Abstract
Introduction
Excess body weight is associated with a state of low-grade chronic inflammation and alterations of the gut microbiome. Powdered meal replacements (PMR) have been shown to be an effective strategy for weight management; however, their effect on inflammation and the gut microbiome remains unclear. The aim of this 12-week randomised control clinical trial is to investigate the effects of PMR consumption, here given as a soy-yoghurt-honey formula, on inflammation, gut microbiome and overall metabolism in individuals with excessive body weight.
Methods and analysis
Healthy adults with excess body weight (n=88) are being recruited and randomly assigned to one of the following groups: (1) Control group (CON): maintaining usual diet for 12 weeks, or (2) PMR group: replacing morning and afternoon snacks daily with a PMR for 12 weeks. Participants are asked to maintain body weight throughout the study and fill out a journal with information about PMR consumption, body weight, food intake, appetite sensations and medications. Three study visits are required: baseline, week 6 and week 12. Outcome measures include systemic inflammatory biomarkers, gut microbiome composition, metabolic blood markers, host energy metabolism, body composition, appetite sensations and host gene expression profile.
Ethics and dissemination
This research protocol was approved by the University of Alberta Ethics Board (Pro00070712) and adheres to the Canadian Tri-Council Policy statement on the use of human participants in research. Procedures and potential risks are fully discussed with participants. Study findings will be disseminated in peer-reviewed journals, conference presentations and social media.
Trial registration number
Keywords: nutrition & dietetics, microbiology, immunology, obesity
STRENGTHS AND LIMITATIONS OF THIS STUDY.
The randomised controlled clinical trial design, coupled with regular assessments and follow-up sessions, as well as a comprehensive range of evaluated outcomes, effectively reduces biases and confounding factors.
Cutting edge technology, such as the metabolic chamber and dual-energy X-ray absorptiometry, enables precise outcome measures.
The exploratory multiomics approach, incorporating gut microbiome, gene expression and genetic polymorphisms, supports the progress of precision nutrition by generating hypothesis.
A primary limitation of the study is the absence of a placebo group and the fact it is not not double-blinded.
Since the gut microbiome analysis depends on faecal samples, it might not fully represent changes in the gut microbiome composition occurring in more proximal parts of the gastrointestinal tract.
Introduction
Excess body weight can be defined as a body mass index (BMI)≥25.0 kg/m2,1 which encompasses both the overweight and obesity categories.2 This condition has been associated with a state of systemic low-grade chronic inflammation, which is characterised by a persistent activation of immune and non-immune cells and production of cytokines, chemokines and acute phase proteins.3 4 Those inflammatory biomarkers include interleukins (IL), such as IL-6 and IL-8, tumour necrosis factor-α (TNF-α), and C-reactive protein (CRP).3 Systemic low-grade chronic inflammation causes tissue and organ damage, which can, in turn, lead to the onset and progression of chronic diseases, such as diabetes mellitus, cancer, metabolic syndrome and cardiovascular diseases.3
In individuals with excessive body weight, the state of systemic low-grade chronic inflammation can be mediated by increased adiposity, as well as by mechanisms through the gut microbiota.3 Increased adipocyte size (ie, hypertrophy) is associated with cellular dysfunction and distress.5 6 Hypertrophic adipocytes secrete an increased number of pro-inflammatory chemokines, such as TNF-α, IL-6, IL-8 and monocyte chemoattractant protein 1.4–6 The increased size of adipocytes and cytokine production lead to adipose tissue hypoxia and death, as well as local and systemic inflammation.4–6
Excess body weight is associated with altered gut microbiome composition and reduced microbiome diversity, which might cause metabolic aberrations and enrich for opportunistic pathogens (eg, at the epithelial interface) that contribute to inflammation.7 8 Individuals with excessive body weight usually present with altered gut permeability, which elevates systemic levels of endotoxins (ie, lipopolysaccharides).3 When in the bloodstream, lipopolysaccharides bind to toll-like receptor 4 leading to activation of nuclear factor kappa B and consequently production of pro-inflammatory cytokines, including IL-6 and TNF-α.8
Considering the numerous negative health outcomes associated with excess body weight, much effort has been made to develop effective weight management strategies. Among those are meal replacements, which are food products fortified with vitamins and minerals used to replace one or more meals per day. Meal replacements are commonly used in association with calorie restriction. Research has shown that the consumption of meal replacements leads to greater weight loss when compared with reduced-calorie diets alone.9 10 Improvement in metabolic parameters is generally observed with weight loss, including improvement in glucose metabolism, reduction of triacylglycerol, low-density lipoprotein cholesterol (LDL-C), systolic blood pressure9 11 12 and the inflammatory markers CRP and IL-6.13 Considering the positive health effects of weight loss in individuals with excessive body weight14–16 and the beneficial health effects of meal replacements,9 11 it is important to differentiate the effects of meal replacements from that of weight loss on overall health, which have not been investigated so far. Therefore, the aim of this study is to compare the effects of a 12-week consumption of a powdered meal replacement (PMR group), given as a soy-yoghurt-honey formula,17 versus usual diet (control group, CON) on inflammation, gut microbiome, overall metabolic health, gene expression profile and genetic background in individuals with excessive body weight who are in weight maintenance.
Methods
Study design and ethical procedures
This study is a randomised, controlled, parallel group, clinical trial conducted at the Human Nutrition Research Unit (HNRU), University of Alberta (Edmonton, AB, Canada). The study is an Investigator Initialized Trial sponsored by the Almased Wellness Comp., Bienenbüttel, Germany. The corresponding research protocol fulfils the requirements of the Standard Protocol Items: Recommendations for Interventional Trials checklist.18 This research protocol was approved by the University of Alberta Health Research Ethics Board (HREB, identifier Pro00070712) and complies with the standards established by the Canadian Tri-Council Policy statement on the use of human participants in research. Procedures and potential risks involved in the study are discussed with participants prior to obtaining informed consent (online supplemental material). This protocol is registered on ClinicalTrials.gov, and recruitment started in April 2019 and is expected to finish in November 2023 (table 1).
Table 1.
WHO trial registration data set
| Data category | Information |
| Primary registry and trial identifying number | ClinicalTrials.gov NCT03235804 |
| Date of registration in primary registry | 1 August 2017 |
| Secondary identifying numbers | University of Alberta Research Ethics Board # Pro00070712 |
| Source(s) of monetary or material support | Almased Wellness-GmbH (Bienenbüttel, Germany) |
| Primary sponsor | Almased Wellness-GmbH (Bienenbüttel, Germany) |
| Secondary sponsor(s) | N/A |
| Contact for public queries | Dr Carla Prado+1 (780) 492–9555 carla.prado@ualberta.ca and Jens Walter+353 (0)21 490–1773 jenswalter@ucc.ie |
| Contact for scientific queries | Dr Carla Prado+1 (780) 492–9555 carla.prado@ualberta.ca and Jens Walter+353 (0)21 490–1773 jenswalter@ucc.ie |
| Public title | The impact of a powdered meal replacement on metabolism and gut microbiota (Premium Study) |
| Scientific title | The impact of a powdered meal replacement on metabolism and gut microbiota: a 12-week study in individuals with excessive body weight (The PREMIUM Study) |
| Countries of recruitment | Canada |
| Health condition(s) or problem(s) studied | Overweight and obesity |
| Intervention(s) | Powdered meal replacement |
| Key inclusion and exclusion criteria | Inclusion criteria: (1) female/male aged 18–50 years; (2) non-smoker; (3) body mass index between 25 and 37 kg/m²; (4) weight stable; (5) fat mass ≥20% for males and ≥25% for females; (6) stable physical activity level. Exclusion criteria: (1) diagnosis of chronic diseases or acute infections; (2) taking any medication that may alter study outcomes; (3) taking prebiotics and probiotics; (4) use of antibiotics in the past 2 months; (5) pregnancy or lactation. |
| Study type | Randomised controlled trial |
| Date of first enrolment | 1 April 2019 |
| Sample size | 88 |
| Recruitment status | Actively recruiting |
| Primary outcome(s) | Interleukin-6 |
| Key secondary outcomes | Gut microbiota |
| Ethics review | University of Alberta Research Ethics Board # Pro00070712 |
| Completion date | N/A |
| Summary results | N/A |
| Individual participant data sharing statement | De-identified data will be shared with the participant on completion of the study (publication) |
bmjopen-2022-070027supp001.pdf (455.9KB, pdf)
Outcome measures
The primary study outcome is to compare changes in IL-6 concentration over time (within groups) between the PMR and CON groups. Secondary outcome is to examine shifts in gut microbiome composition, assessed by relative abundances of amplicon sequence variant (ASV) over time (within groups) between the PMR and CON groups. Exploratory outcomes include:
Remaining gut microbiome diversity indices and relative abundances of bacteria at different taxonomic levels (ie, phylum, family and genus) over time (within groups) between the PMR and CON groups.
Change in markers of systemic inflammation (high-sensitivity CRP (hs-CRP), IL-8 and TNF-α) and immune modulation (IL-10) over time (within groups) between the PMR and CON groups.
Change in concentrations of metabolic blood markers (glucose, insulin, total cholesterol, LDL-C, high-density lipoprotein cholesterol (HDL-C), triglycerides, peptide tyrosine–tyrosine (PYY), glucagon-like peptide-1 (GLP-1), ghrelin, adiponectin, leptin, free glycerol, free fatty acids and thyroid stimulating hormone (TSH)) over time (within groups) between the PMR and CON groups.
Change in resting energy expenditure (REE) and respiratory exchange ratio over time (within groups) between the PMR and CON groups.
Change in body composition (fat mass (FM) and lean soft tissue (LST)) over time (within groups) between the PMR and CON groups.
Change in appetite sensations (hunger, satiety, fullness and prospective food consumption) over time within the PMR group.
Differences in the responses to the intervention according to genetic polymorphisms over time.
Changes in inflammation and excess body weight-related gene expression profile over time (within groups) between the PMR and CON groups.
Research participants
Inclusion criteria are as follows: male or female; non-smoker; between 18 and 50 years of age; BMI between 25.0 and 37.0 kg/m2; with a stable body weight 6 months prior to study initiation (ie, variation <5 kg); FM≥20% for males and ≥25% for females; willingness to maintain stable physical activity level throughout the study; and females must use effective birth control methods.
Exclusion criteria includes participation in >3 hours per week of vigorous physical activity; pregnancy or lactation; diagnosis of any chronic or acute diseases (except for excess body weight); use of any medication that impacts study outcomes, except for antidepressants, anxiolytic and/or thyroid replacement therapy in a stable dose 3 months prior to study initiation and throughout the study period; use of antibiotics 2 months prior to study initiation; use of protein supplements 1 month prior to study initiation; allergy to PMR ingredients (soy, honey and yoghurt); allergy or intolerance to soy, gluten and/or lactose; following a vegetarian, vegan or any other restrictive dietary pattern; claustrophobia; or being unable to comprehend and complete the required questionnaires. Participants consuming supplements or food items that contain prebiotics or probiotics (eg, kefir or kombucha) before being enrolled in the study will be asked to discontinue the use of these products and wait 1 month before starting the study. The use of other nutritional supplements, such as multivitamins and vitamin D3 will be allowed if on a stable dose.
Recruitment, randomisation and intervention
Study advertisement is done using flyers displayed at the University of Alberta campuses, surrounding communities, other post-secondary education institutions in Edmonton (AB, Canada) and healthcare centres in the city. The study is also advertised in University of Alberta email lists, newspapers, classroom presentations and on social media (eg, Kijiji, Facebook and Twitter). Additionally, a personalised website (premium.ualberta.ca) was created.
Individuals interested in being part of the study will be invited to attend a screening visit at the HNRU. This visit will include anthropometric measurements (ie, height, weight and waist circumference), body composition assessment (bioelectrical impedance analysis (BIA)), blood tests (ie, creatinine, estimated glomerular filtration rate (eGFR), albumin, aspartate transaminase (AST), alanine transaminase (ALT), sodium, potassium, chloride and TSH), review of medical history and completion of a Physical Activity Questionnaire. Although glucose, insulin or lipid panel tests are not conducted during the screening visit, individuals who exhibit symptoms of or are taking medications for chronic diseases (eg, diabetes, hypertension and dyslipidaemia) are deemed ineligible for participation.
If deemed eligible, participants are randomly assigned into either the CON or PMR group. Randomisation is stratified by sex using a Microsoft Office Excel spreadsheet. To guarantee impartial allocation of participants to the groups, a study team member created a list of random numbers and assigned them to each group using the website Randomization.com (http://www.jerrydallal.com/random/randomize.htm) with the method of randomly permuted blocks. The list of random numbers is concealed, and a second investigator subsequently follows the predetermined order of numbers and assigns participants to their respective groups based on the order of their screening. Although the investigator has access to the randomisation list, they do not refer to it when assigning participants to their respective groups.
Participants assigned to the CON group are asked to maintain their usual diet for 12 weeks. The ones in the PMR group are asked to replace their morning and afternoon snacks using a PMR (Almased Wellness Comp., Bienenbüttel, Germany) and otherwise maintain their usual diet for 12 weeks. Each snack is replaced by 50 g of powder mixed with 250 mL of water. The nutritional information of the meal replacement is displayed in table 2.
Table 2.
Nutritional information of the tested soy-honey-yoghurt formula, a powdered meal replacement (PMR)
| Nutrient | 50 g of product (PMR) |
| Calories (kcal) | 180 |
| Total fat (g) | 1.0 |
| Saturated fat (g) | 0.5 |
| Trans fat (g) | 0 |
| Polyunsaturated fat (g) | 0.1 |
| Monounsaturated fat (g) | 0.4 |
| Cholesterol (mg) | 3 |
| Total carbohydrates (g) | 15 |
| Dietary fibre (g) | 0.5 |
| Sugars (g) | 15 |
| Protein (g) | 27 |
| Sodium (mg) | 340 |
| Potassium (mg) | 500 |
| Vitamin A (IU) | 794 |
| Vitamin C (mg) | 16 |
| Vitamin E (IU) | 6 |
| Thiamin (vitamin B1) (mg) | 5 |
| Riboflavin (vitamin B2) (mg) | 6 |
| Vitamin B6 (mg) | 7 |
| Calcium (mg) | 215 |
| Iron (mg) | 4.9 |
Experimental protocol
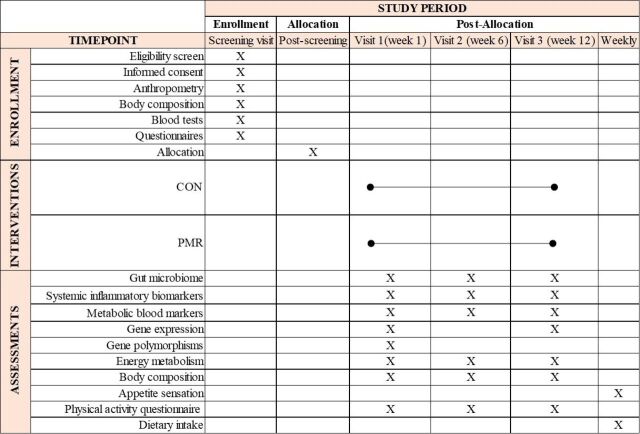
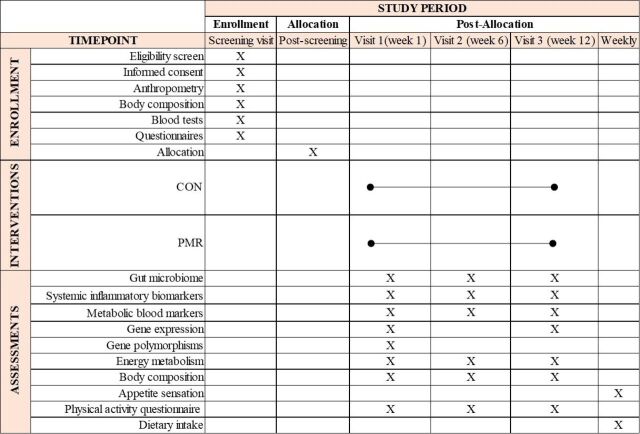
The study design is illustrated in figure 1. The schedule of enrolment, interventions and assessments are shown in figure 2. Following the screening visit and randomisation process, enrolled participants are invited to attend three study visits: baseline, week 6 and week 12. Assessments during each of these visits include: 1-hour resting metabolic rate, blood draw, body composition and Physical Activity Questionnaire. They additionally receive stool collection kits and instructions for faecal sample collection. During the baseline visit, participants receive a scale and a journal to record the following information daily: body weight, date and time of meal replacement intake (PMR group only) and medication intake (if any). They are also asked to record on a weekly basis a 24 hours dietary recall (both groups) and fill out appetite sensation questionnaires (PMR group only). Instructions on how to fill out the journal and dietary records are given. Additionally, participants assigned to the PMR group receive 84 packages of the PMR during visits at baseline and week 6, as well as instructions on how to prepare it. Those assigned to the PMR group start consuming the supplement the day after the first stool sample collection. Study materials (study journal and scale) are returned on week 12.
Figure 1.
Experimental protocol. CON control group, PMR powdered meal replacement group.
Figure 2.
Schedule of enrolment, interventions and assessments (Standard Protocol Items: Recommendations for Interventional Trials figure). CON control group, PMR powdered meal replacement group.
A member of the study team contacts participants weekly to verify adherence to the dietary intervention and potential adverse events. Their body weight is also discussed at that time. If a body weight change greater than ±2% of their initial body weight is noticed, a nutrition consult with a registered dietitian is scheduled to provide instructions on how to increase or decrease food intake and physical activity levels to return to baseline body weight.
Anthropometry and body composition
At the screening visit, anthropometric measurements are taken twice and the average is used for data analysis. Height is measured using a digital stadiometer (235 Heightronic, Concepts, Quick Medical, Snoqualmie, Washington, USA) to the nearest 0.1 cm. Body weight is measured to the nearest 0.1 kg using a calibrated digital scale (Health-o-meter Professional Remote Display, Sunbeam Products, Florida, USA). Waist circumference is measured using a measuring tape at the level of the participant’s belly button, as per standard procedure.19
A digital scale (HD-314 Tanita Corporation, Tokyo, Japan) is provided to participants during the baseline visit, which is returned at the study completion. Body weight is recorded daily in the morning in a fasting state, and with an empty bladder.
Body composition is assessed using dual energy X-ray absorptiometry (DXA, GE Lunar iDXA, General Electric Company, Madison, USA), air displacement plethysmography (ADP, Bod Pod 1SB-060M, Life Measurement Instruments, Concord, California, USA) and BIA (Seca mBCA525, Seca GmbH & Co, Hamburg, Germany). A number of techniques is being used to explore potential changes in body composition using multicompartment modelling: DXA for bone mineral content, BIA for total body water, ADP for body density, which is used to calculate the remaining compartment: adipose tissue and residues (ie, dry LST).20 21
Resting energy expenditure
REE is assessed by indirect calorimetry using an open-circuit metabolic chamber, which measures the volume of oxygen (O2) and carbon dioxide (CO2) from participant’s respiration. Participants lie down in a relaxed position without falling asleep and breathe normally for 60 min. Mixed air with the expired CO2 is drawn from the chamber at a constant flow rate (60±2 L/min) while fresh air with constant O2 is passively drawn into the chamber. The first 30 min of the test are considered time for acclimatisation and hence removed from analysis. Gas exchange (volume of CO2 and O2) is analysed minute-by-minute by the Advance Optima AO2000 Series CO2 analyser (ABB Automation GmbH, Frankfurt, Germany) and the OXYMAT 6 O2 analyser (Siemens AG, Munich, Germany). Data is transferred from those analysers to a computer (Acer Aspire AM3910-E3122, Acer, New Taipei City, Taiwan) via the National Instruments NI USB-6221 device (National Instruments Corporation, Austin, Texas, USA) using the PMCSS Software V.1.8 (Pennington Metabolic Chamber Software Suite, Pennington Biomedical Research Center, Los Angeles, USA). REE (kcal/day) is calculated using the average kcal/min multiplied by 1440.
Blood analysis
Blood is sampled from participants by venipuncture after an overnight fast during the screening visit and at baseline, week 6 and week 12. Evaluated biomarkers are listed in table 3.
Table 3.
Blood parameters, sample and laboratory responsible for blood analysis
| Parameter | Screening | Baseline | Week 6 | Week 12 | Sample | Laboratory |
| Albumin | x | Serum | External | |||
| Creatinine/eGFR | x | Serum | External | |||
| ALT | x | Serum | External | |||
| AST | x | Serum | External | |||
| Electrolytes* | x | Serum | External | |||
| TSH | x | x | x | x | Serum | External |
| hs-CRP | x | x | x | Serum | External | |
| Glucose | x | x | x | Serum | External | |
| Lipid panel† | x | x | x | Serum | External | |
| Free glycerol | x | x | x | Serum | On-site | |
| Free fatty acids | x | x | x | Serum | On-site | |
| Interleukins | x | x | x | Plasma | On-site | |
| TNF-α | x | x | x | Plasma | On-site | |
| Insulin | x | x | x | Plasma | On-site | |
| Leptin | x | x | x | Plasma | On-site | |
| Adiponectin | x | x | x | Plasma | On-site | |
| PYY | x | x | x | Plasma | On-site | |
| GLP-1 | x | x | x | Plasma | On-site | |
| Ghrelin | x | x | x | Plasma | On-site | |
| Polymorphisms | x | Whole blood | On-site | |||
| Gene expression | x | x | Whole blood | On-site |
*Electrolytes include chloride, sodium and potassium.
†Lipid panel include triglycerides, total cholesterol, LDL-C and HDL-C.
‡Interleukins include IL-6, IL-8 and IL-10.
ALT, alanine aminotransferase; AST, aspartate aminotransferase; eGFR, estimated glomerular filtration rate; GLP-1, glucagon like peptide; HDL-C, high-density lipoprotein cholesterol; hs-CRP, high-sensitivity C reactive protein; IL, interleukin; LDL-C, low-density lipoprotein cholesterol; PYY, peptide tyrosine–tyrosine; TNF-α, tumour necrosis factor-α; TSH, thyroid stimulating hormone.
Blood samples are collected using BD Vacutainer tubes (Becton, Dickinson and Company, Franklin Lakes, New Jersey, USA). Tubes containing silica and a polymer are used for serum separation, tubes containing K2-EDTA are used for plasma separation and tubes containing K2-EDTA and protease inhibitors (dipotassium and tacrine, BD P800) are used for GLP-1 and ghrelin analysis.
Creatinine, eGFR, albumin, AST, ALT, sodium, potassium, chloride and TSH are analysed by an external laboratory (DynaLIFE Medical Labs, Edmonton, AB, Canada) at the screening visit prior to enrolment. Glucose, lipid panel (triglycerides, total cholesterol, LDL-C and HDL-C), TSH and hs-CRP will be analysed by DynaLIFE Medical Labs (Edmonton, AB, Canada). IL-6, IL-8, IL-10, TNF-α, insulin, PYY, GLP-1, ghrelin, adiponectin, leptin, free glycerol and free fatty acids will be analysed in our laboratory (University of Alberta, AB, Canada). IL-6, IL-8, IL-10, TNF-α, insulin, PYY, GLP-1, ghrelin, adiponectin and leptin will be analysed by electrochemiluminescence immunoassay (Meso Scale Discovery, Maryland, USA).
An additional blood draw is requested the day following each study visit for hs-CRP analysis due to this being a sensitive marker which can vary substantially within hours of collection for several reasons.22 Therefore, the average of CRP measured on two consecutive days will be taken in case they are similar. If a participant is in an infectious state (ie, CRP>10 mg/L or significant changes between the 2 days measurement) the highest value will be excluded from analysis.
For gene expression profile, RNAs will be sequenced at baseline and week 12. Whole blood (500 µL) is aliquoted into an RNase-free microfuge tube and added 1.3 mL of RNAlater stabilisation solution (Thermo Fisher Scientific, Waltham, Massachusetts, USA). Total RNA will be extracted from whole blood using the RiboPure Blood Kit (Thermo Fisher Scientific, Waltham, USA). The RNA purity will be determined by measuring the 260/280 nm ratio (ideal ratio ~2.0) and the 260/230 nm ratio (ideal ratio 2.0–2.2) using a spectrophotometer. The quality of RNA samples will be evaluated prior to library preparation for RNA sequencing (RNA-seq), using a Bioanalyzer and an RNA Integrity Number ≥7 will be accepted. Samples of high purity and quality RNA will be prepared with the TruSeq RNA Sample Prep kit (Illumina, San Diego, USA). The sequencing will be performed by an external company using the platform Illumina HiSeq 4000 (Illumina, San Diego, USA), in the paired-end mode, in which the two ends will be sequenced with a length of 100 base pairs (bp) (2×100 bp). At the end, two files in the ‘fastq’ format will be generated for each of the evaluated samples.
We will select a set of candidate genes that are known to be involved in the regulation of inflammation and/or excess body weight and are differentially expressed after the intervention. From these genes, we will analyse the most extensively studied polymorphisms. The reasoning behind this approach is that genetic variations in these candidate genes could potentially impact the expression and/or function of the proteins they encode, ultimately influencing the response to the intervention. Genetic polymorphisms will be analysed at baseline. Genomic DNA (gDNA) will be extracted with the QIAamp DNA Micro Kit (Qiagen, Hilden, Germany) from leucocytes in peripheral blood. The gDNA purity will be verified in a spectrophotometer at 260 and 280 nm. The samples will be considered of good quality if the ratio between absorbances is between 1.7 and 2.0. The gDNA concentration will be measured on a fluorimeter. For genotyping, a customised Infinium Global Screening Array-24+v3.0 Kit (Illumina, San Diego, USA) will be used.
Faecal sample collection and gut microbiome sequencing
A total of three faecal samples are collected at baseline, week 6 and week 12. Faecal samples are either collected at the HNRU the day of the study visits, or at home, kept at room temperature, and delivered to the HNRU as soon as possible. During the baseline visit, participants are instructed on how to collect faecal samples using the provided collection kits. The faecal collection tubes (DNA/RNA Shield, Zymo Research, Irvine, California, USA) preserve nucleic acids in the sample and maintain stability at room temperature. Once delivered to the laboratory, the faecal sample tubes are frozen at −80°C until processing and analysis.
The microbial DNA will be extracted from all samples including positive and negative controls, using QIAamp Fast DNA Stool Mini Kit as previously described,23 packed with dried-ice and shipped to University of Minnesota Genomic Center (Minnesota, USA) for sequencing. Shipping will adhere to the regulation of the Environment, Health and Safety Department, University of Alberta. MiSeq Illumina technology (300 bp pair-end) will be used to sequence 16S ribosomal RNA targeting V5–V6 region to characterise the faecal microbiome composition using primer pair 784F [5’-RGGATTAGATACCC −3’] and 1064R [5′-CGACRRCCATGCANCACCT-3′].
Physical Activity Questionnaire
The Godin-Shephard leisure-time Physical Activity Questionnaire will be completed at baseline, week 6 and week 12 to estimate physical activity levels.24 25 In this questionnaire, participants answer how often they perform strenuous, moderate and light exercise for more than 15 min in 1 week. A physical activity score is calculated based on intensity=(9×strenuous)+(5×moderate)+(3×light).25 26 This will be used to classify participants as insufficiently active (<14 units), moderately active (≥14 and <24 units) or active (≥24 units).26
Dietary intake
The dietary intake will be assessed using the online Automated Self-Administered 24-hour Recall (ASA24) Canada.27 A paper-based version is available per individual participant and is returned to the study team weekly by email or fax. Dietary information is entered in ASA24 to ensure consistency. Three 24 hours recalls are completed at weeks 1, 6 and 12 (2 weekdays and 1 weekend day) and one 24 hours recall per week on the remaining weeks of the study period (1 weekday). Energy, macronutrients and micronutrients intake will be obtained using ASA24 automated coding based on the amount of each food consumed.
Appetite sensations
To assess how the PMR affects appetite, participants assigned to the PMR group rate their appetite sensations using the study journals once a week and at five time points: (1) immediately after waking up/fasting, (2) immediately before the morning PMR consumption, (3) 30 min after the morning PMR consumption, (4) immediately before the afternoon PMR consumption and (5) 30 min after the afternoon PMR consumption. Hunger, satiety, fullness and prospective food consumption will be assessed using a paper-and-pen 100 mm Visual Analogue Scale.28 They are instructed to make a single vertical mark between two anchors to indicate the intensity of their subjective states regarding each element, on a scale from 0 to 100 mm. The following questions are asked: How hungry do you feel? (I am not hungry at all—I have never been more hungry); How satisfied do you feel? (I am completely empty—I cannot eat another bite); How full do you feel? (not at all full—totally full); How much do you think you can eat? (nothing at all—a lot).
Adherence and withdraw/discontinuation
Participants are immediately withdrawn from the study if they: (1) have significant variation in body weight (>±3% of baseline body weight29) that does not return to baseline 2 weeks after the nutrition consult; (2) become pregnant; (3) start or change medications or supplement intake listed in the eligibility criteria; (4) no longer meet the inclusion criteria. Participants assigned to the PMR group are asked to return all supplement bags (empty or not) to the visits on week 6 and 12. These are weighted, and participants are excluded from the study if the PMR have not been consumed twice daily during the 12 weeks or if there is >20% of product left inside the bags. In addition, participants can withdraw from the study at any time.
Statistical analyses
Sample size estimate
A total of 74 participants (37 in each group) will be needed to detect a medium effect size (ES) of 0.669. The ES was calculated based on a previously published study,30 in which the mean per cent change in IL-6 from baseline to 12 months was −6.76±36.95 pg/mL in a group receiving soy protein versus 17.62±35.92 pg/mL in the CON. Accounting for a 20% attrition rate, the total sample size of 88 participants (44 in each group) will have a power of 80% with a significance level of 5%. The sample size calculation was done using G*Power V.3.1.9.2.
Data analysis
Normality of the study variables will be assessed by the Shapiro-Wilk W-test. By inspecting boxplots, values >1.5 box-lengths from the edge of the box will be considered as outliers and may be excluded from analysis. Differences between groups of nominal variables will be analysed by Pearson’s χ2 test or Fisher’s exact test. Both group effect and time effect will be analysed using a two-way mixed analysis of variance (ANOVA) or analysis of covariance (ANCOVA) as appropriate. Assumption of homogeneity of variances will be tested using Levene’s test of equality of variances. Correlation between variables will be assessed by Pearson’s correlation. If significant correlations between nutrients and energy intake are noticed, the residual method will be applied in order to describe the relationship between aspects of food intake and biochemical characteristics independent of energy intake.31 All analyses will be performed using IBM SPSS Statistics V.24 (International Business Machines Corporation), considering a critical significance value of 5%, unless otherwise stated.
Regarding genetic polymorphisms analysis, adherence to the Hardy-Weinberg equilibrium will be checked using the χ2 test. The R package ‘argyle’ will be used to analyse the genotype data and assess the potential impact on the responses to the intervention.32 To verify whether the results differ among the genotypes, the dominant model (major allele x heterozygous+minor allele) will be applied. ES will be assessed by Cohen’s d-test and multivariate analysis (MANOVA) will be applied, including time and genotype as the two independent variables. For post hoc analysis, the Steel Dwass test (p<0.05) will be applied. Statistical analysis of the gene expression profile will include the evaluation of the quality of the sequences with the FastQC tool. The Trimmomatic software33 will be used to remove low-quality strings and adapters. Then, the libraries will be evaluated again in the FastQC software, for proper verification. The RNA-seq data will be subjected to analysis by RNA-seq using the protocol described in Trapnell et al .34 The functional annotation of differentially expressed genes will be carried out through the GeneOntology platform (http://geneontology.org). Analyses to identify differentially expressed metabolic pathways will be performed using the fgsea package of the R software.
For the gut microbiome analysis, raw sequencing data will be undergone multiple quality control steps including primer removal, trimming, chimaera removal as previously described.35 36 Sequences will be classified using classify-sklearn algorithm37 against Silva database V.138 generated for given primers by RESCRIPt.38 To decontamination, non-target sequences will be removed such as mitochondria, chloroplast and archaea. Possible contamination detected by positive and negative controls and sequences with raw count <3 and present in <10% of samples will be removed. Sample with an extremely low number of reads (<2000 after filtering) will not be considered in microbiome analysis. Filtered ASV table in count data will be converted to relative abundant data for visualisation, then centred log-ratio transformed. The indices of α-diversity (eg, observed species, Shannon, phylogenetic diversity, Inver Simpson) and β-diversity (eg, Bray-Curtis and Aitchison distance) will be calculated using phyloseq39 and microbiome40 R packages. Bray-Curtis distance will be used to generate non-parametric multidimensional scaling ordination plots for β-diversity metrics with scaled and centred results. Stability over time will also be assessed based on Bray-Curtis distances. The adonis2 function in R package vegan will be used for permutational multivariate analysis of variance (PERMANOVA). Aitchison distance will be used for principal component analysis in mapping microbiome and metabolic markers data to exploring multidimensional association.23 False discovery rate (FDR) will be used to adjust p values, and FDR<0.05 will be considered as statistically significant.
Patient and public involvement
None.
Ethics and dissemination
This study is approved by the University of Alberta’s HREB (Pro00070712) and is registered on ClinicalTrials.gov. This research adheres to the standards as set out in the Canadian Tri-Council Policy statement on the use of human participants in research. This study is regulated by Health Canada. Amendments will be submitted to the HREB and Health Canada review and approval prior to implementations. ClinicalTrials.gov will be updated accordingly.
All personal information is kept private, and participation is anonymous. Participants are assigned a study ID, which is kept separated from any personal information collected. A master list with identifiable information and study IDs is cryptographically protected and stored at the HNRU. The study information will be kept for 15 years after the completion of the study. If participants withdraw consent, they are asked for permission to use the data collected until that point; however, if they deny it, their data is destroyed. Absence of answer is considered as permission to use the data. The Quality Management in Clinical Research (QMCR) Department at the University of Alberta is independent of investigators and sponsor. The QMCR is responsible for monitoring the study data and will conduct yearly auditing.
Following data collection, analysis and review of findings, manuscripts will be prepared for submission to peer-reviewed journals and results presented in national and international conferences. Study findings will also be disseminated through social media. Data will be published regardless of outcomes and the University of Alberta retains the right to publish. Authorship eligibility will adhere to the International Committee of Medical Journal Editors’ recommended guidelines.41 As a mandate of completing a registered trial, the results must be published within 12 months of the completion of the trial. Data set and statistical code may be provided on request.
Supplementary Material
Footnotes
JM and CLPO contributed equally.
Contributors: JM, CLPO, AMA, AB, AMS, LM, CC, SG, CR, NKN, PDC, JW and CMP were involved in the design of the study. JM, CLPO, AMS, JW, and CMP wrote the study protocol. JM, CLPO, AMA, AB, AMS, LM, CC, SG, CR, NKN, PDC, JW and CMP participated in drafting and revising the manuscript. JM, CLPO, AMA, AB, AMS, LM, CC, SG, CR, NKN, PDC, JW and CMP read and approved the final manuscript.
Funding: This was an investigator-initiated trial supported by Almased Wellness-GmbH (Bienenbüttel, Germany). Per contractual agreement, the funder has had no role in the study design and implementation, writing of the manuscript and decision to submit the article for publication. Some of the infrastructure used in the project was funded by the Canadian Foundation for Innovation John R Evans Leaders Fund (Project # 34115). CMP is supported by a Campus Alberta Innovates Program Chair in Nutrition, Food and Health. CLPO is supported by the Mitacs Accelerate International (Mitacs, Canada) in partnership with Almased USA Inc. JM was supported by the Alberta Diabetes Institute.
Competing interests: In addition to what is noted under ‘Funding’, CLPO reports receiving honoraria and/or paid consultancy from Abbott and AMRA Medical Inc. outside the scope of this work. AB received research support for their departments and consultant or speakers’ honoraria from the Almased-Wellness-GmbH. AMS reports receiving honoraria and/or paid consultancy from Novo Nordisk, Johnson & Johnson, Boehringer Ingelheim and Xeno Biosciences outside the scope of this work. PDC is inventor on patent applications dealing with the use of specific bacteria and components in the treatment of different diseases. PDC was co-founder of The Akkermansia Company SA and of Enterosys S.A. JW has received research funding and consulting fees from industry sources involved in the manufacture and marketing of dietary fibres, prebiotics and probiotics. JW is further a co-owner of Synbiotics Health, a developer of synbiotic products. CMP reports receiving honoraria and/or paid consultancy from Abbott Nutrition, Nutricia, Nestle Health Science, Fresenius Kabi, Pfizer, and AMRA medical outside the scope of this work. Other authors declare no conflict of interest.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
Pro00070712
References
- 1. Sung H, Siegel RL, Torre LA, et al. Global patterns in excess body weight and the associated cancer burden. CA Cancer J Clin 2019;69:88–112. 10.3322/caac.21499 [DOI] [PubMed] [Google Scholar]
- 2. World Health Organization . Obesity and overweight. World Health Organization Website.. 2021. Available: https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight
- 3. Furman D, Campisi J, Verdin E, et al. Chronic inflammation in the etiology of disease across the life span. Nat Med 2019;25:1822–32. 10.1038/s41591-019-0675-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Monteiro R, Azevedo I. Chronic inflammation in obesity and the metabolic syndrome. Mediators Inflamm 2010;2010:289645. 10.1155/2010/289645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Karczewski J, Śledzińska E, Baturo A, et al. Obesity and inflammation. Eur Cytokine Netw 2018;29:83–94. 10.1684/ecn.2018.0415 [DOI] [PubMed] [Google Scholar]
- 6. Longo M, Zatterale F, Naderi J, et al. Adipose tissue dysfunction as determinant of Dbesity-associated metabolic complications. Int J Mol Sci 2019;20:2358. 10.3390/ijms20092358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vallianou N, Stratigou T, Christodoulatos GS, et al. Understanding the role of the gut Microbiome and microbial metabolites in obesity and obesity-associated metabolic disorders: Current evidence and perspectives. Curr Obes Rep 2019;8:317–32. 10.1007/s13679-019-00352-2 [DOI] [PubMed] [Google Scholar]
- 8. Boulangé CL, Neves AL, Chilloux J, et al. Impact of the gut Microbiota on inflammation, obesity, and metabolic disease. Genome Med 2016;8:42.:42. 10.1186/s13073-016-0303-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Heymsfield SB, van Mierlo CAJ, van der Knaap HCM, et al. Weight management using a meal replacement strategy: meta and pooling analysis from six studies. Int J Obes 2003;27:537–49. 10.1038/sj.ijo.0802258 [DOI] [PubMed] [Google Scholar]
- 10. Astbury NM, Piernas C, Hartmann-Boyce J, et al. A systematic review and meta-analysis of the effectiveness of meal replacements for weight loss. Obes Rev 2019;20:569–87. 10.1111/obr.12816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kempf K, Schloot NC, Gärtner B, et al. Meal replacement reduces insulin requirement, Hba1C and weight long-term in type 2 diabetes patients with >100 U insulin per day. J Hum Nutr Diet 2014;27 Suppl 2:21–7. 10.1111/jhn.12145 [DOI] [PubMed] [Google Scholar]
- 12. Halle M, Röhling M, Banzer W, et al. Meal replacement by formula diet reduces weight more than a lifestyle intervention alone in patients with overweight or obesity and accompanied cardiovascular risk factors—the ACOORH trial. Eur J Clin Nutr 2021;75:661–9. 10.1038/s41430-020-00783-4 [DOI] [PubMed] [Google Scholar]
- 13. Kempf K, Röhling M, Banzer W, et al. High-protein, low-Glycaemic meal replacement decreases fasting insulin and inflammation markers—A 12-month Subanalysis of the ACOORH trial. Nutrients 2021;13:1433. 10.3390/nu13051433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kolotkin RL, Andersen JR. A systematic review of reviews: exploring the relationship between obesity, weight loss and health-related quality of life. Clin Obes 2017;7:273–89. 10.1111/cob.12203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Forsythe LK, Wallace JMW, Livingstone MBE. Obesity and inflammation: the effects of weight loss. Nutr Res Rev 2008;21:117–33. 10.1017/S0954422408138732 [DOI] [PubMed] [Google Scholar]
- 16. Case CC, Jones PH, Nelson K, et al. Impact of weight loss on the metabolic syndrome. Diabetes Obes Metab 2002;4:407–14. 10.1046/j.1463-1326.2002.00236.x [DOI] [PubMed] [Google Scholar]
- 17. Berg A, McCarthy HD. A soy-Yoghurt-honey product as a therapeutic functional food: mode of action and narrative review. Heliyon 2022;8:e11011. 10.1016/j.heliyon.2022.e11011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chan A-W, Tetzlaff JM, Altman DG, et al. SPIRIT 2013 statement: defining standard protocol items for clinical trials. Ann Intern Med 2013;158:200–7. 10.7326/0003-4819-158-3-201302050-00583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lohman TG, Roche AF, Martorell R. Anthropometric standardization reference manual: human Kinetics books. 1988.
- 20. Siervo M, Jebb SA. Body composition assessment: theory into practice: introduction of Multicompartment models. IEEE Eng Med Biol Mag 2010;29:48–59. 10.1109/MEMB.2009.935471 [DOI] [PubMed] [Google Scholar]
- 21. Fosbøl MØ, Zerahn B. Contemporary methods of body composition measurement. Clin Physiol Funct Imaging 2015;35:81–97. 10.1111/cpf.12152 [DOI] [PubMed] [Google Scholar]
- 22. Pearson TA, Mensah GA, Alexander RW, et al. Markers of inflammation and cardiovascular disease. Circulation 2003;107:499–511. 10.1161/01.CIR.0000052939.59093.45 [DOI] [PubMed] [Google Scholar]
- 23. Nguyen NK, Deehan EC, Zhang Z, et al. Gut Microbiota modulation with long-chain corn bran Arabinoxylan in adults with overweight and obesity is linked to an individualized temporal increase in fecal propionate. Microbiome 2020;8:118. 10.1186/s40168-020-00887-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Godin G, Shephard R. A simple method to assess exercise behavior in the community. Canadian J Appl Sport Sci 1985;10:141–6. [PubMed] [Google Scholar]
- 25. Godin leisure-time exercise questionnaire. Med Sci Sports Exerc 1997;29:36–8. 10.1097/00005768-199706001-00009 [DOI] [Google Scholar]
- 26. Godin G. The Godin-Shephard leisure-time physical activity questionnaire. Health Fitness J Canada 2011;4:18–22. [Google Scholar]
- 27. National Cancer Institute . Asa24 automated self-administered 24-hour recall, epidemiology and Genomics research program, division of cancer control and population sciences. 2015. Available: http://epi.grants.cancer.gov/asa24
- 28. Flint A, Raben A, Blundell J, et al. Reproducibility, power and validity of visual analogue scales in assessment of appetite sensations in single test meal studies. Int J Obes 2000;24:38–48. 10.1038/sj.ijo.0801083 [DOI] [PubMed] [Google Scholar]
- 29. Stevens J, Truesdale KP, McClain JE, et al. The definition of weight maintenance. Int J Obes 2006;30:391–9. 10.1038/sj.ijo.0803175 [DOI] [PubMed] [Google Scholar]
- 30. Mangano KM, Hutchins-Wiese HL, Kenny AM, et al. Soy proteins and Isoflavones reduce Interleukin-6 but not serum lipids in older women: a randomized controlled trial. Nutr Res 2013;33:1026–33. 10.1016/j.nutres.2013.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Willett WC, Howe GR, Kushi LH. Adjustment for total energy intake in epidemiologic studies. Am J Clin Nutr 1997;65:1220S–1228S. 10.1093/ajcn/65.4.1220S [DOI] [PubMed] [Google Scholar]
- 32. Morgan AP. Argyle: an R package for analysis of Illumina Genotyping arrays. G3 Genes|Genomes|Genetics. G3 (Bethesda) 2015;6:281–6. 10.1534/g3.115.023739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible Trimmer for Illumina sequence data. Bioinformatics 2014;30:2114–20. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-Seq experiments with Tophat and cufflinks. Nat Protoc 2012;7:562–78. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bolyen E, Rideout JR, Dillon MR, et al. Reproducible, interactive, Scalable and extensible Microbiome data science using QIIME 2. Nat Biotechnol 2019;37:852–7. 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Callahan BJ, McMurdie PJ, Rosen MJ, et al. Dada2: high-resolution sample inference from Illumina Amplicon data. Nat Methods 2016;13:581–3. 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in python. J Mach Learn Res 2011;12:2825–30. [Google Scholar]
- 38. Robeson MS, O’Rourke DR, Kaehler BD, et al. Rescript: reproducible sequence Taxonomy reference database management. PLOS Comput Biol 2021;17:e1009581. 10.1371/journal.pcbi.1009581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McMurdie PJ, Holmes S. An R package for reproducible interactive analysis and graphics of Microbiome census data. PLoS ONE 2013;8:e61217. 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lahti L, Shetty S, Turaga N, et al. Tools for Microbiome analysis in R. version. 2017.
- 41. International Committee of medical Journal editors . Defining the role of authors and contributors. 2017. Available: https://www.icmje.org/recommendations/browse/roles-and-responsibilities/defining-the-role-of-authors-and-contributors.html
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjopen-2022-070027supp001.pdf (455.9KB, pdf)