Abstract
The herpes simplex virus type 1 (HSV-1) capsid shell is composed of four major polypeptides, VP5, VP19c, VP23, and VP26. VP26, a 12-kDa polypeptide, is associated with the tips of the capsid hexons formed by VP5. Mature capsids form upon angularization of the shell of short-lived, fragile spherical precursors termed procapsids. The cold sensitivity and short-lived nature of the procapsid have made its isolation and biochemical analysis difficult, and it remains unclear whether procapsids contain bound VP26 or whether VP26 is recruited following shell angularization. By indirect immunocytochemical analysis of virally expressed VP26 and by direct visualization of a transiently expressed VP26-green fluorescent protein fusion, we show that VP26 fails to specifically localize to intranuclear procapsids accumulated following incubation of the temperature-sensitive HSV mutant tsProt.A under nonpermissive conditions. However, following a downshift to the permissive temperature, which allows procapsid maturation to proceed, VP26 was seen to concentrate at intranuclear sites which also contained epitopes specific to mature, angularized capsids. Like the formation of these epitopes, the association of VP26 with maturing capsids was blocked in a reversible fashion by the depletion of intracellular ATP. We conclude that unlike the other major capsid shell proteins, VP26 is recruited in an ATP-dependent fashion after procapsid maturation begins.
The major components of the herpes simplex virus (HSV) capsid shell are the virally encoded polypeptides VP5, VP19c, VP23, and VP26 (1, 6, 17, 18, 28). VP5 forms the hexons and pentons of the capsid shell (24, 39) connected by a triplex linker complex composed of VP19c and VP23 (24, 37). VP26 is a 12-kDa polypeptide encoded by the UL35 gene (8, 18) which sits at the tip of each copy of VP5 when present in hexons but not in pentons (1, 42, 44, 45). VP26 is the only major capsid shell constituent not essential for viral replication in tissue culture, but its absence does result in reduced yields of infectious virus in mouse trigeminal ganglia (9). Within unpackaged capsids are scaffold polypeptides, principally VP22a and also the less abundant proteins VP21 and VP24, the products of autoproteolysis by the UL26-encoded protease Pra (8, 15, 19, 20, 25).
The first stage in capsid assembly is thought to be the aggregation of VP5–pre-VP22a and VP5-Pra complexes with each other and with preassembled triplexes to form a spherical structure termed the procapsid or large cored B capsid, analogous to the prohead of double-stranded DNA bacteriophage (21, 23, 26, 34–38). Soon after assembly, the procapsid undergoes a dramatic structural transformation in which the surface shell angularizes to the mature polyhedral form. Angularization is accompanied by the formation of mature hexons and pentons, the display of several new VP5 epitopes (5, 11, 16, 39), and proteolytic cleavage of the interior scaffolds by the Pra polypeptide (2, 8, 13–15, 19, 20, 25, 27, 30). The angular mature capsid is considerably more stable than the fragile, cold-sensitive, thin-walled procapsid (21, 37).
An unresolved question in the pathway of capsid assembly is the time of recruitment of VP26. It is unclear whether VP26 assembles onto VP5 hexons in procapsids or instead is recruited during the conformational changes which accompany shell transformation. It has to date proven impossible to test whether VP26 is present in the HSV procapsid, since the instability and low abundance of these structures has made their isolation and biochemical characterization difficult. Moreover, much of what is known about HSV capsid structure and assembly has come from in vivo and in vitro analyses using baculovirus systems expressing various combinations of the major capsid proteins (21–23, 34, 36–38) but omitting VP26 because of its nonessential role. Purified VP26 (42) or VP26 extracted from wild-type capsids (1, 17, 20) can associate with VP26-null angular capsids, suggesting that at least in vitro, VP26 does not need to assemble with procapsids in order to ensure incorporation. Interestingly, VP26 is incapable of entering cell nuclei when expressed in the absence of other viral proteins but can be localized to the nucleus by coexpression with VP5 and preVP22a (29), suggesting that VP26 might be incorporated directly into the procapsid as part of a VP26-VP5-scaffold precursor complex. This, however, raises the question of how VP26 is excluded from assembly into pentons (42, 44).
For several years our laboratory has made use of the HSV mutant tsProt.A to better understand the process of HSV assembly. tsProt.A carries a temperature-sensitive lesion in the UL26 gene such that at the nonpermissive temperature of 39°C, the UL26-encoded protease Pra is inactive, and tsProt.A-infected cells accumulate nuclear procapsids (5, 11, 26, 31). Following a downshift to the permissive temperature of 31°C, these procapsids angularize, package DNA, and give rise to infectious particles in a single, synchronized wave (5). This assay system therefore enables us to control procapsid maturation in vivo and hence to examine only those events which accompany HSV assembly and egress. Using this system, we have previously demonstrated that DNA packaging, but not angularization of the capsid shell, is inhibited when accumulated procapsids are allowed to mature in the absence of normal levels of cellular ATP (4, 7). Unexpectedly, however, epitopes recognized by the anti-VP5 monoclonal antibodies 8F5 and 5C, previously thought to be characteristic of angularized capsids, failed to form under these conditions. Since these epitopes, like VP26, reside exclusively on VP5 hexons (5, 11, 16, 39), we speculated that ATP depletion might affect some aspect of VP26-hexon interaction (7).
In the present study we tested this hypothesis by examining the intracellular distribution of VP26 both during the accumulation of procapsids and also following their angularization. We report that virally encoded VP26 and a VP26-green fluorescent protein (GFP) fusion remain diffuse in the nucleus and cytoplasm when tsProt.A is maintained under nonpermissive conditions but relocalize to 8F5- and 5C-reactive nuclear punctate structures when cells are shifted down to the permissive conditions. Interestingly, the kinetics of VP26-GFP recruitment closely follows that of 5C and 8F5 epitope generation, and ATP depletion prior to temperature downshift blocks VP26 relocalization. We conclude that VP26 is not assembled onto HSV procapsids but instead is recruited in an ATP-dependent manner during angularization. This recruitment is closely correlated with display of the 8F5 and 5C epitopes.
MATERIALS AND METHODS
Cells and viruses.
COS cells were grown in Dulbecco modified Eagle's medium supplemented with 1% penicillin-streptomycin and 10% fetal calf serum (GIBCO Laboratories). COS cells were transiently transfected by the DEAE-dextran method as previously described (41). HSV strain tsProt.A was grown as previously described (5). Cells were depleted of ATP by using a mixture of 25 mM 2-deoxyglucose and 25 mM sodium azide (32, 40, 43), as in our earlier studies (7).
Construction of a VP26-GFP expression plasmid.
The UL35 gene encoding VP26 was amplified by PCR from the genome of HSV strain SC16, using DNA purified from infected Vero cells as the template. Oligonucleotide 5′ GCGCGCAAGCTTTGATGGCCGTCCCGCAATTTCAC anneals with the first 21 nucleotides of the UL35 ORF and introduces an upstream HindIII restriction site (underlined) to facilitate subsequent cloning of the PCR product. Oligonucleotide 5′ CCGTCCTGGATCCGGGGTCCCGGGCGTCGAAG is complementary to the final 20 nucleotides of the UL35 gene and introduces a BamHI restriction site (underlined). The PCR product was cloned between the HindIII and BamHI sites of vector EGFP-C1 (Clontech), creating a translational fusion between UL35 and the carboxy-terminal coding region of the GFP gene.
Immunocytochemistry.
Transfected or untransfected COS cells were grown on glass coverslips and, as appropriate, infected with HSV strain tsProt.A at a multiplicity of 10 for 1 h at 37°C. The medium was replaced, infection was allowed to continue at 39 or 31°C as required, and then cells were rinsed in phosphate-buffered saline (PBS) and fixed in PBS–2% paraformaldehyde for 15 min. Samples were permeabilized in PBS–0.1% Triton X-100 for 10 min and then incubated for 30 min with blocking buffer (20% newborn calf serum, PBS) and for 1 h with the appropriate primary antibody. After four 5-min washes with PBS, cells were incubated for 1 h with fluorescein isothiocyanate (FITC)- or Texas red-conjugated secondary antibodies (Southern Biotechnology Associates) as appropriate, washed with PBS, and mounted by using ProLong antifade reagent (Molecular Probes, Inc.). Specimens were examined in a Bio-Rad MRC 600 scanning laser confocal microscope.
Isolation of angularized capsids.
Infected cells were incubated under appropriate conditions of temperature and ATP depletion and then washed in PBS; all subsequent procedures were carried out at 4°C. Cells were collected by scraping in distilled water; then Triton X-100 added to a final concentration of 0.5%. Following overnight incubation on ice, the mixture was sonicated and subjected to a 1,500 × g clearing spin for 10 min. The supernatant was collected; then the pellet was resuspended in TNE (500 mM NaCl, 1 mM EDTA, 10 mM Tris-Cl [pH 8.0]), sonicated, and centrifuged as before. After collection of the supernatant, the pellet was subjected to one further round of TNE extraction, and the three supernatants were combined. The pooled supernatants were layered on top of a 35% (wt/vol) sucrose–10 mM Tris-Cl (pH 8.0) cushion and then spun at 25,000 × g in a Beckman TLS55 swinging-bucket rotor for 75 min. The resulting pellet was resuspended in PBS and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12% gel) and Western blotting using the anti-VP26 antiserum NC-7 (6).
RESULTS
VP26 localizes to sites of capsid maturation during synchronized HSV assembly.
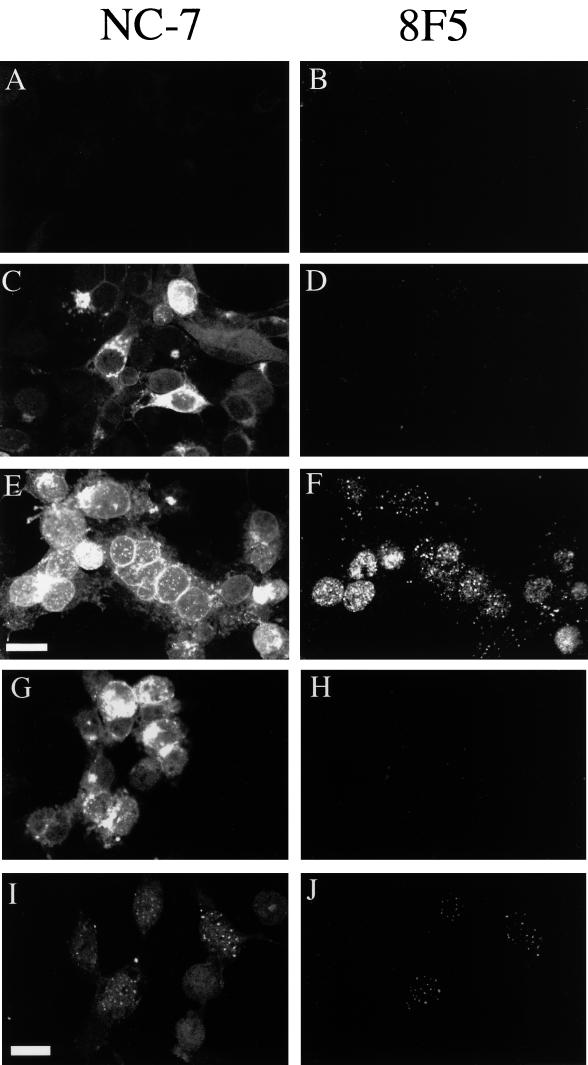
To examine the intracellular localization of VP26 during capsid assembly, we used the experimental approach summarized in Fig. 1. Following infection of COS cells with the HSV mutant tsProt.A, cells were incubated at the nonpermissive temperature of 39°C to enable viral gene expression, DNA replication, and accumulation of a population of immature procapsids. Cells were then downshifted to 31°C to enable procapsids to mature in a single, synchronized wave (5). Figure 2 shows the intracellular localization of VP26 (detected with a polyclonal anti-VP26 antiserum [6]) and of the mature capsid-specific VP5 epitope 8F5 (5, 11, 39). As previously demonstrated (5), mock-infected cells (Fig. 2B) or infected cells accumulating procapsids (Fig. 2D) showed little reactivity to 8F5. In contrast, following the downshift to 31°C, 8F5 reactivity was readily detectable in a punctate nuclear pattern (Fig. 2F), which may correspond to sites at which aggregated procapsids angularize (5, 26, 31). In cells accumulating procapsids, VP26 was found in both nucleus and cytoplasm; however, some cells exhibited particularly bright staining in one or the other location (Fig. 2C). Although staining was rather faint in some cells, it was clearly above the background level of staining in uninfected cells (Fig. 2A). Following the downshift to 31°C, some of the VP26 relocalized to intranuclear punctate structures (Fig. 2E). When the patterns of VP26 and 8F5 staining were colored red and green, respectively, and then merged (Fig. 3A), all of the punctate VP26 staining appeared yellow, showing that the punctate VP26-containing structures are closely associated with sites of 8F5 reactivity. It is important to note that the reverse is not necessarily true: since some VP26 remains diffusely distributed throughout the whole of the interior of the nucleus, it would be expected that all 8F5-reactive structures would appear yellow in a merged image, even if VP26 did not concentrate in the vicinity of the 8F5 reactivity. We conclude from these data that nuclear VP26 preferentially localizes only to those sites at which procapsids are maturing and does not concentrate at intranuclear sites when cells accumulate immature procapsids.
FIG. 1.
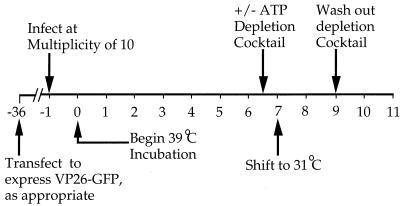
General experimental design. Numbers below the line correspond to time after infection with HSV tsProt.A, in hours. Where necessary, cells were transfected to transiently express a VP26-GFP fusion protein 36 h prior to infection. An ATP depletion cocktail was added or omitted after 6.5 h infection at the nonpermissive temperature of 39°C. After 7 h at 39°C, cells were downshifted to 31°C for a further 2 h. In some experiments, the ATP depletion cocktail was then washed away and incubation continued at 31°C for a further 2 h. At particular times, cells were fixed for indirect immunocytochemistry or collected for Western blot analysis as described in the text.
FIG. 2.
Intracellular localization of VP26 and mature nucleocapsids during synchronized capsid maturation. COS cells were mock infected (A and B) or infected with tsProt.A (C to J) and then incubated as depicted in Fig. 1. Samples were fixed after 7 h at 39°C (C and D) or after an additional 2 h at 31°C in the absence (A, B, E, and F) or presence (G and H) of an ATP depletion cocktail. Panels I and J represent cells which were depleted of ATP as in panels G and H, but the depletion cocktail was washed away and cells were allowed to recover for a further 2 h at 31°C. Fixed cells were immunostained with the anti-VP26 rabbit antiserum NC-7 or the anti-VP5 mouse monoclonal antibody 8F5 as indicated; they were then stained with appropriate secondary antibodies and viewed in the Texas red (A, C, E, G, and I) or FITC (B, D, F, H, and J) channel of a laser scanning confocal microscope. Scale bars in panels E and I represent 30 μm.
FIG. 3.
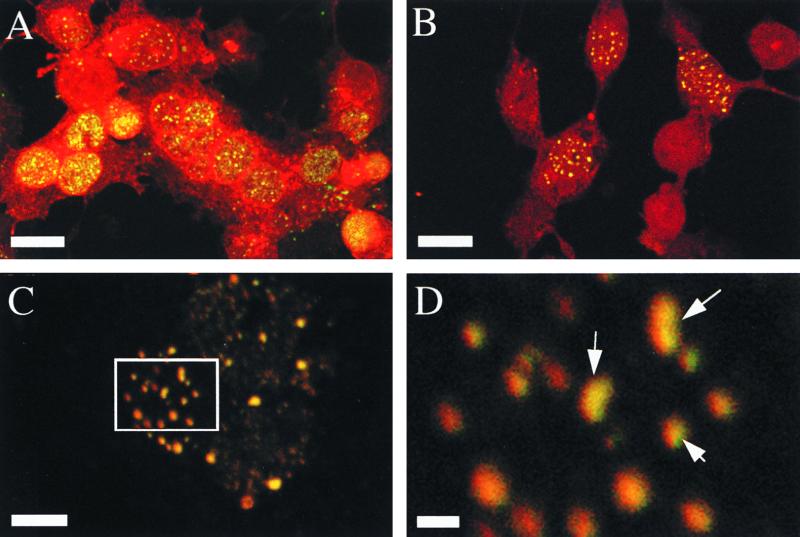
Intranuclear punctate VP26 colocalizes with mature capsids. VP26 staining is represented in red, and 8F5 reactivity (VP5 in mature capsids) is shown in green. (A) Merge of Fig. 2E and F; (B) merge of Fig. 2I and J; (C) higher magnification of a nucleus from the same experiment shown in panel B; (D) fourfold magnification of the boxed region in panel C. Scale bars represent 30 μm (A and B), 10 μm (C) and 2 μm (D). Arrows in panel D indicate structures which appear to contain subdomains reactive to one or the other antibody or to both.
Formation of punctate VP26-containing structures is reversibly inhibited by ATP depletion.
We have previously shown that mature capsid-specific epitopes fail to form under conditions of ATP depletion and have hypothesized that this reflects an ATP dependence in VP26 recruitment (7). To test whether ATP depletion affects VP26 association with maturing capsids, we added an ATP depletion cocktail 0.5 h before the downshift, as depicted in Fig. 1. Figure 2H shows that as expected, the 8F5 epitope fails to form under these conditions and, consistent with our hypothesis, that VP26 remained in a diffuse nuclear and cytoplasmic distribution (Fig. 2G). When the ATP depletion mixture was washed away and cells were allowed to recover for an additional 2 h at 31°C, the 8F5 epitope formed and VP26 relocalized to a punctate nuclear pattern (Fig. 2I and J). A merge of Fig. 2I and J suggests that VP26 and 8F5 reactivity localized to the same structures (Fig. 3B). Figures 3C and D show a higher magnification merge of the nucleus of another infected cell incubated under exactly the same conditions as those shown in Fig. 3B. Note the extensive, (although not complete; see arrows in Fig. 3D) overlap of red (anti-VP26) and green (8F5) reactivity in these intranuclear structures.
VP26-GFP colocalizes with tsProt.A capsids only after release of the temperature block.
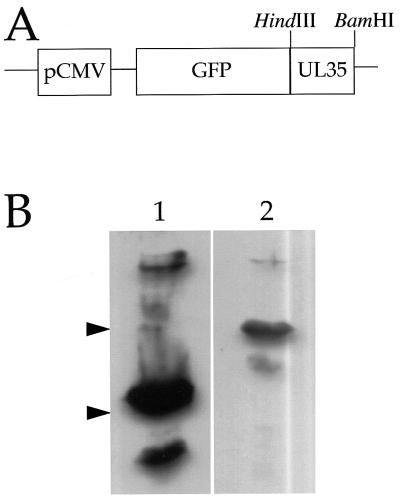
To further investigate the relocalization of VP26 during capsid assembly, and to test our earlier observations by independent means, we prepared a transient expression plasmid in which the gene encoding GFP (3) was fused to the amino-terminal coding region of the VP26 gene under the control of the constitutive cytomegalovirus immediate-early promoter (Fig. 4A). When transfected into COS cells, a VP26-GFP fusion protein of the expected size was detected by Western blotting with an anti-GFP monoclonal antibody (Fig. 4B). We anticipated that this GFP fusion protein would provide a convenient means of visualizing VP26, since a similar fusion protein can be efficiently incorporated into HSV capsids in vivo (10). Furthermore, a VP26–glutathione S-transferase chimera is still able to bind capsids in vitro (42).
FIG. 4.
(A) Partial structure of a VP26-GFP expression plasmid. The VP26 gene UL35 was fused in frame to the carboxy-terminal coding region of the GFP gene as described in Materials and Methods. The BamHI and HindIII restriction sites used for cloning and the cytomegalovirus immediate-early promoter (pCMV) are indicated. (B) Expression of a VP26-GFP fusion in transiently transfected COS cells. COS cells were transfected and after 48 h were collected, subjected to SDS-PAGE (10% gel), Western blotted, and probed with an anti-GFP monoclonal antibody (Clontech). Lane 1, COS cells transfected to express GFP alone; lane 2, COS cells transfected with the VP26-GFP expression plasmid. The mobilities of size markers of 38 and 26 kDa are indicated by arrowheads at the left. The predicted molecular masses of GFP and VP26-GFP are approximately 28 and 40 kDa, respectively.
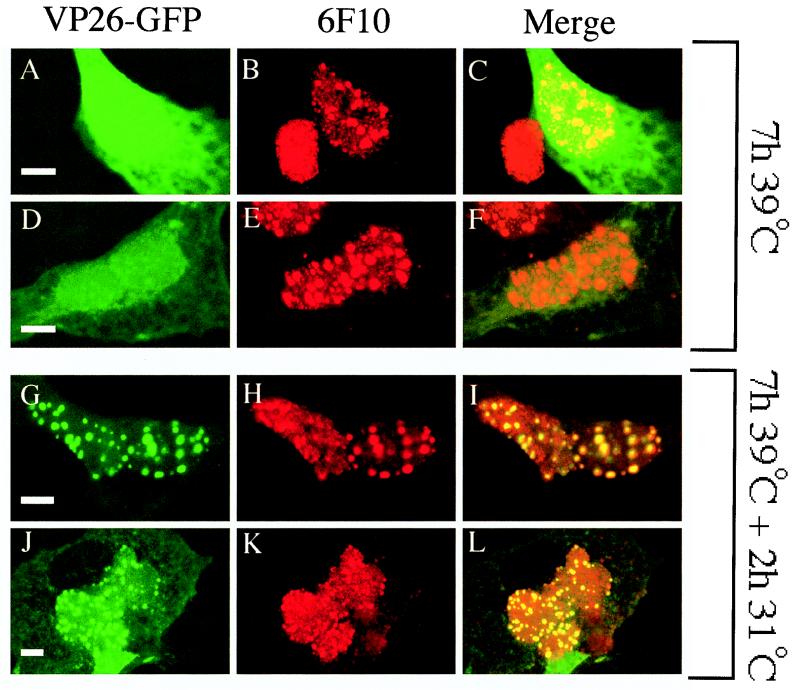
COS cells were transfected to express VP26-GFP and subjected to the procedure depicted in Fig. 1. They were then immunostained with the anti-VP5 monoclonal antibody 6F10. The 6F10 epitope is displayed by VP5 when present in both procapsids and mature capsids (21, 33) and thus enables immunocytochemical detection of procapsids in tsProt.A cells maintained under nonpermissive conditions (5). As previously demonstrated (5), infected cell nuclei contained large 6F10-reactive punctate structures (Fig. 5B, E, H, and K). In contrast, VP26-GFP exhibited a diffuse nuclear and cytoplasmic localization under nonpermissive conditions (Fig. 5A and D), similar to GFP when expressed without fusion to VP26 (see Fig. 7). However, like virally encoded VP26, when cells were shifted to permissive conditions for 2 h to permit procapsid maturation, a substantial fraction of the VP26-GFP relocalized to a punctate staining pattern (Fig. 5G and J). A merge of the red and green images reveals that all of the punctate green VP26-GFP fluorescence colocalized with red punctate 6F10 antigen (Fig. 5I and L). As stated above, it is unsurprising that all of the punctate 6F10 immunostaining appeared to colocalize with VP26-GFP in unshifted cells (Fig. 5C and F) since the latter polypeptide is evenly distributed throughout the interior of the nucleus under these conditions.
FIG. 5.
VP26-GFP colocalizes with maturing capsids but not accumulated procapsids. COS cells were transfected to express VP26-GFP and then infected with tsProt.A and incubated for 7 h at 39°C (A to F) or 7 h at 39°C followed by 2 h at 31°C (G to L). Fixed cells were immunostained with anti-VP5 monoclonal antibody 6F10 and a Texas red-conjugated anti-mouse secondary antibody and then viewed in the Texas red (B, E, H, and K) or FITC (A, D, G, and J) channel of a laser scanning confocal microscope. Panels C, F, I, and L represent merged images. All scale bars represent 10 μm.
FIG. 7.
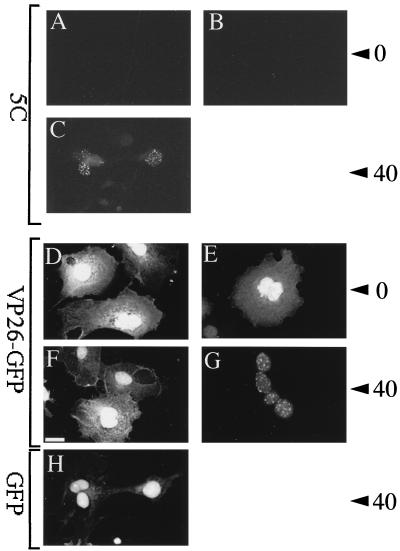
Kinetics of VP26-GFP relocalization and 5C epitope generation. COS cells were transfected to express VP26-GFP (A to G) or GFP alone (H), infected with tsProt.A (B, C, E, G, and H) or mock infected (A, D, and F), and incubated for 7 h at 39°C. Cells were then fixed immediately (A, B, D, and E) or incubated for a further 40 min at 31°C (C, F, G, and H) and either immunostained with monoclonal antibody 5C (A to C) or viewed for direct GFP fluorescence (D to H) as indicated at the left. Numbers at the right refer to 0 or 40 min of incubation at 31°C prior to fixation. The scale bar in panel F represents 30 μm.
We note that there were some differences in the distribution of VP26-GFP compared with wild-type, endogenous VP26. Although VP26-GFP and VP26 were present in both the nucleus and cytoplasm, VP26-GFP appeared to be more abundant in the nucleus and did not exhibit the juxtanuclear staining pattern sometimes seen for VP26 (Fig. 2C and E).
ATP depletion blocks VP26-GFP assembly following release of the temperature block.
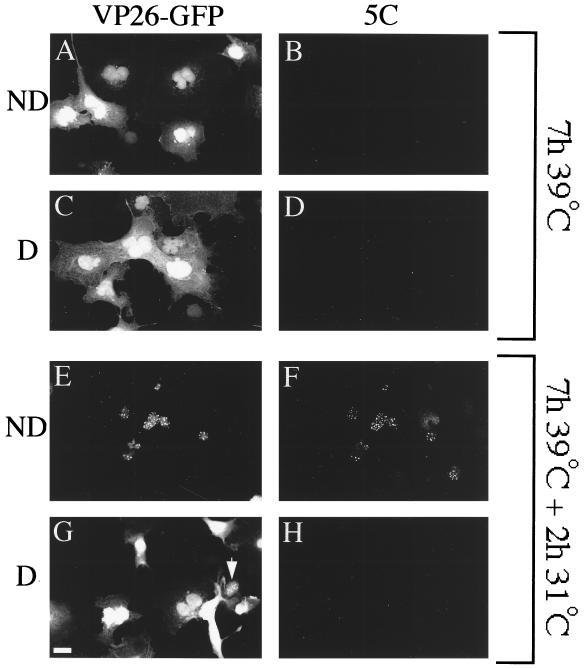
The intracellular distribution of VP26-GFP during capsid maturation was examined in mock-depleted and ATP-depleted cells as depicted in Fig. 1. Figure 6A to D shows that as expected, cells accumulating procapsids failed to display the mature capsid-specific epitope 5C and contained diffuse nuclear and cytoplasmic VP26-GFP. The same result was obtained whether cells had been depleted of ATP (Fig. 6C and D) or not (Fig. 6A and B). Following incubation for 2 h at 31°C, the 5C epitope was expressed and VP26-GFP relocalized to a punctate nuclear pattern in control cells (Fig. 6E and F) but not depleted cells (Fig. 6G and H). The punctate VP26-GFP in panel Fig. 6E colocalized with the 5C antigen in a merged image (data not shown). We obtained similar results with antibody 8F5; however, VP26-GFP-expressing cells exhibited much weaker 8F5 reactivity than untransfected cells, and it was difficult to image reproducibly (see Discussion). We conclude that like virally encoded VP26, VP26-GFP appears to colocalize with mature capsid-specific epitopes in an ATP-dependent manner.
FIG. 6.
An experiment similar to that in Fig. 5 was performed except that cells were treated with an ATP depletion cocktail 0.5 h before the temperature downshift (depleted [D]; C, D, G, and H) or mock treated (not depleted [ND]; A, B, E, and F) and then fixed at the time of temperature downshift (A to D) or following a 2-h incubation at 31°C (E to H). Fixed cells were immunostained with the anti-VP5 monoclonal antibody 5C and a Texas red-conjugated secondary antibody. Fields of cells were then visualized in the FITC (A, C, E, and G) or Texas red (B, D, F, and H) channel. Arrowhead in panel G indicates a rare nucleus exhibiting faint punctate VP26-GFP staining despite depletion of intracellular ATP. The scale bar in panel G represents 30 μm.
Comparing the kinetics of VP26-GFP recruitment with that of hexon maturation.
To further test the relationship between mature VP5 epitope display and the relocalization of VP26, we examined whether the rate of recruitment of VP26-GFP onto maturing capsids was similar to the rate at which the 8F5 and 5C epitopes are generated. Figure 7 shows that the rate of recruitment of VP26-GFP onto capsids closely follows the kinetics of generation of the 5C epitope (39). Although very faint 5C immunoreactivity was occasionally visible after 30 min of incubation at 31°C, the earliest reproducibly visible 5C reactivity occurred after 40 min (Fig. 7C), as previously shown for the epitope 8F5 (5). The relocalization of VP26-GFP showed identical kinetics, with the first punctate VP26-GFP staining visible as early as 40 min after the downshift (Fig. 7G). As additional controls, we tested the intracellular localization of GFP following the temperature shift in infected cells and of VP26-GFP in uninfected cells. In both cases, GFP-dependent fluorescence remained diffuse in the nucleus and cytoplasm (Fig. 7H and F, respectively).
VP26 shows ATP-dependent binding to mature HSV capsids.
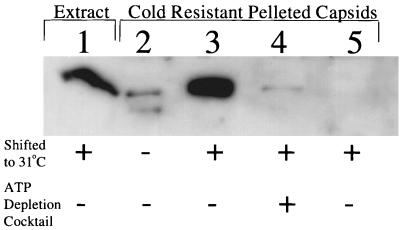
Does ATP-dependent colocalization of VP26 with maturing HSV capsids correspond to stable recruitment of VP26 onto the hexons of maturing capsids, or does it simply reflect their close proximity within the nucleus? To test this, tsProt.A-infected COS cells were incubated under various conditions and then chilled to disrupt the cold-sensitive procapsids; extracts were prepared, and then capsids were pelleted and subjected to SDS-PAGE and Western blotting for VP26. We have previously shown that angularized capsids, but not procapsids, are pelleted under these conditions (7). Figure 8 shows that VP26 could be pelleted with capsids following 31°C incubation under normal conditions (lane 3) but not after ATP depletion (lane 4). Pelleting of VP26 was mature capsid dependent, since far lower levels of VP26 were recovered when capsids were not allowed to mature to the cold-resistant angular form (lane 2). We conclude that when ATP is depleted, VP26 does not stably associate with the angularized, cold-resistant mature capsid. The ATP dependence of VP26-capsid binding therefore correlates with the ATP-dependent relocalization of VP26 to the VP5-containing nuclear punctate structures and is consistent with the possibility that they correspond to the same event in capsid maturation.
FIG. 8.
VP26 can be pelleted in a capsid-dependent manner only when capsids angularize in the presence of normal levels of ATP. Cells were infected with tsProt.A (lanes 1 to 4) or mock infected (lane 5) and then harvested after incubation for 7 h at 39°C (lane 2) or after a further 2-h incubation at 31°C in the absence (lanes 1, 3, and 5) or presence (lane 4) of an ATP depletion cocktail. Cell extracts were then either subjected to SDS-PAGE (12.5% gel) without further treatment (lane 1) or first chilled and solubilized in Triton X-100, after which capsids were washed and collected by pelleting through a Sucrose cushion (lanes 2 to 5). The gel was Western blotted and probed with anti-VP26 antiserum. The autoradiograph was deliberately overexposed to show the low level of capsid-independent VP26 pelleting (lane 2).
DISCUSSION
We previously speculated (7) that the conformational changes which lead to exposure of the mature VP5 hexon-specific epitopes 8F5 and 5C might be driven by, or alternatively be responsible for, the recruitment of VP26 to the tips of VP5 hexons. Here we have shown that under our conditions, VP26 does indeed appear to concentrate only in the vicinity of capsids following their angularization and that this relocalization shows kinetics and ATP dependence similar to those of 5C and 8F5 epitope formation. Our findings lead to the prediction that when techniques are available for the purification of procapsids, these structures will be found to lack the VP26 polypeptide. If this conclusion is correct, why are VP5 and preVP22a capable of relocalizing VP26 from the cytoplasm to the nucleus in transfection assays (29)? Perhaps in the context of a normal viral infection, proteins other than VP5 are responsible for VP26 nuclear sorting. Alternatively, as suggested by Wingfield and colleagues (42), the VP5-VP26 complex might dissociate once within the nucleus, leaving VP5 free to assemble into the procapsid.
Although virally encoded VP26 and a VP26-GFP fusion protein showed similar ATP-dependent relocalization, and although a VP26-GFP fusion similar to this one is known to be incorporated normally into infectious HSV particles (10), we observed differences in the behavior of the two polypeptides. Whereas VP26-GFP (like GFP alone) appeared to concentrate predominantly in the nucleus, endogenous VP26 was found either equally in both nucleus and cytoplasm or concentrated in one location or the other. The different VP26 distributions could reflect the stage of the cell cycle at which the cell was infected or the multiplicity of infection, and its significance for capsid assembly remains unclear. Finally, the quantitative redistribution of VP26-GFP from a nucleoplasmic and cytoplasmic location to punctate nuclear structures was surprising (compare Fig. 6A and E). This suggests that sufficient VP26-binding sites exist for the recruitment of all cellular VP26-GFP, even in the presence of the endogenous, virally encoded VP26. Alternatively, perhaps VP26-GFP associates with some other, very abundant component of the VP5-containing punctate nuclear structures before a smaller subset binds to the mature capsid hexons. This possibility is consistent with the observation that the punctate nuclear structures appear to consist of subdomains which react with either the VP26 antibody, the 8F5 antibody, or both (Fig. 3D, arrows). Further analysis will be required to determine the significance of this observation for the process of capsid maturation.
The relationship between VP26 binding and 5C/8F5 epitope formation remains obscure, since our data do not allow us to conclude whether VP26 binding causes or is a consequence of 8F5/5C epitope formation. To discriminate between these two models, it will be necessary to test whether the angularized capsids formed by VP26-null virions (9) react with the 8F5 and 5C antibodies. Interestingly, although we found it straightforward to visualize 5C and VP26-GFP reactivity in the same nuclei (Fig. 6E and F), we found that 8F5 reactivity was much weaker in VP26-GFP-expressing cells than in untransfected or GFP-expressing cells (data not shown). Perhaps the GFP portion of the VP26-GFP polypeptide lies close to the 8F5 (but not 5C) epitope and interferes with antibody binding.
Our results show that in addition to its role in DNA packaging (7), ATP is required for assembly of the peripheral capsid subunit VP26 onto the hexons of angularized capsids. It remains unclear why ATP should be required for VP26 recruitment and for mature VP5 epitopes to form. One possibility is that ATP-requiring chaperones drive these late events in capsid maturation, as has been suggested for other animal virus capsids (12, 40). Alternatively, ATP may be used to modify VP26 by phosphorylation, a process which could affect VP26-capsid association (17). Nevertheless, VP26 recruitment onto mature capsids clearly does not require ATP in vitro (42). A further possibility is that ATP is required for the entry of VP26 into the nucleus. However, nuclei appear to contain easily detectable levels of both the VP26 and VP26-GFP proteins at the time of shift to permissive conditions (Fig. 2 and 5). Finally, perhaps intranuclear trafficking of VP26 to the sites of capsid maturation is itself an energy-requiring process. Future experiments will attempt to distinguish among these possibilities.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant AI38265 (to D.W.W.) and by NIH training grant T32 AI07506 (to J.H.I.C.). Core support was provided by NIH Cancer Center grant P30-CA13330.
We thank Lily Huang for technical assistance and gratefully acknowledge the Analytical Imaging Facility of the Albert Einstein College of Medicine for help with confocal microscopy. The monoclonal anti-VP5 antibodies 5C and 8F5 were kindly provided by Jay Brown and Bill Newcomb, and the polyclonal anti-VP26 antiserum was kindly provided by Roselyn Eisenberg and Gary Cohen. We thank Amy Sheaffer, Daniel Tenney, and Sandra Weller for helpful discussions.
REFERENCES
- 1.Booy F P, Trus B L, Newcomb W W, Brown J C, Conway J F, Steven A C. Finding a needle in a haystack: detection of a small protein (the 12-kDa VP26) in a large complex (the 200-MDa capsid of herpes simplex virus) Proc Natl Acad Sci USA. 1994;91:5652–5656. doi: 10.1073/pnas.91.12.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braun D K, Roizman B, Pereira L. Characterization of posttranslational products of herpes simplex virus gene 35 proteins binding to the surfaces of full capsids but not empty capsids. J Virol. 1984;49:142–153. doi: 10.1128/jvi.49.1.142-153.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 4.Church G A, Dasgupta A, Wilson D W. Herpes simplex virus DNA packaging without measurable DNA synthesis. J Virol. 1998;72:2745–2751. doi: 10.1128/jvi.72.4.2745-2751.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Church G A, Wilson D W. Study of herpes simplex virus maturation during a synchronous wave of assembly. J Virol. 1997;71:3603–3612. doi: 10.1128/jvi.71.5.3603-3612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen G H, Ponce de Leon M, Diggelmann H, Lawrence W C, Vernon S K, Eisenberg R J. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J Virol. 1980;34:521–531. doi: 10.1128/jvi.34.2.521-531.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dasgupta A, Wilson D W. ATP depletion blocks herpes simplex virus DNA packaging and capsid maturation. J Virol. 1999;73:2006–2015. doi: 10.1128/jvi.73.3.2006-2015.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davison M D, Rixon F J, Davison A J. Identification of genes encoding two capsid proteins (VP24 and VP26) of herpes simplex virus type 1. J Gen Virol. 1992;73:2709–2713. doi: 10.1099/0022-1317-73-10-2709. [DOI] [PubMed] [Google Scholar]
- 9.Desai P, DeLuca N A, Person S. Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology. 1998;247:115–124. doi: 10.1006/viro.1998.9230. [DOI] [PubMed] [Google Scholar]
- 10.Desai P, Person S. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J Virol. 1998;72:7563–7568. doi: 10.1128/jvi.72.9.7563-7568.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao M, Matusick-Kumar L, Hurlburt W, DiTusa S F, Newcomb W W, Brown J C, McCann P J, Deckman I, Colonno R J. The protease of herpes simplex virus type 1 is essential for functional capsid formation and viral growth. J Virol. 1994;68:3702–3712. doi: 10.1128/jvi.68.6.3702-3712.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lingappa J R, Hill R L, Wong M L, Hegde R S. A multistep, ATP-dependent pathway for assembly of human immunodeficiency virus capsids in a cell-free system. J Cell Biol. 1997;136:567–581. doi: 10.1083/jcb.136.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu F, Roizman B. Differentiation of multiple domains in the herpes simplex virus 1 protease encoded by the UL26 gene. Proc Natl Acad Sci USA. 1992;89:2076–2080. doi: 10.1073/pnas.89.6.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu F, Roizman B. Characterization of the protease and other products of amino-terminus-proximal cleavage of the herpes simplex virus 1 UL26 protein. J Virol. 1993;67:1300–1309. doi: 10.1128/jvi.67.3.1300-1309.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu F Y, Roizman B. The herpes simplex virus 1 gene encoding a protease also contains within its coding domain the gene encoding the more abundant substrate. J Virol. 1991;65:5149–5156. doi: 10.1128/jvi.65.10.5149-5156.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matusick-Kumar L, Hurlburt W, Weinheimer S P, Newcomb W W, Brown J C, Gao M. Phenotype of the herpes simplex virus type 1 protease substrate ICP35 mutant virus. J Virol. 1994;68:5384–5394. doi: 10.1128/jvi.68.9.5384-5394.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McNabb D S, Courtney R J. Posttranslational modification and subcellular localization of the p12 capsid protein of herpes simplex virus type 1. J Virol. 1992;66:4839–4847. doi: 10.1128/jvi.66.8.4839-4847.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McNabb D S, Courtney R J. Identification and characterization of the herpes simplex virus type 1 virion protein encoded by the UL35 open reading frame. J Virol. 1992;66:2653–2663. doi: 10.1128/jvi.66.5.2653-2663.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newcomb W W, Brown J C. Use of Ar+ plasma etching to localize structural proteins in the capsid of herpes simplex virus type 1. J Virol. 1989;63:4697–4702. doi: 10.1128/jvi.63.11.4697-4702.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newcomb W W, Brown J C. Structure of the herpes simplex virus capsid: effects of extraction with guanidine hydrochloride and partial reconstitution of extracted capsids. J Virol. 1991;65:613–620. doi: 10.1128/jvi.65.2.613-620.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newcomb W W, Homa F L, Thomsen D R, Booy F P, Trus B L, Steven A C, Spencer J V, Brown J C. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J Mol Biol. 1996;263:432–446. doi: 10.1006/jmbi.1996.0587. [DOI] [PubMed] [Google Scholar]
- 22.Newcomb W W, Homa F L, Thomsen D R, Trus B L, Cheng N, Steven A, Booy F, Brown J C. Assembly of the herpes simplex virus procapsid from purified components and identification of small complexes containing the major capsid and scaffolding proteins. J Virol. 1999;73:4239–4250. doi: 10.1128/jvi.73.5.4239-4250.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newcomb W W, Homa F L, Thomsen D R, Ye Z, Brown J C. Cell-free assembly of the herpes simplex virus capsid. J Virol. 1994;68:6059–6063. doi: 10.1128/jvi.68.9.6059-6063.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newcomb W W, Trus B L, Booy F P, Steven A C, Wall J S, Brown J C. Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. J Mol Biol. 1993;232:499–511. doi: 10.1006/jmbi.1993.1406. [DOI] [PubMed] [Google Scholar]
- 25.Person S, Laquerre S, Desai P, Hempel J. Herpes simplex virus type 1 capsid protein, VP21, originates within the UL26 open reading frame. J Gen Virol. 1993;74:2269–2273. doi: 10.1099/0022-1317-74-10-2269. [DOI] [PubMed] [Google Scholar]
- 26.Preston V G, Coates J A, Rixon F J. Identification and characterization of a herpes simplex virus gene product required for encapsidation of virus DNA. J Virol. 1983;45:1056–1064. doi: 10.1128/jvi.45.3.1056-1064.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Preston V G, Rixon F J, McDougall I M, McGregor M, al Kobaisi M F. Processing of the herpes simplex virus assembly protein ICP35 near its carboxy terminal end requires the product of the whole of the UL26 reading frame. Virology. 1992;186:87–98. doi: 10.1016/0042-6822(92)90063-u. [DOI] [PubMed] [Google Scholar]
- 28.Rixon F J. Structure and assembly of herpesviruses. Semin Virol. 1993;4:135–144. [Google Scholar]
- 29.Rixon F J, Addison C, McGregor A, Macnab S J, Nicholson P, Preston V G, Tatman J D. Multiple interactions control the intracellular localization of the herpes simplex virus type 1 capsid proteins. J Gen Virol. 1996;77:2251–2260. doi: 10.1099/0022-1317-77-9-2251. [DOI] [PubMed] [Google Scholar]
- 30.Rixon F J, Cross A M, Addison C, Preston V G. The products of herpes simplex virus type 1 gene UL26 which are involved in DNA packaging are strongly associated with empty but not with full capsids. J Gen Virol. 1988;69:2879–2891. doi: 10.1099/0022-1317-69-11-2879. [DOI] [PubMed] [Google Scholar]
- 31.Rixon F J, McNab D. Packaging-competent capsids of a herpes simplex virus temperature-sensitive mutant have properties similar to those of in vitro-assembled procapsids. J Virol. 1999;73:5714–5721. doi: 10.1128/jvi.73.7.5714-5721.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skiba P J, Zha X, Maxfield F R, Schissel S L, Tabas I. The distal pathway of lipoprotein-induced cholesterol esterification, but not sphingomyelinase-induced cholesterol esterification, is energy-dependent. J Biol Chem. 1996;271:13392–13400. doi: 10.1074/jbc.271.23.13392. [DOI] [PubMed] [Google Scholar]
- 33.Spencer J V, Trus B L, Booy F P, Steven A C, Newcomb W W, Brown J C. Structure of the herpes simplex virus capsid: peptide A862-H880 of the major capsid protein is displayed on the rim of the capsomer protrusions. Virology. 1997;228:229–235. doi: 10.1006/viro.1996.8392. [DOI] [PubMed] [Google Scholar]
- 34.Tatman J D, Preston V G, Nicholson P, Elliott R M, Rixon F J. Assembly of herpes simplex virus type 1 capsids using a panel of recombinant baculoviruses. J Gen Virol. 1994;75:1101–1113. doi: 10.1099/0022-1317-75-5-1101. [DOI] [PubMed] [Google Scholar]
- 35.Thomsen D R, Newcomb W W, Brown J C, Homa F L. Assembly of the herpes simplex virus capsid: requirement for the carboxyl-terminal twenty-five amino acids of the proteins encoded by the UL26 and UL26.5 genes. J Virol. 1995;69:3690–3703. doi: 10.1128/jvi.69.6.3690-3703.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomsen D R, Roof L L, Homa F L. Assembly of herpes simplex virus (HSV) intermediate capsids in insect cells infected with recombinant baculoviruses expressing HSV capsid proteins. J Virol. 1994;68:2442–2457. doi: 10.1128/jvi.68.4.2442-2457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trus B L, Booy F P, Newcomb W W, Brown J C, Homa F L, Thomsen D R, Steven A C. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J Mol Biol. 1996;263:447–462. doi: 10.1016/s0022-2836(96)80018-0. [DOI] [PubMed] [Google Scholar]
- 38.Trus B L, Homa F L, Booy F P, Newcomb W W, Thomsen D R, Cheng N, Brown J C, Steven A C. Herpes simplex virus capsids assembled in insect cells infected with recombinant baculoviruses: structural authenticity and localization of VP26. J Virol. 1995;69:7362–7366. doi: 10.1128/jvi.69.11.7362-7366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trus B L, Newcomb W W, Booy F P, Brown J C, Steven A C. Distinct monoclonal antibodies separately label the hexons or the pentons of herpes simplex virus capsid. Proc Natl Acad Sci USA. 1992;89:11508–11512. doi: 10.1073/pnas.89.23.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weldon R A, Jr, Parker W B, Sakalian M, Hunter E. Type D retrovirus capsid assembly and release are active events requiring ATP. J Virol. 1998;72:3098–3106. doi: 10.1128/jvi.72.4.3098-3106.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson D W, Davis-Poynter N, Minson A C. Mutations in the cytoplasmic tail of herpes simplex virus glycoprotein H suppress cell fusion by a syncytial strain. J Virol. 1994;68:6985–6993. doi: 10.1128/jvi.68.11.6985-6993.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wingfield P T, Stahl S J, Thomsen D R, Homa F L, Booy F P, Trus B L, Steven A C. Hexon-only binding of VP26 reflects differences between the hexon and penton conformations of VP5, the major capsid protein of herpes simplex virus. J Virol. 1997;71:8955–8961. doi: 10.1128/jvi.71.12.8955-8961.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wrobel I, Collins D. Fusion of cationic liposomes with mammalian cells occurs after endocytosis. Biochim Biophys Acta. 1995;1235:296–304. doi: 10.1016/0005-2736(95)80017-a. [DOI] [PubMed] [Google Scholar]
- 44.Zhou Z H, He J, Jakana J, Tatman J D, Rixon F J, Chiu W. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat Struct Biol. 1995;2:1026–1030. doi: 10.1038/nsb1195-1026. [DOI] [PubMed] [Google Scholar]
- 45.Zhou Z H, Prasad B V, Jakana J, Rixon F J, Chiu W. Protein subunit structures in the herpes simplex virus A-capsid determined from 400 kV spot-scan electron cryomicroscopy. J Mol Biol. 1994;242:456–469. doi: 10.1006/jmbi.1994.1594. [DOI] [PubMed] [Google Scholar]