGCCGGAACAGCAGT; gRNA3: CACCGCTGTGGCCTCCGCCCTAGGT) were selected and cloned into the pSpCas9(BB)-2A-GFP plasmid (Addgene plasmid #48138). BU3 NG iPSCs were pretreated with ROCK Inhibitor (Y-27632) for three hours prior to nucleofection using the Lonza P3 Primary Cell 4DNucleofector™ X Kit (Lonza, cat. no. V4XP-3024) and replated for puromycin resistance screening. Clones were passaged and gDNA was isolated for PCR screening using the following primer pairs (Figure S3): 5’ CTGATCCCCTCAGACATTCTCAGGA 3’ (P5) to 5’ GAGCTGCCGCTGCCGGT 3’ (P6) for outside the left homology arm to the tdTomato cassette and 5’ ACTTGTGTAGCGCCAAGTGC 3’ (P3) to 5’ ACACACACTCGCCTCCTGTT 3’ (P4) for within the puromycin resistance cassette to outside the right homology arm. HEK293 cells that had been transfected with the targeting plasmids or not were used as controls. Clones that showed insertion of tdTomato by PCR were sequenced to confirm insertion, and further expanded. The floxed PGK-Puromycin resistance cassette was excised using transient transfection with Cre plasmid (pHAGE2 EF1aL-Cre-IRES-NeoR-W; plasmid map available at www.kottonlab.com) as previously published25 with transient G418 selection of candidate targeted, Cre-excised clones. Mono-allelic targeting and puromycin resistance excision was confirmed by PCR with primers 5’AGGACTCTTGTCCCAAAGGC 3’ (P1) to 5’ CTGGGGTGTGGGGTTAAAGT 3’ (P2) yielding both a 267bp long band for the untargeted allele and a 2290bp long band for the targeted allele with the puromycin cassette excised. Cells were then assessed by G-banding to identify karyotypically normal clones (Cell Line Genetics).
iPSC differentiation into iAT2s (Alveolospheres)
Human iPSC lines (BU3 NG, BU3 NGAT, SPC2-ST-B2) were differentiated into iAT2s as previously described,50 with detailed protocols and characterizations available for free download at www.kottonlab.com. Briefly, the STEMdiff Definitive Endoderm Kit (STEMCELL Technologies, 05110), was used for differentiation into endoderm, which was scored by co-expression of CKIT and CXCR4 by flow cytometry. Cells were then passaged using Gentle Cell Dissociation Reagent, replated onto Matrigel coated plate, and cultured in “DS/SB” media consisting of complete serum free differentiation media (cSFDM)50 with 2uM Dorsomorphin (“DS”; Stemgent) and 10uM SB431542 (“SB”; Tocris) for 72 hours for anteriorization, the first 24 hours being supplemented with 10 uM Y-27632 (Tocris). Media was then changed to cSFDM with 3 uM CHIR99021 (“C”; Tocris), recombinant human BMP4 (10 ng/ml; “B”; R&D Systems), and 100 nM retinoic acid (“Ra”; Sigma-Aldrich) called “CBRa” for lung specification. Between days 14–16 of differentiation, cells were sorted for lung progenitors either using NKX2–1GFP knock-in reporters or using antibody staining for CPM (FUJIFILM) or CD47hi/CD26neg for lines not containing an NKX2–1 reporter. Sorted cells were resuspended in growth-factor reduced Matrigel (Corning 356231) droplets and covered with alveolar differentiation medium, “CK+DCI” containing a base of cSFDM with 3 uM CHIR99021 (“C”), rhKGF (10 ng/m; “K”; R&D Systems), 50 nM dexamethasone (“D”; Sigma-Aldrich), 0.1 mM 8-bromoadenosine 3′,5′ cyclic monophosphate sodium salt (Sigma-Aldrich), and 0.1 mM 3-isobutyl-1-methylxanthine (IBMX; Sigma-Aldrich) (“CI”). 10 uM Y-27632 (Tocris) was supplemented for 72 hours post sort and cells were refed with CK+DCI every 48–72 hours. For iAT2 maintenance, cells were passaged every 10–14 days as single cells as previously described.50 iAT2s at air-liquid interface were plated onto 6.5mm transwell inserts (Corning) coated with hESC qualified Matrigel (Corning, 8774552), as previously published.67,72
Lentiviral and Adenoviral Transduction
For introduction of lentiviral and adenoviral constructs to iAT2s, alveolospheres were dissociated to single cells as with passaging.50 Cells were then incubated for 4 hours at 37C in suspension with virus in CK+DCI supplemented with 10 uM Y-27632 and 5 ug/mL polybrene. WT YAP and YAP5SA lentiviral transduction was performed at a multiplicity of infection (MOI) of 10, as previously published.50 An MOI of 200 was used for infections with Adeno-Cre-GFP virus. Cells were then replated in Matrigel droplets in CK+DCI.
iAT1 differentiation in “L+DCI” medium
iPSCs were first differentiated into iAT2s and passaged as above in 3D cultures in iAT2 medium (CK+DCI) supplemented for the first 3 days after passaging with 10 uM Y-27632 (Tocris). To generate iAT1s, 3 days after passaging iAT2s in 3D, the medium was replaced with “L+DCI”, consisting of [10 uM LATS-IN-1 (“L”; MedChemExpress Cat. No.: HY-138489), 50 nM dexamethasone (“D”; Sigma-Aldrich), 0.1 mM 8-bromoadenosine 3′,5′ cyclic monophosphate sodium salt (Sigma-Aldrich; “C”), and 0.1 mM 3-isobutyl-1-methylxanthine (IBMX; Sigma-Aldrich; “I”)]. Cells were cultured up to 16 days in L+DCI in 3D while monitoring AGERtdTomato expression as detailed in the text.
iAT1 air-liquid interface (ALI) culture
ALI versions of iAT1 cultures were prepared as follows. To prepare “iAT1 ALI P0” cultures, first a single cell suspension of iAT2s was passaged onto 6.5mm transwell inserts (Corning) coated with hESC qualified Matrigel (Corning, 8774552) as previously published,67,72 but switching CK+DCI medium to L+DCI at the time of plating. To prepare “iAT1 ALI P1” cultures, iAT2s in 3D Matrigel culture were first switched to L+DCI medium in 3D for 9 days before being passaged as single cells onto Matrigel coated transwell inserts for continued L+DCI culture (high density = 200k cells/6.5mm insert, medium density = 100k cells/insert, low density = 50k cells/insert). For all ALI culturing conditions, 10 uM Y-27632 (Tocris) was added to L+DCI media for the first 3 days post passaging and then removed. At the same time (day 3), liquid was aspirated from the apical chamber (air lift) to form the air liquid interface (ALI). Cells in ALI cultures were maintained for up to 10 days (7 days post air lift). Accutase (Innovative Cell Technologies) was used to dissociate cells for FACS analysis and single cell RNA sequencing. iAT2 ALI cultures were plated at 200–250k cells per hESC qualified Matrigel-coated 6.5mm transwell insert in CK+DCI as previously published.67
Primary AT2 culture and differentiation
Primary adult AT2 cells, a generous gift of Purushothama Rao Tata (Duke University), were cultured in Serum Free Feeder Free (SFFF) medium in 3D Matrigel as reported.28 For differentiation experiments, cells were passaged as reported and cultured in SFFF for 7 days before medium was switched to either L+DCI or published human serum containing Alveolar Differentiation Medium (ADM).28
Reverse Transcription quantitative PCR (RT-qPCR)
For RT-qPCR, whole well or sorted cells were collected and stored in Qiazol (Qiagen, 79306) prior to RNA isolation using RNeasy Plus Mini Kit according to the manufacturer’s protocol (Qiagen, 74104). cDNA was then synthesized using MultiScribe Reverse™ Transcriptase (ThermoFisher 4311235). A QuantStudio instrument (Applied Biosciences) and predesigned Taqman probes were used and run in a 384-well format for 40 cycles. Relative expression was normalized to an 18S control and fold change over control cells was calculated using 2−ΔΔCt. Where indicated in the text, RNA extracts from adult primary human distal lung tissue explants, the kind gift of Barry Stripp (Cedars Sinai, Los Angeles), were employed as RT-qPCR controls. RNA was isolated via RNeasy Plus Mini Kit following manufacturer’s protocol.
Flow Cytometry and FACS
0.05% trypsin was used to generate single cell suspensions which were resuspended in sort buffer [HBSS (ThermoFisher) with 2% FBS, 10 uM Y-27632 (Tocris)] with live/dead stain [Calcein blue (Life technologies) or DRAQ7 (Abcam)]. Cells were sorted based on reporter expression: NKX2–1GFP and AGERtdTomato for BU3 NGAT, or SFTPCtdTomato for SPC2B2, as indicated in the text. Cell sorting was performed on a Moflo Astrios EQ (Beckman Coulter) and flow cytometry analysis was performed on an LSRII SORP (BD Biosciences) at the Boston University Flow Cytometry Core Facility. For Edu assays, the Click-iT Plus EdU Alexa Fluor 647 Flow Cytometry Assay Kit (Thermo Fisher Scientific) was used with EdU added 24 hours before cell isolation, and cells fixed in 4% paraformaldehyde (PFA) were analyzed on a Stratedigm (S1000EXI) cytometer with post-processing using FlowJo software (BD Biosciences). For all other analyses, live non-fixed cells were sorted or analyzed as indicated in the text.
Immunofluorescence microscopy
Organoids (3D cultured epithelial spheres or clumps) or cells were fixed in 4% PFA for 20 minutes at room temperature. For whole mount staining, organoids were immediately stained in solution and mounted on cavity slides (Elisco). For sectioning, samples were dehydrated via ethanol and paraffin embedded, or dehydrated using sucrose and frozen in OCT. Prior to staining, paraffin sections were rehydrated, and antigen retrieval was performed using heated citrate buffer. For both paraffin- and cryo-embedded sections, permeabilization was performed using 0.1% Triton and blocking was done with 4% Normal Donkey Serum. Samples were incubated in primary antibody overnight at 4C and incubated in secondary antibody with Hoechst for 2 hrs at RT. Primary antibodies used included ProSFTPC (Pro-SPC): Seven Hills WRAB-9337; HTI-56: Terrace Biotech TB-29AHT1–56; ZO-1: Thermo Fisher Cat# 61–7300; and RFP: Rockland 600–401-379; RAGE: R&D systems AF1145, NKX2–1: Abcam ab76013. Confocal images were taken on either a Leica SP5 or Zeiss LSM 710-Live Duo and were processed using Fiji.
For ALI cross sections, cultures were fixed for 2 hours at RT in 2% glutaraldehyde plus 1% paraformaldehyde in 0.1M cacodylate buffer, pH 7.4 then post fixed overnight at 4C in 1.5% osmium tetroxide. After washing, membranes were cut away from the insert, placed in glass vials and block stained in 1.5% uranyl acetate. Samples were dehydrated through graded acetones, infiltrated in a 50:50 mixture of propylene oxide: resin and embedded in Embed 812. 0.5–1um sections were cut and stained with Toluidine blue.
Western Blots
Samples were lysed in Curtis Lysis Buffer (10mM Tris pH8, 150mM NaCl, 1% Na Deoxycholate, 1% NP-40, 1% SDS, 1mM EDTA) + 1X HALT protease inhibitor (78430, Thermo Fisher Scientific). Sample concentration was measured by BCA and equal amounts of protein were used for each sample. 2X Laemmli sample buffer + 2-Mercaptoethanol was added to each sample for a total volume of 20ul. The samples were then heat denatured for 10 minutes at 95 degrees Celsius. Samples were then loaded onto a 10% SDS-PAGE gel and transferred onto a nitrocellulose membrane (162–0115 Biorad). Primary antibodies were diluted 1:100 in 5% milk and added to the blot overnight (16–18hrs) at 4 degrees Celsius with gentle agitation. The following day the blot was washed 3×10 minutes in 1X TBS-T at room temperature with gentle agitation followed by incubation with the HRP secondary antibody (Anti-Rabbi HRP: #7074, Cell Signaling Technologies or Anti-Goat HRP: 705–036-147 Jackson Immuno Research) diluted 1:5000 in 5% milk at room temperature for 1 hour with gentle agitation. The blot was developed using Femto reagents (PI34096, Thermo Fisher Scientific). The blot was then washed and stripped in a stripping buffer (46430, Thermo Fisher Scientific) for 15 minutes at room temperature with gentle agitation before being stained with new antibody. Primary antibodies used were p-YAP (S127) (13008, Cell Signaling Technology), YAP (14074, Cell Signaling Technology), RAGE (AF1145, R&D Systems) and Hexokinase 1 (19662–1-AP, Proteintech).
VEGFA ELISA
To measure secreted VEGFA protein, conditioned medium after 48 hours of exposure to cells was harvested from each sample indicated in the text and figure legends on day 10 of ALI culture. VEGFA ELISA was performed using the Human VEGFA ELISA kit (Abcam) according to manufacturer’s instructions.
Cell surface area calculations
iAT1 ALI p1 cells were grown in L+DCI medium for 10 days, aspirating apical medium on day 3 (air lift) to form an ALI. Day 10 post plating (7 days after air lift) cells were fixed in 4% PFA for 20 minutes at room temperature and stained for tight junction protein ZO-1 (Thermo Fisher Cat# 61–7300). Randomized Z stack images were taken at 20x in 3 different places of each transwell insert for three transwells and imported into ImageJ. Cell outlines denoted by ZO-1 were traced by hand and area was calculated using ImageJ for around 50 cells per image.
Transepithelial Electrical Resistance
To measure TEER, a Millicell ERS-2 Coltohmmeter (Moillipore Sigma, MERS00002) was used. Electrodes were sterilized by dipping in 70% EtOH followed by conditioned cell media. 200uL media was added to the apical chamber of Transwell culture inserts prior to taking measurements. For each sample, readings were taken at 3 locations in the well. TEER was calculated by subtracting the “blank” Matrigel coated well from the mean and multiplying by the tissue culture growth area.
Generation of YAP5SA and WT YAP lentivirus
Coding sequences for YAP5SA and WT YAP were PCR amplified from pLVX-Tight-Puro-3F-YAP-5SA and pLVX-Tight-Puro-3F-YAP, respectively,73 adding NotI and BglII restriction sites for ligation into a pHAGE2-EF1aL-dsRed-UBC-tagBFP-WPRE backbone in place of the dsRed cassette. We have previously published the dual promoter lentiviral vector for dual transgenesis74 and the pHAGE2 version includes a loxP cassette in the 3’LTR as previously published.75 This cloning strategy generated the following loxP containing plasmids for the current project: pHAGE2-EF1aL-YAP5SA-UBC-tagBFP-WPRE and pHAGE2-EF1aL-WTYAP-UBC-tagBFP-WPRE. The YAP5SA coding sequence was also cloned into a pHAGE1 backbone generating pHAGE1-EF1aL-YAP5SA-UBC-GFP-WPRE. Lentiviral packaging and titering protocols (as we have previously published),74 as well as plasmid maps and primer design protocols for cloning into pHAGE lentiviral backbones are all available at www.kottonlab.com.
Single Cell RNA sequencing of iAT2s and iAT1s
For single cell RNA sequencing of iPSC-derived alveolar epithelial cells, cells were dissociated to single cell suspensions using 0.05% trypsin, or Accutase in the case of ALI cultures. Live cells were sorted via DRAQ7 using Moflo Astrios EQ (Beckman Coulter) at the Boston University Flow Cytometry Core Facility. Single Cell RNA sequencing was then preformed using the Chromium Single Cell 3’system according to manufacturers’ instructions (10x Genomics) at the Single Cell Sequencing Core at Boston University Medical Center. Sequencing files were mapped to the human genome reference (GRCh37) supplemented with GFP, tdTomato, and BFP sequences using CellRanger v3.0.2. Seurat v3.2.376 was used for downstream analysis and quality control. After inspection of the quality control metrics, cells with 15% to 35% of mitochondrial content and <800 detected genes were excluded for downstream analyses. In addition, doublets were also excluded for downstream analysis. We normalized and scaled the unique molecular identifier (UMI) counts using the regularized negative binomial regression (SCTransform).77 Following the standard procedure in Seurat’s pipeline, we performed linear dimensionality reduction (principal component analysis) and used the top 20 principal components to compute the unsupervised Uniform Manifold Approximation and Projection (UMAP).78 For clustering of the cells, we used Louvain algorithm79 which were computed at a range of resolutions from 1.5 to 0.05 (more to fewer clusters). Populations were annotated using Louvain Clustering resolution indicated in the text. Cell cycle scores and classifications were done using the Seurat’s cell-cycle scoring and regression method.80 Cluster specific genes were calculated using MAST framework in Seurat wrapper.81 An online Shiny app82 has been established to allow interactive, user-friendly visualizations of gene expression in each population, as follows:
Figure 3 WT YAP, YAP5SA: https://crem-bu.shinyapps.io/scRNAseq_lentiviral_YAP5SA_iAT1_induction/
Figure 6 L+DCI, YAP5SA, CKDCI: https://crem-bu.shinyapps.io/scRNAseq_iAT1_induction_methods/
Figure 6 L+DCI Time Series: https://crem-bu.shinyapps.io/scRNAseq_iAT1_differentiation_time_series/
Figure 7 3D vs ALI: https://crem-bu.shinyapps.io/scRNAseq_iAT1_3D_ALI_comparison/
For RNA velocity, latent time and Partition-based graph abstraction (PAGA) analysis, we used scVelo.83 For comparing scRNA-seq data generated in this study with existing datasets and computing Jaccard index, we used matchSCore2 R package.84
Single Cell Type Order Parameters (scTOP) analysis was performed as previously published for comparisons between published single cell datasets.64
Primary Single Cell RNA sequencing
Tissue preparation and scRNA sequencing
Samples of normal de-identified human lungs from donors who were not matched for lung transplant, were obtained as described previously37 with the following adaptations. For peripheral tissue experiments, a 3 × 2 cm piece of distal lung tissue was obtained, pleura and visible airways/blood vessels were dissected away, mechanically minced into ~2mm pieces, and processed into a single-cell suspension as described.37 After a single-cell suspension was obtained from the proximal or peripheral tissue, CD45+ immune cells were depleted via MACS LS columns using CD45-microbeads (Miltenyi, 130-045-801) with 2 X 106 cells per column to enhance purity and viability. After CD45 depletion, sorted cells were loaded onto a GemCode instrument (10x Genomics, Pleasanton, CA, USA) to generate single-cell barcoded droplets (GEMs) according to the manufacture’s protocol. The resulting libraries were sequenced on an Illumina HiSeq2500 or NovaSeq instrument.
Analysis of scRNA-seq data
Read were aligned and unique molecular identifier (UMI) counts obtained via the Cell Ranger pipeline (10X Genomics). For further processing, integration and downstream analysis, Seurat V3 (PMID: 29608179) was used. Cells with less than 200 genes, greater than 2 Median absolute deviation above the median, and with potential stress signals of greater than 10% mitochondrial reads were removed. The Cell Cycle phase prediction score was calculated using Seurat function CellCycleScoring, and data was normalized and scaled using the SCTransform function and adjusting for cell cycle, percent mitochondria, number of features per cell, and number of UMI per cell. For integration of individual samples, Canonical Correlation Analysis (CCA) method using SCTransform in Seurat V3; the top 3000 variable genes were used to find anchors. Linear dimension reduction was done via PCA, and the number of PCA dimensions was evaluated and selected based on assessment of an ElbowPlot. Data was clustered using the Louvain graph based algorithm in R and cluster resolution was evaluated using R package ‘clustree’. The Uniform Manifold Projection (UMAP) data reduction algorithm was used to project the cells onto two dimensional coordinates. Subsequently canonical marker genes were used to identify cellular compartments (epithelium, endothelium, mesenchymal, and immune populations). Epithelial clusters were subsetted, and clustering and UMAP reduction were repeated. Clusters were then assigned putative cell types based on annotation with canonical marker genes, or from assessment of top cluster-defining genes based on differential expression (using the FindConservedMarkers function in SeuratV3).
Primary human AT1 gene signatures
Differential gene expression analysis was performed using the Seurat function FindAllMarkers using the MAST algorithm and using the RNA assay for UMIs. The following comparison were performed 1) AT1s vs all lung cells, 2) AT1s vs. lung epithelial cells, and 3) AT1s vs AT2s. The top 50 differentially upregulated genes ordered by LogFC (FDR<0.05) were used as AT1 gene signatures (Table S1), with AT1 vs all lung cells being used as the AT1 gene signature scored in Figures 3, 6, and 7. Similarly, an AT2 50-gene signature (vs all lung cells) was generated and scored in Figure 6. AT1 gene signature (and other signatures) are made using module score function of Seurat.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical methods relevant to each figure are denoted in the figure captions. Unpaired student’s t-tests were used to compare two groups while one-way Analysis of Variance (ANOVA) with Tukey multiple comparisons test was used to compare the means between three or more groups. Analysis was performed using GraphPad Prism and p<0.05 was used to determine statistical significance unless otherwise indicated in the text.
Supplementary Material
Highlights.
Transcriptomic analysis of primary human AT1s shows enrichment of YAP/TAZ signaling
Nuclear YAP activation is sufficient to drive an AT1 transcriptomic shift
An AGERtdTomato reporter iPSC line allows tracking and isolation of human iAT1s
iAT1s form a flat epithelial barrier in vitro in air-liquid interface cultures
ACKNOWLEDGMENTS
The authors wish to thank the Kotton, Varelas, and Morrisey Labs for insightful discussions. We thank Yuriy Alekseyev and the BU sequencing core, as well as Brian R. Tilton for flow cytometry. We are grateful to Greg Miller and Marianne James of the Center for Regenerative Medicine (CReM) for maintenance and characterization of iPSCs, supported by NIH grants N01 75N92020C00005, 1UL1TR001430, and U01TR001810. We thank the Tata Lab (Duke University, Durham, NC) for the gift of cultured primary AT2 cells. Schematics were created with BioRender.com. This study was supported by NIH grants 1F31HL158193, T32HL007035, and PCTC_JC_202003 to C.L.B.; a Pulmonary Fibrosis Foundation Award, a Boston University Department of Medicine Career Investment Award, and NIH K08HL163494 to K.-D.A.; R01HL124392 to X.V.; and U01HL134745, U01HL134766, U01HL152976, R01HL095993, and P01HL170952 to D.N.K.
Footnotes
DECLARATION OF INTERESTS
The authors have filed a patent application related to the generation of human alveolar epithelial type I cells from pluripotent stem cells detailed in this manuscript.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.stem.2024.03.017.
REFERENCES
- 1.Weibel ER (2015). On the tricks alveolar epithelial cells play to make a good lung. Am. J. Respir. Crit. Care Med. 191, 504–513. 10.1164/rccm.201409-1663OE. [DOI] [PubMed] [Google Scholar]
- 2.Habermann AC, Gutierrez AJ, Bui LT, Yahn SL, Winters NI, Calvi CL, Peter L, Chung MI, Taylor CJ, Jetter C, et al. (2020). Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci. Adv. 6, eaba1972. 10.1126/sciadv.aba1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kobayashi Y, Tata A, Konkimalla A, Katsura H, Lee RF, Ou J, Banovich NE, Kropski JA, and Tata PR (2020). Persistence of a regeneration-associated, transitional alveolar epithelial cell state in pulmonary fibrosis. Nat. Cell Biol. 22, 934–946. 10.1038/S41556-020-0542-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi J, Park JE, Tsagkogeorga G, Yanagita M, Koo BK, Han N, and Lee JH (2020). Inflammatory Signals Induce AT2 Cell-Derived Damage-Associated Transient Progenitors that Mediate Alveolar Regeneration. Cell Stem Cell 27, 366–382.e7. 10.1016/J.STEM.2020.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans MJ, Cabral LJ, Stephens RJ, and Freeman G (1975). Transformation of alveolar Type 2 cells to Type 1 cells following exposure to NO2. Exp. Mol. Pathol. 22, 142–150. 10.1016/0014-4800(75)90059-3. [DOI] [PubMed] [Google Scholar]
- 6.Adamson IYR, and Bowden DH (1974). The type 2 cell as progenitor of alveolar epithelial regeneration. A cytodynamic study in mice after exposure to oxygen. Lab. Invest. 30, 35–42. [PubMed] [Google Scholar]
- 7.Barkauskas CE, Cronce MJ, Rackley CR, Bowie EJ, Keene DR, Stripp BR, Randell SH, Noble PW, and Hogan BLM (2013). Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 123, 3025–3036. 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zacharias WJ, Frank DB, Zepp JA, Morley MP, Alkhaleel FA, Kong J, Zhou S, Cantu E, and Morrisey EE (2018). Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature 555, 251–255. 10.1038/nature25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rock JR, Barkauskas CE, Cronce MJ, Xue Y, Harris JR, Liang J, Noble PW, and Hogan BLM (2011). Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc. Natl. Acad. Sci. USA 108, E1475–E1483. 10.1073/pnas.1117988108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desai TJ, Brownfield DG, and Krasnow MA (2014). Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature 507, 190–194. 10.1038/nature12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao L, Yee M, and O’Reilly MA (2013). Transdifferentiation of alveolar epithelial type II to type I cells is controlled by opposing TGF-β and BMP signaling. Am. J. Physiol. Lung Cell. Mol. Physiol. 305, L409–L418. 10.1152/ajplung.00032.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riemondy KA, Jansing NL, Jiang P, Redente EF, Gillen AE, Fu R, Miller AJ, Spence JR, Gerber AN, Hesselberth JR, et al. (2019). Single-cell RNA sequencing identifies TGF-β as a key regenerative cue following LPS-induced lung injury. JCI Insight 5, e123637. 10.1172/jci.insight.123637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z, Wu H, Jiang K, Wang Y, Zhang W, Chu Q, Li J, Huang H, Cai T, Ji H, et al. (2016). MAPK-Mediated YAP Activation Controls Mechanical-Tension-Induced Pulmonary Alveolar Regeneration. Cell Rep. 16, 1810–1819. 10.1016/j.celrep.2016.07.020. [DOI] [PubMed] [Google Scholar]
- 14.Nabhan AN, Brownfield DG, Harbury PB, Krasnow MA, and Desai TJ (2018). Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science 359, 1118–1123. 10.1126/science.aam6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung MI, Bujnis M, Barkauskas CE, Kobayashi Y, and Hogan BLM (2018). Niche-mediated BMP/SMAD signaling regulates lung alveolar stem cell proliferation and differentiation. Development 145, dev163014. 10.1242/dev.163014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Little DR, Gerner-Mauro KN, Flodby P, Crandall ED, Borok Z, Akiyama H, Kimura S, Ostrin EJ, and Chen J (2019). Transcriptional control of lung alveolar type 1 cell development and maintenance by NK homeobox 2–1. Proc. Natl. Acad. Sci. USA 116, 20545–20555. 10.1073/pnas.1906663116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borok Z, Lubman RL, Danto SI, Zhang XL, Zabski SM, King LS, Lee DM, Agre P, and Crandall ED (1998). Keratinocyte growth factor modulates alveolar epithelial cell phenotype in vitro: expression of aquaporin 5. Am. J. Respir. Cell Mol. Biol. 18, 554–561. 10.1165/AJRCMB.18.4.2838. [DOI] [PubMed] [Google Scholar]
- 18.Sun T, Huang Z, Zhang H, Posner C, Jia G, Ramalingam TR, Xu M, Brightbill H, Egen JG, Dey A, et al. (2019). TAZ is required for lung alveolar epithelial cell differentiation after injury. JCI Insight 5, e128674. 10.1172/JCI.INSIGHT.128674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Little DR, Lynch AM, Yan Y, Akiyama H, Kimura S, and Chen J (2021). Differential chromatin binding of the lung lineage transcription factor NKX2–1 resolves opposing murine alveolar cell fates in vivo. Nat. Commun. 12, 2509. 10.1038/S41467-021-22817-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Penkala IJ, Liberti DC, Pankin J, Sivakumar A, Kremp MM, Jayachandran S, Katzen J, Leach JP, Windmueller R, Stolz K, et al. (2021). Age-dependent alveolar epithelial plasticity orchestrates lung homeostasis and regeneration. Cell Stem Cell 28, 1775–1789.e5. 10.1016/j.stem.2021.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Edeen K, Manzer R, Chang Y, Wang S, Chen X, Funk CJ, Cosgrove GP, Fang X, and Mason RJ (2007). Differentiated Human Alveolar Epithelial Cells and Reversibility of their Phenotype In vitro. Am. J. Respir. Cell Mol. Biol. 36, 661–668. 10.1165/rcmb.2006-0410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fehrenbach H, Kasper M, Tschernig T, Pan T, Schuh D, Shannon JM, Müller M, and Mason RJ (1999). Keratinocyte growth factor-induced hyperplasia of rat alveolar type II cells in vivo is resolved by differentiation into type I cells and by apoptosis. Eur. Respir. J. 14, 534–544. 10.1034/j.1399-3003.1999.14c10.x. [DOI] [PubMed] [Google Scholar]
- 23.Qiao R, Yan W, Clavijo C, Mehrian-Shai R, Zhong Q, Kim KJ, Ann D, Crandall ED, and Borok Z (2008). Effects of KGF on alveolar epithelial cell transdifferentiation are mediated by JNK signaling. Am. J. Respir. Cell Mol. Biol. 38, 239–246. 10.1165/rcmb.2007-0172OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Danto SI, Shannon JM, Borok Z, Zabski SM, and Crandall ED (1995). Reversible transdifferentiation of alveolar epithelial cells. Am. J. Respir. Cell Mol. Biol. 12, 497–502. 10.1165/ajrcmb.12.5.7742013. [DOI] [PubMed] [Google Scholar]
- 25.Jacob A, Morley M, Hawkins F, McCauley KB, Jean JC, Heins H, Na CL, Weaver TE, Vedaie M, Hurley K, et al. (2017). Differentiation of Human Pluripotent Stem Cells into Functional Lung Alveolar Epithelial Cells. Cell Stem Cell 21, 472–488.e10. 10.1016/j.stem.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alysandratos KD, Garcia-De-Alba C, Yao C, Pessina P, Huang J, Villacorta-Martin C, Hix OT, Minakin K, Burgess CL, Bawa P, et al. (2023). Culture impact on the transcriptomic programs of primary and iPSC-derived human alveolar type 2 cells. JCI Insight 8, e158937. 10.1172/JCI.INSIGHT.158937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamamoto Y, Gotoh S, Korogi Y, Seki M, Konishi S, Ikeo S, Sone N, Nagasaki T, Matsumoto H, Muro S, et al. (2017). Long-term expansion of alveolar stem cells derived from human iPS cells in organoids. Nat. Methods 14, 1097–1106. 10.1038/nmeth.4448. [DOI] [PubMed] [Google Scholar]
- 28.Katsura H, Sontake V, Tata A, Kobayashi Y, Edwards CE, Heaton BE, Konkimalla A, Asakura T, Mikami Y, Fritch EJ, et al. (2020). Human Lung Stem Cell-Based Alveolospheres Provide Insights into SARS-CoV-2-Mediated Interferon Responses and Pneumocyte Dysfunction. Cell Stem Cell 27, 890–904.e8. 10.1016/J.STEM.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kadur Lakshminarasimha Murthy P, Sontake V, Tata A, Kobayashi Y, Macadlo L, Okuda K, Conchola AS, Nakano S, Gregory S, Miller LA, et al. (2022). Human distal lung maps and lineage hierarchies reveal a bipotent progenitor. Nature 604, 111–119. 10.1038/S41586-022-04541-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanagaki S, Ikeo S, Suezawa T, Yamamoto Y, Seki M, Hirai T, Hagiwara M, Suzuki Y, and Gotoh S (2021). Directed induction of alveolar type I cells derived from pluripotent stem cells via Wnt signaling inhibition. Stem Cells 39, 156–169. 10.1002/STEM.3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salahudeen AA, Choi SS, Rustagi A, Zhu J, van Unen V, de la O SM., Flynn RA., Margalef-Català M., Santos AJM., Ju J., et al. (2020). Progenitor identification and SARS-CoV-2 infection in human distal lung organoids. Nature 588, 670–675. 10.1038/s41586-020-3014-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matthay MA, Zemans RL, Zimmerman GA, Arabi YM, Beitler JR, Mercat A, Herridge M, Randolph AG, and Calfee CS (2019). Acute respiratory distress syndrome. Nat. Rev. Dis. Primers 5, 18. 10.1038/S41572-019-0069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strunz M, Simon LM, Ansari M, Kathiriya JJ, Angelidis I, Mayr CH, Tsidiridis G, Lange M, Mattner LF, Yee M, et al. (2020). Alveolar regeneration through a Krt8+ transitional stem cell state that persists in human lung fibrosis. Nat. Commun. 11, 3559. 10.1038/s41467-020-17358-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jain R, Barkauskas CE, Takeda N, Bowie EJ, Wang Q, Padmanabhan A, Manderfield LJ, Li D, Li L, Trivedi CM, et al. (2015). Plasticity of Hopx+ type I alveolar cells to regenerate type II cells in the lung. Nat. Commun. 6, 6727. 10.1038/ncomms7727.Plasticity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chung MI, and Hogan BLM (2018). AGER-CreERT2: A new genetic tool for studying lung alveolar development, homeostasis, and repair. Am. J. Respir. Cell Mol. Biol. 59, 706–712. 10.1165/rcmb.2018-0125OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flodby P, Borok Z, Banfalvi A, Zhou B, Gao D, Minoo P, Ann DK, Morrisey EE, and Crandall ED (2010). Directed Expression of Cre in Alveolar Epithelial Type 1 Cells. Am. J. Respir. Cell Mol. Biol. 43, 173–178. 10.1165/rcmb.2009-0226OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Basil MC, Cardenas-Diaz FL, Kathiriya JJ, Morley MP, Carl J, Brumwell AN, Katzen J, Slovik KJ, Babu A, Zhou S, et al. (2022). Human distal airways contain a multipotent secretory cell that can regenerate alveoli. Nature 604, 120–126. 10.1038/s41586-022-04552-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Treutlein B, Brownfield DG, Wu AR, Neff NF, Mantalas GL, Espinoza FH, Desai TJ, Krasnow MA, and Quake SR (2014). Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 509, 371–375. 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun X, Perl AK, Li R, Bell SM, Sajti E, Kalinichenko VV, Kalin TV, Misra RS, Deshmukh H, Clair G, et al. (2022). A census of the lung: CellCards from LungMAP. Dev. Cell 57, 112–145.e2. 10.1016/J.DEVCEL.2021.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Tang Z, Huang H, Li J, Wang Z, Yu Y, Zhang C, Li J, Dai H, Wang F, et al. (2018). Pulmonary alveolar type I cell population consists of two distinct subtypes that differ in cell fate. Proc. Natl. Acad. Sci. USA 115, 2407–2412. 10.1073/pnas.1719474115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du Y, Kitzmiller JA, Sridharan A, Perl AK, Bridges JP, Misra RS, Pryhuber GS, Mariani TJ, Bhattacharya S, Guo M, et al. (2017). Lung Gene Expression Analysis (LGEA): an integrative web portal for comprehensive gene expression data analysis in lung development. Thorax 72, 481–484. 10.1136/THORAXJNL-2016-209598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ardini-Poleske ME, Clark RF, Ansong C, Carson JP, Corley RA, Deutsch GH, Hagood JS, Kaminski N, Mariani TJ, Potter SS, et al. (2017). LungMAP: The molecular atlas of lung development program. Am. J. Physiol. Lung Cell. Mol. Physiol. 313, L733–L740. 10.1152/ajplung.00139.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frank DB, Penkala IJ, Zepp JA, Sivakumar A, Linares-Saldana R, Zacharias WJ, Stolz KG, Pankin J, Lu M, Wang Q, et al. (2019). Early lineage specification defines alveolar epithelial ontogeny in the murine lung. Proc. Natl. Acad. Sci. USA 116, 4362–4371. 10.1073/pnas.1813952116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gokey JJ, Snowball J, Sridharan A, Sudha P, Kitzmiller JA, Xu Y, and Whitsett JA (2021). YAP regulates alveolar epithelial cell differentiation and AGER via NFIB/KLF5/NKX2–1. iScience 24, 102967. 10.1016/J.ISCI.2021.102967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nantie LB, Young RE, Paltzer WG, Zhang Y, Johnson RL, Verheyden JM, and Sun X (2018). Lats1/2 inactivation reveals Hippo function in alveolar type I cell differentiation during lung transition to air breathing. Development 145, dev163105. 10.1242/dev.163105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Soldt BJ, Qian J, Li J, Tang N, Lu J, and Cardoso WV (2019). Yap and its subcellular localization have distinct compartment-specific roles in the developing lung. Development 146, dev175810. 10.1242/dev.175810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shiraishi K, Shah PP, Morley MP, Loebel C, Santini GT, Katzen J, Basil MC, Lin SM, Planer JD, Cantu E, et al. (2023). Biophysical forces mediated by respiration maintain lung alveolar epithelial cell fate. Cell 186, 1478–1492.e15. 10.1016/J.CELL.2023.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. (2007). Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747–2761. 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alysandratos KD, Russo SJ, Petcherski A, Taddeo EP, Acín-Pérez R, Villacorta-Martin C, Jean JC, Mulugeta S, Rodriguez LR, Blum BC, et al. (2021). Patient-specific iPSCs carrying an SFTPC mutation reveal the intrinsic alveolar epithelial dysfunction at the inception of interstitial lung disease. Cell Rep. 36, 109636. 10.1016/J.CELREP.2021.109636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jacob A, Vedaie M, Roberts DA, Thomas DC, Villacorta-Martin C, Alysandratos KD, Hawkins F, and Kotton DN (2019). Derivation of self-renewing lung alveolar epithelial type II cells from human pluripotent stem cells. Nat. Protoc. 14, 3303–3332. 10.1038/S41596-019-0220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun YL, Hurley K, Villacorta-Martin C, Huang J, Hinds A, Gopalan K, Caballero IS, Russo SJ, Kitzmiller JA, Whitsett JA, et al. (2021). Heterogeneity in Human Induced Pluripotent Stem Cell-derived Alveolar Epithelial Type II Cells Revealed with ABCA3/SFTPC Reporters. Am. J. Respir. Cell Mol. Biol. 65, 442–460. 10.1165/rcmb.2020-0259OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hurley K, Ding J, Villacorta-Martin C, Herriges MJ, Jacob A, Vedaie M, Alysandratos KD, Sun YL, Lin C, Werder RB, et al. (2020). Reconstructed Single-Cell Fate Trajectories Define Lineage Plasticity Windows during Differentiation of Human PSC-Derived Distal Lung Progenitors. Cell Stem Cell 26, 593–608.e8. 10.1016/J.STEM.2019.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jones RC, Karkanias J, Krasnow MA, Pisco AO, Quake SR, Salzman J, Yosef N, Bulthaup B, Brown P, Harper W, et al. (2022). The Tabula Sapiens: A multiple-organ, single-cell transcriptomic atlas of humans. Science 376, eabl4896. 10.1126/science.abl4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hawkins F, Kramer P, Jacob A, Driver I, Thomas DC, Mccauley KB, Skvir N, Crane AM, Kurmann AA, Hollenberg AN, et al. (2017). Prospective isolation of NKX2–1–expressing human lung progenitors derived from pluripotent stem cells. J. Clin. Invest. 127, 2277–2294. 10.1172/JCI89950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kastan N, Gnedeva K, Alisch T, Petelski AA, Huggins DJ, Chiaravalli J, Aharanov A, Shakked A, Tzahor E, Nagiel A, et al. (2021). Small-molecule inhibition of Lats kinases may promote Yap-dependent proliferation in postmitotic mammalian tissues. Nat. Commun. 12, 3100. 10.1038/s41467-021-23395-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kastan NR, Oak S, Liang R, Baxt L, Myers RW, Ginn J, Liverton N, Huggins DJ, Pichardo J, Paul M, et al. (2022). Development of an improved inhibitor of Lats kinases to promote regeneration of mammalian organs. Proc. Natl. Acad. Sci. USA 119, e2206113119. 10.1073/pnas.2206113119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCauley KB, Hawkins F, Serra M, Thomas DC, Jacob A, and Kotton DN (2017). Efficient Derivation of Functional Human Airway Epithelium from Pluripotent Stem Cells via Temporal Regulation of Wnt Signaling. Cell Stem Cell 20, 844–857.e6. 10.1016/j.stem.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y, Xu X, Maglic D, Dill MT, Mojumdar K, Ng PKS, Jeong KJ, Tsang YH, Moreno D, Bhavana VH, et al. (2018). Comprehensive Molecular Characterization of the Hippo Signaling Pathway in Cancer. Cell Rep. 25, 1304–1317.e5. 10.1016/J.CELREP.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adams TS, Schupp JC, Poli S, Ayaub EA, Neumark N, Ahangari F, Chu SG, Raby BA, DeIuliis G, Januszyk M, et al. (2020). Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci. Adv. 6, eaba1983. 10.1126/sciadv.aba1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kathiriya JJ, Wang C, Zhou M, Brumwell A, Cassandras M, Le Saux CJ, Cohen M, Alysandratos KD, Wang B, Wolters P, et al. (2022). Human alveolar type 2 epithelium transdifferentiates into meta-plastic KRT5+ basal cells. Nat. Cell Biol. 24, 10–23. 10.1038/S41556-021-00809-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang P, de Gil de Rubio RG., Hrycaj SM., Gurczynski SJ., Riemondy KA., Moore BB., Omary MB., Ridge KM., and Zemans RL. (2020). Ineffectual Type 2-to-Type 1 Alveolar Epithelial Cell Differentiation in Idiopathic Pulmonary Fibrosis: Persistence of the KRT8hi Transitional State. Am. J. Respir. Crit. Care Med. 201, 1443–1447. 10.1164/rccm.201909-1726LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Negretti NM, Plosa EJ, Benjamin JT, Schuler BA, Habermann AC, Jetter CS, Gulleman P, Bunn C, Hackett AN, Ransom M, et al. (2021). A single-cell atlas of mouse lung development. Development 148, dev199512. 10.1242/DEV.199512/273783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zepp JA, Morley MP, Loebel C, Kremp MM, Chaudhry FN, Basil MC, Leach JP, Liberti DC, Niethamer TK, Ying Y, et al. (2021). Genomic, epigenomic, and biophysical cues controlling the emergence of the lung alveolus. Science 371, eabc3172. 10.1126/science.abc3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yampolskaya M, Herriges MJ, Ikonomou L, Kotton DN, and Mehta P (2023). scTOP: physics-inspired order parameters for cellular identification and visualization. Development 150, dev201873. 10.1242/dev.201873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.He P, Lim K, Sun D, Pett JP, Jeng Q, Polanski K, Dong Z, Bolt L, Richardson L, Mamanova L, et al. (2022). A human fetal lung cell atlas uncovers proximal-distal gradients of differentiation and key regulators of epithelial fates. Cell 185, 4841–4860.e25. 10.1016/J.CELL.2022.11.005. [DOI] [PubMed] [Google Scholar]
- 66.Herriges MJ, Yampolskaya M, Thapa BR, Lindstrom-Vautrin J, Wang F, Huang J, Na CL, Ma L, Montminy MM, Bawa P, et al. (2023). Durable alveolar engraftment of PSC-derived lung epithelial cells into immunocompetent mice. Cell Stem Cell 30, 1217–1234.e7. 10.1016/j.stem.2023.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Abo KM, Sainz de Aja JS, Lindstrom-Vautrin J, Alysandratos KD, Richards A, Garcia-De-Alba C, Huang J, Hix OT, Werder RB, Bullitt E, et al. (2022). Air-liquid interface culture promotes maturation and allows environmental exposure of pluripotent stem cell-derived alveolar epithelium. JCI Insight 7, e155589. 10.1172/JCI.INSIGHT.155589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schneider JP, Wrede C, Hegermann J, Weibel ER, Mühlfeld C, and Ochs M (2019). On the topological complexity of human alveolar epithelial type 1 cells. Am. J. Respir. Crit. Care Med. 199, 1153–1156. 10.1164/rccm.201810-1866LE. [DOI] [PubMed] [Google Scholar]
- 69.Crapo JD, Barry BE, Gehr P, Bachofen M, and Weibel ER (1982). Cell number and cell characteristics of the normal human lung. Am. Rev. Respir. Dis. 126, 332–337. 10.1164/ARRD.1982.126.2.332. [DOI] [PubMed] [Google Scholar]
- 70.Wang F, Ting C, Riemondy KA, Douglas M, Foster K, Patel N, Kaku N, Linsalata A, Nemzek J, Varisco BM, et al. (2023). Regulation of epithelial transitional states in murine and human pulmonary fibrosis. J. Clin. Invest. 133, e165612. 10.1172/JCI165612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Evans MJ, Cabral LJ, Stephens RJ, and Freeman G (1973). Renewal of Alveolar Epithelium in the Rat Following Exposure to NO2. Am. J. Pathol. 70, 175–198. [PMC free article] [PubMed] [Google Scholar]
- 72.Huang J, Hume AJ, Abo KM, Werder RB, Villacorta-Martin C, Alysandratos KD, Beermann M, Lou S-R,C., Lindstrom-Vautrin J., et al. (2020). SARS-CoV-2 Infection of Pluripotent Stem Cell-Derived Human Lung Alveolar Type 2 Cells Elicits a Rapid Epithelial-Intrinsic Inflammatory Response. Cell Stem Cell 27, 962–973.e7. 10.1016/j.stem.2020.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hiemer SE, Szymaniak AD, and Varelas X (2014). The Transcriptional Regulators TAZ and YAP Direct Transforming Growth Factor β-induced Tumorigenic Phenotypes in Breast Cancer Cells. J. Biol. Chem. 289, 13461–13474. 10.1074/jbc.M113.529115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wilson AA, Kwok LW, Hovav AH, Ohle SJ, Little FF, Fine A, and Kotton DN (2008). Sustained expression of alpha1-antitrypsin after transplantation of manipulated hematopoietic stem cells. Am. J. Respir. Cell Mol. Biol. 39, 133–141. 10.1165/rcmb.2007-0133OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sommer CA, Sommer AG, Longmire TA, Christodoulou C, Thomas DD, Gostissa M, Alt FW, Murphy GJ, Kotton DN, and Mostoslavsky G (2010). Excision of reprogramming transgenes improves the differentiation potential of iPS cells generated with a single excisable vector. Stem Cells 28, 64–74. 10.1002/STEM.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21. 10.1016/J.CELL.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hafemeister C, and Satija R (2019). Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296. 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mcinnes L, Healy J, and Melville J (2020). UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. 10.1016/abs/1802.03426. [DOI] [Google Scholar]
- 79.Blondel VD, G JL., Lambiotte R., and Lefebvre E. (2008). Fast unfolding of communities in large networks. J. Stat. Mech. Theor. Exp. 2008, 10008. 10.1088/1742-5468/2008/10/P10008. [DOI] [Google Scholar]
- 80.Tirosh I, Izar B, Prakadan SM, Wadsworth MH, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, et al. (2016). Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196. 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, et al. (2015). MAST: A flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278. 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ouyang JF, Kamaraj US, Cao EY, and Rackham OJL (2021). ShinyCell: simple and sharable visualization of single-cell gene expression data. Bioinformatics 37, 3374–3376. 10.1093/BIOINFORMATICS/BTAB209. [DOI] [PubMed] [Google Scholar]
- 83.Bergen V, Lange M, Peidli S, Wolf FA, and Theis FJ (2020). Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 38, 1408–1414. 10.1038/s41587-020-0591-3. [DOI] [PubMed] [Google Scholar]
- 84.Mereu E, Lafzi A, Moutinho C, Ziegenhain C, McCarthy DJ, Álvarez-Varela A, Batlle E, Sagar, Grün D., Lau JK., et al. (2020). Benchmarking single-cell RNA-sequencing protocols for cell atlas projects. Nat. Biotechnol. 38, 747–755. 10.1038/s41587-020-0469-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell RNA-seq datasets have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Interactive visualizations of these datasets will also be available on the Kotton Lab’s Bioinformatics Portal at http://www.kottonlab.com. This paper does not report original code. Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Rabbit anti-NKX2–1 antibody | Abcam | Cat#ab76013; RRID:AB_1310784 |
| Goat anti-RFP antibody | My Biosource | Cat#MBS448122 |
| Rabbit anti-proSFTPC antibody | Seven Hills | Cat#r76694 |
| Goat anti-RAGE antibody | R&D Systems | Cat#AF1145 |
| Mouse anti-HT1–56 antibody | Terrace Biotech | SKU: TB-29AHT1–56 |
| Rabbit anti-ZO-1 antibody | ThermoFisher | Cat#61–7300; RRID:AB_2533938 |
| Rabbit anti-YAP antibody | Cell Signaling Technology | Cat#14074 |
| Rabbit anti-phospho-YAP antibody | Cell Signaling Technology | Cat#13008 |
| Rabbit anti-Hexokinase1 antibody | Proteintech | Cat#19662–1-AP; RRID:AB_10859778 |
| Alexa Fluor 647 donkey anti-rabbit | Invitrogen | Cat#A31573; RRID:AB_2536183 |
| Alexa Fluor 488 donkey anti-mouse | Invitrogen | Cat#A21202; RRID:AB_141607 |
| Alexa Fluor 647 donkey anti-goat | Invitrogen | Cat#A21447; RRID:AB_2535864 |
| Alexa Fluor 546 donkey anti-goat | Invitrogen | Cat#A11056; RRID:AB_2534103 |
| Alexa Fluor 488 donkey anti-rabbit | Invitrogen | Cat#A21208; RRID:AB_2535794 |
| Alexa Fluor 488 donkey anti-goat | Invitrogen | Cat#A11055; RRID:AB_2534102 |
| Anti-rabbit IgG, HRP-linked antibody | Cell Signaling Technology | Cat#7074 |
| Anti-goat IgG, HRP-linked antibody | Jackson ImmunoRsearch | Cat#705–036-147; RRID:AB_2340392 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| Cre Recombinase Adenovirus | Vector Biolabs | Cat#1700 |
|
| ||
| Biological samples | ||
|
| ||
| Primary Human Lung samples | Gift from Dr. Barry Stripp, Cedars Sinai | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Growth Factor Reduced Matrigel | Corning | Cat#356234 |
| mTeSR 1 complete kit | StemCell Technologies | Cat#85850 |
| DMEM | Gibco | Cat#2414671 |
| IMDM | ThermoFisher | Cat#12440053 |
| Glutamax | Life Technologies | Cat#35050–061 |
| Ham's F12 | Cellgro | Cat#10–080-CV |
| B27 supplement | Invitrogen | Cat#17504–44 |
| N2 supplement | Invitrogen | Cat#17502–048 |
| Primocin | Invivogen | Cat#ANTPM1 |
| 7.5% BSA Fraction V | ThermoFisher | Cat#15260–037 |
| 1-thioglycerol (MTG) | Sigma | Cat#M6145 |
| Ascorbic Acid | Sigma | Cat#A4544 |
| SB431542 | Sigma | Cat#S4317 |
| Y-27632 (ROCK inhibitor) | Tocris | Cat#1254 |
| Growth Factor Reduced 3D Matrigel | Corning | Cat#354230 |
| Dispase | Gibco | Cat#17105–041 |
| Retinoic acid | Sigma | Cat#R2625 |
| 0.05% Trypsin-EDTA | Gibco | Cat#25300062 |
| Fetal bovine serum | Gibco | Cat#16141079 |
| Paraformaldehyde | Electron Microscopy Sciences | Cat#19208 |
| Normal donkey serum | Sigma | Cat#D9663 |
| Antigen Unmasking Solution, Citric Acid Based | Vector Laboratories | Cat#H-3300–250 |
| Hoechst 33342 | Thermo Fisher | Cat#H3570 |
| Triton-X | Sigma | Cat#T9284 |
| Calcein Blue | ThermoFisher | Cat#C1429 |
| DRAQ7 | Abcam | Cat#ab109202 |
| Dorsomorphin | Fisher Scientific | Cat#NC0275327 |
| CHIR99021 | Fisher Scientific | Cat#44–235-0 |
| rhKGF (FGF7) | Fisher Scientific | Cat#251KG050 |
| rhBMP4 | R&D systems | Cat#314-BP |
| rh IL-1beta | Gibco | Cat#200–01B |
| rhFGF18 | Gibco | Cat#100–28 |
| TRULI (LATS-IN-1) | MedChem Express | Cat#HY-138489 |
| TDI-011536 | MedChem Express | Cat#HY-150042 |
| Dexamethasone | Sigma | Cat#D4902 |
| 8-bromoadenosine 30,50-cyclic monophosphate sodium salt (cAMP) | Sigma | Cat#B7880 |
| 3-Isobutyl-1-methylxanthine (IBMX) | Sigma | Cat#I5879 |
| Polybrene | Tocris | Cat#7711 |
| Phalloidin (Texas Red) | Invitrogen | Cat#T7471 |
| Puromycin Dihydrochloride | ThermoFisher Scientific | Cat#A1113802 |
| Geneticin Sulfate | Life Technologies | Cat#11811–023 |
| Dimethyl Sulfoxide (DMSO) | Sigma Aldrich | Cat#D2650 |
| ProLong Diamond Antifade Mountant | Invitrogen | Cat#P36965 |
| West Femto Maximum Sensitivity Substrate | Thermo Scientific | Cat#PI34096 |
| Restore PLUS Western Blot Stripping Buffer | Thermo Scientific | Cat#46430 |
| Human Serum | Sigma-Aldrich | Cat#H6914 |
|
| ||
| Critical commercial assays | ||
|
| ||
| RNeasy Mini Kit | QIAGEN | Cat#74104 |
| QIAzol Lysis Reagent | QIAGEN | Cat#79306 |
| TaqMan Fast Universal PCR Master Mix (2X) | Thermo Fisher | Cat#4364103 |
| High-Capacity cDNA Reverse Transcription Kit | Applied Biosytems | Cat#4368814 |
| P3 Primary Cell 4D-Nucleofector X Kit | Lonza | Cat#V4XP-3032 |
| Human VEGFA ELISA Kit | Abcam | Cat#119566 |
| Click-iT EdU Cell Proliferation Kit | ThermoFisher | Cat#C10340 |
|
| ||
| Deposited data | ||
|
| ||
| scRNAseq data: WT YAP, YAP5SA (Figure 3) | This paper | GEO: GSE221342 |
| scRNAseq data: L+DCI, YAP5SA, CK+DCI (Figure 6) | This paper | GEO: GSE221343 |
| scRNAseq data: L+DCI time series (Figure 6) | This paper | GEO: GSE246243 |
| scRNAseq data: 3D vs ALI (Figure 7) | This paper | GEO: GSE221344 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Human: Donor iPSC line targeted with SFTPCtdTomato (SPC2-ST-B2) | Kotton Lab; Hurley et al.52 | http://stemcellbank.bu.edu |
| Human: Normal donor iPSC line targeted with NKX2–1gfp (BU3 NG) | Kotton Lab; Hawkins et al.54 | http://stemcellbank.bu.edu |
| Human: Normal donor iPSC line targeted with NKX2–1gfp and AGERtdTomato (BU3 NGAT) | This paper | http://stemcellbank.bu.edu |
|
| ||
| Oligonucleotides | ||
|
| ||
| Oligonucleotide primers | Integrated DNA Technologies | N/A |
| P1 (AGER left homology arm) | This paper | 5'AGGACTCTTGTCCCAAAGGC 3' |
| P2 (AGER right homology arm) | This paper | 5'CTGGGGTGTGGGGTTAAAGT 3' |
| P3 (Puromycin resistance Fwd) | This paper | 5'ACTTGTGTAGCGCCAAGTGC 3' |
| P4 (Puromycin resistance Rev) | This paper | 5'ACACACACTCGCCTCCTGTT 3' |
| P5 (tdTomato insert Fwd) | This paper | 5'CTGATCCCCTCAGACATTCTCAGGA 3' |
| P6 (tdTomato insert Rev) | This paper | 5'GAGCTGCCGCTGCCGGT 3' |
|
| ||
| Recombinant DNA | ||
|
| ||
| p2701-AGER-tdTomato | This paper | Addgene 216470 |
| p2702-AGER-gRNA1 | This paper | Addgene216471 |
| p2703-AGER-gRNA2 | This paper | Addgene216472 |
| p2704-AGER-gRNA3 | This paper | Addgene 216473 |
| p2706-pHAGE-EF1aL-YAP5SA-UBC-GFP | This paper | Addgene 216475 |
| p2709-pHAGE2-Ef1aL-YAP5SA-UBC-tagBFP-loxP | This paper | Addgene216478 |
| p2710-pHAGE2-Ef1aL-WTYAP-UBC-tagBFP-loxP | This paper | Addgene 216479 |
|
| ||
| Software and algorithms | ||
|
| ||
| ImageJ | National Institutes of Health | https://imagej.nih.gov/ij/ |
| Prism | GraphPad | https://www.graphpad.com |
| FlowJo | Becton Dickinson & Company | https://flowjo.com/solutions/flowjo |
|
| ||
| Other | ||
|
| ||
| StemDiff Definitive Endoderm Kit | StemCell Technologies | Cat#05110 |
| Gentle Cell Dissociation Reagent | StemCell Technologies | Cat#07174 |
| 0.4μm pore Polyester Membrane Transwell Insert | Corning | Cat#3470 |
| Accutase | Sigma | Cat#A6964 |
| TaqMan probe: 18S | Thermo Fisher | 4318839 |
| TaqMan probe: SFTPC | Thermo Fisher | Hs00161628_m1 |
| TaqMan probe: NAPSA | Thermo Fisher | Hs00362192_m1 |
| TaqMan probe: NKX2–1 | Thermo Fisher | Hs00968940_m1 |
| TaqMan probe: CYR61 | Thermo Fisher | Hs00155479_m1 |
| TaqMan probe: ANKRD1 | Thermo Fisher | Hs00173317_m1 |
| TaqMan probe: AGER | Thermo Fisher | Hs00542584_g1 |
| TaqMan probe: CAV1 | Thermo Fisher | Hs00971716_m1 |
| TaqMan probe: CLIC5 | Thermo Fisher | Hs00213494_m1 |
| TaqMan probe: PDPN | Thermo Fisher | Hs00366766_m1 |
| TaqMan probe: MKI67 | Thermo Fisher | Hs04260396_g1 |
| TaqMan probe: HOPX | Thermo Fisher | Hs05028646_s1 |
| TaqMan probe: SFTPA1 | Thermo Fisher | Hs00831305_s1 |
| TaqMan probe: SNAI1 | Thermo Fisher | Hs00195591_m1 |
| TaqMan probe: TWIST1 | Thermo Fisher | Hs04989912_s1 |
| TaqMan probe: ECAD (CDH1) | Thermo Fisher | Hs01023895_m1 |
| TaqMan probe: TP63 | Thermo Fisher | Hs00978340_m1 |
| TaqMan probe: SCGB3A2 | Thermo Fisher | Hs00369678_m1 |
| TaqMan probe: CTGF | Thermo Fisher | Hs00170014_m1 |
| TaqMan probe: AXIN2 | Thermo Fisher | Hs00610344_m1 |
| TaqMan probe: LEF1 | Thermo Fisher | Hs01547250_m1 |
| TaqMan probe: VEGFA | Thermo Fisher | Hs00900055_m1 |


