Abstract
Aging is a nearly inescapable trait among organisms yet lifespan varies tremendously among different species spanning several orders of magnitude in vertebrates alone. This vast phenotypic diversity is driven by distinct evolutionary trajectories and tradeoffs that are reflected in patterns of diversification and constraint in organismal genomes. Age-specific impacts of selection also shape allele frequencies in populations impacting disease susceptibility and environment-specific mortality risk. Further, the mutational processes that spawn this genetic diversity both in the germline and somatic cells are strongly influenced by age and life history. Here, we discuss recent advances in our understanding of the evolution of aging and lifespan at organismal, population, and cellular scales and highlight the outstanding questions that remain unanswered.
Keywords: aging, lifespan, genetics, evolution, mutation, diversity
Evolutionary theory of aging
Evolutionary theory predicts that aging is an inevitable result of the increased selective impact of genes that influence early-life survival and fecundity compared with genes that act late in life [1–3]. This observation was first explicitly made by Peter Medawar, building off of R.A. Fisher’s introduction of the concept of age-specific reproductive value, which models the age-dependent future genetic contributions of individuals [4]. Indeed, many of the seminal contributions to the evolutionary theory of aging can be traced to the founders of the modern synthesis (reviewed in [5]). Medawar’s mutation-accumulation theory posits that late-acting deleterious alleles are likely to accumulate due to the reduced force of natural selection in older individuals (sometimes referred to as a “selection shadow”). A similar but distinct theory of aging, antagonistic pleiotropy, proposes that alleles that are beneficial early in life, but deleterious late in life, will accumulate. The key difference between these two theories is that in antagonistic pleiotropy aging evolves due to an evolutionary trade-off between the fitness of old and young individuals while in contrast, in mutation accumulation aging-associated alleles are neutral in young individuals. While the relative contributions of antagonistic pleiotropy and mutation accumulation remain unclear, the extensive and extraordinary variation in lifespan among organisms highlights that different evolutionary scenarios and tradeoffs can favor vastly different outcomes of this phenotype. Medawar also noted the relative ambiguity of the term “aging,” which is used to refer to almost any time-dependent change on a biological entity [6]. This is distinct from lifespan which refers to the age of death of an individual, or senescence which refers to biological changes yielding an increased probability of mortality as a function of age. Recently, several technological advances alongside large-scale genetic datasets have propelled renewed excitement about the evolution of aging and the genetics of age structured populations. Here, we highlight recent progress in our understanding of this topic, ranging from new insights into the life histories of diverse organisms to the new approaches enabling us to identify aging-genes and age-associated biological phenomena (Figure 1).
Figure 1:
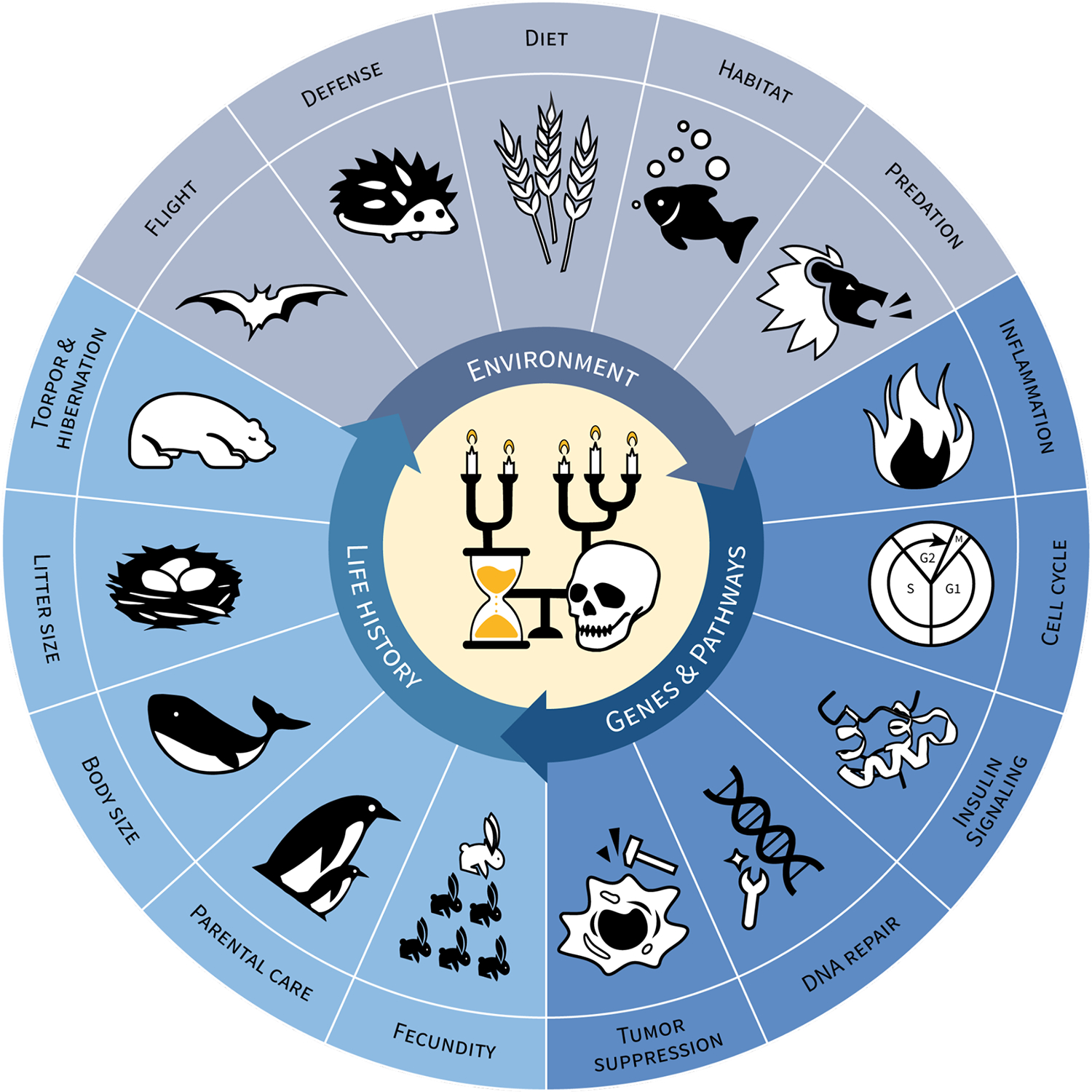
Evolutionary dynamics underlying aging and longevity.
The interplay between genes, life history traits, and environmental factors drives the evolution of aging and lifespan. Highlighted above are several of the key elements in each domain that have known associations with aging and longevity. The iconography in the center of the figure depicts differences in lifespan (candle height) and their evolutionary relationships, in the Vanitas style.
Insights from extreme agers of the animal kingdom
There exists an extraordinary variation in lifespan among organisms on this planet spanning several orders of magnitude in vertebrates alone [7,8]. Such variation provides an exquisite natural dataset in which to probe the evolution of this trait providing insights into the evolutionary trade-offs that constrain and mold this phenotype, as well as the scenarios under which the extremes evolve.
The life history of longevity:
Evolutionary theory predicts that increased lifespan will evolve in the context of low extrinsic mortality, i.e. few environmental threats posing a risk of death. In contrast, short lifespans will evolve in organisms with high extrinsic mortality (for example due to predation). Lifespan, along with many other life history traits, tends to exhibit strong allometric scaling, with larger animals being longer lived. However, even after controlling for body size and phylogenetic relatedness, lifespan exhibits covariation with other life history traits, with the majority of this variance explainable across two independent axes [9,10]. Generation time and age-at-first reproduction co-vary with lifespan along one axis, while the distribution of age-specific mortality and reproduction co-vary along the other. Thus, short lived species tend to be smaller and have shorter generation and maturation times. They also have high rates of iteroparity and many offspring, with highly concentrated mortality risk (e.g. low juvenile survival). Importantly, these life-history correlations mean that extreme lifespan can emerge from indirect selection on a covarying trait.
The relationship between age and mortality is commonly modeled using the Gompertz hazard function (Box 1). This simple empirical model describes the exponential increase in the mortality rate with age. While the increased probability of mortality with age has been considered a universal trait among organisms, recent comparative studies of tortoises [11], and nonavian reptiles and amphibians [9,12] have identified several cases in which the rates of aging are negligibly small, or even negative. Other species that have been suggested to exhibit so-called “negligible senescence” include rockfish [13] and naked mole rats [14]. Many of these species, in particular fish and some reptiles and amphibians, exhibit “indeterminate” growth and fecundity, providing a key clue into the basis of their remarkable longevity: the continual growth and production of offspring throughout lifespan places a strong selective cost on late-acting deleterious mutations. Additionally, in rockfish and many other fishes, fecundity scales disproportionately with body size [15] meaning that larger, older, females will contribute more offspring to the next generation than younger, smaller individuals. Several other correlates of lifespan have been identified including metabolic rate [16,17], temperature and thermoregulatory mode [12], protective phenotypes (e.g. toxins, shells, and flight) [18], and sex. Thus, extreme lifespan evolves across many different phenotypic axes.
Box 1 -. Modeling mortality.
The lifespan of organisms is commonly modeled using the Gompertz-Makeham equation which takes the form of the hazard function:
where m(t), the mortality hazard, represents the instantaneous probability of death at time t and m(t)dt is the fraction of the population that dies from t to t + dt. This equation states that the probability of mortality of an organism can be modeled by an age-dependent component that increases exponentially with age (the Gompertzian component), and an age-independent component (the Makeham component). The parameter G reflects the exponential mortality rate coefficient, or “rate of aging”. A0 is a constant representing the intrinsic vulnerability or initial mortality rate (IMR) while M0 is the age independent mortality rate or extrinsic mortality rate (EMR). Due to the challenge in distinguishing IMR from EMR often a simplified Gompertz equation is used to model mortality in which the M0 term is dropped. The survival function S(t), which represents the probability of an individual surviving until time t, can be derived from the hazard function and takes the form:
More complex mortality functions, such as the Siler model [105], allow mortality to vary more flexibly throughout lifespan quantifying the increased mortality in infants and post-reproductive adults explicitly. The figure below shows mortality and survival curves for several different species, including long-lived humans and ultra long-lived rockfish, as well as a turtle and lizard that exhibit “negative senescence.”
Figure Box 1:
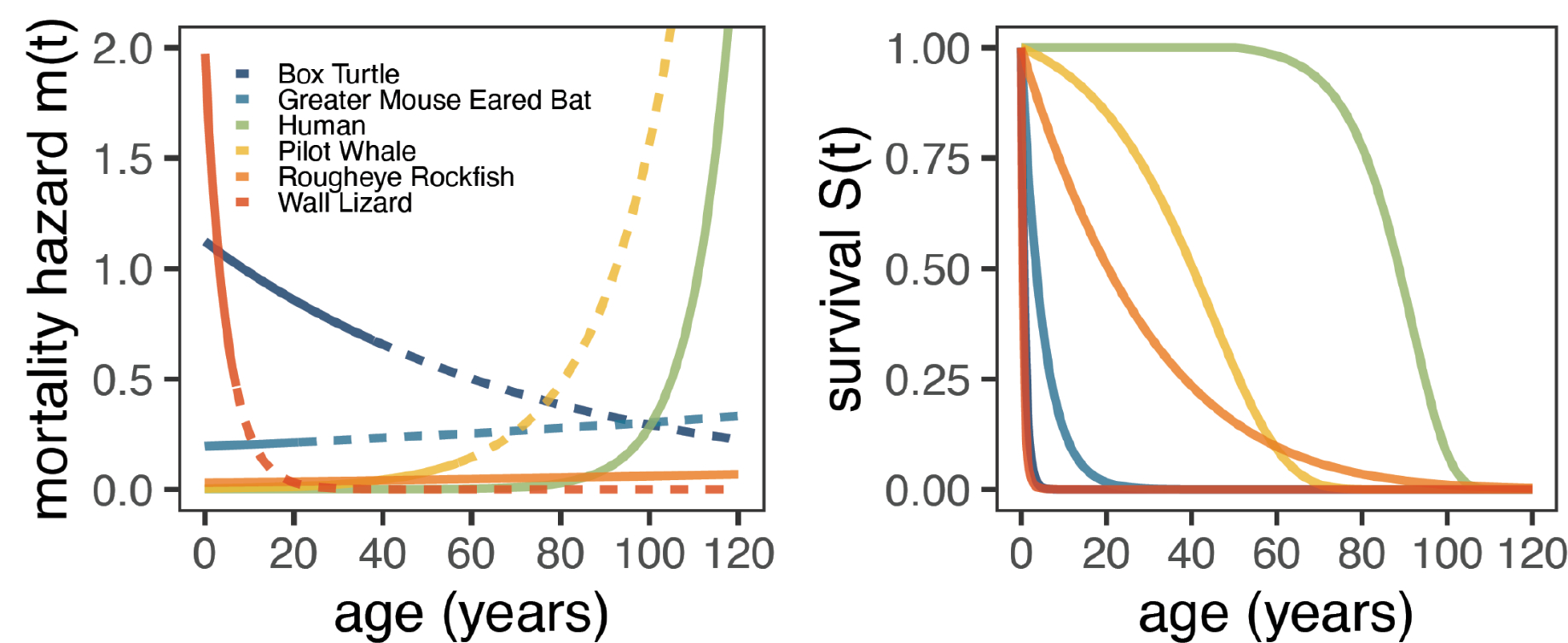
Mortality hazard (instantaneous probability of death) and survival curves for six diverse species. Mortality hazard lines become dotted after the maximum lifespan of the species.
Death and sex (chromosomes):
Extensive sex differences in lifespan are apparent across many taxa (Reviewed in [19]); intriguingly, the direction of this effect consistently favors the homogametic sex (i.e. females in XY systems like mammals, and males in ZW systems like birds). Indeed, a recent comparison of 299 species found that the homogametic sex lived, on average, 17.6% longer than the heterogametic sex [20] and in-depth analyses of mammals [21] and amphibians [22] have shown similar results. One explanation of this phenomenon, the “unguarded X hypothesis,” posits that the recessive mutations on the X (or Z) chromosomes will be exposed only in the heterogametic context. However, population genetics models have suggested that the effect size of the difference in lifespan between the hetero- and homo-gametic sexes is too large to be explained by the unmasking of recessive variation alone [23]. An alternative explanation is that the Y (or Z) chromosome itself becomes toxic with age, due to the derepression and misexpression of repetitive DNA. This hypothesis is supported by recent work in drosophila demonstrating that increased Y chromosome copy number is negatively correlated with lifespan [24]. Further bioinformatic analyses across several taxa provide further support for this hypothesis and find that the relative sizes of the X and Y chromosomes in mammals (though not Z or W in birds) are associated with lifespan [25]. Heterochromatin becomes de-repressed with age in several species, however, the Y chromosome is particularly rich in heterochromatin-repressed repetitive sequences. Such repetitive, heterochromatic sequences have classically been challenging to interrogate due to limitations in sequencing technologies. The advent of several new long reads sequencing approaches will hopefully open these regions to study across species with different lifespans as well as throughout the lifespan of individual organisms.
Comparative genomics of extreme aging
While classical model organisms including yeast, worms, flies, and mice have provided myriad insights into the molecular underpinnings of lifespan variation, novel long read sequencing and genome assembly approaches are empowering comparative genomics-based interrogation of long-lived wild species across the planet that would be otherwise be intractable to study in the lab (Box 2).
Box 2 -. The evolution of comparative genomics.
Long-read sequencing technologies have dramatically reshaped modern approaches to evolutionary genomics. Over the past decade, the rapid improvement of nanopore and PacBio sequencing technologies has enabled remarkable advances in genome assembly. These technologies also allow us to access notoriously challenging regions of the genome, including segmental duplications, telomeres, centromeres, and other repeat-rich regions which can span many megabases. Short read assemblies in contrast are remarkably fragmented, frustrating analyses. Long-read approaches obviate these difficulties by directly sequencing single molecules of HMW (high molecular weight) DNA up to many megabases in length, permitting coverage across the aforementioned regions. Complete, reference quality genomes have historically only been available for a handful of species (e.g. human and mouse) and were the result of massive-scale consortia efforts. In contrast, long read sequencing alongside recent improvements in genome assembly allow individual labs to contribute near-complete genomes. Complete genomes from diverse species enables analysis of previously uncharacterized coding and non-coding sequences. The many structural rearrangements present in genomes can also be assessed for the first time: linking genome structural variants to phenotypes has remained relatively unexplored due to technological shortcomings in short-read assembly. Ultimately, comprehensive comparative genomics across the tree of life requires so-called telomere-to-telomere assemblies which are gapless covering even the most structurally challenging loci. Nevertheless, currently comparative genomics relies on genomes assembled to varying levels of completion which are suited for different comparative genomics analyses (Table Box II).
Table Box II:
A brief overview of reference genome resources for comparative and evolutionary genomics.
| Assembly type | Strengths | Recent resources |
|---|---|---|
| Short-read genomes | Resequencing, population genetics, initial characterizations of genome diversity | Zoonomia [106], Primate Genome Project [107] |
| Long-read genomes | Near-complete analyses of autosomal gene content and diversity, characterization of structural variation, near-complete haplotype assembly, initial characterizations of complex loci (see below) | Vertebrate Genome Project [48]; Human Pangenome Reference Consortium [108] |
| T2T genomes | Complete analyses of autosomal and sex chromosomal gene content, full characterization of complex regions including segmental duplications, centromeres, telomeres, and rDNA repeats | T2T Consortium [82] |
Insights from genome assemblies:
The complete genomes of several extremely long lived non-model species have been generated recently including the bowhead whale (Balaena mysticetus, >200 years) [26]; the giant tortoises Chelonoidis abingdonii and Aldabrachelys gigantea (>100 years) [27,28]; the Asian and African elephant (Loxodonta africana and Elephas maximus, >80 and >65 years respectively) [28]; naked mole rats (Heterocephalus glaber, >35 years) [29]; the Canadian beaver (Castor canadensis, >23 years) [29]; several long-lived rockfish of the Sebastes clade (100–200 years) [13]; immortal cnidarian jellyfish (Turriitopsis dohrnii) [30]; and various bats [31]. Substantially less attention has been paid to the genomes of wild, short-lived species with the critical exception of the short-lived African turquoise killifish (Nothobranchius furzeri, 4–6mo) [32,33], which has emerged as a key model organism of aging and suspended animation [34]. Individual analyses of the genomes of these diverse species have repeatedly highlighted signatures of selection in key aging pathways including insulin signaling, fatty acid metabolism, DNA repair, inflammation, cell cycle, and tumor suppression pathways among others (pathways reviewed in [35–37]). In long lived species these pathways often exhibit signatures of positive selection [13,29,38] highlighting the evolutionary innovations necessary for extreme lifespan. Intriguingly, in short lived killifish evidence points to these critical age-associated pathways being under relaxed constraint [32,33] due to the reduced selective pressure on later-acting genes with roles in maintenance and longevity.
Such signatures are also observed in structural and copy number changes in the genome. Bowhead whales exhibit duplications in genes associated with DNA damage repair and cancer [26]; rockfish exhibit age-associated copy number expansion of butyrophilin genes, which serve an immuno-suppresive function [13]; immortal cnidarian jellyfish exhibit expansions in genes associated with DNA repair and replication, telomere maintenance, oxidative stress, stem cell maintenance and intercellular communication [30]; and bats exhibit an expanded repertoire of APOBEC3 genes, which have antiviral functions and have known mutational signatures [31,39,40]. In contrast, short-lived annual killifish exhibit increased genome sizes attributed to increased mobile element content, potentially facilitated by relaxed selection on genome maintenance in these species [33].
Insights from genome pensembles:
While studies of individual genomes have been highly informative, methods leveraging comparative analyses of phylogenetically diverse taxa with broad variation in a phenotype of interest can also yield unique insights. Broadly speaking, such methods seek to identify correlations between the rate of evolution of a gene or region with the phenotype of interest across taxa. This approach has been successfully used to quantify genetic constraint associated with extended lifespan across the entire mammalian clade [38], longevity among different rockfish species [13,41], and differences in the constraint of both genes and non-coding regions associated with several different phenotypes in other mammals [42–44]. These approaches have highlighted that overall, long-lived species tend to exhibit signatures of increased constraint in many key aging pathways including DNA repair, cell cycle, cell death, insulin signaling and immunity [38]. In contrast, short-lived killifish populations exhibit small effective population sizes and the accumulation of deleterious mutations in similar pathways [45]. Taken together alongside insights from individual genomes it emerges that in general, key aging pathways are under strong purifying selection in long lived taxa with independent distinct innovations observed in select genes in individual species. This is well illustrated in rockfish where several long lived species exhibit signatures of positive selection in DNA repair genes, however different positively selected genes are found in different species [13]. In rockfish these approaches have also been used to dissect apart the direct genetic drivers of extreme lifespan (largely associated with nutrient signaling) from the indirect genetic drivers which acted to increase lifespan by increasing body size (associated with DNA repair and mTOR signaling associated genes).
Despite the power of these comparative approaches they still exhibit some drawbacks, such as a reliance on large multiple sequence alignments (MSAs). While advances have been made in this space [46], whole genome MSAs are still extremely computationally expensive to generate. Furthermore, genetic variation that is not well described by MSAs, such as structural variation, will be ignored. Another important caveat is that poor quality input genomes will likely lead to unreliable inferences and exclude complex and rapidly evolving loci (Box 2). While recent efforts have prioritized broad taxonomical sampling over genome quality [47], future projects such as the Vertebrate Genome Project [48] are focused on constructing high quality resources which will be of substantial value for comparative genomics of extreme phenotypes.
Insights from functional genomics:
Functionally characterizing the genes and pathways identified from comparative genomics studies is essential to fully dissecting the mechanistic underpinnings of differences in lifespan. However, a primary challenge to such work is the intractability of exploring molecular mechanisms in non-model, long-lived organisms (Box 3). Nonetheless, several studies have leveraged cell culture models to explore hypotheses from comparative genomics studies. Work in elephants and their large-bodied relatives (Proboscideans) has identified gene duplications in several tumor suppressor genes [49–52]; two of these genes, LIF6 and TP53-Retrogene9, represent functional retrogenes with the ability to induce apoptosis upon expression [49,52]. In contrast, similar work in the long-lived bowhead whale has demonstrated a preference for repair mechanisms over the elimination of damaged cells. This includes work demonstrating the effectiveness of a cloned retrogene of the cell cycle regulator CDKN2C that serves to enhance cell cycle arrest and cell viability in response to DNA damage [53]; and additional work in bowhead whale cells demonstrating an enhanced DNA damage repair response [54]. Work in long-lived rodents has shown a spectrum of responses to various aging-related stresses, many private to each species [55–58]. These studies highlight the exciting possibilities of using cellular models of diverse and intractable species to understand even highly complex phenotypes such as body size and aging (Box 3).
Box 3 -. Aging in a dish.
While the ultimate tests of aging-related hypotheses and treatment are in vivo, longitudinal studies of putative life- or health-span extensions are costly and complex, if not impossible in exceptionally long lived taxa. Cell culture models have long served as a bridge between the trials of research and the tribulations of clinical trials; yet it is critical that appropriate models are utilized to avoid wasted effort.
Many processes in aging are tissue-specific, and non-cell autonomous. While any cell type can be used in principle for initial exploration of candidate genes and pathways, robust characterization of aging-associated genes and interventions requires more faithful experimental systems. Organoid models and organism-on-a-chip studies have been proposed as a means of resolving this in humans [109]. These approaches involve either ex vivo culturing of organ tissues under controlled conditions; or the establishment of organoids derived from induced pluripotent stem cells. Compared to in vivo studies, ex vivo organoid systems provide an unmatched increase in experimental tractability and a level of control of extrinsic, confounding factors.
Because these approaches require invasive, possibly lethal sampling of target species, an alternative approach is to construct organoid models de novo using stem-cell-based approaches. Specifically, induced pluripotent stem cells (iPSCs) promise a robust system for generating any tissue of interest with minimal non-lethal sampling of individual animals. Both a key strength and limitation of iPSC-based approaches is that epigenetic changes associated with aging are lost during the reprogramming process [110,111]. The generation of iPSCs, however, remains a challenging process outside of key model species such as humans and mice. While many methods exist to transform cells into iPSCs, many of these methods rely or can result in alterations to the genome or to the fundamental cellular biology of the species, and special care must be taken to ensure that the resulting iPSCs replicate the true biology of their original host.
The following table highlights a selection of some of the ongoing work in different long-lived clades of interest using cell culture models, and the progress that has been made towards improving the state-of-the-art of ex vivo aging research.
Table Box III:
A select list of studies leveraging cell culture or organoid models in long-lived clades of interest.
| Species | Cell Types | iPSC protocols | Organoids |
|---|---|---|---|
| Human | Various; see references for reviews | Various; see references for reviews | Various (iPSC, ex vivo); see references for reviews, eg [112] |
| Non-human primate | Various; see references for reviews | Various [113–116] | Various (iPSC, ex vivo); see references for reviews, eg [117] |
| Mouse | Various; see references for reviews | Various; see references for reviews | Various (iPSC, ex vivo); see references for reviews, eg [117] |
| Naked Mole Rat | fibroblasts (various tissues)[55,118–121] | Integrative plasmid; Viral transduction [122–125] | Aorta (ex vivo) [117,126] |
| Elephant | fibroblasts[49,51] | N/A | N/A |
| Whale | Fibroblasts [53,54,127–129] | N/A | N/A |
| Fish and Sharks | Various [130,131] | Viral transformation [132] | Retina (iPSC) [133,134] |
| Turtles | fibroblasts (various tissues)[135–137] | NA | Liver [138] |
| Bats | Fibroblasts (skin, kidney) [139–143] | Integrative plasmid [144,145]; Viral transformation [146] | Trachea (ex vivo) [147], Intestines (ex vivo) [148,149] |
Population genetics of aging
The age-specific impacts of selection:
Increasingly large-scale cohort studies such as the UK Biobank [59] and All of Us Research Program [60] present new opportunities to study human aging from a population genetics perspective. Trends in allele frequencies across age strata have the potential to reveal longitudinal dynamics of variant effects. In humans, both mutation accumulation and antagonistic pleiotropy have been postulated as explanations for the presence of segregating disease-causing alleles. However, studies of the GWAS catalog [60] purporting to have identified evidence of this [61] have suffered from statistical and logical flaws [62]. Taking advantage of GWAS data from more than 175,000 participants, Mostafavi et al identified just a handful of common variants with strong impacts on age-specific mortality [63]. These variants were found at the APOE ε4 and CHRNA3 loci, which predispose to Alzheimer’s disease and smoking behavior respectively. These results highlight the strong impact of purifying selection to purge deleterious alleles, even if they act later in life.
Epidemiological studies have documented an association between reproductive traits, such as age at onset of puberty or first childbirth, on female lifespan [64,65]. By calculating polygenic prediction scores for these traits, Mostafavi et al were able to show that genetic variants that delay puberty and increase age at first birth in mothers are associated with increased lifespan. In contrast, polygenically predicted increases in BMI, cholesterol, and coronary artery disease risk are associated with decreased lifespan.
The observation in humans that purifying selection strongly influences late-acting disease-causing alleles is intriguing given that these alleles should exert their effect well after menopause and peak reproductive ages in women. Indeed, for many late-onset diseases, causal variants tend to be evolutionarily recent and segregate at low frequencies. In an attempt to reconcile these observations with theory Pavard and Coste [66] developed an evolutionary demographic model that accounted for male fertility extending into old age, and the role of familial care by parents or grandparents. This model predicted that variants that cause disease later in life are indeed expected to be under strong purifying selection in many cases due to the impact of post-menopausal parental and grandparental care.
The many complexities inherent to dissecting the evolution and genetics of lifespan have also been highlighted in attempts to estimate the heritability of this trait. While estimates as high as 30% have been reported [67], recent analyses have suggested that these estimates are substantially inflated due to assortative mating, with the true estimated heritability <10% [68]. Indeed, some of the strongest predictors of lifespan are geographic and environmental in nature [69,70]. Together, these data highlight the clear importance of socio-cultural and environmental factors in influencing human lifespan.
Interactions between genes and the environment (GxE) also almost certainly play an important role, although are exceptionally hard to dissect in humans. A recent study in Drosophila identified alleles associated with lifespan under dietary stress (high sugar versus a control) [71]. Remarkably, one third of alleles mediated an environment-specific (GxE) effect on shortening lifespan. These alleles were evolutionary younger and exhibited signatures of selection in the wild. These results provide support for the “evolutionary mismatch hypothesis,” i.e. differences between ancient and modern environments contribute to disease [72]. Humans notably have undergone massive dietary changes over the last several thousand years with the neolithic transition from hunter-gatherer to agricultural sustenance.
Though many late-acting disease-causing genes appear to be under strong purifying selection, in general genes expressed in old age do tend to exhibit signatures of relaxed selection compared to genes expressed early in life. Analyses of gene expression across several mammals and insects have found that the rate of non-synonymous substitution (dn/ds) is significantly higher in genes expressed later in life [73–75]. Late-expressed genes also exhibit more segregating non-synonymous substitutions and have a reduced effective strength of selection (Nes). Similar signatures have been identified in killifish, with genes expressed early in life recapitulating stringencies in selection [33]. To explore these patterns more broadly in human populations, Yamamoto et al developed a statistical model to quantify the proportion of variance in gene expression that is attributable to age or genetics in 948 humans across 27 different tissues [76]. Intriguingly, while indeed the force of purifying selection was stronger on genes expressed earlier in life for the majority of tissues, recapitulating work in other species, several highly proliferative tissues exhibited the opposite trend. These ‘non-Medawarian’ tissues displayed high rates of cancer and age-of-expression associated somatic mutation. The genes responsible for driving this signature were highly enriched for pathways associated with DNA repair, cellular proliferation, differentiation, and cancer. One explanation for these signatures is that these genes are highly pleiotropic with critical roles both in early development as well as later in life in these specific “non-Medwarian” tissues.
Mutation: cause and/or consequence of aging and death
The aging germline:
Mutation is the fundamental source of genetic variation. Observations of an association of advanced paternal age with achondroplasia provided some of the first clues that the germline mutation rate increases with age [77]. Recent advances in sequencing methodologies have allowed us to directly quantify this mutation rate across a multitude of organisms linking aging and several other life history traits to this key biological trait.
Some of the most in depth analyses of the impact of age on mutation rate have been performed in humans. A landmark study of 1,548 pedigrees showed the paternal mutation rate to increase with age at a rate approximately four-times higher than the maternal rate (1.51 versus 0.37 mutations per year) [78]. The types of de-novo mutations from sperm and eggs also differed significantly with several egg-specific mutation hotspots observed. Intriguingly, analyses of almost 10,000 individuals from multiple single-generation families both with and without autism spectrum disorder (ASD) did not find any age-association with the rate of structural variant (SV) formation [79], though SVs were observed at a significantly higher rate in ASD probands and were more likely to be paternal in origin. These results are consistent with recent analyses of macaque parent-offspring trios, which also failed to identify any association of SV formation with age [80]. These observations are however at odds with analyses highlighting significant age-associated increases in instability and fragmentation in sperm [81]. Future studies and new sequencing technologies are required to reconcile the differences in mutations identified between family studies and direct observation of gametes. In particular, de-novo SVs are extremely challenging to identify using short read sequencing. Long-read based approaches, including those leveraging new telomere-to-telomere assemblies [82], may provide more sensitivity in detecting de-novo events (Box 2).
Germline mutation rates also vary substantially by species, reflecting differences in life history and reproductive strategies. A recent hallmark study of 151 trios from different mammals, fish, birds, and reptiles identified an average germline mutation rate of 1.17 × 10e-8 across vertebrates, with higher mutation rates driven primarily by greater parental age at reproduction [83]. However, per-generation mutation rates varied by up to 40-fold among different species. A paternal bias in the mutation rate was observed for mammals and birds, but not reptiles and fishes. Age at maturity and generation time were both positively associated with increased mutation rates, and in mammals the number of offspring per generation was negatively correlated with mutation rate. The mutational spectrum also differed substantially between vertebrate classes with the largest differences being observed in fish in A>C and C>A mutations. Changes in life history can have strong effects on germ-line mutation rates and profiles. One extreme example is in domesticated animals which have been selected for short generation times resulting in exceptionally high per-year mutation rates [83]. In rockfish, species with increased lifespans exhibit more segregating CpG->TpG mutations [13]. This mutational signature is characteristic of spontaneous methylated cytosine deamination, highlighting that shifts in the average generation time of a population can influence the spectrum of segregating genetic variation. Extensive comparisons of the rate and spectrum of de-novo SVs between species have yet to be performed. Together these results highlight the close relationship between the evolution of mutation rates and the evolution of aging.
Somatic mutation:
Somatic mutations accumulate with age in cells throughout the body. Indeed, this accumulation of somatic mutation is considered a “hallmark” of aging [84], though it is not clear to what degree such mutations are a “cause” of aging. Somatic mutations are difficult to measure due to their low frequency and the challenge of distinguishing them from artifactual sequencing errors. Newly developed single-molecule sequencing techniques, such as Nano-seq (duplex sequencing), enable highly accurate somatic mutation calls and to establish somatic mutation rates [85]. Somatic mutation rates tend to exceed their matched germline rates by 1- to 2-orders of magnitude [86–88], however they differ between tissues and cell types [89,90]. This increased somatic mutation rate is consistent with the disposable soma theory, which posits that organisms face a resource trade-off in their investment in germline vs somatic repair [91].
While the vast majority of work on somatic mutations has focused on humans, a recent analysis of intestinal crypts from 16 mammalian species provides several key insights into the evolution of somatic mutation [87]. Endogenous mutational processes were found to dominate the observed mutations, as opposed to environmentally associated mutations, though this trend may differ between cell types and tissues. Across mammals, the mutational signatures observed were largely the same, mirroring results in de-novo germline mutations which only found differences in the mutational spectrum between vertebrate classes. Most strikingly, the somatic mutation rate showed an inverse relationship with lifespan, with 82% of inter-species variation explained by this trait. Variation in body size was surprisingly not associated with somatic mutation rate. Moreover, the end-of-lifespan burden across species varied by only 3-fold regardless of lifespan, supporting the theory that somatic mutation accumulation may be a key contributing factor to lifespan. Together these findings suggest that, at least in some cell types, somatic mutation burden represents a highly accurate estimator of absolute age across species.
Cancer is caused by somatic mutations that activate uncontrolled cell proliferation [92], with cancer risk varying between cell types. Thus, if every cell possesses some intrinsic risk of tumorigenesis, it follows that larger organisms with a greater number of cells should be at a greater risk of developing cancer [93]. This is indeed the case within species: in humans for example, the risk of many different cancer types increases with height [94]. Likewise in dogs, larger breeds have a higher cancer incidence [95]. However, there is no apparent correlation between lifespan, body size, and cancer risk between species [96], a phenomenon referred to as Peto’s paradox [97]. Organisms with longer lifespans should have a similarly increased risk of cancer, as a consequence of increased mutation burden.
A recent study of 110,148 captive individuals across 191 mammalian species did identify a dramatic range in cancer mortality risk (CMR, i.e. the likelihood of dying from cancer) among species [98]. Over 20% of species examined exhibited substantial risk (exceeding 10% CMR) of cancer-related death [98]. However, body mass and lifespan accounted for only 0.78% and 2.94% of cross-species variance in CMR respectively, robustly supporting Peto’s paradox in mammals. This line of evidence, taken alongside findings of an inverse relationship between lifespan and somatic mutation rate, support life history-dependent evolution of cancer mitigation. However, the genetic drivers underpinning increased cancer resistance in larger organisms are still largely unresolved, with recent evidence suggesting a plurality of mechanisms (Box 3).
Concluding Remarks & Future Perspectives
The scope of the field of aging and evolution is extraordinarily broad encompassing a myriad of topics beyond what we have covered here including epigenetic modifications [99–101], telomere attrition [102], other hallmarks of aging [84]. We also choose here to focus primarily on vertebrates, however many of the molecular mechanisms underlying aging are conserved in invertebrate and even in plant models, as described in other reviews [103,104].
Differences in lifespan have evolved countless times throughout the tree of life with longer and shorter lifespans covarying strongly with several distinct phenotypes and environmental conditions. This means that extreme lifespans can emerge indirectly from selection on covarying traits or adaptations to new environments, signatures of which will be left in their genomes. Dissecting the functional impact of selected genes and pathways remains challenging, however cellular and organoid models have emerged as potentially transformative tools to explore these lifespan-extending adaptations.
The age-specific forces of selection are now for the first time being revealed in humans demonstrating that strongly deleterious alleles, even when late-acting, exhibit signatures of purifying selection. However, in general, genes expressed earlier in life exhibit increased constraint compared to those expressed late in life, except in the case of a handful of highly proliferative ‘non-medawarian’ tissues. The cell type-specific patterning of expression timing and constraint have yet to be explored however. Such studies have the potential to reveal “medwarian” and “non-medawarian” patterns in individual cell types. Future profiling of cellular phenotypes throughout lifespan will also enable linking individual cellular phenomena to organism level phenotypes and evolutionary patterns.
The evolution of aging and lifespan is tightly linked to the evolution of mutation rates and processes. While rapid advances have been observed in dissecting single nucleotide mutation, our understanding of both germline and somatic structural variation remains incomplete. As somatic mutation profiling approaches become more cost effective, future studies also have the potential to increase our understanding of cellular aging across taxa. These studies will hopefully provide further insights into the lifespan-associated pathways identified from comparative genomics studies, clarifying their cell-type specific actions.
The proliferation of several genomic technologies has propelled our understanding of the evolution of aging enormously. However, many of these technologies have only recently started being applied at scale to explore fundamental questions of evolutionary biology. The future thus holds unprecedented promise as we begin to explore the full extent of organismal diversity to uncover the many different origins of life history differences on earth.
Highlights.
The extensive variation in lifespan among organisms provides a natural dataset to probe the evolutionary trade-offs that constrain and mold this phenotype across taxa
Several key pathways repeatedly emerge as the targets of selection in comparative genomics of long lived species. Overall, these pathways exhibit increased constraint in long lived species, and reduced constraint in short lived species with distinct signatures of diversifying selection observed in individual taxa.
Large cohort studies demonstrate that even late-acting deleterious alleles appear to be under strong purifying selection in human populations.
Somatic mutation rates scale with lifespan and are significantly higher than matched germline mutation rates.
Cell culture models of extreme agers are beginning to facilitate characterization of key genotype-phenotype interactions in biological aging.
Outstanding questions.
Gene-by-environment (GxE) interactions play an extremely important role in influencing lifespan and other life history traits. However, identifying GxE interactions is challenging even in model organisms, and still underexplored in humans. A major outstanding challenge is the development of both study designs and statistical approaches that are empowered to identify GxE in large human cohorts and to elucidate the impacts of these interactions on human health and lifespan.
The covariance of lifespan with several other life history traits implies that shared genetic pathways shape the biology of aging. Many of these pathways are extremely deeply conserved and essential. While these pathways are discovered repeatedly in comparative genomics, disentangling the many pleiotropic and epistatic effects of perturbations in these pathways remains extremely challenging.
Long read sequencing is unveiling the extensive structural differences that exist within and between species in previously intractable regions such as centromeres, telomeres, and rDNA repeats as well as large structural variants such as inversions and translocations. However, current comparative genomics approaches based on multiple sequence alignments are not empowered to assess this variation. An outstanding question is how these structural rearrangements influence phenotypes. Answering this question will require novel computational and statistical comparative approaches in addition to high quality genomes and phenotype data.
While heterochromatic repeat sequences found on sex chromosomes have been implicated in aging, the longitudinal dynamics of their de-repression in different species and the mechanism of their toxicity is not well understood.
While recent work highlights the tight linkage between the evolution of lifespan and both germline and somatic single nucleotide mutation patterns, our understanding of age-associated somatic and germline structural variation remains incomplete.
Acknowledgements
This work was supported by the National Institute of General Medical Sciences grant R35GM142916 to P.H.S, Vallee Scholars Award to P.H.S, NSF PRFB 2109915 to J.M.V. and NIH NIA T32 AG000266 to J.M.V.
Footnotes
Declaration of interests
The authors declare no competing interests.
References
- 1.Medawar PB (1952) An Unsolved Problem of Biology: An Inaugural Lecture Delivered at University College, London, 6 December, 1951 [Google Scholar]
- 2.Williams GC (1957) Pleiotropy, natural selection, and the evolution of senescence. Evolution 11, 398–411 [Google Scholar]
- 3.Hamilton WD (1966) The moulding of senescence by natural selection. J. Theor. Biol. 12, 12–45 [DOI] [PubMed] [Google Scholar]
- 4.Fisher RA (1930) The genetical theory of natural selection, Clarendon Press [Google Scholar]
- 5.Charlesworth B (2000) Fisher, Medawar, Hamilton and the evolution of aging. Genetics 156, 927–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finch CE (1994) Longevity, Senescence, and the Genome, University of Chicago Press [Google Scholar]
- 7.Nielsen J et al. (2016) Eye lens radiocarbon reveals centuries of longevity in the Greenland shark (Somniosus microcephalus). Science 353, 702–704 [DOI] [PubMed] [Google Scholar]
- 8.Depczynski M and Bellwood DR (2005) Shortest recorded vertebrate lifespan found in a coral reef fish. Curr. Biol. 15, R288–9 [DOI] [PubMed] [Google Scholar]
- 9.Healy K et al. (2019) Animal life history is shaped by the pace of life and the distribution of age-specific mortality and reproduction. Nat Ecol Evol 3, 1217–1224 [DOI] [PubMed] [Google Scholar]
- 10.Healy K et al. (2014) Ecology and mode-of-life explain lifespan variation in birds and mammals. Proc. Biol. Sci. 281, 20140298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.da Silva R et al. (2022) Slow and negligible senescence among testudines challenges evolutionary theories of senescence. Science 376, 1466–1470 [DOI] [PubMed] [Google Scholar]
- 12.Reinke BA et al. (2022) Diverse aging rates in ectothermic tetrapods provide insights for the evolution of aging and longevity. Science 376, 1459–1466 [DOI] [PubMed] [Google Scholar]
- 13.Kolora SRR et al. (2021) Origins and evolution of extreme life span in Pacific Ocean rockfishes. Science 374, 842–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham Ruby J et al. (2018) Naked mole-rat mortality rates defy Gompertzian laws by not increasing with age. DOI: 10.7554/eLife.31157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barneche DR et al. (2018) Fish reproductive-energy output increases disproportionately with body size. Science 360, 642–645 [DOI] [PubMed] [Google Scholar]
- 16.Auer SK et al. (2018) Metabolic rate evolves rapidly and in parallel with the pace of life history Nature Communications, 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinho GM et al. (2022) Hibernation slows epigenetic ageing in yellow-bellied marmots. Nat Ecol Evol 6, 418–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilkinson GS and Adams DM (2019) Recurrent evolution of extreme longevity in bats. Biol. Lett. 15, 20180860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Austad SN and Fischer KE (2016) Sex Differences in Lifespan. Cell Metab. 23, 1022–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xirocostas ZA et al. (2020) The sex with the reduced sex chromosome dies earlier: a comparison across the tree of life. Biol. Lett. 16, 20190867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemaître J-F et al. (2020) Sex differences in adult lifespan and aging rates of mortality across wild mammals. Proc. Natl. Acad. Sci. U. S. A. 117, 8546–8553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cayuela H et al. (2022) Sex-related differences in aging rate are associated with sex chromosome system in amphibians. Evolution 76, 346–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Connallon T et al. (2022) How much does the unguarded X contribute to sex differences in life span? Evol Lett 6, 319–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown EJ et al. (2020) The Y chromosome may contribute to sex-specific ageing in Drosophila. Nat Ecol Evol 4, 853–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sultanova Z et al. (2023) Genetic sex determination, sex chromosome size and sex-specific lifespans across tetrapods. J. Evol. Biol. 36, 480–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keane M et al. (2015) Insights into the evolution of longevity from the bowhead whale genome. Cell Rep. 10, 112–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quesada V et al. (2019) Giant tortoise genomes provide insights into longevity and age-related disease. Nat Ecol Evol 3, 87–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Çilingir FG et al. (2022) Chromosome-level genome assembly for the Aldabra giant tortoise enables insights into the genetic health of a threatened population. Gigascience 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou X et al. (2020) Beaver and Naked Mole Rat Genomes Reveal Common Paths to Longevity. Cell Rep. 32, 107949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pascual-Torner M et al. (2022) Comparative genomics of mortal and immortal cnidarians unveils novel keys behind rejuvenation. Proc. Natl. Acad. Sci. U. S. A. 119, e2118763119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jebb D et al. (2020) Six reference-quality genomes reveal evolution of bat adaptations. Nature 583, 578–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valenzano DR et al. (2015) The African Turquoise Killifish Genome Provides Insights into Evolution and Genetic Architecture of Lifespan. Cell 163, 1539–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui R et al. (2019) Relaxed Selection Limits Lifespan by Increasing Mutation Load. Cell 178, 385–399.e20 [DOI] [PubMed] [Google Scholar]
- 34.Hu C-K et al. (2020) Vertebrate diapause preserves organisms long term through Polycomb complex members. Science 367, 870–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma S and Gladyshev VN (2017) Molecular signatures of longevity: Insights from cross-species comparative studies. Semin. Cell Dev. Biol. 70, 190–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh PP et al. (2019) The Genetics of Aging: A Vertebrate Perspective. Cell 177, 200–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian X et al. (2017) Molecular Mechanisms Determining Lifespan in Short- and Long-Lived Species. Trends Endocrinol. Metab. 28, 722–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kowalczyk A et al. (2020) Pan-mammalian analysis of molecular constraints underlying extended lifespan. Elife 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alexandrov LB et al. (2020) The repertoire of mutational signatures in human cancer. Nature 578, 94–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stavrou S and Ross SR (2015) APOBEC3 Proteins in Viral Immunity. J. Immunol. 195, 4565–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Treaster S et al. (2023) Convergent genomics of longevity in rockfishes highlights the genetics of human life span variation. Sci Adv 9, eadd2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kowalczyk A et al. (2022) Complementary evolution of coding and noncoding sequence underlies mammalian hairlessness. Elife 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Partha R et al. (2017) Subterranean mammals show convergent regression in ocular genes and enhancers, along with adaptation to tunneling. Elife 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kowalczyk A et al. (2019) RERconverge: an R package for associating evolutionary rates with convergent traits. Bioinformatics 35, 4815–4817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willemsen D et al. (2020) Intra-species differences in population size shape life history and genome evolution. DOI: 10.7554/eLife.55794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Armstrong J et al. (2020) Progressive Cactus is a multiple-genome aligner for the thousand-genome era. Nature 587, 246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Christmas MJ et al. (2023) Evolutionary constraint and innovation across hundreds of placental mammals. Science 380, eabn3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rhie A et al. (2021) Towards complete and error-free genome assemblies of all vertebrate species. Nature 592, 737–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vazquez JM et al. (2018) A Zombie LIF Gene in Elephants Is Upregulated by TP53 to Induce Apoptosis in Response to DNA Damage. Cell Rep. 24, 1765–1776 [DOI] [PubMed] [Google Scholar]
- 50.Vazquez JM and Lynch VJ (2021) Pervasive duplication of tumor suppressors in Afrotherians during the evolution of large bodies and reduced cancer risk. Elife 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sulak M et al. (2016) copy number expansion is associated with the evolution of increased body size and an enhanced DNA damage response in elephants. Elife 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Preston AJ et al. (2023) Elephant TP53-RETROGENE 9 induces transcription-independent apoptosis at the mitochondria. Cell Death Discov 9, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vazquez JM et al. (2022) A CDKN2C retroduplication in Bowhead whales is associated with the evolution of extremely long lifespans and alerted cell cycle dynamics bioRxiv, 2022.09.07.506958 [Google Scholar]
- 54.Firsanov D et al. (2023) DNA repair and anti-cancer mechanisms in the longest-living mammal: the bowhead whale [Google Scholar]
- 55.Salmon AB et al. (2008) Fibroblasts from naked mole-rats are resistant to multiple forms of cell injury, but sensitive to peroxide, ultraviolet light, and endoplasmic reticulum stress. J. Gerontol. A Biol. Sci. Med. Sci. 63, 232–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gorbunova V et al. (2012) Cancer resistance in the blind mole rat is mediated by concerted necrotic cell death mechanism. Proc. Natl. Acad. Sci. U. S. A. 109, 19392–19396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gorbunova V et al. (2008) Rodents for comparative aging studies: from mice to beavers. Age 30, 111–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Q et al. (2021) Genomic expansion of Aldh1a1 protects beavers against high metabolic aldehydes from lipid oxidation. Cell Rep. 37, 109965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sudlow C et al. (2015) UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.All of Us Research Program Investigators et al. (2019) The “All of Us” Research Program. N. Engl. J. Med. 381, 668–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rodríguez JA et al. (2017) Antagonistic pleiotropy and mutation accumulation influence human senescence and disease. Nat Ecol Evol 1, 55. [DOI] [PubMed] [Google Scholar]
- 62.Long E and Zhang J (2019) Retesting the influences of mutation accumulation and antagonistic pleiotropy on human senescence and disease Nature ecology & evolution, 3992–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mostafavi H et al. (2017) Identifying genetic variants that affect viability in large cohorts. PLoS Biol. 15, e2002458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith KR et al. (2009) Familial aggregation of survival and late female reproduction. J. Gerontol. A Biol. Sci. Med. Sci. 64, 740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shadyab AH et al. (2017) Maternal Age at Childbirth and Parity as Predictors of Longevity Among Women in the United States: The Women’s Health Initiative. Am. J. Public Health 107, 113–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pavard S and Coste CFD (2021) Evolutionary demographic models reveal the strength of purifying selection on susceptibility alleles to late-onset diseases. Nat Ecol Evol 5, 392–400 [DOI] [PubMed] [Google Scholar]
- 67.Ljungquist B et al. (1998) The effect of genetic factors for longevity: a comparison of identical and fraternal twins in the Swedish Twin Registry. J. Gerontol. A Biol. Sci. Med. Sci. 53, M441–6 [DOI] [PubMed] [Google Scholar]
- 68.Ruby JG et al. (2018) Estimates of the Heritability of Human Longevity Are Substantially Inflated due to Assortative Mating. Genetics 210, 1109–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dwyer-Lindgren L et al. (2017) Inequalities in Life Expectancy Among US Counties, 1980 to 2014: Temporal Trends and Key Drivers. JAMA Intern. Med. 177, 1003–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vierboom YC and Preston SH (2020) Life Beyond 65: Changing Spatial Patterns of Survival at Older Ages in the United States, 2000–2016. J. Gerontol. B Psychol. Sci. Soc. Sci. 75, 1093–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pallares LF et al. (2023) Dietary stress remodels the genetic architecture of lifespan variation in outbred Drosophila. Nat. Genet. 55, 123–129 [DOI] [PubMed] [Google Scholar]
- 72.Corbett S et al. (2018) The transition to modernity and chronic disease: mismatch and natural selection. Nat. Rev. Genet. 19, 419–430 [DOI] [PubMed] [Google Scholar]
- 73.Cheng C and Kirkpatrick M (2021) Molecular evolution and the decline of purifying selection with age. Nat. Commun. 12, 2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Turan ZG et al. (2019) Molecular footprint of Medawar’s mutation accumulation process in mammalian aging. Aging Cell 18, e12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jia K et al. (2018) An analysis of aging-related genes derived from the Genotype-Tissue Expression project (GTEx). Cell Death Discov 4, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yamamoto R et al. (2022) Tissue-specific impacts of aging and genetics on gene expression patterns in humans. Nat. Commun. 13, 5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Penrose LS (1955) Parental age and mutation. Lancet 269, 312–313 [DOI] [PubMed] [Google Scholar]
- 78.Jónsson H et al. (2017) Parental influence on human germline de novo mutations in 1,548 trios from Iceland. Nature 549, 519–522 [DOI] [PubMed] [Google Scholar]
- 79.Belyeu JR et al. (2021) De novo structural mutation rates and gamete-of-origin biases revealed through genome sequencing of 2,396 families. Am. J. Hum. Genet. 108, 597–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thomas GWC et al. (2021) Origins and Long-Term Patterns of Copy-Number Variation in Rhesus Macaques. Mol. Biol. Evol. 38, 1460–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Laurentino S et al. (2020) A germ cell-specific ageing pattern in otherwise healthy men. Aging Cell 19, e13242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nurk S et al. (2022) The complete sequence of a human genome. Science 376, 44–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bergeron LA et al. (2023) Evolution of the germline mutation rate across vertebrates. Nature 615, 285–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.López-Otín C et al. (2023) Hallmarks of aging: An expanding universe. Cell 186, 243–278 [DOI] [PubMed] [Google Scholar]
- 85.Abascal F et al. (2021) Somatic mutation landscapes at single-molecule resolution. Nature 593, 405–410 [DOI] [PubMed] [Google Scholar]
- 86.Milholland B et al. (2017) Differences between germline and somatic mutation rates in humans and mice. Nat. Commun. 8, 15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cagan A et al. (2022) Somatic mutation rates scale with lifespan across mammals. Nature 604, 517–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Moore L et al. (2021) The mutational landscape of human somatic and germline cells. Nature 597, 381–386 [DOI] [PubMed] [Google Scholar]
- 89.Rockweiler NB et al. (2023) The origins and functional effects of postzygotic mutations throughout the human life span. Science 380, eabn7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yizhak K et al. (2019) RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues. Science 364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kirkwood TB (1977) Evolution of ageing. Nature 270, 301–304 [DOI] [PubMed] [Google Scholar]
- 92.Hanahan D and Weinberg RA (2000) The hallmarks of cancer. Cell 100, 57–70 [DOI] [PubMed] [Google Scholar]
- 93.Leroi AM et al. (2003) Cancer selection. Nat. Rev. Cancer 3, 226–231 [DOI] [PubMed] [Google Scholar]
- 94.Green J et al. (2011) Height and cancer incidence in the Million Women Study: prospective cohort, and meta-analysis of prospective studies of height and total cancer risk. Lancet Oncol. 12, 785–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dobson JM (2013) Breed-predispositions to cancer in pedigree dogs. ISRN Vet. Sci. 2013, 941275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Abegglen LM et al. (2015) Potential Mechanisms for Cancer Resistance in Elephants and Comparative Cellular Response to DNA Damage in Humans. JAMA 314, 1850–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Peto R (1977) Origins of human cancer, Cold Spring Harbor Laboratory [Google Scholar]
- 98.Vincze O et al. (2022) Cancer risk across mammals. Nature 601, 263–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lu AT et al. (2023) Universal DNA methylation age across mammalian tissues. Nat Aging DOI: 10.1038/s43587-023-00462-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Moqri M et al. (2022) PRC2 clock: a universal epigenetic biomarker of aging and rejuvenation bioRxiv, 2022.06.03.494609 [Google Scholar]
- 101.Haghani A et al. (2023) DNA methylation networks underlying mammalian traits. Science 381, eabq5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Whittemore K et al. (2019) Telomere shortening rate predicts species life span. Proc. Natl. Acad. Sci. U. S. A. 116, 15122–15127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nooden LD (2012) Senescence and Aging in Plants, Elsevier [Google Scholar]
- 104.Zhang S et al. (2020) Caenorhabditis elegans as a Useful Model for Studying Aging Mutations. Front. Endocrinol. 11, 554994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Siler W (1979) A competing-risk model for animal mortality. Ecology 60, 750–757 [Google Scholar]
- 106.Zoonomia Consortium (2020) A comparative genomics multitool for scientific discovery and conservation. Nature 587, 240–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kuderna LFK et al. (2023) A global catalog of whole-genome diversity from 233 primate species. Science 380, 906–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liao W-W et al. (2023) A draft human pangenome reference. Nature 617, 312–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ferreira JV et al. (2022) Cell Non-autonomous Proteostasis Regulation in Aging and Disease. Front. Neurosci. 16, 878296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mertens J et al. (2018) Aging in a Dish: iPSC-Derived and Directly Induced Neurons for Studying Brain Aging and Age-Related Neurodegenerative Diseases. Annu. Rev. Genet. 52, 271–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Browder KC et al. (2022) In vivo partial reprogramming alters age-associated molecular changes during physiological aging in mice. Nat Aging 2, 243–253 [DOI] [PubMed] [Google Scholar]
- 112.Torrens-Mas M et al. (2021) Organoids: An Emerging Tool to Study Aging Signature across Human Tissues. Modeling Aging with Patient-Derived Organoids. Int. J. Mol. Sci. 22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liu H et al. (2008) Generation of induced pluripotent stem cells from adult rhesus monkey fibroblasts. Cell Stem Cell 3, 587–590 [DOI] [PubMed] [Google Scholar]
- 114.Hong SG et al. (2014) Path to the clinic: assessment of iPSC-based cell therapies in vivo in a nonhuman primate model. Cell Rep. 7, 1298–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pollen AA et al. (2019) Establishing Cerebral Organoids as Models of Human-Specific Brain Evolution. Cell 176, 743–756.e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pavlovic BJ et al. (2018) A Comparative Assessment of Human and Chimpanzee iPSC-derived Cardiomyocytes with Primary Heart Tissues. Sci. Rep. 8, 15312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mitchell SJ et al. (2015) Animal models of aging research: implications for human aging and age-related diseases. Annu Rev Anim Biosci 3, 283–303 [DOI] [PubMed] [Google Scholar]
- 118.Tian X et al. (2013) High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature 499, 346–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hadi F et al. (2020) Transformation of naked mole-rat cells Nature, 583E1–E7 [DOI] [PubMed] [Google Scholar]
- 120.Zhao J et al. (2020) Reply to: Transformation of naked mole-rat cells Nature, 583E8–E13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhao S et al. (2014) High autophagy in the naked mole rat may play a significant role in maintaining good health. Cell. Physiol. Biochem. 33, 321–332 [DOI] [PubMed] [Google Scholar]
- 122.Miura K et al. (2021) Induced Pluripotent Stem Cells from Cancer-Resistant Naked Mole-Rats. Adv. Exp. Med. Biol. 1319, 329–339 [DOI] [PubMed] [Google Scholar]
- 123.Tan L et al. (2017) Naked Mole Rat Cells Have a Stable Epigenome that Resists iPSC Reprogramming. Stem Cell Reports 9, 1721–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Miyawaki S et al. (2016) Tumour resistance in induced pluripotent stem cells derived from naked mole-rats. Nat. Commun. 7, 11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lee S-G et al. (2017) Naked Mole Rat Induced Pluripotent Stem Cells and Their Contribution to Interspecific Chimera. Stem Cell Reports 9, 1706–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Labinskyy N et al. (2006) Comparison of endothelial function, O2-* and H2O2 production, and vascular oxidative stress resistance between the longest-living rodent, the naked mole rat, and mice. Am. J. Physiol. Heart Circ. Physiol. 291, H2698–704 [DOI] [PubMed] [Google Scholar]
- 127.Burkard M et al. (2015) Establishment of the first humpback whale fibroblast cell lines and their application in chemical risk assessment. Aquat. Toxicol. 167, 240–247 [DOI] [PubMed] [Google Scholar]
- 128.Pereiro X et al. (2022) Characteristics of Whale Müller Glia in Primary and Immortalized Cultures. Front. Neurosci. 16, 854278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yajing S et al. (2018) Establishment and characterization of pygmy killer whale (Feresa attenuata) dermal fibroblast cell line. PLoS One 13, e0195128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hightower LE and Renfro JL (1988) Recent applications of fish cell culture to biomedical research. J. Exp. Zool. 248, 290–302 [DOI] [PubMed] [Google Scholar]
- 131.Goswami M et al. (2022) Role and relevance of fish cell lines in advanced in vitro research. Mol. Biol. Rep. 49, 2393–2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Peng L et al. (2019) Generation of Stable Induced Pluripotent Stem-like Cells from Adult Zebra Fish Fibroblasts. Int. J. Biol. Sci. 15, 2340–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Schwarz JS et al. (2015) Value of Organoids from Comparative Epithelia Models. Yale J. Biol. Med. 88, 367–374 [PMC free article] [PubMed] [Google Scholar]
- 134.Zilova L et al. (2021) Fish primary embryonic pluripotent cells assemble into retinal tissue mirroring in vivo early eye development. Elife 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tan F et al. (2010) Validation of an in vitro cytotoxicity test for four heavy metals using cell lines derived from a green sea turtle (Chelonia mydas). Cell Biol. Toxicol. 26, 255–263 [DOI] [PubMed] [Google Scholar]
- 136.Glaberman S et al. (2021) Concurrent Evolution of Antiaging Gene Duplications and Cellular Phenotypes in Long-Lived Turtles. Genome Biol. Evol. 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Martins GS et al. (2016) Cytochemical characteristics of blood cells from Brazilian tortoises (Testudines: Testudinidae). Genet. Mol. Res. 15 [DOI] [PubMed] [Google Scholar]
- 138.Zdyrski C et al. (2023) Characterization of the first turtle organoids: A model for investigating unique adaptations with biomedical potential [Google Scholar]
- 139.Brook CE et al. (2020) Accelerated viral dynamics in bat cell lines, with implications for zoonotic emergence. Elife 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jacquet S et al. (2022) Adaptive duplication and genetic diversification of protein kinase R contribute to the specificity of bat-virus interactions. Sci Adv 8, eadd7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Koh J et al. (2019) ABCB1 protects bat cells from DNA damage induced by genotoxic compounds. Nat. Commun. 10, 2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tarigan R et al. (2021) Distinct interferon response in bat and other mammalian cell lines infected with Pteropine orthoreovirus. Virus Genes 57, 510–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cosby RL et al. (2021) Recurrent evolution of vertebrate transcription factors by transposase capture. Science 371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Aurine N et al. (2021) Reprogrammed Bat Stem Cells as A Model to Study Host-Pathogen Interaction during Infection. Microorganisms 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mo X et al. (2014) Generation and characterization of bat-induced pluripotent stem cells. Theriogenology 82, 283–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Déjosez M et al. (2023) Bat pluripotent stem cells reveal unusual entanglement between host and viruses. Cell 186, 957–974.e28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chan LLY et al. (2023) Generation of self-replicating airway organoids from the cave nectar bat as a model system for studying host-pathogen interactions in the bat airway epithelium. Emerg. Microbes Infect. 12, e2148561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Elbadawy M et al. (2021) Establishment of Intestinal Organoid from and the Susceptibility to Bat-Associated Viruses, SARS-CoV-2 and Pteropine Orthoreovirus. Int. J. Mol. Sci. 22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhou J et al. (2020) Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 26, 1077–1083 [DOI] [PubMed] [Google Scholar]


