Abstract
Introduction
Patients with pediatric low-grade gliomas (pLGGs), the most common primary brain tumors in children, can often benefit from MAPK inhibitor (MAPKi) treatment. However, rapid tumor regrowth, also referred to as rebound growth, may occur once treatment is stopped, constituting a significant clinical challenge.
Methods
Four patient-derived pediatric glioma models were investigated to model rebound growth in vitro based on viable cell counts in response to MAPKi treatment and withdrawal. A multi-omics dataset (RNA sequencing and LC-MS/MS based phospho-/proteomics) was generated to investigate possible rebound-driving mechanisms. Following in vitro validation, putative rebound-driving mechanisms were validated in vivo using the BT-40 orthotopic xenograft model.
Results
Of the tested models, only a BRAFV600E-driven model (BT-40, with additional CDKN2A/Bdel) showed rebound growth upon MAPKi withdrawal. Using this model, we identified a rapid reactivation of the MAPK pathway upon MAPKi withdrawal in vitro, also confirmed in vivo. Furthermore, transient overactivation of key MAPK molecules at transcriptional (e.g. FOS) and phosphorylation (e.g. pMEK) levels, was observed in vitro. Additionally, we detected increased expression and secretion of cytokines (CCL2, CX3CL1, CXCL10 and CCL7) upon MAPKi treatment, maintained during early withdrawal. While increased cytokine expression did not have tumor cell intrinsic effects, presence of these cytokines in conditioned media led to increased attraction of microglia cells in vitro.
Conclusion
Taken together, these data indicate rapid MAPK reactivation upon MAPKi withdrawal as a tumor cell intrinsic rebound-driving mechanism. Furthermore, increased secretion of microglia-recruiting cytokines may play a role in treatment response and rebound growth upon withdrawal, warranting further evaluation.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11060-024-04672-9.
Keywords: Pediatric low-grade glioma, MAPK inhibitor, Rebound growth, Treatment withdrawal, Cytokines, Tumor microenvironment
Introduction
Pediatric low-grade gliomas (pLGG) are the most common primary brain tumors in children [1]. They are a diverse group of WHO grade 1 and grade 2 glial and glioneuronal tumors [2], which typically contain a high proportion of tumor microenvironment (TME) cells, especially microglia/macrophages (30–50%) [3]. While pLGG patients have a good overall prognosis (15-year overall survival 80–90%), progression-free survival is considerably lower (15-year PFS: 55%), in particular in cases of incomplete resection (15-year PFS for non-resected patients: 27%) [4]. Incompletely resected pLGGs are a chronic disease, often requiring multiple lines of therapy, leading to accumulation of therapy-related sequelae [4]. This underscores the need for better and definitive therapies, to improve the quality of life for these patients.
pLGGs are almost exclusively driven by mutually exclusive alterations in different components of the extracellular-regulated kinase/mitogen-activated protein kinase (MAPK) pathway [5]. The most common alterations in pLGGs affect the BRAF gene, and comprise either structural BRAF rearrangements (~ 50% KIAA1549:BRAF fusions) or BRAF point mutations (~ 10% BRAFV600E mutations) [3].
The fact that pLGGs are mainly driven by increased MAPK activation offers the possibility for targeted therapy using different small molecule inhibitors of the MAPK pathway. Several clinical trials investigating the efficacy of these inhibitors have shown promising effects [6]. Two studies assessing the efficacy of the BRAF type I ½ inhibitor [7] dabrafenib, one retrospective analysis [8] and one phase I/II clinical trial [9], showed an objective response rate (ORR) of 80% [8] and 44% [9] respectively. Furthermore, the combination of dabrafenib and trametinib was recently FDA-approved for the first-line treatment of BRAFV600E-driven pLGGs [10], as it showed an improved overall response (OR, defined as best overall complete or partial response; 47%), compared to standard-of-care (SOC) chemotherapy (OR: 11%), as well as less toxicity [11].
However, despite the efficacy of MAPKi treatment, a subset of patients experiences fast tumor regrowth upon treatment stop [8, 12]. This rapid tumor regrowth is also referred to as rebound growth, and constitutes a significant clinical challenge. Rebound growth is characterized by a rapid tumor regrowth (≥25%) within three months after treatment stop (Patricia O’Hare et al., Neuro Oncol., under review; e.g. median time to progression after dabrafenib treatment: 2.3 months) [8], and in some patients faster tumor growth is observed after MAPKi treatment compared to before treatment (clinical observation). While tumor regrowth is also observed in patients after chemotherapy treatment, time to progression is considered to be shorter in bona fide rebound (Patricia O’Hare et al., Neuro Oncol., under review). Interestingly, tumors showing rebound growth remain sensitive to the initial MAPKi treatment [8, 13], suggesting that acquired resistance mechanisms are not responsible for the observed pattern.
In the present study, we show that, in a BRAFV600E-driven model with co-occurring CDKN2A/B deletion, rebound growth after MAPKi withdrawal is associated with a fast reactivation of the MAPK pathway. Additionally, our results indicate a possible involvement of MAPKi-induced cytokine expression on the TME, specifically microglia cells.
Materials and methods
Cell culture
BT-40 cells, a kind gift from Prof. Peter Houghton, were culture in RPMI containing L-glutamine (cat. no 21,875,034, ThermoFisher Scientific) and 10% FCS (cat. no. F7524, Sigma Aldrich). DKFZ-BT66, DKFZ-BT308 and DKFZ-BT314 were cultured as described previously [14, 15] and used in proliferating and senescent state. As described previously, to culture cells in proliferating state oncogene-induced senescence, otherwise observed in these cells, is inhibited through the induction of the SV40-TAg [14, 15]. Further details on this system and the pipeline used to establish these models are described in detail in the original publications [14–16].
The microglia cell line HMC3 was purchased from the ATCC and cultured in MEM with L-glutamine (cat. no. 31,095,029) supplemented with 10% FCS, non-essential amino acids (cat. no. 11,140,035, ThermoFisher Scientific) and sodium pyruvate (cat. no. 11,360,039, ThermoFisher Scientific).
If not otherwise indicated, BT-40, DKFZ-BT308 and DKFZ-BT314 were seeded for experiments two days prior to experiment start in complete media (containing all supplements) and upon experiment start switched to minimum media (MM).
For further details see Supplementary materials and methods.
Drug treatments and withdrawal
All inhibitors used for in vitro experiments are listed in Table S3. For details on drug concentration choice and the withdrawal procedure see Supplementary materials and methods.
Metabolic activity assay for IC50 determination
Metabolic activity was measured using CellTiter-Glo 2.0 (cat. no. G9241, Promega) according to the manufacturer’s instructions. For further details see Supplementary materials and methods.
Cell counting for growth curve analysis
Cells were counted using the Vi-CELL XR (Beckman Coulter; Software v2.03) using the settings described in Table S1. For further details see Supplementary materials and methods.
Cell cycle analysis by flow cytometry
Cell cycle analysis by flow cytometry was performed as described previously [17].
RNA isolation, cDNA synthesis and quantitative reverse transcription real-time PCR (RT-qPCR)
qPCR was performed as described previously [18]. Primers were purchased from Qiagen or Invitrogen (Table S5). For further details see Supplementary materials and methods.
Protein extraction and immunoblotting
Antibodies used are listed in Table S6. For further details see Supplementary materials and methods.
RNA sequencing and data processing
2 × 100 bp paired-end sequencing was performed on the Illumina NovaSeq 6000 according to the manufacturer’s protocol. For details see Supplementary materials and methods.
LC-MS/MS proteomics and phosphoproteomics data generation and processing
Briefly, proteins were cleaved using Trypsin/Lys-C mix (cat. no. V5072, Promega) and either cleaned up using self-made SDB-PRS stage tips in case of proteomics or enriched for phospho-peptides using the High-Select™ TiO2 Phosphopeptide Enrichment Kit (cat. no. A32993, ThermoFisher Scientific) in case of phospho-proteomics analysis. For details on sample processing, data generation and processing see Supplementary materials and methods.
Luminex-based multiplex assay
The Luminex-based multiplex assay was performed using the Bio-Plex 200 System (Bio-Rad) with the Bio-Plex Pro Reagent Kit 3 (cat. no. 171,304,090 M, Bio-Rad) using the Bio-Plex Pro HuCSP standard (cat. no. 12,007,919, Bio-Rad) and the Bio-Plex Pro Human Chemokine Standards (cat. no. 171DK0001, Bio-Rad) according to the manufacturer’s instructions. Custom detection antibody multiplexes were used as indicated in Table S7. For details see Supplementary materials and methods.
Kinase phosphorylation array
Proteome Profiler Human Phospho-Kinase Array Kit (ARY003C, R&D systems) was used according to the manufacturer’s instructions using 400 µg of protein lysate per array set. Arrays were visualized with Amersham ECL Prime Western Blotting Detection Reagent (cat. no. RPN2232, GE Healthcare Dharmacon) using the Azure c400 imaging system (Azure Biosystems). Quantification was done using ImageJ (v2.9.0).
Stimulation with recombinant cytokines
Cells were treated for 1 h with 100 ng/ml of recombinant human CCL2 (cat. no. 300-04, PeproTech), CX3CL1 (cat. no. 300 − 31, PeproTech), CXCL10 (cat. no. 300 − 12, PeproTech), CCL7 (cat. no. 300 − 17, PeproTech) or the combination of all four.
Treatment with neutralizing antibodies
For cell count experiments and western blot analysis, cells were treated with antibodies neutralizing CCL2 (0.5 µg/ml; cat. no. MAB279, R&D systems), CX3CL1 (0.25 µg/ml; cat. no. MAB3652, R&D systems), CXCL10 (0.25 µg/mL; cat. no. MAB266, R&D systems) and CCL7 (0.1ng/ml; cat. no. MAB282, R&D systems) or mouse IgG (cat. no. MAB002, R&D systems).
Conditioned media collection for transwell assay
For conditioned media collection, BT-40 were treated for five days, with media changes on day 2 and day 4 of treatment, and on day 5 treatment withdrawal was performed. CM (containing 2% FCS) was collected after five days of treatment (24 h after the last media change), and 24 h after treatment withdrawal. After collection, CM was centrifuged for 5 min at 1200 rpm and filtered using a 0.22 µM sterile filter (cat. no. SLGS033, Sigma Aldrich).
Transwell migration assay
Transwell migration assays were performed using a 24-well transwell chamber (insert: 8 µM pore size; cat. no. 353,097, Corning; companion plate: 353,504, Corning). Detailed experimental procedures are described in the Supplementary materials and methods.
Transwell co-culture
HMC3 cells were switched to RPMI containing 10% FCS upon seeding in the upper transwell chamber (6-well format, 0.4 µM pore size; cat. no. 353,493, Corning). Two days after seeding, transwell inserts were added into wells containing BT-40 (6-well companion plate, cat. no. 353,502, Corning) and media was switched to MM (RPMI, 2% FCS) for the whole transwell chamber.
BT40 xenograft in vivo model
All mouse procedures were performed under license (PP8308129), following UK Home Office Animals (Scientific Procedures) Act 1986 and local institutional guidelines (UCL ethical review committee) and ARRIVE guidelines. For details on experimental procedures see Supplementary materials and methods.
Statistics
Statistical analysis was either performed using GraphPad Prism (v8.0.2) or R Studio (R version 4.2.2). Statistical tests performed and number of independent biological replicates for each experiment are specified in the respective figure legends. Significance was defined as p-value/adj-p-value ≤ 0.05 if not otherwise indicated. For details see Supplementary materials and methods.
Results
Establishment of an in vitro MAPKi withdrawal rebound model
We tested several patient-derived in vitro models with the aim of establishing a MAPKi withdrawal rebound model fulfilling the following criteria: (1) response upon treatment (i.e. decreased cell proliferation or increased cell death); (2) cell regrowth upon treatment withdrawal; (3) cell regrowth upon MAPKi withdrawal faster than after chemotherapy withdrawal; optionally (4) faster cell proliferation after MAPKi withdrawal compared to untreated cells. Drug concentrations were chosen based on effect in vitro (Fig. S1, for details see Supplementary materials and methods section) and are within clinically relevant concentrations (Table S4).
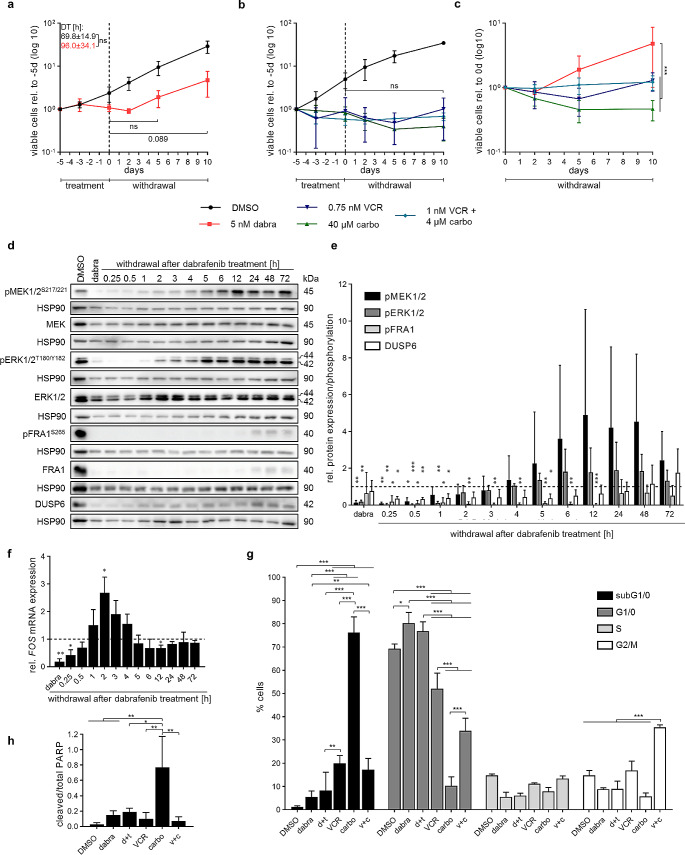
Treatment of pilocytic astrocytoma (PA)-derived KIAA1549:BRAF fusion- (DKFZ-BT66, DKFZ-BT308) or BRAFV600E-driven cells (DKFZ-BT314) in proliferating state (induced by SV40-TAg) or senescent state (no SV40-TAg induction) did not reduce viable cell numbers compared to untreated cells (Fig. S2). Due to the lack of effect on viable cell numbers observed during treatment (i.e. rebound model criteria 1), presence or absence of rebound growth upon treatment withdrawal could not be assessed, making these models unsuitable to study the rebound growth. In contrast, treatment of the pleomorphic xanthoastrocytoma (PXA)-like cell line BT-40 (derived from a juvenile PA, without further genetic modification) [19], driven by BRAFV600E and co-occurring CDKN2A/B deletion, with 5 nM dabrafenib induced a decrease of cell proliferation (Fig. 1a). Upon treatment withdrawal, cells started to re-proliferate after two days, at a similar rate as control cells (DT control:69.8 ± 14.9 h vs. DT after: 96.0 ± 34.1 h), with increased cell numbers at 10 days after withdrawal, hence fulfilling rebound model criteria 2 but not the optional criteria 4. Similar patterns were observed upon dabrafenib and trametinib combination treatment and withdrawal (Fig. S3a). Cells treated with chemotherapy (vincristine (VCR) and carboplatin), showed no cell regrowth upon withdrawal in the timeframe investigated (Fig. 1b). Accordingly, cell regrowth upon dabrafenib treatment withdrawal was significantly increased compared to SOC chemotherapy treatment (Fig. 1c, i.e. rebound model criteria 3), thereby mimicking the effect that is observed in patients and making BT-40 a suitable model to study the rebound growth.
Fig. 1.
Development and characterization of an in vitro rebound model using BT-40 (BRAFV600E, CDKN2A/Bdel). (a-b) Viable cell counts during treatment and withdrawal with 5 nM dabrafenib (dabra) (a) or 0.75 nM vincristine (VCR), 40 µM carboplatin (carbo) and 1 nM VCR and 4µM carbo (b). Dashed line indicates withdrawal timepoint. Viable cell counts are normalized to treatment start (-5d). Data is shown on a logarithmic scale (base 10) as mean ± SD (n = 3 independent biological replicates). Doubling time (DT) was calculated from two to ten days for each biological replicate (n = 3) and is indicated in hours as mean ± SD. Unpaired two-sided t-test; ns: not significant. (c) Viable cell counts during withdrawal of cells pretreated with 5 nM dabrafenib (dabra), 0.75 nM vincristine (VCR), 40 µM carboplatin (carbo) or 1 nM VCR and 4 µM carbo for five days; viable cell counts are normalized to the withdrawal timepoint (five days of treatment) for each condition. Data is shown on a logarithmic scale (base 10) as mean ± SD (n = 3 independent biological). Two-way ANOVA, Bonferroni post-hoc test, *** adj-p-value ≤ 0.001; no indication: not significant. (d-e) Western blot analysis of MAPK activity markers after five days treatment with 5 nM dabrafenib (dabra) followed by treatment withdrawal. Blots shown are representative of three independent biological replicates (d). Quantification (e) is relative to solvent control (DMSO; dashed line) and shown as mean ± SD (n = 3 independent biological replicates). One-sample t-test, *p-value ≤ 0.05 **p-value ≤ 0.01 ***p-value ≤ 0.001; no indication: not significant. (f) RT-qPCR analysis of FOS gene expression after five days treatment with 5 nM dabrafenib (dabra) followed by treatment withdrawal. Quantification is relative to solvent control (DMSO; dashed line) and shown as mean ± SD (n = 3 independent biological replicates). One-sample t-test, *p-value ≤ 0.05 **p-value ≤ 0.01 ***p-value ≤ 0.001; no indication: not significant. (g) Cell cycle analysis using FACS after treatment for five days with 5nM dabrafenib (dabra), 2.7nM dabrafenib and 0.3nM trametinib (d + t), 0.75nM vincristine (VCR), 40µM carboplatin (carbo) or 1nM vincristine and 4µM carboplatin (v + c). Percentage of single cell population in the different cell cycle phases are shown as mean ± SD (n = 3 independent biological replicates). One-way ANOVA, Tukey post-hoc test, * adj-p-value ≤ 0.05 ** adj-p-value ≤ 0.01 *** adj-p-value ≤ 0.001; no indication: not significant. (h) Western blot quantification (of blots in Figure S2b) showing cleaved PARP relative to full-length PARP after treatment for five days with 5 nM dabrafenib (dabra), 2.7 nM dabrafenib and 0.3 nM trametinib (d + t), 0.75 nM vincristine (VCR), 40 µM carboplatin (carbo) or 1 nM vincristine and 4 µM carboplatin (v + c). Data is shown as mean ± SD (n = 3 independent biological replicates). One-way ANOVA, Tukey post-hoc test, *adj-p-value ≤ 0.05 **adj-p-value ≤ 0.01; no indication: not significant
Characterization of the BT-40 in vitro rebound model
Molecular analysis of MAPK pathway activity in BT-40 showed near complete inhibition of the pathway after five days treatment with 5 nM dabrafenib (Fig. 1d-f). Cell cycle analysis (Fig. 1g) indicated arrest in G1/0-phase upon MAPKi treatment, without significant increase of PARP cleavage (Fig. 1h, Fig. S3b). Upon dabrafenib withdrawal, MEK and ERK phosphorylation returned to control levels after five hours (Fig. 1d-e), indicating a rapid reactivation of the pathway. MEK phosphorylation further transiently increased above control levels (Fig. 1d-e). Furthermore, DUSP6, downstream of ERK activation [20], was re-expressed at later withdrawal timepoints (24–72 h). FOS gene expression, an immediate target gene of ERK [21], showed a 2.5-fold increase compared to baseline two hours after withdrawal (Fig. 1f). While reactivation occurred slightly later (6 h) after combined dabrafenib and trametinib withdrawal, overall MAPK reactivation patterns were comparable (Fig. S3c-e). Importantly, DMSO withdrawal did not increase MAPK pathway activity (Fig. S3f-h), indicating the effect observed is not an artefact of the withdrawal procedure.
In contrast, as expected, SOC chemotherapy treatment and withdrawal did not induce consistent changes in the MAPK pathway, except for fluctuations in FOS gene expression observed after VCR withdrawal (Fig. S3i-n). Cell cycle analysis (Fig. 1g) showed an increase in the sub-G1 population in all chemotherapy treatments, indicating cell death. VCR and carboplatin combination additionally led to an increased G2/M population. Accordingly, PARP cleavage, was observed with all SOC drugs but only showed a significant increase upon carboplatin treatment (Fig. 1h, Fig. S2b), in line with the strongest induction of subG0/1 (Fig. 1g).
Taken together, these data indicate that MAPKi treatment induces cell cycle arrest with no significant cytotoxic effects, followed by a rapid reactivation of the pathway specifically upon MAPKi withdrawal.
Multi-omics analysis reveals dynamic changes in MAPK pathway activation, cytokine expression and AKT signaling upon dabrafenib treatment and withdrawal
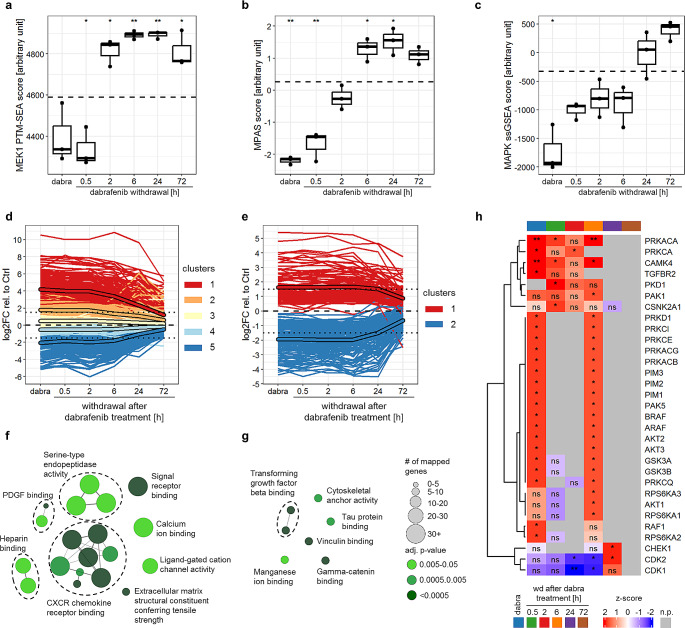
To investigate the underlying molecular mechanisms of regrowth after MAPKi withdrawal, we generated a multi-omics dataset from our BT-40 rebound model upon dabrafenib treatment and withdrawal, using RNA sequencing, LC-MS/MS based proteomics and phosphoproteomics.
Decreased MAPK activity, measured using omics-specific signatures, was observed upon dabrafenib treatment (Fig. 2a-c). Upon treatment withdrawal, MAPK reactivation was observed within hours to one day (protein phosphorylation: 2 h, gene expression: 6 h, protein expression: 24 h) (Fig. 2a-c). Additionally, overactivation of the pathway during dabrafenib withdrawal was observed on the level of gene expression and protein phosphorylation (Fig. 2a-b), in line with our aforementioned data.
Fig. 2.
Multi-omics analysis reveals dynamic changes in MAPK pathway activation, cytokine expression and AKT signaling upon dabrafenib treatment and withdrawal. (a-c) MAPK pathway activity scores after five days treatment with 5 nM dabrafenib followed by withdrawal measured using the MEK1 PTM-SEA score [54] for phosphoproteomics data (a), the MPAS score [55] for RNAseq data (b) and a proteomics-based MAPK ssGSEA-score [56] for proteomics data (c). Boxplots depict the median, first and third quartiles. Whiskers extend from the hinge to the largest/smallest value no further than 1.5 *IQR from the hinge (where IQR is the interquartile range). Dashed line indicates the mean of the solvent control (five days DMSO). Two-tailed unpaired t-test, *p-value ≤ 0.05 **p-value ≤ 0.01 no indication: not significant (n = 3 independent biological replicates). (d-e) Longitudinal k-means clustering of differentially expressed genes (a) and proteins (e) after five days treatment with 5 nM dabrafenib followed by withdrawal relative to solvent control (five days DMSO; dashed line). Only genes and proteins with an adjusted p-value < 0.01 for at least one timepoint were included in the analysis. Unframed lines represent single genes or proteins, framed lines show clusters mean. Dotted line: log2FC = +/-1.5 (n = 3 independent biological replicates). (f-g) GO-term enrichment analysis of upregulated genes (clusters 1 and 2 from panel d, cluster-mean ≥ 1.5) (f), and proteins (cluster 1 from panel e, cluster-mean ≥ 1.5) (g). Only terms with significant enrichment (adj. p-value ≤ 0.05) are shown. GO-term groups are defined by overlapping genes and named based on the GO-Term with highest percentage of mapped genes. (h) Kinase-substrate-enrichment-analysis (KSEA) of differentially regulated phosphopeptides relative to solvent control after five days treatment with 5 nM dabrafenib (dabra) followed by withdrawal (wd). Only phospho-peptides with a significant regulation (adj. p-value < 0.01 at a given timepoint) were included in the analysis. *adj-p-value ≤ 0.05 **adj-p-value ≤ 0.01; ns: not significant; n.p.= no prediction (n = 3 independent biological replicates)
Longitudinal k-means clustering (kml) of differentially regulated genes, proteins and phospho-peptides showed that the majority of differential gene and protein expression or phosphorylation was induced during dabrafenib treatment, and maintained at least until 24 h of withdrawal (mean of highest differentially regulated clusters ≥ 1.5 or ≤ -1.5; Fig. 2d-e, Fig. S4a) before reaching baseline levels (= untreated control) again. Within the upregulated genes (kml-clusters 1 and 2, cluster-mean ≥ 1.5) and proteins (kml-cluster 1, cluster-mean ≥ 1.5), a significant enrichment of GO-terms related to cytokine signaling and TGF-beta signaling, respectively, was observed (Fig. 2f-g, Table S8, Table S9). Furthermore, KSEA analysis of significantly regulated phospho-peptides (adj. p-value < 0.01 at a given time-point) suggested increased activity of several signaling pathways (TGF-beta, AKT, JAK/STAT indicated by downstream effectors PIM and PAK) upon dabrafenib treatment and early withdrawal (up to 6 h; Fig. 2h).
On the other hand, decreased cell cycle activity was indicated by GO-term analysis of downregulated genes (kml-clusters 4 and 5, cluster mean ≤ -1.5) and proteins (kml-cluster 2, cluster mean ≤ -1.5) (Fig. S4b-c, Table S10, Table S11) as well as by KSEA showing decreased CDK activity (Fig. 2h). This is in line with cell cycle arrest upon treatment (Fig. 1g) and decreased cell proliferation seen until two days of withdrawal (Fig. 1a).
Taken together, these data indicate that MAPKi treatment induced the upregulation of cytokines and activates several signaling pathways (TGF-beta, AKT and JAK/STAT), potentially participating in tumor rebound upon withdrawal. Treatment withdrawal on the other hand led to the reactivation of the MAPK pathway and cell cycle related pathways, while still maintaining some of the MAPKi treatment induced changes.
Cytokine expression and increased AKT activity are independent mechanisms without a tumor cell intrinsic effect on rebound growth
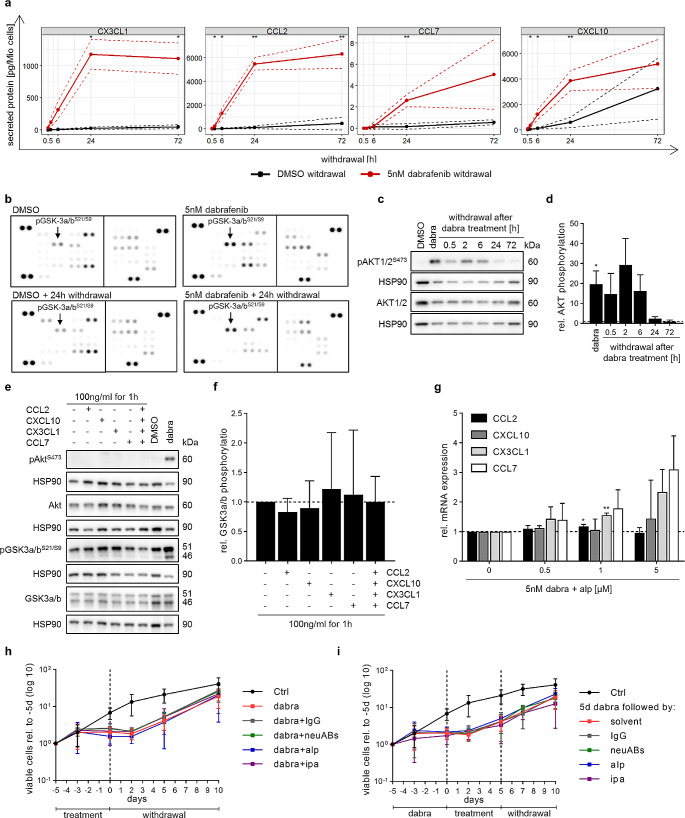
We next investigated the secretion of cytokines showing increased gene expression (21 cytokines; log2FC > 2, padj < 0.01 at 6 h of withdrawal; Fig. S4d) using a commercially available Luminex-based multiplex assay (covering 16/21 cytokines). Cytokine secretion was increased upon dabrafenib treatment and maintained upon withdrawal (Fig. S5a), with 11/16 cytokines maintaining increased secretion with a > 2-fold increase compared to DMSO withdrawal (AUC-log2FC > 1; Table S12). Of these eleven, secretion of CCL2, CX3CL1, CCL7 and CXCL10 was significantly higher 24 h after dabrafenib withdrawal compared to DMSO, one day before cells are proliferating again (Fig. 3a, Fig. S5b).
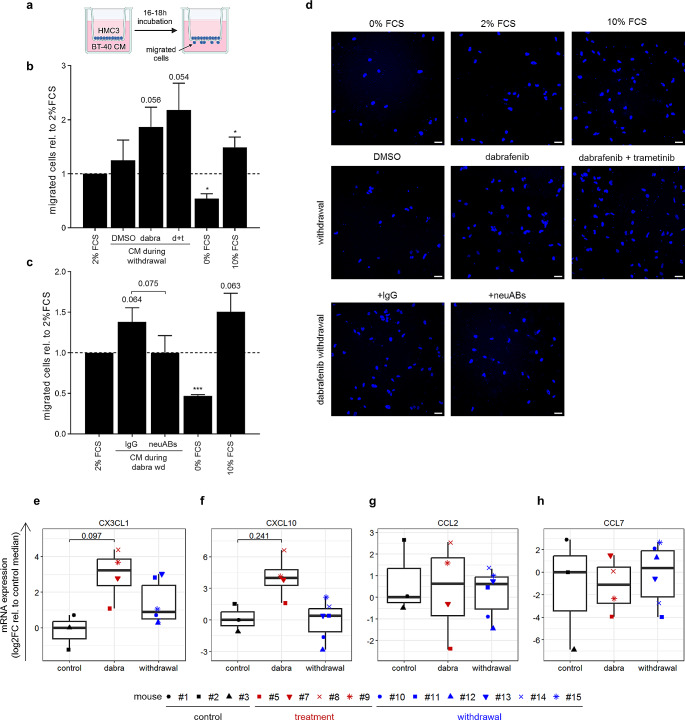
Fig. 3.
Cytokine expression and increased AKT activity are independent mechanisms without a tumor cell intrinsic effect on rebound growth. (a) Luminex-based multiplex assay results showing cytokine secretion during withdrawal after treatment for five days with either DMSO (solvent control) or 5 nM dabrafenib. Data is shown as mean ± SD (n = 3 independent biological replicates). Two-tailed unpaired t-test, *p-value ≤ 0.05 **p-value ≤ 0.01; no indication: not significant. (b) Kinase phosphorylation array of samples treated for five days with 5 nM dabrafenib or DMSO (solvent control) followed by 24 h withdrawal (wd). Arrays consist of two membranes, each target is detected in technical duplicates. Images shown are representatives of two biological replicates. (c-d) Western blot analysis of AKT phosphorylation after five days treatment with 5 nM dabrafenib (dabra) followed by up to 72 h withdrawal. Blots shown are representative of three independent biological replicates (c). Quantification (d) is relative to solvent control (DMSO; dashed line) and shown as mean ± SD (n = 3 independent biological replicates). One-sample t-test, * p-value ≤ 0.05; no indication: not significant. (e-f) Western blot analysis of AKT activity markers after treatment with recombinant cytokines. Treatment for five days with DMSO or 5 nM dabrafenib (dabra) served as negative and positive control for western blots (e). Blots shown are representative of three biological replicates (e). Quantification (f) was done relative to untreated samples (dashed line) and is shown as mean ± SD (n = 3 independent biological replicates). One-sample t-test; no indication: not significant. (g) RT-qPCR analysis of chemokine gene expression after five days treatment with 5 nM dabrafenib (dabra) alone or in combination with varying concentrations of alpelisib (alp). Quantification was done relative to dabrafenib only treatment (0; dashed line) and is shown as mean ± SD (n = 3 independent biological replicates). One-sample t-test, *p-value ≤ 0.05 **p-value ≤ 0.01; no indication: not significant. (h) Viable cell counts during treatment with 5nM dabrafenib (dabra) alone or in combination with a combination of antibodies neutralizing CCL2 (0.5 µg/mL), CX3CL1 (0.25 µg/mL), CXCL10 (0.25 µg/mL) and CCL7 (0.1 ng/mL) (neuABs), IgG control (1 µg/mL), 5 µM alpelisib (alp) or 1 µM ipatasertib (ipa) followed by ten days of withdrawal. Dashed line indicates withdrawal timepoint. Viable cell counts are normalized to treatment start (-5d). Data is shown on a logarithmic scale (base 10) as mean ± SD of at least three biological replicates. (i) Viable cell counts during treatment with 5 nM dabrafenib (dabra) followed by withdrawal. During withdrawal cells were either untreated (= solvent) or treated for five days with a combination of antibodies neutralizing CCL2 (0.5 µg/mL), CX3CL1 (0.25 µg/mL), CXCL10 (0.25 µg/mL) and CCL7 (0.1 ng/mL) (neuABs), IgG control (1 µg/mL), 5 µM alpelisib (alp) or 1 µM ipatasertib (ipa), followed by five days of withdrawal. Dashed lines indicate withdrawal timepoints. Viable cell counts are normalized to treatment start (-5d). Data is shown on a logarithmic scale (base 10) as mean ± SD of at least three biological replicates
To validate the differential regulation of the aforementioned pathways, a phosphorylation kinase array and western blot analysis was used. Increased AKT activity (GSK3-a/b and AKT phosphorylation) upon treatment and withdrawal was confirmed (Fig. 3b-d, Fig. S6c), but no increased TGF-beta or JAK/STAT signaling was observed (Fig. S6a-c).
Of note, increased expression of CCL2, CX3CL1, CCL7 and CXCL10, and increased AKT activity, were also observed for dabrafenib and trametinib combination treatment and withdrawal (Fig. S6d-f).
We next investigated a possible interplay between CCL2, CX3CL1, CCL7 and CXCL10 expression and AKT activation. While some cytokine receptors were expressed in BT-40 (Fig. S7a), neither stimulation with recombinant cytokines (Fig. 3e-f) nor inhibition of cytokines using neutralizing antibodies (neuABs, Fig. S7b-c) affected AKT activity. Furthermore, inhibition of AKT activity using the PI3K inhibitor alpelisib did not suppress dabrafenib-induced cytokine expression (Fig. 3g, Fig. S7d-e). Overall, this indicates independence of MAPKi-induced cytokine expression from AKT signaling in our model.
While both cytokines and AKT signaling independently can be survival- and growth-promoting, neither cytokine inhibition by neuABs nor AKT inhibition by alpelisib (PI3Ki) or ipatasertib (AKTi) during MAPKi treatment or withdrawal reduced viable cell counts compared to MAPK inhibition only (Fig. 3h-i, Fig. S7d-j).
Taken together these data indicate the absence of a tumor cell intrinsic growth-promoting effect of the cytokines or AKT signaling during MAPKi treatment or withdrawal, and suggests that MAPK pathway reactivation is the natural driver of the intrinsic tumor rebound mechanism in our rebound model.
In vivo validation of MAPK pathway reactivation during rebound growth
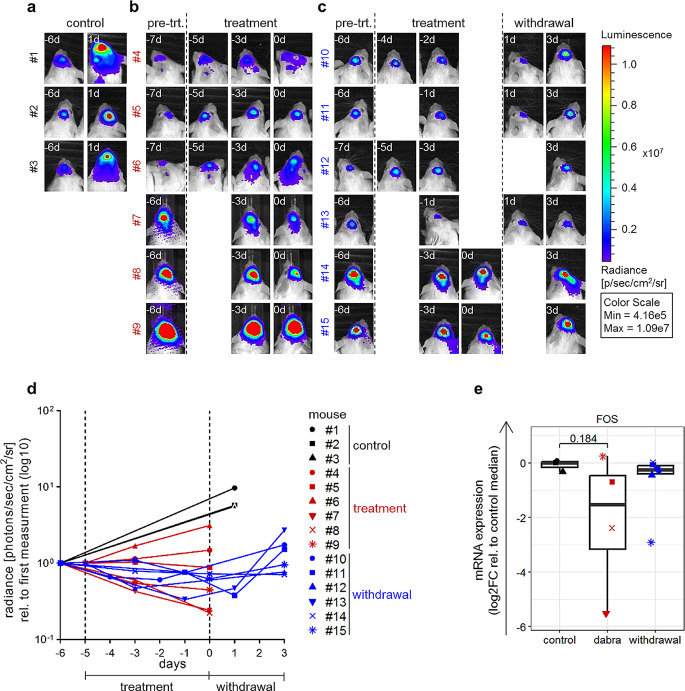
To investigate the MAPK pathway reactivation upon treatment withdrawal in vivo, BT-40 cells were engrafted orthotopically in NOD scid gamma (NSG) mice to generate samples for molecular analysis (gene expression) during treatment and withdrawal. As shown by bioluminescent imaging, used to control for treatment effect, 100 mg/kg dabrafenib treatment stabilized tumor growth within six days of treatment in 10/12 mice (non-responding mice were excluded from further analysis). Upon treatment stop, a trend for increased tumor size was observed during the first three days of withdrawal and before harvesting of tumors in 2/6 mice, suggesting the possibility of rebound growth in our in vivo model (Fig. 4a-d).
Fig. 4.
MAPK reactivation upon dabrafenib treatment and withdrawal in BT-40 orthotopic xenograft model. (a-d) NSG mice carrying BT-40 xenograft tumors were treated with 100 mg/kg dabrafenib (treatment; six doses, once daily) followed by three days of withdrawal. Tumor growth was measure by bioluminescence imaging and is shown for each individual animal as radiance in photons/s/cm²/steradian (d). Dashed lines indicate treatment start and treatment stop. Bioluminescence images (a-c) of untreated animals (control, n = 3; a), animals undergoing treatment (n = 6; b) and animals undergoing treatment followed by withdrawal (n = 6; c); pre-trt. = before treatment start. (e) RT-qPCR analysis of FOS expression in BT-40 xenograft tumors after six days treatment with dabrafenib (dabra; 100 mg/kg, six doses, once daily) followed by three days of withdrawal (withdrawal). Samples from mice showing tumor progression (#4, #6; panel d) during dabrafenib treatment were excluded from the analysis. Quantification was done relative to the median of untreated samples (control). FOS was undetected in two samples (#7, #8), for these samples Ct values were set to 40 (max. number of cycles). Boxplots depict the median, first and third quartiles. Whiskers extend from the hinge to the largest/smallest value no further than 1.5 *IQR from the hinge (where IQR is the interquartile range). Two-tailed unpaired t-test; no indication: not significant (control: n = 3 mice, dabra: n = 4 mice, withdrawal: n = 6 mice)
Gene expression analysis of tumor samples showed decreased FOS expression during treatment and re-expression during treatment withdrawal compared to untreated controls (Fig. 4e), in line with MAPK pathway regulation patterns observed in vitro.
MAPKi-induced cytokines attract microglia in a paracrine fashion
Since the cytokines had no tumor cell intrinsic effects, we hypothesized that they might have tumor extrinsic properties, that could further increase tumor growth (i.e. optional rebound model criteria 4), which was not observed in BT-40 monoculture. As cytokines, in particular CCL2 and CX3CL1, have been described to attract microglia cells [22–24], we investigated microglia attraction using conditioned media (CM; Fig. 5a). Indeed, increased migration of HMC3 cells, with a trend towards statistical significance, was observed using CM collected from BT-40 cells during MAPKi withdrawal (Fig. 5b, d), which was reversed by the addition of a combination of antibodies neutralizing CCL2, CX3CL1, CXCL10 and CCL7 (Fig. 5c-d). Additionally, we could show that microglia cells support BT-40 growth (Fig. S8), suggesting that microglia cells recruited upon treatment withdrawal could increase tumor growth (i.e. optional tumor rebound criteria 4). Importantly, a trend for increased mRNA expression of CX3CL1 and CXCL10 in tumor cells was also observed upon dabrafenib treatment in vivo (Fig. 5e-h), suggesting that such a recruitment could take place in vivo. Further investigation in pLGG PDX-models in immunocompetent mice, yet to be established [25], will be necessary to validate these findings.
Fig. 5.
Increased cytokine secretion upon MAPKi treatment and withdrawal induces increased microglia attraction. (a) Schematic of transwell migration assay setup to investigate migration of HMC3 cells towards conditioned media (CM) collected from BT-40. (b) Transwell migration assay of HMC3 cells towards conditioned media (CM) collected from BT-40 cells after 24 h of withdrawal after five days treatment with DMSO (solvent control), 5nM dabrafenib (d) or 2.7 nM dabrafenib and 0.3 nM trametinib (d + t). 2% FCS serves as baseline control as CM contains 2% FCS, 0% FCS as negative control and 10% FCS as positive control. Quantification is shown as mean ± SD (n = 3 independent biological replicates; 2 technical duplicates per condition; 10–12 randomly distributed images were quantified per transwell) relative to 2% FCS. One-sample t-test, * p-value ≤ 0.05; no indication: not significant. (c) Transwell migration assay of HMC3 cells towards CM collected 24 h after dabrafenib withdrawal (dabra wd) containing either antibodies neutralizing CCL2 (1 µg/mL), CX3CL1 (0.5 µg/mL), CXCL10 (0.5 µg/mL) and CCL7 (0.2 ng/mL) (neuABs) or IgG (2 µg/mL). 2% FCS serves as baseline control as CM contains 2% FCS, 0% FCS as negative control and 10% FCS as positive control. Quantification is shown as mean ± SD (n = 3 independent biological replicates; 2 technical duplicates per condition; 10–12 randomly distributed images were quantified per transwell) relative to 2% FCS. One-sample t-test and two-tailed unpaired t-test (comparing IgG to neuABs), *p-value ≤ 0.05 ***p-value ≤ 0.001; no indication: not significant. (d) Representative fluorescence images showing HMC3 migrated through the transwell, nuclei were stained with DAPI. Scale bar = 50 µM. (e-h) RT-qPCR analysis of CX3CL1 (e), CXCL10 (f), CCL2 (g) and CCL7 (h) expression in BT-40 xenograft tumors after six days treatment with dabrafenib (dabra; 100 mg/kg, six doses, once daily) followed by three days of withdrawal (withdrawal). Samples from mice showing tumor progression (#4, #6; Fig. 4d) during dabrafenib treatment were excluded from the analysis. Quantification was done relative to the median of untreated samples (control). CX3CL1 was undetected in one sample (#2), CCL2 was undetected in two samples (#2, #7) and CCL7 was undetected in six samples (#2, #3, #5, #7, #8, #11), for these samples Ct values were set to 40 (max. number of cycles). Boxplots depict the median, first and third quartiles. Whiskers extend from the hinge to the largest/smallest value no further than 1.5 * IQR from the hinge (where IQR is the interquartile range). Two-tailed unpaired t-test; no indication: not significant (control: n = 3 mice, dabra: n = 4 mice, withdrawal: n = 6 mice)
Of note, increased microglia attraction was also observed using conditioned media collected during MAPKi treatment (Fig. S9). The role of microglia in the treatment period remains to be elucidated.
Taken together, these data suggest a possible tumor cell extrinsic, paracrine mechanism of MAPKi-induced cytokine secretion involving microglia cells, warranting further investigation using more complex models.
Discussion
Rebound growth of pLGGs after MAPKi treatment remains a clinical challenge and is not well understood. Both prediction as well as prevention of rebound growth need a deeper molecular and biological understanding of the underlying mechanisms driving it.
We successfully modeled the rebound growth of pediatric glioma upon MAPKi withdrawal in vitro using BT-40 (BRAFV600E, CDKN2A/Bdel). However, in additional pLGG models driven by KIAA1549:BRAF fusion (DKFZ-BT66, DKFZ-BT308) or BRAFV600E mutation (DKFZ-BT314) we did not observe an effect on viable cell counts during treatment, in line with previous data [14, 15, 26]. This may be explained by inhibition of p53 and pRB through expression of SV40-TAg [14, 27, 28] in the proliferating cells and by the biology of senescent cells, which commonly do not undergo apoptosis [15, 26, 29]. Due to the lack of effect during MAPKi treatment in these models, the presence or absence of rebound growth could not be assessed and these models could not be further used in this study. Nonetheless, our BT-40 rebound model represents an important pediatric glioma subgroup, as patients with this genetic background have the poorest prognosis [30]. Whether our findings in BT-40 also apply to pLGG entities with different genetic backgrounds needs to be further validated in future studies using more suitable models, including non-rebounding models as additional control.
Using our BT-40 rebound model, we observed rapid reactivation of the MAPK pathway upon MAPKi withdrawal. As rebound growth in pLGG is not associated with MAPKi resistance [8, 13], a MAPK-dependent driving mechanism is plausible and we assume that the fast MAPK reactivation plays a role in the rebound growth. In addition to a rapid reactivation, we observed a transient overshooting activation of the pathway, which has also been observed in other MAPK-driven tumor entities [31–33]. Interestingly, in our model, MAPK overactivation was most prominently observed on the level of MEK phosphorylation and MAPK target gene expression (FOS, MPAS score). Substantial pERK rebound, as observed in other studies and associated with reduced proliferation and cell death in MAPKi addiction models [31–33], was not observed, likely due to the parallel re-expression of DUSP6, which negatively regulates ERK activity [34]. Taken together, this indicates a potential vulnerability to DUSP6 inhibitors, as targeting of this phosphatase may induce apoptosis due to overshooting MAPK activity [35, 36].
In addition to a fast reactivation of the MAPK pathway, we observed upregulation of AKT signaling, which is commonly observed upon MAPKi in other entities [37–39] and combination of MAPKi and PI3K/AKT/mTORi was shown to have beneficial effects in different tumor entities [40–42], including pLGGs [43]. In contrast to what was previously described for BT-40 [43], inhibition of AKT signaling in combination with MAPKi did not further decrease viable cell counts compared to MAPKi only, and most importantly it did not affect rebound growth in our setting. This could be explained by the fact that the synergistic activity of mTORCi and MEKi involved effects mediated by the microenvironment (i.e. angiogenesis) [43], which is not reflected in our in vitro setting.
Our analysis furthermore revealed increased expression and secretion of several cytokines, in particular CCL2, CX3CL1, CXCL10 and CCL7, upon dabrafenib treatment and withdrawal in vitro, with a trend for increased expression of CX3CL1 and CXCL10 also in vivo. While cytokines have been described to have direct growth-promoting effects on tumor cells [44–47], in our rebound model, inhibition of these cytokines had no effect on BT-40 viability or proliferation, indicating that these cytokines have no tumor cell intrinsic effect. However, we were able to demonstrate a tendency for increased attraction of microglia cells, which make up the majority of the TME in pLGG tumors [3]. In line with previous work demonstrating the role of cytokines in immune cell attraction [22–24, 48–51], a trend for decreased microglia attraction was observed upon inhibition of CCL2, CX3CL1, CXCL10 and CCL7 in our setting.
Increased microglia attraction by the tumor cells upon MAPKi withdrawal observed in vitro could possibly contribute to a neuroradiologically growing tumor mass, as observed on MRI. Furthermore, we observed growth-promoting effects of microglia on BT-40 cells in vitro. In line, a study using BRAF fusion-driven neural stem cells showed the importance of microglia recruitment for tumor formation in vivo [52]. This suggests a potential tumor growth-promoting role of microglia cells (attracted either during MAPKi treatment and/or immediately after withdrawal while cytokines are still increased), possibly enhancing tumor growth after MAPKi withdrawal and leading to faster growth after compared to before treatment. Furthermore, attraction of microglia cells may have implications during MAPKi treatment, as microglia infiltration in pLGG tumors correlated with a high predictive MAPK inhibitor sensitivity score (MSS) [53]. Taken together, these data indicate a possible complex role of microglia in response to MAPKi treatment and rebound growth after treatment withdrawal in pLGG. The exact role of microglia cells and how their activation status (i.e. tumor-promoting or -suppressive) may be influenced by MAPKi treatment and withdrawal needs further investigation, for which more complex models, such as organoids containing immune cells or immunocompetent mouse models [25], will be necessary.
In summary, we have identified the MAPK pathway as a tumor cell intrinsic rebound driving mechanism in pediatric glioma cells with BRAFV600E mutation and CDKN2A/Bdel. In addition, MAPKi withdrawal was associated with the expression and secretion of microglia-recruiting cytokines, which may contribute to rebound growth in a paracrine manner. Further validation of our findings in patient samples will be necessary. Such samples remain to date extremely scarce, since re-challenging with MAPKi is currently favored to surgery to treat rebounding patients (clinical communication). Nonetheless, our data opens the question whether further development of different MAPK inhibitors for the treatment of pediatric glioma is enough to sufficiently improve patient outcome, as the MAPK pathway will most likely be reactivated with any inhibitor upon treatment stop. Therefore, the exploration of more effective treatment strategies, possibly involving modulation of the tumor immune microenvironment, suggested to play a role by our data, are warranted to improve patient’s quality of life.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Isabel Büdenbender, Carina Müller, Anna Laudat, Johanna Bauer and Daniela Kuhn for their excellent technical support and Natja Sachse, Elisabeth Hillier and Nicola Herrmann-Wichmann for excellent administrative assistance. We thank Marta Emperador Melero for her scientific input. We thank the NGS Core Facility, German Cancer Research Center (DKFZ), for providing excellent RNA sequencing services, the Omics IT and Data Management Core Facility (ODCF), German Cancer Research Center (DKFZ), for providing excellent support with the RNAseq data pre-processing as well as the Light Microscopy Facility at the German Cancer Research Center (DKFZ) and the Flow Cytometry Core Facility at the German Cancer Research Center (DKFZ) for their support during this study. Illustrations were created with BioRender.com.
Author contributions
D.K., R.S. and T.M. conceived the ideas and designed the study. D.K., L.C., R.G., M.L., Y.L., P.B., H.H., J.R., K.K.M., S.Ha. and R.S. performed experiments and data analysis. D.F. and S.He. performed data analysis. R.G., F.S., D.V., J.Z., G.V., D.P., J.R., K.K.M., K.W.P., M.R., S.P., S.M.P., I.O., D.T.W.J., S.Ha., T.B., J.P.M.-B. and O.W. provided critical input on the project and experimental design. A.vD., S.P. and D.T.W.J. provided resources. O.W. and T.M. acquired funding. All authors contributed to writing and reviewing the manuscript.
Funding
D.T.W.J., O.W. and T.M. were supported by The Everest Centre for Low-grade Paediatric Brain Tumours (GN-000707, The Brain Tumour Charity, UK). T.M., O.W. and T.B. were supported by DKTK JF Upgrade Next Gen LOGGIC (B310-JF-LOGGIC-MDE). J.P.M.-B. was funded by Cancer Research UK (C54322/A27727), the Brain Tumour Charity (EVEREST (GN-000382) and GN-000522) and National Institutes of Health Research Biomedical Research Center at the Great Ormond Street Hospital for Children NHS Foundation Trust, and the University College London. S.Ha. and T.B. were also supported by the German Research Foundation (DFG) funded SFB 1479-project, ID: 441891347 (Junior Fellowship (S.Ha.); P14 (T.B.)) and project HA 8638/1–1 (S.Ha). I.O. was supported by Deutsche Krebshilfe (#70113843).
Open Access funding enabled and organized by Projekt DEAL.
Data availability
The RNA sequencing data (GSE249718) generated in this study is available from the GEO platform. The LC-MS/MS proteomics and phosphoproteomics data (PXD048277) generated in this study are available from the PRIDE database.
Declarations
Conflict of interest
T.M. has received research grants from Biomed Valley Discoveries, Inc. and Day One Biopharmaceuticals. O.W is on the advisory board with Novartis and has received research grants from Biomed Valley Discoveries, Inc., Day One Biopharmaceuticals and PreComb. IO received research grants from PreComb.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Till Milde, Email: t.milde@kitz-heidelberg.de.
Romain Sigaud, Email: r.sigaud@kitz-heidelberg.de.
References
- 1.Ostrom QT, Price M, Ryan K, et al. CBTRUS Statistical Report: Pediatric Brain Tumor Foundation Childhood and adolescent primary brain and other Central Nervous System tumors diagnosed in the United States in 2014–2018. Neuro Oncol. 2022;24:iii1–iii38. doi: 10.1093/neuonc/noac161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis DN, Perry A, Wesseling P, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 2021;23:1231–1251. doi: 10.1093/neuonc/noab106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Milde T, Rodriguez FJ, Barnholtz-Sloan JS, et al. Reimagining pilocytic astrocytomas in the context of pediatric low-grade gliomas. Neuro Oncol. 2021 doi: 10.1093/neuonc/noab138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armstrong GT, Conklin HM, Huang S, et al. Survival and long-term health and cognitive outcomes after low-grade glioma. Neuro Oncol. 2011;13:223–234. doi: 10.1093/neuonc/noq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones DTW, Bandopadhayay P, Jabado N. The Power of Human Cancer Genetics as revealed by Low-Grade Gliomas. Annu Rev Genet. 2019;53:483–503. doi: 10.1146/annurev-genet-120417-031642. [DOI] [PubMed] [Google Scholar]
- 6.Manoharan N, Liu KX, Mueller S, et al. Pediatric low-grade glioma: targeted therapeutics and clinical trials in the molecular era. Neoplasia (United States) 2023;36:100857. doi: 10.1016/j.neo.2022.100857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karoulia Z, Gavathiotis E, Poulikakos PI. New perspectives for targeting RAF kinase in human cancer. Nat Rev Cancer. 2017;17:676–691. doi: 10.1038/nrc.2017.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nobre L, Zapotocky M, Ramaswamy V et al (2020) Outcomes of BRAF V600E pediatric Gliomas treated with targeted BRAF inhibition. JCO Precis Oncol 561–571. 10.1200/po.19.00298 [DOI] [PMC free article] [PubMed]
- 9.Hargrave DR, Bouffet E, Tabori U, et al. Efficacy and safety of Dabrafenib in Pediatric patients with BRAF V600 mutation-positive relapsed or refractory low-Grade Glioma: results from a phase I/IIa study. Clin Cancer Res. 2019;25:7303–7311. doi: 10.1158/1078-0432.CCR-19-2177. [DOI] [PubMed] [Google Scholar]
- 10.Barbato MI, Nashed J, Bradford D, et al. FDA approval Summary: Dabrafenib in combination with trametinib for BRAF V600E mutation-positive low-grade glioma. Clin Cancer Res. 2023;30:OF1–OF6. doi: 10.1158/1078-0432.CCR-23-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouffet E, Hansford JR, Garrè ML, et al. Dabrafenib plus Trametinib in Pediatric Glioma with BRAF V600 mutations. N Engl J Med. 2023;389:1108–1120. doi: 10.1056/NEJMoa2303815. [DOI] [PubMed] [Google Scholar]
- 12.Tsai JW, Choi JJ, Ouaalam H et al (2023) Integrated response analysis of pediatric low-grade gliomas during and after targeted therapy treatment. Neuro-Oncology Adv 5. 10.1093/noajnl/vdac182 [DOI] [PMC free article] [PubMed]
- 13.Selt F, van Tilburg CM, Bison B, et al. Response to trametinib treatment in progressive pediatric low-grade glioma patients. J Neurooncol. 2020;149:499–510. doi: 10.1007/s11060-020-03640-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Selt F, Hohloch J, Hielscher T, et al. Establishment and application of a novel patient-derived KIAA1549: BRAF-driven pediatric pilocytic astrocytoma model for preclinical drug testing. Oncotarget. 2017;8:11460–11479. doi: 10.18632/oncotarget.14004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selt F, Sigaud R, Valinciute G, et al. BH3 mimetics targeting BCL-XL impact the senescent compartment of pilocytic astrocytoma. Neuro Oncol. 2023;25:735–747. doi: 10.1093/neuonc/noac199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Selt F, El Damaty A, Schuhmann MU, et al. Generation of patient-derived pediatric pilocytic astrocytoma in-vitro models using SV40 large T: evaluation of a modeling workflow. J Neurooncol. 2023;165:467–478. doi: 10.1007/s11060-023-04500-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valinciute G, Ecker J, Selt F, et al. Class I HDAC inhibitor entinostat synergizes with PLK1 inhibitors in MYC-amplified medulloblastoma cells. J Neurooncol. 2023;163:143–158. doi: 10.1007/s11060-023-04319-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milde T, Oehme I, Korshunov A, et al. HDAC5 and HDAC9 in medulloblastoma: novel markers for risk stratification and role in tumor cell growth. Clin Cancer Res. 2010;16:3240–3252. doi: 10.1158/1078-0432.CCR-10-0395. [DOI] [PubMed] [Google Scholar]
- 19.Kolb EA, Gorlick R, Houghton PJ, et al. Initial testing (stage 1) of AZD6244 (ARRY-142886) by the pediatric preclinical testing program. Pediatr Blood Cancer. 2010;55:668–677. doi: 10.1002/pbc.22576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ram A, Murphy D, DeCuzzi N, et al. A guide to ERK dynamics, part 2: downstream decoding. Biochem J. 2023;480:1909–1928. doi: 10.1042/BCJ20230277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy LO, Smith S, Chen RH, et al. Molecular, interpretation of ERK signal duration by immediate early gene products. Nat Cell Biol. 2002;4:556–564. doi: 10.1038/ncb822. [DOI] [PubMed] [Google Scholar]
- 22.He M, Dong H, Huang Y, et al. Astrocyte-derived CCL2 is associated with m1 activation and recruitment of cultured microglial cells. Cell Physiol Biochem. 2016;38:859–870. doi: 10.1159/000443040. [DOI] [PubMed] [Google Scholar]
- 23.Hulshof S, van Haastert ES, Kuipers HF, et al. CX3CL1 and CX3CR1 expression in human brain tissue: Noninflammatory Control versus multiple sclerosis. J Neuropathol Exp Neurol. 2003;62:899–907. doi: 10.1093/jnen/62.9.899. [DOI] [PubMed] [Google Scholar]
- 24.Held-Feindt J, Hattermann K, Müerköster SS, et al. CX3CR1 promotes recruitment of human glioma-infiltrating microglia/macrophages (GIMs) Exp Cell Res. 2010;316:1553–1566. doi: 10.1016/j.yexcr.2010.02.018. [DOI] [PubMed] [Google Scholar]
- 25.Milde T, Fangusaro J, Fisher MJ, et al. Optimizing preclinical pediatric low-grade glioma models for meaningful clinical translation. Neuro Oncol. 2023;25:1920–1931. doi: 10.1093/neuonc/noad125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buhl JL, Selt F, Hielscher T, et al. The senescence-associated secretory phenotype mediates Oncogene-induced senescence in Pediatric Pilocytic Astrocytoma. Clin Cancer Res. 2019;25:1851–1866. doi: 10.1158/1078-0432.CCR-18-1965. [DOI] [PubMed] [Google Scholar]
- 27.Ahuja D, Sáenz-Robles MT, Pipas JM. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene. 2005;24:7729–7745. doi: 10.1038/sj.onc.1209046. [DOI] [PubMed] [Google Scholar]
- 28.Usta D, Sigaud R, Buhl JL, et al. A cell-based MAPK reporter assay reveals synergistic MAPK pathway activity suppression by MAPK inhibitor combination in BRAF-driven pediatric low-grade glioma cells. Mol Cancer Ther. 2020;19:1736–1750. doi: 10.1158/1535-7163.MCT-19-1021. [DOI] [PubMed] [Google Scholar]
- 29.Soto-Gamez A, Quax WJ, Demaria M. Regulation of Survival Networks in senescent cells: from mechanisms to interventions. J Mol Biol. 2019;431:2629–2643. doi: 10.1016/j.jmb.2019.05.036. [DOI] [PubMed] [Google Scholar]
- 30.Lassaletta A, Zapotocky M, Mistry M, et al. Therapeutic and prognostic implications of BRAF V600E in Pediatric Low-Grade Gliomas. J Clin Oncol. 2017;35:2934–2941. doi: 10.1200/JCO.2016.71.8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leung GP, Feng T, Sigoillot FD, et al. Hyperactivation of MAPK signaling is deleterious to RAS/RAF-mutant Melanoma. Mol Cancer Res. 2019;17:199–211. doi: 10.1158/1541-7786.MCR-18-0327. [DOI] [PubMed] [Google Scholar]
- 32.Chiou LW, Chan CH, Jhuang YL, et al. DNA replication stress and mitotic catastrophe mediate sotorasib addiction in KRASG12C-mutant cancer. J Biomed Sci. 2023;30:50. doi: 10.1186/s12929-023-00940-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farnsworth DA, Inoue Y, Johnson FD, et al. MEK inhibitor resistance in lung adenocarcinoma is associated with addiction to sustained ERK suppression. npj Precis Oncol. 2022;6:1–15. doi: 10.1038/s41698-022-00328-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pratilas CA, Taylor BS, Ye Q, et al. V600EBRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci U S A. 2009;106:4519–4524. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ito T, Young MJ, Li R, et al. Paralog knockout profiling identifies DUSP4 and DUSP6 as a digenic dependence in MAPK pathway-driven cancers. Nat Genet. 2021;53:1664–1672. doi: 10.1038/s41588-021-00967-z. [DOI] [PubMed] [Google Scholar]
- 36.Ecker V, Brandmeier L, Stumpf M, et al. Negative feedback regulation of MAPK signaling is an important driver of chronic lymphocytic leukemia progression. Cell Rep. 2023;42:113017. doi: 10.1016/j.celrep.2023.113017. [DOI] [PubMed] [Google Scholar]
- 37.Corrales E, Levit-Zerdoun E, Metzger P, et al. PI3K/AKT signaling allows for MAPK/ERK pathway independency mediating dedifferentiation-driven treatment resistance in melanoma. Cell Commun Signal. 2022;20:187. doi: 10.1186/s12964-022-00989-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Obenauf AC, Zou Y, Ji AL, et al. Therapy-induced tumour secretomes promote resistance and tumour progression. Nature. 2015;520:368–372. doi: 10.1038/nature14336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi H, Hugo W, Kong X, et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov. 2014;4:80–93. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi H, Kong X, Ribas A, Lo RS. Combinatorial treatments that overcome PDGFRβ-driven resistance of melanoma cells to V600EB-RAF inhibition. Cancer Res. 2011;71:5067–5074. doi: 10.1158/0008-5472.CAN-11-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steinbrunn T, Stühmer T, Sayehli C, et al. Combined targeting of MEK/MAPK and PI3K/Akt signalling in multiple myeloma. Br J Haematol. 2012;159:430–440. doi: 10.1111/bjh.12039. [DOI] [PubMed] [Google Scholar]
- 42.Schreck KC, Allen AN, Wang J, Pratilas CA (2020) Combination MEK and mTOR inhibitor therapy is active in models of glioblastoma. 10.1093/noajnl/vdaa138. Neuro-Oncology Adv 2: [DOI] [PMC free article] [PubMed]
- 43.Arnold A, Yuan M, Price A, et al. Synergistic activity of mTORC1/2 kinase and MEK inhibitors suppresses pediatric low-grade glioma tumorigenicity and vascularity. Neuro Oncol. 2019 doi: 10.1093/neuonc/noz230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y, Cai Y, Liu L et al (2018) Crucial biological functions of CCL7 in cancer. 10.7717/peerj.4928. PeerJ 2018: [DOI] [PMC free article] [PubMed]
- 45.Liu M, Guo S, Stiles JK. The emerging role of CXCL10 in cancer. Oncol Lett. 2011;2:583–589. doi: 10.3892/ol.2011.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu M, Wang Y, Xia R et al (2021) Role of the CCL2-CCR2 signalling axis in cancer: mechanisms and therapeutic targeting. Cell Prolif. 54 [DOI] [PMC free article] [PubMed]
- 47.Conroy MJ, Lysaght J (2020) CX3CL1 Signaling in the Tumor Microenvironment. Advances in Experimental Medicine and Biology. Springer, pp 1–12 [DOI] [PubMed]
- 48.Clarner T, Janssen K, Nellessen L et al (2015) CXCL10 triggers early microglial activation in the Cuprizone Model. 194:3400–3413 [DOI] [PubMed]
- 49.Wetzel K, Menten P, Opdënakker G, et al. Transduction of human MCP-3 by a parvoviral vector induces leukocyte infiltration and reduces growth of human cervical carcinoma cell xenografts. J Gene Med. 2001;3:326–337. doi: 10.1002/jgm.191. [DOI] [PubMed] [Google Scholar]
- 50.Tsou CL, Peters W, Si Y, et al. Critical roles for CCR2 and MCP-3 in monocyte mobilization from bone marrow and recruitment to inflammatory sites. J Clin Invest. 2007;117:902–909. doi: 10.1172/JCI29919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoneyama H, Narumi S, Zhang Y, et al. Pivotal role of dendritic cell-derived CXCL10 in the retention of T helper cell 1 lymphocytes in secondary lymph nodes. J Exp Med. 2002;195:1257–1266. doi: 10.1084/jem.20011983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen R, Keoni C, Waker CA, et al. KIAA1549-BRAF expression establishes a permissive Tumor Microenvironment through NFκB-Mediated CCL2 production. Neoplasia (United States) 2019;21:52–60. doi: 10.1016/j.neo.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sigaud R, Albert TK, Hess C et al (2023) MAPK inhibitor sensitivity scores predict sensitivity driven by the immune infiltration in pediatric low-grade gliomas. Nat Commun 14. 10.1038/s41467-023-40235-8 [DOI] [PMC free article] [PubMed]
- 54.Krug K, Mertins P, Zhang B, et al. A curated resource for Phosphosite-specific signature analysis. Mol Cell Proteom. 2019;18:576–593. doi: 10.1074/mcp.TIR118.000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wagle M-C, Kirouac D, Klijn C, et al. A transcriptional MAPK pathway activity score (MPAS) is a clinically relevant biomarker in multiple cancer types. npj Precis Oncol. 2018;2:7. doi: 10.1038/s41698-018-0051-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maxwell MJ, Arnold A, Sweeney H et al (2021) Unbiased proteomic and phosphoproteomic analysis identifies response signatures and novel susceptibilities after combined MEK and mTOR inhibition in BRAFV600E mutant glioma. Mol Cell Proteom 20. 10.1016/j.mcpro.2021.100123 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing data (GSE249718) generated in this study is available from the GEO platform. The LC-MS/MS proteomics and phosphoproteomics data (PXD048277) generated in this study are available from the PRIDE database.