Abstract
Unlike all the other Rift Valley fever virus strains (Bunyaviridae, Phlebovirus) studied so far, clone 13, a naturally attenuated virus, does not form the filaments composed of the NSs nonstructural protein in the nuclei of infected cells (R. Muller, J. F. Saluzzo, N. Lopez, T. Drier, M. Turell, J. Smith, and M. Bouloy, Am. J. Trop. Med. Hyg. 53:405–411, 1995). This defect is correlated with a large in-frame deletion in the NSs coding region of the S segment of the tripartite genome. Here, we show that the truncated NSs protein of clone 13 is expressed and remains in the cytoplasm, where it is degraded rapidly by the proteasome. Through the analysis of reassortants between clone 13 and a virulent strain, we localized the marker(s) of attenuation in the S segment of this attenuated virus. This result raises questions regarding the role of NSs in pathogenesis and highlights, for the first time in the Bunyaviridae family, a major role of the S segment in virulence and attenuation, possibly associated with a defect in the nonstructural protein.
Rift Valley fever (RVF) virus is an arthropod-borne virus which periodically causes epidemics and epizootics in sub-Saharan countries of Africa and in Egypt (for a review, see reference 17). The most recent outbreaks occurred in 1997 and 1998 in eastern (Kenya, Somalia, and Uganda) and western (Mauritania) Africa (references 1 and 21 and references therein). In humans, infection provokes a wide range of clinical symptoms from benign fever to encephalitis, retinitis, and fatal hepatitis associated with hemorrhages. Among young animals, lambs, calves, and kids are severely affected and die from acute hepatitis. In adults, the symptoms are less pronounced but teratogenic and abortogenic effects are frequent in pregnant animals. Mice, hamsters, and some strains of rats are laboratory animal models for the study of RVF pathogenesis since they are highly sensitive to virulent strains and develop hepatitis or encephalitis when inoculated by peripheral routes (17).
Rift Valley fever virus belongs to the Phlebovirus genus of the Bunyaviridae family. Like all the members of the family, it possesses a single-stranded tripartite RNA genome composed of large (L), medium (M), and small (S) segments (for a review, see reference 23). The L and M segments are of negative polarity and code, respectively, for the L RNA-dependent RNA polymerase and for a polyprotein precursor cleaved to generate the envelope glycoproteins G1 and G2 and two nonstructural proteins, 14K and 78K. The S segment utilizes an ambisense strategy and codes for two proteins: the nucleoprotein N and the nonstructural protein NSs. These proteins are translated from two individual mRNAs of opposite polarities. The mRNA synthesizing the N protein is complementary to the genomic sense molecule, whereas the mRNA synthesizing the NSs protein is of genomic polarity. The role of the nonstructural proteins, for any member of the family, is still undetermined. Except in clone 13, the NSs proteins of all the RVF strains analyzed so far form filamentous structures in the nuclei of infected cells (16). The absence of nuclear NSs-associated filament in cells infected with clone 13 was correlated with a large internal deletion of the NSs open reading frame. This defect, which affects 70% of the coding sequence (i.e., 549 nucleotides) and conserves in frame the N and C termini of the protein, was not found to affect the ability of clone 13 to replicate to high titers in mammalian and mosquito cells as well as in the mosquito vector Culex pipiens. In addition, this virus was shown to be avirulent for mice. These observations raise the issues of the necessity of NSs in the viral cycle and its possible role in viral pathogenesis.
In this study, we extended the analysis of the truncated NSs protein which was not detected in our previous work and analyzed the genetic determinants of the attenuation of clone 13 through the production of reassortants with the virulent Egyptian strain ZH548.
Analysis of the clone 13 NSs gene product.
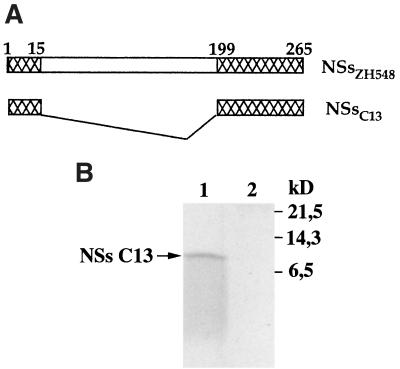
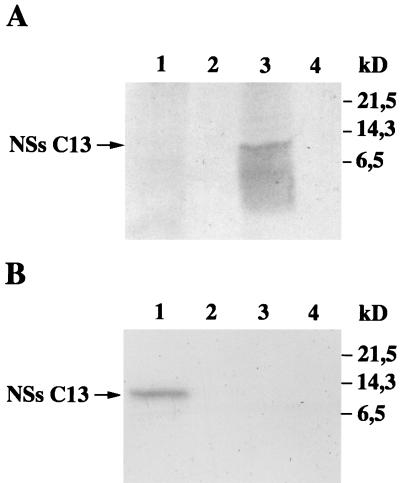
Compared to other virulent or attenuated RVF virus strains, the deletion in the S segment of clone 13 removed 183 of the 265 amino acids composing the complete protein and conserved the 67 C-terminal amino acids in frame with the 15 N-terminal amino acids (Fig. 1A). In our previous study, this polypeptide of 82 amino acids, which was expected to have a molecular mass of 9,056 Da and a pI of 5.8, was not detected, either by immunoprecipitation or by immunofluorescence assays of clone 13-infected cells with several sources of antibodies, a hyperimmune mouse ascitic fluid to an RVF virus virulent strain, the NSs-specific monoclonal antibody RB1-3C3, and monospecific polyclonal antibodies against the purified 31-kDa NSs protein expressed in RVF virus-infected cells (16). Since Northern blot analysis indicated that NSs-specific mRNAs were present in the polysomal fraction in amounts at least as abundant as in MP12-infected cells (not shown), the absence of NSs polypeptide in clone 13-infected cells was interpreted to be due to (i) the inability of the mRNA to be translated, (ii) the absence or low reactivity of antibodies to the epitopes present in the truncated NSs protein, and (iii) the rapid degradation of the polypeptide in infected cells. To investigate further these hypotheses, we tested antibodies prepared against the baculovirus-expressed NSs protein of MP12 (29) and constructed a recombinant Semliki Forest virus (SFV) replicon expressing the NSs of clone 13 (NSsC13), which was used as a control for protein expression. These recently produced antibodies were used thruout this study. The SFV replicon was chosen because of its capacity to overexpress foreign proteins (15). Thus, the sequence coding for the NSs protein of clone 13 was amplified by PCR with pBS-NSsC13 as a template and the oligodeoxynucleotides NSFG5′ and NSFAG3′ as described previously (29). After digestion with BglII, the DNA fragment was ligated into the BamHI-cleaved pSFV-1 plasmid (15). The protocols to synthesize the SFV-NSsC13 replicon and to produce suicidal SFV particles by cotransfecting BHK21 cells with helper 2 RNA has already been described (29). When BSR cells were infected with SFV-NSsC13, labeled with [35S]methionine and [35S]cysteine (Promix; Amersham), and analyzed in 17.5% polyacrylamide gels, no clone 13-specific NSs polypeptide was visible over the cellular background (not shown). This was in contrast to what occurred with the 31-kDa protein observed in cells infected with SFV-NSsMP12, which was visualized as an intense and sharply defined band over the cellular proteins (29). However, when the extract was immunoprecipitated with the NSs-specific antibodies by using protein A-Sepharose (Pharmacia) as described previously (29), synthesis of truncated NSsC13 was clearly revealed but the polypeptide did not migrate as a distinct band (Fig. 1B, lane 1). The small amount of protein and the smearing pattern of migration suggested that the polypeptide had undergone degradation.
FIG. 1.
(A) Schematic representation of the NSs open reading frame in the S segment of clone 13 and ZH548 RVF virus strains. Numbers represent the amino acid positions. (B) Expression of NSsC13 via recombinant SFV-NSsC13. BSR cells were infected with SFV-NSsC13 at an MOI of 5 and labeled with 100 μCi of [35S]methionine and [35S]cysteine per ml in methionine-deficient medium for 2 h from 22 to 24 h p.i. (lane 1). Mock-infected cells were run as a control (lane 2). Proteins from total cell extracts were immunoprecipitated with an immune mouse ascitic fluid prepared against the baculovirus expressed-NSs of the MP12 strain (29) and analyzed in a sodium dodecyl sulfate–17.5% polyacrylamide gel.
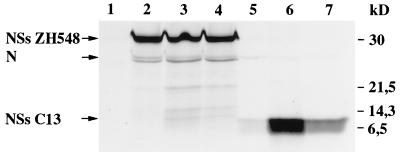
Since, in many cases, degradation of proteins occurs through the proteasome pathway, we tested the effect of two inhibitors of the proteasome. In cells infected with clone 13 that were treated with a 20 μM concentration of the aldehyde peptide MG132 (synthesized at the Pasteur Institute) or 10 μM lactacystin (from Biomol Research Laboratories, Plymouth Meeting, Pa.) and labeled with [35S]methionine and [35S]cysteine for 2 h from 8 to 10 h postinfection (p.i.), immunoprecipitation with the NSs-specific polyclonal antibodies showed the presence of the NSs protein, which migrated as a broad band at the expected position (Fig. 2, lanes 6 and 7). In the absence of the inhibitor (lane 5), trace amounts of the 9-kDa protein were detected only when labeling was performed relatively late in infection, i.e., not before 8 h p.i. It should be noted that, in contrast to the truncated protein of clone 13, the full-length NSs protein expressed in cells infected with ZH548 is not sensitive to degradation since treatment with MG132 or lactacystin did not increase the amount of protein recovered after immunoprecipitation (Fig. 2, lanes 2 to 4). As was already observed during infection of Vero cells with MP12, the antibodies to NSs coimmunoprecipitated small amounts of the nucleoprotein due to interactions between the two proteins (F. Yadani, unpublished data) as well as other minor proteins, probably of cellular origin, which migrated slightly more slowly than NSsC13 and were visible in cells infected with ZH548 and treated with the inhibitors (lanes 3 and 4).
FIG. 2.
Effect of proteasome inhibitor on the expression of NSs. Vero cells infected with ZH548 (lanes 2 to 4) or clone 13 (lanes 5 to 7) or mock infected (lane 1) were untreated (lanes 2 and 5) or treated with 20 μM MG132 (lanes 3 and 6) or 10 μM lactacystin (lanes 4 and 7) and labeled from 8 to 10 h p.i. with 200 μCi of a mixture of [35S]methionine and [35S]cysteine per ml. Proteins from total cellular extracts were immunoprecipitated with anti-NSs antibodies and analyzed in a sodium dodecyl sulfate–17.5% polyacrylamide gel. The positions of the viral proteins NSs and N (on the left) and the molecular mass markers (on the right) are indicated. In this experiment, the N protein migrated as a doublet, a situation observed occasionally.
To determine the cellular localization of the NSsC13 protein, clone 13-infected cells were fractionated into cytoplasmic and nuclear extracts after being labeled with [35S]methionine and [35S]cysteine from 6 to 8 h p.i. in the presence of MG132. Polyacrylamide gel analysis of the immunoprecipitation products indicated that most, if not all, of the NSs protein remained in the cytoplasm (Fig. 3A). Of note is that, in spite of the presence of the inhibitor, the NSsC13 protein appeared degraded, suggesting that pathways other than the proteasome pathway may be used or that inhibition is incomplete. Confirming the cytoplasmic localization of the protein, immunofluorescence assays using the NSs-specific antibodies and fluorescein-labeled sheep anti-mouse immunoglobulin G antibodies (Byosis, Compiègne, France) revealed a faint fluorescence localized in the cytoplasm and no staining in the nuclei (Fig. 4A). In this experiment, cells were not treated with the proteasome inhibitor but were fixed at 24 h p.i., a time sufficient for NSs to accumulate. Interestingly, cytoplasmic fluorescence was found in the vicinity of the nucleus, a pattern not observed in the control cells infected with ZH548, in which nuclear filaments as well as a diffuse cytoplasmic staining were visible (Fig. 4B). It is noteworthy that two factors, the source of antibodies and the method of cell fixation, were different from those used previously (16). Although the carboxy terminus was found to be essential for the filament formation in MP12-infected cells (29), such a structure was not observed in clone 13-infected cells, in spite of the presence of the sequence in the molecule. This absence can be explained by the fact that NSs is rapidly degraded by the proteasome and/or that the region corresponding to the deletion contains other elements involved in filament formation, including signals for nuclear migration. Work is in progress to determine the mechanism(s) involved in filament formation.
FIG. 3.
Cellular localization of NSsC13. Vero cells infected with clone 13 at an MOI of 5 (lanes 1 and 2 in panel A and lanes 3 and 4 in panel B) or mock infected (lanes 3 to 4 in panel A and lanes 1 and 2 in panel B) were treated with 20 μM MG132 and labeled for 2 h from 6 to 8 h p.i. with 360 μCi of a mixture of [35S]methionine and [35S]cysteine per ml in methionine-deficient medium (A) or with 250 μCi of [32P]orthophosphate per ml (B). Cytoplasmic (lanes 1 and 3) and nuclear (lanes 2 and 4) extracts were prepared, and proteins from them were immunoprecipitated with anti-NSs antibodies and analyzed in a sodium dodecyl sulfate–17.5% polyacrylamide gel.
FIG. 4.
Vero cells infected with clone 13 (A) or ZH548 (B) or left uninfected (C) were collected at 24 hours p.i., treated with formaldehyde and then Triton X-100, and stained with antibodies to NSs. Complexes were revealed with fluorescein-labeled sheep anti-mouse immunoglobulin G antibodies.
Finally, NSs was described as a phosphoprotein (25), and we recently mapped the phosphorylation sites of the MP12 NSs protein to two serine residues located in the carboxy terminus at positions 252 and 256 (13a). Since this region is conserved in NSs of clone 13, we analyzed the protein after labeling infected cells with [32P]orthophosphate in the presence of MG132. Immunoprecipitation and polyacrylamide gel electrophoresis indicated that 32P was incorporated into the protein (Fig. 3B). Two-dimensional analysis of CNBr cleavage peptides resulted in two phosphopeptides with a migration pattern similar to that obtained with the MP12 strain (not shown). This result strongly suggests that the same sites, i.e., serines 252 and 256, are phosphorylated in NSsC13. In contrast to the protein labeled with 35S (Fig. 3A), the 32P band was relatively distinct (Fig. 3B), which could be explained by cleavage of the carboxy terminus, which would have left the polypeptide unlabeled.
Selection of reassortants.
Reassortants between the attenuated RVF MP12 strain and a virulent strain isolated in Senegal were useful in determining the molecular basis of attenuation (22). Exchange of one segment of the virulent strain by the corresponding segment of MP12 resulted in a virus attenuated for mice, demonstrating that each of the three segments of MP12 carries attenuating mutations and that attenuation of the RVF virus is under polygenic control. However, in our hands the Senegalese strain was not stable in Vero cell cultures and became attenuated after a few passages. Thus, strain ZH548, isolated from a human patient during the Egyptian outbreak in 1977, was chosen as a partner for the coinfection because it retained a high level of virulence after 16 passages in tissue culture (4). We verified that after five cycles of plaque purification and passages in Vero cells, the subclones were as virulent for mice as the uncloned virus; the 50% lethal doses were similar (5 to 10 PFU), and the survival times (5 to 5.5 days) were not significantly different. Therefore, one of these clones was used for reassortment in an experiment in which Vero cells were coinfected with clone 13 and ZH548 at a multiplicity of infection (MOI) of 5 and 1 PFU per cell, respectively. The extracellular virus was harvested at 72 h p.i., and plaques were assayed as described previously (2). The parental origins of the segments were determined after reverse transcription-PCR amplification and sequencing of specific regions in the NSs (positions 31 to 841 in the genomic sense S RNA), G2 (positions 772 to 1580 in the antigenomic sense M RNA), and L (positions 4440 to 4651 in the antigenomic L RNA) coding regions, where the two strains differ (16, 21, 28). Reactions for reverse transcription-PCRs and sequencing and the sequences of the primers were described previously (21). All the PCR products were sequenced completely, but no mutation was found.
Among 30 plaque-purified viruses, the eight expected genotypes were identified (Table 1). A few plaques which contained the two parental copies of the S segment were not analyzed further, leaving open the questions of whether two individual particles coinfected cells or whether one particle contains a diploid genome as has already been reported (20, 27). One representative of each genotype was plaque purified three times or until homogeneity and analyzed for its properties in cell culture as well as in mice.
TABLE 1.
Reassortants between ZH548 and clone 13
| Genotypea | No. of representatives | Plaque | Mortality of mice inoculated with:
|
|||
|---|---|---|---|---|---|---|
| 104 PFU
|
105 PFU
|
|||||
| No. dead/no. inoculated (%) | Survivalb (day) | No. dead/no. inoculated (%) | Survival (day) | |||
| C/C/C | 2 | R407.11 | 0/16 (0) | 0/8 (0) | ||
| Z/C/C | 1 | R407.3 | 0/16 (0) | 0/8 (0) | ||
| C/Z/C | 3 | R403 | 0/16 (0) | 0/8 (0) | ||
| Z/Z/C | 7 | R406 | 1c/16 (6.25) | 0/8 (0) | ||
| C/C/Z | 1 | R414 | 16/16 (100) | 4.3 ± 1.5 | 8/8 (100) | 3 ± 0 |
| Z/C/Z | 2 | R413 | 16/16 (100) | 4.6 ± 1.0 | 8/8 (100) | 4 ± 1.8 |
| C/Z/Z | 1 | R422 | 16/16 (100) | 5.0 ± 0 | 8/8 (100) | 3.75 ± 1.2 |
| Z/Z/Z | 6 | R405 | 16/16 (100) | 5.1 ± 0.3 | 8/8 (100) | 4.6 ± 1.2 |
| Clone 13 | 0/16 (0) | 2d/16 (12.5) | ||||
| ZH548 | 16/16 (100) | 5.1 ± 1.3 | 16/16 (100) | 4.75 ± 1.0 | ||
The parental origins of segments L, M, and S in that order are indicated by Z and C for ZH548 and clone 13, respectively.
Values are means ± standard deviations.
Death occurred at day 7 p.i.
Death occurred at day 15 p.i.
Reassortants with the S segment of clone 13 do not plaque in MRC5 and have an increased ability to induce persistence.
Proteins from cells infected with each reassortant were analyzed after being labeled for 2 h from 8 to 10 h p.i. in the presence or absence of MG132 and being immunoprecipitated with NSs-specific antibodies. The pattern of migration of NSs correlated with the genotype determined by sequencing. In the experiment carried out in the presence of MG132 (Fig. 5), NSs derived from clone 13 was recovered in larger amounts than in the absence of inhibitor (not shown), indicating that degradation of the truncated NSs occurred but was not linked to a particular combination of L and M segments.
FIG. 5.
Expression of the NSs protein in Vero cells infected with clone 13 or ZH548 and in reassortants obtained after coinfection with these two strains. Cells were treated with 20 μM MG132 and labeled with 200 μCi of a mixture of [35S]methionine and [35S]cysteine per ml from 8 to 10 h p.i. Proteins were immunoprecipitated with anti-NSs antibodies and analyzed in a sodium dodecyl sulfate–17.5% polyacrylamide gel. The positions of the viral proteins (on the left) and the molecular mass markers (on the right) are indicated. N, nucleoprotein. The parental origins of segments L, M, and S (in that order) are indicated by C for clone 13 and Z for ZH548.
Like clone 13 and ZH548, all the reassortants grew to similar titers in Vero cells (1 × 107 to 5 × 107 PFU per ml). Clone 13 exhibits a particular property in human embryonic MRC5 cells by not forming plaques (16). Plaquing of the reassortant viruses in MRC5 clearly showed that the capacity of the virus to form plaques was associated with the parental origin of the S segment (not shown).
Furthermore, these two strains exhibited different cytopathic effects in Vero cells: in cultures infected with ZH548, most of the cells were killed within 72 h, whereas in cultures infected with clone 13, a significant proportion of cells survived the infection and became persistently infected, still expressing viral antigens after numerous passages (2). Although establishment of persistenly infected cells was difficult to measure, we observed that, like clone 13, all the reassortants containing the S segment of clone 13 were able to infect Vero cells and led to a persistent infection (not shown).
The S segment contains markers for attenuation.
As already reported by Peters and Linthicum, all the strains of mice are sensitive to RVF virus infection by the peripheral route and develop hepatitis (17). Thus, the pathogenicity of the reassortants and parent strains was assayed in 4- to 6-week-old outbred Swiss mice (OF1; IFFA-CREDO, Les Oncins, France) by inoculating intraperitoneally 104 or 105 PFU into groups of eight mice and observing them for 21 days or until death occurred (Table 1). Except for the two mice inoculated with 105 PFU of clone 13, which died at day 15 exhibiting neurological disorders and paralysis, the rest of the animals inoculated with clone 13 or with the reassortants containing the clone 13 S segment survived the infection and did not present any clinical symptoms. As to the death of the mouse inoculated with 104 PFU of R406, we do not know whether it was fortuitous or resulted from RVF virus, since a survival of 7 days seemed longer than usual for hepatitis and too short for encephalitis and since inoculation with a higher dose (105 PFU) did not provoke any deleterious effect (Table 1). All the mice surviving at day 21 p.i. were bled, and their sera were shown to contain RVF virus-specific antibodies, strongly suggesting that the virus did replicate in mice. The different combinations of segments did not seem to affect significantly the level of antibody response, the enzyme-linked immunosorbent assay titers being at least 1:12,800 and 1:1,000 in neutralization.
After inoculation with ZH548 and with the reassortants which contain the S segment of ZH548, all the mice died within an average survival time which was approximately the same for each group of mice, with inoculation with higher doses of virus accelerating death (Table 1). Histopathological examinations revealed an acute hepatitis (M. Huerre, not shown). Of note is that reassortant R414, which possesses the L and M segments of clone 13 and the S segment of ZH548 (genotype C/C/Z [see Table 1]), was found to be at least as virulent for mice as ZH548, with the 50% lethal doses of the two viruses being estimated to be in the same range (less than 10 PFU). These results indicate that no attenuation marker is present in the L and M segments of clone 13.
Reassortment between virulent and avirulent strains and monoclonal antibody-resistant variants has been used to study the molecular mechanism of virulence of the bunyavirus La Crosse (for reviews, see references 10 and 18). Virulence was shown to be under polygenic control, with the major determinants for neurovirulence and neuroinvasiveness located in the L and M segments, respectively (6, 7, 11). Here, we describe for the first time the localization of a major attenuation marker within the S segment. This raises the question of whether the attenuating mutation corresponds to the deletion in the NSs protein. Since a system of reverse genetics to introduce specific mutations in the Rift Valley fever virus genome is not yet available, the question cannot be answered. Comparison of the sequences of the clone 13 and ZH548 S segments indicated that, besides the deletion in the NSs gene, there is only one amino acid change (glycine versus glutamic acid) at position 159 in the N protein sequence and that there are six nucleotide changes in the intergenic region but none in the 5′ and 3′ noncoding regions (16, 28). Although the role of the changes in the noncoding sequences and the point mutation in the N gene could not be excluded, it is tempting to speculate on the role of the deletion in the NSs protein in attenuation and cytopathic properties.
Interestingly, the NSs protein of clone 13 is extremely unstable and rapidly degraded by the proteasome. Degradation of the RNA polymerase nsP4 of Sindbis virus by the N-end rule pathway was presumed to regulate the viral life cycle (5). Similarly, the proteasome was shown to play a role in picornavirus and human immunodeficiency virus infections by degrading the 3C proteases of poliovirus and hepatitis A virus (9) or several virion components (24). In the case of RVF virus, NSs is normally quite stable, as documented here with ZH548, and the situation observed for clone 13 is exceptional. In fact, the presence of the protein in the nucleus may be an alternative to taking away molecules which are not necessary in the cytoplasm, where replication and virion formation occur. Since we do not know the role of NSs protein in the RVF virus life cycle, the biological significance of the degradation of the clone 13 NSs protein remains unclear, but this observation supports the speculation that NSs is an accessory protein in tissue culture. Recently, a number of RNA nonstructural virus proteins were found to be nonessential for growth in tissue culture: NS2 of the human and bovine respiratory syncytial viruses (3, 26), the C protein of vesicular stomatitis virus (14), the SH protein of simian virus 5 (12), and the reovirus ς1s protein (19). It could well be that, although it is nonessential in tissue culture, NSs represents a major determinant in the outcome of infection in mammals. Similar observations were reported for the V protein of Sendai virus (13) and the nonstructural C protein of measles virus, which is required for efficient replication in human peripheral blood cells (8). It would therefore be of interest to address the question of the role of NSs in the pathogenesis, tropism, and virulence of RVF virus.
Acknowledgments
We thank Daniel Coudrier for help with animal work, M. Huerre and H. Khun for histopathological work, and C. Prehaud and F. Yadani for helpful discussions.
A.K. was supported by a fellowship from the Ministère de l'Education Nationale du Grand Duché du Luxembourg.
REFERENCES
- 1.Anonymous. An outbreak of Rift Valley fever, Eastern Africa, 1997–98. Weekly Epidemiol Rep. 1998;73:105–112. [PubMed] [Google Scholar]
- 2.Billecocq A, Vialat P, Bouloy M. Persistent infection of mammalian cells by Rift Valley fever virus. J Gen Virol. 1996;77:3053–3062. doi: 10.1099/0022-1317-77-12-3053. [DOI] [PubMed] [Google Scholar]
- 3.Buchholz U J, Finke S, Conzelmann K K. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol. 1999;73:251–259. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caplen H, Peters C J, Bishop D H. Mutagen-directed attenuation of Rift Valley fever virus as a method for vaccine development. J Gen Virol. 1985;66:2271–2277. doi: 10.1099/0022-1317-66-10-2271. [DOI] [PubMed] [Google Scholar]
- 5.de Groot R J, Rumenapf T, Kuhn R J, Strauss E G, Strauss J H. Sindbis virus RNA polymerase is degraded by the N-end rule pathway. Proc Natl Acad Sci USA. 1991;88:8967–8971. doi: 10.1073/pnas.88.20.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Endres M J, Griot C, Gonzalez-Scarano F, Nathanson N. Neuroattenuation of an avirulent bunyavirus variant maps to the L RNA segment. J Virol. 1991;65:5465–5470. doi: 10.1128/jvi.65.10.5465-5470.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Endres M J, Valsamakis A, Gonzalez-Scarano F, Nathanson N. Neuroattenuated bunyavirus variant: derivation, characterization, and revertant clones. J Virol. 1990;64:1927–1933. doi: 10.1128/jvi.64.5.1927-1933.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Escoffier C, Manie S, Vincent S, Muller C P, Billeter M, Gerlier D. Nonstructural C protein is required for efficient measles virus replication in human peripheral blood cells. J Virol. 1999;73:1695–1698. doi: 10.1128/jvi.73.2.1695-1698.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gladding R L, Haas A L, Gronros D L, Lawson T G. Evaluation of the susceptibility of the 3C proteases of hepatitis A virus and poliovirus to degradation by the ubiquitin-mediated proteolytic system. Biochem Biophys Res Commun. 1997;238:119–125. doi: 10.1006/bbrc.1997.7251. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez-Scarano F, Nathanson N. Bunyaviridae. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. New York, N.Y: Raven Press; 1996. pp. 1473–1504. [Google Scholar]
- 11.Griot C, Pekosz A, Lukac D, Scherer S S, Stillmock K, Schmeidler D, Endres M J, Gonzalez-Scarano F, Nathanson N. Polygenic control of neuroinvasiveness in California serogroup bunyaviruses. J Virol. 1993;67:3861–3867. doi: 10.1128/jvi.67.7.3861-3867.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He B, Leser G P, Paterson R G, Lamb R A. The paramyxovirus SV5 small hydrophobic (SH) protein is not essential for virus growth in tissue culture cells. Virology. 1998;250:30–40. doi: 10.1006/viro.1998.9354. [DOI] [PubMed] [Google Scholar]
- 13.Kato A, Kiyotani K, Sakai Y, Yoshida T, Nagai Y. The paramyxovirus, Sendai virus, V protein encodes a luxury function required for viral pathogenesis. EMBO J. 1997;16:578–587. doi: 10.1093/emboj/16.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.Kohl A, di Bartolo V, Bouloy M. The Rift Valley fever virus nonstructural protein NSs is phosphorylated at serine residues located in casein kinase II consensus motifs in the carboxy terminus. Virology. 1999;263:517–525. doi: 10.1006/viro.1999.9978. [DOI] [PubMed] [Google Scholar]
- 14.Kretzschmar E, Peluso R, Schnell M J, Whitt M A, Rose J K. Normal replication of vesicular stomatitis virus without C proteins. Virology. 1996;216:309–316. doi: 10.1006/viro.1996.0066. [DOI] [PubMed] [Google Scholar]
- 15.Liljestrom P, Garoff H. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Bio/Technology. 1991;9:1356–1361. doi: 10.1038/nbt1291-1356. [DOI] [PubMed] [Google Scholar]
- 16.Muller R, Saluzzo J F, Lopez N, Dreier T, Turell M, Smith J, Bouloy M. Characterization of clone 13, a naturally attenuated avirulent isolate of Rift Valley fever virus, which is altered in the small segment. Am J Trop Med Hyg. 1995;53:405–411. doi: 10.4269/ajtmh.1995.53.405. [DOI] [PubMed] [Google Scholar]
- 17.Peters C J, Linthicum K J. Rift Valley fever. In: Beran G W, Steele J H, editors. Handbook of zoonoses, section B. Viral. 2nd ed. Boca Raton, Fla: CRC Press; 1994. pp. 125–138. [Google Scholar]
- 18.Pringle C R. Genetics and genome segment reassortment. In: Elliott R M, editor. The viruses. The Bunyaviridae. New York, N.Y: Plenum Press; 1996. pp. 189–226. [Google Scholar]
- 19.Rodgers S E, Connolly J L, Chappell J D, Dermody T S. Reovirus growth in cell culture does not require the full complement of viral proteins: identification of a ς1s-null mutant. J Virol. 1998;72:8597–8604. doi: 10.1128/jvi.72.11.8597-8604.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodriguez L L, Owens J H, Peters C J, Nichol S T. Genetic reassortment among viruses causing hantavirus pulmonary syndrome. Virology. 1998;242:99–106. doi: 10.1006/viro.1997.8990. [DOI] [PubMed] [Google Scholar]
- 21.Sall A A, Zanotto P M de A, Sene O K, Zeller H G, Digoutte J P, Thiongane Y, Bouloy M. Genetic reassortment of Rift Valley fever virus in nature. J Virol. 1999;73:8196–8200. doi: 10.1128/jvi.73.10.8196-8200.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saluzzo J F, Smith J F. Use of reassortant viruses to map attenuating and temperature-sensitive mutations of the Rift Valley fever virus MP-12 vaccine. Vaccine. 1990;8:369–375. doi: 10.1016/0264-410x(90)90096-5. [DOI] [PubMed] [Google Scholar]
- 23.Schmaljohn C. Bunyaviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. New York, N.Y: Raven Press; 1996. pp. 1447–1471. [Google Scholar]
- 24.Schwartz O, Marechal V, Friguet B, Arenzana-Seisdedos F, Heard J M. Antiviral activity of the proteasome on incoming human immunodeficiency virus type 1. J Virol. 1998;72:3845–3850. doi: 10.1128/jvi.72.5.3845-3850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Struthers J K, Swanepoel R, Shepherd S P. Protein synthesis in Rift Valley fever virus infected cells. Virology. 1984;134:118–124. doi: 10.1016/0042-6822(84)90277-0. [DOI] [PubMed] [Google Scholar]
- 26.Teng M N, Collins P L. Altered growth characteristics of recombinant respiratory syncytial viruses which do not produce NS2 protein. J Virol. 1999;73:466–473. doi: 10.1128/jvi.73.1.466-473.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Urquidi V, Bishop D H L. Non-random reassortment between the tripartite RNA genomes of La Crosse and snowshoe hare viruses. J Gen Virol. 1992;73:2255–2265. doi: 10.1099/0022-1317-73-9-2255. [DOI] [PubMed] [Google Scholar]
- 28.Vialat P, Muller R, Vu T H, Prehaud C, Bouloy M. Mapping of the mutations present in the genome of the Rift Valley fever virus attenuated MP12 strain and their putative role in attenuation. Virus Res. 1997;52:43–50. doi: 10.1016/s0168-1702(97)00097-x. [DOI] [PubMed] [Google Scholar]
- 29.Yadani F-Z, Kohl A, Préhaud C, Billecocq A, Bouloy M. The carboxy-terminal acidic domain of Rift Valley fever virus NSs protein is essential for the formation of filamentous structures but not for the nuclear localization of the protein. J Virol. 1999;73:5018–5025. doi: 10.1128/jvi.73.6.5018-5025.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]