Abstract
Mucosal-associated invariant T (MAIT) cells are innate-like T cells that recognize bacterial riboflavin–based metabolites as activating antigens. Although MAIT cells are found in tissues, it is unknown whether any host tissue–derived antigens exist. Here, we report that a sulfated bile acid, cholic acid 7-sulfate (CA7S), binds the nonclassical MHC class I protein MR1 and is recognized by MAIT cells. CA7S is a host-derived metabolite whose levels were reduced by more than 98% in germ-free mice. Deletion of the sulfotransferase 2a family of enzymes (Sult2a1–8) responsible for CA7S synthesis reduced the number of thymic MAIT cells in mice. Moreover, recognition of CA7S induced MAIT cell survival and the expression of a homeostatic gene signature. By contrast, recognition of a previously described foreign antigen, 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil (5-OP-RU), drove MAIT cell proliferation and the expression of inflammatory genes. Thus, CA7S is an endogenous antigen for MAIT cells, which promotes their development and function.
INTRODUCTION
Mucosal-associated invariant T (MAIT) cells are innate-like T cells that recognize riboflavin-based metabolites, such as 5-OP-RU, presented by major histocompatibility complex (MHC) class I-related protein (MR1) as foreign antigens through semi-invariant T cell receptors (TCRs) (1–5). Although conventional T cells recognize self-derived weak antigens for selection and maintenance, microbiome-derived metabolites mediate the development of MAIT cells, as evidenced by the fact that MAIT cell numbers are decreased in germ-free mice (6, 7). Because the microbiome has a strong effect on endogenous metabolite levels (8), it is possible that host-derived antigen(s) may also contribute to MAIT cell development.
In many mammalian species, MAIT cells are most abundant in the liver (9, 10) and are mainly localized in the hepatic sinusoid around bile ducts (11–13). In contrast to circulating MAIT cells, liver MAIT cells exhibit an activated phenotype despite being in a nonproliferating state (11, 14, 15). This suggests that weak MR1 ligand(s) may be expressed in these specific regions. MAIT cell ligands of this type have not yet been identified, although previous studies have suggested the presence of self-derived ligands for MR1 (16–18).
Here, we report that sulfated bile acid is presented by MR1 and recognized by MAIT TCRs to promote MAIT cell homeostasis. Deletion of enzyme required for its synthesis in mice impaired MAIT cell development.
RESULTS
Cholic acid 7-sulfate is a MAIT cell ligand presented by MR1
Not all MR1 ligands act as agonistic antigens to MAIT cells (4, 19, 20). To simultaneously evaluate MR1 binding and antigen activity, we established a nuclear factor of activated T-cells (NFAT)–green fluorescent protein (GFP) reporter T cell line expressing both MR1 and a MAIT TCR (fig. S1A). A known MAIT cell antigen, 5-OP-RU, induced the dose-dependent activation of these reporter cells (fig. S1B) and marginal up-regulation of MR1 on the cell surface (fig. S1B). By contrast, a nonstimulatory MR1 ligand, acetyl-6-formylpterin (Ac-6-FP), potently enhanced MR1 surface expression (fig. S1B) as previously reported (21, 22) without inducing GFP expression. Thus, the reporter cells can function as both antigen-presenting cells and responder T cells.
To search for host-derived ligand(s) for MAIT cells, we used this cell line to screen fractionated intestinal extracts from specific pathogen–free (SPF) mice. Among the 100 fractions separated by reversed-phase column chromatography (table S1A), three major peaks were detected around fractions #11, #42, and #84 (Fig. 1A). To determine which peak corresponded to a known microbe-derived antigen, we subjected 5-OP-RU to the same column separation process (Fig. 1B). The activities of fractions #11 and #42 were again observed (Fig. 1B) corresponding to the expected retention of 5-OP-RU and its related lumazine derivatives, respectively (5).
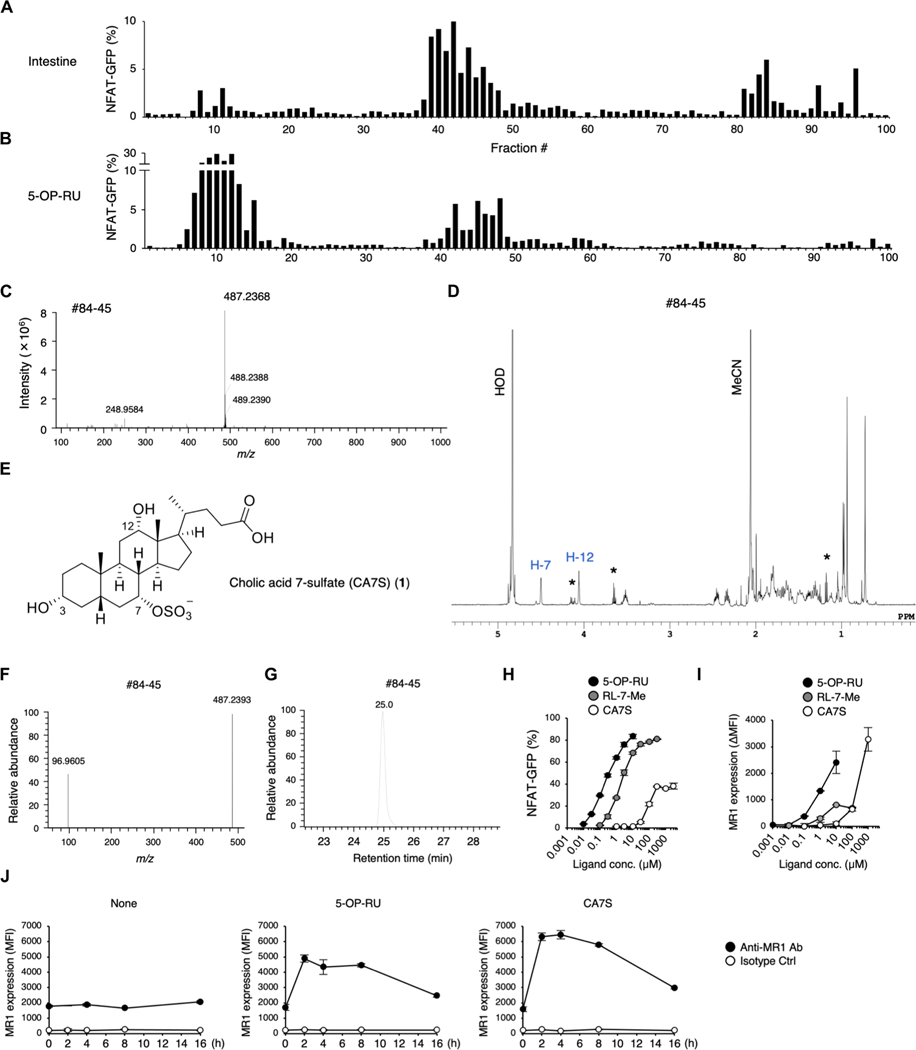
Fig. 1. Purification and structural determination of a ligand for MAIT cells.
(A and B) Screening of MAIT cell agonists from mouse intestine. NFAT-GFP reporter cells expressing MAIT TCR and MR1 were stimulated with HPLC-separated fractions from SPF mouse intestine (A) and freshly prepared 5-OP-RU as a control (B) for 16 to 20 hours and analyzed by flow cytometry. (C and D) HRMS (C) and 1H NMR spectra (600 MHz, D2O) (D) of fraction #84–45 from SPF mice intestine. In (D), impurities are denoted by asterisks. (E) Chemical structure of CA7S. (F and G) HRMS/MS spectra (F) and extracted ion chromatogram (G) of fraction #84–45. (H and I) NFAT-GFP reporter cells were stimulated with 5-OP-RU, RL-7-Me, and CA7S. Percentages of GFP+ cells (H) and MR1 expression (I) were analyzed at 20 and 6 hours after stimulation, respectively. MR1 surface expression is presented as mean fluorescence intensity (MFI) values of stimulated cells subtracted by those of vehicle-treated unstimulated cells (ΔMFI). (J) MR1 surface expression at 0, 2, 4, 8, and 16 hours after stimulation with vehicle control (None), 5-OP-RU, or CA7S. (H to J) Data are presented as the means ± SD of triplicate assays and are representative of more than two independent experiments.
To determine the chemical structure of the activating molecule in fraction #84, we further purified this fraction by hydrophilic interaction chromatography and collected a single fraction (designated as #84–45) (table S1B and fig. S1C). The activity was nuclease and protease resistant, suggesting a metabolite(s) other than polypeptides or oligonucleic acids (fig. S1D). High-resolution mass spectrometry (HRMS) analysis revealed a major peak at mass/charge ratio (m/z) 487.2368 in negative ion mode, suggesting a small compound (Fig. 1C). We next performed one-dimensional (1D)– and 2D–nuclear magnetic resonance (NMR) spectroscopic analysis (Fig. 1D and fig. S2, A to D) and showed that the compound is a cholic acid (CA) analog with a functional group attached to the hydroxy group at the C7 position (fig. S2E). The molecular formula estimated from the main peak at m/z 487.2368 was C24H40O8S [M − H]−, suggesting that the major component of the fraction was cholic acid 7-sulfate (CA7S) (Fig. 1E). HRMS/MS analysis confirmed the presence of a sulfate group (Fig. 1F). We next synthesized CA7S to authenticate the assignment (fig. S3A). The 1H and 13C NMR spectra of fraction #84–45 agreed with those of the synthetic CA7S (Fig. 1D and fig. S2, A, F, and G), and liquid chromatography HRMS (LC/HRMS) analysis showed identical retention times for fraction #84–45 and the CA7S standard (Fig. 1, F and G, and fig. S2, H and I). Synthetic CA7S activated reporter cells expressing a MAIT TCR, but the efficacy and potency were weaker than known antigens (Fig. 1H). Because absolute concentrations of unstable 5-OP-RU converted from 5-amino-6-d-ribitylaminouracil (5-A-RU) cannot be estimated precisely (23), more-stable synthetic ribityllumazine, 7-methyl-8-d-ribityllumazine (RL-7-Me), was used for comparison (24). The activity of CA7S was approximately 5% of RL-7-Me, indicating that CA7S is a weak agonist to MAIT cells (fig. S1E). Synthetic CA7S also elevated cell-surface MR1 expression (Fig. 1, I and J). CA7S was recognized by several different MAIT TCRs derived from mouse (m) and human (h) on m/h MR1 (fig. S1F). Thus, CA7S appears to be a ligand for MAIT cells presented by MR1.
CA7S is present in various SPF mouse tissues
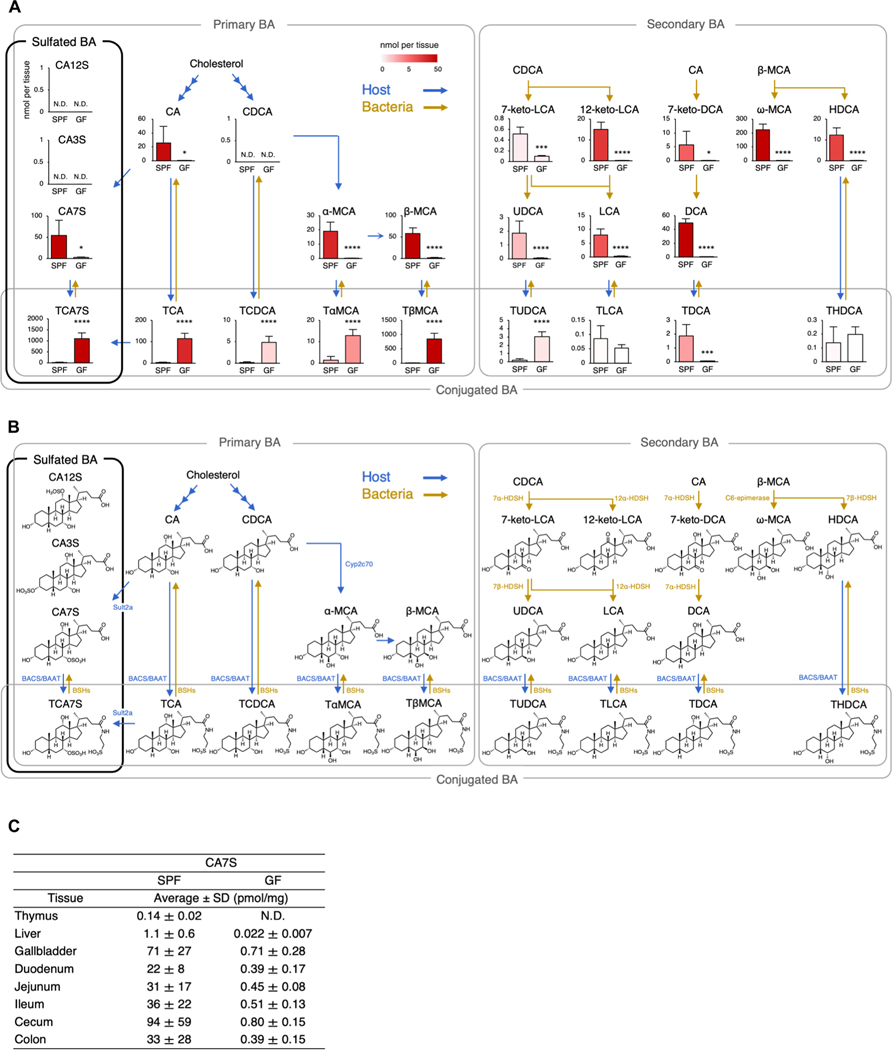
CA7S is a bile acid metabolite biosynthesized by host sulfotransferase 2a (Fig. 2, A and B) (25). Because bile acid metabolism is largely influenced by symbiotic bacteria, we next examined the effect of microbiota on intestinal bile acid metabolites, including CA7S. To assess this, we analyzed intestines from SPF and germ-free (GF) mice using targeted metabolomics (data S1). The concentration of many bile acid derivatives were affected by symbiotic bacteria (Fig. 2A) that mediate the deconjugation/dehydroxylation of bile acids (Fig. 2B) (26–28). Among these, CA7S were decreased by more than 98% under GF conditions (Fig. 2A), presumably due to the lack of deconjugation of tauro CA7S (TCA7S) by symbiotic bacteria (Fig. 2A) (29), because TCA7S was increased in GF mice (Fig. 2A).
Fig. 2. Decrease in the level of CA7S in GF mice.
(A) Quantification of bile acid metabolites in the cecum and cecal contents from SPF and GF mice. Blue and yellow arrows indicate host- and bacteria-mediated enzymatic responses, respectively. N.D., not detected (<0.010 nmol per tissue). (B) Metabolic pathway map of bile acids in mice, related to (A). Sult, sulfotransferase; HDSH, hydroxysteroid dehydrogenase; BAAT, bile acid–CoA:amino acid N-acyltransferase; BACS, bile acid–CoA synthetase; Cyp, cytochrome P450; BSHs, bile salt hydrolases. (C) Tissue distribution of CA7S in various tissues of SPF and GF mice. N.D., not detected (<0.010 pmol/mg). (A and C) Data are presented as the means ± SD from experiments with three or more mice per group and are representative of two independent experiments. *P < 0.05, ***P < 0.005, and ****P < 0.001 by two-tailed, unpaired Student’s t tests with Benjamini-Hochberg correction.
In addition to the intestines, CA7S was detected in the gallbladders, livers, and thymi of SPF mice. CA7S levels were detectable, but lower, in all GF tissues examined (Fig. 2C). LC-MS/MS analyses confirmed that other regioisomers of CA7S, such as CA3S and CA12S, could not be detected in mouse tissue (table S2) as previously reported (25). Thus, CA7S is a host-derived bile acid metabolite, but its abundance is largely dependent on symbiotic bacteria.
Cholic acid sulfate is a unique agonistic bile acid metabolite
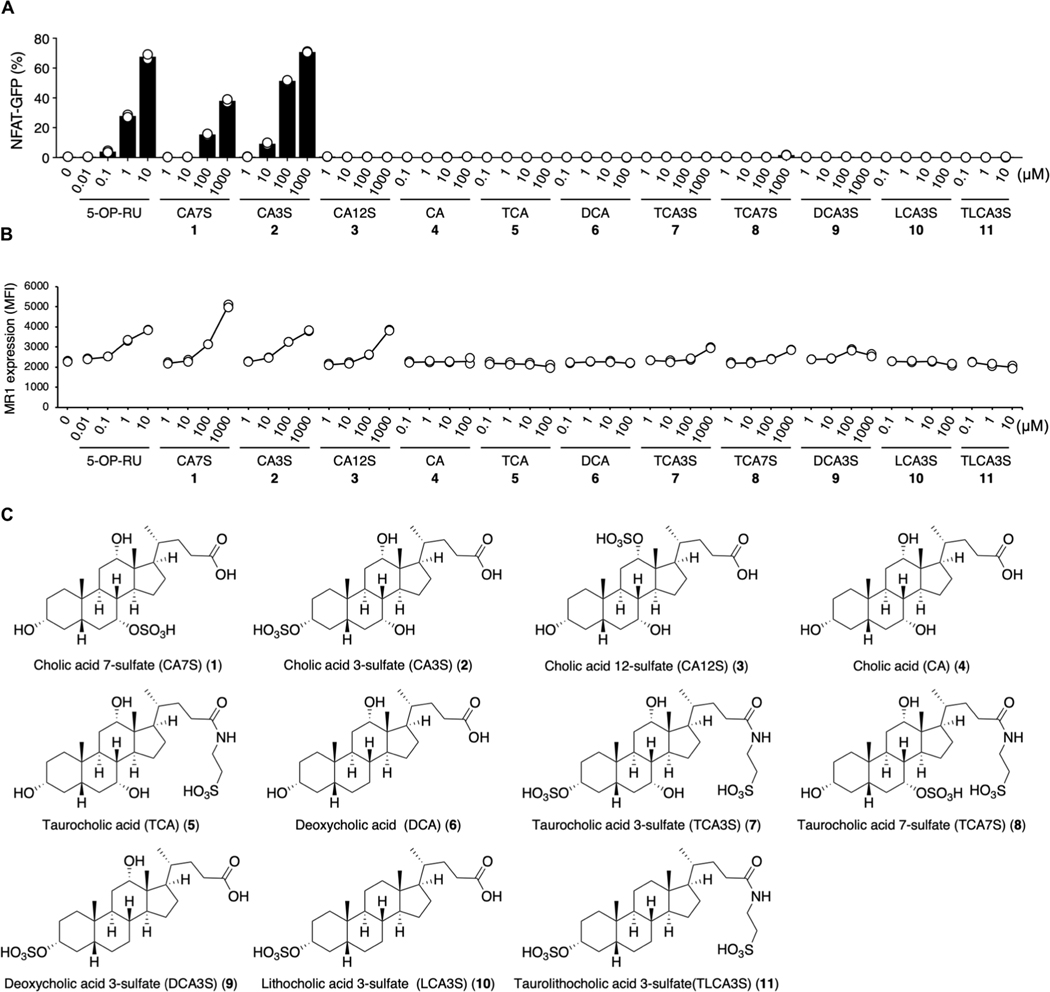
CA7S is biosynthesized by sulfate conjugation of CA, which is a major component of bile acid released into the intestine from the gallbladder and which aids absorption of dietary lipids through micellization (30). However, CA itself did not activate reporter cells expressing MAIT TCR (Fig. 3, A and C), suggesting that the sulfate group was required for agonistic function.
Fig. 3. Structure-activity relationship of bile acid metabolites for MAIT cell activation.
(A and B) MAIT TCR activation assays using NFAT-GFP reporter cells expressing mouse MAIT TCR and MR1. Reporter cells were stimulated with vehicle control, 5-OP-RU, and analogs of CA7S (1); cholic acid 3-sulfate (CA3S: 2), cholic acid 12-sulfate (CA12S: 3), cholic acid (CA: 4), taurocholic acid (TCA: 5), deoxycholic acid (DCA: 6), taurocholic acid 3-sulfate (TCA3S: 7), taurocholic acid 7-sulfate (TCA7S: 8), deoxycholic acid 3-sulfate (DCA3S: 9), lithocholic acid 3-sulfate (LCA3S: 10), and taurolithocholic acid 3-sulfate (TLCA3S: 11). Because of their inherent cell toxicity as surfactants, several bile acids were added at lower concentrations (4, 5, 6, 10, and 11). Some water-insoluble bile acids (4, 5, 6, and 10) were coated on plates as described (81). Compound 11 was added as dimethyl sulfoxide solution because of its insolubility in both water and organic solvents. NFAT-GFP (A) and MR1 (B) expressions were evaluated at 20 and 6 hours after stimulation, respectively. (C) Structural formula of bile acid analogs related to (A) and (B). (A and B) Data are presented as individual values of duplicate assays and are representative of more than two independent experiments.
In addition to the hydroxy group at position 7 (7-OH) of CA, 3-OH and 12-OH groups are also potential sulfation sites. We therefore also synthesized these isomers (fig. S3A). CA3S, which is present in some mammalian species (25), but not CA12S, activated MAIT reporter cells in the presence of MR1 (Fig. 3, A and C). By contrast, all three CA sulfate forms (CA3S, CA7S, and CA12S) up-regulated the cell-surface expression of MR1 to a similar extent (Fig. 3, B and C), suggesting that CA sulfates may have the ability to bind to MR1.
Because primary mouse bile acids are often conjugated with taurine, we next synthesized taurine-conjugated forms of CA3S (TCA3S) and CA7S (TCA7S) (fig. S3B) and examined the effect of C24 modification on activity. Neither TCA, TCA3S, or TCA7S showed any substantial activity either as an antigen or an MR1 ligand (Fig. 3, A to C). Thus, this amino acid conjugation of CA, which incorporates the addition of a hydrophilic group at position 24, abolishes any MR1-binding capability.
Primary bile acids are further dehydroxylated by symbiotic bacteria, producing secondary bile acids. However, the sulfated forms of secondary bile acids, such as deoxy CA3S (DCA3S), litho CA3S (LCA3S), and taurolitho CA3S (TLCA3S) did not have activity in reporter cells (Fig. 3, A and C). Thus, the sulfation of the 3- or 7-OH of CA is the key structural feature for MAIT cell activation.
CA7S is presented by MR1 in a manner similar to bacterial ligands
We next investigated the mode of CA7S presentation by MR1. MAIT TCR recognition of CA7S was inhibited by blocking anti-MR1 antibodies and Ac-6-FP (21) in a dose-dependent manner (Fig. 4, A and B), implying that CA7S is presented by MR1 in a conventional manner (4, 5) despite lacking a carbonyl group that could form a Schiff base with MR1. We therefore assessed the possible MR1-binding mode by introducing mutations in MR1. Most mutations affecting 5-OP-RU binding to MR1 also affected CA7S binding, consistent with CA7S and 5-OP-RU potentially binding to a similar region of MR1 (Fig. 4C). We next assessed the competitive ligand binding to MR1 of CA7S in the presence of the fluorescent ligand, JYM20, which has been reported to bind in the A′ pocket of MR1 (31). CA7S outcompeted JYM20 with a similar potency to that of the reported ligand, diclofenac (DCF) (19) (Fig. 4D). TCRα Y95 was also essential for the recognition of CA7S, as has also been reported for 5-OP-RU (Fig. 4E) (5), although this mutant TCR could still respond normally to anti-TCRβ monoclonal antibody stimulation (fig. S4). Thus, although CA7S differs in both size and structure from other reported MR1 ligands (4, 5, 18, 19), it is presented by the MR1 A′ pocket and is recognized by MAIT TCRs.
Fig. 4. Binding mode of CA7S to MR1 and MAIT TCR.
(A and B) Inhibition assay of 5-OP-RU (0.5 μM) and CA7S (500 μM) by anti-MR1 Ab (26.5) (A) or Ac-6-FP (B). Percentages of GFP+ cells were shown. (C) Effect of hMR1 mutations (Y7A, R9A, K43A, Y62A, L66A, W69A, M72A, R79A, R94A, W156A, and W164A) on the recognition of ligands. Cells expressing hMR1 mutants and a MAIT TCR were stimulated with vehicle control, 5-OP-RU, or CA7S and analyzed by flow cytometry after 6 hours. MR1 expression was shown as MFI of anti-MR1 staining subtracted by isotype control. (D) MR1-restricted ligand affinities (IC50) determined by FP assay (31). Left: Titration curves of strong MR1 binders (5-OP-RU and Ac-6-FP), moderate MR1 binder (RL-6-Me-7-OH), weak MR1 binders (CA7S, CA3S, and DCF), and MR1 nonbinding substances [epigallocatechin gallate (EGCG) and NLV peptide] are displayed. Tetrahydroxy bile acid (THBA) was used as a bile acid control with similar hydrophilicity to CA7S and CA3S. The table represents a summary of IC50 for all investigated compounds. (E) Effect of MAIT TCRα mutation on ligand recognition. Reporter cells transfected with vector alone (Mock), MAIT TCRβ together with WT MAIT TCRα (WT), or mutant TCRα (Y95F) were stimulated with vehicle control, 5-OP-RU, or CA7S and analyzed after 20 hours. (C and D) Data are presented as the means ± SD of triplicate assays. (A, B, and E) Data are presented as individual values of duplicate assays. All data are representative of at least two independent experiments. NB, no binding.
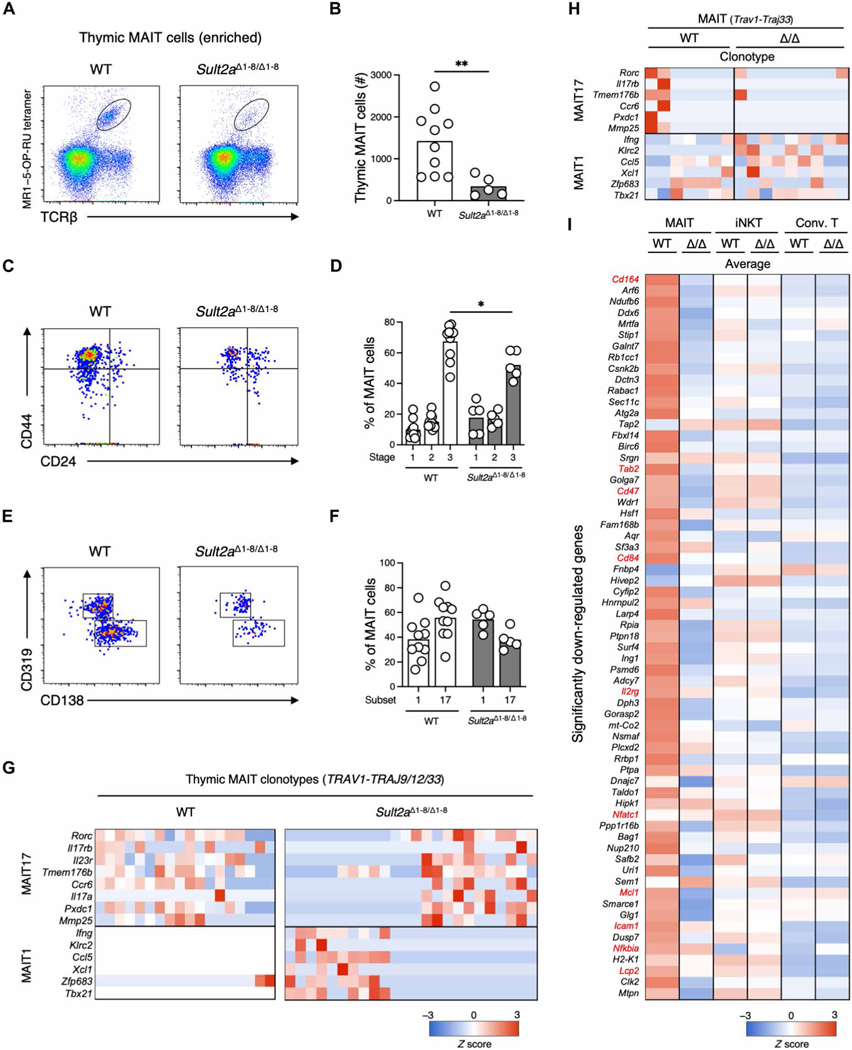
Thymic MAIT cell numbers are reduced in Sult2a-deficient mice
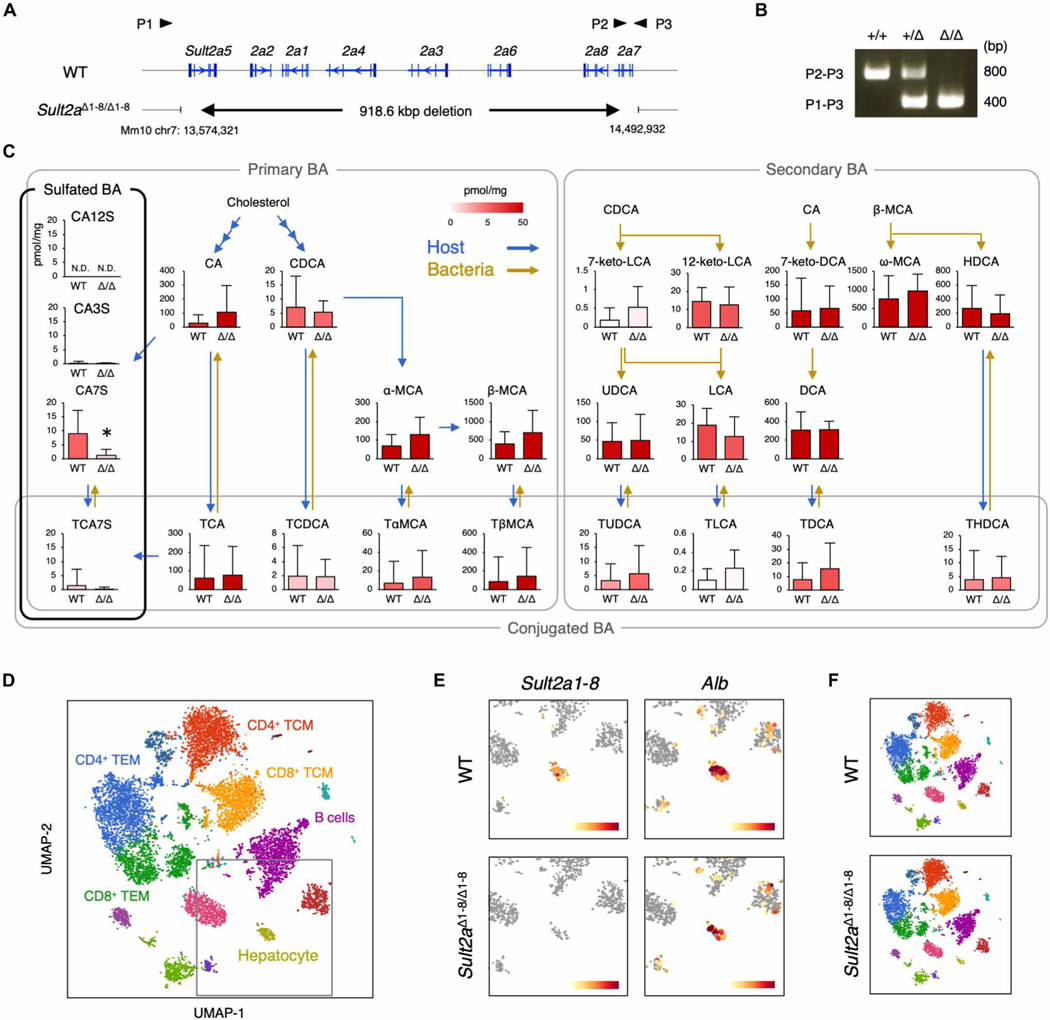
CA7S is biosynthesized by the SULT2A family of enzymes, which incorporate a sulfate group into CA (32). Because eight isoforms of SULT2A (Sult2a1-8) exist in a cluster on mouse chromosome 7A1, we generated mice lacking all eight isoforms using the CRISPR-Cas9 system (Sult2aΔ1–8/Δ1–8 mice) (Fig. 5, A and B). Targeted metabolomics analyses revealed that CA7S was significantly decreased in Sult2aΔ1–8/Δ1–8 mice, whereas other bile acid metabolites were unchanged (Fig. 5C). Nevertheless, Sult2aΔ1–8/Δ1–8 mice were viable and showed no macroscopic abnormalities, suggesting that CA7S is not a vital metabolite in mice. Because SULT2A is abundantly expressed in the liver (33, 34), we confirmed the lack of expression of all Sult2a isoforms in hepatocytes from Sult2aΔ1–8/Δ1–8 mice via single-cell RNA sequencing (scRNA-seq) (Fig. 5, D and E). However, hepatocyte and resident immune cell clusters were similar in wild-type (WT) and Sult2a-deficient mice (Fig. 5F). Thus, CA7S deficiency does not grossly affect the development of hepatocytes and adjacent immune cells, allowing us to use Sult2aΔ1–8/Δ1–8 mice to investigate the contribution of CA7S to MAIT cell development and function.
Fig. 5. Generation of Sult2aΔ1–8/Δ1–8 mice.
(A) Gene-targeting strategy for Sult2aΔ1–8/Δ1–8 mice. P1, P2, and P3 indicate primers used for genomic PCR. (B) Genotyping PCR of Sult2a+/+, Sult2a+/Δ1–8, and Sult2aΔ1–8/Δ1–8 mice. (C) Quantification of bile acid metabolites in feces from WT and Sult2aΔ1–8/Δ1–8 mice. Blue and yellow arrows indicate host- and bacteria-mediated enzymatic responses, respectively. (D) UMAP projection based on sc-RNA-seq of density-fractionated liver cell suspensions from WT and Sult2aΔ1–8/Δ1–8 mice. Erythrocytes expressing Hbb-bs above 1.8 were excluded in the process of analysis. (E) Expression of Sult2a1–8 (left) and Alb (right) in the hepatocyte clusters derived from WT (top) and Sult2aΔ1–8/Δ1–8 (bottom) mice. (F) UMAP projection of WT (top) and Sult2aΔ1–8/Δ1–8 (bottom) mice. (C) Data are presented as the means ± SD from experiment with six or more mice per group. (D to F) Data are from experiments with three mice per group. (C) A direct product of SULT2A, CA7S, was analyzed by two-tailed, unpaired Student’s t tests (*P < 0.05). For all other bile acid metabolites, P values were adjusted with Benjamini-Hochberg correction (FDR < 0.05).
A significant decrease in the number of MR1–5-OP-RU tetramer+ (MR1–5-OP-RU tet+) thymic MAIT cells was observed in Sult2aΔ1–8/Δ1–8 mice compared with littermate controls (Fig. 6, A and B). Thymic MAIT cells can be divided into three developmental stages. The most mature stage (stage 3) was dominant in WT mice (35), whereas this stage was significantly less frequent in Sult2aΔ1–8/Δ1–8 mice (Fig. 6, C and D). Mature thymic MAIT cells skew to a MAIT17 subset, interleukin-17 (IL-17)–producing MAIT cells, which requires more signaling for development (36), whereas MAIT17 was less dominant in Sult2aΔ1–8/Δ1–8 mice, as assessed by surface marker expression (Fig. 6, E and F). scTCR-RNA-seq analyses of WT and Sult2aΔ1–8/Δ1–8 thymocytes expressing MAIT clonotypes (Trav1-Traj9/12/33) also supported the reduction of MAIT17 (Fig. 6G). Thus, thymic MAIT cell development is impaired under CA7S-deficient conditions.
Fig. 6. Impaired MAIT cell development in Sult2aΔ1–8/Δ1–8 mice.
(A to F) Flow cytometry analysis of MAIT cells among MR1–5-OP-RU tetramer-enriched thymocytes from 3- to 4-week-old WT and Sult2aΔ1–8/Δ1–8 mice. (A) Representative flow cytometry dot plots of MR1–5-OP-RU tet+TCRβ+ MAIT cells in thymocytes. (B) Absolute number of thymic MAIT cells in WT and Sult2aΔ1–8/Δ1–8 mice. (C) Representative CD44 and CD24 expression in MAIT cells from WT (left) and Sult2aΔ1–8/Δ1–8 (right) mice. (D) Frequency of stage 1 (CD44−CD24+), stage 2 (CD44−CD24−), and stage 3 (CD44+CD24−) MAIT cells in WT and Sult2aΔ1–8/Δ1–8 mice. (E) Representative CD319 and CD138 expression in CD44+CD24− MAIT cells in WT (left) and Sult2aΔ1–8/Δ1–8 (right) mice. (F) Frequency of MAIT17 cells (CD138+) and MAIT1 cells (CD319+) in WT and Sult2aΔ1–8/Δ1–8 mice. (G and H) Heatmap of mRNA expression of MAIT1 and 17 signature genes in canonical MAIT cells (Trav1-Traj9/12/33) in the (G) thymi and (H) livers of WT and Sult2aΔ1–8/Δ1–8 mice. (I) Heatmap of mRNA expression in canonical MAIT cells (Trav1-Traj9/12/33), canonical iNKT cells (Trav11-Traj18), and conventional T cells in the liver of WT and Sult2aΔ1–8/Δ1–8 mice. The genes significantly down-regulated in Trav1-expressing cells from Sult2aΔ1–8/Δ1–8 mice (P < 0.05) compared with those from WT mice are shown. The gene expression levels are shown as average values. Data are from experiments with three or more mice per group. *P < 0.05 and **P < 0.01 by two-tailed, unpaired Student’s t tests or one-way ANOVA followed by Tukey’s multiple comparison test.
Because MAIT cells are highly abundant in the livers of many mammalian species (9, 10, 37), we also examined MR1–5-OP-RU tet+ mature MAIT cells in this organ. The frequency of liver MAIT cells was not significantly different in Sult2aΔ1–8/Δ1–8 mice (fig. S5), and most MAIT cell clones in the livers of both WT and Sult2aΔ1–8/Δ1–8 mice expressed previously reported MAIT1 signature genes (38) (Fig. 6H). However, genes significantly down-regulated in Sult2aΔ1–8/Δ1–8 mice compared with WT mice included those involved in TCR signaling and costimulation (e.g., Lcp2 encoding SLP-76, Cd84 encoding Slamf5, Nfatc1, Nfkbia, and Tab2) (38–40) and cell adhesion (Icam1 and Cd164) (38). The common gamma chain (γc) (Il2rg) [a component of IL-7R–mediated licensing of MAIT cells (14)], Bcl-2 family proteins (Mcl1) (41), and other anti-apoptotic proteins (Bag1, Birc6, and Rb1cc1) (42–45) were also down-regulated, suggesting that liver MAIT cells in the absence of SULT2A may not receive sufficient signals to support their homeostasis (Fig. 6I). By contrast, these genes were not down-regulated in iNKT and conventional T cells in the livers of Sult2aΔ1–8/Δ1–8 mice (Fig. 6I). Thus, CA7S specifically contributes to MAIT cell homeostasis.
CA7S contributes to the maintenance of canonical human MAIT cells
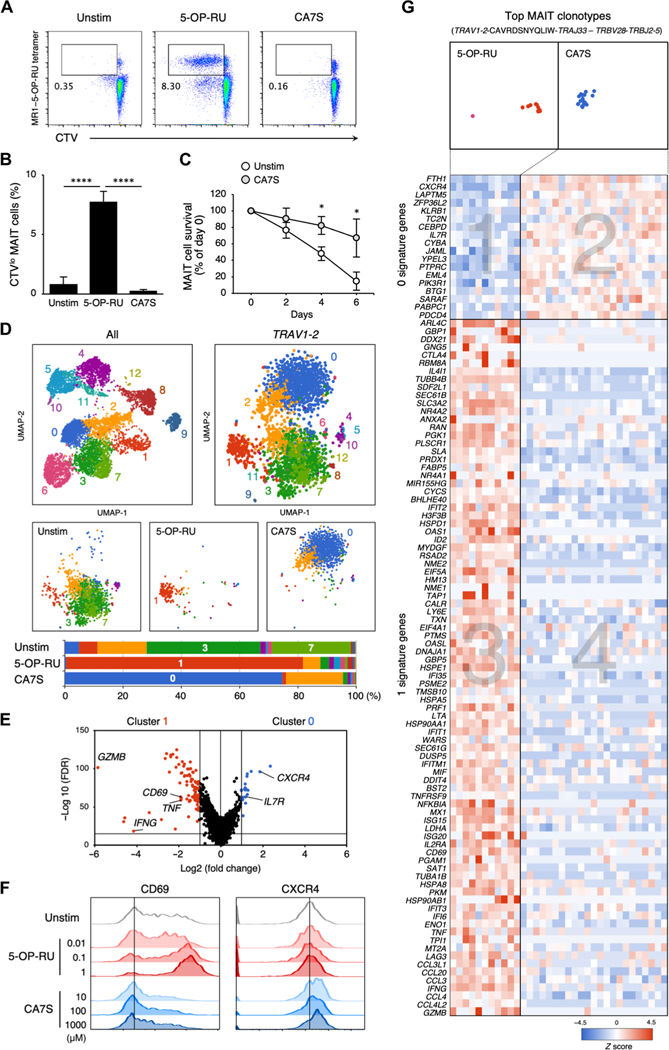
To confirm whether CA7S acts on primary human MAIT cells, we stimulated peripheral blood mononuclear cells (PBMCs) with CA7S. To compare the functional differences for each agonist, we stimulated cells with minimum effective concentrations of 5-OP-RU (10 μM) and CA7S (1000 μM) based on their reporter cell activity. As assessed by CellTrace violet (CTV) dilution, 5-OP-RU induced cell division of MR1–5-OP-RU tet+ MAIT cells, whereas CA7S did not (Fig. 7, A and B). However, the number of MAIT cells cultured in the presence of CA7S was four times greater than that of control cultures without CA7S at day 6 (Fig. 7C). 5-OP-RU, but not CA7S, increased the surface expression of CD161 (KLRB1) during culture (fig. S6A), suggesting that these ligands induced different responses via the MAIT TCR. To test this hypothesis, we compared gene expression signatures using scTCR-RNA-seq of MAIT cells that were either unstimulated (Unstim) or stimulated with 5-OP-RU or CA7S for 1 day. Uniform manifold approximation and projection (UMAP) clustering analysis of MAIT cells (TRAV1–2+) demonstrated that gene expression signatures were distinct depending on the ligand. 5-OP-RU mainly induced cluster 1, whereas cluster 0 was enriched upon stimulation with CA7S. Both were distinct from the Unstim signature (clusters 3 and 7) (Fig. 7D). Comparison of clusters 0 and 1 revealed that distinct genes were up-regulated upon stimulation with 5-OP-RU versus CA7S (Fig. 7E). CD69, GZMB, IFNG, and TNF, for example, were induced by 5-OP-RU (46, 47), whereas CXCR4 and IL7R were selectively induced upon CA7S stimulation (Fig. 7E). CA7S induced surface expression of CXCR4, but not CD69, in the peripheral MAIT cells (Fig. 7F). Gene ontology enrichment analysis also supported this differential gene expression, because 5-OP-RU up-regulated immune activation–related genes as previously reported (table S3) (46–48), whereas CA7S induced genes involved in wound healing (top enrichment score, P = 0.002) and negative regulation of immune activation (second and third enrichment score, P = 0.002) (table S3). This differential regulation was also observed within the most frequent clonotypes expressing the canonical MAIT TCR pair (TRAV1–2-CAVRDSNYQLIW-TRAJ33–TRBV28-TRBJ2–5) (Fig. 7G). 5-OP-RU preferentially induced genes in area 3 rather than area 1, whereas CA7S-induced genes were included in area 2 rather than area 4 (Fig. 7G). Thus, CA7S appears to trigger different signals than those induced by 5-OP-RU through the same MAIT TCR. We also performed scTCR-RNA-seq after long-term stimulation with CA7S and CA3S, which are both present in humans. MAIT cells that survived in the presence of CA7S/3S expressed canonical MAIT TCRs, such as TRAV1–2-TRAJ33/12/20 paired with TRBV6–4/6–1/20–1, which were also enriched upon 5-OP-RU stimulation (fig. S6B), suggesting that CA7S/3S is a ligand for canonical MAIT cells. However, differential gene expression signatures were also retained, because CA7S/3S induced homeostatic genes, such as IL7R (14, 49), KLF2 (50), and TCF7 (36, 51, 52), which were not induced by 5-OP-RU (fig. S6, C and D). Thus, sulfated bile acid is a host-derived ligand distinct from the microbe-derived antigen 5-OP-RU, which contributes to the development and survival of canonical MAIT cells.
Fig. 7. Responses of human MAIT cell to CA7S.
(A and B) Human PBMCs were labeled with CTV and stimulated with vehicle control (Unstim), 5-OP-RU (10 μM), or CA7S (1000 μM) in the absence of cytokines on day 6. Representative flow cytometry dot plots of proliferating CD3+MR1–5-OP-RU tet+CTVlo MAIT cells (A). Proportion of MR1–5-OP-RU tet+CTVlo MAIT cells in CD3+-gated cells (B). (C) Survival ratio of MAIT cells stimulated with 1000 μM CA7S (CA7S) or left unstimulated (Un-stim). (D and E) Sc-RNA-seq of CD3+CD161+MR1–5-OP-RU tet+ MAIT cells stimulated with vehicle control (Unstim), 5-OP-RU (10 μM), or CA7S (1000 μM) for 24 hours. UMAP based on mRNA expression was performed on all isolated cells (top left). UMAPs of Trav1–2+ cells (top right) were separately shown for different stimuli (Unstim, 5-OP-RU, and CA7S) (middle). Proportions of the clusters in Trav1–2+ cells of each group are shown as bar graphs (bottom) (D). Volcano plot of mRNA expression comparing the characteristic clusters of 5-OP-RU stimulation (cluster 1) and CA7S stimulation (cluster 0) (E). (F) Surface expression of CD69 and CXCR4 after CA7S stimulation. PBMCs were stimulated with indicated concentrations of 5-OP-RU (red) or CA7S (blue), and surface expression of CD69 and CXCR4 within CD3+CD161+MR1–5-OP-RU tet+ MAIT cells was determined at day 9. (G) Heatmap of mRNA expression in the most frequent MAIT clonotypes (TRAV1–2-CAVRDSNYQLIW-TRAJ33–TRBV28-TRBJ2–5). The genes with differential expression between clusters 1 and 0 in (E) are shown. (B and C) Data are presented as the means ± SD of triplicate assays, and (A), (B), (C), and (E) are representative of more than three independent experiments. *P < 0.05 and ****P < 0.001 by two-tailed, unpaired Student’s t tests with Benjamini-Hochberg correction or one-way ANOVA followed by Tukey’s multiple comparison test.
DISCUSSION
In the bile acid metabolic pathway, sulfation contributes to the excretion of excess bile acids through the gastrointestinal tract for bile acid homeostasis (25). However, the “active” physiological roles of CA7S have not been fully understood (29, 53). The current study suggests that an abundant by-product of one physiological process can be utilized for the regulation of other physiological processes. CA sulfates are mainly produced in the liver and small intestine, both of which express SULT2A (33, 34, 54) and where MAIT cells are enriched in humans (55), suggesting that this metabolite may play an important role in MAIT cell homeostasis in these tissues. Although there are some limitations to exploring the effect of CA7S deficiency using laboratory mice in which MAIT cells are rare, further analyses across multiple species or use of MAIT-enriched mice (56–58) may help clarify this issue. Because common genetic polymorphisms for human SULT2A1 result in the reduction of its enzymatic activity (59, 60), future work should investigate whether there is any association between MAIT cell development/function and the presence of these polymorphisms.
Impairment of thymic MAIT cells, particularly stage 3, in Sult2aΔ1–8/Δ1–8 mice suggests that CA7S plays a preferential role in MAIT cell maturation. The reduction of CA7S in GF thymi may explain impaired thymic MAIT cell development in addition to the lack of 5-OP-RU (6, 7). Although CA7S was detected in the thymus, it is currently unknown how this metabolite comes to be located in this lymphoid organ. In GF mice (3, 6, 7, 35), SPF Sult2a-deficient mice (Fig. 6B), and even GF Sult2a-deficient mice (fig. S7), a few thymic MAIT cells were detected. It is therefore possible that other unknown endogenous ligand(s) may also contribute to the early development of thymic MAIT cells. An alternative but not mutually exclusive hypothesis is that an intrinsic binding affinity of MR1 to MAIT TCRs (21) may also play a role in thymic selection in a ligand-independent manner.
Conventional T cells are positively selected within the thymus by moderate interaction with weak-affinity peptides (61). In the periphery, these weak peptides cannot induce activation but contribute to the maintenance or homeostasis of mature T cells (52, 62, 63). CA7S also induced homeostatic gene expression in the peripheral MAIT cells, although it did not trigger inflammatory responses. The role of CA7S in liver MAIT cell function is not clearly understood, but we observed a difference in gene expression in Sult2aΔ1–8/Δ1–8 mice. CA7S may be involved in tissue residency, local survival, or functional priming/licensing within the tissue. One of the CA7S-induced molecules, CXCR4, contributes to the residency of MAIT cells in the liver (64), although further investigation is warranted. Liver MAIT cells respond to biliary epithelial cells (BECs) in an MR1-dependent manner (11). Thus, it is possible that hepatic cells, such as BECs, liver sinusoidal endothelial cells (LSECs), or hepatocytes may act as a niche for MAIT cells by presenting bile acid metabolites on MR1, as MAIT cells are localized in the liver sinusoid around bile ducts (9). Alternatively, as BECs are normally separated from MAIT cells in the sinusoid under steady-state conditions, MAIT cells may recognize CAS presented by BECs when the LSEC barrier is breached by trauma or infection, therefore serving as a possible damage sensor leading to tissue repair (7, 46–48). Considering their unique tissue distribution, MAIT cells may need to be resistant to bile acid stresses, and MAIT cells do express high levels of the ATP-binding cassette subfamily (37), also called multidrug resistance 1 polypeptide, which is known to protect T cells in the ileum from bile acid–driven stresses (65).
CA7S may accumulate in disease settings such as cholestasis-related inflammation, because bile sulfation is required for excretion of excessive bile (25, 66). Correlation between MAIT cell activation and primary sclerosing cholangitis or primary biliary cholangitis has been reported (67, 68). Whether excessive CA7S is involved in the development of such inflammatory diseases through the dysregulation of MAIT cells is an important area for future research.
Mammalian species lacking MR1 lose TRAV1, suggesting the coevolution of the MR1–MAIT cell system (69). Some such species also lack Sult2a homologs (table S4, A and B). Both MR1 and MAIT cells are absent in fish and amphibians (70). These animals use larger C27 bile alcohols as major bile salts (71), and these are unlikely to bind the MR1 pocket even if sulfated. Further phylogenetic studies will be needed to validate a possible link between sulfated bile acids and the MR1–MAIT system.
In conclusion, this study extends previous work underscoring the structural diversity of ligands presented by MR1 as compared with other MHC class I–related molecules (72). To monitor a wide variety of exogenous and endogenous metabolites, mammals may have acquired a “promiscuity” in presenting ligands with various structures and affinities (4, 18–20, 73) on MR1, which has a certain intrinsic affinity to MAIT TCRs (21). Further identification of unexpected structures of natural ligands therefore is likely possible and may contribute to further understanding of MAIT cell physiology.
MATERIALS AND METHODS
Study design
The goal of this study is to identify and characterize host-derived MAIT cell ligand(s). To enable this end, we established a platform using column chromatography–based separation and mass spectrometry together with a sensitive reporter assay. To determine the chemical structure of the ligand, we used NMR spectroscopy and chemical synthesis. We established mice lacking a particular bile acid metabolite using mouse genetics and metabolomics. Sample sizes were determined on the basis of previous studies and pilot experiments in our laboratory and are indicated in the figure legends. All experiments were performed three times unless otherwise indicated in the figure legends. Mice of the same sex and similar ages were randomly assigned to each experimental group. For in vivo experiments, the investigators were blinded to the genotype of the mouse samples. None of the data was excluded from our analysis.
Mice
Sult2aΔ1–8/Δ1–8 mice were generated using the CRISPR-Cas9 system, as previously described (74). In brief, Cas9 mRNA and sgRNAs were microinjected into fertilized embryos of BDF1 mice. Homozygous KO mice were born from a heterozygous intercross. All mice were genotyped 2 to 4 weeks after birth using PCR with specific primers (P1, 5′-ACCTGGAAAGACTAATACTTGCC-3′; P2, 5′-CCCCACAGAGACAGACCAAT-3′; and P3, 5′-CAAATGATCTCTCAATGAGTTCAC-3′). Sult2aΔ1–8/Δ1–8 mice were used for phenotypic analyses in 3- to 8-week-old mice with WT littermate controls (Sult2a+/+ or Sult2a+/Δ1–8). Germ-free mice (C57BL/6NJcl [Gf]) were purchased from CLEA Japan Inc. and analyzed in the same weeks of age compared with SPF mice. Germ-free Sult2aΔ1–8/Δ1–8 mice were generated by in vitro fertilization maintained in vinyl isolators within the facility in CLEA Japan. All animal protocols were approved by the committee of Ethics on Animal Experiment, Research Institute for Microbial Diseases, Osaka University (Biken-AP-R03–17-0).
Reagents
5-OP-RU for the cellular assay was prepared with 5-A-RU (Toronto Research Chemical) and methylglyoxal (Sigma-Aldrich) as previously described (5). Ac-6-FP (catalog no. 11.418) was purchased from Schircks Laboratories. 2-Hydroxy-5-methoxybenzaldehyde (HMB) (catalog no. 146862), DCF (catalog no. D6899), and epigallocatechin gallate (EGCG) (CAS no. 93894) were supplied by Sigma-Aldrich. For FP assays, 5-OP-RU (75), JYM20 (76), and RL-6-Me-7-OH (4) were synthesized as previously described. The native cytomegalovirus MHC-I–restricted peptide (NLV) (catalog no. 181329) was synthesized by GL Biochem. TCA3S, LCA3S, and TLCA3S were purchased from Cayman. DCA3S and THBA were purchased from Avanti. CA and TCA were purchased from Nacalai Tesque. DCA was purchased from FUJIFILM Wako. Synthetic CA7S, CA3S, CA12S, TCA3S, TCA7S, and RL-7-Me were prepared as described in Supplementary Methods. Anti-human/mouse/rat MR1 (26.5), anti-mouse CD3 (2C11), anti-mouse CD19 (6D5), anti-mouse TCRβ (H57–597), anti-mouse CD45 (30-F11), anti-mouse CD319 (4G2), anti-mouse CD138 (281–2), anti-mouse CD24 (M1/69), mouse IgG2a, κ isotype control (MOPC-173), Armenian hamster immunoglobulin G (IgG) isotype control (HTK888), anti-human CD3 (HIT3a), anti-human CD161 (HP-3G10), anti-human CD69 (FN50), anti-human CXCR4 (12G5), anti-mouse CD16/32 (93), and anti-human TotalSeq-C Hashtags (LNH-94; 2M2) antibodies were purchased from BioLegend. Anti-mouse CD44 (IM7) was from BD Biosciences. Dead cells were stained with propidium iodide (PI) (Sigma-Aldrich) or 7-amino-actinomycin D (7AAD) (BioLegend). Mouse or human MR1 tetramers loaded with 5-OP-RU or 6-FP conjugated to PE or APC were provided by the NIH tetramer core facility (Emory University) (5).
Cells
TCRα- and TCRβ-chain cDNA sequences were cloned into retroviral vector pMX-IRES-rat CD2. TCRαβ sequences for mouse MAIT TCR #1 clonotype (TRAV1-CAVRDSNYQLIW-TRAJ33–TRBV13–3-TRBJ2–1) and mouse MAIT TCR #2 clonotype (TRAV1-CAVRDSNYQLIW-TRAJ33–TRBV13–3-TRBJ2–7) were from previous reports (3, 77). αβTCR sequences for human MAIT TCR #1 clonotype (TRAV1–2-CAVKDSNYQLIW-TRAJ33–TRBV6–2-TRBJ2–2) were from Protein Data Bank (4L4T). Human and mouse MR1s were cloned into retroviral vector pMX-IRES-human CD8 (78). These plasmids were cotransfected into Phoenix packaging cells using PEI MAX (Polysciences). Supernatant containing retroviruses was used for infection (79) into TCR-deficient mouse T cell hybridoma with an NFAT-GFP reporter gene (80). MR1 and TCRα mutants were constructed by site-directed mutagenesis using KOD-FX (TOYOBO) following the manufacturer’s instruction.
Ligand extraction and purification
Mouse large intestines were frozen in liquid nitrogen. They were then crushed and extracted with 6 ml of double-distilled water. The supernatant was ultrafiltered (<100 kDa) and fractionated by HPLC (JASCO LC-NetII/ADC) using COSMOSIL PBr 4.6 mm I.D. by 250 mm (Nacalai Tesque) and TSK gel Amide-80 5 μm 4.6 mm I.D. by 250 mm (TOSOH Bioscience) as described in fig. S1 (C and D). Purified samples were dried using a freeze-dryer (EYELA FDU-1200).
Reporter cell assay
For the stimulation of reporter cells, tissue-extracted samples and synthetic compounds were dissolved in water and added to each well of the 96-well plate in the absence of methylglyoxal. Some water-insoluble samples were dissolved in ethanol or chloroform/methanol (2:1, vol/vol) and coated on plates as previously described (81). NFAT-GFP reporter cells (1 × 104 to 3 × 104 cells/100 μl per well) were cultured for 6 to 20 hours at 37°C and analyzed for GFP and MR1 expression by flow cytometry. Tissue-extracted fractions were treated with DNase I (Roche) or trypsin (Promega) overnight at 37°C before 5-OP-RU and CA7S stimulation in fig. S1F. For the inhibition assay, anti-MR1 antibody (0.001 to 1 μg/ml) and Ac-6-FP (0.1 to 100 μM) were added to reporter cells for 1 hour before the stimulation with 5-OP-RU and CA7S.
Human studies
The institutional review boards of Osaka University (approval number 29–4-10) approved blood draw protocols for healthy individuals. The research was performed in accordance with all relevant guidelines and regulations.
Stimulation of PBMCs
To prepare PBMCs, whole blood was collected in heparin-coated tubes and centrifuged to separate the cellular fraction and plasma using lymphocyte separation solution (d = 1.077) (Nacalai Tesque). PBMCs were labeled with CTV (Invitrogen) according to the manufacturer’s instructions. PBMCs were cultured with 5-OP-RU, CA3S, and CA7S in the absence of cytokines in RPMI 1640 (Sigma-Aldrich) supplemented with 10% fetal bovine serum, penicillin (100 U/ml; Sigma-Aldrich), streptomycin (100 μg/ml; MP Biomedicals), and 50 μM 2-mercaptoethanol (Nacalai Tesque). At the indicated days after stimulation, PBMCs were analyzed by flow cytometry (Attune NxT flow cytometer, Thermo Fisher Scientific) and CD3+CD161+MR1–5-OP-RU tetramer+ cells were sorted by the SH800S cell sorter (Sony Biotechnology) for sc-TCR-RNA-seq analyses. For survival assays, 1 × 106 PBMCs were left untreated or treated with 1 mM CA7S for 6 days in 150-μl cultures using 96-well round-bottom plates. At day 0, 150 μl of culture suspension was analyzed by flow cytometry, and the number of living (PI-negative) CD3+CD161+MR1–5-OP-RU-tetramer+ MAIT cells was defined as 100%. Every 2 days, the same staining was performed, and the survival ratio was calculated as a percentage against day 0.
Preparation of MAIT cells from mouse tissues
Thymi were dissociated by homogenizer to make single-cell suspensions. Single-cell suspensions of thymocyte were stained with APC-mouse MR1–5-OP-RU tetramer (NIH Tetramer Core Facility, Emory University Vaccine Center, Atlanta, USA). These were enriched by anti-APC microbeads according to the manufacturer’s instructions (Miltenyi Biotec). Livers were dissociated over a 40-μm nylon mesh. Liver mononuclear cells were isolated by density gradient centrifugation on a discontinuous 40 to 80% Percoll (Cytiva) gradient.
Single-cell–based transcriptome and TCR repertoire analysis
Single-cell transcriptome and TCR repertoire analysis were performed using Chromium Controller (10x Genomics) according to the manufacturer’s instructions as previously described (82). Gene expression-based clustering was performed using the Seurat R package [v3.1; (83)]. Cells with a mitochondrial content >10% and cells with <200 or >4000 genes detected were considered outliers (dying cells and empty droplets and doublets, respectively) and filtered out. The cluster analysis based on UMAP plot, volcano plot, and differential expression analysis were performed by BBrowser (BioTuring).
NMR spectroscopy
Fraction #84–45 was dissolved in 750 μl of D2O containing approximately 0.1% (v/v) MeCN as an internal standard (IS). NMR spectra were acquired on a JEOL ECZ600R spectrometer and were referenced internally according to residual solvent signals (HOD, MeCN). Signals characteristic of CA analogs were observed (three methyl protons at δ 0.73, 0.93, and 0.97 ppm; two methylene protons at δ 2.32 and 2.45 ppm; three methine protons at δ 3.52, 4.07, and 4.51 ppm in the 1H NMR spectrum and a carbonyl carbon at δ 180.8 ppm in the 13C NMR spectrum, Fig. 1D and fig. S2A). Through analyses including 2D-NMR (fig. S2, B to E), two peaks at δ 4.07 and 4.51 ppm in the 1H NMR spectrum were assigned to methine protons H-12 and H-7, respectively. The H-7 signal appeared more deshielded than predicted, indicating that a functional group is linked to the hydroxy group located at the C7 position of the CA skeleton.
LC-MS/MS quantitative analysis
Bile acids were extracted from each of the mouse tissue samples by 1 ml of extraction solvent (ice-cold methanol) after supplementing deuterium-labeled IS compounds (d4-CA; d4-CDCA; d4-TCA; d4-TCDCA; d4-LCA; d4-DCA; d4-GDCA; and d5-TLCA). Samples were mixed vigorously by vortex for 1 min followed by 5 min of sonication. The samples were centrifuged at 16,000g for 5 min at 4°C. The supernatant (800 μl) was then collected into clean tubes. For the targeted quantitative analysis of bile acids (data S1), the supernatant was diluted with methanol according to the abundance of bile acid contained in each tissue to prepare an analytical sample. A triple-quadrupole mass spectrometer equipped with an electrospray ionization (ESI) ion source (LCMS-8060; Shimadzu Corporation) was used in multiple reaction monitoring (MRM) mode. The conditions for the LC-MS/MS analysis were as follows: column, metal-free PEEK-coated InertSustain C18 (2.1 × 150 mm; particle size, 3 μm; GL Sciences Inc.); column temperature, 50°C; flow rate, 0.3 ml/min; mobile phase, water/acetonitrile/acetic acid (3:1:0.05, v/v/v) (A) and methanol/isopropanol/acetic acid (1:19:0.05, v/v/v) (B); gradient curve, 0% B at 0 min, 38% B at 17 min, 100% B at 25 min, 100% B at 35 min, 0% B at 35.1 min, and 0% B at 50 min; injection volume, 5 μl; mass analysis mode, negative ion mode; electrospray voltage, −3.0 kV; nebulizer gas flow rate, 2.0 liters/min; drying gas flow rate, 10.0 liters/min; desolvation temperature, 250°C; heat block temperature, 400°C; and detector voltage, 2.16 kV. The MRM mode and a dwell time of 10 ms per channel were used. Other optimized MRM parameters for bile acids are shown in data S1. The absolute contents of bile acids were calculated based on the calibration curves. The MRM calibration curves were generated from the triplicate analyses of these standard solutions using the chromatographic peak area of each analyte to that of the IS.
MR1 protein expression and purification
Recombinant human MR1 was produced in vitro as previously described (19, 84). Briefly, DNA encoding the extracellular domains of the MR1 heavy chain and β2m was transformed into Escherichia coli BL21(DE3). The proteins were subsequently overexpressed and purified as inclusion bodies. Refolding of MR1-HMB was performed via addition of 80 μM HMB, 120 mg of denatured inclusion body MR1 heavy chain, and 60 mg of β2m protein in 1 liter of refold buffer consisting of 0.1 M tris, pH 8.5, 2.5 M urea, 0.4 M l-arginine, 2 mM Na-EDTA, 0.5 mM oxidized glutathione, and 5 mM reduced glutathione for 16 hours at 4°C. The refolding solution was dialyzed against 10 mM tris, pH 8.0. The MR1-HMB complex was purified via size exclusion (S200 Superdex 16/600, GE Healthcare) and then dialyzed against 10 mM bis-tris propane, pH 6.5, and 150 mM NaCl at 4°C for 16 hours. The resultant MR1-empty was buffer exchanged to 10 mM tris, pH 8.0, and purified using anion exchange (HiTrap Q HP, GE Healthcare) chromatography as previously described (31).
Fluorescence polarization inhibition assay
Bile acids (CA7S, CA3S, and THBA) were each solubilized in ultra-pure water. The remainder of the ligands were dissolved in dimethyl sulfoxide (Sigma-Aldrich). Fluorescence polarization (FP) assays were conducted with serial dilutions of each ligand starting at a maximum concentration of 50 mM for CA7S, CA3S, and THBA; 20 mM DCF; 5 μM Ac-6-FP; 500 nM 5-OP-RU; 500 μM RL-6-Me-7-O; 5 mM EGCG; and 1 mM NLV peptide. Each compound was incubated with the previously reported synthetic fluorescent MR1-specific ligand, JYM20 (10 nM), and human MR1-empty (100 nM) in assay buffer (25 mM Hepes, pH 7.5, 150 mM NaCl, and 5 mM Na-EDTA) (31). FP was measured after 24 hours at 25°C using the PHERAstar microplate reader (BMG LabTech) as previously reported (31). Ligand binding curves were graphed as a sigmoidal concentration response curve using Prism Version 9.3.0 (GraphPad Software Inc.). Median inhibitory concentration (IC50) values for binding affinity were calculated at the ligand concentration required for 50% of inhibition for JYM20 binding to MR1-empty.
Nontargeted LC-HRMS/MS analysis
Nontargeted LC-HRMS/MS analyses were performed using a Nexera X2 UHPLC system (Shimadzu Co.) coupled with a Q Exactive high-performance benchtop quadrupole Orbitrap mass spectrometer (Thermo Fisher Scientific Inc.). LC analytical conditions were identical to those used in the targeted bile acid analysis method using LC-MS/MS. The full scanning HRMS analysis conditions were as follows: polarity, positive and negative ionization; sheath gas flow rate, 50 arb; auxiliary (Aux) gas flow rate, 10 arb; spray voltage for positive ion mode, 4.0 kV; spray voltage for negative ion mode, −3.0 kV; capillary temperature, 250°C; S-lens level, 60; heater temperature, 400°C; mass resolution, 70,000; automatic gain control (AGC) target, 3 × 106; maximum injection time, 200 ms; and scan range, 150 to 1500 (m/z). The conditions for data-dependent MS2 (dd-MS2) were as follows: mass resolution, 17,500; AGC target, 5 × 104; trap fill time, 80 ms; isolation width, ± 1.2 Da; fixed first mass, m/z 50; stepped normalized collision energy, 10, 30, and 45 eV; intensity threshold of precursor ions for dd-MS2 analysis, 1 × 104; apex trigger, 2 to 4 s; and dynamic exclusion, 2 s. The Compound Discoverer ver. 3.0 (Thermo Fisher Scientific Inc.) was used for HRMS data processing. Infusion analysis was performed using an ion trap/time-of-flight HRMS fitted with an ESI ion source (Shimadzu). The HRMS operating conditions were as follows: polarity, negative; electrospray voltage, −3.5 kV; curve desolvation line temperature, 200°C; heat block temperature, 200°C; nebulizing gas (N2) flow, 1.5 liters/min; drying gas (N2) pressure, 0.1 MPa; scan range, m/z 100 to 1500.
Statistical analysis
An unpaired two-tailed Student’s t test or one-way analysis of variance (ANOVA) with the Tukey’s multiple comparison test were performed for the statistical analyses using GraphPadPrism (Version 9.1.0, GraphPad Software Inc.). Asterisks denote the level of statistical significance (*P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001). P values were adjusted for multiple comparisons using the Benjamini-Hochberg method (FDR < 0.05) using Graph-Pad Prism version 9.1.0.
Supplementary Material
Acknowledgments:
We thank Y. Takeuchi, D. Motooka, Y-C. Liu, D. Okuzaki, X. Lu, A. Hidaka, C. Motozono, M. Yasuda, A. Tanaka, Y. Harima, T. Kondo, M. Goto, and M. Matsuda for experimental support and C. Schutt for discussion. The MR1 tetramer technology was developed jointly by J. McCluskey, J.R., and D.P.F., and the material was produced by the National Institutes of Health Tetramer Core Facility as permitted to be distributed by the University of Melbourne.
Funding:
This research was supported by AMED [JP21gm0910010, JP223fa627002 and JP21ak0101070 (S.Y.)], JSPS KAKENHI [JP20H00505, JP22H05183 (S.Y.), JP20K06938, JP20H04773 (S.I.), JP22H05185 (Y.I.), and JP21J22863 (E. Ito)], JST-Mirai Program [JPMJMI20G1 (Y.I.)], and Chugai Pharmaceuticals. This research was also supported by the Platform Project for Supporting Drug Discovery and Life Science Research [Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)] from AMED under grant number JP21am0101119 (support number 2774). This research was also supported by the Australian Research Council (ARC) [DP220102401 (J.R.) and CE200100012 (D.P.F.)], the National Institutes of Health (NIH) [RO1 AI148407-01A1(J.R. and D.P.F.)] and NHMRC investigator grants [2008981 (J.R.) and 2009551 (D.P.F.)]. W.A. was supported by Australian ARC Discovery Early Career Researcher Award (DECRA) fellowships (DE220101491).
Footnotes
Competing interests: J.R., J.Y.W.M., and D.P.F. are coinventors on patents describing MR1 tetramers and MR1-ligand complexes. All other authors declare that they have no competing financial interests.
Data and materials availability: Single-cell–based transcriptome data and bulk TCR-sequencing data have been deposited in Gene Expression Omnibus datasets under accession numbers GSE203394, GSE204917, and GSE228498. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
REFERENCES AND NOTES
- 1.Godfrey DI, Koay HF, McCluskey J, Gherardin NA, The biology and functional importance of MAIT cells. Nat. Immunol 20, 1110–1128 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Legoux F, Salou M, Lantz O, MAIT cell development and functions: The microbial connection. Immunity 53, 710–723 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Treiner E, Duban L, Bahram S, Radosavljevic M, Wanner V, Tilloy F, Affaticati P, Gilfillan S, Lantz O, Addendum: Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature 423, 1018 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, Bhati M, Chen Z, Kostenko L, Reantragoon R, Williamson NA, Purcell AW, Dudek NL, McConville MJ, O’Hair RAJ, Khairallah GN, Godfrey DI, Fairlie DP, Rossjohn J, McCluskey J, MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 491, 717–723 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Corbett AJ, Eckle SBG, Birkinshaw RW, Liu L, Patel O, Mahony J, Chen Z, Reantragoon R, Meehan B, Cao H, Williamson NA, Strugnell RA, Van Sinderen D, Mak JYW, Fairlie DP, Kjer-Nielsen L, Rossjohn J, McCluskey J, T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature 509, 361–365 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Legoux F, Bellet D, Daviaud C, El Morr Y, Darbois A, Niort K, Procopio E, Salou M, Gilet J, Ryffel B, Balvay A, Foussier A, Sarkis M, El Marjou A, Schmidt F, Rabot S, Lantz O et al. , Science 366, 494–499 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Constantinides MG, Link VM, Tamoutounour S, Wong AC, Perez-Chaparro PJ, Han SJ, Chen YE, Li K, Farhat S, Weckel A, Krishnamurthy SR, Vujkovic-Cvijin I, Linehan JL, Bouladoux N, Merrill ED, Roy S, Cua DJ, Adams EJ, Bhandoola A, Scharschmidt TC, Aubé J, Fischbach MA, Belkaid Y, MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science 366, eaax6624 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blacher E, Levy M, Tatirovsky E, Elinav E, Microbiome-modulated metabolites at the interface of host immunity. J. Immunol 198, 572–580 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Kurioka A, Walker LJ, Klenerman P, Willberg CB, MAIT cells: New guardians of the liver. Clin. Transl. Immunol 5, e98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edmans MD, Connelley TK, Jayaraman S, Vrettou C, Vordermeier M, Mak JYW, Liu L, Fairlie DP, Maze EA, Chrun T, Klenerman P, Eckle SBG, Tchilian E, Benedictus L, Identification and phenotype of MAIT cells in cattle and their response to bacterial infections. Front. Immunol 12, 627173 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeffery HC, Van Wilgenburg B, Kurioka A, Parekh K, Stirling K, Roberts S, Dutton EE, Hunter S, Geh D, Braitch MK, Rajanayagam J, Iqbal T, Pinkney T, Brown R, Withers DR, Adams DH, Klenerman P, Oo YH, Biliary epithelium and liver B cells exposed to bacteria activate intrahepatic MAIT cells through MR1. J. Hepatol 64, 1118–1127 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.von Seth E, Zimmer CL, Reuterwall-Hansson M, Barakat A, Arnelo U, Bergquist A, Ivarsson MA, Björkström NK, Primary sclerosing cholangitis leads to dysfunction and loss of MAIT cells. Eur. J. Immunol 48, 1997–2004 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Lett MJ, Mehta H, Keogh A, Jaeger T, Jacquet M, Powell K, Meier MA, Fofana I, Melhem H, Vosbeck J, Cathomas G, Heigl A, Heim MH, Burri E, Mertz KD, Niess JH, Kollmar O, Zech CJ, Ivanek R, Duthaler U, Klenerman P, Stroka D, Sinnreich MF, Stimulatory MAIT cell antigens reach the circulation and are efficiently metabolised and presented by human liver cells. Gut 71, 2526–2538 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang X-Z, Jo J, Tan AT, Sandalova E, Chia A, Tan KC, Lee KH, Gehring AJ, De Libero G, Bertoletti A, IL-7 licenses activation of human liver intrasinusoidal mucosal-associated invariant T cells. J. Immunol 190, 3142–3152 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Lamichhane R, Munro F, Harrop TWR, de la Harpe SM, Dearden PK, Vernall AJ, McCall JL, Ussher JE, Human liver-derived MAIT cells differ from blood MAIT cells in their metabolism and response to TCR-independent activation. Eur. J. Immunol 51, 879–892 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Lepore M, Kalinichenko A, Calogero S, Kumar P, Paleja B, Schmaler M, Narang V, Zolezzi F, Poidinger M, Mori L, de Libero G, Functionally diverse human T cells recognize non-microbial antigens presented by MR1. eLife 6, e24476 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crowther MD, Dolton G, Legut M, Caillaud ME, Lloyd A, Attaf M, Galloway SAE, Rius C, Farrell CP, Szomolay B, Ager A, Parker AL, Fuller A, Donia M, McCluskey J, Rossjohn J, Svane IM, Phillips JD, Sewell AK, Genome-wide CRISPR–Cas9 screening reveals ubiquitous T cell cancer targeting via the monomorphic MHC class I-related protein MR1. Nat. Immunol 21, 178–185 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young MH, U’Ren L, Huang S, Mallevaey T, Scott-Browne J, Crawford F, Lantz O, Hansen TH, Kappler J, Marrack P, Gapin L, MAIT cell recognition of MR1 on bacterially infected and uninfected Cells. PLOS ONE 8, e53789 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keller AN, Eckle SBG, Xu W, Liu L, Hughes VA, Mak JYW, Meehan BS, Pediongco T, Birkinshaw RW, Chen Z, Wang H, D’Souza C, Kjer-Nielsen L, Gherardin NA, Godfrey DI, Kostenko L, Corbett AJ, Purcell AW, Fairlie DP, McCluskey J, Rossjohn J, Drugs and drug-like molecules can modulate the function of mucosal-associated invariant T cells. Nat. Immunol 18, 402–411 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Harriff MJ, McMurtrey C, Froyd CA, Jin H, Cansler M, Null M, Worley A, Meermeier EW, Swarbrick G, Nilsen A, Lewinsohn DA, Hildebrand W, Adams EJ, Lewinsohn DM, MR1 displays the microbial metabolome driving selective MR1-restricted T cell receptor usage. Sci. Immunol 3, eaao2556 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eckle SBG, Birkinshaw RW, Kostenko L, Corbett AJ, McWilliam HEG, Reantragoon R, Chen Z, Gherardin NA, Beddoe T, Liu L, Patel O, Meehan B, Fairlie DP, Villadangos JA, Godfrey DI, Kjer-Nielsen L, McCluskey J, Rossjohn J, A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells. J. Exp. Med 211, 1585–1600 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McWilliam HEG, Eckle SBG, Theodossis A, Liu L, Chen Z, Wubben JM, Fairlie DP, Strugnell RA, Mintern JD, McCluskey J, Rossjohn J, Villadangos JA, The intracellular pathway for the presentation of Vitamin B-related antigens by the antigen-presenting molecule MR1. Nat. Immunol 17, 531–537 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Mak JYW, Xu W, Reid RC, Corbett AJ, Meehan BS, Wang H, Chen Z, Rossjohn J, McCluskey J, Liu L, Fairlie DP, Stabilizing short-lived Schiff base derivatives of 5-aminouracils that activate mucosal-associated invariant T cells. Nat. Commun 8, 14599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuoka T, Motozono C, Hattori A, Kakeya H, Yamasaki S, Oishi S, Ohno H, Inuki S, The effects of 5-OP-RU stereochemistry on its stability and MAIT-MR1 axis. Chembiochem 22, 672–678 (2021). [DOI] [PubMed] [Google Scholar]
- 25.Alnouti Y, Bile acid sulfation: A pathway of bile acid elimination and detoxification. Toxicol. Sci 108, 225–246 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Funabashi M, Grove TL, Wang M, Varma Y, McFadden ME, Brown LC, Guo C, Higginbottom S, Almo SC, Fischbach MA, A metabolic pathway for bile acid dehydroxylation by the gut microbiome. Nature 582, 566–570 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones BV, Begley M, Hill C, Gahan CGM, Marchesi JR, Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl. Acad. Sci. U.S.A. 105, 13580–13585 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choucair I, Nemet I, Li L, Cole MA, Skye SM, Kirsop JD, Fischbach MA, Gogonea V, Mark Brown J, Wilson Tang WH, Hazen SL, Quantification of bile acids: A mass spectrometry platform for studying gut microbe connection to metabolic diseases. J. Lipid Res. 61, 159–177 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaudhari SN, Luo JN, Harris DA, Aliakbarian H, Yao L, Paik D, Subramaniam R, Adhikari AA, Vernon AH, Kiliç A, Weiss ST, Huh JR, Sheu EG, Devlin AS, A microbial metabolite remodels the gut-liver axis following bariatric surgery. Cell Host Microbe 29, 408–424.e7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russell DW, Fifty years of advances in bile acid synthesis and metabolism. J. Lipid Res. 50, S120–S125 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang CJH, Awad W, Liu L, Mak JYW, Veerapen N, Illing PT, Purcell AW, Eckle SBG, Mccluskey J, Besra GS, Fairlie DP, Rossjohn J, Nours JL, Quantitative affinity measurement of small molecule ligand binding to major histocompatibility complex class-I-related protein 1 MR1. J. Biol. Chem 298, 102714 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang J, Bathena SP, Tong J, Roth M, Hagenbuch B, Alnouti Y, Kinetic analysis of bile acid sulfation by stably expressed human sulfotransferase 2A1 (SULT2A1). Xenobiotica 40, 184–194 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Alnouti Y, Klaassen CD, Tissue distribution and ontogeny of sulfotransferase enzymes in mice. Toxicol. Sci 93, 242–255 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Feng L, Yuen YL, Xu J, Liu X, Chan MYC, Wang K, Fong WP, Cheung WT, Lee SST, Identification and characterization of a novel PPARα-regulated and 7α-hydroxyl bile acid-preferring cytosolic sulfotransferase mL-STL (Sult2a8). J. Lipid Res. 58, 1114–1131 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koay HF, Gherardin NA, Enders A, Loh L, Mackay LK, Almeida CF, Russ BE, Nold-Petry CA, Nold MF, Bedoui S, Chen Z, Corbett AJ, Eckle SBG, Meehan B, D’Udekem Y, Konstantinov IE, Lappas M, Liu L, Goodnow CC, Fairlie DP, Rossjohn J, Chong MM, Kedzierska K, Berzins SP, Belz GT, McCluskey J, Uldrich AP, Godfrey DI, Pellicci DG, A three-stage intrathymic development pathway for the mucosal-associated invariant T cell lineage. Nat. Immunol 17, 1300–1311 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Koay HF, Su S, Amann-Zalcenstein D, Daley SR, Comerford I, Miosge L, Whyte CE, Konstantinov IE, D’Udekem Y, Baldwin T, Hickey PF, Berzins SP, Mak JYW, Sontani Y, Roots CM, Sidwell T, Kallies A, Chen Z, Nüssing S, Kedzierska K, Mackay LK, McColl SR, Deenick EK, Fairlie DP, McCluskey J, Goodnow CC, Ritchie ME, Belz GT, Naik SH, Pellicci DG, Godfrey DI, A divergent transcriptional landscape underpins the development and functional branching of MAIT cells. Sci. Immunol 4, eaay6039 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dusseaux M, Martin E, Serriari N, Péguillet I, Premel V, Louis D, Milder M, Le Bourhis L, Soudais C, Treiner E, Lantz O, Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood 117, 1250–1259 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Salou M, Legoux F, Gilet J, Darbois A, Du Halgouet A, Alonso R, Richer W, Goubet AG, Daviaud C, Menger L, Procopio E, Premel V, Lantz O, A common transcriptomic program acquired in the thymus defines tissue residency of MAIT and NKT subsets. J. Exp. Med 216, 133–151 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dias J, Leeansyah E, Sandberg JK, Multiple layers of heterogeneity and subset diversity in human MAIT cell responses to distinct microorganisms and to innate cytokines. Proc. Natl. Acad. Sci. U.S.A 114, E5434–E5443 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flament H, Rouland M, Beaudoin L, Toubal A, Bertrand L, Lebourgeois S, Rousseau C, Soulard P, Gouda Z, Cagninacci L, Monteiro AC, Hurtado-Nedelec M, Luce S, Bailly K, Andrieu M, Saintpierre B, Letourneur F, Jouan Y, Si-Tahar M, Baranek T, Paget C, Boitard C, Vallet-Pichard A, Gautier JF, Ajzenberg N, Terrier B, Pène F, Ghosn J, Lescure X, Yazdanpanah Y, Visseaux B, Descamps D, Timsit JF, Monteiro RC, Lehuen A, Outcome of SARS-CoV-2 infection is linked to MAIT cell activation and cytotoxicity. Nat. Immunol 22, 322–335 (2021). [DOI] [PubMed] [Google Scholar]
- 41.Dzhagalov I, Dunkle A, He Y-W, The anti-apoptotic Bcl-2 family member Mcl-1 promotes T lymphocyte survival at multiple stages. J. Immunol 181, 521–528 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takayama S, Sato T, Krajewski S, Kochel K, Irie S, Milian JA, Reed JC, Cloning and functional analysis of BAG-1: A novel Bcl-2-binding protein with anti-cell death activity. Cell 80, 279–284 (1995). [DOI] [PubMed] [Google Scholar]
- 43.Low CG, Luk ISU, Lin D, Fazli L, Yang K, Xu Y, Gleave M, Gout PW, Wang Y, BIRC6 protein, an inhibitor of apoptosis: Role in survival of human prostate cancer cells. PLOS ONE 8, e55837 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xia H, Wang W, Crespo J, Kryczek I, Li W, Wei S, Bian Z, Maj T, He M, Liu RJ, He Y, Rattan R, Munkarah A, Guan JL, Zou W, Suppression of FIP200 and autophagy by tumor-derived lactate promotes naïve T cell apoptosis and affects tumor immunity. Sci. Immunol 2, eaan4631 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Young TM, Reyes C, Pasnikowski E, Castanaro C, Wong C, Decker CE, Chiu J, Song H, Wei Y, Bai Y, Zambrowicz B, Thurston G, Daly C, Autophagy protects tumors from T cell-mediated cytotoxicity via inhibition of TNFα-induced apoptosis. Sci. Immunol 5, eabb9561 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Hinks TSC, Marchi E, Jabeen M, Olshansky M, Kurioka A, Pediongco TJ, Meehan BS, Kostenko L, Turner SJ, Corbett AJ, Chen Z, Klenerman P, McCluskey J, Activation and in vivo evolution of the MAIT cell transcriptome in mice and humans reveals tissue repair functionality. Cell Rep. 28, 3249–3262.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leng T, Akther HD, Hackstein CP, Powell K, King T, Friedrich M, Christoforidou Z, McCuaig S, Neyazi M, Arancibia-Cárcamo CV, Hagel J, Powrie F, Peres RS, Millar V, Ebner D, Lamichhane R, Ussher J, Hinks TSC, Marchi E, Willberg C, Klenerman P, TCR and inflammatory signals tune human MAIT cells to exert specific tissue repair and effector functions. Cell Rep. 28, 3077–3091.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lamichhane R, Schneider M, de la Harpe SM, Harrop TWR, Hannaway RF, Dearden PK, Kirman JR, Tyndall JDA, Vernall AJ, Ussher JE, TCR- or cytokine-activated CD8+ mucosal-associated invariant T cells are rapid polyfunctional effectors that can coordinate immune responses. Cell Rep. 28, 3061–3076.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Willing A, Jäger J, Reinhardt S, Kursawe N, Friese MA, Production of IL-17 by MAIT cells is increased in multiple sclerosis and is associated with IL-7 receptor expression. J. Immunol 200, 974–982 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Odumade OA, Weinreich MA, Jameson SC, Hogquist KA, Krüppel-like factor 2 regulates trafficking and homeostasis of γδ T cells. J. Immunol 184, 6060–6066 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mielke LA, Liao Y, Clemens EB, Firth MA, Duckworth B, Huang Q, Almeida FF, Chopin M, Koay HF, Bell CA, Hediyeh-Zadeh S, Park SL, Raghu D, Choi J, Putoczki TL, Hodgkin PD, Franks AE, Mackay LK, Godfrey DI, Davis MJ, Xue HH, Bryant VL, Kedzierska K, Shi W, Belz GT, TCF-1 limits the formation of Tc17 cells via repression of the MAF–RORγt axis. J. Exp. Med 216, 1682–1699 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pais Ferreira D, Silva JG, Wyss T, Fuertes Marraco SA, Scarpellino L, Charmoy M, Maas R, Siddiqui I, Tang L, Joyce JA, Delorenzi M, Luther SA, Speiser DE, Held W, Central memory CD8+ T cells derive from stem-like Tcf7hi effector cells in the absence of cytotoxic differentiation. Immunity 53, 985–1000.e11 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Chaudhari SN, Harris DA, Aliakbarian H, Luo JN, Henke MT, Subramaniam R, Vernon AH, Tavakkoli A, Sheu EG, Devlin AS, Bariatric surgery reveals a gut-restricted TGR5 agonist with anti-diabetic effects. Nat. Chem. Biol 17, 20–29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Teubner W, Meinl W, Florian S, Kretzschmar M, Glatt H, Identification and localization of soluble sulfotransferases in the human gastrointestinal tract. Biochem. J 404, 207–215 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Provine NM, Klenerman P, MAIT cells in health and disease. Annu. Rev. Immunol 38, 203–228 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Martin E, Treiner E, Duban L, Guerri L, Laude H, Toly C, Premel V, Devys A, Moura IC, Tilloy F, Cherif S, Vera G, Latour S, Soudais C, Lantz O, Stepwise development of MAIT cells in mouse and human. PLoS Biol. 7, e54 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cui Y, Franciszkiewicz K, Mburu YK, Mondot S, Le Bourhis L, Premel V, Martin E, Kachaner A, Duban L, Ingersoll MA, Rabot S, Jaubert J, De Villartay JP, Soudais C, Lantz O, Mucosal-associated invariant T cell-rich congenic mouse strain allows functional evaluation. J. Clin. Invest 125, 4171–4185 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wakao H, Yoshikiyo K, Koshimizu U, Furukawa T, Enomoto K, Matsunaga T, Tanaka T, Yasutomi Y, Yamada T, Minakami H, Tanaka J, Oda A, Sasaki T, Wakao R, Lantz O, Udagawa T, Sekiya Y, Higuchi K, Harada N, Nishimura K, Ohtaka M, Nakanishi M, Fujita H, Expansion of functional human mucosal-associated invariant T cells via reprogramming to pluripotency and redifferentiation. Cell Stem Cell 12, 546–558 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Thomae BA, Eckloff BW, Freimuth RR, Wieben ED, Weinshilboum RM, Human sulfotransferase SULT2A1 pharmacogenetics: Genotype-to-phenotype studies. Pharmacogenomics J. 2, 48–56 (2002). [DOI] [PubMed] [Google Scholar]
- 60.Gohal SA, Rasool MI, Bairam AF, Alatwi ES, Alherz FA, Abunnaja MS, El Daibani AA, Kurogi K, Liu MC, Effects of genetic polymorphisms on the sulfation of doxorubicin by human SULT1C4 allozymes. J. Biochem 170, 419–426 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR, T cell receptor antagonist peptides induce positive selection. Cell 76, 17–27 (1994). [DOI] [PubMed] [Google Scholar]
- 62.Tanchot C, Lemonnier FA, Pérarnau B, Freitas AA, Rocha B, Differential requirements for survival and proliferation of CD8 naïve or memory T cells. Science 276, 2057–2062 (1997). [DOI] [PubMed] [Google Scholar]
- 63.Viret C, Wong FS, Janeway CA Jr., Designing and maintaining the mature TCR repertoire. Immunity 10, 559–568 (1999). [DOI] [PubMed] [Google Scholar]
- 64.Chen Z, Liu S, He C, Sun J, Wang L, Chen H, Zhang F, CXCL12-CXCR4-mediated chemotaxis supports accumulation of mucosal-associated invariant T cells into the liver of patients with PBC. Front. Immunol 12, 578548 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cao W, Kayama H, Chen ML, Delmas A, Sun A, Kim SY, Rangarajan ES, McKevitt K, Beck AP, Jackson CB, Crynen G, Oikonomopoulos A, Lacey PN, Martinez GJ, Izard T, Lorenz RG, Rodriguez-Palacios A, Cominelli F, Abreu MT, Hommes DW, Koralov SB, Takeda K, Sundrud MS, The xenobiotic transporter Mdr1 enforces T cell homeostasis in the presence of intestinal bile acids. Immunity 47, 1182–1196.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Corbett CL, Bartholomew TC, Billing BH, Summerfield JA, Urinary excretion of bile acids in cholestasis: Evidence for renal tubular secretion in man. Clin. Sci. 61, 773–780 (1981). [DOI] [PubMed] [Google Scholar]
- 67.Valestrand L, Zheng F, Hansen SH, Øgaard J, Hov JR, Björkström NK, Karlsen TH, Jiang X, Melum E, Bile from patients with primary sclerosing cholangitis contains mucosal-associated invariant T-cell antigens. Am. J. Pathol 192, 629–641 (2022). [DOI] [PubMed] [Google Scholar]
- 68.Jiang X, Lian M, Li Y, Zhang W, Wang Q, Wei Y, Zhang J, Chen W, Xiao X, Miao Q, Bian Z, Qiu D, Fang J, Ansari AA, Leung PSC, Coppel RL, Tang R, Gershwin ME, Ma X, The immunobiology of mucosal-associated invariant T cell (MAIT) function in primary biliary cholangitis: Regulation by cholic acid-induced interleukin-7. J. Autoimmun 90, 64–75 (2018). [DOI] [PubMed] [Google Scholar]
- 69.Boudinot P, Mondot S, Jouneau L, Teyton L, Lefranc MP, Lantz O, Restricting nonclassical MHC genes coevolve with TRAV genes used by innate-like T cells in mammals. Proc. Natl. Acad. Sci. U.S.A. 113, E2983–E2992 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsukamoto K, Deakin JE, Graves JAM, Hashimoto K, Exceptionally high conservation of the MHC class I-related gene, MR1, among mammals. Immunogenetics 65, 115–124 (2013). [DOI] [PubMed] [Google Scholar]
- 71.Hofmann AF, Hagey LR, Krasowski MD, Bile salts of vertebrates: Structural variation and possible evolutionary significance. J. Lipid Res. 51, 226–246 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Awad W, Le Nours J, Kjer-Nielsen L, McCluskey J, Rossjohn J, Mucosal-associated invariant T cell receptor recognition of small molecules presented by MR1. Immunol. Cell Biol. 96, 588–597 (2018). [DOI] [PubMed] [Google Scholar]
- 73.Salio M, Awad W, Veerapen N, Gonzalez-Lopez C, Kulicke C, Waithe D, Martens AWJ, Lewinsohn DM, Hobrath JV, Cox LR, Rossjohn J, Besra GS, Cerundolo V, Ligand-dependent downregulation of MR1 cell surface expression. Proc. Natl. Acad. Sci. U.S.A. 117, 10465–10475 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xue W, Chen S, Yin H, Tammela T, Papagiannakopoulos T, Joshi NS, Cai W, Yang G, Bronson R, Crowley DG, Zhang F, Anderson DG, Sharp PA, Jacks T, CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature 514, 380–384 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mak JYW, Liu L, Fairlie DP, Chemical modulators of mucosal associated invariant T cells. Acc. Chem. Res 54, 3462–3475 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McWilliam HEG, Mak JYW, Awad W, Zorkau M, Cruz-Gomez S, Lim HJ, Yan Y, Wormald S, Dagley LF, Eckle SBG, Corbett AJ, Liu H, Li S, Reddiex SJJ, Mintern JD, Liu L, McCluskey J, Rossjohn J, Fairlie DP, Villadangos JA, Endoplasmic reticulum chaperones stabilize ligand-receptive MR1 molecules for efficient presentation of metabolite antigens. Proc. Natl. Acad. Sci. U.S.A. 117, 24974–24985 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tilloy F, Treiner E, Park SH, Garcia C, Lemonnier F, De La Salle H, Bendelac A, Bonneville M, Lantz O, An invariant T cell receptor α chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted α/β T cell subpopulation in mammals. J. Exp. Med 189, 1907–1921 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yamasaki S, Ishikawa E, Sakuma M, Ogata K, Sakata-Sogawa K, Hiroshima M, Wiest DL, Tokunaga M, Saito T, Mechanistic basis of pre-T cell receptor-mediated autonomous signaling critical for thymocyte development. Nat. Immunol 7, 67–75 (2006). [DOI] [PubMed] [Google Scholar]
- 79.Yamasaki S, Ishikawa E, Sakuma M, Hara H, Ogata K, Saito T, Mincle is an ITAM-coupled activating receptor that senses damaged cells. Nat. Immunol 9, 1179–1188 (2008). [DOI] [PubMed] [Google Scholar]
- 80.Matsumoto Y, Kishida K, Matsumoto M, Matsuoka S, Kohyama M, Suenaga T, Arase H, A TCR-like antibody against a proinsulin-containing fusion peptide ameliorates type 1 diabetes in NOD mice. Biochem. Biophys. Res. Commun 534, 680–686 (2021). [DOI] [PubMed] [Google Scholar]
- 81.Ishikawa E, Ishikawa T, Morita YS, Toyonaga K, Yamada H, Takeuchi O, Kinoshita T, Akira S, Yoshikai Y, Yamasaki S, Direct recognition of the mycobacterial glycolipid, trehalose dimycolate, by C-type lectin Mincle. J. Exp. Med 206, 2879–2888 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lu X, Hosono Y, Nagae M, Ishizuka S, Ishikawa E, Motooka D, Ozaki Y, Sax N, Maeda Y, Kato Y, Morita T, Shinnakasu R, Inoue T, Onodera T, Matsumura T, Shinkai M, Sato T, Nakamura S, Mori S, Kanda T, Nakayama EE, Shioda T, Kurosaki T, Takeda K, Kumanogoh A, Arase H, Nakagami H, Yamashita K, Takahashi Y, Yamasaki S, Identification of conserved SARS-CoV-2 spike epitopes that expand public cTfh clonotypes in mild COVID-19 patients. J. Exp. Med 218, e20211327 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hafemeister C, Satija R, Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Awad W, Ler GJM, Xu W, Keller AN, Mak JYW, Lim XY, Liu L, Eckle SBG, Le Nours J, McCluskey J, Corbett AJ, Fairlie DP, Rossjohn J, The molecular basis underpinning the potency and specificity of MAIT cell antigens. Nat. Immunol 21, 400–411 (2020). [DOI] [PubMed] [Google Scholar]
- 85.Goto J, Kato H, Hasegawa F, Nambara T, Synthesis of monosulfates of unconjugated and conjugated bile acids. Chem. Pharm. Bull 27, 1402–1411 (1979). [Google Scholar]
- 86.Thakare R, Gao H, Kosa RE, Bi YA, Varma MVS, Cerny MA, Sharma R, Kuhn M, Huang B, Liu Y, Yu A, Walker GS, Niosi M, Tremaine L, Alnouti Y, Rodrigues AD, Leveraging of rifampicin-dosed cynomolgus monkeys to identify bile acid 3-O-sulfate conjugates as potential novel biomarkers for organic anion-transporting polypeptides. Drug Metab. Dispos 45, 721–733 (2017). [DOI] [PubMed] [Google Scholar]
- 87.Nonappa UM, First chemical synthesis, aggregation behavior and cholesterol solubilization properties of pythocholic acid and 16α-hydroxycholic acid. European J. Org. Chem 2007, 3331–3336 (2007). [Google Scholar]
- 88.Del Amo V, Siracusa L, Markidis T, Baragaña B, Bhattarai KM, Galobardes M, Naredo G, Pérez-Payán MN, Davis AP, Differentially-protected steroidal triamines; scaffolds with potential for medicinal, supramolecular, and combinatorial chemistry. Org. Biomol. Chem 2, 3320–3328 (2004). [DOI] [PubMed] [Google Scholar]
- 89.Kakiyama G, Muto A, Shimada M, Mano N, Goto J, Hofmann AF, Iida T, Chemical synthesis of 3β-sulfooxy-7β-hydroxy-24-nor-5-cholenoic acid: An internal standard for mass spectrometric analysis of the abnormal Δ5-bile acids occurring in Niemann-Pick disease. Steroids 74, 766–772 (2009). [DOI] [PubMed] [Google Scholar]
- 90.Festa C, Renga B, D’Amore C, Sepe V, Finamore C, De Marino S, Carino A, Cipriani S, Monti MC, Zampella A, Fiorucci S, Exploitation of cholane scaffold for the discovery of potent and selective farnesoid X receptor (FXR) and G-protein coupled bile acid receptor 1 (GP-BAR1) ligands. J. Med. Chem 57, 8477–8495 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.