Summary
The size of the human head is highly heritable, but genetic drivers of its variation within the general population remain unmapped. We perform a genome-wide association study on head size (N = 80,890) and identify 67 genetic loci, of which 50 are novel. Neuroimaging studies show that 17 variants affect specific brain areas, but most have widespread effects. Gene set enrichment is observed for various cancers and the p53, Wnt, and ErbB signaling pathways. Genes harboring lead variants are enriched for macrocephaly syndrome genes (37-fold) and high-fidelity cancer genes (9-fold), which is not seen for human height variants. Head size variants are also near genes preferentially expressed in intermediate progenitor cells, neural cells linked to evolutionary brain expansion. Our results indicate that genes regulating early brain and cranial growth incline to neoplasia later in life, irrespective of height. This warrants investigation of clinical implications of the link between head size and cancer.
Keywords: genetics, genome-wide association study, head size, intracranial volume, head circumference, cancer, meta-analysis
Graphical abstract
Highlights
-
•
Knol, Poot, et al. identify 67 genetic loci associated with human head size
-
•
Genes harboring or near head size genetic variants enrich for macrocephaly genes
-
•
Head size genetic variants preferentially locate to cancer genes and pathways
-
•
Further research is needed on the potential link between head size and cancer risk
Knol, Poot, et al. identify 67 loci for human head size in a genome-wide association study. Genes harboring the lead variants enrich for cancer genes and pathways, which was not seen for height variants. These findings suggest a potential link between a larger head and a higher cancer risk.
Introduction
The size of the human head, measured by head circumference or intracranial volume, correlates closely with brain size. Head size is determined by growth in the first years of life and is largely completed by 6 years of age, whereas the rest of the body typically grows until early adulthood.1 Head size is highly genetically determined, ranging from near 90% during childhood to 75% during adulthood.2 Rare genetic syndromes have revealed individual genes strongly affecting head size.3 Nevertheless, genetic determinants of its variation within the general population are still poorly characterized, with no coherent and well-supported picture of associated biological pathways.
A previous genome-wide association study (GWAS) on 47,000 individuals identified 18 genetic loci for intracranial volume,4 while another GWAS on head size in 46,000 children and adults identified 17 loci for head size including low-frequency variants in TP53.5 Here, we increased the sample size to a total GWAS discovery sample size of 80,890 individuals, and validated the results in an independent sample of 25,088 individuals. Our GWAS analyses show strong enrichment for genes and multiple pathways involved in cancer, macrocephaly genes, and show preferential expression of genes near variants in intermediate progenitor cells.
Results
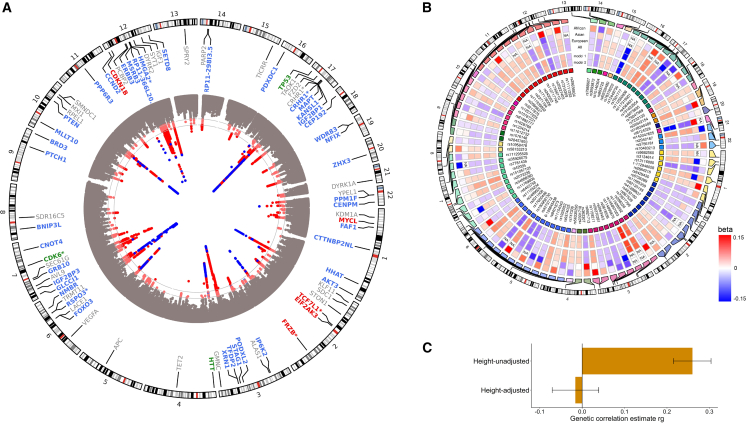
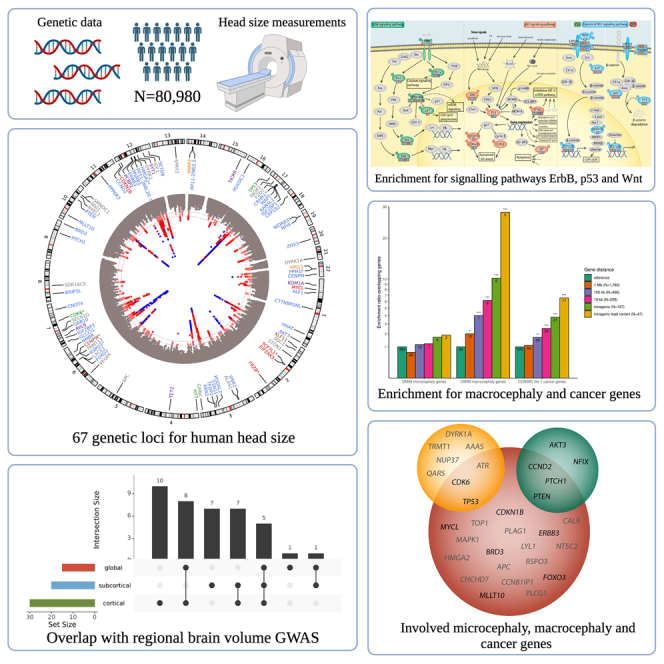
We performed a meta-analysis of GWASs for head size, as proxied by intracranial volume from brain imaging, or head circumference (Tables S1–S3 and S4; STAR Methods). Compared with previous efforts,5,6 we nearly doubled the sample size (N = 80,890), in majority from European ancestry (N = 75,309). We identified 90 independent genetic variants in 67 loci associated with human head size in the European sample (Figure 1A; Table S6. Lead genetic variants and their effects on human head size in samples of different ethnicities, with and without adjustment for height. Related to Figure 1B, Table S7. Genome-wide significant genetic variants and variants in linkage disequilibrium (r2>0.6), including functional annotations. Related to Figure 1A and 1B, Table S8. Lead variants of the combined discovery and validation GWAS meta-analysis, related to Figure 1A and 1B; Data S1, S2, and S3), of which 50 loci were novel. Although the results showed some bias (linkage disequilibrium [LD] score regression intercept 1.056; Table S5), the identified variants remained genome-wide significant after correction for this amount of bias. Most variants (N = 48) showed consistent directions of association among the European, African (N = 1,356), and Asian (N = 4,225) ancestry samples (Figure 1B; Table S6), suggesting population-specific genetic effects on head size in these loci. Since we had limited non-European samples, we also tested the combined effect of the lead variants, which showed positive associations in African and East Asian ancestry samples (βAfrican = 0.34, confidence interval [CI] 0.08–0.60; βEast Asian = 0.40, CI 0.24–0.57). In the European validation sample (N = 25,088), 20 of the 89 lead variants were associated with head size at a Bonferroni significance level (p < 5.6 × 10−4) and 54 at a nominal significance level, while all lead variants showed the same direction of effect. In the UK Biobank validation sample (N = 23,046), the 89 available lead variants together explained 2.3% of the phenotypic head size variance. A meta-analysis combining the European discovery and validation sample (N = 101,241) identified 102 genomic loci with 126 lead variants (Table S8), of which 60 loci overlapped with the 67 genomic loci identified by the discovery meta-analysis.
Figure 1.
Genome-wide association studies on human head size
(A) Circos Manhattan plot of the European ancestry head size GWAS, with gray lines corresponding to genome-wide significant (p < 5 × 10−8) or sub-significant (p < 1 × 10−6) p value thresholds. Known variants are in blue, novel ones in red. For each lead variant, the nearest gene is presented, with the color corresponding to its position to the lead variant: exonic (red), 3′-UTR (green), intronic (blue), intergenic including up- and downstream, exonic and intronic non-coding RNA (gray). Nearest genes for more than one locus are denoted with an asterisk (∗).
(B) Circos heatmap showing the betas of lead variants in African, Asian, and European ancestry meta-analyses, as well as the transancestral meta-analysis. Differences between the height-unadjusted (model 1) and -adjusted (model 2) meta-analysis are also shown.
(C) Bar plot of the genetic correlation coefficient (ρgenetic) of the height-unadjusted and -adjusted head size GWAS with the height GWAS, with their accompanying 95% confidence intervals.
Head-specific growth vs. general growth
We investigated whether variants affecting head size are specific for growth of the human head or are driven, at least in part, by an effect on human body height. Accordingly, we performed a height-adjusted head size GWAS (N = 50,424). The genetic correlation between head size and height (ρgenetic = 0.26, p = 2.1 × 10−30) disappeared in this model (ρgenetic = −0.02, p = 0.58) (Figure 1C), confirming the removal of height-associated effects. Importantly, there was no significant reduction for any of the lead variants’ effect sizes with head size (Table S6). We further explored the effect of these variants on the size of other body parts using area measures obtained from bone density scans (N = 3,313). As expected, a polygenic score of the lead variants was associated with the skull area, even after adjusting for height (p = 2.1 × 10−12). One lead genetic variant (rs12277225) was significantly associated with the L1-L4 spine area (p = 1.3 × 10−5), but the other lead variants did not affect bone area measures of arm, leg, and spine (Table S9). Altogether, this indicates that the effect of the identified variants on head size is predominantly head-specific.
Regional brain volumetric effects
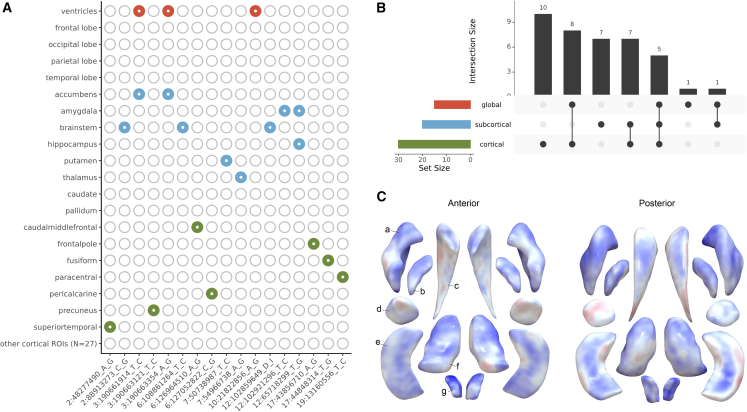
Head size may reflect growth of specific brain regions. Indeed, 15 lead genetic variants or variants in LD (r2 > 0.6) from 12 genetic loci were previously reported to affect volumes of subregions of the brain (Figure 2A; Table S10). We screened all loci previously associated with these regional brain volumes, and found 16 of those 132 loci significantly related with head size after multiple testing correction (Table S11). To determine if the current findings can be localized to specific brain regions, we investigated the 90 independent head size variants in relation to more fine-grained measures of brain morphometry—corrected for head size—in 22,145 individuals (Figure 2B; Table S12). Thirty-nine variants were associated with one or multiple cortical, subcortical, and global brain regions of which 17 variants were preferentially associated with one or two specific cortical or subcortical regions. For example, rs111939932, an intronic variant in PCBP2, is associated with nucleus accumbens volume and is an expression quantitative trait locus (eQTL) for several genes, including ATP5G2 in the nucleus accumbens and basal ganglia. Further analysis revealed its localized effects on this structure’s shape (Figure 2C; Table S13). In the largest GWAS on nucleus accumbens volume,7 this variant was nominally significant (p = 0.02), showing the improved power of our current study to identify novel brain morphometry loci. For the other 51 variants there was no apparent association with particular brain regions. Overall, these results suggest that most head size variants affect generalized brain or cranial growth, while a minority influence regional brain growth.
Figure 2.
Genetic loci for head size and effects on regional brain volumes
(A) Heatmap showing head size loci that overlap with previously identified loci for global brain volumes (red), subcortical volumes (blue), and cortical region of interest volumes (green).
(B) UpSet plot of associations between head size lead variants and brain volumes. Intersection size corresponds to the frequency of the combination depicted below the bar. Set size corresponds to the frequency of associations with one of the brain volume categories (i.e., global, subcortical, or cortical).
(C) Plot showing the subcortical shape analysis of rs111939932 using log Jacobian determinants. Colors correspond to t values, with positive associations depicted in blue, and negative ones in red. Letters point to different subcortical structures: a, putamen; b, pallidum; c, caudate; d, amygdala; e, hippocampus; f, thalamus; g, accumbens.
Genetic correlation with neuropsychiatric traits
Genetic correlation analyses with neuropsychiatric traits have been conducted previously.5,6 We replicated positive genetic correlations with cognitive functioning and Parkinson’s disease, also when only including new samples (Figure S1; Table S14). The replicated correlation with Parkinson’s disease provides independent evidence for the proposed brain overgrowth hypothesis in this disorder.8 Novel genetic correlations were found with multiple psychiatric traits; negative correlations with attention-deficit hyperactivity disorder (ρgenetic = −0.18, p = 4.5 × 10−7), insomnia (ρgenetic = −0.19, p = 1.8 × 10−5), major depressive disorder (ρgenetic = −0.11, p = 2.6 × 10−4), and neuroticism (ρgenetic = −0.11, p = 5.4 × 10−4) (Figure S1; Table S14). Since psychiatric disorders themselves are genetically correlated, incorporating head size and other brain anatomy traits could aid in disentangling underlying genetic factors.
Pathway analysis
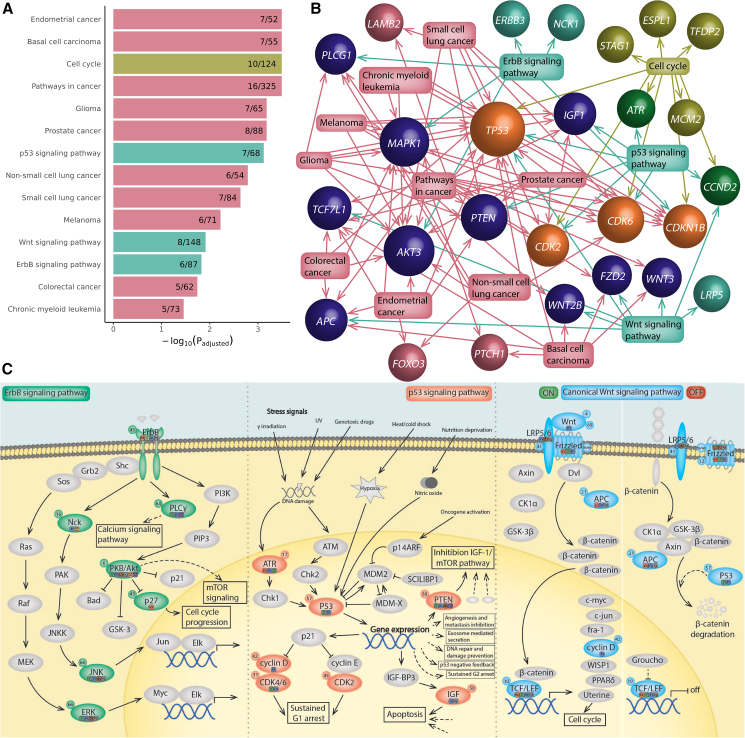
To obtain novel insights into the biological mechanisms underlying head size variation, we performed a gene set enrichment analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG)9 gene sets and found 14 to be significantly enriched (Figure 3A; Table S15). Nine of those gene sets represent different cancer types that substantially overlap between each other and share underlying biological pathways (Figure 3B). The remaining gene sets represent the p53, Wnt, and ErbB signaling pathways, all involved in tumorigenesis including in the abovementioned cancer types.10 Remarkably, lead variants in our GWAS were predominantly intragenic for the seven genes in the p53 pathway, eight genes in the Wnt pathway, and six genes in the ErbB-EGFR pathway (Figure 3C), suggesting that modulation of these pathways plays an important role in head size variation.
Figure 3.
Gene sets enriched in human head size loci
(A) Bar plots presenting enriched KEGG gene sets. –log10 of adjusted p value and proportion of nearby genes overlapping with the gene set are presented. Cancer gene sets are depicted in pink, cell growth and death gene sets in yellow-green, and signal transduction gene sets in turquoise.
(B) Network graph showing enriched KEGG gene sets and their included genes near genetic lead variants. Gene sets are shown in squares with arrows to overlapping genes. Colors correspond to gene set categories: only cancer gene sets (pink), only cell growth and death gene sets (yellow-green), only signal transduction gene sets (turquoise), cancer gene sets and cell growth and death gene sets (dark blue), cell growth and death and signal transduction gene sets (green), or all three gene set categories (orange). Sphere size corresponds to the number of gene sets linked to that gene.
(C) Schematic overview of enriched signaling pathways with proteins encoded by genes near (<10 kb) identified genetic loci. Proteins encoded by these genes are colored (green, ErbB pathway; red, p53 pathway; blue, Wnt pathway), other proteins are depicted in gray. Circles next to protein names provide the locus number of the encoding gene. Locations of lead variants and variants in LD (r2 > 0.6) are shown in squares next to the proteins: exonic (e; red), 3′-UTR (3′; green), 5′-UTR (5; light green), intronic (i; blue), intergenic including up- and downstream, exonic and intronic non-coding RNA (g; gray). For Frizzled, not only FZD2 but also FRZB is taken into consideration.
The p53 signaling pathway showed the strongest enrichment (padjusted = 7.6 × 10−4) (Figure 3A; Table S15). Tumor suppressor protein p53, encoded by TP53, is activated by different stress signals to regulate the cell cycle and apoptosis. Our lead signal in this locus was TP53 3′-UTR variant rs78378222 with predicted deleterious effects (CADD = 15.93), which was identified previously.5 Three other genes in this pathway (ATR, CDK6, and PTEN) also contained 3′-UTR or exonic variants in LD (r2 > 0.6) with lead variants. Identified genes act in cell-cycle arrest and cellular senescence (CDK6, CDK2, and CCND2), apoptosis (IGF1), or inhibition of the insulin growth factor (IGF)-1/mammalian target of rapamycin (mTOR) pathway (PTEN), suggesting comprehensive involvement of the p53 signaling pathway in head growth. This finding is in line with evidence that p53 signaling regulates both normal and malignant neural stem cell populations.11,12,13
The Wnt signaling pathway has links to carcinogenesis and the developing and adult central nervous system,14,15 as well as to bone development including cranial growth.16 Of the eight overlapping genes, three contained exonic or 3′-UTR variants in LD (r2 > 0.6) with identified lead variants (APC, TP53, and TCF7L1). Wnt signaling pathway gene FRZB, not annotated in KEGG, also contained exonic and 3′-UTR variants. In total, 1,948 genetic variants in LD with the identified lead variants (r2 > 0.6), including 35 exonic variants, are eQTLs for WNT3 in 27 different tissues including the cerebellar hemispheres. In addition, various exonic, 3′-UTR and 5′-UTR variants in LD with the lead variants are eQTLs for TCF7L1 in brain tissues. These observations suggest that variants in this pathway affect brain and cranial growth in the human population.
The ErbB pathway (padjusted = 0.014, Figure 3A), also known as the EGFR signaling pathway, has six overlapping genes near head size variants, which are involved in calcium signaling (PLCG1), MAPK signaling (NCK1 and MAPK1), and PI3K-AKT signaling (ERBB3, AKT3, and CDKN1B). In addition, five genetic variants are eQTLs for EGFR in the cerebellum. Interestingly, both AKT3 and CDKN1B are linked to clinical head size syndromes and cancer risk17,18,19,20 and contain, respectively, 3′-UTR variants and an exonic variant that reach genome-wide significance. ErbB signaling is involved in neurodevelopment,21,22,23 making it a plausible pathway involved in head size variation.
Since the above signaling pathways also have universal roles in cell growth, we determined their enrichment in the height GWAS. We found that only the Wnt signaling pathway was significantly enriched in the height GWAS (padjusted = 0.038), suggesting that the p53 and ErbB signaling pathways are more specifically involved in head growth rather than generalized body growth.
Functional prioritization using gene expression
Using a transcriptome-wide association study (TWAS), we identified 156 head size-associated variants functioning as eQTLs, regulating the expression of 112 genes (eGenes) in relevant tissue types (Table S16). Genomic overlap with additional gene-regulatory and epigenetic features provides evidence for 67 eQTLs regulating the expression of 58 eGenes (RegulomeDB probability score >0.5), including AKT3 in brain tissue and TCF7L1 in the cerebellum—part of the ErbB and Wnt pathway, respectively. In addition, 22 eGenes were suggested to be regulated by 22 splicing QTLs (sQTLs), including AKT3. The omnibus test revealed a shared effect for 80 eGenes across the tested gene expression panels (Table S17), including WNT3, AKT3, and EGFR.
Enrichment of Mendelian head size genes and cancer genes
Target genes of GWAS variants are often close to the lead variant.24 Accordingly, we determined the enrichment of different categories of genes located nearby head size variants, stratified by their distance (Table S18).
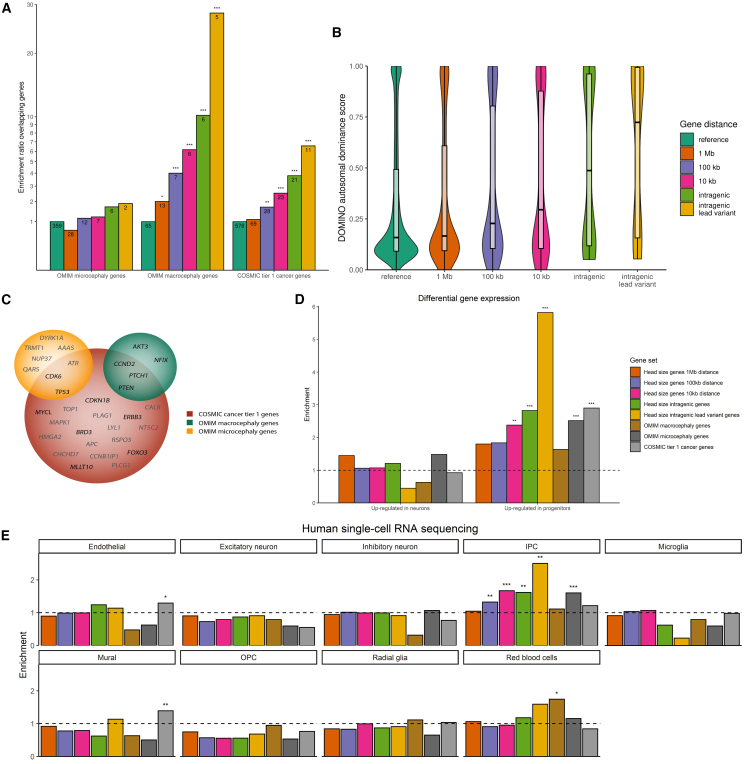
First, we investigated genes mutated in OMIM syndromes associated with abnormal head size, i.e., macrocephaly or microcephaly (Tables S19 and S20). We found increasing enrichment for macrocephaly genes with decreasing distance to the lead variants, culminating in a 37-fold enrichment of macrocephaly genes in genes containing an intragenic lead variant (Figure 4A). In contrast, microcephaly genes were not enriched with shorter distance from lead variants. The striking enrichment of macrocephaly genes did not change in the height-adjusted head size GWAS (Table S21). Furthermore, there was only a modest enrichment for macrocephaly genes in the height GWAS, even for the top 67 loci (i.e., the same number of loci as our GWAS; Table S21). Macrocephaly syndrome genes with intragenic lead variants include AKT3, PTCH1, PTEN, CCND2, and NFIX (Table S19). We conclude that common genetic variants near genes associated with macrocephaly syndromes, but not microcephaly syndromes, contribute to variation in head size in the general population. Our GWAS of head size may therefore identify novel macrocephaly genes. Accordingly, a patient with intellectual disability25 presented with macrocephaly and a mutation in TICRR, a gene for which a lead variant and variants in LD were eQTLs in 12 different tissues. TICRR acts in initiation of DNA replication and interacts with CDK2,26 a gene nearby another lead variant. TICRR is therefore an interesting candidate macrocephaly syndrome gene.
Figure 4.
Gene enrichments stratified by distance from head size lead variants
(A) Enrichment of OMIM macro- and microcephaly genes and COSMIC tier 1 genes near identified genetic loci. Depicted are enrichments of genes within 1 Mb (orange), 100 kb (purple), or 10 kb (pink) of identified genetic loci, genes with intragenic genetic variants (light green) and genes with intragenic genetic lead variants (yellow) in comparison with genes in the reference genome (dark green). ∗p < 0.05; ∗∗p < 0.0125 (0.05/4); ∗∗∗p < 0.0025 (0.05/4/5).
(B) Violin plots showing DOMINO autosomal dominance scores of different gene sets. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
(C) Venn diagram showing genes within 10 kb of genetic loci that overlap with OMIM microcephaly genes (yellow) or macrocephaly genes (green) or COSMIC cancer tier 1 genes (red). Genes with intragenic lead variants are depicted in black, others in gray.
(D) Bar plot showing enrichments of gene sets for genes differentially expressed in neurons and progenitors. ∗p < 0.05; ∗∗p < 0.025 (0.05/2); ∗∗∗p < 0.003 (0.05/2/8).
(E) Bar plots showing enrichments of gene sets for the various cell types in the human cortical brain using single-cell RNA-sequencing data. ∗p < 0.05; ∗∗FDR < 0.05; ∗∗∗p < 0.0007 (0.05/9/8).
We determined whether cancer genes are enriched close to lead variants (Figure 4A). Indeed, there was a 9-fold enrichment for high-fidelity cancer genes (first-tier COSMIC27) among genes with an intragenic lead variant, which persisted after height adjustment (Table S21). There was only a modest enrichment of cancer genes close to height GWAS variants, providing additional evidence that cancer-related genes are specifically relevant for head size variation.
At a variant-level, no genetic correlation was found with GWAS meta-analyses of various cancer types28,29,30,31 (Table S22).
Autosomal dominance score
We did not observe a significant enrichment for microcephaly genes (Figure 4A). This may be due to differences between the micro- and macrocephaly gene sets. Macrocephaly typically results from mutations with an autosomal dominant inheritance pattern (64.6%, Table S19), whereas microcephaly predominantly involves mutations with an autosomal recessive inheritance pattern (72.3%, Table S20). We observed a profound increase for genes with a predicted dominant inheritance pattern closer to our lead variants (Figure 4B). However, neither dominant nor recessive microcephaly genes were enriched (Table S21) and the predominant recessive inheritance patterns of microcephaly genes could not explain their lack of enrichment. An alternative explanation is that microcephaly syndromes are more clinically heterogeneous and the underlying mechanisms are less specific to brain and cranial growth.
Gain of function and loss of function
The overlap among macrocephaly genes, microcephaly genes, and cancer genes is shown in Figure 4C. Macrocephaly-associated genes were more enriched for high-fidelity cancer genes than microcephaly-associated genes (enrichment ratio 12.9 vs. 3.2, Table S21). We therefore investigated whether the same mutation type, i.e., gain of function or loss of function, causes both macrocephaly syndromes as a germline mutation but also associate with cancer as somatic mutations. We found that this was the case for the vast majority of macrocephaly-associated genes with a defined role in cancer (37 of 41 genes, Table S19), i.e., the same type of mutation associated with both macrocephaly and cancer. Moreover, germline mutations in 14 of these 37 genes, including our GWAS genes PTEN, PTCH1, and SUFU, are associated with a syndrome or condition with a suggested cancer predisposition (Table S19). Our GWAS data and these observations may therefore suggest that subtle up-regulation of oncogenes and oncogenic pathways or down-regulation of tumor suppressor genes and pathways increases head size in the general population.
Brain cell expression
As neural progenitors are the actively dividing cells in the developing brain, their expressed genes may explain the observed genetic variants for head size.32 Indeed, genes at or near the head size loci were enriched in differentially expressed neural progenitor cell genes (Figure 4D; Table S23). Subsequently, we looked at a single-cell RNA-sequencing (scRNA-seq) dataset from cell types in the human cortex.33,34 Intriguingly, we find that genes close to head size variants are strongly enriched for genes preferentially expressed in intermediate progenitor cells (IPCs) (Figure 4E; Tables S24 and S25; Figure S2). Increased proliferation of IPCs in a primate-specific area of the brain, the outer region subventricular zone, is believed to be responsible for the evolutionary expansion of the human brain.35,36 This suggests that genetic variation regulating the proliferation or neuronal differentiation of IPCs plays an important role in determining human head size. Indeed, Wnt pathway genes, p53 pathway genes, and PTCH1, SUFU, and NFIX, which we find near genetic variants determining head size, are examples of regulators of IPCs.37,38,39,40,41,42,43 To understand which type of variants influence head size, we performed a partitioned heritability analysis that classifies variants into categories based on functional elements. We found an enrichment for variants in the regulatory elements of both neural progenitors and their neuronal progenies (enrichmentprogenitors = 12.7, p = 8.3 × 10−4; enrichmentneurons = 16.1, p = 3.7 × 10−4).
Finally, we assessed whether a similar pattern was seen for the Catalog of Somatic Mutations in Cancer (COSMIC) first-tier cancer genes. Indeed, our differential gene expression analysis dataset indeed showed an enrichment of cancer genes in the genes specific for neural progenitors (enrichment = 2.9, p = 1.7 × 10−6, Table S23). However, no significant enrichment was found for IPCs using the scRNA-seq data.
Discussion
Here we performed the largest head size GWAS to date and found that associated genetic variants significantly locate to cancer genes and cancer-associated pathways. Genes near head size variants were enriched for high-fidelity cancer genes even after adjustment for height, suggesting a specific association of head growth with cancer, rather than general growth. Germline mutations in multiple macrocephaly syndrome genes are known to be an increased cancer risk, including PTEN (Cowden syndrome) and PTCH1 (Gorlin syndrome) (Table S19). Our GWAS was performed in the general population, which prompts the question of whether the link between head size and cancer extends beyond rare genetic syndromes.
Previous meta-analyses of prospective observational studies found associations between adult height and increased risk for various forms of cancer.44 Similarly, head circumference at birth has previously been positively associated with brain cancer during childhood,45 and with different types of cancer later in life including stomach cancer and breast cancer,46 with stronger associations than for respectively birth weight or birth length. The correlation between head size at birth and breast cancer later in life was further supported by a pooled analysis of 32 studies,47 but not by another prospective cohort study.48 Our study provides further evidence for this link between head size and cancer.
The abovementioned observational studies together with our genetic results suggest that early growth rather than later adolescent growth may be associated with neoplasia, since cranial growth is completed around the sixth year of age, whereas height is primarily determined by peri-pubertal growth. Head size at birth and its growth during early infancy in relation to cancer risk therefore deserves further studies to identify potential underlying pathophysiological mechanisms and its potential clinical implications.45,49,50
Limitations of the study
Although this study suggests an association between head growth and cancer, further studies are needed to investigate whether head size is causally related to cancer development. In our study, we were not able to account for environmental factors such as socio-economic status and diet, especially during childhood, which would be important to adjust for in future studies. In addition, the clinical implications of the findings of our study need to be investigated, for example if patients with clinical macrocephaly syndromes need to be screened for cancer more extensively.
Consortia
The members of the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium are Philippe Amouyel, Konstantinos Arfanakis, Benjamin S. Aribisala, Mark E. Bastin, Ganesh Chauhan, Christopher Chen, Ching-Yu Cheng, Philip L. de Jager, Ian J. Deary, Debra A. Fleischman, Rebecca F. Gottesman, Vilmundur Gudnason, Saima Hilal, Edith Hofer, Deborah Janowitz, J. Wouter Jukema, David C.M. Liewald, Lorna M. Lopez, Oscar Lopez, Michelle Luciano, Oliver Martinez, Wiro J. Niessen, Paul Nyquist, Jerome I. Rotter, Tatjana Rundek, Ralph L. Sacco, Helena Schmidt, Henning Tiemeier, Stella Trompet, Jeroen van der Grond, Henry Völzke, Joanna M. Wardlaw, Lisa Yanek, and Jingyun Yang.
The members of the Enhancing NeuroImaging Genetics through Meta-Analysis (ENIGMA) Consortium are Ingrid Agartz, Saud Alhusaini, Laura Almasy, David Ames, Katrin Amunts, Ole A. Andreassen, Nicola Armstrong, Manon Bernard, John Blangero, Laura M.E. Blanken, Marco P. Boks, Dorret I. Boomsma, Adam M. Brickman, Henry Brodaty, Randy L. Buckner, Jan K. Buitelaar, Dara M. Cannon, Vaughan J. Carr, Stanley V. Catts, M. Mallar Chakravarty, Qiang Chen, Christopher R.K. Ching, Aiden Corvin, Benedicto Crespo-Facorro, Joanne E. Curran, Gareth E. Davies, Eco J.C. de Geus, Greig I. de Zubicaray, Anouk den Braber, Sylvane Desrivières, Allissa Dillman, Srdjan Djurovic, Wayne C. Drevets, Ravi Duggirala, Stefan Ehrlich, Susanne Erk, Thomas Espeseth, Iryna O. Fedko, Guillén Fernández, Simon E. Fisher, Tatiana M. Foroud, Tian Ge, Sudheer Giddaluru, David C. Glahn, Aaron L. Goldman, Robert C. Green, Corina U. Greven, Oliver Grimm, Narelle K. Hansell, Catharina A. Hartman, Ryota Hashimoto, Andreas Heinz, Frans Henskens, Derrek P. Hibar, Beng-Choon Ho, Pieter J. Hoekstra, Avram J. Holmes, Martine Hoogman, Jouke-Jan Hottenga, Hilleke E. Hulshoff Pol, Assen Jablensky, Mark Jenkinson, Tianye Jia, Karl-Heinz Jöckel, Erik G. Jönsson, Sungeun Kim, Marieke Klein, Peter Kochunov, John B. Kwok, Stephen M. Lawrie, Stephanie Le Hellard, Hervé Lemaître, Carmel Loughland, Andre F. Marquand, Nicholas G. Martin, Jean-Luc Martinot, Mar Matarin, Daniel H. Mathalon, Karen A. Mather, Venkata S. Mattay, Colm McDonald, Francis J. McMahon, Katie L. McMahon, Rebekah E. McWhirter, Patrizia Mecocci, Ingrid Melle, Andreas Meyer-Lindenberg, Patricia T. Michie, Yuri Milaneschi, Derek W. Morris, Bryan Mowry, Kwangsik Nho, Thomas E. Nichols, Markus N. Nöthen, Rene L. Olvera, Jaap Oosterlaan, Roel A. Ophoff, Massimo Pandolfo, Christos Pantelis, Irene Pappa, Brenda Penninx, G. Bruce Pike, Paul E. Rasser, Miguel E. Rentería, Simone Reppermund, Marcella Rietschel, Shannon L. Risacher, Nina Romanczuk-Seiferth, Emma Jane Rose, Perminder S. Sachdev, Philipp G. Sämann, Andrew J. Saykin, Ulrich Schall, Peter R. Schofield, Sara Schramm, Gunter Schumann, Rodney Scott, Li Shen, Sanjay M. Sisodiya, Hilkka Soininen, Emma Sprooten, Velandai Srikanth, Vidar M. Steen, Lachlan T. Strike, Anbupalam Thalamuthu, Arthur W. Toga, Paul Tooney, Diana Tordesillas-Gutiérrez, Jessica A. Turner, Maria del C. Valdés Hernández, Dennis van der Meer, Nic J.A. Van der Wee, Neeltje E.M. Van Haren, Dennis van 't Ent, Dick J. Veltman, Henrik Walter, Daniel R. Weinberger, Michael W. Weiner, Wei Wen, Lars T. Westlye, Eric Westman, Anderson M. Winkler, Girma Woldehawariat, Margaret J. Wright, and Jingqin Wu.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Genome-wide association study summary statistics | CHARGE dbGaP and http://enigma.ini.usc.edu/research/download-enigma-gwas-results | phs000930 (dbGaP accession number) |
| Software and algorithms | ||
| EasyQC | Winkler et al.51 | Software - Universität Regensburg (uni-regensburg.de) |
| METAL | Willer et al.52 | METAL Documentation - Genome Analysis Wiki (umich.edu) |
| LD score regression | Bulik-Sullivan et al.53 | GitHub - bulik/ldsc: LD Score Regression (LDSC) |
| LocusZoom | Pruim et al.54 | LocusZoom - Create Plots of Genetic Data |
| FUMA GWAS | Watanabe et al.55 | Functional Mapping and Annotation of Genome-wide association studies (ctglab.nl) |
| TWAS-Fusion | Gusev et al.56 | TWAS/FUSION (gusevlab.org) |
| DOMINO | Quinodoz et al.57 | Domino (iob.ch) |
| Other | ||
| OMIM database | Amberger et al.58 | Home - OMIM |
| Cortical organoids’ scRNA-seq data | Bhaduri et al.34 | https://organoidreportcard.cells.ucsc.edu |
| e/sQTLs, and allele-specific expression in cultured primary human neural progenitors and their sorted neuronal progeny | Aygün et al.32 | https://bitbucket.org/steinlabunc/expression_splicing_qtls_public/src/master/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Hieab H.H. Adams (Hieab.Adams@radboudumc.nl).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The genome-wide summary statistics that support the findings of this study will be made available through the CHARGE dbGaP (accession number phs000930) and ENIGMA (http://enigma.ini.usc.edu/research/download-enigma-gwas-results) websites.
No previously unreported custom computer code or mathematical algorithm was used to generate results central to the conclusions.
Any additional information required to re-analyse the data reported in this work paper is available from the lead contact upon request.
Experimental model and subject details
Study population
Most studies participate in the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE)59 or the Enhancing NeuroImaging Genetics through Meta-Analysis (ENIGMA)60 consortium. We also included the results of the most recent head circumference GWAS.5 A complete overview of the included studies is shown in Table S1 and their population characteristics are presented in Table S2. Each contributing study was approved by their institutional review boards or local ethical committees. Written informed consent was obtained from all study participants.
Genotyping
Genotyping of individuals was performed on commercially available arrays, and imputed to 1000 Genomes (1KG) or Haplotype Reference Consortium (HRC) imputation panels (Table S3). Quality control was performed using the EasyQC software.51 In each study, genetic variants with an imputation quality r2 below 0.3 and a minor allele frequency (MAF) below 0.001 were excluded. Additionally, variants were filtered on study level requiring .
Phenotyping
Different methods were used to measure human head size across studies. Briefly, either head circumference was measured, or intracranial volume was measured on computed tomography (CT) or magnetic resonance imaging (MRI) scans. In total, human head size was measured using intracranial volume measured on CT or MRI scans in respectively 1,283 and 84,171 individuals, and using head circumference in 20,524 individuals (Table S4). These measures have previously shown to be phenotypically and genetically correlated,.5,6,61 Genetic correlations between our MRI scans and head circumference measurements was 0.75. Together, this allowed us to perform a combined meta-analyse of different measures of head size.
Method details
Genome-wide association studies
GWAS were performed for each study adjusted for age, age2 (if significant), gender, eigenstrat PC1-4 (if significant), study-specific adjustments and case-control status (if applicable). In a second model, additional adjustment for height were made. The METAL software52 was used to perform a sample size weighted Z score meta-analysis. After meta-analysis, genetic variants available in less than 5,000 individuals were excluded. Comparable betas were derived using the formula as was done previously.62 Genomic inflation and polygenic heterogeneity were assessed using the LD score regression software53 by comparing the genomic control inflation factor and the LD score regression intercept (Table S5).
GWAS meta-analyses were performed separately for African, Asian and European samples. We also performed a transancestral meta-analysis. Since the analyses in non-European samples were underpowered, we additionally used an inverse-variance weighted method to test the combined effects of the lead variants in the non-European samples. This analysis was performed using the gtx package as implemented in R.
Functional annotations
Regional association plots were made with the LocusZoom software.54 The Functional Mapping and Annotation of Genome-Wide Association Studies (FUMA GWAS) platform55 was used to derive the independent genomic loci and genetic lead variants, and to functionally annotate the identified genetic variants. Additionally, enrichment for KEGG9 biological pathways was assessed for genes located nearby the identified genetic loci using the default options in FUMA, using hypergeometric tests. Genotype-Tissue Expression (GTEx) v7 was used to identify expression quantitative trait loci (eQTL) for the lead genetic variants and variants in LD (r2 > 0.6).
We performed a transcriptome-wide association study (TWAS) using the association statistics from the European-only head size GWAS summary statistics and weights from 21 publicly available gene expression reference panels. We focused on the gene expression weights from blood (Young Finns Study, YFS), arterial (GTEx), brain (GTEx, CommonMind Consortium (CMC)) and peripheral nerve tissues (GTEx). Precomputed SNP-expression weights in the 1-Mb window were obtained for each gene in the reference panel, including the highly-tissue specific splicing QTL (sQTL) information on gene isoforms in the dorsolateral prefrontal cortex (DLPFC, CMC). Using the SNP-expression weights, SNP-trait effect estimates and the SNP correlation matrix, we used the TWAS-Fusion56 to estimate the association statistic between the predicted expression and head size (TWAS Z score). Transcriptome-wide significant genes (eGenes) and the corresponding QTLs (eQTLs) were determined using Bonferroni correction in each reference panel, based on the average number of features (4,320 genes) tested across all the reference panels.56 Finally, using a prior association probability of 1.1 × 10−5 and colocalization analysis (COLOC)63 for each locus we estimated the posterior probability of a shared causal variant (PP4>0.75) between the gene expression and trait association. eGene regions with eQTLs not reaching genome-wide significance in the head size GWAS were considered putatively novel TWAS signals. Furthermore, functional validation of the eGenes was performed by integrating eQTL with the functional genomics feature from the RegulomeDB.64 A RegulomeDB probability score greater than 0.5 and closer to 1 indicates the likelihood of the eQTL having a gene-regulatory role. Finally, accounting for pairwise correlation between the gene expression features we conducted the multiple degree of freedom omnibus analysis, to test for the shared effect of eGenes across the different gene reference panels. A significance threshold of p < 3.48 × 10−6 accounting for the number of genes (N = 14,385) tested was used to identify significant eGenes in the omnibus test.
Effects on anthropomorphic measures and regional brain volumes
The LD score regression software53,65 was used to assess genetic correlations with adult height,66 for both the height-unadjusted and height-adjusted model.
Dual-energy X-ray absorptiometry (DXA) measurements of the UK Biobank imaging subsample (N = 3,313) were used to examine the effect of the identified lead variants on anthropometric measures across the body, i.e., bone area of the arms, legs, pelvis, ribs, spine, trunk and vertebrae L1-L4. In these analyses values more than three standard deviations from the mean were considered outliers and removed from the analyses. We adjusted for age, age,2 gender and principal components (model 1), and additionally for height (model 2) to correct for an overall growth effect.
To investigate the effects of the identified variants for head size on growth in specific brain regions, we investigated the overlap between the identified loci for head size and previous genome-wide association studies (GWASs) on brain volumes.7,67,68,69,70 We also analyzed the associations between the identified lead genetic variants and global volumes (i.e., four brain lobes and lateral ventricle volumes), subcortical volumes (i.e., volumes of eight subcortical structures) and cortical volumes (i.e., volumes of 34 cortical regions of interest) in the UK Biobank (N = 22,145). Volumes were derived using the FreeSurfer 6.0 software. Values more than 3.5 standard deviations away from the mean were considered outliers and removed from the analysis. In the first model, we adjusted for age, age,2 gender and principal components, and in the second model additionally for intracranial volume.
Additionally, we took the lead variants specifically associated with one or two subcortical volumes, and investigated their effects on the shape of seven subcortical structures, i.e., amygdala, caudate nucleus, hippocampus, nucleus accumbens, pallidum, putamen and thalamus. The radial distances and log Jacobian determinants were derived using the ENIGMA-Shape package (http://enigma.usc.edu/ongoing/enigma-shape-analysis/). Volumetric outliers more than 3.5 standard deviations from the mean were removed from the analysis.
We performed 10,000 permutations to define the number of independent DXA, brain volumetric and subcortical shape outcomes. We used this number to define our multiple testing adjusted p value thresholds for significance, i.e., 0.05/(number of independent outcomes x number of lead genetic variants).
Genetic correlations
We investigated the genetic correlations with neuropsychiatric traits using the LD score regression software.53,65 Genetic correlation analyses were performed for educational attainment,71 general cognitive function,72 all stroke,73 Alzheimer’s disease,74 frontotemporal dementia,75 Parkinson’s disease,76 anorexia nervosa,77 attention-deficit hyperactivity disorder,78 autism spectrum disorder,79 bipolar disorder,80 extraversion,81 insomnia,82 major depressive disorder,83 neuroticism,84 obsessive compulsive disorder85 and schizophrenia.80 Analyses were performed in the entire GWAS dataset as well as in the GWAS set with newly included studies in comparison to the intracranial volume GWAS performed by Adams et al.6
We also performed genetic correlation analyses for publicly available cancer GWAS, namely for breast cancer,28 ovarian cancer29 and prostate cancer.30 To obtain information on more cancer types, we additionally included GWAS of cancer registries from the UK Biobank and Kaiser Permanente Genetic Epidemiology Research on Adult Health and Aging (GERA).31 Of those, we excluded cancer types with less than 1,000 cases, which left the following cancer types to be analyzed: bladder cancer (Ncases = 2,242), breast cancer (Ncases = 17,881), cervical cancer (Ncases = 6,563), colon cancer (Ncases = 3,793), endometrial cancer (Ncases = 2,037), esophageal/gastric cancer (Ncases = 1,091), kidney cancer (Ncases = 1,338), lung cancer (Ncases = 2,485), malignant melanoma (Ncases = 6,777), non-Hodgkin’s lymphoma (Ncases = 2,400), prostate cancer (Ncases = 10,792) and rectal cancer (Ncases = 2,091). Genetic correlations with oral cavity/pharyngeal cancer (Ncases = 1,223) and ovarian cancer (Ncases = 1,259) could not be calculated due to low heritability estimates.
Enrichment analyses
We performed enrichment analyses of different gene sets: genes within 1 Mb, 100 kb or 10 kb of the identified genetic loci, genes within 10 kb of the identified genetic loci with intragenic genetic variants, and genes within 10 kb of the identified genetic loci with intragenic genetic lead variants. As a reference, we used the rest of the protein-coding genome.
First, the Online Mendelian Inheritance in Man (OMIM) database58 was used to retrieve information on genes related to heritable phenotypes affecting head size (Tables S19 and S20). Second, the COSMIC database27 was used to extract Tier 1 cancer genes. Taking the rest of the genome as our reference gene set, we calculated the enrichment of these macrocephaly, microcephaly and cancer genes in the abovementioned gene sets.
Lastly, DOMINO,57 a previously developed machine learning tool, was used to assess if the genes in the different gene sets were more often predicted to harbor dominant changes in comparison with genes in the rest of the genome.
Mean autosomal dominance scores were compared with the reference genome using a Mann-Whitney test. Differences in the proportions for the OMIM macro- and microcephaly genes, intellectual disability genes and COSMIC genes were calculated using a Pearson’s χ2 test.
We performed these analyses for the head size height-unadjusted GWAS results, but also the GWAS in the subset of studies for which height was available, the height-adjusted GWAS and the height GWAS.66 For comparison, we also selected the top 67 loci for the height GWAS, so the results were not driven by a difference in the number of associated loci.
Experimental datasets of brain cells
To assess whether the identified genes in the current study are enriched for genes differentially expressed in human progenitors versus neurons, we utilized differential gene expression data of those cell lines, derived from a previously published sample population (Ndonor = 85 in progenitors and Ndonor = 74 in neurons).32 Using genes with at least 10 counts in more than 5% of the cell-type specific donors in either cell-type (resulting in 16,172 protein-coding genes out of 28,785 genes in total), we performed a paired differential gene expression analysis with design matrix: model.matrix(∼ CellType + as.factor(DonorID) + RIN, data) as described previously,32 using the limma R package.86 We detected 1,095/1,420 protein genes upregulated in progenitors/neurons, respectively, for abs(logFC) > 1.5 and adjusted p value < 0.05. Performing a hypergeometric test, we evaluated if multiple protein-coding gene sets: head size gene sets with different distances from the lead variants, OMIM macrocephaly and microcephaly genes, and COSMIC tier 1 cancer genes are enriched among the protein-coding genes upregulated in progenitors or neurons.
Using a different approach, scRNA-seq data were used to investigate whether our genes of interest were enriched for genes specific for certain cortical brain cell types. Specifically, scRNA-seq data from the developing human cortex (gestational week 6–22, more than 189,000 cells) were used to identify the top 10% of genes specific for a certain cell type.34 Using this data, we first performed LD score regression53 based enrichment analyses of the head size GWAS summary statistics, as previously described.33,87 Gene specificity was defined as the ratio of expression of a gene in a cell type by the total expression of that gene in all cell types. In parallel, we again tested the enrichment of various gene sets: head size gene sets with different distances from the lead variants, OMIM macrocephaly and microcephaly genes, and COSMIC tier 1 cancer genes, with the top 10% of cell specific genes for each cell type using hypergeometric tests. FDR correction was used to correct for the multiple gene sets tested for enrichment in each cell type.
To determine if regulatory elements of neural progenitors are enriched for the heritability of head size, we performed partitioned heritability analyses53,88 using chromatin accessibility profiles from a population of 76 primary human neural progenitor cells and 61 of their differentiated neuronal progenies, as was done previously.89
Quantification and statistical analysis
Please see the statistical analyses and software in method details.
Acknowledgments
Acknowledgments are provided for the studies that contributed new samples in addition to samples in previous efforts.5,6
The TWAS analysis in this study is supported by the following funding resource: P30AG066546 (South Texas Alzheimer’s Disease Research Center).
The 1000BRAINS study was funded by the Institute of Neuroscience and Medicine, Research Center Juelich, Germany. We thank the Heinz Nixdorf Foundation (Germany) for the generous support of the Heinz Nixdorf Recall Study on which 1000BRAINS is based. We also thank the scientists and the study staff of the Heinz Nixdorf Recall Study and 1000BRAINS. Funding was also granted by the Initiative and Networking Fund of the Helmholtz Association (S. Caspers) and the European Union’s Horizon 2020 Research and Innovation Programme under grant agreements 785907 (Human Brain Project SGA2; K. Amunts, S. Caspers, and S. Cichon) and 945539 (Human Brain Project SGA3; K. Amunts, S. Caspers, and S. Cichon).
The Three-City (3C) Study (Bordeaux and Dijon) is conducted under a partnership agreement between the Institut National de la Santé et de la Recherche Médicale (INSERM), the Institut de Santé Publique et Développement of the Victor Segalen Bordeaux 2 University, and Sanofi-Aventis. The Fondation pour la Recherche Médicale funded the preparation and initiation of the study. The 3C Study is also supported by the Caisse Nationale Maladie des Travailleurs Salariés, Direction Générale de la Santé, Mutuelle Générale de l’Education Nationale, Institut de la Longévité, Regional Governments of Aquitaine and Bourgogne, Fondation de France, Ministry of Research-INSERM Program “Cohortes et collections de données biologiques,” French National Research Agency COGINUT (ANR-06-PNRA-005), the Fondation Plan Alzheimer (FCS 2009–2012), and the Caisse Nationale pour la Solidarité et l’Autonomie (CNSA). This project has received funding from the European Union’s Horizon 2020 Research and Innovation Programme under grant agreement nos. 643417 and 640643, the French National Research Agency (ANR) - France 2030: ANR-18-RHUS-0002 and ANR-23-IAHU-0001, and the University of Bordeaux Initiative of Excellence (IdEX). Part of the computations were performed at the Bordeaux Bioinformatics Center (CBiB), the University of Bordeaux, and the CREDIM (Centre de Ressource et Développement en Informatique Médicale) at University of Bordeaux, on a server infrastructure supported by the Fondation Claude Pompidou. The project is supported through the following funding organizations under the aegis of JPND (www.jpnd.eu; BRIDGET project): Australia, National Health and Medical Research Council; Austria, Federal Ministry of Science, Research and Economy; Canada, Canadian Institutes of Health Research; France, French National Research Agency; Germany, Federal Ministry of Education and Research; Netherlands, The Netherlands Organisation for Health Research and Development; and United Kingdom, Medical Research Council.
The Atherosclerosis Risk in Communities (ARIC) study has been funded in whole or in part with federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, Department of Health and Human Services (contract numbers HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700004I, and HHSN268201700005I), R01HL087641 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. The authors thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by grant number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. This project was supported in part by National Institute of Neurological Disorders and Stroke grant NS087541.
Data and samples were collected by the Australian Schizophrenia Research Bank (ASRB), supported by the Australian NHMRC, the Pratt Foundation, Ramsay Health Care, and the Viertel Charitable Foundation. The ASRB was also supported by the Schizophrenia Research Institute (Australia), utilizing infrastructure funding from NSW Health and the Macquarie Group Foundation. DNA analysis was supported by the Neurobehavioral Genetics Unit, using funding from NSW Health. M.J.C. was supported by an NHMRC Senior Research Fellowship (1121474), and M.J.C. and M.J.G. were supported by NHMRC project grants 1147644 and 1051672.
The authors are deeply indebted to Gael Jobard, Marc Joliot, Emmanuel Mellet, Laurent Petit, and Laure Zago for their contribution to the design, acquisition, and analyses of the BIL&GIN. A.C.-C. was funded by a grant to C.F. from the Netherlands Organization for Scientific Research (NWO) (054-15-101) and B.M. and F.C. by a grant from the French National Research Agency (ANR) (grant no. 15-HBPR-0001-03) as part of the FLAG-ERA consortium project “MULTI-LATERAL,” a partner project to the European Union’s Flagship Human Brain Project.
The Coronary Artery Risk Development in Young Adults Study (CARDIA) is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with the University of Alabama at Birmingham (75N92023D00002 and 75N92023D00005), Northwestern University (75N92023D00004), University of Minnesota (75N92023D00006), and Kaiser Foundation Research Institute (75N92023D00003). CARDIA was also partially supported by the Intramural Research Program of the National Institute on Aging (NIA) and an intra-agency agreement between NIA and NHLBI (AG0005). This manuscript has been reviewed by CARDIA for scientific content.
The CROMIS-2 ICH study is funded by the Stroke Association and British Heart Foundation. Funding for genotyping was provided by the UCLH/UCL National Institute for Health Research (NIHR) Biomedical Research Centre.
The Diabetes Heart Study (DHS) was supported in part by the National Institutes of Health through R01 HL67348, R01 HL092301, R01 NS058700, R01 NS075107, and R01 AG058921 and the General Clinical Research Center at Wake Forest School of Medicine (M01 RR07122 and F32 HL085989). The authors thank the investigators, staff, and participants of the DHS for their valuable contributions.
The Duke Neurogenetics Study (DNS) was supported by Duke University as well as National Institutes of Health grants R01DA033369 and R01DA031579. R.A., A.R.K., and A.R.H. received further support from National Institutes of Health grant R01AG049789.
We are grateful to all the participants of the Epidemiological Prevention Study Zoetermeer (EPOZ). We would like to thank Dr. Ir. Natalie Terzikhan for imputing the genetic data.
The Erasmus Stroke Study (ESS) was supported by Stroke Research Foundation and Erasmus MC MRACE grants.
This work was supported by the National Center for Research Resources at the National Institutes of Health (grant numbers NIH 1 U24 RR021992 [Function Biomedical Informatics Research Network] and NIH 1 U24 RR025736-01 [Biomedical Informatics Research Network Coordinating Center]), the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health through grant UL1 TR000153, and the National Institutes of Health through 5R01MH094524 and P20GM103472. J.M.F. was funded by the Veterans Administration (1IK6CX002519).
This work was supported by the NHLBI’s Framingham Heart Study (contracts N01-HC-25195, HHSN268201500001I, and 75N92019D00031) and its contract with Affymetrix, Inc., for genotyping services (contract no. N02-HL-6-4278). A portion of this research utilized the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. This study was also supported by grants from the National Institute of Aging (R01s AG033040, AG033193, AG054076, AG049607, AG008122, AG016495, U01-AG049505, AG052409, AG058589, and RF1AG059421) and the National Institute of Neurological Disorders and Stroke (R01-NS017950). We would like to thank the dedication of the Framingham Study participants as well as the Framingham Study team, especially investigators and staff from the Neurology group, for their contributions to data collection. C.D. is supported by the Alzheimer’s Disease Center (P30 AG 010129) and by National Institute on Aging grants R01AG054076 and P30 AG072972. The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the NHLBI, the National Institutes of Health, or the US Department of Health and Human Services.
The Generation R Study was supported by ZonMw (TOP project 91211021 [T.W.]), the Sophia Foundation (grant S18-20 [R.L.M.]), and the European Union’s Horizon 2020 Research and Innovation Programme (no. 733206 LifeCycle Project [S. Lamballais], no. 848158 EarlyCause Project [C.A.M.C.], and Marie Skłodowska-Curie grant agreement no. 707404 [C.A.M.C.]). The Generation R Study is conducted by the Erasmus Medical Center in close collaboration with the School of Law and Faculty of Social Sciences of the Erasmus University Rotterdam; the Municipal Health Service Rotterdam Area, Rotterdam; the Rotterdam Homecare Foundation, Rotterdam; and the Stichting Trombosedienst & Artsenlaboratorium Rijnmond (STAR-MDC), Rotterdam. We gratefully acknowledge the contribution of children and parents, general practitioners, hospitals, midwives, and pharmacies in Rotterdam. The general design of the Generation R Study is made possible by financial support from the Erasmus Medical Center, Rotterdam; the Erasmus University Rotterdam; the Netherlands Organization for Health Research and Development (ZonMw); the Netherlands Organisation for Scientific Research (NWO); the Ministry of Health, Welfare and Sport; and the Ministry of Youth and Families.
The Trøndelag Health Study (HUNT) is a collaboration between HUNT Research Center (Faculty of Medicine and Health Sciences, NTNU – Norwegian University of Science and Technology), Nord-Trøndelag County Council, Central Norway Health Authority, and the Norwegian Institute of Public Health. HUNT MRI was funded by the liaison committee between the Central Norway Regional Health Authority and the Norwegian University of Science and Technology as well as the Norwegian National Advisory Unit for functional magnetic resonance imaging (MRI).
The Institute of Mental Health (IMH) study was supported by research grants from the National Healthcare Group, Singapore (SIG/05004 and SIG/05028), and the Singapore Bioimaging Consortium (RP C-009/2006) research grants awarded to K. Sim.
The Internet-based Students HeAlth Research Enterprise (i-Share) study is conducted by the Universities of Bordeaux and Versailles Saint-Quentin-en-Yvelines (France). The i-Share study has received funding from the French National Agency (Agence Nationale de la Recherche [ANR]) via the “Investissements d’Avenir” program (grant number ANR-10-COHO-05) and from the University of Bordeaux Initiative of Exellence (IdEX). This project has also received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation Programme under grant agreement no. 640643. The 3C Study is conducted under a partnership agreement among the Institut National de la Santé et de la Recherche Médicale (INSERM), the University of Bordeaux, and Sanofi-Aventis. The Fondation pour la Recherche Médicale funded the preparation and initiation of the study. S. Debette has received investigator-initiated research funding from the French National Research Agency (ANR) and from the Fondation Leducq. This work was supported by the National Foundation for Alzheimer’s Disease and Related Disorders, the Institut Pasteur de Lille, the LabEx DISTALZ, and the Centre National de Génotypage. This project is an EU Joint Program - Neurodegenerative Disease Research (JPND) project. The project is supported through the following funding organizations under the aegis of JPND (www.jpnd.eu): Australia, National Health and Medical Research Council; Austria, Federal Ministry of Science, Research and Economy; Canada, Canadian Institutes of Health Research; France, French National Research Agency; Germany, Federal Ministry of Education and Research; Netherlands, The Netherlands Organisation for Health Research and Development; and United Kingdom, Medical Research Council. In addition, S. Debette is supported by a grant overseen by the French National Research Agency (ANR) as part of the Investment for the Future Program ANR-18-RHUS-0002. Part of the computations were performed at the Bordeaux Bioinformatics Center (CBiB), University of Bordeaux, and at the CREDIM at University of Bordeaux, on a server infrastructure supported by the Fondation Claude Pompidou.
LIFE-Adult is supported by LIFE – Leipzig Research Center for Civilization Diseases, an organizational unit affiliated to the Medical Faculty of the University of Leipzig. LIFE is funded by means of the European Union, European Regional Development Fund (ERDF) and by funds of the Free State of Saxony within the framework of the excellence initiative (project numbers 713–241202, 713–241202, 14505/2470, and 14575/2470). We thank all participants and Kerstin Wirkner, Ulrike Scharrer, Katrin Arelin, and everyone involved in MRI data acquisition and analysis.
The Nagahama Prospective Genome Cohort for Comprehensive Human Bioscience is grateful to the Nagahama City Office and the nonprofit organization Zeroji Club for their help in conducting the study. This project is supported by operational funds of Kyoto University and the Top Global University Project of the Ministry of Education, Culture, Sports, Science and Technology (MEXT) in Japan. We also receive Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science and research grants from the Japan Agency for Medical Research and Development for the Practical Research Project for Rare/Intractable Diseases and the Comprehensive Research on Aging and Health Science for Dementia R&D. M.-G.D. received a grant from the Fondation Bettencourt Schueller.
As part of the Poznan MS study, M.A.P. reported receiving grants from the Polish National Science Centre: 2011/01/D/NZ4/05801.
The Rotterdam Study (RS) is funded by Erasmus Medical Center and Erasmus University, Rotterdam; the Netherlands Organization for Health Research and Development (ZonMw); the Research Institute for Diseases in the Elderly (RIDE); the Ministry of Education, Culture and Science; the Ministry for Health, Welfare and Sports; the European Commission (DG XII); and the Municipality of Rotterdam. The authors are grateful to the study participants, the staff from the Rotterdam Study, and the participating general practitioners and pharmacists. The generation and management of GWAS genotype data for the Rotterdam Study (RS I, RS II, and RS III) were executed by the Human Genotyping Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, Rotterdam, the Netherlands. The GWAS datasets are supported by the Netherlands Organisation of Scientific Research NWO Investments (no. 175.010.2005.011, 911-03-012); the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC; the Research Institute for Diseases in the Elderly (014-93-015; RIDE2); the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO); and Netherlands Consortium for Healthy Aging (NCHA) project no. 050-060-810. We thank Pascal Arp, Mila Jhamai, Marijn Verkerk, Lizbeth Herrera, Marjolein Peters, and Carolina Medina-Gomez for their help in creating the GWAS database and Karol Estrada, Yurii Aulchenko, and Carolina Medina-Gomez for the creation and analysis of imputed data. H.H.H.A. is supported by ZonMw grant number 916.19.151.
Study of Health in Pomerania - TREND (SHIP-TREND) is part of the Community Medicine Research net of the University of Greifswald, Germany, which is funded by the Federal Ministry of Education and Research (grant nos. 01ZZ9603, 01ZZ0103, and 01ZZ0403), the Ministry of Cultural Affairs, and the Social Ministry of the Federal State of Mecklenburg-West Pomerania. MRI scans and genome-wide SNP typing in SHIP-TREND have been supported by a joint grant from Siemens Healthineers, Erlangen, Germany and the Federal State of Mecklenburg-West Pomerania. The University of Greifswald is a member of the Caché Campus program of the InterSystems GmbH.
The Canadian Institutes of Health Research and the Heart and Stroke Foundation of Canada fund the Saguenay Youth Study (SYS). Computations were performed on the GPC supercomputer at the SciNet HPC Consortium. SciNet is funded by the Canada Foundation for Innovation under the auspices of Compute Canada, the Government of Ontario, Ontario Research Fund - Research Excellence, and the University of Toronto.
This research has been conducted using the UK Biobank Resource under application number 23509.
Vitamin D Intervention in Infants (VIDI) is supported by the Finnish Medical Foundation, the Academy of Finland, the Sigrid Jusélius Foundation, the Swedish Research Council, the Novo Nordisk Foundation, Finska Läkaresällskapet, and the Folkhälsan Research Foundation. We want to thank Dr. Helena Hauta-alus, Dr. Elisa Holmlund-Suila, Dr. Saara Valkama, and Dr. Jenni Rosendahl for their contribution in acquiring the data.
Author contributions
Performed statistical analysis: M.J.K., R.A.P., T.E.E., C.L.S., A.M., S.v.d.A., M.-G.D., X.J., I.C.H., S. Lamballais, M.A.P., C.E.L., A.C.-C., T.G.M.v.E., C.S.R., J.S., M. Scholz, A.K., G.H.Y.L., R.A., J.R.A., F.-C.H., A.R.A., M.L., A. Tsuchida, M.W.A.T., N.A., Y.P., D.L., M. Sargurupremraj, F.B., R.N.B., S. Dalvie, M.J.G., S.H., N.J., Y.K., A.R.K., S. Li, K.L., S.E.M., S.N., N.D.P., Y.Q., W.R.R., G.V.R., S.Y.S., W.V.W., K.W., M.J.C., C.-L.C., and H.H.H.A. Acquired data: T.E.E., I.C.H., D.H.K.v.D.-N., M.A.P., T.G.M.v.E., A.K.H., A.S.B., F.B., J.C.B., D.B., R.N.B., R.B., S. Caspers, G.C., C.A.M.C., J.-F.D., C.D., M.E.-C., J.M.F., B.F., B.I.F., N.F., C.H., P.H., G.H., M.K.I., C.R.J., C.J., Y.K., A.R.K., W.T.L., F. Macciardi, O.M., B.M., S.E.M., S. Miyamoto, S. Moebus, T.H.M., R.M., T.W.M., M.N., Z.P., A.P., R.S., P.J.S., K. Setoh, S. Sidney, B.S.P., J.L.S., Y.T., A. Teumer, A.U., A.v.d.L., M.W.V., D.J.W., B.G.W., A.V.W., Q.Y., K.Y., H.G.B., Q.L.G., K. Sim, D.J.S., D.W.B., M.J.C., A.R.H., S.A., A.V., T.P., S. Cichon, V.D.C., F.C., L.J.L., T.W., P.J.K., H.H., M.F., F. Matsuda, H.J.G., M.A.I., S. Debette, P.M.T., S. Seshadri, and H.H.H.A. Drafted the manuscript: M.J.K., R.A.P., and H.H.H.A. Revised the manuscript for important intellectual content: M.J.K., R.A.P., T.E.E., C.L.S., A.M., S.v.d.A., M.-G.D., X.J., I.C.H., D.H.K.v.D.-N., S. Lamballais, M.A.P., C.E.L., A.C.-C., T.G.M.v.E., C.S.R., J.S., M. Scholz, A.K.H., A.K., G.H.Y.L., R.A., J.R.A., F.-C.H., A.R.A., M.L., A. Tsuchida, M.W.A.T., N.A., Y.P., D.L., M. Sargurupremraj, A.S.B., F.B., J.C.B., D.B., R.N.B., R.B., S. Caspers, G.C., C.A.M.C., S. Dalvie, J.-F.D., C.D., M.E.-C., J.M.F., B.F., B.I.F., N.F., M.J.G., S.H., C.H., P.H., G.H., M.K.I., C.R.J., N.J., C.J., Y.K., A.R.K., S. Li, K.L., W.T.L., F. Macciardi, O.M., B.M., S.E.M., S. Miyamoto, S. Moebus, T.H.M., R.M., T.W.M., M.N., S.N., N.D.P., Z.P., A.P., Y.Q., W.R.R., G.V.R., R.S., P.J.S., K. Setoh, C.Y.S., S. Seshadri, B.S.P., J.L.S., Y.T., A. Teumer, A.U., A.v.d.L., M.W.V., D.J.W., B.G.W., A.V.W., K.W., Q.Y., K.Y., H.G.B., Q.L.G., K. Sim, D.J.S., D.W.B., M.J.C., A.R.H., C.-L.C., S.A., A.V., T.P., S. Cichon, V.D.C., F.C., L.J.L., T.W., P.J.K., H.H., M.F., F. Matsuda, H.J.G., M.A.I., S. Debette, P.M.T., S. Seshadri, and H.H.H.A.
Declaration of interests
H.H. and I.C.H. received funding from Alzheimer’s Research UK and the Dunhill Medical Trust Foundation. M.A.P. reported receiving grants and personal and travel fees from Roche, Novartis, Merck, and Biogen outside the submitted work. M. Scholz receives funding from Pfizer Inc. for a project not related to this research. C.D. serves as a consultant of Novartis Pharmaceuticals. B.F. has received educational speaking fees from Medice. N.J. and P.M.T. are MPIs of a research grant from Biogen Inc. for work unrelated to the contents of this manuscript. D.J.W. received funding from the Stroke Foundation/British Heart Foundation. D.J.S. has received consultancy honoraria from Discovery Vitality, Johnson & Johnson, Kanna, L’Oreal, Lundbeck, Orion, Sanofi, Servier, Takeda, and Vistagen. H.H. received funding from MRC, Wellcome Trust, and NIHR UCLH BRC. H.J.G. has received travel grants and speaker’s honoraria from Fresenius Medical Care, Neuraxpharm, and Janssen Cilag as well as research funding from Fresenius Medical Care.
Published: May 3, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2024.101529.
Contributor Information
Hieab H.H. Adams, Email: hieab.adams@radboudumc.nl.
The Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium:
Philippe Amouyel, Konstantinos Arfanakis, Benjamin S. Aribisala, Mark E. Bastin, Ganesh Chauhan, Christopher Chen, Ching-Yu Cheng, Philip L. de Jager, Ian J. Deary, Debra A. Fleischman, Rebecca F. Gottesman, Vilmundur Gudnason, Saima Hilal, Edith Hofer, Deborah Janowitz, J. Wouter Jukema, David C.M. Liewald, Lorna M. Lopez, Oscar Lopez, Michelle Luciano, Oliver Martinez, Wiro J. Niessen, Paul Nyquist, Jerome I. Rotter, Tatjana Rundek, Ralph L. Sacco, Helena Schmidt, Henning Tiemeier, Stella Trompet, Jeroen van der Grond, Henry Völzke, Joanna M. Wardlaw, Lisa Yanek, and Jingyun Yang
The Enhancing NeuroImaging Genetics through Meta-Analysis (ENIGMA) Consortium:
Ingrid Agartz, Saud Alhusaini, Laura Almasy, David Ames, Katrin Amunts, Ole A. Andreassen, Nicola Armstrong, Manon Bernard, John Blangero, Laura M.E. Blanken, Marco P. Boks, Dorret I. Boomsma, Adam M. Brickman, Henry Brodaty, Randy L. Buckner, Jan K. Buitelaar, Dara M. Cannon, Vaughan J. Carr, Stanley V. Catts, M. Mallar Chakravarty, Qiang Chen, Christopher R.K. Ching, Aiden Corvin, Benedicto Crespo-Facorro, Joanne E. Curran, Gareth E. Davies, Eco J.C. de Geus, Greig I. de Zubicaray, Anouk den Braber, Sylvane Desrivières, Allissa Dillman, Srdjan Djurovic, Wayne C. Drevets, Ravi Duggirala, Stefan Ehrlich, Susanne Erk, Thomas Espeseth, Iryna O. Fedko, Guillén Fernández, Simon E. Fisher, Tatiana M. Foroud, Tian Ge, Sudheer Giddaluru, David C. Glahn, Aaron L. Goldman, Robert C. Green, Corina U. Greven, Oliver Grimm, Narelle K. Hansell, Catharina A. Hartman, Ryota Hashimoto, Andreas Heinz, Frans Henskens, Derrek P. Hibar, Beng-Choon Ho, Pieter J. Hoekstra, Avram J. Holmes, Martine Hoogman, Jouke-Jan Hottenga, Hilleke E. Hulshoff Pol, Assen Jablensky, Mark Jenkinson, Tianye Jia, Karl-Heinz Jöckel, Erik G. Jönsson, Sungeun Kim, Marieke Klein, Peter Kochunov, John B. Kwok, Stephen M. Lawrie, Stephanie Le Hellard, Hervé Lemaître, Carmel Loughland, Andre F. Marquand, Nicholas G. Martin, Jean-Luc Martinot, Mar Matarin, Daniel H. Mathalon, Karen A. Mather, Venkata S. Mattay, Colm McDonald, Francis J. McMahon, Katie L. McMahon, Rebekah E, McWhirter, Patrizia Mecocci, Ingrid Melle, Andreas Meyer-Lindenberg, Patricia T. Michie, Yuri Milaneschi, Derek W. Morris, Bryan Mowry, Kwangsik Nho, Thomas E. Nichols, Markus N. Nöthen, Rene L. Olvera, Jaap Oosterlaan, Roel A. Ophoff, Massimo Pandolfo, Christos Pantelis, Irene Pappa, Brenda Penninx, G. Bruce Pike, Paul E. Rasser, Miguel E. Rentería, Simone Reppermund, Marcella Rietschel, Shannon L. Risacher, Nina Romanczuk-Seiferth, Emma Jane Rose, Perminder S. Sachdev, Philipp G. Sämann, Andrew J. Saykin, Ulrich Schall, Peter R. Schofield, Sara Schramm, Gunter Schumann, Rodney Scott, Li Shen, Sanjay M. Sisodiya, Hilkka Soininen, Emma Sprooten, Velandai Srikanth, Vidar M. Steen, Lachlan T. Strike, Anbupalam Thalamuthu, Arthur W. Toga, Paul Tooney, Diana Tordesillas-Gutiérrez, Jessica A. Turner, Maria del C. Valdés Hernández, Dennis van der Meer, Nic J.A. Van der Wee, Neeltje E.M. Van Haren, Dennis van 't Ent, Dick J. Veltman, Henrik Walter, Daniel R. Weinberger, Michael W. Weiner, Wei Wen, Lars T. Westlye, Eric Westman, Anderson M. Winkler, Girma Woldehawariat, Margaret J. Wright, and Jingqin Wu
Supplemental information
References
- 1.Dekaban A.S. Changes in brain weights during the span of human life: relation of brain weights to body heights and body weights. Ann. Neurol. 1978;4:345–356. doi: 10.1002/ana.410040410. [DOI] [PubMed] [Google Scholar]
- 2.Smit D.J.A., Luciano M., Bartels M., van Beijsterveldt C.E.M., Wright M.J., Hansell N.K., Brunner H.G., Estourgie-van Burk G.F., de Geus E.J.C., Martin N.G., Boomsma D.I. Heritability of head size in Dutch and Australian twin families at ages 0-50 years. Twin Res. Hum. Genet. 2010;13:370–380. doi: 10.1375/twin.13.4.370. [DOI] [PubMed] [Google Scholar]
- 3.Pirozzi F., Nelson B., Mirzaa G. From microcephaly to megalencephaly: determinants of brain size. Dialogues Clin. Neurosci. 2018;20:267–282. doi: 10.31887/DCNS.2018.20.4/gmirzaa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jansen P.R., Nagel M., Watanabe K., Wei Y., Savage J.E., de Leeuw C.A., van den Heuvel M.P., van der Sluis S., Posthuma D. Genome-wide meta-analysis of brain volume identifies genomic loci and genes shared with intelligence. Nat. Commun. 2020;11:5606. doi: 10.1038/s41467-020-19378-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haworth S., Shapland C.Y., Hayward C., Prins B.P., Felix J.F., Medina-Gomez C., Rivadeneira F., Wang C., Ahluwalia T.S., Vrijheid M., et al. Low-frequency variation in TP53 has large effects on head circumference and intracranial volume. Nat. Commun. 2019;10:357. doi: 10.1038/s41467-018-07863-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adams H.H.H., Hibar D.P., Chouraki V., Stein J.L., Nyquist P.A., Rentería M.E., Trompet S., Arias-Vasquez A., Seshadri S., Desrivières S., et al. Novel genetic loci underlying human intracranial volume identified through genome-wide association. Nat. Neurosci. 2016;19:1569–1582. doi: 10.1038/nn.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Satizabal C.L., Adams H.H.H., Hibar D.P., White C.C., Knol M.J., Stein J.L., Scholz M., Sargurupremraj M., Jahanshad N., Roshchupkin G.V., et al. Genetic architecture of subcortical brain structures in 38,851 individuals. Nat. Genet. 2019;51:1624–1636. doi: 10.1038/s41588-019-0511-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krabbe K., Karlsborg M., Hansen A., Werdelin L., Mehlsen J., Larsson H.B.W., Paulson O.B. Increased intracranial volume in Parkinson's disease. J. Neurol. Sci. 2005;239:45–52. doi: 10.1016/j.jns.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 9.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanchez-Vega F., Mina M., Armenia J., Chatila W.K., Luna A., La K.C., Dimitriadoy S., Liu D.L., Kantheti H.S., Saghafinia S., et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell. 2018;173:321–337.e10. doi: 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng H., Ying H., Yan H., Kimmelman A.C., Hiller D.J., Chen A.-J., Perry S.R., Tonon G., Chu G.C., Ding Z., et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meletis K., Wirta V., Hede S.-M., Nistér M., Lundeberg J., Frisén J. p53 suppresses the self-renewal of adult neural stem cells. Development. 2006;133:363–369. doi: 10.1242/dev.02208. [DOI] [PubMed] [Google Scholar]
- 13.Stecca B., Ruiz i Altaba A. A GLI1-p53 inhibitory loop controls neural stem cell and tumour cell numbers. EMBO J. 2009;28:663–676. doi: 10.1038/emboj.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inestrosa N.C., Varela-Nallar L. Wnt signalling in neuronal differentiation and development. Cell Tissue Res. 2015;359:215–223. doi: 10.1007/s00441-014-1996-4. [DOI] [PubMed] [Google Scholar]
- 15.Chenn A., Walsh C.A. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- 16.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 17.Grey W., Izatt L., Sahraoui W., Ng Y.M., Ogilvie C., Hulse A., Tse E., Holic R., Yu V. Deficiency of the cyclin-dependent kinase inhibitor, CDKN1B, results in overgrowth and neurodevelopmental delay. Hum. Mutat. 2013;34:864–868. doi: 10.1002/humu.22314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wasserman J.D., Tomlinson G.E., Druker H., Kamihara J., Kohlmann W.K., Kratz C.P., Nathanson K.L., Pajtler K.W., Parareda A., Rednam S.P., et al. Multiple Endocrine Neoplasia and Hyperparathyroid-Jaw Tumor Syndromes: Clinical Features, Genetics, and Surveillance Recommendations in Childhood. Clin. Cancer Res. 2017;23:e123–e132. doi: 10.1158/1078-0432.CCR-17-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alcantara D., Timms A.E., Gripp K., Baker L., Park K., Collins S., Cheng C., Stewart F., Mehta S.G., Saggar A., et al. Mutations of AKT3 are associated with a wide spectrum of developmental disorders including extreme megalencephaly. Brain. 2017;140:2610–2622. doi: 10.1093/brain/awx203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davies M.A., Stemke-Hale K., Tellez C., Calderone T.L., Deng W., Prieto V.G., Lazar A.J.F., Gershenwald J.E., Mills G.B. A novel AKT3 mutation in melanoma tumours and cell lines. Br. J. Cancer. 2008;99:1265–1268. doi: 10.1038/sj.bjc.6604637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mei L., Nave K.A. Neuregulin-ERBB signaling in the nervous system and neuropsychiatric diseases. Neuron. 2014;83:27–49. doi: 10.1016/j.neuron.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aguirre A., Dupree J.L., Mangin J.M., Gallo V. A functional role for EGFR signaling in myelination and remyelination. Nat. Neurosci. 2007;10:990–1002. doi: 10.1038/nn1938. [DOI] [PubMed] [Google Scholar]
- 23.Kataria H., Alizadeh A., Karimi-Abdolrezaee S. Neuregulin-1/ErbB network: An emerging modulator of nervous system injury and repair. Prog. Neurobiol. 2019;180 doi: 10.1016/j.pneurobio.2019.101643. [DOI] [PubMed] [Google Scholar]
- 24.Brodie A., Azaria J.R., Ofran Y. How far from the SNP may the causative genes be? Nucleic Acids Res. 2016;44:6046–6054. doi: 10.1093/nar/gkw500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lelieveld S.H., Reijnders M.R.F., Pfundt R., Yntema H.G., Kamsteeg E.J., de Vries P., de Vries B.B.A., Willemsen M.H., Kleefstra T., Löhner K., et al. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat. Neurosci. 2016;19:1194–1196. doi: 10.1038/nn.4352. [DOI] [PubMed] [Google Scholar]
- 26.Kumagai A., Shevchenko A., Shevchenko A., Dunphy W.G. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell. 2010;140:349–359. doi: 10.1016/j.cell.2009.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sondka Z., Bamford S., Cole C.G., Ward S.A., Dunham I., Forbes S.A. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat. Rev. Cancer. 2018;18:696–705. doi: 10.1038/s41568-018-0060-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H., Ahearn T.U., Lecarpentier J., Barnes D., Beesley J., Qi G., Jiang X., O'Mara T.A., Zhao N., Bolla M.K., et al. Genome-wide association study identifies 32 novel breast cancer susceptibility loci from overall and subtype-specific analyses. Nat. Genet. 2020;52:572–581. doi: 10.1038/s41588-020-0609-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phelan C.M., Kuchenbaecker K.B., Tyrer J.P., Kar S.P., Lawrenson K., Winham S.J., Dennis J., Pirie A., Riggan M.J., Chornokur G., et al. Identification of 12 new susceptibility loci for different histotypes of epithelial ovarian cancer. Nat. Genet. 2017;49:680–691. doi: 10.1038/ng.3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schumacher F.R., Al Olama A.A., Berndt S.I., Benlloch S., Ahmed M., Saunders E.J., Dadaev T., Leongamornlert D., Anokian E., Cieza-Borrella C., et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat. Genet. 2018;50:928–936. doi: 10.1038/s41588-018-0142-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rashkin S.R., Graff R.E., Kachuri L., Thai K.K., Alexeeff S.E., Blatchins M.A., Cavazos T.B., Corley D.A., Emami N.C., Hoffman J.D., et al. Pan-cancer study detects genetic risk variants and shared genetic basis in two large cohorts. Nat. Commun. 2020;11:4423. doi: 10.1038/s41467-020-18246-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aygün N., Elwell A.L., Liang D., Lafferty M.J., Cheek K.E., Courtney K.P., Mory J., Hadden-Ford E., Krupa O., de la Torre-Ubieta L., et al. Genetic effects on brain traits impact cell-type specific gene regulation during neurogenesis. bioRxiv. 2021 doi: 10.1101/2020.10.21.349019. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skene N.G., Bryois J., Bakken T.E., Breen G., Crowley J.J., Gaspar H.A., Giusti-Rodriguez P., Hodge R.D., Miller J.A., Muñoz-Manchado A.B., et al. Genetic identification of brain cell types underlying schizophrenia. Nat. Genet. 2018;50:825–833. doi: 10.1038/s41588-018-0129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhaduri A., Andrews M.G., Mancia Leon W., Jung D., Shin D., Allen D., Jung D., Schmunk G., Haeussler M., Salma J., et al. Cell stress in cortical organoids impairs molecular subtype specification. Nature. 2020;578:142–148. doi: 10.1038/s41586-020-1962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hansen D.V., Lui J.H., Parker P.R.L., Kriegstein A.R. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- 36.Cárdenas A., Villalba A., de Juan Romero C., Picó E., Kyrousi C., Tzika A.C., Tessier-Lavigne M., Ma L., Drukker M., Cappello S., Borrell V. Evolution of Cortical Neurogenesis in Amniotes Controlled by Robo Signaling Levels. Cell. 2018;174:590–606.e21. doi: 10.1016/j.cell.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munji R.N., Choe Y., Li G., Siegenthaler J.A., Pleasure S.J. Wnt signaling regulates neuronal differentiation of cortical intermediate progenitors. J. Neurosci. 2011;31:1676–1687. doi: 10.1523/JNEUROSCI.5404-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chodelkova O., Masek J., Korinek V., Kozmik Z., Machon O. Tcf7L2 is essential for neurogenesis in the developing mouse neocortex. Neural Dev. 2018;13:8. doi: 10.1186/s13064-018-0107-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pollock A., Bian S., Zhang C., Chen Z., Sun T. Growth of the developing cerebral cortex is controlled by microRNA-7 through the p53 pathway. Cell Rep. 2014;7:1184–1196. doi: 10.1016/j.celrep.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glickstein S.B., Monaghan J.A., Koeller H.B., Jones T.K., Ross M.E. Cyclin D2 is critical for intermediate progenitor cell proliferation in the embryonic cortex. J. Neurosci. 2009;29:9614–9624. doi: 10.1523/JNEUROSCI.2284-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Antonelli F., Casciati A., Tanori M., Tanno B., Linares-Vidal M.V., Serra N., Bellés M., Pannicelli A., Saran A., Pazzaglia S. Alterations in Morphology and Adult Neurogenesis in the Dentate Gyrus of Patched1 Heterozygous Mice. Front. Mol. Neurosci. 2018;11:168. doi: 10.3389/fnmol.2018.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yabut O.R., Ng H.X., Fernandez G., Yoon K., Kuhn J., Pleasure S.J. Loss of Suppressor of Fused in Mid-Corticogenesis Leads to the Expansion of Intermediate Progenitors. J. Dev. Biol. 2016;4 doi: 10.3390/jdb4040029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harris L., Zalucki O., Gobius I., McDonald H., Osinki J., Harvey T.J., Essebier A., Vidovic D., Gladwyn-Ng I., Burne T.H., et al. Transcriptional regulation of intermediate progenitor cell generation during hippocampal development. Development. 2016;143:4620–4630. doi: 10.1242/dev.140681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Green J., Cairns B.J., Casabonne D., Wright F.L., Reeves G., Beral V., Million Women Study collaborators Height and cancer incidence in the Million Women Study: prospective cohort, and meta-analysis of prospective studies of height and total cancer risk. Lancet Oncol. 2011;12:785–794. doi: 10.1016/S1470-2045(11)70154-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samuelsen S.O., Bakketeig L.S., Tretli S., Johannesen T.B., Magnus P. Head circumference at birth and risk of brain cancer in childhood: a population-based study. Lancet Oncol. 2006;7:39–42. doi: 10.1016/S1470-2045(05)70470-8. [DOI] [PubMed] [Google Scholar]
- 46.McCormack V.A., dos Santos Silva I., Koupil I., Leon D.A., Lithell H.O. Birth characteristics and adult cancer incidence: Swedish cohort of over 11,000 men and women. Int. J. Cancer. 2005;115:611–617. doi: 10.1002/ijc.20915. [DOI] [PubMed] [Google Scholar]
- 47.dos Santos Silva I., De Stavola B., McCormack V., Collaborative Group on Pre-Natal Risk Factors and Subsequent Risk of Breast Cancer Birth size and breast cancer risk: re-analysis of individual participant data from 32 studies. PLoS Med. 2008;5:e193. doi: 10.1371/journal.pmed.0050193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sandvei M.S., Lagiou P., Romundstad P.R., Trichopoulos D., Vatten L.J. Size at birth and risk of breast cancer: update from a prospective population-based study. Eur. J. Epidemiol. 2015;30:485–492. doi: 10.1007/s10654-015-0045-2. [DOI] [PubMed] [Google Scholar]
- 49.McCormack V.A., dos Santos Silva I., De Stavola B.L., Mohsen R., Leon D.A., Lithell H.O. Fetal growth and subsequent risk of breast cancer: results from long term follow up of Swedish cohort. BMJ. 2003;326:248. doi: 10.1136/bmj.326.7383.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vatten L.J., Nilsen T.I.L., Tretli S., Trichopoulos D., Romundstad P.R. Size at birth and risk of breast cancer: prospective population-based study. Int. J. Cancer. 2005;114:461–464. doi: 10.1002/ijc.20726. [DOI] [PubMed] [Google Scholar]
- 51.Winkler T.W., Day F.R., Croteau-Chonka D.C., Wood A.R., Locke A.E., Mägi R., Ferreira T., Fall T., Graff M., Justice A.E., et al. Quality control and conduct of genome-wide association meta-analyses. Nat. Protoc. 2014;9:1192–1212. doi: 10.1038/nprot.2014.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bulik-Sullivan B.K., Loh P.R., Finucane H.K., Ripke S., Yang J., Schizophrenia Working Group of the Psychiatric Genomics Consortium. Patterson N., Daly M.J., Price A.L., Neale B.M. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watanabe K., Taskesen E., van Bochoven A., Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gusev A., Ko A., Shi H., Bhatia G., Chung W., Penninx B.W.J.H., Jansen R., de Geus E.J.C., Boomsma D.I., Wright F.A., et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 2016;48:245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quinodoz M., Royer-Bertrand B., Cisarova K., Di Gioia S.A., Superti-Furga A., Rivolta C. DOMINO: Using Machine Learning to Predict Genes Associated with Dominant Disorders. Am. J. Hum. Genet. 2017;101:623–629. doi: 10.1016/j.ajhg.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Amberger J.S., Bocchini C.A., Schiettecatte F., Scott A.F., Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM(R)), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Psaty B.M., O'Donnell C.J., Gudnason V., Lunetta K.L., Folsom A.R., Rotter J.I., Uitterlinden A.G., Harris T.B., Witteman J.C.M., Boerwinkle E., CHARGE Consortium Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ. Cardiovasc. Genet. 2009;2:73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thompson P.M., Stein J.L., Medland S.E., Hibar D.P., Vasquez A.A., Renteria M.E., Toro R., Jahanshad N., Schumann G., Franke B., et al. The ENIGMA Consortium: large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging Behav. 2014;8:153–182. doi: 10.1007/s11682-013-9269-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jorgensen J.B., Paridon E., Quaade F. The correlation between external cranial volume and brain volume. Am. J. Phys. Anthropol. 1961;19:317–320. doi: 10.1002/ajpa.1330190402. [DOI] [PubMed] [Google Scholar]
- 62.Chauhan G., Adams H.H.H., Bis J.C., Weinstein G., Yu L., Töglhofer A.M., Smith A.V., van der Lee S.J., Gottesman R.F., Thomson R., et al. Association of Alzheimer's disease GWAS loci with MRI markers of brain aging. Neurobiol. Aging. 2015;36:1765.e7–1765.e16. doi: 10.1016/j.neurobiolaging.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Giambartolomei C., Vukcevic D., Schadt E.E., Franke L., Hingorani A.D., Wallace C., Plagnol V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dong S., Boyle A.P. Predicting functional variants in enhancer and promoter elements using RegulomeDB. Hum. Mutat. 2019;40:1292–1298. doi: 10.1002/humu.23791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bulik-Sullivan B., Finucane H.K., Anttila V., Gusev A., Day F.R., Loh P.R., ReproGen Consortium. Psychiatric Genomics Consortium. Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3. Duncan L., et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yengo L., Sidorenko J., Kemper K.E., Zheng Z., Wood A.R., Weedon M.N., Frayling T.M., Hirschhorn J., Yang J., Visscher P.M., GIANT Consortium Meta-analysis of genome-wide association studies for height and body mass index in approximately 700000 individuals of European ancestry. Hum. Mol. Genet. 2018;27:3641–3649. doi: 10.1093/hmg/ddy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van der Lee S.J., Knol M.J., Chauhan G., Satizabal C.L., Smith A.V., Hofer E., Bis J.C., Hibar D.P., Hilal S., van den Akker E.B., et al. A genome-wide association study identifies genetic loci associated with specific lobar brain volumes. Commun. Biol. 2019;2:285. doi: 10.1038/s42003-019-0537-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vojinovic D., Adams H.H., Jian X., Yang Q., Smith A.V., Bis J.C., Teumer A., Scholz M., Armstrong N.J., Hofer E., et al. Genome-wide association study of 23,500 individuals identifies 7 loci associated with brain ventricular volume. Nat. Commun. 2018;9:3945. doi: 10.1038/s41467-018-06234-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hofer E., Roshchupkin G.V., Adams H.H.H., Knol M.J., Lin H., Li S., Zare H., Ahmad S., Armstrong N.J., Satizabal C.L., et al. Genetic Determinants of Cortical Structure (Thickness, Surface Area and Volumes) among Disease Free Adults in the CHARGE Consortium. bioRxiv. 2019 doi: 10.1101/409649. Preprint at. [DOI] [Google Scholar]
- 70.Hibar D.P., Adams H.H.H., Jahanshad N., Chauhan G., Stein J.L., Hofer E., Renteria M.E., Bis J.C., Arias-Vasquez A., Ikram M.K., et al. Novel genetic loci associated with hippocampal volume. Nat. Commun. 2017;8 doi: 10.1038/ncomms13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee J.J., Wedow R., Okbay A., Kong E., Maghzian O., Zacher M., Nguyen-Viet T.A., Bowers P., Sidorenko J., Karlsson Linnér R., et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet. 2018;50:1112–1121. doi: 10.1038/s41588-018-0147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Davies G., Lam M., Harris S.E., Trampush J.W., Luciano M., Hill W.D., Hagenaars S.P., Ritchie S.J., Marioni R.E., Fawns-Ritchie C., et al. Study of 300,486 individuals identifies 148 independent genetic loci influencing general cognitive function. Nat. Commun. 2018;9:2098. doi: 10.1038/s41467-018-04362-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Malik R., Chauhan G., Traylor M., Sargurupremraj M., Okada Y., Mishra A., Rutten-Jacobs L., Giese A.K., van der Laan S.W., Gretarsdottir S., et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat. Genet. 2018;50:524–537. doi: 10.1038/s41588-018-0058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lambert J.C., Ibrahim-Verbaas C.A., Harold D., Naj A.C., Sims R., Bellenguez C., DeStafano A.L., Bis J.C., Beecham G.W., Grenier-Boley B., et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat. Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ferrari R., Hernandez D.G., Nalls M.A., Rohrer J.D., Ramasamy A., Kwok J.B.J., Dobson-Stone C., Brooks W.S., Schofield P.R., Halliday G.M., et al. Frontotemporal dementia and its subtypes: a genome-wide association study. Lancet Neurol. 2014;13:686–699. doi: 10.1016/S1474-4422(14)70065-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nalls M.A., Blauwendraat C., Vallerga C.L., Heilbron K., Bandres-Ciga S., Chang D., Tan M., Kia D.A., Noyce A.J., Xue A., et al. Expanding Parkinson’s disease genetics: novel risk loci, genomic context, causal insights and heritable risk. bioRxiv. 2019 doi: 10.1101/388165. Preprint at. [DOI] [Google Scholar]
- 77.Duncan L., Yilmaz Z., Gaspar H., Walters R., Goldstein J., Anttila V., Bulik-Sullivan B., Ripke S., Eating Disorders Working Group of the Psychiatric Genomics Consortium. Thornton L., et al. Significant Locus and Metabolic Genetic Correlations Revealed in Genome-Wide Association Study of Anorexia Nervosa. Am. J. Psychiatr. 2017;174:850–858. doi: 10.1176/appi.ajp.2017.16121402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Demontis D., Walters R.K., Martin J., Mattheisen M., Als T.D., Agerbo E., Baldursson G., Belliveau R., Bybjerg-Grauholm J., Bækvad-Hansen M., et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 2019;51:63–75. doi: 10.1038/s41588-018-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Grove J., Ripke S., Als T.D., Mattheisen M., Walters R.K., Won H., Pallesen J., Agerbo E., Andreassen O.A., Anney R., et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 2019;51:431–444. doi: 10.1038/s41588-019-0344-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium Genomic Dissection of Bipolar Disorder and Schizophrenia, Including 28 Subphenotypes. Cell. 2018;173:1705–1715.e16. doi: 10.1016/j.cell.2018.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.van den Berg S.M., de Moor M.H.M., Verweij K.J.H., Krueger R.F., Luciano M., Arias Vasquez A., Matteson L.K., Derringer J., Esko T., Amin N., et al. Meta-analysis of Genome-Wide Association Studies for Extraversion: Findings from the Genetics of Personality Consortium. Behav. Genet. 2016;46:170–182. doi: 10.1007/s10519-015-9735-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hammerschlag A.R., Stringer S., de Leeuw C.A., Sniekers S., Taskesen E., Watanabe K., Blanken T.F., Dekker K., Te Lindert B.H.W., Wassing R., et al. Genome-wide association analysis of insomnia complaints identifies risk genes and genetic overlap with psychiatric and metabolic traits. Nat. Genet. 2017;49:1584–1592. doi: 10.1038/ng.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wray N.R., Ripke S., Mattheisen M., Trzaskowski M., Byrne E.M., Abdellaoui A., Adams M.J., Agerbo E., Air T.M., Andlauer T.M.F., et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 2018;50:668–681. doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Turley P., Walters R.K., Maghzian O., Okbay A., Lee J.J., Fontana M.A., Nguyen-Viet T.A., Wedow R., Zacher M., Furlotte N.A., et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat. Genet. 2018;50:229–237. doi: 10.1038/s41588-017-0009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.International Obsessive Compulsive Disorder Foundation Genetics Collaborative (IOCDF-GC) and OCD Collaborative Genetics Association Studies (OCGAS) Revealing the complex genetic architecture of obsessive-compulsive disorder using meta-analysis. Mol. Psychiatr. 2018;23:1181–1188. doi: 10.1038/mp.2017.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bryois J., Skene N.G., Hansen T.F., Kogelman L.J.A., Watson H.J., Liu Z., Eating Disorders Working Group of the Psychiatric Genomics Consortium. International Headache Genetics Consortium. 23andMe Research Team. Brueggeman L., et al. Genetic identification of cell types underlying brain complex traits yields insights into the etiology of Parkinson's disease. Nat. Genet. 2020;52:482–493. doi: 10.1038/s41588-020-0610-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Finucane H.K., Bulik-Sullivan B., Gusev A., Trynka G., Reshef Y., Loh P.R., Anttila V., Xu H., Zang C., Farh K., et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liang D., Elwell A.L., Aygün N., Lafferty M.J., Krupa O., Cheek K.E., Courtney K.P., Yusupova M., Garrett M.E., Ashley-Koch A., et al. Cell-type specific effects of genetic variation on chromatin accessibility during human neuronal differentiation. bioRxiv. 2020 doi: 10.1101/2020.01.13.904862. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome-wide summary statistics that support the findings of this study will be made available through the CHARGE dbGaP (accession number phs000930) and ENIGMA (http://enigma.ini.usc.edu/research/download-enigma-gwas-results) websites.
No previously unreported custom computer code or mathematical algorithm was used to generate results central to the conclusions.
Any additional information required to re-analyse the data reported in this work paper is available from the lead contact upon request.