Abstract
Among 34 Epstein-Barr virus isolates from nonimmunocompromised Chinese donors, we identified three intertypic recombinants with type 1 sequences at the EBNA2 locus and type 2 sequences at some or all of the EBNA3A, -3B, and -3C loci. These appear to have arisen from independent, evolutionarily recent recombination events; such events may be commoner in nonimmunocompromised populations than hitherto imagined.
The classification of Epstein-Barr virus (EBV) strains into types 1 and 2 is based on linked polymorphisms in the EBV nuclear antigen 2 (EBNA2) gene, situated in the BamHI Y region of the genome, and in the EBNA3A, -3B, and -3C genes, tandemly arranged over 40 kb away in the BamHI E region (2, 10, 20). The two virus types are biologically indistinguishable in almost all respects (18) and coexist within all human populations studied to date (1, 3, 11, 22, 30, 33, 34). In most populations type 1 strains appear the more prevalent, with only a minority of individuals carrying type 2 (1, 11, 15, 30, 34); coinfection with both types appears to be relatively rare, except in certain immunocompromised groups (5, 16, 21, 32). Interestingly, almost all of the >300 EBV isolates analyzed to date from various geographic areas are either uniformly type 1 or uniformly type 2 at all four type-specific loci (6, 11, 13, 29, 30). The only exceptions in the literature are a single intertypic recombinant isolated from a healthy donor from New Guinea (6), an area where type 1 and type 2 viruses show almost equal prevalence (3), and two recombinants isolated from human immunodeficiency virus-positive Caucasian homosexuals (31), an immunocompromised group in which type 1-type 2 coinfection is unusually common (25, 32) and in which high levels of virus coreplication in vivo predispose to recombination events (26, 27). These various findings imply that type 1 and type 2 EBVs, though coexistent in human populations, have followed largely separate evolutionary paths with little impact from intertypic recombination. The present work focuses on virus isolates from southern China, an area which attracts interest because of the unusually high prevalence of an EBV-associated tumor, nasopharyngeal carcinoma (NPC) (18). The limited evidence available from normal donor-derived isolates and from the analysis of NPC biopsies suggests that type 1 strains are predominant in Chinese populations, with overt type 2 virus infection at the same low levels reported for the Western world (7, 9, 14, 34). Superimposing our results on this pattern, we now report an unexpectedly high frequency of intertypic recombinants.
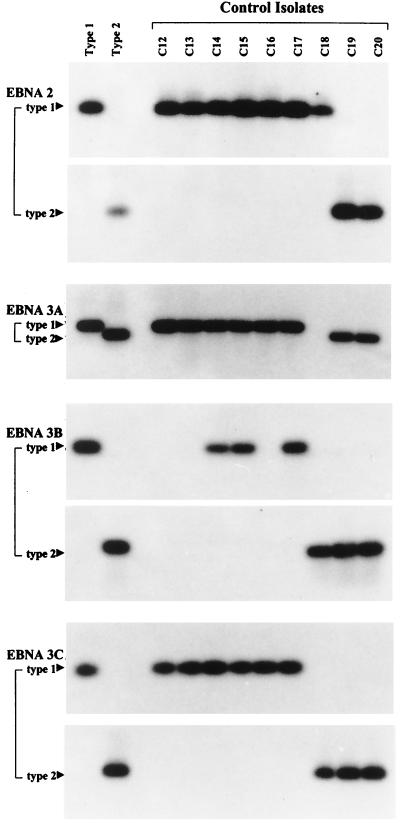
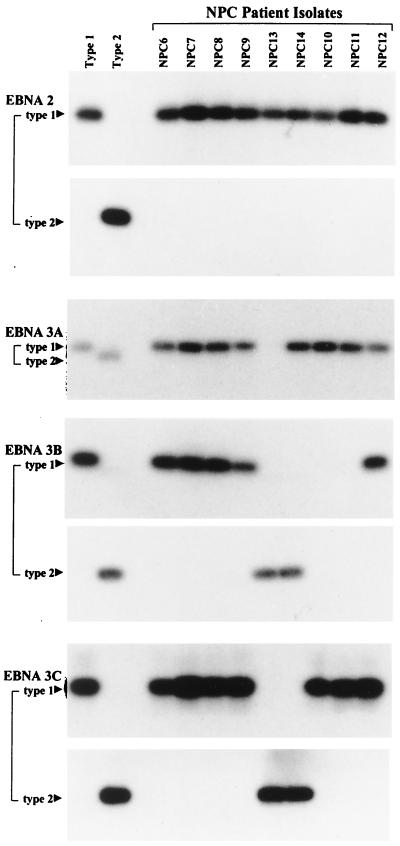
Resident EBV strains were isolated from the blood lymphocytes and/or throat washings of 34 ethnic southern Chinese donors by virus rescue in vitro as EBV-transformed lymphoblastoid cell lines. The methods used (31) were optimized to generate multiple independent isolates per donor and to minimize bias towards type 1 strains, which are generally more efficient than type 2 in in vitro transformation assays (19); where multiple isolates were obtained, all isolates from one donor gave the same pattern of results. Twenty of these donors were healthy individuals and 14 were patients with recently diagnosed NPC in whom we could detect no marked shift in the EBV-host balance or impairment of virus-specific T-cell responses by standard virological and immunological assays (31). In 29 of these 34 donors the resident EBV strain was identified as type 1 at all four polymorphic loci. Healthy control donor isolates C14, C15, and C17 (Fig. 1) and NPC patient isolates NPC 6 to 9 and 12 (Fig. 2) illustrate typical cases where type 1 signals were obtained at EBNA2, -3A, -3B, and -3C by using the standard PCR typing protocols established in earlier work (20). However, a subset of Chinese type 1 viruses (C12, C13, and C16 in Fig. 1 and NPC 10 and 11 in Fig. 2) failed to give a signal at EBNA3B when the standard typing protocol was used. In each case this reflected the presence of two sequence changes relative to the standard B95.8 type 1 sequence (T→G at nucleotide 97414 and A→G at nucleotide 97417) within the region detected by the standard type 1-specific probe; otherwise, the EBNA3B amplification product followed the classical type 1 sequence (data not shown). Only 2 of 34 isolates, from healthy donors C19 and C20 (Fig. 1), were uniformly type 2 at all loci, in line with the comparative rarity of type 2 viruses observed in other surveys of Chinese EBV strains (7, 9, 14, 34). Surprisingly, this left 3 of 34 isolates (C18 [Fig. 1] and NPC 13 and NPC 14 [Fig. 2]) which were immediately recognizable as intertypic recombinants. Thus, isolates C18 and NPC 13 gave a type 1 signal at EBNA2, no signal at EBNA3A, and a type 2 signal at EBNA3B and -3C; isolate NPC 14 was type 1 at EBNA2 and -3A and type 2 at EBNA3B and -3C. Two blood isolates from donor C18, 30 blood and throat washing isolates from NPC patient 13, and 7 blood and throat washing isolates from NPC patient 14 were available; every one had the relevant recombinant structure (data not shown).
FIG. 1.
Genomic typing of representative Chinese healthy control donor isolates C12 to C20 at the EBNA2, -3A, -3B, and -3C type-specific loci using standard PCR protocols. Amplifications were carried out with a common 5′ primer and type-specific 3′ primers in the case of EBNA2 and common 5′ and 3′ primers in the cases of EBNA3A, -3B, and -3C; the products were detected with type-specific probes in the cases of EBNA2, -3B, and -3C and a common probe in the case of EBNA3A (20). All assays included reference DNA samples from the prototype 1 B95.8 cell line and the prototype 2 AG876 cell line.
FIG. 2.
Genomic typing of representative Chinese NPC patient-derived isolates NPC 6-14 at the EBNA2, -3A, -3B, and -3C type-specific loci. PCR protocols and reference DNA samples were as in Fig. 1.
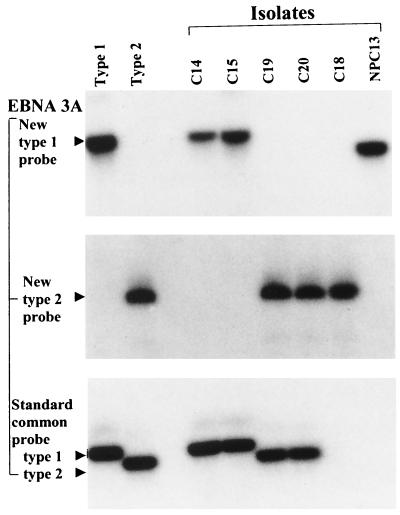
To resolve the EBNA3A status of isolate C18, we employed new primer-probe combinations to amplify a fragment of the EBNA3A sequence larger than that used in the standard assay. This involved the 5′ common primer 5′-GAGACGGCACAGGCTTGGAAT-3′ (B95.8 coordinates 93276 to 93296) and the 3′ common primer 5′-CCTCAGCACGCAAACGAGCCAG-3′ (B95.8 coordinates 94152 to 94130, inclusive); the type 1-specific probe was 5′-GCCCCCTGTATCTCCAGG-3′ (B95.8 coordinates 93767 to 93784) and the type 2-specific probe was 5′-CCCTTAGGGGACCAACT-3′ (AG876 sequence colinear with B95.8 coordinates 93762 to 93766 and 93782 to 93793 and spanning a type 2-specific 15-bp deletion equivalent to B95.8 coordinates 93767 to 93781 [20]). As illustrated in Fig. 3, this assay showed a type 2-specific signal at the EBNA3A locus of C18; the failure to amplify such a product in the original assay proved to be due to a single point mutation in the annealing site for the original type-common 3′ primer (data not shown). Sequencing of C18 confirmed maintenance of a type 1 sequence throughout the EBNA2 gene and of a type 2 sequence throughout EBNA3A. Isolate C18 therefore has a type 1 EBNA2–type 2 EBNA3A, -3B, -3C recombinant structure similar to that seen for the three other recombinants described in the literature (6, 31). In such cases the position of recombination must lie somewhere within the 40-kbp region between the EBNA2 and EBNA3A loci.
FIG. 3.
Further analysis of EBNA3A gene type using new common 5′ and 3′ primers and probing with a new type 1-specific probe or with a new type 2-specific probe compared to using the standard common primers and common probe at this locus. Note that of the two isolates which could not be typed at EBNA3A by the standard protocol, C18 was identified as type 2 and NPC 13 as type 1 (but with a smaller PCR product). Parallel results are shown for the reference B95.8 (type 1) and AG876 (type 2) viruses and for standard Chinese type 1 (C14 and C15) and type 2 (C19 and C20) isolates.
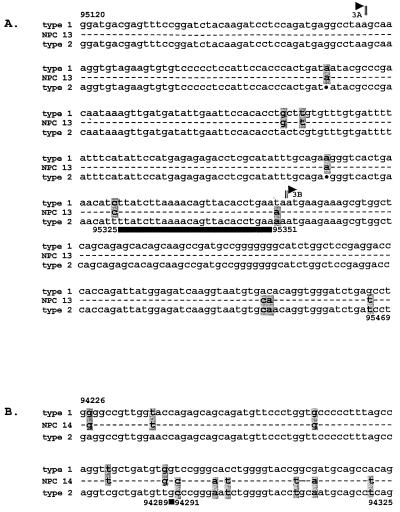
The NPC 13 isolate, which also failed to produce an EBNA3A signal in the original screening, was reanalyzed as described above and yielded a PCR product which hybridized to the type 1-specific probe but was significantly smaller than the expected 877-bp type 1 product (Fig. 3). Sequencing revealed a 228-bp deletion in the EBNA3A gene of NPC 13, but the amplified sequence was clearly type 1 on both sides of the deletion, with <1% nucleotide divergence from the B95.8 prototype in an area where the AG876 type 2 sequence shows >10% divergence (20). Interestingly the deletion removes codons 402 to 477 of the EBNA3A sequence (including one of the primer binding sites used in the original PCR typing assay) but leaves the reading frame intact; we confirmed by immunoblotting that the lymphoblastoid cell lines carrying this virus did indeed express the relevant truncated EBNA3A protein (data not shown). We then went on to identify the position of recombination in NPC 13 by sequencing between the type 1-specific amplified region in EBNA3A and the type 2-specific amplified region in EBNA3B. As shown in Fig. 4A, type 1 signature sequences were maintained beyond the end of the last EBNA3A coding exon (BERF1) through to at least nucleotide 95325 in the intervening sequence; thereafter, the next signature nucleotide at 95351 assumes a type 2 signature which was maintained into the first exon of EBNA3B (BERF2a). A similar analysis of isolate NPC 14, which the original screening had classified as type 1 EBNA2, -3A–type 2 EBNA3B, -3C, identified its point of recombination even more precisely. As shown in Fig. 4B, the EBNA3A gene maintained a type 1 signature downstream of the typing locus to nucleotide 94289 (EBNA3A codon 653) but switched to a type 2 signature from nucleotide 94291 (EBNA3A codon 654) onwards.
FIG. 4.
Genomic sequences in the regions of intertypic recombination. (A) Sequence of the NPC 13 isolate showing the end of the EBNA3A open reading frame, the intervening noncoding sequence, and the beginning of the EBNA3B open reading frame relative to the standard type 1 (B95.8 coordinates 95120 to 95469) and type 2 (AG876) virus sequences. Dashes represent sequences conserved in NPC 13, B95.8, and AG876. At positions where B95.8 and AG876 diverge, the identity of the relevant nucleotide in NPC 13 is shown and its adherence to the type 1 or type 2 consensus is indicated by shading. Dots represent deletions in AG876 relative to B95.8. The black horizontal bar denotes the region within which recombination has occurred in NPC 13. (B) Sequence of the NPC 14 isolate within the EBNA3A gene, shown as in panel A, relative to the standard type 1 (B95.8 coordinates 94226 to 94325) and type 2 (AG876) virus sequences. The black box identifies the site of recombination in NPC 14.
The EBNA2, -3A, -3B, and -3C genotypes of all 34 Chinese viral isolates are summarized in Table 1. Arranged alongside in correct genomic order are the results at two other informative polymorphisms where, in the present panel of isolates, allelic frequencies proved to be influenced by virus type. Thus, at the EBNA1 locus (6.5 kbp downstream of the EBNA3C gene) we sequenced codons 475 to 535 of the EBNA1 C terminus, where a number of allelic variants have been identified among EBV strains from different geographic areas and/or disease states (4, 8, 12, 13, 23, 28). Of the 12 type 1 Chinese strains analyzed for this region, one (C1) followed the prototype B95.8 sequence, designated as the A allele from the codon corresponding to the alanine residue as signature amino acid 487 (4, 13). All the rest showed six amino acid changes relative to B95.8 (487 A→V, 499 D→E, 502 T→N, 524 T→I, 528 I→V, and 533 L→I); this is the same V-allele sequence that others have reported as preferentially associated with NPC (12) but which is clearly a geographically related polymorphism present in many Chinese isolates from healthy donors as well as from NPC patients. Interestingly, both type 2 Chinese isolates showed a different EBNA1 sequence variation (476 P→Q, 487 A→T, 492 S→C, and 524 T→I) characteristic of the T allele present in many Caucasian and African isolates (13). When the analysis was extended to the latent membrane protein 1 (LMP1) locus 58 kbp downstream of EBNA1, 26 of 29 type 1 Chinese strains showed the 30-bp deletion spanning LMP1 codons 343 to 352, a polymorphism known to be common among Chinese isolates (17). Nine representative type 1 viruses with this deletion all had the same LMP1 codon 318 to 386 sequence, with six coding changes relative to B95.8 characterizing the Ch′ LMP1 allele (322 Q→N, 334 Q→R, 335 G→D, 338 L→S, and 366 S→T [9, 24]). Of the 3 of 29 type 1 viruses that did not have deletions at the LMP1 locus, one (C1) was entirely B95.8-like in this region, while the other two (C4 and C13) showed another pattern of changes, here designated Ch" (331 G→Q, 338 L→P, 344 G→D, 355 G→A, 366 S→T [9, 24]). Once again, however, the two type 2 viruses were distinct; both had no deletion at the LMP1 locus but showed a hitherto-unreported pattern of sequence changes (322 Q→E, 334 Q→R, 338 L→S, 352 H→R, 354 G→D, 355 G→V, and 366 S→T), here designated Ch‴.
TABLE 1.
Summary of virus genotype analysisa
| Isolate | Type
|
C terminus
|
||||
|---|---|---|---|---|---|---|
| EBNA2 | EBNA3A | EBNA3B | EBNA3C | EBNA1 | LMP1 | |
| C1 | 1 | 1 | 1 | 1 | A | Non-del, B95 |
| C2 | 1 | 1 | 1 | 1 | Del | |
| C3 | 1 | 1 | 1 | 1 | Del | |
| C5 | 1 | 1 | 1 | 1 | Del | |
| C6 | 1 | 1 | 1 | 1 | Del | |
| C7 | 1 | 1 | 1 | 1 | Del | |
| C8 | 1 | 1 | 1 | 1 | Del | |
| C9 | 1 | 1 | 1 | 1 | Del | |
| C10 | 1 | 1 | 1 | 1 | Del | |
| C11 | 1 | 1 | 1 | 1 | Del | |
| C12 | 1 | 1 | 1 | 1 | V | Del, Ch′ |
| C14 | 1 | 1 | 1 | 1 | V | Del, Ch′ |
| C15 | 1 | 1 | 1 | 1 | V | Del, Ch′ |
| C16 | 1 | 1 | 1 | 1 | V | Del, Ch′ |
| C17 | 1 | 1 | 1 | 1 | V | Del, Ch′ |
| NPC 1 | 1 | 1 | 1 | 1 | V | Del, Ch′ |
| NPC 2 | 1 | 1 | 1 | 1 | V | Del, Ch′ |
| NPC 3 | 1 | 1 | 1 | 1 | V | Del, Ch′ |
| NPC 4 | 1 | 1 | 1 | 1 | V | Del, Ch′ |
| NPC 5 | 1 | 1 | 1 | 1 | Del | |
| NPC 6 | 1 | 1 | 1 | 1 | Del | |
| NPC 7 | 1 | 1 | 1 | 1 | Del | |
| NPC 8 | 1 | 1 | 1 | 1 | Del | |
| NPC 9 | 1 | 1 | 1 | 1 | Del | |
| NPC 10 | 1 | 1 | 1 | 1 | Del | |
| NPC 11 | 1 | 1 | 1 | 1 | Del | |
| NPC 12 | 1 | 1 | 1 | 1 | Del | |
| C4 | 1 | 1 | 1 | 1 | V | Non-del, Ch" |
| C13 | 1 | 1 | 1 | 1 | V | Non-del, Ch" |
| NPC 13 | 1 | 1 | 2 | 2 | V | Del, Ch′ |
| NPC 14 | 1 | 1/2 | 2 | 2 | V | Non-del, Ch" |
| C18 | 1 | 2 | 2 | 2 | T | Non-del, Ch‴ |
| C19 | 2 | 2 | 2 | 2 | T | Non-del, Ch‴ |
| C20 | 2 | 2 | 2 | 2 | T | Non-del, Ch‴ |
Boldface denotes intertypic recombinants. Del, deletion containing; Non-del, non-deletion containing.
Though the numbers of isolates studied were small, there clearly was a tendency among Chinese virus strains for EBNA1 and LMP1 alleles to segregate with virus type. It was therefore interesting to analyze the recombinant virus genomes in that context. The C18 recombinant carried both an EBNA1 allele (T) and an LMP1 allele (nondeletion containing, Ch‴) that were otherwise unique to type 2 Chinese isolates; this suggests that C18 carries a substantial genomic fragment (at least from EBNA3A to LMP1) of type 2 viral origin. In the cases of the NPC 13 and NPC 14 recombinants, both carried an EBNA1 allele (V) and LMP1 alleles (deletion-containing Ch′ or non-deletion-containing Ch‴) only seen here among type 1 Chinese viruses; this suggests that in these cases the second recombination locus must lie in the 6.5 kbp between EBNA3C and EBNA1.
The unique structures of the three intertypic recombinants indicate that these viruses have arisen from separate recombination events. Furthermore these events appear to be evolutionarily quite recent since the recombinants each carry contemporary Chinese type 1 or type 2 sequences at the EBNA1 and LMP1 loci and not the divergent sequences one might expect had these viruses been descendants of much earlier founder recombinations. Though the C18, NPC 13, and NPC 14 viruses could have arisen within the individual donors themselves, multiple virus isolates from these individuals failed to reveal evidence of type 1 or type 2 parental strains. We consider it more likely that these viruses represent recombinant strains that are circulating in the Chinese population. All three recombinants retain B-cell transforming activity, and, at least in the two cases where the isolates came from throat washings (NPC 13 and 14), the viruses are clearly replication competent; based on our current understanding of EBV biology, these are two key requirements for successful EBV infection and transmission in the wild (18). The existence of intertypic recombinants in the southern Chinese population, and their relative frequency (3 of 34) among the isolates studied, was quite unexpected and in sharp contrast to recent experience with >250 European virus isolates (11, 13, 29, 30, 31). More studies will be required to determine the generality of the present findings. However the work implies that, at least in the southern Chinese population, type 1-type 2 coinfection may be commoner than first thought and that intertypic recombination is having a significant impact on the range of EBV strains in circulation.
Acknowledgments
This work was supported by the Cancer Research Campaign, United Kingdom, and by the Medical Research Council in the form of a Clinical Research Fellowship to R.S.M. and a Career Development Award to S.P.L.
We are grateful to Deborah Williams for excellent secretarial help.
REFERENCES
- 1.Abdel-Hamid M, Chen J J, Constantine N, Massoud M, Raab-Traub N. EBV strain variation: geographical distribution and relation to disease state. Virology. 1992;190:168–175. doi: 10.1016/0042-6822(92)91202-6. [DOI] [PubMed] [Google Scholar]
- 2.Adldinger H K, Delius H, Freese U K, Clarke J, Bornkamm G W. A putative transforming gene of Jijoye virus differs from that of Epstein-Barr virus prototypes. Virology. 1985;141:221–234. doi: 10.1016/0042-6822(85)90253-3. [DOI] [PubMed] [Google Scholar]
- 3.Aitken C, Sengupta S K, Aedes C, Moss D J, Sculley T B. Heterogeneity within the Epstein-Barr virus nuclear antigen 2 gene in different strains of Epstein-Barr virus. J Gen Virol. 1994;75:95–100. doi: 10.1099/0022-1317-75-1-95. [DOI] [PubMed] [Google Scholar]
- 4.Bhatia K, Raj A, Gutierrez M I, Judde J-G, Spangler G, Venkatesh H, Magrath I T. Variation in the sequence of Epstein-Barr virus nuclear antigen 1 in normal peripheral blood lymphocytes and in Burkitt's lymphomas. Oncogene. 1996;13:177–181. [PubMed] [Google Scholar]
- 5.Buisson M, Morand P, Genoulaz O, Bourgeat M-J, Micoud M, Seigneurin J-M. Changes in the dominant Epstein-Barr virus type during human immunodeficiency virus infection. J Gen Virol. 1994;75:431–437. doi: 10.1099/0022-1317-75-2-431. [DOI] [PubMed] [Google Scholar]
- 6.Burrows J M, Khanna R, Sculley T B, Alpers M P, Moss D J, Burrows S R. Identification of a naturally occurring recombinant Epstein-Barr virus isolate from New Guinea that encodes both type 1 and type 2 nuclear antigen sequences. J Virol. 1996;70:4829–4833. doi: 10.1128/jvi.70.7.4829-4833.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X Y, Pepper S D, Arrand J R. Prevalence of the A type and B type of Epstein-Barr virus DNA in nasopharyngeal carcinoma biopsies from Southern China. J Gen Virol. 1992;73:463–466. doi: 10.1099/0022-1317-73-2-463. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y Y, Chang K L, Chen W G, Shibata D, Hayashi K, Weiss L M. Epstein-Barr virus-associated nuclear antigen-1 carboxy-terminal gene sequences in Japanese and American patients with gastric carcinoma. Lab Invest. 1998;78:877–882. [PubMed] [Google Scholar]
- 9.Cheung S-T, Leung S-F, Lo K-W, Chiu K W, Tam J S L, Fok T F, Johnson P J, Lee J C K, Huang D P. Specific latent membrane protein 1 gene sequences in type 1 and type 2 Epstein-Barr virus from nasopharyngeal carcinoma in Hong Kong. Int J Cancer. 1998;76:399–406. doi: 10.1002/(sici)1097-0215(19980504)76:3<399::aid-ijc18>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 10.Dambaugh T, Hennessy K, Chamnankit L, Kieff E. U2 region of Epstein-Barr virus DNA may encode Epstein-Barr nuclear antigen 2. Proc Natl Acad Sci USA. 1984;81:7632–7636. doi: 10.1073/pnas.81.23.7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gratama J W, Oosterveer M A P, Weimar W, Sintnicolaas K, Sizoo W, Bolhuis R L H, Ernberg I. Detection of multiple ‘Ebnotypes’ in individual Epstein-Barr virus carriers following lymphocyte transformation by virus derived from peripheral blood and oropharynx. J Gen Virol. 1994;75:85–94. doi: 10.1099/0022-1317-75-1-85. [DOI] [PubMed] [Google Scholar]
- 12.Gutierrez M I, Raj A, Spangler G, Sharma A, Hussain A, Judde J-G, Tsao S W, Yuen P W, Joab I, Magrath I T, Bhatia K. Sequence variations in EBNA1 may dictate restriction of tissue distribution of Epstein-Barr virus in normal and tumour cells. J Gen Virol. 1997;78:1663–1670. doi: 10.1099/0022-1317-78-7-1663. [DOI] [PubMed] [Google Scholar]
- 13.Habeshaw G, Yao Q-Y, Bell A I, Morton D, Rickinson A B. Epstein-Barr virus nuclear antigen 1 sequences in endemic and sporadic Burkitt's lymphoma reflect virus strains prevalent in different geographic areas. J Virol. 1999;73:965–975. doi: 10.1128/jvi.73.2.965-975.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khanim F, Yao Q Y, Niedobitek G, Sihota S, Rickinson A B, Young L S. Analysis of Epstein-Barr virus gene polymorphisms in geographically distinct virus isolates of normal donor and tumour origin. Blood. 1996;88:3562–3568. [PubMed] [Google Scholar]
- 15.Kunimoto M, Tamura S, Tabata T, Yoshie O. One-step typing of Epstein-Barr virus by polymerase chain reaction: predominance of type 1 virus in Japan. J Gen Virol. 1992;73:455–461. doi: 10.1099/0022-1317-73-2-455. [DOI] [PubMed] [Google Scholar]
- 16.Luxton J C, Williams A, Weller I, Crawford D H. Epstein-Barr virus infection of HIV-seropositive individuals is transiently suppressed by high dose acyclovir treatment. AIDS. 1993;7:1337–1343. doi: 10.1097/00002030-199310000-00006. [DOI] [PubMed] [Google Scholar]
- 17.Miller W E, Edwards R H, Walling D M, Raab-Traub N. Sequence variation in the Epstein-Barr virus latent membrane protein 1. J Gen Virol. 1994;75:2729–2740. doi: 10.1099/0022-1317-75-10-2729. [DOI] [PubMed] [Google Scholar]
- 18.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2397–2446. [Google Scholar]
- 19.Rickinson A B, Young L S, Rowe M. Influence of the Epstein-Barr virus nuclear antigen EBNA2 on the growth phenotype of virus-transformed B cells. J Virol. 1987;61:1310–1317. doi: 10.1128/jvi.61.5.1310-1317.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sample J, Young L, Martin B, Chatman T, Kieff E, Rickinson A. Epstein-Barr virus type 1 (EBV-1) and 2 (EBV-2) differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J Virol. 1990;64:4084–4092. doi: 10.1128/jvi.64.9.4084-4092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sculley T B, Apolloni A, Hurren L, Moss D J, Cooper D A. Co-infection with A- and B-type Epstein-Barr virus in human immunodeficiency virus-positive subjects. J Infect Dis. 1990;162:643–648. doi: 10.1093/infdis/162.3.642. [DOI] [PubMed] [Google Scholar]
- 22.Sixbey J W, Shirley P, Chesney P J, Buntin D M, Resnick L. Detection of a second widespread strain of Epstein-Barr virus. Lancet. 1989;ii:761–765. doi: 10.1016/s0140-6736(89)90829-5. [DOI] [PubMed] [Google Scholar]
- 23.Snudden D K, Smith P R, Lai D, Ng M H, Griffin B. Alterations in the structure of the EBV nuclear antigen, EBNA1, in epithelial cell tumours. Oncogene. 1995;10:1545–1552. [PubMed] [Google Scholar]
- 24.Sung N S, Edwards R H, Seillier-Moiseiwitsch F, Perkins A G, Zeng Y, Raab-Traub N. EBV strain variation in nasopharyngeal carcinoma from the endemic and non-endemic regions of China. Int J Cancer. 1998;76:207–215. doi: 10.1002/(sici)1097-0215(19980413)76:2<207::aid-ijc7>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 25.Walling D M, Edmistan S N, Sixbey J W, Abdel-Hamid M, Resnick L, Raab-Traub N. Co-infection of multiple strains of the Epstein-Barr virus in human immunodeficiency virus-associated hairy leukoplakia. Proc Natl Acad Sci USA. 1992;89:6560–6564. doi: 10.1073/pnas.89.14.6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walling D M, Perkins A G, Webster-Cyriaque J, Resnick L, Raab-Traub N. The Epstein-Barr virus EBNA-2 gene in oral hairy leukoplakia: strain variation, genetic recombination, and transcriptional expression. J Virol. 1994;68:7918–7926. doi: 10.1128/jvi.68.12.7918-7926.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walling D M, Raab-Traub N. Epstein-Barr virus intrastrain recombination in oral hairy leukoplakia. J Virol. 1994;68:7909–7917. doi: 10.1128/jvi.68.12.7909-7917.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wrightham M N, Stewart J P, Janjua N J, Pepper S D V, Sample C, Rooney C M, Arrand J R. Antigenic and sequence variation in the C-terminal unique domain of the Epstein-Barr virus nuclear antigen EBNA-1. Virology. 1995;208:521–530. doi: 10.1006/viro.1995.1183. [DOI] [PubMed] [Google Scholar]
- 29.Yao Q-Y, Croom-Carter D S G, Tierney R J, Habeshaw G, Wilde J T, Hill F G H, Conlon C, Rickinson A B. Epidemiology of infection with Epstein-Barr virus types 1 and 2: lessons from the study of a T cell immunocompromised hemophiliac cohort. J Virol. 1998;72:4352–4363. doi: 10.1128/jvi.72.5.4352-4363.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao Q Y, Rowe M, Martin B, Young L S, Rickinson A B. The Epstein-Barr virus carrier state: dominance of a single growth-transforming isolate in the blood and in the oropharynx of healthy virus carriers. J Gen Virol. 1991;72:1579–1590. doi: 10.1099/0022-1317-72-7-1579. [DOI] [PubMed] [Google Scholar]
- 31.Yao Q Y, Tierney R J, Croom-Carter D, Cooper G M, Ellis C J, Rowe M, Rickinson A B. Isolation of intertypic recombinants of Epstein-Barr virus from T cell-immunocompromised individuals. J Virol. 1996;70:4895–4903. doi: 10.1128/jvi.70.8.4895-4903.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yao Q Y, Tierney R J, Croom-Carter D, Dukers D, Cooper G M, Ellis C J, Rowe M, Rickinson A B. Frequency of multiple Epstein-Barr virus infections in T cell-immunocompromised individuals. J Virol. 1996;70:4884–4894. doi: 10.1128/jvi.70.8.4884-4894.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young L S, Yao Q Y, Rooney C M, Sculley T B, Moss D J, Rupani H, Laux G, Bornkamm G W, Rickinson A B. New type B isolates of Epstein-Barr virus from Burkitt's lymphoma and from normal individuals in endemic areas. J Gen Virol. 1987;68:2853–2862. doi: 10.1099/0022-1317-68-11-2853. [DOI] [PubMed] [Google Scholar]
- 34.Zimber U, Adldinger H K, Lenoir G M, Vuillaume M, Knebel-Doeberitz M V, Laux G, Desgranges C, Wittman P, Freese U K, Schneider U, Bornkamm G W. Geographic prevalence of two types of Epstein-Barr virus. Virology. 1986;154:56–66. doi: 10.1016/0042-6822(86)90429-0. [DOI] [PubMed] [Google Scholar]