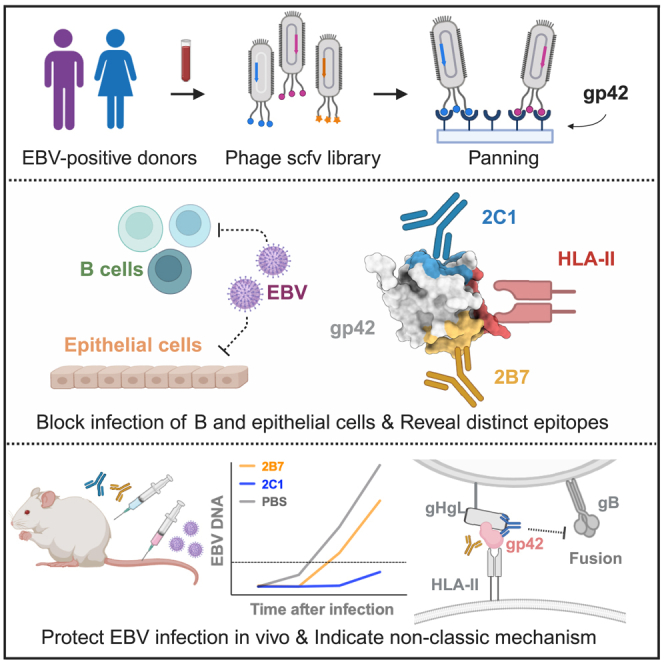
Summary
Epstein-Barr virus (EBV) is linked to various malignancies and autoimmune diseases, posing a significant global health challenge due to the lack of specific treatments or vaccines. Despite its crucial role in EBV infection in B cells, the mechanisms of the glycoprotein gp42 remain elusive. In this study, we construct an antibody phage library from 100 EBV-positive individuals, leading to the identification of two human monoclonal antibodies, 2B7 and 2C1. These antibodies effectively neutralize EBV infection in vitro and in vivo while preserving gp42’s interaction with the human leukocyte antigen class II (HLA-II) receptor. Structural analysis unveils their distinct binding epitopes on gp42, different from the HLA-II binding site. Furthermore, both 2B7 and 2C1 demonstrate potent neutralization of EBV infection in HLA-II-positive epithelial cells, expanding our understanding of gp42’s role. Overall, this study introduces two human anti-gp42 antibodies with potential implications for developing EBV vaccines targeting gp42 epitopes, addressing a critical gap in EBV research.
Keywords: Epstein-Barr virus, human neutralizing antibody, gp42, fusion machinery, monoclonal antibody, epitope, sites of vulnerability, vaccine
Graphical abstract
Highlights
-
•
Human anti-gp42 mAbs can neutralize EBV infection in both B and epithelial cells
-
•
2B7 and 2C1 target different gp42 epitopes, distinct from the HLA-II binding site
-
•
High prevalence of 2C1-like antibodies among individuals infected with EBV
-
•
2C1 has shown significant protection against EBV infection in humanized mice models
Epstein-Barr virus presents a significant global health challenge. Zhao et al. isolate human anti-gp42 mAbs, 2B7 and 2C1, which neutralize EBV in B and epithelial cells by targeting distinct epitopes, demonstrating non-classic neutralizing mechanisms. In humanized mice, mAb 2C1 provides significant protection against EBV infection, showcasing its promising therapeutic potential.
Introduction
More than 90% of individuals are infected with the Epstein-Barr virus (EBV).1 It is linked to various human diseases, including autoimmune disorders such as multiple sclerosis, as well as nasopharyngeal carcinoma (NPC) and B cell malignancies in immunocompromised individuals.2,3,4,5 For instance, individuals undergoing organ transplantation or those with HIV/AIDS are particularly susceptible to EBV-related lymphoproliferative disorders.6 Therefore, developing effective vaccines and targeted drugs against EBV is crucial for public health.
However, despite decades of research, there are currently no available EBV vaccines or specific anti-EBV treatments.7,8,9 This is primarily due to our limited understanding of the mechanisms underlying EBV infection and persistence. Additionally, we have limited knowledge regarding the requirements for protection against EBV, including the specificity and strength of the immune response needed. Neutralizing antibodies have a pivotal role in unraveling the mechanisms of EBV infection, holding promise for effective treatment and vaccine development.10,11,12,13 Therefore, the development of neutralizing antibodies is an urgent priority in order to enhance our understanding of the infection mechanism and to create effective vaccines and targeted drugs against EBV.
Individuals infected with EBV present a diverse repertoire of potent neutralizing antibodies that target EBV-encoded proteins and exhibit effective potency to inhibit both B cell and epithelial cell infection in vitro.14 Nevertheless, the precise range of epitopes and the protection mechanism against EBV infection of these antibodies remain poorly understood. During infection, EBV targets host cells through a complex process involving multiple viral envelope glycoproteins, including gH, gL, gp42, gB, and gp350/220.15 While glycoproteins gH, gL, and gB are required for fusion in both B and epithelial cells, there is a need for an extra glycoprotein gp42 for B cell infection, which combines with gH/gL to form a gH/gL/gp42 complex.16 The N terminus of gp42 binds gH/gL with high affinity, while the C terminus of gp42 interacts with human leukocyte antigen class II (HLA-II), triggering fusion initiated by the interaction between the gH/gL/gp42-HLA-II complex and subsequent gB conformational change.17 A recent finding has shown that incorporating gp42 into gHgL-based subunit vaccines enhances the neutralizing activity and fusion inhibition efficacy of the immune sera, indicating the importance of a gp42-elicited immune response for protection against EBV.18 In addition, gp42 contributes to viral tropism switching.19 Epithelial cell-derived EBV with a high level of gp42 tends to infect B cells, while B cell-derived EBV expressing low levels of gp42 is more likely to infect epithelial cells.19 However, the precise functions of gp42 and the impact of anti-gp42 immune response on both B and epithelial cell infections remain unclear.
The previously reported murine antibody F-2-1 targeting gp42 has been shown to disrupt the binding of HLA-II and gp42. It hinders the fusion of EBV and B cells, exhibiting B cell-neutralizing, but not epithelial cell-neutralizing, potential.20,21,22,23 Monoclonal antibodies (mAbs) targeting the N terminus of gp42 have been developed in rabbits, but these antibodies present no obvious neutralization effects.24 Recently, 5E3, A10, and 4C12, recognizing the C-type lectin domain of gp42, have been isolated from animals immunized with gp42 or gH/gL/gp42 and shown to possess neutralizing activity in B cells.25,26,27 However, to date, there is no report of human antibodies against gp42.
Here, to better understand the role of gp42 in EBV infection, we established a phage-displayed antibody library using peripheral blood samples from EBV-positive volunteers. Through screening, we identified two high-affinity antibodies, 2B7 and 2C1, that target distinct epitopes other than the HLA-II binding site on gp42.
Results
Isolation and identification of high-affinity anti-gp42 antibodies
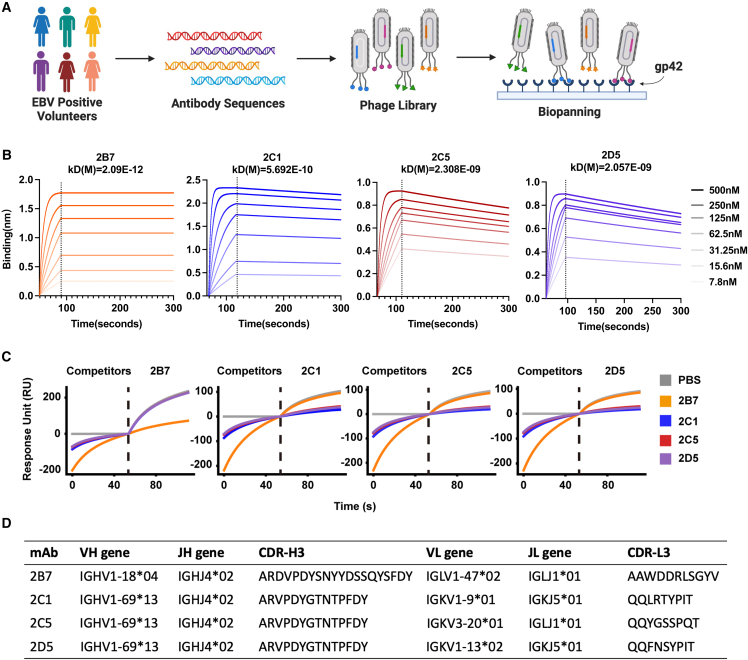
We collected peripheral blood samples from 100 EBV-positive volunteers (Figure 1A). The peripheral blood mononuclear cells of the volunteers were separated for cDNA construction (Figure S1A). To capture the diversity of both heavy (H)- and light (L)-chain sequences, we employed a human immunoglobulin G (IgG) primer set to amplify the variable regions of the H and L chains (vH and vL, respectively) from the cDNA (Figure S1B). Subsequently, we constructed the vL-vH sequence library by linking the amplified sequences together through overlap PCR, incorporating a flexible linker. A phage library with a diversity of 2.9 × 109 was then constructed using the pComb3XSS phage vector (Figures S1C and S1D). The gp42 protein was purified from eukaryotic cells (293F) and used as bait protein for phage screening (Figures 1A and S1E). Following three rounds of screening, the titer of phages binding to the gp42 protein exhibited a 134-fold increase compared to the titer observed with the control (Figure S1F). Finally, 96 single clones were selected to verify the relative affinity of each phage clone for the gp42 protein using ELISA (Figure S1G).
Figure 1.
Isolation and identification of anti-gp42 antibodies
(A) Schematic representation of the phage library generation and screening process. Peripheral blood samples were collected from EBV-positive volunteers. DNA fragments encoding the variable region of the B cell receptor domain were amplified and incorporated into the phage genomes. Following three rounds of screening on immobilized gp42, phages bound to gp42 were isolated and subjected to sequencing.
(B) Measurement of binding kinetics by bio-layer interferometry (BLI) assay for purified 2B7, 2C1, 2C5, and 2D5. KD represents the equilibrium dissociation constant. The biotin-conjugated gp42 protein was immobilized on SA sensors. The range of antibody concentrations was from 500 to 7.8 nM.
(C) Competition assay of purified antibodies (2B7, 2C1, 2C5, and 2D5) using surface plasmon resonance (SPR).
(D) Antibody characteristics of 2B7, 2C1, 2C5, and 2D5. CDRs are defined using the IMGT numbering scheme.28
See also Figure S1.
Sequencing analysis identified four distinct antibody sequences: 2B7, 2C1, 2C5, and 2D5 (Figure S1G). Notably, 2C1, 2C5, and 2D5 shared a very similar H-chain sequence but different L-chain sequences (Figure S1H). To further investigate these antibodies, their sequences were engineered into the human IgG1 eukaryotic expression vector, and the full-length antibodies were expressed. Among the identified antibodies, 2B7 and 2C1 exhibited the highest apparent affinity, with KD values of 2.09 × 10−12 and 5.692 × 10−10 M, respectively, as determined by bio-layer interferometry (BLI) analysis (Figure 1B). Additionally, the binding affinities of 2B7 and 2C1 Fabs to gp42 were assessed. The binding affinity of 2B7 Fab to gp42 is 11.24 nM, while that of 2C1 Fab was measured at 1.89 nM, as depicted in Figure S1I. In competition assays, it was observed that 2B7 did not compete with 2C1, 2C5, or 2D5, suggesting that they bind to distinct epitopes (Figure 1C). However, 2C1, 2C5, and 2D5 antibodies exhibited complete competition with each other, indicating their binding at the same epitope (Figure 1C). The third complementarity-determining region (CDR3) sequences and Ig gene assignments are listed in Figure 1D. Given the superior affinity demonstrated by 2C1, which shares the same CDR3 of the H chain (CDR-H3) and epitope with 2C5 and 2D5, we selected 2C1 and 2B7 antibodies for further investigation.
2B7 and 2C1 inhibit EBV infection in B and HLA-II-positive epithelial cells
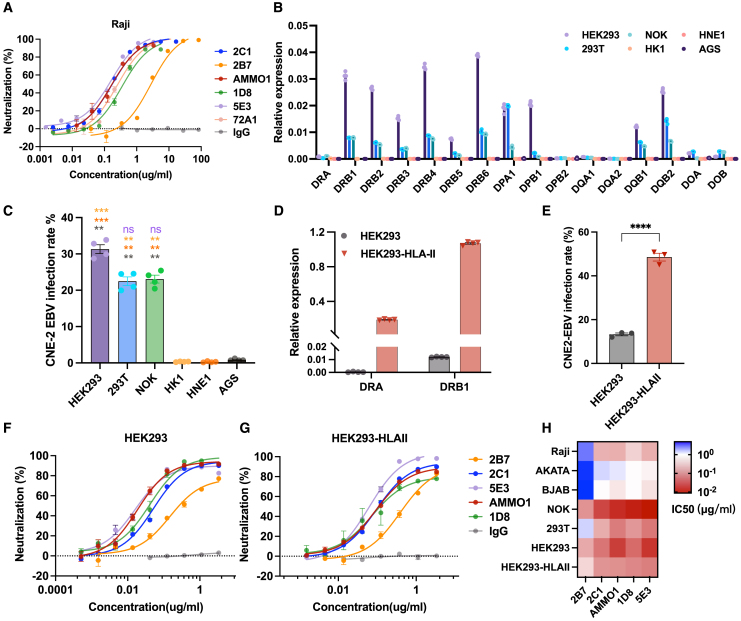
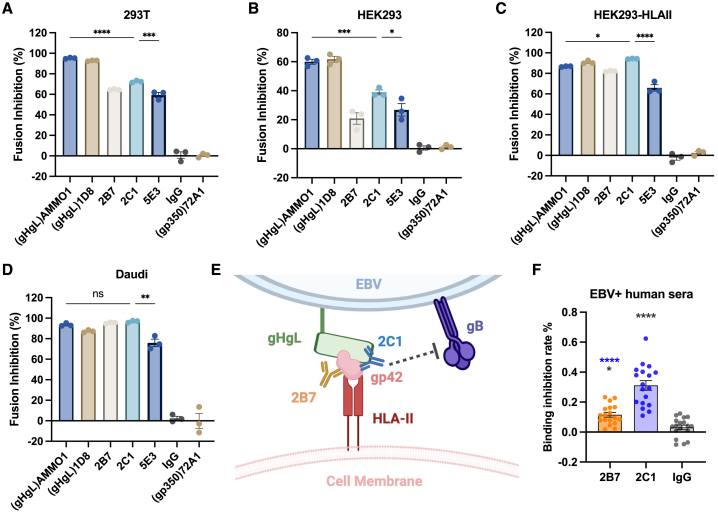
Evaluation of the neutralization potency of 2B7 and 2C1 against EBV infection was initially conducted in B cells (Figures 2A, S2A, and S2B). 2C1 displayed a potent Raji B cell neutralization with an half maximal inhibitory concentration (IC50) value of 0.171 μg/mL, which was comparable to the neutralization ability of the anti-gp350 antibody 72A1 (IC50 = 0.243 μg/mL) (Figure 2A). Given the essential role of gp42 in EBV infection within B cells, which rely on HLA-II-mediated viral entry,17 we extended our investigation to explore whether 2B7 and 2C1 exhibit neutralization effects on HLA-II-positive epithelial cells. To investigate this, we first assessed the HLA-II expression across six epithelial cell lines (HEK293, 293T, NOK, HK1, HNE1, AGS) and observed relatively elevated HLA-II levels in HEK293, 293T, and NOK epithelial cells (Figure 2B). Interestingly, HEK293, 293T, and NOK cells were susceptible to efficient infection by epithelial-derived CNE2-EBV, which is supposed to express a high level of gp42,19 but the HK1, HNE1, and AGS cells are resistant to CNE2-EBV infection (Figure 2C). To further study the impact of HLA-II expression on the infection efficacy of CNE2-EBV in epithelial cells, we established a stable HEK293 cell line expressing HLA-II (HEK293-HLA-II) (Figure 2D). Subsequently, we compared the infection rates of CNE2-EBV in HEK293-HLA-II cells with those in wild-type HEK293 cells. This revealed that the infection rate of CNE2-EBV in HEK293-HLA-II cells was three times higher than in wild-type HEK293 cells, suggesting that HLA-II expression enhances the susceptibility of epithelial cells to CNE2-EBV infection (Figure 2E).
Figure 2.
Neutralization effect of anti-gp42 antibodies in B and epithelial cells
(A) The neutralization effect of 2B7, 2C1, 5E3, AMMO1 (anti-gH/gL), 1D8 (anti-gH/gL), 72A1 (anti-gp350), and control IgG was assessed against EBV infection in Raji B cells (n = 3).
(B) qPCR analysis of human leukocyte antigen class II (HLA-II) gene expression in various epithelial cell lines. The expression levels are presented as relative fold change to the GAPDH gene (n = 4).
(C) CNE2-EBV infection rate in different epithelial cell lines. CNE2-EBV was added to each cell in equal amounts (n = 4).
(D) qPCR analysis of HLA-II gene expression in HEK293 cells and HEK293 cells overexpressing HLA-II (HEK293-HLA-II). The expression levels are presented as relative fold change to the GAPDH gene (n = 4).
(E) CNE2-EBV infection rate in HEK293 cells and HEK293-HLA-II. CNE2-EBV was added to each cell in equal amounts (n = 3).
(F and G) The neutralizing activities of mAbs against CNE2-EBV infection in HEK293 cells (F) or HEK293-HLA-II cells (G) (n = 3).
(H) The IC50 values of mAbs in different B and epithelial cell lines (n = 3).
Significance was calculated using one-way ANOVA followed by Tukey’s multiple comparisons (C) and two-tailed unpaired Student’s t test (E). Error bars represent ±SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
See also Figure S2.
Furthermore, we found that both 2B7 and 2C1 exhibited the capacity to inhibit CNE2-EBV infection in HEK293, HEK293-HLA-II, NOK, and 293T epithelial cells (Figures 1F, 1G, S2C, and S2D). The IC50 values of 2B7 and 2C1 across various B and epithelial cell lines are shown in Figure 2H. In addition, to investigate potential differences in HLA expression among cultured cells, we assessed HLA-II expression in various batches of HEK293 and 293T cells. We observed slight variations between batches and noted a positive correlation between the infection rate across different batches of cells and the expression levels of HLA-II (Figures S2E–S2H). Furthermore, the transfection process did not affect the rates of EBV infection or the neutralizing efficiency of mAbs in HEK293 cells (Figures S2I and S2J). It is noteworthy that these antibodies can neutralize B cell infection against both epithelial-derived CNE2-EBV and B cell-derived AKATA-EBV. However, their epithelial neutralization effect was observed when infected only with CNE2-EBV but not with AKATA-EBV (Figures S2K and S2L).
Cryo-EM structure of the EBV gH/gL/gp42 and 2C1 Fab complex
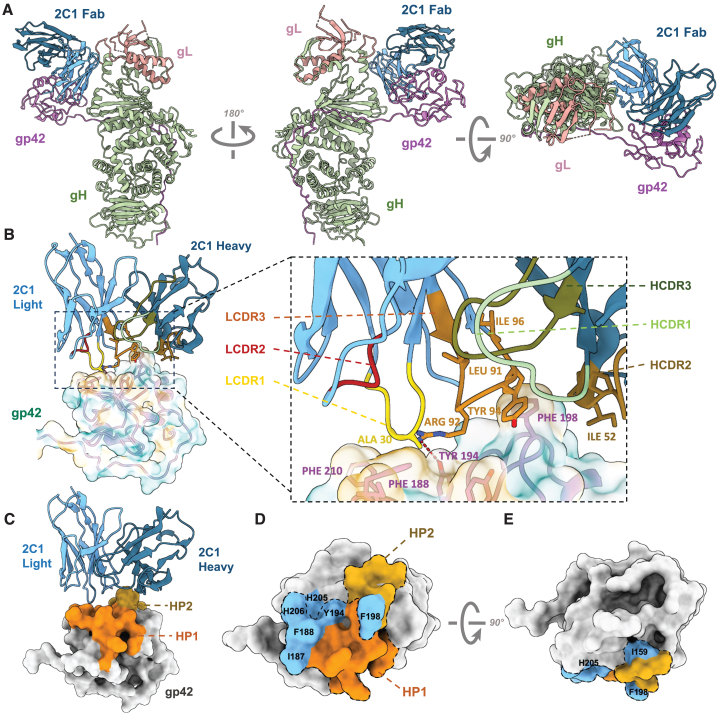
To provide insight into the structural basis for the neutralizing potency of 2C1, we used cryoelectron microscopy (cryo-EM) to elucidate the structure of recombinant gH/gL/gp42 in complex with the Fab fragments of 2C1 at resolutions of 3.3 Å (Figure 3A). Detailed information regarding data collection and EM analysis is provided in the STAR Methods and supplemental information (Figures S3A–S3H; Table S1). The resolution of the gp42/Fab interface was 3.4 Å by local refinement. 2C1 Fab forms a binding interaction with the gH/gL/gp42 protein, covering a total buried surface area of 382.7 Å2. This interaction engages both the vH (99.3 Å2) and vL (283.4 Å2) sequences to recognize gp42. The IMGT numbering system is used to designate the numbering of all antibody residues and CDR loops. Fab 2C1 uses CDR L1 and CDR L3 for its binding with gp42 (Figure 3B). The primary interaction site of the antibody predominantly involves the hydrophobic patches, which are reported as the gp42 canonical ligand binding site (Figures 3C–3E and S3I).29 The region consists mainly of hydrophobic and aromatic residues, which encompass two tyrosines (at positions 185 and 194) and three phenylalanines (at positions 188, 198, and 210), along with a cluster of aliphatic residues (Ile159, Ile187, Val201, and Pro213) within gp42. These residues created a shallow and broad groove above the HLA-DR1 binding site of gp42. Lys197 and Phe198 of gp42 form extensive aromatic residue interactions with the interface of the H chain and L chain, which may play an important role in hydrophobic interaction. These amino acids could provide the conformational signal for triggering viral entry.29 To identify key residues crucial for antibody binding, we introduced a range of single amino acid mutations to the interacting sites on gp42. Given that the hydrophobic patches are believed to interact with 2C1, hydrophobic amino acids were mutated to either alanine or hydrophilic amino acids with similar side-chain lengths (Figure S4). All mutant variants were successfully expressed and purified. Subsequently, we evaluated the impact of these mutations on 2C1 binding using BLI. Among them, mutations in residues I159, I187, F188, Y194, F198, H205, and H206 were found to affect the binding between 2C1 and gp42 (Figure S4). Their positions are indicated in Figures 3C–3E (highlighted in blue). Notably, mutating F198 almost completely abolishes the binding of 2C1 to gp42. Mutations in Y194, I159, and F188 strongly affected the dissociation affinity of 2C1 (Figure S4). The mutation results indicate the prominent role of hydrophobic interactions in the binding between 2C1 and gp42. Furthermore, the key residue Y194 forms a hydrogen bond with R92L (Figure 3B).
Figure 3.
Cryo-EM structure of EBV gH/gL/gp42-2C1 Fab and mutation verification
(A) Structure of EBV gH/gL/gp42-2C1 Fab complex. gH, gL, gp42, 2C1 Fab heavy chain, and 2C1 Fab light chain are colored green, salmon, purple, dark blue, and light blue, respectively.
(B) Structure model and zoomed-in views of the binding site of 2C1 Fab (ribbon representation). The CDRs of the vH (HCDR1-3) and the CDRs of the vL (LCDR1-3) are shown in different colors. The side chains of residues that form the hydrophobic surface (yellow: hydrophobic amino acids, blue: hydrophilic amino acids) of gp42 are displayed.
(C) Structure model of 2C1 Fab (ribbon representation) with EBV gp42 (surface representation). Hydrophobic patches (HPs) are indicated.
(D and E) Different views of overlap between primary functional amino acids and HPs. Residues Ile159, Ile187, Phe188, Tyr194, Phe198, His205, and His206 of the HPs are indicated in blue.
See also Figures S3–S5 and Table S1.
Given the high degree of similarity in the H chains among 2C1, 2C5, and 2D5, we employed tFold-Ab to generate models for these Fabs.30 Despite the less conserved CDR sequences in the L chain compared to the H chain, they demonstrated similar hydrophobic surfaces (Figures S5A and S5B). An analysis of the density map highlighted that F198 inserts into the hydrophobic pocket of 2C1, which is constituted by conserved residues including W47H, I51H, I52H, K63H, and P95L. These residues are common across 2C1, 2C5, and 2D5 (Figures 3B, S5C, and S5D). Examination of the effects of gp42 mutations on 2C5 and 2D5 revealed a similar sensitivity to mutations that influence 2C1 binding (Figures S4C, S5E, and S5F). This suggests that these antibodies are capable of interacting with the hydrophobic pockets of gp42 due to their analogous hydrophobic surfaces. In contrast, 2B7 was not impacted by any of these mutations, implying that the binding mechanism of 2B7 involves distinct residues (Figure S5G).
Additionally, we observed that the gH/gL/gp42 conformation is different compared to the previously reported structure 5T1D.31 In our structure, the two arms of the gH/gL/gp42-2C1 Fab complex demonstrate angles of ∼66° relative to each other, while the angle is ∼33° in 5T1D (Figures S6A and S6B). Earlier research has shown that gH/gL/gp42 complexes can adopt two conformational states: a "closed" state where gH/gL and gp42-HLA-II molecules are closely aligned in one configuration and a more heterogeneous "open" state in the other configuration (Figures S6C and S6D).32 In the closed state, the two arms of the gH/gL/gp42 complex have angles of ∼20° relative to each other, while in the open state, the angles range from ∼40° to 115° relative to each other.32 This indicates that the position of gp42 is relatively flexible in relation to gH/gL and that the change in angle is probably influenced by the gp42 binding ligand.
2B7 and 2C1 recognize two distinct epitopes on gp42
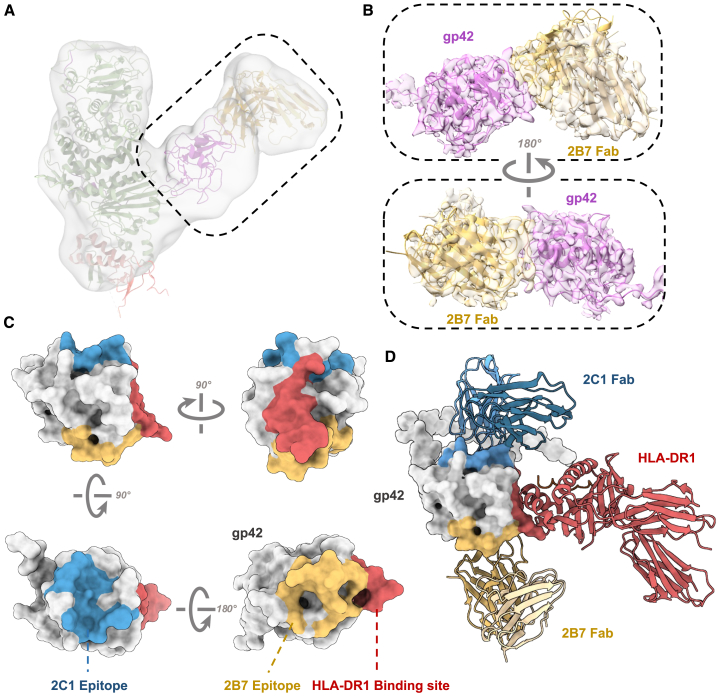
We attempted to obtain the 2B7-gH/gL/gp42 complex structure (Figures S6E and S6F). However, we encountered challenges due to orientation preferences that hindered acquiring the atom model, so we fit the model of the gH/gL/gp42 complex (PDB: 5T1D)31 along with the predicted structure of the 2B7 Fab into a low-pass-filtered map (Figures 4A and 4B). This fitting approach allowed us to discern that the epitope of 2B7 is distinct from the interface where gp42 interacts with HLA-DR1 (Figures 4C and 4D).29
Figure 4.
2B7 and 2C1 antibodies reveal two distinct sites on gp42
(A) The semi-transparent cryo-EM maps of the EBV gH/gL/gp42-2B7 Fab complex. The model structure of EBV gH/gL/gp42/2B7 Fab is aligned with the low-pass-filtered electron density as a reference.
(B) Zoomed-in views of a semi-transparent map superimposed with the atomic models generated by crystal diffraction.
(C) The 2B7 epitope (yellow), 2C1 epitopes (blue), and HLA-DR1 binding site (red) are mapped on gp42 (gray).
(D) Structure model of gp42 (gray) in complex with HLA-DR1 (red), 2C1 Fab (blue), and 2B7 Fab (yellow).
See also Figure S6.
Previously, HLA-II was identified as the EBV gp42 receptor mediating EBV infection of B cells.23 The complex structure of the HLA-II ligand-binding domain with gp42 has been elucidated.29 A comparison of this structure with the structures of the gH/gL/gp42-2C1 Fab and gH/gL/gp42-2B7 Fab complexes reveals that the epitopes of both antibodies are unique and distinct from the HLA-II binding site (data not shown). Additionally, we compared the recently reported antibody 5E3, which competes with HLA-II25 (Figure S6G). To verify whether the antibodies compete with HLA-II for gp42 binding, we purified the ectodomain of HLA-DR1 (Figure S6H). Our findings indicate that unlike 5E3, neither 2B7 nor 2C1 inhibits the binding of HLA-DR1 to gp42 or the gH/gL/gp42 complex (Figures S6I–6L), consistent with the structural analysis. To our surprise, the binding of 2B7 even promotes the binding ability of HLA-DR1 to gp42 and the stability of the HLA-DR1-gp42 complex (Figures S6I and S6J). These findings indicate that the interaction between gp42 and 2B7 might induce a certain extent of conformational change in gp42, which resulted in the increased affinity between gp42 and HLA-DR1.
Furthermore, we investigated whether the presence of antibodies could affect the binding of EphA2 (receptor of EBV gH/gL) and the gH/gL/gp42 complex.33,34 Our findings suggest that the interaction between the gH/gL/gp42 complex and 2B7 or 2C1 does not hinder its ability to interact with the gH/gL receptor EphA2 (Figure S6M).
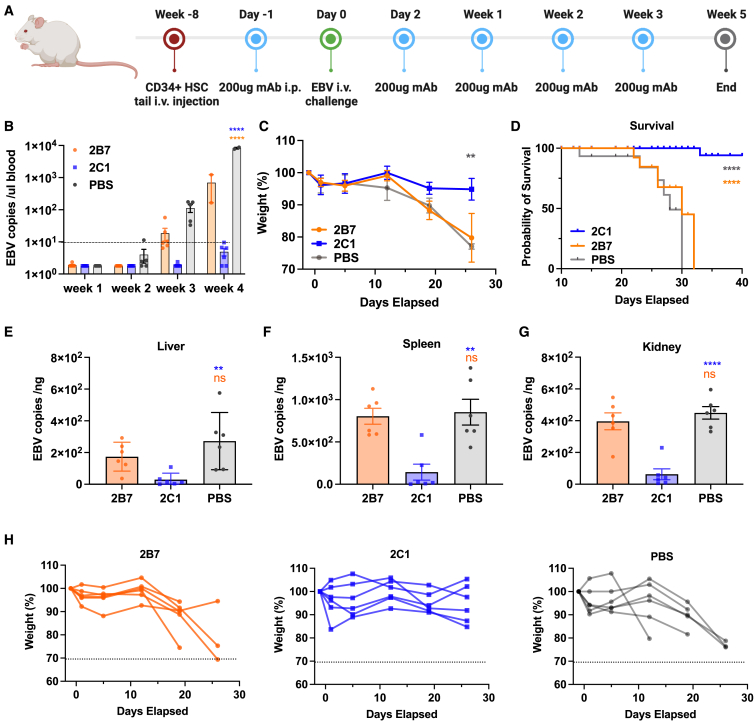
2C1 protects against EBV challenge and EBV-induced lymphoma in humanized mice
To evaluate the in vivo protective efficacy of 2B7 and 2C1, we performed an EBV challenge assay in humanized mice. The NOD-Prkdcnull interleukin-2 receptor γnull mice were engrafted with human CD34+ stem cells. After an 8-week maturation period, the mice were injected with anti-gp42 antibodies and infected with EBV. Each humanized mouse (six per group) received an intraperitoneal administration of 200 μg (in 200 μL PBS) of either 2B7 or 2C1 or 200 μL PBS as negative control. One day later, the mice were intravenously injected with EBV equivalent to 2.5 × 106 green Raji units. Following that, all animals were given 200 μg of the respective antibodies on the second, seventh, fourteenth, and twenty-first days (Figure 5A). To assess the suitability of our dosing regimen, we evaluated serum antibody levels in BALB/c and NSG mice following comparable dosing schedules. The findings revealed that, on average, the concentrations of serum antibodies in mice were similar to those of gp42-specific antibodies in individuals infected with EBV (Figures S7A–S7C).
Figure 5.
2C1 provides effective protection against EBV infection in humanized mice
(A) Timeline represents the engraftment of CD34+ human hematopoietic stem cells, the administration of antibodies, and the EBV challenge. Intraperitoneal administration of 200 μg 2C1 (n = 6), 200 μg 2B7 (n = 6), or 200 μL PBS (negative control, n = 6) was performed in mice for a total of five doses. Following the initial dose, the mice were intravenously challenged with CNE2-EBV.
(B) EBV DNA copies in the peripheral blood of each group of mice following the EBV infection. Each data point represents an individual mouse, with the dotted line indicating the detection limit.
(C) The body weight changes of mice in the 2C1, 2B7, and PBS groups were monitored.
(D) The survival curve illustrates the outcomes of mice from each group following the EBV challenge.
(E–G) EBV DNA copies in the liver (E), spleen (F), and kidney (G) were quantified using real-time qPCR.
(H) The body weight of each mouse in the 2B7, 2C1, or PBS group is indicated.
Significance was calculated using two-way ANOVA followed by Dunnett’s multiple comparisons (B and C), log-rank test (D), or one-way ANOVA followed by Tukey’s multiple comparisons (E–G). Error bars, mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
Two weeks after the EBV challenge, we observed detectable levels of EBV DNA in all mice of the PBS group, with levels increasing rapidly thereafter (Figure 5B). By the third week, overt EBV DNA was detectable in all mice of the 2B7 and PBS groups, while only trace amounts were found in one mouse in the 2C1 group (Figure 5B). In the fourth week, the average EBV DNA copy numbers in the surviving mice of the 2B7 and PBS groups exceeded 100 and 1,000 copies/μL, respectively (Figure 5B). In contrast, EBV DNA copy numbers of mice in the 2C1 remained below 10 copies/μL, notably lower than the copy numbers observed in the 2B7 or PBS group (Figure 5B). Moreover, mice treated with 2B7 or PBS experienced significant weight loss starting from the second week post-challenge (Figure 5C), and all mice in the 2B7 and PBS groups succumbed to the infection within 5 weeks (Figure 5D). Mice treated with 2B7 exhibited similar survival rates and body weight loss compared to the mice treated with PBS (Figures 5C and 5D). In contrast, mice treated with 2C1 demonstrated stable body weight and higher survival rates, with five mice (83.3%) surviving after 6 weeks (Figures 5C and 5D). While 2B7 did not exhibit notable protection in terms of survival rate and body weight, mice treated with 2B7 had lower EBV copy numbers than those treated with PBS. These results indicate that 2B7 has a weaker ability to inhibit EBV infection in vivo (Figure 5B). Notably, the 2C1-treated group exhibited significantly lower EBV DNA copy numbers in the spleen, kidney, and liver in comparison to the 2B7- and PBS-treated mice, indicating a stronger inhibitory effect of 2C1 on EBV infection (Figures 5E–5G). It is worth noting that the spleen generally exhibits higher EBV copy numbers than the liver and kidneys (Figures 5E–5G). The body weight of each mouse is shown in Figure 5H.
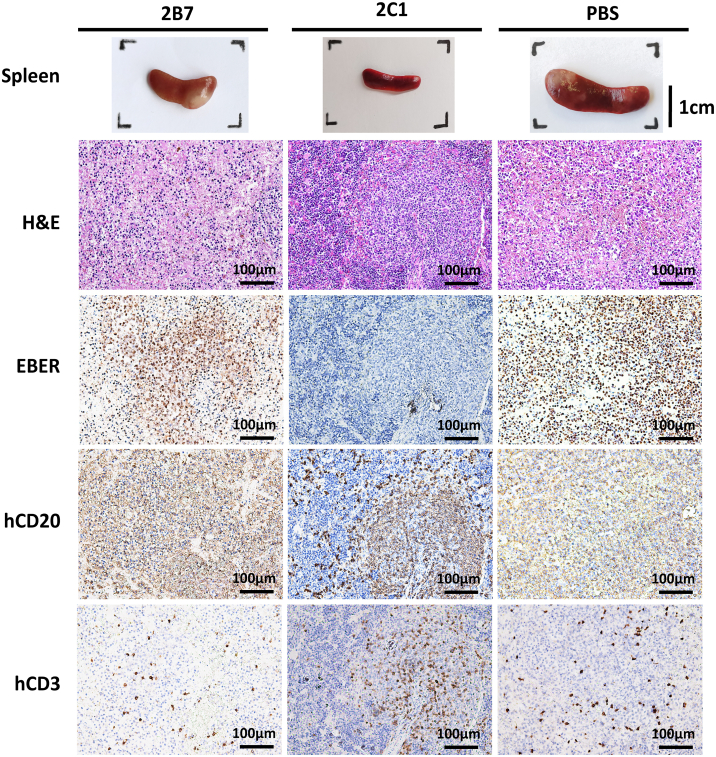
We next performed a pathological analysis in EBV-induced lymphomas during autopsies. All mice treated with 2B7 or PBS showed enlarged spleens with pale tumors on the surface. In contrast, most mice (5/6) in the 2C1 group had normal spleens without visible tumors (Figure 6). We proceeded with histopathological analysis of spleen sections using in situ hybridization for EBV-encoded RNA (EBER), immunohistochemistry for hCD20 and hCD3, and H&E staining. The 2B7- and PBS-treated mice had typical large B cell lymphoma in the white pulp area, with strong positivity for EBER staining and infiltrated by substantial hCD20+ B cells. The lymphomas in the mice treated with 2B7 and PBS disrupted the integrity of the tissue structure. In contrast, in the 2C1 group, the overall structure of the spleen remained intact, with immune cell infiltration primarily concentrated in the white pulp area rather than being diffusely distributed. Additionally, compared to the 2B7 and PBS groups, the 2C1 group exhibited a lower number of cells positive for EBER (Figure 6). Taken together, these findings demonstrate that the 2C1 antibody reduces EBV replication and lymphomagenesis in the spleen compared to the 2B7 and PBS groups, which explains the protective effects of 2C1 against lethal EBV infection in vivo.
Figure 6.
2C1 prevents EBV-induced lymphoma and inhibits viral replication in EBV-challenged mice
Representative images of spleens and splenic sections stained for hematoxylin and eosin (H&E), in situ hybridization targeting EBV-encoded RNA (EBER), and human CD20 and CD3 (hCD20 and hCD3).
2B7 and 2C1 inhibit cell fusion and 2C1-like antibodies are prevalent in human sera
To gain deeper insights, we investigated the effects of 2B7 and 2C1 on the fusion process in both B and epithelial cells using virus-free fusion assays. Our findings revealed that both 2B7 and 2C1 exerted robust inhibition on B cell fusion, with 2C1 showing superior effectiveness, consistent with its potent neutralization capacity (Figure 7A). However, in the epithelial fusion assay, the fusion-inhibitory potency of 2B7 and 2C1 was comparatively modest and weaker than that of gH/gL antibody AMMO1 in 293T and HEK293 cells (Figures 7B and 7C). This observation is consistent with previous research suggesting that gp42 may not play a pivotal role in the epithelial fusion process, and epithelial cell fusion can occur independently of gp42 presence.15
Figure 7.
Inhibition of cell fusion and high prevalence of 2C1-like antibodies in EBV-positive human sera
(A–D) The fusion inhibition effect of AMMO1 (anti-gH/gL), 1D8 (anti-gH/gL), 72A1 (anti-gp350), 2B7, 2C1, and IgG control was assessed using Daudi B cells (A), 293T cells (B), HEK293 cells (C), and HEK293-HLA-II cells (D). The percentage of fusion inhibition was calculated using the formula ((RLU_positive − RLU_mAb)/RLU_positive × 100%). RLU, relative luminescence units. n = 3.
(E) Potential mechanism of 2B7 and 2C1 neutralization. The interaction between gp42 antibodies and the gH/gL/gp42-HLA-II complex may impede the subsequent triggering of gB conformational changes and the membrane fusion process.
(F) Competitive inhibition of anti-gp42 antibodies in EBV-positive human sera by 2B7, 2C1, and IgG control was assessed using SPR (n = 18). See also Figure S7.
Error bars represent ± SEM. Significance was calculated using one-way ANOVA followed by Tukey’s multiple comparisons. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
Surprisingly, we observed a significant enhancement in the fusion-inhibitory potency of 2B7 and 2C1 in HEK293-HLA-II cells compared to HEK293 cells, with 2C1 demonstrating stronger inhibition than the gH/gL antibody AMMO1 (Figure 7D). This suggests that the fusion-inhibitory ability of 2B7 and 2C1 is related to the HLA-II expression levels. Moreover, the fusion inhibitory potency of 2C1 in both B cells and epithelial cells surpassed that of 5E3 (Figures 7A–7D). This suggests that 2C1, targeting gp42 hydrophobic patches, could more effectively inhibit the cell fusion process compared to blocking the HLA-II binding site. Furthermore, considering that gp42 is not needed for EBV binding,16 we hypothesize that the neutralization effect of 2B7 and 2C1 arises from their ability to disrupt the structure of the gH/gL/gp42-HLA-II complex, which is crucial for triggering the conformational change of gB and subsequent membrane fusion17 (Figure 7E).
To determine the prevalence of antibodies resembling 2B7 or 2C1 induced by natural EBV infections, we collected serum samples from 40 individuals who tested positive for EBV. We then used surface plasmon resonance to examine the binding of these serum samples to gp42. Among the collected sera, 18 showed significant binding to gp42, and these samples were subsequently used in the binding inhibition assay. The control IgG displayed a nonspecific inhibition of approximately 3.1% (±1.54%, SEM) of serum antibodies binding to gp42. In contrast, both 2B7 and 2C1 exhibited stronger inhibition, blocking approximately 11.5% (±1.55%, SEM) and 31.1% (±3.21%, SEM) of serum antibodies from binding to gp42, respectively (Figure 7F). The stronger blocking capacity of 2C1 suggests a higher prevalence of 2C1-like antibodies, indicating the widespread presence of antibodies targeting similar epitopes in individuals infected with EBV.
In addition, to further characterize the 18 individuals who tested positive for anti-gp42 antibodies in comparison to the other 22 individuals who tested negative, we collected and analyzed additional data. These data include EBV DNA copy numbers, early antigen IgA (EA-IgA), viral capsid antigen IgA (VCA-IgA), viral bZIP transcription factor IgA (Zta-IgA) (signal-to-cut-off, SCO), EBV nuclear antigen 1 IgA (EBNA1-IgA) (cut-off index, COI), EBNA1-IgG (U/mL), R transactivator IgG (Rta-IgG), and the age of the EBV carriers. We compared these parameters between individuals with positive anti-gp42 (18/40) and those with negative anti-gp42 (22/40). The results showed that lower levels of EA-IgA, VCA-IgA, and Rta-IgG were observed in anti-gp42-negative carriers compared to their positive counterparts (Figures S7D–S7K).
Discussion
EBV gp42 is a membrane protein crucial for facilitating the fusion and entry of EBV into B cells.35 The gp42 protein forms a stable heterotrimer with gH/gL, playing a pivotal role in HLA-II-dependent B cell entry.15 In our research, we successfully identified two anti-gp42 human antibodies, 2B7 and 2C1, which demonstrate potent neutralization of EBV infection in both B and HLA-II-positive epithelial cells. Neutralizing antibodies function by binding vulnerable epitopes of viral proteins. Consequently, identifying the specific epitopes they recognize can provide valuable insights into the mechanisms of virus infection, offering guidance for the rational design of vaccines. Strnad et al. isolated the mouse-derived anti-gp42 antibody F-2-1, which inhibits B cell infection by interrupting the binding of HLA-II with gp42.21,23 Junping et al. generated nine mAbs in mice against the N-terminal region of gp42, targeting the 44–61 or 67–81 aa, but none exhibited neutralization ability.24 Recently, 1A7, 6G7, and 5E3 were isolated from rabbits and showed neutralizing abilities in B cells.25,26 The epitopes of 1A7 and 6G7 were predicted through chimeric gp42 construct binding: 1A7 binds to gp42 residues 149–163, partially overlapping with the HLA-II binding site, while the 6G7 epitope mapped in gp42 174–183 aa, distinct from the HLA-II binding site.26 However, their epitopes were not determined by structural analysis. The structure of 5E3 in complex with gH/gL/gp42 was recently reported, showing a large overlapping region with the HLA-II binding site.25
Our structural data indicate that the epitopes recognized by 2B7 and 2C1 are distinct from the HLA-II binding site and the estimated epitopes of antibodies reported previously. This suggests that the neutralizing effects of 2B7 and 2C1 are not mediated by directly competing with the binding of gp42 and HLA-II. Instead, these antibodies may inhibit infection by hindering the fusion process after HLA-II binding.16 Specifically, the hydrophobic pocket where 2C1 binds is crucial for the EBV fusion process that occurs downstream of receptor binding.32 Studies have demonstrated that mutations within this pocket impede fusion without affecting gp42 binding to HLA-II or gHgL, indicating its functional importance in viral fusion.36,37 In a study by Karthik et al., altering the gp42 residue I159 effectively blocked membrane fusion.32 Intriguingly, this residue is one of the seven critical amino acids involved in the binding of 2C1 to gp42. This suggests that 2C1 may achieve neutralization by occupying the hydrophobic pocket and interfering with the fusion process by perturbing the overall structure and conformation of the gH/gL/gp42-HLA-II complex.17 On the other hand, 2B7 does not target this pocket and exhibits a less potent neutralization and fusion-inhibition effect compared to 2C1 despite its higher affinity for gp42. This indicates that while the binding of 2B7 might influence the gH/gL/gp42-HLA-II-mediated fusion process to some extent, its epitope is not as vulnerable as that of 2C1.
It is worth noting that both 2C1 and 2B7 antibodies displayed potent neutralization of EBV infection in both B and epithelial cells. However, their neutralization effect on epithelial cells appears exclusive to epithelial cell-derived CNE2-EBV, which expresses a high level of gp42.19 This suggests that the inhibition of epithelial cell infection by these two antibodies is dependent on the virion gp42. Moreover, the susceptibility of HEK293, 293T, and NOK cells38 (as they exhibit relatively high HLA-II expression) to CNE2-EBV infection, along with the significantly increased CNE2-EBV infection in cells overexpressing HLA-II (HEK293-HLA-II), strongly suggests that CNE2-EBV can infect epithelial cells via an HLA-II-dependent pathway.39 In addition, the fusion-inhibition effect of 2B7 and 2C1 was notably enhanced in HEK293-HLA-II cells, suggesting that these antibodies exert their inhibitory effect by targeting the HLA-II-dependent fusion pathway.
While the precise mechanisms underlying the EBV neutralization effect of 2B7 and 2C1 remain unclear, the binding of antibodies, particularly 2C1, to gp42 could potentially alter the gH/gL/gp42-HLA-II structure, thereby impeding subsequent membrane fusion processes. Furthermore, our serum inhibition assay indicates a widespread prevalence of 2C1-like antibodies in EBV-infected individuals, suggesting that a similar neutralizing mechanism is commonly employed in physiological contexts.
Prior research has established the inhibitory role of gp42 in the infection of epithelial cells, which do not express HLA-II constitutively.19,40 However, upregulation of HLA-II has been observed in tumor cells of NPC,41,42,43 and the nasopharyngeal epithelium is prone to exposure to epithelial-derived, gp42-rich EBV.44,45 This suggests that the neutralizing effect of the anti-gp42 antibodies on epithelial cells may play a physiological role.
In addition, our findings reveal higher levels of EA-IgA, VCA-IgA, and Rta-IgG in individuals lacking anti-gp42 antibodies compared to those who test positive. Notably, the EA-IgA, VCA-IgA, and Rta-IgG titers are elevated in patients with NPC and are commonly used for NPC screening and early diagnosis.46 Moreover, elevated serum levels of EBV EA-IgA and VCA-IgA have been linked to poor treatment response and prognosis in patients with extranodal natural killer/T cell lymphoma.47 These findings suggest that the presence of gp42-specific antibodies may contribute to better control of EBV reactivation and the related diseases.
Moreover, our unpublished data based on a population study show that the presence of serum gp42 IgG is a protective factor against the risk of developing NPC (X.W. Kong, unpublished data). Notably, individuals with NPC contain lower levels of gp42 IgG in their serum compared to control groups. These results further support that gp42 plays a pivotal role in nasopharyngeal epithelium infection and emphasize its potential as a valuable vaccine target to reduce the risk of EBV infection and NPC. However, the precise binding and entry mechanism of EBV and the specific role of gp42 in B and epithelial cell infection remain unclear, and further investigation is warranted to fully elucidate the relationship between gp42-specific antibodies, EBV infection, and disease control.
Finally, in humanized mice, 2C1 demonstrated significant protection against EBV infection. In contrast, although 2B7 reduced EBV copy numbers, it did not extend the mice’s survival. This may be due to our use of a challenge with 2.5 × 106 Raji units of CNE2-EBV, which exceeds the viral dosage employed in previous studies.11,13 As a result, the mice had relatively short survival times, and the excessive EBV exposure may have overshadowed the protective effect of antibodies with weaker neutralizing capabilities. In addition, the mouse protection data revealed that the 2B7 and 2C1 mAbs were unable to completely prevent viral infection, as evidenced by the detection of the virus in all animal groups after 4 weeks. This could be attributed to several factors, including excessive EBV exposure or the potential inability of the mAbs to achieve complete neutralization of the virus in this experimental setting. Antibodies have been shown to exert various effects beyond direct neutralization, such as limiting viral dissemination or modulating host immune responses.48 Future studies employing additional experimental approaches, such as in vitro viral entry assays or in vivo imaging techniques, could provide further insights into the precise mechanisms by which these antibodies impact viral infection dynamics.
In conclusion, we have successfully isolated the neutralizing human antibodies targeting EBV gp42. Structural analysis revealed two distinct epitopes on gp42, suggesting that anti-gp42 antibodies could hinder the fusion process without competing with HLA-II binding and holding promise for further mechanistic investigations. In addition, 2C1 identifies a previously unknown vulnerable site on gp42, which holds significant implications for the future design of gp42-based EBV vaccines. Furthermore, we demonstrate that 2C1 effectively protects against EBV infecting humanized mice, which can be combined with other EBV antibodies (e.g., AMMO1, 1D8) to develop antibody-based strategies for preventing or treating EBV infection.
Limitations of the study
Humanized mice provide a valuable tool for assessing antibody protection against EBV infection. However, the model has inherent limitations, including differences in immune responses compared to humans and potential variability in tissue tropism. Additionally, the lack of an animal model for EBV-associated malignancies hinders the assessment of antibodies in preventing and mitigating those diseases. Therefore, the development of new animal models is essential for future research. Moreover, while our study illuminates the neutralizing actions of antibodies, the detailed mechanisms through which these antibodies disrupt the fusion process remain partially understood. Further exploration through structural studies or functional assays could unveil the precise molecular interactions at play and inform the creation of more focused therapeutic approaches against EBV infection.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| AMMO1 | Snijder et al.12 | N/A |
| 1D8 | Zhu et al.13 | N/A |
| 72A1 | Hoffman et al.49 | N/A |
| 5E3 | Hong et al.25 | N/A |
| 2B7 | This paper | N/A |
| 2C1 | This paper | N/A |
| 2C5 | This paper | N/A |
| 2D5 | This paper | N/A |
| 2G4 | Audet et al.50 | N/A |
| anti-M13 HRP antibody | NBbiolab | Cat #S004H |
| anti-hCD3 antibody | VENTANA | Cat#790-4341 |
| anti-hCD20 antibody | VENTANA | Cat#760–2531 |
| anti-human IgG HRP antibody | Abcam | Cat# ab97160; RRID:AB_10688252 |
| Goat anti-human IgG | Shuangliu Zhenglong Biochem Lab | Cat#H0111-6 |
| Bacterial and virus strains | ||
| EBV Akata GFP | Zhu et al.13 | N/A |
| EBV CNE2 GFP | Zhang et al.51 | N/A |
| E.coli: TG1 strain | AlpVHHs | Cat# P006 |
| Helper phage: M13KO7 | NEB | Cat# N0315S |
| Chemicals, peptides, and recombinant proteins | ||
| Sodium butyrate | Sigma Aldrich | Cat# V900464 |
| 12-O-tetradecanoylphorbol-13-acetate | Beyotime | Cat# S1819 |
| BsmBI-v2 | NEB | Cat# R0739L |
| Vaccinia Capping Enzyme | Novoprotein | Cat# GMP-M062 |
| 2′-O-Methyltransferase | Novoprotein | Cat# GMP-M072 |
| Critical commercial assays | ||
| Ni Sepharose excel | Cytiva | Cat#17371202 |
| rProtein A Sepharose Fast Flow resin | Cytiva | Cat#17127906 |
| GoScript™ Reverse Transcription System | Promega | Cat# A5001 |
| T7 High Yield RNA Transcription kit | Novoprotein | Cat# E131 |
| Tissue DNA kit | Omega | Cat# D3396-02 |
| EBER detection kit | ZSGB-BIO | Cat# ISH-7001–100 |
| ChamQ Universal SYBR qPCR Master Mix | Vazyme Biotech | Cat# Q711-02 |
| Superdex 200 Increase 10/300 GL | Cytiva | Cat# 28990944 |
| EZ-Link Sulfo-NHS-Biotin | ThermoFisher | Cat#21338 |
| Dual-Glo luciferase assay system | Promega | Cat# E2940 |
| Blood DNA Extraction Kit | TIANGEN | Cat #DP304-03 |
| Standard EBV genome | BDS biotech | Cat#BDS-BW-087 |
| Streptavidin (SA) sensors | ForteBio, Pall LLC | Cat# 18-5019 |
| Protein A sensors | ForteBio, Pall LLC | Cat# 18-5010 |
| Deposited data | ||
| CryoEM Maps of gH/gL/gp42-2C1 complex | This paper | EMD-37249 |
| CryoEM Maps of gH/gL/gp42-2B7 complex | This paper | N/A |
| Atomic Model of gH/gL/gp42-2C1 complex | This paper | PDB 8KHR |
| Original Data | This paper | Research Data Deposit: https://www.researchdata.org.cn/; RDDB2024979584 |
| Atomic Model of gH/gL/gp42/E1D1 complex | Sathiyamoorthy et al.31 | PDB: 5T1D |
| Atomic Model of EBV gH/gL complex | Matsuura et al.52 | PDB: 3PHF |
| Atomic Model of gp42/HLA-DR1 complex | Mullen et al.29 | PDB: 1KG0 |
| Experimental models: Cell lines | ||
| CNE2-EBV | Zhang et al.53 | N/A |
| 293T | ATCC | CRL-3216 |
| 293F | Thermofisher | Cat# R79007 |
| NOK | Piboonniyom et al.54 | N/A |
| HK1 | Huang et al.55 | N/A |
| HNE1 | Zhan et al.56 | N/A |
| AGS | ATCC | CRL-1739 |
| HEK293 | ATCC | CRL-1573 |
| HEK293-HLAII | This paper | N/A |
| Raji | ATCC | CCL-86 |
| Bjab | Klein et al.57 | N/A |
| Daudi | ATCC | CCL-213 |
| Akata-EBV | Molesworth et al.58 | N/A |
| Akata | Wang et al.59 | N/A |
| Experimental models: Organisms/strains | ||
| NOD-Prkdcscid IL2Rγnull mice (NPG®) | Beijing Vitalstar Biotech Co., Ltd. | N/A |
| Oligonucleotides | ||
| Forward primer specific for EBV BALF5 gene: 5′- CCCAACACTCCACCACACC -3′ | This paper | N/A |
| Reverse primer specific for EBV BALF5 gene: 5′- TCTTAGGAGCTGTCCGAGGG -3′ | This paper | N/A |
| Primers for HLA-II qPCR assay, See Table S1 | This paper | N/A |
| Recombinant DNA | ||
| pCAGGS-gH | Hann et al.60 | N/A |
| pCAGGS-gL | Hann et al.60 | N/A |
| pCAGGS-gB | Hann et al.60 | N/A |
| pCAGGS-gp42 | This paper | N/A |
| pCAG-T7 | Okuma et al.61 | N/A |
| pT7EMCluc | Okuma et al.61 | N/A |
| pcDNA3.1-gL-Linker-gH-His | Zhu et al.13 | N/A |
| pcDNA3.1-gL-Linker-gH | This paper | N/A |
| pCRC8400-gp42(34-223aa)-His | This paper | N/A |
| pLVX-HLA-DRA1 | This paper | N/A |
| pLVX-HLA-DRB1 | This paper | N/A |
| pcDNA3.1-HLA-DRA1∗01:01 | Walker et al.62 | N/A |
| pcDNA3.1-HLA-DRB1∗04:01 | Walker et al.62 | N/A |
| pcDNA3.1-EphA2 | Zhu et al.13 | N/A |
| pUC57-T7-T7 polymerase-poly(A) | This paper | N/A |
| Pcomb3XSS | Andris-Widhopf et al.63 | Addgene Plasmid, #63890; RRID: Addgene_63890 |
| Software and algorithms | ||
| GraphPad Prism version 8 | Graphpad | https://www.graphpad.com |
| ChimeraX | Goddard et al.64 | https://www.cgl.ucsf.edu/chimerax/ |
| Chimera | Pettersen et al.65 | https://www.cgl.ucsf.edu/chimera/ |
| cryoSPARC v3 | Punjani et al.66 | https://cryosparc.com |
| Coot | Emsley et al.67 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Phenix | Adams et al.68 | http://phenix-online.org |
| MolProbity | Chen et al.69 | http://molprobity.biochem.duke.edu/ |
| Alpha Fold2 | Jumper et al.70 | https://alphafold.ebi.ac.uk/ |
| PyMOL | Schrödinger, Inc.71 | https://pymol.org/ |
| Biorender | Biorender | http://www.biorender.com |
| Other | ||
| Biacore™ 8K | Cytiva | https://www.cytivalifesciences.com/en/us/shop/protein-analysis/spr-label-freeanalysis/spr-systems/biacore-8k-p-05540 |
| ÄKTA pure™ 25M | Cytiva | https://www.cytivalifesciences.com/en/us/shop/chromatography/chromatography-systems/akta-pure-p-05844 |
| Octet RED96 | Forte'Bio, Pall LLC | https://www.sartorius.com/en/products/biolayer-interferometry |
| CytoFLEX | Beckman Coulter | https://www.beckman.com/flow-cytometry/research-flow-cytometers/cytoflex |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Mu-Sheng Zeng (zengmsh@sysucc.org.cn).
Materials availability
For all requests concerning resources and reagents, please contact the lead contact author. All reagents, including antibodies, proteins, plasmids, and viruses, will be made available upon request after the completion of a Material Transfer Agreement for non-commercial usage.
Data and code availability
-
•
The cryo-EM map of the EBV gH/gL/gp42-2C1 Fab has been submitted to the Electron Microscopy DataBank (EMDB:EMD-37249). The atomic coordinates of the complex have been submitted and are available in the Protein Data Bank (PDB:8KHR).
-
•
This paper does not report the original code.
-
•
The original data has been uploaded to Research Data Deposit: https://www.researchdata.org.cn/ with accession number RDDB2024979584. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Cell lines
Cell lines were maintained in a humidified atmosphere at 37°C with 5% CO2. NOK cells, HEK293T cells, HEK293 cells, and CNE2 cells were cultured in DMEM supplemented with 10% FBS. BJAB B cells, Raji B cells, AGS cells, and EBV-negative AKATA (AKATA-) cells were cultured in RPMI 1640 medium (Gibco) supplemented with 10% FBS. 293F cells were maintained with SMM 293-TII Expression Medium (Sino Biological). For the establishment of HLA-DR over-expressing cell lines, lentiviruses expressing full-length HLA-DRA1 and HLA-DRB1, along with Puromycin and Neomycin (G418) selection markers, were prepared using 293T cell lines. HEK293 cells were infected with the HLA-DRA1 lentivirus. Then, 48 h post-infection, Puromycin was added and maintained for two weeks. Subsequently, these cells were infected with the HLA-DRB1 lentivirus. Again, 48 h post-infection, Puromycin was replaced with G418 for another two weeks. Overexpression of HLA-DRA1 and HLA-DRB1 was confirmed by Western blot analysis. 100 mg/mL streptomycin and 100 U/mL penicillin were added to all cell culture media.
Human specimens
Peripheral blood mononuclear cell (PBMC) samples were collected from 100 EBV-positive participants in 2020 and 2021 in Guangdong, China. Sera from EBV-positive individuals (n = 40) were collected from Sun Yat-sen University Cancer Center in 2022 and 2023 in Guangdong, China. The sample size was determined both by empirical and clinical availability. Samples were not artificially assigned to experimental or control groups. Instead, the grouping in Figures S7D–S7K was determined solely by the detection results of the anti-gp42 antibody. Ethical approval for the use of human specimens was granted by the Ethics Review Committee of Sun Yat-sen University Cancer Center (SYSUCC, G2022-145-01), and the study adhered to the principles outlined in the Declaration of Helsinki. All participants involved in human research provided written informed consent.
Mice
Animal experiments involving infectious EBV were conducted within the animal biosafety level 2 facilities at Zhongshan School of Medicine, following approval by the Sun Yat-sen University Committee on the Animal Experiments Ethics (SYSUCC, L102012021020P). The animal studies strictly followed the ARRIVE guidelines (Animal Research: Reporting of In Vivo Experiments). NOD-Prkdcscid IL2Rγnull mice (NPG) were employed for generating humanized mice acquired from Beijing Vitalstar Biotechnology Co., Ltd., and they were kept in a specific pathogen-free (SPF) environment. All mice were housed under controlled conditions with a temperature of 25 ± 2°C, humidity maintained at 50%, and a 12-h light-dark cycle.
Method details
Scfv library construction and panning procedure
Pcomb3XSS vector was a gift from Carlos Barbas (RRID: Addgene_63890), used to construct the scfv phage library as described by previous studies.63,72,73 In brief, fragments encoding single-chain variable fragments (scFv) were obtained through PCR amplification from PBMC cDNA. These fragments were then ligated into the Pcomb3XSS vector and transformed into E coli (TG1 strain). The diversity of the library was estimated to encompass approximately 2.9 × 109 distinct clones. Subsequently, this library underwent panning against purified gp42 antigen. Immuno tubes (Nunc-Immuno MaxiSorp, Thermo Scientific) were incubated with 1mL 10 μg/mL gp42 protein (dissolved in PBS) overnight at 4°C. Following this, 2 mL of a 3% bovine serum albumin (BSA) solution (w/v, dissolved in PBS) was used for the subsequent blocking by incubating the tube at 37°C for 1 h. Subsequently, the tubes were washed with PBS–0.1% Tween 20 (PBST) five times, and the scfv phage library (dissolved in the blocking buffer) was added to the tubes, with approximately 1011 phage particles in each tube. The plate underwent incubation for 1 h at 37°C, followed by 20 washes with PBST to eliminate unbound phages. To elute the bound phages, 500 μL of elution buffer (containing 0.25 mg/mL trypsin) was added to each well and incubated at 37°C for 10 min. The eluted phage was mixed with log-phase E. coli TG1 cells and cultivated in 2×YT medium, which was supplemented with 100 μg/mL ampicillin. Once the medium reached an OD600 of 0.5, helper phage M13KO7 (NEB, N0315S) was added at a multiplicity of infection of 20:1 and allowed to incubate at 37°C for an additional 30 min. Subsequently, kanamycin was added at a final concentration of 50 μg/mL, and the cultures were incubated overnight at 30°C with shaking. This panning protocol was repeated for two to three rounds and individually amplified phage clones were assessed for their capacity to bind to gp42 using Enzyme-linked immunosorbent assay (ELISA).74 Briefly, individual phage clones are generated in a 96-well format and incubated onto a 96-well plate coated with gp42. Following incubation and washing, the remaining phages on the plate were detected using an anti-M13 HRP antibody (NBbiolab, Cat #S004H) and read using a BioTek Epoch microplate spectrophotometer.
Plasmids
The EBV protein genes were amplified from the EBV M81 strain (GenBank: KF373730.1). The ectodomain of gp42 (34–223 aa), gH(aa 19–678)/gL(24–137 aa) and EphA2(28–530 aa) were inserted into a pCDNA3.1 vector, with an N-terminal CD5 signal peptide and a C-terminal 6× His tag. The gH and gL were linked with (G4S)3 linker. A gH/gL construct lacking the His tag was employed to purify the gH/gL/gp42 complex. Plasmids encoding the soluble form of HLA-DR1 (HLA-DRA1∗01:01/DRB1∗04:01) were constructed following a previous report.62 pCAGGS-gB, pCAGGS-gH, pCAGGS-gL, pCAG-T7, and pT7EMCLuc were generously given by Professor R. Longnecker (Northwestern University) and Wolfgang Hammerschmidt (Helmholtz Zentrum München). The pUC57 plasmid was engineered to contain the mRNA template encoding T7 RNA polymerase. This construct includes a T7 promoter sequence, a 5′ untranslated region (UTR) at the N-terminal, a 3' UTR, and a 120-poly-A tail at the C-terminal, denoted as pUC57-T7-T7 polymerase-poly(A). Separate primer pairs were utilized to obtain the variable domain of antibody heavy chain and light chain (vH and vL, respectively). PCR products were cloned into antibody expression vectors with human IgG1 constant regions. Sanger sequencing was conducted to verify the recombinant plasmids.
Soluble protein purification
Soluble gp42, gH/gL, gH/gL/gp42, HLA-DR1, EphA2, recombinant antibodies, and Fabs were purified by 293F protein expression system cultured with chemically defined medium (UP1000, Union Bio) under CO2 supplied shaking incubator.13,26 The gH/gL/gp42 complex was purified by co-transfect 293F with soluble gp42(with his tag) and soluble gH/gL(no tag) plasmids. Six to seven days post-transfection, the supernatant was collected, filtered through a 0.22um filter, and purified with Ni Sepharose Excel (17371202, Cytiva).75 Eluate was further purified by Superdex 200 Increase 10/300 GL (28990944, Cytiva). Recombinant antibodies were purified by passing the supernatant through rProtein A Sepharose Fast Flow resin (17127906, Cytiva), followed by PBS washing and subsequent elution with 15 mL of 0.2M glycine (pH 3.0). The elution buffer was immediately neutralized with 2 mL of 1M Tris HCl (pH 8.0). After purification, the antibodies were dialyzed into PBS. Purified proteins underwent additional purification through size exclusion chromatography using ÄKTA pure 25M (Cytiva Life Sciences). Protein concentration was determined by NanoDrop One Spectrophotometer (Thermo Scientific). After purification, the recombinant protein was aliquoted, rapidly frozen using liquid nitrogen, and then stored at −80°C for further use.
Recombinant protein biotinylation
Purified HLA-DR1, gp42, and gH/gL/gp42 were biotinylated at a theoretical 3:1 biotin/protein ratio. This biotinylation process was carried out using EZ-Link Sulfo-NHS-Biotin (ThermoFisher Cat#21338) for 30 min at 37oC. The sample underwent three consecutive rounds of buffer replacement with PBS using MilliporeSigma Amicon Ultra-2 Centrifugal Filter Units to eliminate excess unbound biotin.
Bio-layer interferometry (BLI)
Interactions between purified gp42 or gH/gL/gp42 proteins with antibodies and gp42 with HLA-DR1 were assessed using an Octet RED96 instrument (Forte'Bio, Pall LLC). The purified gp42 or gH/gL/gp42 proteins were biotinylated and immobilized on streptavidin (SA) sensors (ForteBio, Pall LLC). The sensors were then equilibrated in KB buffer and subjected to target protein association and subsequent dissociation in KB buffer (0.1% BSA [w/v] and 0.02% Tween 20[v/v] in PBS). In each experiment, a sensor loaded with biotinylated protein but immersed in KB buffer during association and dissociation served as a background reference. For competition binding assays, 10ug/ml biotinylated gp42 or biotinylated gH/gL/gp42 was immobilized onto the streptavidin sensors (ForteBio, Pall LLC). KB buffer was used to establish the baseline interference. Subsequently, the sensors were associated with 500nM 2C1, 100nM 2B7, or KB buffer (blank), followed by equilibrium in KB buffer and subsequent association with 500 nM 2C1, 100 nM 2B7, or 10 μM HLA-DR1. Dissociation was carried out with KB buffer. One sensor loaded with antigens was placed in the KB buffer as a reference during both the association and dissociation steps. This reference sensor was used to subtract background signals. For measuring 2C1 affinities to variant mutations of gp42, Protein A sensors (ForteBio, Pall LLC) were employed to capture 10ug/ml 2C1. As analytes, 1000 nM gp42 with amino acid substitutions was utilized. All proteins subjected to BLI analysis were diluted in KB buffer.
Surface plasmon resonance (SPR)
SPR was performed with Biacore 8K equipment (Cytiva). Soluble gp42 was covalently coupled onto the flow cell 2 of a CM5 chip, and the reference flow cells (flow cell 1) were not modified. Purified antibodies were diluted using a running buffer containing 150 mM NaCl, 100 mM HEPES-NaOH (pH 7.4), 30 mM EDTA, and 0.5% TWEEN 20 to the indicated concentration. In a multi-cycle kinetics experiment, the interaction between gp42 and antibodies was assessed by introducing antibodies at decreasing concentrations with an injection rate of 30 μL/min (analyte contact time was set to 60 s) at 25°C. Following each reaction, 50 mM NaOH was injected to regenerate the chip. Kinetics were fitted by Biacore Evaluation Software 3.0.12.15655. To perform competitive analysis with SPR, antibodies were injected sequentially without regeneration in between (contact time was set to 60 s). To evaluate the strength of competition, reaction units (RUs) of a specific antibody pair were subtracted from the RUs of a specific control pair (running buffer followed by antibodies). To assess the competition binding of 2B7 and 2C1 with antibodies in human serum, we determined the corrected RUs of a serum sample after 2B7, 2C1, and IgG contact (mAb_RU). The corrected RUs (Control_RU) were obtained by subtracting the RUs of a corresponding control pair, which involved a running buffer followed by the corresponding serum sample. The binding inhibition rate was calculated as the percentages of the change in response units (ΔRUs): ΔRUs % = (Control_RU - mAb_RU)/Control_RU x 100%. The ΔRUs % served as the indicators of the strength of competition, with higher ΔRUs % indicating stronger competition.
Virus production
CNE2-EBV was produced from CNE2 epithelial cells infected with EBV-GFP. Upon reaching 90% density within a 10 cm dish, NaB (sodium butyrate) and TPA (12-O-tetradecanoylphorbol-13-acetate) were added to the medium to induce the release of EBV virus. After 12 h, the medium was replaced. Under sterile conditions, the culture supernatant was collected to isolate and purify the virus. The supernatant was directly aspirated and centrifugated, followed by filtration through a 0.45 μm filter. Subsequently, the virus underwent a 100-fold concentration through high-speed centrifugation at 50,000 g, after which it was resuspended with RPMI 1640 medium (free of FBS) and stored at −80°C until use. AKATA-EBV was produced as described previously.13 Briefly, Akata-EBV-GFP cells were suspended in RPMI 1640 medium and treated with goat anti-human IgG (Shuangliu Zhenglong Biochem Lab Cat#H0111-6) for 6h to induce Epstein-Barr virus (EBV) production. After incubation, Akata-EBV-GFP cells were cultured in RPMI 1640 with 5% FBS for another 72h, and the virus-containing supernatant was collected, filtered, and concentrated as CNE2-EBV.
Neutralization assay
Before the neutralization assay, the virus was titrated on various cell lines. For B cell neutralization, antibodies were added in serial dilutions to 96-well plates, each containing 60 μL RPMI 1640. Subsequently, each well was added with 20 μL of CNE2-EBV-GFP (in RPMI 1640), followed by an incubation at 37°C for 2 h. Then,130 μL of fresh 10% FBS RPMI 1640 containing 1 × 104 B cells (Raji, Akata, or BJAB B cells) were added into each well. Incubation then continued at 37°C for 48 h. For epithelial cell neutralization, the process is similar except that the resulting mixture was added to 96-well plates containing 8 x 103 epithelial cells per well. 2G450 was consistently used as the IgG control throughout the study. Subsequent incubation at 37°C for 48 h followed. Flow cytometry was used to determine the percentage of GFP-positive cells. Uninfected cells served as negative controls, while cells infected by EBV without antibodies were used as positive controls. The percentage of neutralization was defined as [1 - (infection rate of cells in antibody-added well/infection rate of the positive control)] × 100%. Data were analyzed and calculated using non-linear regression analysis to determine the IC50 value.
Virus-free fusion inhibition assay
Effector cells (293T cells) were transfected with pCAGGS-gB, pCAGGS-gH, pCAGGS-gL, pCAGGS-gp42, and pT7EMCLuc (2 μg for each plasmid). 24 h later, effector cells (2 × 105 cells/test) were incubated with 2.5 μg of the respective antibodies (AMMO1, 1D8, 2B7, 2C1, or IgG control) per test at 37°C for 30 min. Then, the effector cells were mixed with either target epithelial cells (293T, HEK293, or HEK293-HLAII cells) or target Daudi B cells (2 × 105 cells/test). Target epithelial cells were transfected with a plasmid encoding T7 polymerase (pCAG-T7). For transfection of Daudi B cells, mRNA encoding T7 polymerase was synthesized in vitro and introduced into Daudi B cells via electroporation, following previously described protocols.76 Briefly, Daudi cells (7.5 × 106) were electroporated 50 μg of mRNA (encoding T7 polymerase) in 250 μL of RPMI1640.
Electroporation was conducted in 4-mm cuvettes at 190 V and a capacitance of 975 μF, utilizing a Bio-Rad electroporation apparatus. The mixed cells were cultured in a 12-well plate for an additional 24 h. After this incubation, the culture medium was aspirated, and the cell luciferase activity was determined, which can be detected only if cell-cell fusion has occurred. Specifically, the luciferase activity was measured using the Dual-Glo luciferase assay system (Promega) according to the manufacturer’s instructions and detected with a Bio-Tek microplate reader. The relative fusion activity was calculated as the ratio of luciferase activity in cells incubated with mAbs to that with the IgG control.
Generation of IVT mRNA
To generate IVT mRNA, the pUC57-T7-T7 polymerase-poly(A) plasmid containing the T7 promoter, 5′-UTR, open reading frame (ORF) encoding T7 polymerase, 3′-UTR, and a 120-polyA tail was linearized using BsmBI-v2 restriction enzyme (NEB, R0739L) and served as template, as previously described.77 Briefly, The T7 High Yield RNA Transcription kit (Novoprotein, E131) was used for the synthesis of uncapped mRNA transcripts. Subsequently, the cap structure was added using Vaccinia Capping Enzyme (Novoprotein, GMP-M062) and mRNA Cap 2′-O-Methyltransferase (Novoprotein, GMP-M072) in a single step. mRNA was purified with LiCl, eluted in RNase-free water, and stored at −80°C until use.
Animal experiments
To generate humanized mice, human CD34+ cells, extracted from umbilical cord blood with a purity exceeding 90% (provided by Beijing Novay biotech), were intravenously injected into the tail 48 h post a single intraperitoneal Busulfan injection at a dosage of 20 mg per kilogram of body weight. Evaluation of human CD45+ cell presence in peripheral blood, using flow cytometry, was conducted at both 4- and 8-week intervals following engraftment. Mice were administered 200 μg of monoclonal antibodies (2B7 or 2C1) or 200 μL of PBS intraperitoneally on days −1, 2, 7, 14, and 21. The challenge involved an intravenous tail injection of CNE2-EBV (3 x 106 Green Raji Units) on day 0. Peripheral blood was collected once a week for four weeks following the EBV challenge. Mice were euthanized with either a weight loss greater than 30%, inability to take food or water, or at week 6.
H&E, IHC staining, and in situ hybridization
Tissue samples underwent fixation in 10% formalin, followed by embedding in paraffin and sectioning onto slides. Consecutive slides were used for staining with H&E, hCD3 (anti-hCD3 antibody, VENTANA Cat#790–4341), hCD20 (anti-hCD20 antibody, VENTANA Cat#760–2531), and EBERs. hCD20 and hCD3 antibodies were used at a 1:200 dilution. To detect EBV-encoded RNAs (EBERs), in situ hybridization was carried out with an EBER probe (EBER detection kit, ZSGB-BIO, Cat#ISH-7001–100), following the instructions provided by the manufacturer.
Quantitative real-time PCR (qPCR)
For assessing EBV DNA copy numbers, EBV DNA was extracted from mouse peripheral blood utilizing a Blood DNA Extraction Kit (TIANGEN, China, Cat #DP304-03). Quantification of EBV DNA copy numbers followed established protocols.13 In brief, the EBV DNA copy numbers were assessed using Roche Light Cycler 480 with TaqMan BamHI probes (sense: 5′-CCCAACACTCCACCACACC-3′; antisense: 5′-TCTTAGGAGCTGTCCGAGGG-3′). A standard EBV genome (BDS biotech Cat#BDS-BW-087) served as a control for accurate copy number quantification.
To assess the expression of the human leukocyte antigen (HLA)-II, cellular RNA was extracted utilizing TRIzol Reagent (Thermo Fisher Scientific) according to the manufacturer’s guidelines. The concentration and purity of the extracted RNA were assessed using a spectrophotometer (NanoDrop, Thermo Fisher Scientific). Subsequently, complementary DNA (cDNA) was generated from the extracted RNA utilizing a reverse transcription kit (Promega GoScript Reverse Transcription System) following the manufacturer’s instruction. QPCR was performed using the SYBR-Green PCR kit (ChamQ Universal SYBR qPCR Master Mix, Vazyme Biotech Co., Ltd., Nanjing, China), and signal detection was carried out using the Roche LightCycler 480 II instrument. The primers used for the HLA-II genes are listed in Table S2.
Negative-stain electron microscopy
For negative-stain electron microscopy imaging of gH/gL/gp42-Fab, the protein samples were diluted to approximately 0.02 mg/mL in PBS buffer. Formvar and carbon-coated 300 mesh copper grids (Electron Microscopy China) were used, and they were glow discharged immediately before use. 5 μL of the sample was applied onto the grid and left for 1 min, after which it was blotted away with Whatman No. 1 filter paper. Subsequently, the grids were stained with 10 μL of 0.75% (w/v) uranyl acetate for 30 s, then blotting away the excess stain solution. Grids were checked on the FEI Talos L120C G2 transmission electron microscope, and images were acquired with a Gatan Ceta 16M 4 K × 4 K CMOS camera.
Cryo-EM sample preparation and data collection
Purified samples were checked by negative staining using the Talos L120C G2 (Thermo Fisher Scientific) at 120 kV. To image ghglgp42 -Fab by cryo-EM, 4 μL of 0.5 mg/mL sample was loaded onto a freshly glow-discharged (60 s at 15 mA). Plunge freezing was performed on Quantifoil Cu R1.2/1.3 300 mesh grids using a Vitrobot Mark IV system (ThermoFisher Scientific Inc.) with 0-blot force, 7 s blot time, 100% humidity, and 8°C. A 300 kV Titan Krios microscope (Thermo Fisher Scientific Inc.) equipped with a BioContinuum Imaging Filter (Gatan Inc) was used to collect the cryo-EM data. Images were captured on a K3 Summit direct detection camera (Gatan Inc) using EPU software and GIF Quantum energy filter for the automated acquisition of images. The images were captured at a nominal magnification of 105,000× in super-resolution mode, resulting in a calibrated pixel size of 0.855 Å. Each exposure was dose-fractionated into 32 frames to reach a total dose of 50 e−/Å2. The micrographs had a final defocus range ranging from −2.0 to −1.0 μm.
Cryo-EM data processing
All image processing was done in cryoSPARC (v3.3.2).66 Raw movie frames were subjected to dose-weighting and correction for beam-induced drift using the Patch motion correction (multi) integrated into the cryoSPARC software. The parameters for the contrast transfer function (CTF) were estimated using CTFFIND4.78 Particles were first picked using blob picker. Particle images were obtained using a box size of 384 pixels for two-dimensional classification. From these, we selected high-quality 2D class averages that served as templates for training in Topaz.79 These class averages represented projections from different orientations. Then, we used Topaz extract to pick particles and subjected them to another round of 2D classification. After ab initio model building and homogeneous refinement, heterogeneous refinement was carried out. Particles from the specific class were employed to produce the final map through nonuniform (NU) refinement with a global resolution of 3.3 Å. Local refinement focusing on the interface with the mask could reconstitute the structure at a 3.4 Å resolution. A local resolution estimate was conducted using cryoSPARC, and the reported resolutions were evaluated based on the gold-standard Fourier Shell Correlation (FSC) criterion of 0.143.
Model building, refinement, and validation
The structure of the gH/gL/gp42 (PDB: 5T1D) in complex with a fab (predicted by Alpha Fold270) was docked into the cryo-EM density maps using CHIMERA.65 Subsequently, these models underwent manual adjustments for local fitting in COOT.67 The refinement of the models against the corresponding maps in real space was carried out using PHENIX,68 with the application of secondary structural restraints and Ramachandran restraints. The quality of each model’s stereochemistry was evaluated using MolProbity.69 Structural visualizations were generated utilizing PyMOL,71 Chimera, or ChimeraX.64,65 Table S1 and Figure S4 showed detailed statistics on model refinement and validation.
Quantification and statistical analysis
The sample size (n) varied for different assays and is specified in figure legends. Mean values ±standard error of the mean (SEM) was presented. One-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons test was utilized in the comparison among more than two groups. two-way ANOVA followed by Dunnett’s multiple comparisons test was employed for analyzing EBV copy number and weight change in humanized mice. The log rank (Mantel-Cox) test was used to evaluate survival curves. Detailed descriptions of statistical methods for each experiment are provided in figure legends. Significance was indicated as ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, and non-significant (ns) when p > 0.05. Data analysis and p-value calculations were performed using GraphPad Prism software (8.0).
Additional resources
No additional resources.
Acknowledgments
This work was supported by grants from the National Key Research and Development Program of China (2022YFC3400900), the National Natural Science Foundation of China (82030046), the Program for Guangdong Introducing Innovative and Entrepreneurial Teams (2019BT02Y198), and the Guangdong Science and Technology Department (2020B1212030004). We thank Professor Ning-Shao Xia (Xiamen University, China) for kindly providing the anti-gp42 antibody 5E3. We thank Professor Lin-Qi Zhang (Tsinghua University, China) for kindly providing the anti-gp350 antibody 72A1. We thank Prof. Richard Longnecker (Northwestern University, USA) for kindly providing the plasmids pCAGGS-T7, pCAGGS-gH, pCAGGS-gL, pCAGGS-gB, and pT7EMCLuc. The cartoons in the graphical abstract and Figures 1A, 5A, and 7E were created with BioRender.com.
Author contributions
Conceptualization, M.-S.Z., Z.L., X.-W.K., and G.-X.Z.; investigation, G.-X.Z., X.-Y.F., X.-W.K., G.-L.B., T.L., C.X., and S.-X.L.; resources, M.-S.Z., Z.L., X.-W.K., and Y.W.; formal analysis, G.-X.Z., X.-Y.F., G.-L.B., X.-W.K., and S.-J.-B.C.; writing – original draft, G.-X.Z. and X.-Y.F.; writing – review & editing, M.-S.Z., Z.L., X.-W.K., C.S., C.X., and N.M.; funding acquisition, M.-S.Z.; supervision, M.-S.Z. and Z.L.
Declaration of interests
The authors declare that patent applications have been filed covering 2B7 and 2C1 (patent application number: 2B7: CN202310261915.7, M.-S.Z., G.-X.Z., C.X., and G.-K.F. are the inventors; 2C1: CN202310261961.7, M.-S.Z., G.-X.Z., G.-L.B., and G.-K.F. are the inventors). The patent applicant is Sun Yat-sen University Cancer Center.
Declaration of generative AI and AI-assisted technologies in the writing process
During the preparation of this work, the authors used ChatGPT3.5 in order to check the grammar and improve clarity. After using this tool/service, the authors reviewed and edited the content as needed and took full responsibility for the content of the publication.
Published: May 21, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2024.101573.
Contributor Information
Xiang-Wei Kong, Email: kongxw8@mail.sysu.edu.cn.
Zheng Liu, Email: liuz3@sustech.edu.cn.
Mu-Sheng Zeng, Email: zengmsh@sysucc.org.cn.
Supplemental information
References
- 1.Dunmire S.K., Verghese P.S., Balfour H.H. Primary Epstein-Barr virus infection. J. Clin. Virol. 2018;102:84–92. doi: 10.1016/j.jcv.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Liu S.-X., Zhao G.-X., Lin R.-B., Zeng M.-S., Zhong Q. Classifying the tumor microenvironment to stratify nasopharyngeal carcinoma patients. Ann. Nasopharynx Cancer. 2022;6:8. doi: 10.21037/anpc-21-10. [DOI] [Google Scholar]
- 3.Crombie J.L., LaCasce A.S. Epstein Barr Virus Associated B-Cell Lymphomas and Iatrogenic Lymphoproliferative Disorders. Front. Oncol. 2019;9:109. doi: 10.3389/fonc.2019.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bjornevik K., Cortese M., Healy B.C., Kuhle J., Mina M.J., Leng Y., Elledge S.J., Niebuhr D.W., Scher A.I., Munger K.L., Ascherio A. Longitudinal analysis reveals high prevalence of Epstein-Barr virus associated with multiple sclerosis. Science. 2022;375:296–301. doi: 10.1126/science.abj8222. [DOI] [PubMed] [Google Scholar]
- 5.Young L.S., Dawson C.W. Epstein-Barr virus and nasopharyngeal carcinoma. Chin. J. Cancer. 2014;33:581–590. doi: 10.5732/cjc.014.10197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burns D.M., Chaganti S. Epstein-Barr virus-associated lymphoproliferative disorders in immunosuppressed patients. Annals of Lymphoma 5. Ann. Lymphoma. 2021;5:24. doi: 10.21037/aol-20-42. [DOI] [Google Scholar]
- 7.Sun C., Kang Y.-F., Fang X.-Y., Liu Y.-N., Bu G.-L., Wang A.-J., Li Y., Zhu Q.-Y., Zhang H., Xie C., et al. A gB nanoparticle vaccine elicits a protective neutralizing antibody response against EBV. Cell Host Microbe. 2023;31:1882–1897.e10. doi: 10.1016/j.chom.2023.09.011. [DOI] [PubMed] [Google Scholar]
- 8.Zhu Q.-Y., Zhao G.-X., Li Y., Talakatta G., Mai H.-Q., Le Q.-T., Young L.S., Zeng M.-S. Advances in pathogenesis and precision medicine for nasopharyngeal carcinoma. MedComm. 2021;2:175–206. doi: 10.1002/mco2.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai J., Zhang B., Li Y., Zhu W., Akihisa T., Li W., Kikuchi T., Liu W., Feng F., Zhang J. Prophylactic and Therapeutic EBV Vaccines: Major Scientific Obstacles, Historical Progress, and Future Direction. Vaccines (Basel) 2021;9:1290. doi: 10.3390/vaccines9111290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie C., Zhong L.-Y., Bu G.-L., Zhao G.-X., Yuan B.-Y., Liu Y.-T., Sun C., Zeng M.-S. Anti-EBV antibodies: Roles in diagnosis, pathogenesis, and antiviral therapy. J. Med. Virol. 2023;95 doi: 10.1002/jmv.28793. [DOI] [PubMed] [Google Scholar]
- 11.Chen W.-H., Kim J., Bu W., Board N.L., Tsybovsky Y., Wang Y., Hostal A., Andrews S.F., Gillespie R.A., Choe M., et al. Epstein-Barr virus gH/gL has multiple sites of vulnerability for virus neutralization and fusion inhibition. Immunity. 2022;55:2135–2148.e6. doi: 10.1016/j.immuni.2022.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snijder J., Ortego M.S., Weidle C., Stuart A.B., Gray M.D., McElrath M.J., Pancera M., Veesler D., McGuire A.T. An antibody targeting the fusion machinery neutralizes dual-tropic infection and defines a site of vulnerability on Epstein-Barr virus. Immunity. 2018;48:799–811.e9. doi: 10.1016/j.immuni.2018.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Q.-Y., Shan S., Yu J., Peng S.-Y., Sun C., Zuo Y., Zhong L.-Y., Yan S.-M., Zhang X., Yang Z., et al. A potent and protective human neutralizing antibody targeting a novel vulnerable site of Epstein-Barr virus. Nat. Commun. 2021;12:6624. doi: 10.1038/s41467-021-26912-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Q.-Y., Kong X.-W., Sun C., Xie S.-H., Hildesheim A., Cao S.-M., Zeng M.-S. Association between Antibody Responses to Epstein-Barr Virus Glycoproteins, Neutralization of Infectivity, and the Risk of Nasopharyngeal Carcinoma. mSphere. 2020;5:e00901-20. doi: 10.1128/mSphere.00901-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirschner A.N., Omerović J., Popov B., Longnecker R., Jardetzky T.S. Soluble Epstein-Barr Virus Glycoproteins gH, gL, and gp42 Form a 1:1:1 Stable Complex That Acts Like Soluble gp42 in B-Cell Fusion but Not in Epithelial Cell Fusion. J. Virol. 2006;80:9444–9454. doi: 10.1128/JVI.00572-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X., Hutt-Fletcher L.M. Epstein-Barr Virus Lacking Glycoprotein gp42 Can Bind to B Cells but Is Not Able To Infect. J. Virol. 1998;72:158–163. doi: 10.1128/jvi.72.1.158-163.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sathiyamoorthy K., Jiang J., Möhl B.S., Chen J., Zhou Z.H., Longnecker R., Jardetzky T.S. Inhibition of EBV-mediated membrane fusion by anti-gHgL antibodies. Proc. Natl. Acad. Sci. USA. 2017;114:E8703–E8710. doi: 10.1073/pnas.1704661114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bu W., Joyce M.G., Nguyen H., Banh D.V., Aguilar F., Tariq Z., Yap M.L., Tsujimura Y., Gillespie R.A., Tsybovsky Y., et al. Immunization with Components of the Viral Fusion Apparatus Elicits Antibodies That Neutralize Epstein-Barr Virus in B Cells and Epithelial Cells. Immunity. 2019;50:1305–1316.e6. doi: 10.1016/j.immuni.2019.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borza C.M., Hutt-Fletcher L.M. Alternate replication in B cells and epithelial cells switches tropism of Epstein–Barr virus. Nat. Med. 2002;8:594–599. doi: 10.1038/nm0602-594. [DOI] [PubMed] [Google Scholar]
- 20.Li Q., Turk S.M., Hutt-Fletcher L.M. The Epstein-Barr virus (EBV) BZLF2 gene product associates with the gH and gL homologs of EBV and carries an epitope critical to infection of B cells but not of epithelial cells. J. Virol. 1995;69:3987–3994. doi: 10.1128/JVI.69.7.3987-3994.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strnad B.C., Schuster T., Klein R., Hopkins R.F., Witmer T., Neubauer R.H., Rabin H. Production and characterization of monoclonal antibodies against the Epstein-Barr virus membrane antigen. J. Virol. 1982;41:258–264. doi: 10.1128/jvi.41.1.258-264.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ressing M.E., van Leeuwen D., Verreck F.A.W., Keating S., Gomez R., Franken K.L.M.C., Ottenhoff T.H.M., Spriggs M., Schumacher T.N., Hutt-Fletcher L.M., et al. Epstein-Barr Virus gp42 Is Posttranslationally Modified To Produce Soluble gp42 That Mediates HLA Class II Immune Evasion. J. Virol. 2005;79:841–852. doi: 10.1128/JVI.79.2.841-852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Q., Spriggs M.K., Kovats S., Turk S.M., Comeau M.R., Nepom B., Hutt-Fletcher L.M. Epstein-Barr virus uses HLA class II as a cofactor for infection of B lymphocytes. J. Virol. 1997;71:4657–4662. doi: 10.1128/jvi.71.6.4657-4662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong J., Wei D., Wu Q., Zhong L., Chen K., Huang Y., Zhang W., Chen J., Xia N., Zhang X., Chen Y. Antibody Generation and Immunogenicity Analysis of EBV gp42 N-Terminal Region. Viruses. 2021;13:2380. doi: 10.3390/v13122380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong J., Zhong L., Liu L., Wu Q., Zhang W., Chen K., Wei D., Sun H., Zhou X., Zhang X., et al. Non-overlapping epitopes on the gHgL-gp42 complex for the rational design of a triple-antibody cocktail against EBV infection. Cell Rep. Med. 2023;4 doi: 10.1016/j.xcrm.2023.101296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Q., Zhong L., Wei D., Zhang W., Hong J., Kang Y., Chen K., Huang Y., Zheng Q., Xu M., et al. Neutralizing antibodies against EBV gp42 show potent in vivo protection and define novel epitopes. Emerg. Microbes Infect. 2023;12 doi: 10.1080/22221751.2023.2245920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bu W., Kumar A., Board N.L., Kim J., Dowdell K., Zhang S., Lei Y., Hostal A., Krogmann T., Wang Y., et al. Epstein-Barr virus gp42 antibodies reveal sites of vulnerability for receptor binding and fusion to B cells. Immunity. 2024;57:559–573.e6. doi: 10.1016/j.immuni.2024.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lefranc M.-P. In: Encyclopedia of Systems Biology. Dubitzky W., Wolkenhauer O., Cho K.-H., Yokota H., editors. Springer; 2013. Complementarity Determining Region (CDR-IMGT) pp. 451–453. [DOI] [Google Scholar]
- 29.Mullen M.M., Haan K.M., Longnecker R., Jardetzky T.S. Structure of the Epstein-Barr virus gp42 protein bound to the MHC class II receptor HLA-DR1. Mol. Cell. 2002;9:375–385. doi: 10.1016/s1097-2765(02)00465-3. [DOI] [PubMed] [Google Scholar]
- 30.Wu J., Wu F., Jiang B., Liu W., Zhao P. tFold-Ab: Fast and Accurate Antibody Structure Prediction without Sequence Homologs. bioRxiv. 2022 doi: 10.1101/2022.11.10.515918. Preprint at. [DOI] [Google Scholar]
- 31.Sathiyamoorthy K., Hu Y.X., Möhl B.S., Chen J., Longnecker R., Jardetzky T.S. Structural basis for Epstein–Barr virus host cell tropism mediated by gp42 and gHgL entry glycoproteins. Nat. Commun. 2016;7 doi: 10.1038/ncomms13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sathiyamoorthy K., Jiang J., Hu Y.X., Rowe C.L., Möhl B.S., Chen J., Jiang W., Mellins E.D., Longnecker R., Zhou Z.H., Jardetzky T.S. Assembly and Architecture of the EBV B Cell Entry Triggering Complex. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J., Sathiyamoorthy K., Zhang X., Schaller S., Perez White B.E., Jardetzky T.S., Longnecker R. Ephrin receptor A2 is a functional entry receptor for Epstein-Barr virus. Nat. Microbiol. 2018;3:172–180. doi: 10.1038/s41564-017-0081-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H., Li Y., Wang H.-B., Zhang A., Chen M.-L., Fang Z.-X., Dong X.-D., Li S.-B., Du Y., Xiong D., et al. Ephrin receptor A2 is an epithelial cell receptor for Epstein-Barr virus entry. Nat. Microbiol. 2018;3:1–8. doi: 10.1038/s41564-017-0080-8. [DOI] [PubMed] [Google Scholar]
- 35.Shaw P.L., Kirschner A.N., Jardetzky T.S., Longnecker R. Characteristics of Epstein-Barr Virus Envelope Protein gp42. Virus Gene. 2010;40:307–319. doi: 10.1007/s11262-010-0455-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silva A.L., Omerovic J., Jardetzky T.S., Longnecker R. Mutational analyses of Epstein-Barr virus glycoprotein 42 reveal functional domains not involved in receptor binding but required for membrane fusion. J. Virol. 2004;78:5946–5956. doi: 10.1128/JVI.78.11.5946-5956.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirschner A.N., Sorem J., Longnecker R., Jardetzky T.S. Structure of Epstein-Barr Virus glycoprotein 42 suggests a mechanism for triggering receptor-activated virus entry. Structure. 2009;17:223–233. doi: 10.1016/j.str.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eichelberg M.R., Welch R., Guidry J.T., Ali A., Ohashi M., Makielski K.R., McChesney K., Van Sciver N., Lambert P.F., Keleș S., et al. Epstein-Barr Virus Infection Promotes Epithelial Cell Growth by Attenuating Differentiation-Dependent Exit from the Cell Cycle. mBio. 2019;10 doi: 10.1128/mBio.01332-19. e013322–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borza C.M., Morgan A.J., Turk S.M., Hutt-Fletcher L.M. Use of gHgL for Attachment of Epstein-Barr Virus to Epithelial Cells Compromises Infection. J. Virol. 2004;78:5007–5014. doi: 10.1128/JVI.78.10.5007-5014.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chesnokova L.S., Nishimura S.L., Hutt-Fletcher L.M. Fusion of epithelial cells by Epstein–Barr virus proteins is triggered by binding of viral glycoproteins gHgL to integrins αvβ6 or αvβ8. Proc. Natl. Acad. Sci. USA. 2009;106:20464–20469. doi: 10.1073/pnas.0907508106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin S., Li R., Chen M.-Y., Yu C., Tang L.-Q., Liu Y.-M., Li J.-P., Liu Y.-N., Luo Y.-L., Zhao Y., et al. Single-cell transcriptomic analysis defines the interplay between tumor cells, viral infection, and the microenvironment in nasopharyngeal carcinoma. Cell Res. 2020;30:950–965. doi: 10.1038/s41422-020-00402-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas J.A., Iliescu V., Crawford D.H., Ellouz R., Cammoun M., de-Thé G. Expression of HLA-DR antigens in nasopharyngeal carcinoma: An immunohistological analysis of the tumour cells and infiltrating lymphocytes. Int. J. Cancer. 1984;33:813–819. doi: 10.1002/ijc.2910330616. [DOI] [PubMed] [Google Scholar]
- 43.Busson P., Braham K., Clausse B., Tursz T. Constitutive expression of HLA class II antigens on EBV positive malignant cells from nasopharyngeal carcinoma: possible involvement in T cell infiltration. Cancer Detect. Prev. 1988;12:363–368. [PubMed] [Google Scholar]
- 44.Haan K.M., Kwok W.W., Longnecker R., Speck P. Epstein-Barr Virus Entry Utilizing HLA-DP or HLA-DQ as a Coreceptor. J. Virol. 2000;74:2451–2454. doi: 10.1128/jvi.74.5.2451-2454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang R., Scott R.S., Hutt-Fletcher L.M. Epstein-Barr Virus Shed in Saliva Is High in B-Cell-Tropic Glycoprotein gp42. J. Virol. 2006;80:7281–7283. doi: 10.1128/JVI.00497-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lian M. Combining Epstein–Barr virus antibodies for early detection of nasopharyngeal carcinoma: A meta-analysis. Auris Nasus Larynx. 2023;50:430–439. doi: 10.1016/j.anl.2022.09.010. [DOI] [PubMed] [Google Scholar]
- 47.Huang Y., Rao H., Yan S., Wang F., Wu Q., Feng Y., Zhang Y. Serum EBV EA-IgA and VCA-IgA antibodies can be used for risk group stratification and prognostic prediction in extranodal NK/T cell lymphoma: 24-year experience at a single institution. Ann. Hematol. 2017;96:1331–1342. doi: 10.1007/s00277-017-3013-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pelegrin M., Naranjo-Gomez M., Piechaczyk M. Antiviral Monoclonal Antibodies: Can They Be More Than Simple Neutralizing Agents? Trends Microbiol. 2015;23:653–665. doi: 10.1016/j.tim.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoffman G.J., Lazarowitz S.G., Hayward S.D. Monoclonal antibody against a 250,000-dalton glycoprotein of Epstein-Barr virus identifies a membrane antigen and a neutralizing antigen. Proc. Natl. Acad. Sci. USA. 1980;77:2979–2983. doi: 10.1073/pnas.77.5.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Audet J., Wong G., Wang H., Lu G., Gao G.F., Kobinger G., Qiu X. Molecular Characterization of the Monoclonal Antibodies Composing ZMAb: A Protective Cocktail Against Ebola Virus. Sci. Rep. 2014;4:6881. doi: 10.1038/srep06881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Protective anti-gB neutralizing antibodies targeting two vulnerable sites for EBV-cell membrane fusion. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2202371119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matsuura H., Kirschner A.N., Longnecker R., Jardetzky T.S. Crystal structure of the Epstein-Barr virus (EBV) glycoprotein H/glycoprotein L (gH/gL) complex. Proc. Natl. Acad. Sci. USA. 2010;107:22641–22646. doi: 10.1073/pnas.1011806108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang H.-J., Tian J., Qi X.-K., Xiang T., He G.-P., Zhang H., Yu X., Zhang X., Zhao B., Feng Q.-S., et al. Epstein-Barr virus activates F-box protein FBXO2 to limit viral infectivity by targeting glycoprotein B for degradation. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Piboonniyom S.o., Duensing S., Swilling N.W., Hasskarl J., Hinds P.W., Münger K. Abrogation of the retinoblastoma tumor suppressor checkpoint during keratinocyte immortalization is not sufficient for induction of centrosome-mediated genomic instability. Cancer Res. 2003;63:476–483. [PubMed] [Google Scholar]
- 55.Huang D.P., Ho J.H., Poon Y.F., Chew E.C., Saw D., Lui M., Li C.L., Mak L.S., Lai S.H., Lau W.H. Establishment of a cell line (NPC/HK1) from a differentiated squamous carcinoma of the nasopharynx. Int. J. Cancer. 1980;26:127–132. doi: 10.1002/ijc.2910260202. [DOI] [PubMed] [Google Scholar]
- 56.Zhan F., Jiang N., Cao L., Deng L., Tan G., Zhou M., Xie Y., Li G. [Primary study of differentially expressed cDNA sequences in cell line HNE1 of human nasopharyngeal carcinoma by cDNA representational difference analysis] Zhonghua. Yi. Xue. Yi. Chuan. Xue. Za. Zhi. 1998;15:341–344. https://pubmed.ncbi.nlm.nih.gov/9845762/ [PubMed] [Google Scholar]
- 57.Klein G., Lindahl T., Jondal M., Leibold W., Menézes J., Nilsson K., Sundström C. Continuous lymphoid cell lines with characteristics of B cells (bone-marrow-derived), lacking the Epstein-Barr virus genome and derived from three human lymphomas. Proc. Natl. Acad. Sci. USA. 1974;71:3283–3286. doi: 10.1073/pnas.71.8.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Molesworth S.J., Lake C.M., Borza C.M., Turk S.M., Hutt-Fletcher L.M. Epstein-Barr virus gH is essential for penetration of B cells but also plays a role in attachment of virus to epithelial cells. J. Virol. 2000;74:6324–6332. doi: 10.1128/jvi.74.14.6324-6332.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang H.-B., Zhang H., Zhang J.-P., Li Y., Zhao B., Feng G.-K., Du Y., Xiong D., Zhong Q., Liu W.-L., et al. Neuropilin 1 is an entry factor that promotes EBV infection of nasopharyngeal epithelial cells. Nat. Commun. 2015;6:6240. doi: 10.1038/ncomms7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haan K.M., Lee S.K., Longnecker R. Different functional domains in the cytoplasmic tail of glycoprotein B are involved in Epstein-Barr virus-induced membrane fusion. Virology. 2001;290:106–114. doi: 10.1006/viro.2001.1141. [DOI] [PubMed] [Google Scholar]
- 61.Okuma K., Nakamura M., Nakano S., Niho Y., Matsuura Y. Host range of human T-cell leukemia virus type I analyzed by a cell fusion-dependent reporter gene activation assay. Virology. 1999;254:235–244. doi: 10.1006/viro.1998.9530. [DOI] [PubMed] [Google Scholar]
- 62.Walker L.E., Reisfeld R.A. A high yield purification procedure for alpha and beta chains of HLA-DR antigens. J. Biol. Chem. 1982;257:7940–7943. doi: 10.1016/S0021-9258(18)34275-3. [DOI] [PubMed] [Google Scholar]
- 63.Andris-Widhopf J., Rader C., Steinberger P., Fuller R., Barbas C.F. Methods for the generation of chicken monoclonal antibody fragments by phage display. J. Immunol. Methods. 2000;242:159–181. doi: 10.1016/s0022-1759(00)00221-0. [DOI] [PubMed] [Google Scholar]
- 64.Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein. Sci. 2018;27:14–25. doi: 10.1002/pro.3235. https://pubmed.ncbi.nlm.nih.gov/28710774/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 66.Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. https://www.nature.com/articles/nmeth.4169 [DOI] [PubMed] [Google Scholar]
- 67.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 68.Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.-W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen V.B., Arendall W.B., Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC. https://www.sciepub.com/reference/159710.
- 72.Nzuma R.M., Liu F., Grant I.R. Generation and characterization of a novel recombinant scFv antibody specific for Campylobacter jejuni. Appl. Microbiol. Biotechnol. 2018;102:4873–4885. doi: 10.1007/s00253-018-8949-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Frei J.C., Lai J.R. Protein and Antibody Engineering by Phage Display. Methods Enzymol. 2016;580:45–87. doi: 10.1016/bs.mie.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee C.M.Y., Iorno N., Sierro F., Christ D. Selection of human antibody fragments by phage display. Nat. Protoc. 2007;2:3001–3008. doi: 10.1038/nprot.2007.448. [DOI] [PubMed] [Google Scholar]
- 75.Sun C., Yang J.-W., Xie C., Fang X.-Y., Bu G.-L., Zhao G.-X., Dai D.-L., Liu Z., Zeng M.-S. The structure of HSV-1 gB bound to a potent neutralizing antibody reveals a conservative antigenic domain across herpesviruses. hLife. 2024;2:141–146. doi: 10.1016/j.hlife.2023.12.004. [DOI] [Google Scholar]
- 76.Kalla M., Göbel C., Hammerschmidt W. The Lytic Phase of Epstein-Barr Virus Requires a Viral Genome with 5-Methylcytosine Residues in CpG Sites. J. Virol. 2012;86:447–458. doi: 10.1128/JVI.06314-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao G.-X., Bu G.-L., Liu G.-F., Kong X.-W., Sun C., Li Z.-Q., Dai D.-L., Sun H.-X., Kang Y.-F., Feng G.-K., et al. mRNA-based Vaccines Targeting the T-cell Epitope-rich Domain of Epstein Barr Virus Latent Proteins Elicit Robust Anti-Tumor Immunity in Mice. Adv. Sci. 2023;10 doi: 10.1002/advs.202302116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rohou A., Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bepler T., Morin A., Rapp M., Brasch J., Shapiro L., Noble A.J., Berger B. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods. 2019;16:1153–1160. doi: 10.1038/s41592-019-0575-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The cryo-EM map of the EBV gH/gL/gp42-2C1 Fab has been submitted to the Electron Microscopy DataBank (EMDB:EMD-37249). The atomic coordinates of the complex have been submitted and are available in the Protein Data Bank (PDB:8KHR).
-
•
This paper does not report the original code.
-
•
The original data has been uploaded to Research Data Deposit: https://www.researchdata.org.cn/ with accession number RDDB2024979584. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.