Abstract
Levels of virus in the plasma are closely related to the pathogenicity of human immunodeficiency virus type 1 (HIV-1). HIV-2 is much less pathogenic than HIV-1, and infection with HIV-2 leads to significantly lower plasma viral load. To identify the source of this difference, we measured both viral RNA and proviral DNA in matched samples from 34 HIV-2-infected individuals. Nearly half had undetectable viral RNA loads (<100 copies/ml), but levels of proviral DNA were relatively high and confirmed that quantities of provirus in HIV-1 and HIV-2 infection were similar. Overall, HIV-2 proviral DNA load did not correlate with viral RNA load, and higher viral RNA load was associated with increased production of plasma virus from the proviral template. These results suggest that low viral load in HIV-2 infection is due to decreased rates of viral production, rather than differences in target cell infectivity.
Human immunodeficiency virus type 2 (HIV-2) is less pathogenic than HIV-1. Rates of both heterosexual transmission and perinatal transmission are much lower than those for HIV-1 (1, 2, 20). These differences are reflected in the different patterns of the HIV-1 and HIV-2 epidemics; while HIV-1 has spread virtually worldwide over the past 20 years, HIV-2 has largely been confined to West Africa (18). Rates of disease development are also much lower than for HIV-1, and more than 95% of infected individuals followed for at least 8 years fit a clinical definition of long-term nonprogression (17, 22).
HIV-1 infection in vivo is now known to be a highly dynamic process, even during the clinically latent period. Studies have demonstrated high rates of viral replication and great variation in the levels of virus found in the peripheral blood, measured as plasma viral load (17, 36). The pathogenesis of HIV-1 is closely related to plasma viral load; differences in viral load have been clearly associated with differences in rates of disease progression (23, 24, 26, 32). Levels of proviral DNA in infected cells have also been related to disease, though this appears to be a weaker association than that for plasma RNA (8, 9, 34).
We have recently reported that viral load, as measured by levels of viral RNA in the plasma, is much lower in HIV-2- infected individuals than in HIV-1-infected individuals, despite similarities in age at infection and time infected (29). This suggests that viral replication is associated with the difference in pathogenicity of the two viruses. However, levels of proviral DNA in HIV-2 infection are, surprisingly, similar to those found in HIV-1 infection, even when adjusted for clinical status (3, 7, 30). The conjunction of these two findings suggests that HIV-2 may replicate less actively than HIV-1. To test this hypothesis, we have examined the relationship between levels of proviral DNA and viral RNA in matched samples from HIV-2-infected individuals. This has resulted in a fuller characterization of viral production in HIV-2 infection that can be useful for understanding the viral dynamics involved in the attenuated clinical phenotype of HIV-2.
Blood was collected from 34 HIV-2 seropositive individuals in a cohort of registered female sex workers in Dakar, Senegal. The epidemiologic and clinical aspects of HIV-1 and HIV-2 infection in the cohort have been previously described (19). All women had given informed consent prior to enrollment, and none had received antiretroviral therapy. CD4+ cell counts were obtained for 18 of the 34 women at the time of sample collection. Levels ranged from 324 to 1,221 CD4+ cells/mm3, with a mean of 672 CD4+ cells/mm3; one individual had AIDS at the time of sample collection. Serostatus was determined by immunoblotting whole-virus lysates of HIV-1 and HIV-2. Serodiagnostic criteria have been previously described (19). The samples were collected in EDTA-containing Vacutainer tubes, and the peripheral blood mononuclear cells (PBMCs) were separated by Ficoll-Hypaque (Cappel, Aurora, Ohio), after which the plasma was stored at −70°C within 6 h of collection. PBMCs from the same separation were either lysed immediately at 56°C with 100 μg of proteinase K per ml in 1× sodium chloride-Tris-EDTA with 1% sodium dodecyl sulfate or stored in liquid nitrogen and processed in Boston, Mass.
HIV-2 viral RNA load was measured as described previously (29). Virions were pelleted from plasma and lysed with a guanidinium isothiocyanate solution. An internal control (IC) RNA was prepared by in vitro transcription and was added to samples during the purification process. The IC contained the same conserved primer binding sites as the HIV-2 samples, but was 25 nucleotides longer, enabling us to distinguish the sample and IC amplicons by size. The purified RNA was amplified with a one-step reverse transcriptase (RT)-PCR kit (rTth EZ kit; PE Biosystems, Foster City, Calif.) and primers designed to amplify a 200-bp fragment of HIV-2 gag. One of the primers (OG63) was labeled with a fluorescent dye, and the reaction product was denatured and processed on an ABI 373XL automated sequencer. The intensity of the fluorescence from each of the two products (sample and IC) was recorded with Gene-Scan software (ABI, Foster City, Calif.). The sample copy number was calculated as the ratio of fluorescence of the two products multiplied by the number of copies of the IC RNA per RT-PCR mixture (1,000) and adjusted for the volume of sample processed (200 μl). Previous studies have shown that the assay was linear over a 4-log range, and the limit of detection was 100 copies/ml (29).
The range of plasma RNA levels in the 34 women was <100 copies/ml (the limit of sensitivity of the assay) to 227,000 copies/ml (Fig. 1); the highest level was found in the individual diagnosed with AIDS. The median level of HIV-2 RNA for the group was 189 copies/ml (Table 1). Levels of HIV-2 RNA were below the limit of detection in 15 of 34 individuals (44%), consistent with what we have previously reported in this population (29). Levels of viral RNA and CD4+ cells were inversely correlated (ρ = −0.62, P < 0.01).
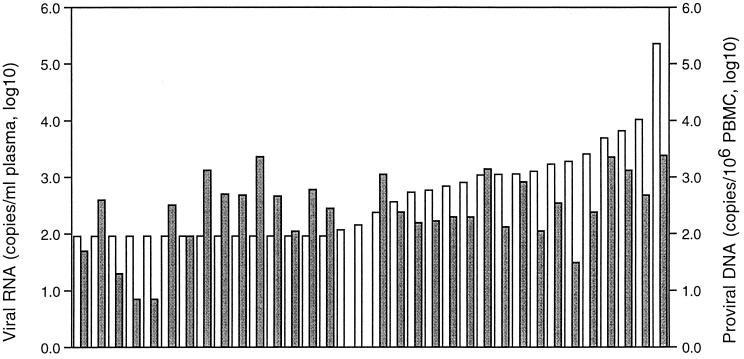
FIG. 1.
Levels of plasma viral RNA and proviral DNA in matched samples. Light bars, RNA measurements; dark bars, proviral DNA measurements.
TABLE 1.
HIV-2 viral RNA and proviral DNA levels from matched samples
| Nucleic acid (n = 34) | Measured levels
|
|
|---|---|---|
| Median | 25th–75th percentile | |
| Viral RNA (copies/ml plasma) | 189 | <100–1,124 |
| Proviral DNA (copies/106 PBMCs) | 259 | 110–600 |
| RNA <100 copies/ml (n = 15) | 356 | 50–501 |
| RNA 100–999 copies/ml (n = 8) | 182 | 79–217 |
| RNA ≥1,000 copies/ml (n = 11) | 479 | 131–1,395 |
| Viral RNA-to-proviral DNA ratio | 2.8 | 0.3–10.7 |
| RNA <100 copies/ml (n = 15) | 0.3 | 0.2–1.8 |
| RNA 100–999 copies/ml (n = 8) | 3.5 | 2.5–60.1a |
| RNA ≥1,000 copies/ml (n = 11) | 8.5 | 2.2–21.6a |
Significantly differs from those with less than 100 copies/ml RNA (P < 0.05).
To measure levels of provirus in the PBMCs, DNA was extracted with phenol-chloroform-isoamyl alcohol, followed by ethanol precipitation. The concentration of the purified DNA was calculated by measuring the optical density at 260 nm, and the purity was calculated by comparison with the optical density at 280 nm; all samples used in the study had a ratio of 1.6 to 2.0. As negative controls, samples from seronegative individuals were processed and tested along with those from HIV-2-infected individuals. A quantitative assay for HIV-2 proviral DNA was established in our laboratory, and we have previously reported results obtained with this technique (30). The assay used a nested PCR to amplify the same portion of the gag gene of HIV-2 as in the RNA assay; results were obtained by comparison of the signal strength of the products from the sample and an IC DNA template amplified in the same tube. For this study we employed a nonradioactive format, replacing the 32P label for the OG63 primer in the second round with the fluorescent dye 6-FAM, processed the samples with an ABI 373XL sequencer, and analyzed the results with Gene-Scan software (ABI). Standard curves generated by the two methods demonstrated that the nonradioactive method led to equivalent results (data not shown).
Levels of HIV-2 proviral DNA ranged from 7 to 2,295 DNA copies per 106 PBMCs, with a median of 259 copies/106 PBMCs (Table 1). There were two samples in which no DNA was detected despite repeated testing; both contained amplifiable DNA as determined by a control PCR amplification of the human β-globin gene (data not shown). For each of these, another DNA sample, collected within 3 months of the sample in question, was tested; one was also negative, and the other had 67 copies/106 PBMCs. For purposes of analysis, the negative samples were assigned values of 1 copy/106 PBMCs. Levels of HIV-2 provirus observed among these women were similar to those found in other studies of HIV-2-infected individuals. Ariyoshi et al. found a median level of 355 copies/106 PBMCs in a community-based study of HIV-2-infected individuals (3), and Berry et al. reported a mean of 17 to 446 copies/106 PBMCs, depending on the level of CD4+ cells (6). We previously reported that HIV-2-infected individuals with CD4+ cell counts greater than 400 had a median proviral load of 64 copies/105 CD4+ cells, which corresponded to 270 copies/106 PBMCs (30). Viral RNA levels are 30-fold less in HIV-2 infection compared to HIV-1 infection (29). In contrast, the levels of HIV-2 proviral DNA reported here are similar to those found in studies of HIV-1 infection, in which median levels of HIV-1 proviral DNA ranged from 105 to 400 copies/106 PBMCs in individuals without AIDS (4, 9, 34).
Levels of proviral DNA, as measured in this study, were not related to CD4+ cell counts (ρ = −0.03, P = 0.90). Most, but not all, studies of HIV-2 proviral load have found an inverse association with CD4+ cell counts (3, 7, 15, 30). Correlations between proviral load and CD4+ cell counts are in part dependent on the denominator used in calculating proviral levels; such an association is weaker when PBMCs rather than CD4+ cells are used as the denominator and most obvious at lower CD4+ cell counts (13). In this study, it is possible that the inclusion of more individuals with lower CD4+ cell counts might have allowed an association of provirus and CD4+ cell count to be detected.
The matched results are presented in Fig. 1, ordered according to the level of plasma viral RNA measured in samples from the study participants. Overall, there was no significant correlation between levels of viral RNA and proviral DNA. There was a trend among those with detectable levels of RNA (ρ = 0.61, P < 0.05), but this relationship disappeared when confined to those with detectable DNA as well. Importantly, the large fraction of HIV-2-infected individuals with fewer than 100 copies/ml of plasma did not have lower levels of proviral DNA than those with more than 100 copies/ml; the median in each group was 320 and 238 copies/106 PBMCs, respectively (P = 0.72). When individuals were further stratified by 10-fold increases in plasma viral load, even those with more than 1,000 copies/ml did not have significantly higher levels of provirus (Table 1). Consequently, the ratio of the RNA and DNA values, which ranged from 0.04 to 141, reflected the change in RNA: there was a significant increase in this relative measure across the three groups, from a median of 0.3 in those with undetectable levels of RNA to 8.5 in the group with greater than 1,000 copies/ml of viral RNA (Table 1). This suggests that increases in viral RNA load may be related to increased expression by proviral DNA templates. To determine if relative rates of viral production were related to HIV-2 pathogenesis, we compared the RNA-to-DNA ratio to the level of CD4+ cells. The mean ratio, based on the normally distributed log transformation of the data, was 8.7 among those with less than 500 CD4+ cells/mm3 and 0.8 among those with higher levels of CD4+ cells, and the ratio of RNA to DNA was significantly correlated with CD4+ cell counts (ρ = −0.57, P < 0.05) (Fig. 2).
FIG. 2.
Ratio of viral RNA and proviral DNA as a function of CD4+ cell count. ρ = −0.57, P < 0.05 (Spearman's rank correlation).
Plasma viral load represents the net balance of the production of virions from infected cells and clearance of virions from the bloodstream. It is formally possible that differences in the RNA-to-DNA ratio are due to changes in the rate of viral clearance, but in vivo studies of HIV-1 viral dynamics have found clearance rates to be constant across a wide range of conditions and levels of viral replication (28). Higher levels of replicating virus appear to be, at least in part, a function of increased levels of expression from the integrated provirus, rather than solely due to the presence of more templates for transcription of the viral genomic RNA. It is possible that the effect is due to the increased release of virions from infected cells or a shift in splicing patterns from multiply spliced to the genomic unspliced RNA needed for production of virions. It has been proposed that a predominantly multiply spliced pattern of viral gene expression is associated with nonprogression of HIV-1 infection; however, this remains controversial (5, 12, 25, 35). Studies of HIV-2 cellular transcripts would be necessary to determine if this might play a role in HIV-2 infection.
In theory, an optimal measure of the relative rates of virus expression might be to measure viral mRNA levels in the same cells that are used to quantify proviral load. However, since viral replication is ultimately dependent on the production of infectious viral particles, measurement of plasma viral RNA may be more relevant for determining in vivo pathogenesis. Furthermore, studies of HIV-1 have found the expected correlation between the levels of full-length unspliced viral mRNAs and plasma RNA. Nonetheless, such measurements, or quantification of proviral DNA as a function of blood volume rather than PBMCs, would yield absolute measures of the amount of virus produced per infected cell. The ratio reported in this study, while not amenable to such calculations, does allow for relative measures of viral expression of both HIV-1 and HIV-2, among studies using similar parameters.
Nearly half of the HIV-2-infected individuals have a nearly quiescent infection, and the paired levels of viral RNA and proviral DNA differ dramatically from those found in HIV-1 infection, where RNA-to-DNA ratios are only rarely less than 1.0 and are above 10.0 in the majority of individuals (4, 27, 34). It is not known if these represent infections that are truly latent at the molecular level; we have been able to detect viral RNA in some individuals with less than 100 copies/ml by a qualitative RT-PCR (S. Popper, unpublished data), suggesting that virus may be produced at very low levels and that the proviruses present in these individuals are not defective.
Productive infection by both HIV-1 and HIV-2 is dependent on continued activation of infected target cells (14); lower production of virus in HIV-2-infected cells may reflect a lower activation state in those cells or that HIV-2 is less responsive to such activation. The long terminal repeat (LTR) of both HIV-1 and HIV-2 regulates the expression of the proviral DNA in response to cellular transactivation signals. The HIV-2 LTR differs from that of HIV-1, in both the number and identity of enhancer elements (11). As a consequence, they are likely to be less responsive to transcription factors present in activated T cells. Hannibal et al. demonstrated that the HIV-2 LTR does not respond as well as the HIV-1 LTR to tumor necrosis factor alpha (16). Similar results were obtained in experiments measuring viral replication (21, 33). There are also potential differences in the activation signals present in HIV-1- and HIV-2-infected cells. Sekigawa et al. found that recombinant HIV-2 envelope glycoprotein was superior to the HIV-1 envelope in its ability to stimulate production of higher levels of gamma interferon and interleukin 16, both of which inhibit viral replication, and lower levels of interleukin 4, which stimulates viral replication (31).
The implications of these in vitro findings for HIV infection in vivo are largely unknown, though there is one report that suggests levels of tumor necrosis factor alpha are lower in HIV-2-infected individuals than in those infected with HIV-1 (10). Such results might indicate that inadequate activation is responsible for low plasma viral load in HIV-2 infection, despite levels of DNA template that result in much higher levels of viral RNA in HIV-1 infection. The results of this study suggest that viral load is related to increased activation and expression from proviral DNA. Comparative studies of both viral and host factors that may affect expression will be useful for understanding the determinants of HIV-2 pathogenesis and the differences between HIV-1 and HIV-2.
Acknowledgments
We thank Chris Mullins, Khady Diop, and M. Lamine Diaw for technical assistance and Jean-Louis Sankalé for helpful suggestions.
This work was supported by grants from the U.S. Army Medical Research and Material Command (17-95-C-5005 and NIH NO1-AI-35173-123). A.D.S. and A.G.-N. are Fogarty International Fellows.
REFERENCES
- 1.Adjorlolo-Johnson G, Decock K M, Ekpini E, Vetter K M, Sibailly T, Brattegaard K, Yavo D, Doorly R, Whitaker J P, Kestens L, Ou C Y, George J R, Gayle H D. Prospective comparison of mother-to-child transmission of HIV-1 and HIV-2 in Abidjan, Ivory Coast. JAMA. 1994;272:462–466. [PubMed] [Google Scholar]
- 2.Andreasson P A, Dias F, Naucler A, Andersson S, Biberfeld G. A prospective study of vertical transmission of HIV-2 in Bissau, Guinea-Bissau. AIDS. 1993;7:989–993. [PubMed] [Google Scholar]
- 3.Ariyoshi K, Berry N, Wilkins A, Ricard D, Aaby P, Naucler A, Ngom P T, Jobe O, Jaffar S, Dias F, Tedder R S, Whittle H. A community-based study of human immunodeficiency virus type 2 provirus load in a rural village in West Africa. J Infect Dis. 1996;173:245–248. doi: 10.1093/infdis/173.1.245. [DOI] [PubMed] [Google Scholar]
- 4.Bagnarelli P, Menzo S, Valenza A, Manzin A, Giacca M, Ancarani F, Scalise G, Varaldo P E, Clementi M. Molecular profile of human immunodeficiency virus type 1 infection in symptomless patients and in patients with AIDS. J Virol. 1992;66:7328–7335. doi: 10.1128/jvi.66.12.7328-7335.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bagnarelli P, Valenza A, Menzo S, Sampaolesi R, Varaldo P E, Butini L, Montroni M, Perno C F, Aquaro S, Mathez D, Leibowitch J, Balotta C, Clementi M. Dynamics and modulation of human immunodeficiency virus type 1 transcripts in vitro and in vivo. J Virol. 1996;70:7603–7613. doi: 10.1128/jvi.70.11.7603-7613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berry N, Ariyoshi K, Jaffar S, Sabally S, Corrah T, Tedder R, Whittle H. Low peripheral blood viral HIV-2 RNA in individuals with high CD4 percentage differentiates HIV-2 from HIV-1 infection. J Hum Virol. 1998;1:457–468. [PubMed] [Google Scholar]
- 7.Berry N, Ariyoshi K, Jobe O, Ngum P T, Corrah T, Wilkins A, Whittle H, Tedder R. HIV type 2 proviral load measured by quantitative polymerase chain reaction correlates with CD4+ lymphopenia in HIV type 2-infected individuals. AIDS Res Hum Retrovir. 1994;10:1031–1037. doi: 10.1089/aid.1994.10.1031. [DOI] [PubMed] [Google Scholar]
- 8.Bruisten S M, Frissen P H J, Vanswieten P, Harrigan P R, Kinghorn I, Larder B, Weigel H M, Devries E, Regez R M, Henrichs J H, Koot M, Huisman J G. Prospective longitudinal analysis of viral load and surrogate markers in relation to clinical progression in HIV type 1-infected persons. AIDS Res Hum Retrovir. 1997;13:327–335. doi: 10.1089/aid.1997.13.327. [DOI] [PubMed] [Google Scholar]
- 9.Chevret S, Kirstetter M, Mariotti M, Lefrere F, Frottier J, Lefrere J J. Provirus copy number to predict disease progression in asymptomatic human immunodeficiency virus type 1 infection. J Infect Dis. 1994;169:882–885. doi: 10.1093/infdis/169.4.882. [DOI] [PubMed] [Google Scholar]
- 10.Chollet-Martin S, Simon F, Matheron S, Joseph C A, Elbim C, Gougerotpocidalo M A. Comparison of plasma cytokine levels in African patients with HIV-1 and HIV-2 infection. AIDS. 1994;8:879–884. doi: 10.1097/00002030-199407000-00003. [DOI] [PubMed] [Google Scholar]
- 11.Clark N M, Hannibal M C, Markovitz D M. The peri-kappa B site mediates human immunodeficiency virus type 2 enhancer activation in monocytes but not in T cells. J Virol. 1995;69:4854–4862. doi: 10.1128/jvi.69.8.4854-4862.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Comar M, Simonelli C, Zanussi S, Depaoli P, Vaccher E, Tirelli U, Giacca M. Dynamics of HIV-1 mRNA expression in patients with long-term nonprogressive HIV-1 infection. J Clin Investig. 1997;100:893–903. doi: 10.1172/JCI119605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cone R W, Gowland P, Opravil M, Grob P, Ledergerber B. Levels of HIV-infected peripheral blood cells remain stable throughout the natural history of HIV-1 infection. AIDS. 1998;12:2253–2260. doi: 10.1097/00002030-199817000-00005. [DOI] [PubMed] [Google Scholar]
- 14.Finzi D, Siliciano R F. Viral dynamics in HIV-1 infection. Cell. 1998;93:665–671. doi: 10.1016/s0092-8674(00)81427-0. [DOI] [PubMed] [Google Scholar]
- 15.Gomes P, Taveira N C, Pereira J M, Antunes F, Ferreira M O S, Lourenco M H. Quantitation of human immunodeficiency virus type 2 DNA in peripheral blood mononuclear cells by using a quantitative-competitive PCR assay. J Clin Microbiol. 1999;37:453–456. doi: 10.1128/jcm.37.2.453-456.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hannibal M C, Markovitz D M, Clark N, Nabel G J. Differential activation of human immunodeficiency virus type-1 and type-2 transcription by specific T-cell activation signals. J Virol. 1993;67:5035–5040. doi: 10.1128/jvi.67.8.5035-5040.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 18.Kanki P. Epidemiology and natural history of human immunodeficiency virus type 2. In: DeVita V T Jr, Hellman S, Rosenberg S A, editors. AIDS: biology, diagnosis, treatment and prevention. 4th ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1997. pp. 127–135. [Google Scholar]
- 19.Kanki P, M'Boup S, Marlink R, Travers K, Hsieh C C, Gueye A, Boye C, Sankale J L, Donnelly C, Leisenring W, et al. Prevalence and risk determinants of human immunodeficiency virus type 2 (HIV-2) and human immunodeficiency virus type 1 (HIV-1) in West African female prostitutes. Am J Epidemiol. 1992;136:895–907. doi: 10.1093/aje/136.7.895. [DOI] [PubMed] [Google Scholar]
- 20.Kanki P J, Travers K U, Mboup S, Hsieh C C, Marlink R G, Guèye-Ndiaye A, Siby T, Thior I, Hernandezavila M, Sankale J L, Ndoye I, Essex M E. Slower heterosexual spread of HIV-2 than HIV-1. Lancet. 1994;343:943–946. doi: 10.1016/s0140-6736(94)90065-5. [DOI] [PubMed] [Google Scholar]
- 21.Markovitz D M, Hannibal M, Perez V L, Gauntt C, Folks T M, Nabel G J. Differential regulation of human immunodeficiency viruses (HIVs): a specific regulatory element in HIV-2 responds to stimulation of the T-cell antigen receptor. Proc Natl Acad Sci USA. 1990;87:9098–9102. doi: 10.1073/pnas.87.23.9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marlink R, Kanki P, Thior I, Travers K, Eisen G, Siby T, Traore I, Hsieh C C, Dia M C, Gueye E, Hellinger J, Guèye-Ndiaye A, Sankale J L, Ndoye I, Mboup S, Essex M. Reduced rate of disease development after HIV-2 infection as compared to HIV-1. Science. 1994;265:1587–1590. doi: 10.1126/science.7915856. [DOI] [PubMed] [Google Scholar]
- 23.Mellors J W, Kingsley L A, Rinaldo C R, Jr, Todd J A, Hoo B S, Kokka R P, Gupta P. Quantitation of HIV-1 RNA in plasma predicts outcome after seroconversion. Ann Intern Med. 1995;122:573–579. doi: 10.7326/0003-4819-122-8-199504150-00003. [DOI] [PubMed] [Google Scholar]
- 24.Mellors J W, Rinaldo C R, Jr, Gupta P, White R M, Todd J A, Kingsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 25.Michael N L, Mo T, Merzouki A, Oshaughnessy M, Oster C, Burke D S, Redfield R R, Birx D L, Cassol S A. Human immunodeficiency virus type 1 cellular RNA load and splicing patterns predict disease progression in a longitudinally studied cohort. J Virol. 1995;69:1868–1877. doi: 10.1128/jvi.69.3.1868-1877.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Brien W A, Hartigan P M, Martin D, Esinhart J, Hill A, Benoit S, Rubin M, Simberkoff M S, Hamilton J D. Changes in plasma HIV-1 RNA and CD4+ lymphocyte counts and the risk of progression to AIDS. Veterans Affairs Cooperative Study Group on AIDS. N Engl J Med. 1996;334:426–431. doi: 10.1056/NEJM199602153340703. [DOI] [PubMed] [Google Scholar]
- 27.Paxton W B, Coombs R W, McElrath M J, Keefer M C, Hughes J, Sinangil F, Chernoff D, Demeter L, Williams B, Corey L. Longitudinal analysis of quantitative virologic measures in human immunodeficiency virus-infected subjects with greater-than-or-equal-to-400 CD4 lymphocytes—implications for applying measurements to individual patients. J Infect Dis. 1997;175:247–254. doi: 10.1093/infdis/175.2.247. [DOI] [PubMed] [Google Scholar]
- 28.Perelson A S, Neumann A U, Markowitz M, Leonard J M, Ho D D. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 29.Popper S, Dieng-Sarr A, Guèye-Ndiaye A, Essex M, Mboup S, Kanki P. Lower HIV-2 viral load reflects the difference in pathogenicity of HIV-1 and HIV-2. J Infect Dis. 1999;180:1116–1121. doi: 10.1086/315010. [DOI] [PubMed] [Google Scholar]
- 30.Sarr A D, Popper S, Thior I, Hamel D, Sankale J, Siby T, Marlink R, Essex M, Mboup S, Kanki P. Relationship between HIV-2 proviral load and CD4+ lymphocyte count differs in HIV monotypic and dual infections. J Hum Virol. 1999;2:45–51. [PubMed] [Google Scholar]
- 31.Sekigawa I, Kaneko H, Neoh L P, Takedahirokawa N, Akimoto H, Hishikawa T, Hashimoto H, Hirose S, Yamamoto N, Kaneko Y. Differences of HIV envelope protein between HIV-1 and HIV-2—possible relation to the lower virulence of HIV-2. Viral Immunol. 1998;11:1–8. doi: 10.1089/vim.1998.11.1. [DOI] [PubMed] [Google Scholar]
- 32.Stein D S, Lyles R H, Graham N M, Tassoni C J, Margolick J B, Phair J P, Rinaldo C, Detels R, Saah A, Bilello J. Predicting clinical progression or death in subjects with early-stage human immunodeficiency virus (HIV) infection: a comparative analysis of quantification of HIV RNA, soluble tumor necrosis factor type II receptors, neopterin, and beta2-microglobulin. Multicenter AIDS Cohort Study. J Infect Dis. 1997;176:1161–1167. doi: 10.1086/514108. [DOI] [PubMed] [Google Scholar]
- 33.Tong-Starksen S E, Welsh T M, Peterlin B M. Differences in transcriptional enhancers of HIV-1 and HIV-2. Response to T cell activation signals. J Immunol. 1990;145:4348–4354. [PubMed] [Google Scholar]
- 34.Verhofstede C, Reniers S, Vanwanzeele F, Plum J. Evaluation of proviral copy number and plasma RNA level as early indicators of progression in HIV-1 infection—correlation with virological and immunological markers of disease. AIDS. 1994;8:1421–1427. doi: 10.1097/00002030-199410000-00008. [DOI] [PubMed] [Google Scholar]
- 35.Vesanen M, Markowitz M, Cao Y Z, Ho D D, Saksela K. Human immunodeficiency virus type-1 mRNA splicing pattern in infected persons is determined by the proportion of newly infected cells. Virology. 1997;236:104–109. doi: 10.1006/viro.1997.8718. [DOI] [PubMed] [Google Scholar]
- 36.Wei X P, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, Saag M S, Shaw G M. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]