Abstract
We have demonstrated that intracellular forms of NOTCH1 transactivate two major Epstein-Barr virus (EBV) latent promoters, the LMP1 and Cp1 promoters in an EBV-negative B-cell line, BJAB. Truncated intracellular NOTCH1 associated with the nuclear membrane (ΔE) transactivates the LMP1 promoter fivefold; however, the intranucleus localized form of NOTCH1 (ICN) transactivates this promoter approximately twofold in chloroamphenicol acetyltransferase (CAT) reporter assays in BJAB cells. Additionally, ΔE activated the major Cp1 promoter 12-fold, whereas the ICN form of NOTCH1 activates at only about half that level when compared to that of ΔE membrane-bound NOTCH1. This result differs from previously observed data, where intracellular NOTCH1 bound to the nuclear membrane, ΔE, and nucleus-localized NOTCH1, ICN, all had similar levels of activation in 293 cells. This suggests distinct transcriptional activities in different cell types. Moreover, in Jurkat cells, a T-cell line, intranucleus localized NOTCH1 molecules demonstrated a repressive activity against the two EBV major latent promoters. Only ΔE activated the Cp1 and LMP1 promoters at a level slightly above background, whereas intranucleus localized NOTCH1 ICN, or the form of NOTCH1 lacking the ankyrin repeats, ΔETAR, surprisingly resulted in the repression of these promoters in Jurkat cells. Similarly, another truncated form of NOTCH1, referred to as ICNW, which contains the tryptophan residue W1767 within one of the RBP-Jκ interacting domains, repressed the LMP1 promoter approximately twofold. Further analysis of the truncated NOTCH1 molecules on the LMP1 promoter element, lacking the two RBP-Jκ binding sites, suggests that repression in Jurkat cells may be affected by the presence of the two RBP-Jκ binding sites. These studies indicate that intracellular NOTCH1 can activate the EBV major latent promoters in BJAB cells. However, in Jurkat cells, intracellular truncated forms of NOTCH1 lacking the RBP-Jκ binding sites repress these EBV latent promoters. Only the membrane-bound form of NOTCH1, ΔE, activated the EBV major latent promoters in Jurkat cells, albeit at a lower level than that seen in BJAB cells. Our data suggest that EBNA2 and truncated intracellular nuclear localized forms of NOTCH1 may be functionally similar in their interactions with RBP-Jκ; however, these molecules may have distinctly different transcriptional partners in BJAB and Jurkat cells. Moreover, these truncated NOTCH1 molecules may not represent the normal processed forms of NOTCH1 in cells and may exhibit dominant negative phenotypes in the absence of the required posttranslational modifications. Further investigations are necessary to determine the similarity and differences occurring with intracellular NOTCH1 in other B- and T-cell lines.
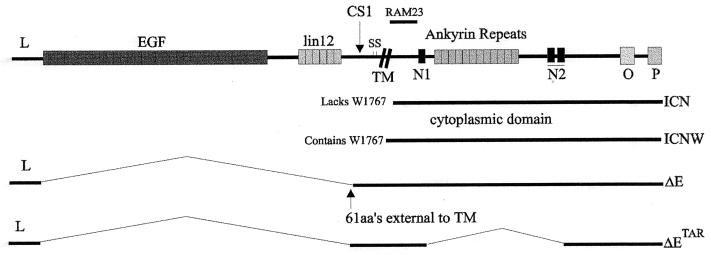
Human NOTCH1 is associated with T-cell acute lymphoblastic leukemia/lymphoma, with a recurrent translocation occurring between chromosomes 7 and 9 at locus q34;q34.3 and with homology to Drosophila melanogaster NOTCH1 (7). The wild-type (wt) human NOTCH1 is expressed in most tissues but is present in high levels in the thymus and brain. It encodes a 2,555-amino-acid type I transmembrane receptor protein larger than 300 kDa with a domain architecture similar to that of the Drosophila homologs (Fig. 1) (6). The chromosomal translocation results in overexpression of the intracellular 3′ end of the NOTCH1 gene. The truncated molecules lacking the external epidermal growth factor-like and lin-12-like repeats range from approximately 100 to 125 kDa in size (1, 2, 6). Truncated forms of intracellular NOTCH1 vary in their intracellular localization. The ΔE form contains the transmembrane domain of the NOTCH1 molecule, lacks most of the extracellular domain, and is localized to the nuclear membrane. The ICN form of NOTCH1 has most of the amino-terminal end, to amino acid 14 of the intracellular domain, deleted and localizes to the nucleus of cells (2, 3). Another truncated NOTCH1 molecule, ICNW, contains the tryptophan residue W1767 important for binding to RBP-Jκ (see Fig. 1 for a schematic diagram of the different truncated forms of intracellular activated NOTCH1) (3, 26). In transient transfections, activated NOTCH1 molecules transactivate the Epstein-Barr virus (EBV) latent Cp1 promoter to similar levels in 293 cells and have similar oncogenic properties in mouse bone marrow cells (3, 20).
FIG. 1.
Schematic diagram showing the full-length NOTCH1 protein with the identified extracellular and intracellular domains. The epidermal growth factor-like repeats and the lin-12-like repeats are positioned extracellular to the transmembrane domain. Two cysteine repeats are potentially involved in the formation of disulfide bridges. The cdc10 (ankyrin) repeats, the two nuclear localization signals (N1 and N2), and the PEST (P) and OPA (O) sequences are intracellular domains (6). The ΔE construct contains the leader peptide, the transmembrane domain, and the intracellular region of NOTCH1 and localizes to the nuclear membrane. The ICN construct lacks the leader peptide and the transmembrane region and localizes to the nucleus. The ΔETAR construct contains the same sequence as ΔE but lacks the ankyrin repeats (3). ICNW contains the entire intracellular domain, including W1767 that is crucial for RBP-Jκ association (3).
RBP-Jκ, the human homolog of the Drosophila Suppressor of Hairless (SuH), is a known transcriptional repressor capable of regulating transcription through numerous cellular and viral promoters by binding to its cognate sequence at these promoter sites (4, 7, 9, 15, 19, 30). The activation of these promoters requires interactions with other cellular and viral transcription factors (24, 25, 27–29). The EBV transcription factor EBNA2 is a potent activator of transcription and is tethered to these promoters by its interaction with RBP-Jκ (9, 15, 33). Another cellular molecule capable of activating viral and cellular promoters through RBP-Jκ is NOTCH1 (3, 12, 13). The interaction of RBP-Jκ with NOTCH1 occurs through two regions. One region is immediately downstream of the transmembrane domain (referred to as the RAM23 domain) and contains the essential tryptophan residue at amino acid position 1767; the other region lies within the ankyrin repeats (see Fig. 1) (3, 26). Constructs without the critical W1767 domain activate promoter elements as efficiently as constructs with the entire cytoplasmic domain intact (3, 13), suggesting that the association with RBP-Jκ can occur without the presence of W1767 within the RAM23 domain. The second region of intracellular NOTCH1 without the ankyrin repeats, ΔETAR, activates at a rate approximately threefold less than NOTCH1 with intact ankyrin repeats but without the W1767 residue. This indicates a requirement for the ankyrin repeats to achieve maximum transactivation of the RBP-Jκ-responsive promoters (3).
The EBV transactivator EBNA2 is incapable of binding to DNA by itself and requires association with RBP-Jκ for targeting and activating promoter elements (9, 17). EBNA2 is essential for EBV immortalization of human primary B lymphocytes, and the association of RBP-Jκ and EBNA2 is a critical component of the immortalization process (5, 8, 32). Viral recombinants mutated in the RBP-Jκ binding site of EBNA2 do not transform human primary B cells in vitro (5, 8, 32). In addition, EBNA2 also associates with numerous components of the basic transcriptional machinery, including PU.1 and AML1 (14, 24, 25). However, its interaction with RBP-Jκ is essential for EBV-induced immortalization of primary B lymphocytes (32).
The similarities between EBNA2 and NOTCH1 are primarily due to their respective involvement in induction of B- and T-cell proliferation and transactivation of cellular and viral promoters through their association with RBP-Jκ (3, 12, 23). Is EBNA2 a functional viral homolog to NOTCH1, usurping the role of NOTCH1 in the immortalization of primary B cells? The association of EBNA2 and NOTCH1 with RBP-Jκ prompted us to compare the activity seen in 293 cells to that in T and B cells. This may provide an explanation as to why EBV can efficiently immortalize human primary B cells and, to a lesser extent, human primary T cells (16, 21). Specifically, we were interested in whether or not the activated intracellular forms of NOTCH1 can transactivate the EBV major latent Cp1 promoter in B and T cells at levels similar to those in previous studies of this promoter in 293 cells. Our studies showed that the intracellular form of NOTCH1, ΔE, associated with the nuclear membrane and activated the EBV Cp1 and LMP1 promoter elements. However, mutants lacking any of the RBP-Jκ binding sites, ICN and ΔETAR, were less potent in their transactivation activity, as determined by transient chloramphenicol acetyltransferase (CAT) reporter assays in BJAB cells.
In Jurkat cells, the nucleus-localized, intracellular form of NOTCH1, ICN, had a surprising effect in repressing the Cp1 and −512/+40 LMP1 promoters. Only ΔE, localized to the nuclear membrane, activated these promoters in Jurkat cells. We tested this observation on a larger LMP1 promoter element lacking the RBP-Jκ binding sites (18). Our observation suggests that these NOTCH1 molecules require the interaction with RBP-Jκ for activation. However, the demonstrated repressive activity may be dependent on interactions with RBP-Jκ, as well as other cellular factors in the transcriptional milieu, expressed in Jurkat cells but not in BJAB cells.
MATERIALS AND METHODS
Cell lines.
Cell culture reagents were obtained from Gibco-BRL Life Technologies. All cell lines were cultured at 37°C under 5% CO2. The Jurkat cell line (obtained from David Gutsch) is a clonal human lymphoblastic T-cell line, and the BJAB cell line (obtained from Elliott Kieff) is an EBV-negative Burkitt's lymphoma cell line. These cell lines were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum purchased from Gemini Bio-Products, Inc. All media were supplemented with gentamicin (20 μg/ml) (Gemini Bio-Products), penicillin-streptomycin (Gibco-BRL) at concentrations of 5 U/ml and 5 μg/ml, respectively, and glutamine at a final concentration of 2 mM.
Expression constructs.
The cDNA constructs NOTCH1 (codons 1 to 2555), ΔE (codons 1 to 22, fused to codons 1673 to 2555), ΔETAR (a subclone of ΔE with a deletion removing the ankyrin repeats; codons 1858 to 2206), and ICN (codons 1770 to 2555) were cloned into pCDNA3 vector and were described previously (20). ICNW, also cloned in pCDNA3, contains the entire intracellular domain of NOTCH1 and includes the W1767 residue important for RBP-Jκ binding (codons 1761 to 2555) (3). All constructs were checked for expression of the appropriate truncated NOTCH1 molecules. The EBNA2 construct is a cDNA construct carrying the entire EBNA2 open reading frame under the control of the simian virus 40 promoter element in the pSG5 vector and has been described previously (29). The multimerized Cp1 CAT reporter construct was obtained from Paul Ling and Diane Hayward (17). The −512/+40 LMP1 CAT reporter construct was obtained from Elliott Kieff (31). The −2350 wt and −2350-Jκ (lacking the RBP-Jκ binding sites) CAT reporter constructs were a gift from Clare Sample (18).
CAT assays.
The various NOTCH1 cDNAs and EBNA2 were used in cotransfections of BJAB and Jurkat cells along with CAT reporter constructs containing a promoter element from the EBV major latent promoters, the −512/+40 LMP1 promoter element (31), or the EBNA major latent Cp1 promoter (17). An LMP1 promoter, −2350, with and without the RBP-Jκ binding sites GTGGGAA (18) was also used in transient reporter assays in Jurkat cells.
For the efficient transfection of Jurkat and BJAB cells, cells were cultured in medium diluted to a density of 250,000 cells per ml and incubated at 37°C and 5% CO2 until they achieved a density of 800,000 to 1 million cells per ml. The medium was then aspirated and the cells were resuspended in fresh medium and further incubated for 24 h. Cells were transfected more efficiently when more than 95% of the cells were actively dividing. Ten million Jurkat cells were electroporated at 260 V and 1,000 μF in 400 μl of medium and were then resuspended in 10 ml of medium followed by incubation for 22 h. BJAB cells were electroporated similar to Jurkat cells, except for the voltage, which was changed to 210 V and 975 μF. Ten micrograms of CAT reporter construct with 20 μg of the various expression constructs were mixed with β-galactosidase expression construct and empty vector (to normalize the total amount of DNA per transfection) in 400 μl of medium. Cells were resuspended in the DNA mixture and allowed to sit for 10 min in a 0.4-μm gap electroporation cuvette at room temperature before electroporation.
All transfections were done with the cDNA encoding β-galactosidase driven by the glucokinase housekeeping promoter as an internal control. Cell extracts were routinely prepared 22 h posttransfection and were analyzed for β-galactosidase and CAT activity according to previously described protocols (22). CAT activity was determined with respect to β-galactosidase activity and quantitated by arbitrary counts on a Molecular Dynamics PhosphorImager system. All transfections were repeated multiple times, and the results were plotted as a mean of multiple experiments.
Immunolocalization.
Transfected cells were harvested and washed in phosphate-buffered saline (PBS). Cells were spread on slides and fixed with methanol-acetone (1:1) for 15 min at −20°C. Cells were incubated with NOTCH1 rabbit polyclonal antibody at a 1:500 dilution, followed by goat anti-rabbit antibody linked to fluorescein isothiocyanate (1:1,000) in the presence of 0.1% Triton X-100, as described previously (3). Microscopy was performed with an Olympus epifluorescence microscope and captured with a charge-coupled device cooled digital camera with the Esprit software program. All images were mounted with the Coreldraw8 software program.
Western blotting.
Transfected cells were collected and washed once in PBS. One million cells were then harvested, resuspended in lysis buffer, fractionated by electrophoresis on a sodium dodecyl sulfate–6% polyacrylamide gel, and then transferred to a 0.45-μm nitrocellulose membrane. All buffers were supplemented with 2 mM sodium thioglycolate. Membranes were then incubated with rabbit anti-NOTCH1 polyclonal serum at a 1:100 dilution in PBS for 24 h at 4°C, washed, and incubated with goat anti-rabbit horseradish peroxidase secondary antibody at a 1:5,000 dilution. The signals were detected by using standard chemiluminescence protocols from the manufacturer.
RESULTS
Intracellular forms of human NOTCH1 activate two EBV major latent promoters in B cells.
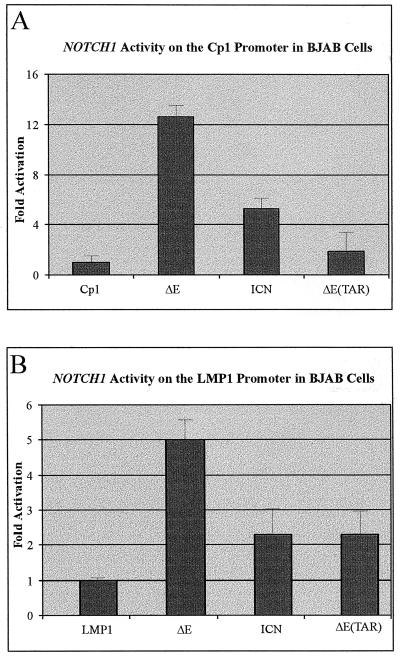
In an effort to determine the ability of activated intracellular forms of NOTCH1 to activate latent EBV promoters in B cells, we performed transient CAT reporter assays with two EBV major latent promoters, the LMP1 −512/+40 and Cp1 promoter regions containing RBP-Jκ binding sites in an EBV-negative Burkitt's lymphoma cell line, BJAB. Characterization of the ability of the different intracellular forms of NOTCH1 to activate the LMP1 promoter containing the RBP-Jκ binding sites showed a distinct difference between nucleus-localized NOTCH1, ICN, and the membrane-associated form of NOTCH1, ΔE. The ΔE construct activated the promoter at a level approximately twofold higher than the ICN expression construct. The ΔETAR construct, lacking the ankyrin repeats, activated the promoter to an extent similar to that of ICN, indicating that since the ΔE construct is the strongest activator, the ankyrin repeats and the RAM23 domain which contains the critical W1767 amino acid deleted in ΔETAR and ICN, respectively, may provide similar or equivalent levels of activation through interaction with RBP-Jκ (Fig. 2B).
FIG. 2.
Intracellular forms of NOTCH1 activate the major latent EBV promoters in BJAB cells. The CAT gene was used as the reporter for activity on the Cp1 (A) and −512/+40 LMP1 (B) promoter regions. Ten micrograms of each reporter construct and 20 μg (each) of the constructs cloned into pCDNA3.1 were transfected into BJAB cells to measure the level of transactivation of each reporter construct. Levels labelled LMP1 and Cp1 indicate transfection with reporter alone for basal activity. PCDNA3.1 vector DNA was used to normalize the amount of DNA in each transfection, and 2.5 μg of an expression plasmid containing the β-galactosidase gene was used as an internal control for transfection. Activity was counted on a Molecular Dynamics PhosphorImager and values are expressed as arbitrary units.
Expectedly, the NOTCH1 ΔE construct activated the −512/+40 LMP1 promoter construct, albeit to a lesser extent than that seen on the multimerized Cp1 promoter in BJAB cells (Fig. 2) and previously seen in 293 cells (3). However, it was surprising to find that the NOTCH1 ICN construct did not activate the promoter to an extent similar to that seen in the same report about 293 cells where NOTCH1 ΔE and ICN had similar activation levels (3). It is possible that both regions of NOTCH1 capable of associating with RBP-Jκ have additive transactivation activity on this LMP1 promoter element. These results indicate that although similar activation was seen on other major latent EBV promoters (e.g., the Cp1 promoter), it is possible that the RAM23 region, which contains the critical W1767 amino acid, has some additive effects on the ability of the intracellular forms of NOTCH1 to activate viral and cellular promoters. Comparably, the NOTCH1 ΔETAR construct, lacking the ankyrin repeats, has similar activities on the −512/+40 LMP1 promoter, indicating that the two regions important for the association with RBP-Jκ each provide similar levels of transactivation activity for the NOTCH1 molecules in BJAB cells (Fig. 2A).
Intracellular NOTCH1 molecules lacking either of the RBP-Jκ interacting domains repress major EBV promoters in Jurkat cells.
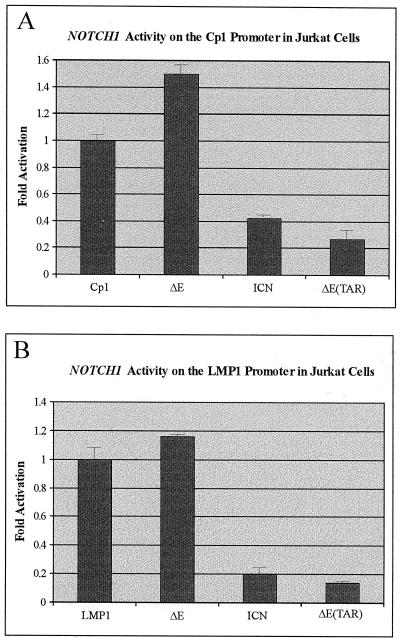
We decided to test these constructs in Jurkat cells, to compare the results obtained in BJAB cells. Previous transactivation experiments with the oncogenic forms of human NOTCH1 were done not in T or B cells but in epithelial cells. Therefore, it was critical to demonstrate whether or not the activation seen in BJAB cells was similar to that seen in a T-cell line. In these studies, we demonstrated that both NOTCH1 ICN and ΔETAR lacking the RBP-Jκ interacting regions at the critical W1767 and ankyrin repeats, respectively, repressed the reporter activity from both the Cp1 and the −512/+40 LMP1 promoters (Fig. 3). The ICN form of NOTCH1 repressed activity approximately 2.5- and 5-fold on the Cp1 and −512/+40 LMP1 promoters, respectively (Fig. 3). The NOTCH1 ΔETAR construct repressed the basal promoter activity in Jurkat cells about fourfold and eightfold on the Cp1 and −512/+40 LMP1 promoter elements, respectively (Fig. 3). Intracellular NOTCH1, ΔE, localized to the nuclear membrane and transactivated the Cp1 promoter approximately 1.5-fold and the −512/+40 LMP1 promoter approximately 1.2-fold (Fig. 3).
FIG. 3.
Intracellular nucleus-localized forms of NOTCH1 repress the major latent EBV promoter elements in the Jurkat cells. Transient CAT reporter assays with the EBV Cp1 promoter (A) and transient transactivation activity on the EBV −512/+40 LMP1 promoter (B) are shown. Cells were transfected with equivalent amounts of DNA, including 10 μg of each reporter plasmid and 20 μg (each) of the various NOTCH1 constructs. The total amount of DNA was normalized by adding vector DNA. Levels labelled LMP1 and Cp1 represent transfection with reporter alone to determine basal levels of activation. The transfected cells were harvested after 22 h and CAT assays were done as described previously (3, 22). Data were collected on a Molecular Dynamics PhosphorImager and values are expressed as arbitrary units.
The results of these transient CAT reporter assays in Jurkat cells were surprising and demonstrated that intracellular forms of NOTCH1, localized to the nucleus lacking the RBP-Jκ binding RAM23 domain that lacked the critical W1767 amino acid, function as repressors of transcription in Jurkat cells. Additionally, the construct lacking the ankyrin repeats which contains the second RBP-Jκ interacting domain, ΔETAR (1, 3, 13), had a greater repressive effect, as demonstrated on both the Cp1 and −512/+40 LMP1 promoters (Fig. 3). The increase in transactivation activity of ΔE when bound to the nuclear membrane indicates that interaction with RBP-Jκ is important for activation. However, the levels seen were much less than those observed in BJAB cells. This suggests that RBP-Jκ may not be the only factor required for the maximum activation of the promoters and that other transcriptional factors may be critical for intracellular NOTCH1 activity on these promoters. However, these factors may not be expressed in the Jurkat cells, resulting in a reduced level of transactivation activity. It is possible that this observed effect, functionally similar to a dominant negative phenotype, could be specific to Jurkat cells. It would be important to perform these experiments with other T-cell lines. However, transfection of other T-cell lines is difficult and so far we have not been able to efficiently transfect other T cells.
These experiments suggest that in Jurkat cells, intracellular activated forms of NOTCH1 that localize to the nucleus are repressive on RBP-Jκ-responsive promoters. However, intracellular forms of NOTCH1 which localize to the nuclear membrane do not function as dominant negative molecules and may be processed in ways similar to wt-activated NOTCH1. We therefore decided to determine if the source of activation or repression by the intracellular forms of NOTCH1 on EBV major latent promoters is through interaction with RBP-Jκ at the promoters.
Intracellular forms of NOTCH1 repress the −2350 LMP1 major latent promoter lacking the RBP-Jκ binding sites in ways similar to that seen on the −512/+40 LMP1 promoter in Jurkat cells.
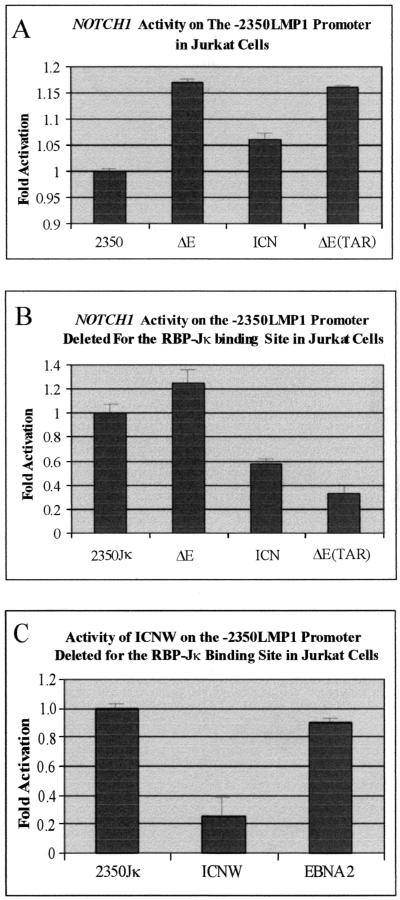
To define the basis of this repression by the intracellular forms of NOTCH1, we used a larger region of the EBV major promoter, −2350 LMP1, lacking the RBP-Jκ binding sites at positions −298 and −223 of the LMP1 promoter element (18). In these experiments, marginal activation was observed on the wild-type promoter with intact RBP-Jκ binding sites (Fig. 4A). The NOTCH1 constructs ΔE, ICN, and ΔETAR all activated this larger −2350 LMP1 promoter at a level only slightly above baseline in Jurkat cells (see Fig. 5A). This was consistent in numerous experiments, suggesting that the larger promoter region may contain additional binding sites for other transcription factors not present in the −512/+40 LMP1 promoter element. These additional factors recruited to this larger promoter resulted in the derepression of the dominant negative activity in Jurkat cells. However, the repression was again seen with the same −2350 LMP1 promoter that lacked both of the cognate RBP-Jκ binding sites (see Fig. 4B). The NOTCH1 ΔE construct transactivated this promoter slightly above baseline, whereas ICN repressed activity approximately twofold and ΔETAR repressed it approximately threefold (see Fig. 4B), levels similar to that seen with the −512/+40 LMP1 promoter. ICNW had about a fivefold reduction of reporter activity. As expected, EBNA2 demonstrated little or no effect on this promoter lacking RBP-Jκ binding sites in Jurkat cells (Fig. 4C). These results suggest that RBP-Jκ is necessary for activation of the EBV major LMP1 promoter in B cells and is critical for interactions with transcription factors or activators for transactivation. However, deletion of the binding sites results in a dominant negative phenotype in Jurkat cells. Promoters that lack the RBP-Jκ binding sites may also be repressed, due to interactions of the intracellular forms of NOTCH1 with other transcription factors at the promoter.
FIG. 4.
Intracellular forms of NOTCH1 repress the larger −2350 LMP1 promoter element lacking the RBP-Jκ binding sites in Jurkat cells. Activity on the larger wild-type −2350 LMP1 promoter element (A) and activity of intracellular NOTCH1 on the −2350 (lacking the RBP-Jκ binding site) promoter construct (B) are shown. The effect of ICNW and EBNA2 on the −2350 LMP1 element lacking the RBP-Jκ binding sites (C) is shown. Ten million Jurkat cells were transfected with the appropriate amounts of DNA, and the cells were harvested after 22 h. Lysates were then used for CAT reporter assays, and the results were obtained with a PhosphorImager from Molecular Dynamics. Only ΔE transactivated this LMP1 promoter, lacking RBP-Jκ binding sites, slightly above background. All other forms of NOTCH1 repressed this promoter in this assay.
FIG. 5.
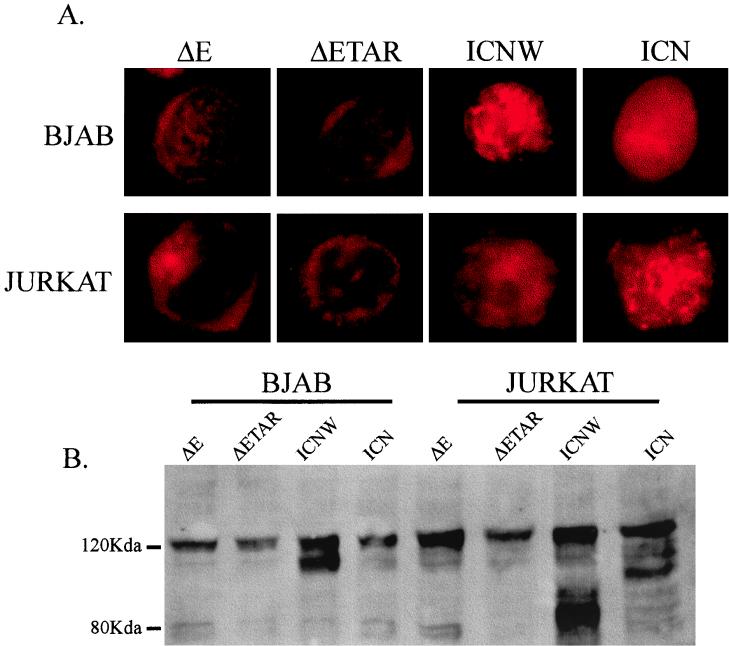
Immunolocalization of activated forms of NOTCH1 in BJAB and Jurkat cells. As expected, ΔE constructs localize predominantly to the nuclear membrane in Jurkat and BJAB cells, whereas ICNW and ICN localize mostly to the nucleus (A). The upper section of panel A shows the immunofluorescence of transfected BJAB cells with the different intracellular NOTCH1 constructs. Note the preferential membrane localization of the ΔE constructs compared to the ICN construct, which predominantly localizes to the nucleus. The lower section shows the transfected Jurkat cells with the forms of activated NOTCH1 and shows similar localization of ΔE to the membrane and ICNW and ICN to the nucleus. A Western blot analysis of the transfected BJAB and Jurkat cells with the different NOTCH1 constructs is shown (B). Transfected cells were harvested, and 500,000 cells were then collected, washed in PBS, lysed, and fractionated by electrophoresis on a sodium dodecyl sulfate–6% polyacrylamide gel. The fractionated proteins were then transferred to nitrocellulose and then probed with rabbit polyclonal antibody against the cytoplasmic domain of NOTCH1. Note the expression of the different forms of NOTCH1 in all lanes.
Oncogenic forms of NOTCH1 that lack the RBP-Jκ binding sites and repress transcription in Jurkat cells are expressed in transfected cells and localize predominantly to the nuclear membrane and nucleus.
Previous experiments with 293 cells indicated activation of transcription with the RBP-Jκ binding element of Cp1 by the ΔE, ICNW, and ICN forms of activated NOTCH1 and not by full-length NOTCH1 (3). This suggests that truncation of the extracellular domain of NOTCH1 leads to transcriptional activation occurring through RBP-Jκ. It is clear that most of the ΔE signal localizes to the nuclear membrane, demonstrating that activation of RBP-Jκ-responsive promoters does not require that the activated molecules have predominant access to the nucleus. However, it is possible that undetected amounts of ΔE can be truncated from the cleavage site and be released from the membrane into the nucleus, activating transcription. We were curious, based on our results with Jurkat cells, whether or not the intracellular truncated forms of NOTCH1, capable of repressing transcription, localize to regions of the cell similar to those seen in 293 cells (3). In transient transfections of the intracellular forms of NOTCH1, we demonstrated that ΔE localizes predominantly to the nuclear membrane, as shown by staining that rims the nucleus, whereas ICNW and ICN preferentially show intranuclear localization in a fashion similar to that seen previously in 293 cells (Fig. 5A) (3). Western blot analysis with rabbit polyclonal antibody against the cytoplasmic region of NOTCH1 also indicated that the truncated forms of NOTCH1 were also efficiently expressed in Jurkat cells (Fig. 5B). These data indicate that oncogenic forms of NOTCH1 that function as dominant negative molecules in Jurkat cells are localized to regions of the nucleus similar to those seen in 293 cells (3). Moreover, these same truncated NOTCH1 molecules function as activators of transcription in BJAB cells, although the levels of activation for the forms of truncated NOTCH1 which localize to the nucleus are less than that for membrane-localized ΔE. These results suggest different patterns of transcriptional regulation relative to NOTCH1 signaling in 293, B, and T cells. A more thorough analysis of these reporter assays in multiple B- and T-cell lines will be important in providing a general mechanism of activation or repression in these cell types.
DISCUSSION
Previous studies investigating the ability of intracellular NOTCH1 to activate RBP-Jκ responsive promoters indicated that the oncogenic forms of NOTCH1 can activate the Cp1 major EBNA promoter in 293 cells, the TP1 promoter in COS7 cells, and a targeted GAL4 promoter in HeLa cells (3, 13, 23). Most of these studies suggest that the EBV transactivator EBNA2 can substitute for these oncogenic forms of NOTCH1 in transformation of B cells induced by EBV (11, 23). We decided to determine the ability of the intracellular, oncogenic forms of NOTCH1 to transactivate the EBV major latent promoters in B cells, the predominant cell type immortalized by EBV and in T cells typically associated with T-cell acute lymphoblastic leukemia/lymphoma and increased expression of intracellular activated NOTCH1 molecules (6, 8, 20). Transient CAT reporter assays were used to investigate the activity of intracellular forms of NOTCH1 on the major EBV −512/+40 LMP1 and Cp1 promoters in BJAB and Jurkat cells. We report a distinct difference in the activities of the NOTCH1 molecules when the results of transient CAT reporter assays are compared. Our results indicate that the intracellular forms of NOTCH1 repress the EBV major latent promoters in Jurkat cells. In 293 cells, these NOTCH1 molecules had similar levels of activation; however, the level of activation of the EBV promoters was reduced in BJAB cells, except for the ΔE, which localizes to the nuclear membrane (3). The level of activation obtained in cells under wt conditions on the EBV genome is expected to be less than that obtained with the multimerized Cp1 promoter, and it became important to compare the results to other normal promoter elements (like LMP1) in these transient CAT reporter assays.
Specifically, the ICN construct differed in its levels of activity on the −512/+40 promoter in BJAB cells. These results indicate that the region 5′ to the ankyrin repeats, including the RAM23 domain that contains one of the RBP-Jκ interaction domains and the critical tryptophan residue W1767, is important for increased transcriptional activity in BJAB cells. This domain did not show any major difference from other forms of intracellular NOTCH1 described in a previous report on 293 cells (3). Therefore, our results with the ICN construct are distinct from but similar to results with the ΔETAR form of intracellular NOTCH1 lacking the ankyrin repeats. The −512/+40 LMP1 promoter contains two RBP-Jκ binding sites, in contrast to the Cp1 promoter reporter construct (17). In other experiments with a hexamerized TP1 promoter reporter construct, the intracellular domain of NOTCH1 was more potent than the EBV transactivator EBNA2. However, both constructs activated to similar extents on the cellular HES1 CAT promoter reporter construct (23). This could be another case of promoter specificity that is not easily explained by transactivation data. Cumulatively, these data suggest that there are specific factors interacting with the different intracellular forms of NOTCH1 at the different promoters, resulting in variations in the levels of activity at these promoters.
The ability of EBV to infect and immortalize human primary B cells and the ability of the virus to utilize major cellular signaling pathways are of important consequence in understanding basic mechanisms of virus-host interactions as they relate to DNA tumor viruses. In these studies, we investigated the correlation between a transformed Burkitt's lymphoma cell line, BJAB, and Jurkat cells. EBNA2 or intracellular NOTCH1 molecules associate with RBP-Jκ in cells activating viral and cellular promoters containing RBP-Jκ binding sites (23, 34). It is possible that the transformation processes for B and T cells are similar in nature and that one of the keys to this process is the interaction with the ubiquitous cellular transcription factor RBP-Jκ. RBP-Jκ interaction with EBNA2 is essential for EBV immortalization of human primary B cells, and the overexpression of intracellular NOTCH1 is crucial for T-cell transformation (32). These events increase the normal activity of the RBP-Jκ-associated promoters, which under normal conditions are repressed (10). Both intracellular NOTCH1 and EBNA2 can activate promoters through interaction with RBP-Jκ in 293, HeLa, and COS7 cells (10, 11).
Intracellular forms of NOTCH1 are oncogenic in T-cell progenitors (20). The interaction of intracellular forms of NOTCH1 with RBP-Jκ may contribute in part to the oncogenic nature of these molecules, and other cellular interactions may have important roles in the initiation and maintenance of the oncogenic state leading to tumor development. Moreover, the interaction of RBP-Jκ with a number of EBV latent proteins crucial for EBV transformation of human primary B cells, resulting in derepression and/or regulation of promoters, may be critical points in the disregulation of the normal activity of RBP-Jκ, leading to development of the neoplastic state. Viral proteins are known to function in the disruption of normal cellular processes. Therefore, the interaction of RBP-Jκ with EBNA2 and the intracellular form of NOTCH1 could be important for the transformation of human primary B and T cells, respectively, leading to increased cell proliferation.
Oncogenic forms of NOTCH1 can activate the EBV major latent promoter, Cp1, in 293 cells (3). Therefore, we wanted to determine if NOTCH1 can activate another major latent promoter, the −512/+40 LMP1 promoter. Our results were consistent with ΔE activating the latent promoter; however, ICN activated at a level twofold lower than ΔE. This was unexpected, as ΔE, ICN, and ICNW contain the critical tryptophan at codon 1767 (the RAM23 domain) and all three transactivated the Cp1 promoter to similar levels in 293 cells (3). Additionally, ΔE activated the LMP1 promoter five- to sixfold above baseline, whereas ICN did not, having only about half the level of activation on the same promoter in BJAB cells. The activation was similar to the level of activation seen with ΔETAR, which had only a moderate level of activation on the multimerized Cp1 promoter in 293 cells (3). This suggests that ICN and ΔETAR had relatively similar binding affinities for RBP-Jκ and that interaction with other basal transcription factors is important for transactivation. However, these results clearly demonstrate that there are differences in the level of activation in these different viral promoters in BJAB and 293 cells. The differences seen between ΔE and ICN on the Cp1 and −512/+40 LMP1 promoters in BJAB cells indicate that ΔE, associated with the nuclear membrane, may obtain the necessary modifications or association with other factors required for efficient activation of these viral promoter elements, whereas ICN and ΔETAR may not.
The ability of EBNA2 to compete with intracellular forms of NOTCH1 indicates that they are interacting with RBP-Jκ on similar domains (23). This creates a scenario whereby EBNA2 can usurp the association of intracellular NOTCH1 with RBP-Jκ interacting at promoters. This makes it possible for EBV to impart its influence on normal cell transcriptional control, driving the proliferation of infected primary B lymphocytes. Molecular genetics and recombinant virus studies demonstrated that a mutation in the two WW residues in EBNA2 that are essential for binding to RBP-Jκ results in a null virus for primary B-cell transformation (32). Recent studies by Sakai and colleagues showed that EBNA2 and the intracellular cytoplasmic domain of NOTCH1 transactivate cellular HES1 and another EBV promoter, TP1, in COS7 cells (23). One of the questions raised by these studies is the possibility that there are differences in the interactions between intracellular NOTCH1 and RBP-Jκ in BJAB cells, compared to 293 or COS7 cells. These differences may be due to the available cell-specific transcription factors recruited to the promoters. Therefore, variations in the degree of activation of these viral promoters containing RBP-Jκ binding sites in BJAB, 293, and COS7 cells are possible, based on the available transcription factors. These related transformation events in lymphocytes may be due to the transcription factor milieu and can provide an explanation for why EBV predominantly transforms primary B cells but not primary T cells (16).
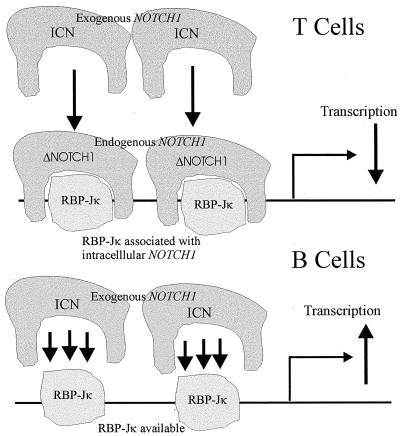
Based on our data, we propose that in B cells, oncogenic intracellular NOTCH1 molecules interact with RBP-Jκ quite efficiently when transfected into BJAB cells, due to an available pool of RBP-Jκ. However, in Jurkat cells the available pool of RBP-Jκ is sequestered by endogenous activated NOTCH1 molecules. Therefore, the addition of exogenous NOTCH1 molecules in large amounts through transient transfections results in competition for the available pool of RBP-Jκ. Thus, exogenous intracellular NOTCH1 constructs act as dominant negative molecules in a T-cell background (see Fig. 6). Additionally, we have shown in separate experiments that little or no endogenous NOTCH1 is associated with RBP-Jκ in B cells, whereas in T cells, the associated NOTCH1 is easily detected with RBP-Jκ (E. Robertson, unpublished observations). Thus, our observation that intracellular NOTCH1 constructs act as dominant negative mutants in Jurkat cells may be a relevant factor important for signaling in B and T cells through the NOTCH1 pathway.
FIG. 6.
Schema of the potential interactions of RBP-Jκ and endogenous and exogenous forms of activated NOTCH1. In BJAB cells, RBP-Jκ is available for interaction with the transfected forms of NOTCH1, therefore activating the promoters. In Jurkat cells, the available RBP-Jκ is sequestered with endogenous activated forms of NOTCH1 and competes with the transfected NOTCH1 molecules, resulting in a dominant negative phenotype that is seen by repression of the EBV latent promoters.
The differences seen in Jurkat cells, where only ΔE had any transactivation activity on the latent EBV promoters, compared to the repressive effects of ICN and ΔETAR, are surprising and once again reflect changes in transcriptional activity of these intracellular forms of NOTCH1 in these cell lines. The fact that deletion of RBP-Jκ interacting domains in ICN and ΔETAR results in repression of the viral promoters suggests that this interaction is critical for the derepression of these promoters. Additionally, deletion of the RBP-Jκ binding sites in the LMP1 promoter also resulted in similar repressive activities by intracellular forms of NOTCH1. This suggests that the interactions of RBP-Jκ with NOTCH1 at the promoter are important for the activation of these major EBV latent promoters in transient CAT reporter assays in BJAB and Jurkat cells. Most of the data so far propose a view that EBNA2 can substitute for NOTCH1 in the activation of promoters in the NOTCH1 signaling pathway (13, 23). We will be investigating these transcriptional activities in other B- and T-cell lines to determine if these differences are specific to BJAB and Jurkat cells. We cannot yet say that this is a general phenomenon in B and T cells, as we have only investigated one cell line. We hope to successfully transfect other T-cell lines in future experiments. Further studies of these and other potential pathways through which EBNA2 and NOTCH1 associate may lead to a greater understanding of the complexity of the resulting transformation processes.
ACKNOWLEDGMENTS
We thank Clare Sample for providing the −2350 LMP1 (with and without the RBP-Jκ binding sites) promoter CAT reporter constructs and Jeffrey Sklar for the NOTCH1 expression constructs. We also thank Elliott Kieff for the −512/+40 LMP1 promoter CAT reporter construct and his thoughtful suggestions. We thank Diane Hayward and Paul Ling for providing the multimerized Cp1 promoter CAT reporter construct.
This work was supported by grants to E.R. from the American Heart Association and a Public Health Service grant from NCI, CA072150. E.R. is scholar of the Leukemia Society of America. M.C. is a fellow of the Lady Tata Memorial Trust.
REFERENCES
- 1.Artavanis-Tsakonas S, Matsuno K, Fortini M E. Notch signaling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 2.Aster J, Pear W, Hasserjian R, Erba H, Davi F, Luo B, Scott M, Baltimore D, Sklar J. Functional analysis of the TAN-1 gene, a human homolog of Drosophila notch. Cold Spring Harbor Symp Quant Biol. 1994;59:125–136. doi: 10.1101/sqb.1994.059.01.016. [DOI] [PubMed] [Google Scholar]
- 3.Aster J C, Robertson E S, Hasserjian R P, Turner J R, Kieff E, Sklar J. Oncogenic forms of NOTCH1 lacking either the primary binding site for RBP-Jkappa or nuclear localization sequences retain the ability to associate with RBP-Jkappa and activate transcription. J Biol Chem. 1997;272:11336–11343. doi: 10.1074/jbc.272.17.11336. [DOI] [PubMed] [Google Scholar]
- 4.Bailey A M, Posakony J W. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 1995;9:2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- 5.Cohen J I, Wang F, Mannick J, Kieff E. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc Natl Acad Sci USA. 1989;86:9558–9562. doi: 10.1073/pnas.86.23.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellisen L W, Bird J, West D C, Soreng A L, Reynolds T C, Smith S D, Sklar J. TAN-1, the human homolog of the Drosophila Notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 7.Fortini M E, Artavanis-Tsakonas S. The suppressor of hairless protein participates in Notch receptor signaling. Cell. 1994;79:273–282. doi: 10.1016/0092-8674(94)90196-1. [DOI] [PubMed] [Google Scholar]
- 8.Hammerschmidt W, Sugden B. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature. 1989;340:393–397. doi: 10.1038/340393a0. [DOI] [PubMed] [Google Scholar]
- 9.Henkel T, Ling P D, Hayward S D, Peterson M G. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein Jkappa. Science. 1994;265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 10.Hsieh J J, Hayward S D. Masking of the CBF1/RBPJ kappa transcriptional repression domain by the Epstein-Barr virus EBNA2. Science. 1995;268:560–563. doi: 10.1126/science.7725102. [DOI] [PubMed] [Google Scholar]
- 11.Hsieh J J D, Henkel T, Salmon P, Robey E, Peterson M G, Hayward S D. Truncated mammalian Notch1 activates CBF1/RBPJκ-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol Cell Biol. 1996;16:952–959. doi: 10.1128/mcb.16.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsieh J J, Nofziger D E, Weinmaster G, Hayward S D. Epstein-Barr virus immortalization: Notch2 interacts with CBF1 and blocks differentiation. J Virol. 1997;71:1938–1945. doi: 10.1128/jvi.71.3.1938-1945.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarriault S, Brou C, Logeat F, Schroeter E H, Kopan R, Israel A. Signaling downstream of activated mNotch. Nature (London) 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 14.Johannsen E, Koh E, Mosialos G, Tong X, Kieff E, Grossman S R. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 2 transactivation is mediated by Jκ and PU.1. J Virol. 1995;69:253–262. doi: 10.1128/jvi.69.1.253-262.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kempkes B, Zimber-Strobl U, Eissner G, Pawlita M, Falk M, Hammerschmidt W, Bornkamm G W. Epstein-Barr virus nuclear antigen 2 (EBNA2)-oestrogen receptor fusion proteins complement the EBNA2-deficient Epstein-Barr virus strain P3HR1 in transformation of primary B cells but suppress growth of human B cell lymphoma lines. J Gen Virol. 1996;77:227–237. doi: 10.1099/0022-1317-77-2-227. [DOI] [PubMed] [Google Scholar]
- 16.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2346–2396. [Google Scholar]
- 17.Ling P D, Rawlins D R, Hayward S D. The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc Natl Acad Sci USA. 1993;90:9237–9241. doi: 10.1073/pnas.90.20.9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marshall D, Sample C. Epstein-Barr virus nuclear antigen 3C is a transcriptional regulator. J Virol. 1995;69:3624–3630. doi: 10.1128/jvi.69.6.3624-3630.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsunami N, Hamaguchi Y, Yamamoto Y, Kuze K, Kangawa K, Matsuo H, Kawaichi M, Honjo T. A protein binding to the Jκ recombination sequence of immunoglobulin genes contains a sequence related to the integrase motif. Nature (London) 1989;342:934–937. doi: 10.1038/342934a0. [DOI] [PubMed] [Google Scholar]
- 20.Pear W S, Aster J C, Scott M L, Hasserjian R P, Soffer B, Sklar J, Baltimore D. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med. 1996;183:2283–2291. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rickinson A A, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2397–2446. [Google Scholar]
- 22.Robertson E S, Grossman S, Johannsen E, Miller C, Lin J, Tomkinson B, Kieff E. Epstein-Barr virus nuclear protein 3C modulates transcription through interaction with the sequence-specific DNA-binding protein Jκ. J Virol. 1995;69:3108–3116. doi: 10.1128/jvi.69.5.3108-3116.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakai T, Taniguchi Y, Tamura K, Minoguchi S, Fukuhara T, Strobl L J, Zimber-Strobl U, Bornkamm G W, Honjo T. Functional replacement of the intracellular region of the Notch1 receptor by Epstein-Barr virus nuclear antigen 2. J Virol. 1998;72:6034–6039. doi: 10.1128/jvi.72.7.6034-6039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sjoblom A, Jansson A, Yang W, Lain S, Nilsson T, Rymo L. PU box-binding transcription factors and a POU domain protein cooperate in the Epstein-Barr virus (EBV) nuclear antigen 2-induced transactivation of the EBV latent membrane protein 1 promoter. J Gen Virol. 1995;76:2679–2692. doi: 10.1099/0022-1317-76-11-2679. [DOI] [PubMed] [Google Scholar]
- 25.Sjoblom A, Yang W, Palmqvist L, Jansson A, Rymo L. An ATF/CRE element mediates both EBNA2-dependent and EBNA2-independent activation of the Epstein-Barr virus LMP1 gene promoter. J Virol. 1998;72:1365–1376. doi: 10.1128/jvi.72.2.1365-1376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H) Curr Biol. 1995;5:1416–1423. doi: 10.1016/s0960-9822(95)00279-x. [DOI] [PubMed] [Google Scholar]
- 27.Tong X, Drapkin R, Reinberg D, Kieff E. The 62- and 80-kDa subunits of transcription factor IIH mediate the interaction with Epstein-Barr virus nuclear protein 2. Proc Natl Acad Sci USA. 1995;92:3259–3263. doi: 10.1073/pnas.92.8.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tong X, Drapkin R, Yalamanchili R, Mosialos G, Kieff E. The Epstein-Barr virus nuclear protein 2 acidic domain forms a complex with a novel cellular coactivator that can interact with TFIIE. Mol Cell Biol. 1995;15:4735–4744. doi: 10.1128/mcb.15.9.4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tong X, Wang F, Thut C J, Kieff E. The Epstein-Barr virus nuclear protein 2 acidic domain can interact with TFIIB, TAF40, and RPA70 but not with TATA-binding protein. J Virol. 1995;69:585–588. doi: 10.1128/jvi.69.1.585-588.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waltzer L, Logeat F, Brou C, Israel A, Sergeant A, Manet E. The human Jκ recombination signal sequence binding protein (RBP-Jκ) targets the Epstein-Barr virus EBNA2 protein to its DNA responsive elements. EMBO J. 1994;13:5633–5638. doi: 10.1002/j.1460-2075.1994.tb06901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang F, Tsang S F, Kurilla M G, Cohen J I, Kieff E. Epstein-Barr virus nuclear antigen 2 transactivates latent membrane protein LMP1. J Virol. 1990;64:3407–3416. doi: 10.1128/jvi.64.7.3407-3416.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yalamanchili R, Tong X, Grossman S, Johannsen E, Mosialos G, Kieff E. Genetic and biochemical evidence that EBNA 2 interaction with a 63-kDa cellular GTG-binding protein is essential for B lymphocyte growth transformation by EBV. Virology. 1994;204:634–641. doi: 10.1006/viro.1994.1578. [DOI] [PubMed] [Google Scholar]
- 33.Zimber-Strobl U, Kremmer E, Grasser F, Marschall G, Laux G, Bornkamm G W. The Epstein-Barr virus nuclear antigen 2 interacts with an EBNA2 responsive cis-element of the terminal protein 1 gene promoter. EMBO J. 1993;12:167–175. doi: 10.1002/j.1460-2075.1993.tb05642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zimber-Strobl U, Strobl L J, Meitinger C, Hinrichs R, Sakai T, Furukawa T, Honjo T, Bornkamm G W. Epstein-Barr virus nuclear antigen 2 exerts its transactivating function through interaction with recombination signal binding protein RBP-J kappa, the homologue of Drosophila suppressor of Hairless. EMBO J. 1994;13:4973–4982. doi: 10.1002/j.1460-2075.1994.tb06824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]