Abstract
The Cre-lox system is an indispensable tool in neuroscience research for targeting gene deletions to specific cellular populations. Here we assess the utility of several transgenic Cre lines, along with a viral approach, for targeting cerebellar Purkinje cells (PCs) in mice. Using a combination of a fluorescent reporter line (Ai14) to indicate Cre-mediated recombination and a floxed Dystroglycan line (Dag1flox), we show that reporter expression does not always align precisely with loss of protein. The commonly used Pcp2Cre line exhibits a gradual mosaic pattern of Cre recombination in PCs from Postnatal Day 7 (P7) to P14, while loss of Dag1 protein is not complete until P30. Ptf1aCre drives recombination in precursor cells that give rise to GABAergic neurons in the embryonic cerebellum, including PCs and molecular layer interneurons. However, due to its transient expression in precursors, Ptf1aCre results in stochastic loss of Dag1 protein in these neurons. NestinCre, which is often described as a “pan-neuronal” Cre line for the central nervous system, does not drive Cre-mediated recombination in PCs. We identify a Calb1Cre line that drives efficient and complete recombination in embryonic PCs, resulting in loss of Dag1 protein before the period of synaptogenesis. AAV8-mediated delivery of Cre at P0 results in gradual transduction of PCs during the second postnatal week, with loss of Dag1 protein not reaching appreciable levels until P35. These results characterize several tools for targeting conditional deletions in cerebellar PCs at different developmental stages and illustrate the importance of validating the loss of protein following recombination.
Keywords: AAV, cerebellum, conditional knock-out, cre, dystroglycan, Purkinje cell
Significance Statement
The development of Cre lines for targeting gene deletions to defined cellular populations has led to important discoveries in neuroscience. As with any tool, there are inherent limitations that must be carefully considered. Here we describe several Cre lines available for targeting cerebellar Purkinje cells at various developmental stages. We use the combination of a Cre-dependent fluorescent reporter line and conditional deletion of the synaptic scaffolding molecule Dystroglycan as an example to highlight the potential disconnect between the presence of a fluorescent reporter and the loss of protein.
Introduction
Over the past 35 years, researchers have used transgenic mouse models to define the molecular pathways involved in nervous system development and function. This began with the generation of the first gene knock-out mouse lines in 1989 (Joyner et al., 1989; Koller et al., 1989; Schwartzberg et al., 1989; Zijlstra et al., 1989; Navabpour et al., 2020). While the use of knock-out mice has led to many fundamental discoveries, the constitutive nature of gene deletion can have inherent limitations. These can include early developmental phenotypes that prevent analysis of a gene's function at later stages, as well as phenotypes that arise from cellular populations other than the ones being directly studied. This limitation was circumvented by the development of conditional knock-outs that enable control over when and where a specific gene is manipulated (Gu et al., 1994; Tsien, Chen, et al., 1996; Tsien, Huerta, et al., 1996; Luo et al., 2020). Spatiotemporal control of gene deletion using the Cre-lox system requires two components: (1) a “floxed” allele of the target gene that incorporates 34 base pair loxP recognition sites flanking a critical genomic region of the target gene and (2) a driver line that expresses Cre recombinase under the control of regulatory elements that confer cellular and/or temporal specificity. When Cre is expressed in an animal homozygous for the floxed allele, Cre drives recombination of the loxP sites, deleting the intervening genomic region. Importantly, this deletion only occurs in cells expressing Cre, leaving the floxed allele intact in Cre-negative cells. For example, one of the original Cre driver lines used regulatory elements of CamKIIα to drive recombination in specific excitatory neuron populations beginning in the third postnatal week (Tsien, Chen, et al., 1996). This allowed for the investigation of the role of NMDAR1 (GluN1) in hippocampal plasticity and spatial learning when crossed to a NMDAR1 floxed line, which is lethal when deleted constitutively (Li et al., 1994; Tsien, Huerta, et al., 1996). Similar conditional genetics approaches using different recombinase/recognition sites have been developed, including Flp-FRT (Dymecki and Tomasiewicz, 1998; Park et al., 2011) and Dre-rox (Anastassiadis et al., 2009). The availability of multiple orthogonal recombination systems allows for intersectional approaches that require the coexpression of multiple recombinases to affect gene expression, giving rise to increased cellular specificity.
An important consideration when using conditional deletion is the fidelity of recombination and efficiency of deletion, which is often assayed by crossing the Cre line to a Cre-dependent reporter line. These reporter lines typically express a fluorescent (tdTomato, EGFP) or enzymatic (LacZ, Alkaline Phosphatase) gene from “safe harbor” loci in the genome that are accessible to Cre-mediated recombination. The reporter is preceded by a “lox-stop-lox” (LSL) cassette that prevents constitutive expression in the absence of Cre. However, these cassettes can be highly sensitive to low levels or transient expression of Cre and therefore can be an insufficient proxy for the deletion of a target floxed allele (Luo et al., 2020). It is therefore imperative to validate the expected deletion of the target allele directly by assaying mRNA to show loss of the expected transcript in Cre-positive cells. However, due to differences in the rate of protein turnover, loss of mRNA may not precisely recapitulate the timing of functional protein loss. Therefore, evaluating the loss of protein should define the gold standard for verifying a conditional deletion.
One particular challenge in using conditional deletion strategies for studying developmental events like synapse formation is identifying lines that express Cre early in development in a cell-type–specific manner. For example, Parvalbumin (PV)-positive interneurons in the hippocampus are widely studied and commonly targeted by several PVCre lines (Hippenmeyer et al., 2005; Madisen et al., 2010). However, Cre expression does not initiate in these cells until Postnatal Day 10 (P10)–P12 (de Lecea et al., 1995), which is after they begin the process of synaptogenesis ∼P6 (Doischer et al., 2008). Therefore, the PVCre line can be used to examine the role of genes in synaptic maintenance and/or function, but not initial synapse formation. Several lines express Cre early in the development of cells that give rise to the hippocampal PV+ interneuron population (Dlx5/6Cre, GAD67Cre, Nkx2.1Cre), but these lines target multiple interneuron populations (Potter et al., 2009; Taniguchi et al., 2011).
In this study, we examine the efficiency of several Cre lines in driving deletion of the synaptic cell adhesion protein Dystroglycan (Dag1) from cerebellar Purkinje cells (PCs). We show that the widely used Pcp2Cre line is insufficient for driving loss of Dag1 protein before the period of synaptogenesis. We show that Ptf1aCre drives recombination of a tdTomato reporter allele in all embryonic GABAergic neurons in the cerebellum yet results in stochastic loss of Dag1 protein from PCs. We identify a Calb1-IRES-Cre-D (Calb1Cre) line that drives specific and robust recombination in PCs by P0, resulting in loss of Dag1 protein prior to synaptogenesis. Finally, we show that AAV-mediated delivery of Cre results in gradual transduction of PCs with a lag between reporter expression and loss of Dag1 protein on the order of weeks.
Materials and Methods
Animal husbandry
All animals were housed and cared for by the Department of Comparative Medicine at Oregon Health and Science University, an Association for Assessment and Accreditation of Laboratory Animal Care-accredited institution. Animal procedures were approved by Oregon Health and Science University's Institutional Animal Care and Use Committee (Protocol No. TR02_IP00000539) and adhered to the NIH Guide for the Care and Use of Laboratory Animals. Animal facilities are regulated for temperature and humidity and maintained on a 12 h light–dark cycle (lights on 6A.M.–6P.M.); animals are provided with 24 h veterinary care and food and water ad libitum. Animals were group housed whenever possible and provided with environmental enrichment in the form of extra crinkle paper and a red plastic shelter. Experiments were performed between Zeitgeber Time (ZT) 0 and ZT12. Animals older than P6 were killed by administration of CO2 and animals <P6 were killed by rapid decapitation. Mice of both sexes were used for all experiments.
Mouse strains and genotyping
The day of birth was designated P0. Ages of mice used for each analysis are indicated in the figure and figure legends. Mice were maintained on a C57BL/6 background. Dag1+/− mice were generated by crossing a male Dag1flox/flox mouse to a female Sox2Cre mouse to generate germline Dag1Δ/+ mice, hereafter referred to as Dag1+/− mice; Dag1+/− offspring lacking the Sox2Cre allele were thereafter maintained as heterozygotes (Vincent and Robertson, 2003). For all Dag1 conditional crosses, a Cre-positive Dag1+/− breeder was crossed to a Dag1flox/flox;tdT/tdT breeder to generate Cre-positive Dag1flox/+;tdT/+ controls and Cre-positive Dag1flox/−;tdT/+ conditional knock-outs. Genomic DNA extracted from toe or tail samples using the HotSHOT method (Truett et al., 2000) was used to genotype animals. Primers for genotyping can be found on The Jackson Laboratory webpage or originating article (Table 1). Dag1+/− mice were genotyped with the following primers: CGAACACTGAGTTCATCC (forward) and CAACTGCTGCATCTCTAC (reverse). For each mouse strain, littermate controls were used for comparison with mutant mice.
Table 1.
Mouse strains
| Common name | Strain name | Reference | Stock no. |
|---|---|---|---|
| BAC-Pcp2-IRES-Cre | B6.Cg-Tg(Pcp2-cre)3555Jdhu/J | Zhang et al. (2004) | 010536 |
| Ai14(RCL-tdT)-D | B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J | Madisen et al. (2010) | 007914 |
| Dag1flox | B6.129(Cg)-Dag1tm2.1Kcam/J | Cohn et al. (2002) | 009652 |
| Nestin-Cre | B6.Cg-Tg(Nes-cre)1Kln/J | Tronche et al. (1999) | 003771 |
| p48-Cre | Ptf1atm1(cre)Hnak/RschJ | Nakhai et al. (2007) | 023329 |
| Calb1-IRES2-Cre-D | B6;129S-Calb1tm2.1(cre)Hze/J | Daigle et al. (2018) | 028532 |
| B6NJ.Sox2-Cre | B6N.Cg-Edil3Tg(Sox2-cre)1Amc/J | Hayashi et al. (2002) | 014094 |
Intracerebroventricular virus injection
A borosilicate capillary glass (World Precision Instruments, catalog #1B150F-4) was pulled into a fine tip and then beveled to an angled point. The capillary was then filled with AAV8-CMV.III.eGFP-Cre.WPRE.SV40 with a titer of 1.00 × 1013 (Penn Vector Core, University of Pennsylvania) diluted 1:10 in phosphate-buffered saline (PBS) with Fast Green FCF (Thermo Fisher Scientific, catalog #BP123-10) for visualization (1.00 × 1012 final titer after dilution). P0 mice were deeply anesthetized through indirect exposure to ice until they were unresponsive to light touch. An ethanol wipe was used to sterilize the skin of each pup prior to injection. The capillary tip was manually guided into the lateral cerebral ventricle, and a Toohey Spritzer Pressure System IIe (Toohey) delivered a 30 psi pulse for 30 msec. The process was repeated in the other hemisphere. Pups were returned to their home cage after they have recovered on a warm pad and are mobile.
Perfusions and tissue preparation
Brains from mice younger than P21 were dissected and drop fixed in 5 ml of 4% paraformaldehyde (PFA) in PBS overnight for 18–24 h at 4°C. Mice P21 and older were deeply anesthetized using CO2 and transcardially perfused with ice-cold 0.1 M PBS followed by 15 ml of ice-cold 4% PFA in PBS. After perfusion, brains were postfixed in 4% PFA for 30 min at room temperature. Brains were rinsed with PBS, embedded in 4% low-melt agarose (Thermo Fisher Scientific, catalog #16520100), and sectioned at 70 µm using a vibratome (VT1200S, Leica Microsystems) into 24-well plates containing 1 ml of 0.1 M PBS with 0.02% sodium azide.
Immunohistochemistry
Free-floating vibratome sections (70 µm) were briefly rinsed with PBS and then blocked for 1 h in PBS containing 0.2% Triton X-100 (PBST) plus 2% normal goat or donkey serum. For staining of Dystroglycan synaptic puncta, an antigen retrieval step was performed prior to the blocking step: sections were incubated in sodium citrate solution, pH 6.0 (10 mM sodium citrate, 0.05% Tween-20), for 12 min at 95°C in a water bath followed by 12 min at room temperature. Free-floating sections were incubated with primary antibodies (Table 2) diluted in blocking solution at 4°C for 48–72 h. Sections were then washed with PBS three times for 20 min each. Sections were then incubated with a cocktail of secondary antibodies (1:500, Alexa Fluor 488, 546, 647) in blocking solution containing Hoechst 33342 (1:10,000, Life Technologies, catalog #H3570) overnight at room temperature followed by three final washes with PBS. Finally, sections were mounted on Fisher Superfrost Plus microscope slides (Thermo Fisher Scientific, catalog #12-550-15) using Fluoromount-G (SouthernBiotech, catalog #0100-01), covered with no. 1 coverslip glass (Thermo Fisher Scientific, catalog #12-541-025), and sealed using nail polish.
Table 2.
Primary antibodies used for immunohistochemistry
| Target | Host species | Dilution | Source | Catalog # | RRID |
|---|---|---|---|---|---|
| α-Dystroglycan (IIH6C4) | Mouse | 1:250 | Millipore Sigma | 05-593 | AB_309828 |
| tdTomato | Goat | 1:1,000 | Biorbyt | orb182397 | AB_2687917 |
| Calbindin | Chicken | 1:1,000 | Boster Bio | M03047-2 | AB_2936235 |
| Calbindin | Rabbit | 1:1,000 | Swant | CB38a | AB_10000340 |
| PV | Goat | 1:1,000 | Swant | PVG213 | AB_2721207 |
Microscopy
Imaging was performed on either a Zeiss Axio Imager M2 fluorescence upright microscope equipped with an Apotome.2 module or a Zeiss LSM 980 laser scanning confocal build around a motorized Zeiss Axio Observer Z1 inverted microscope with a Piezo stage. The Zeiss Axio Imager M2 uses a metal halide light source (HXP 200 C), Axiocam 506 mono camera, and 10×/0.3 NA EC Plan-Neofluar, 20×/0.8 NA Plan-Apochromat objectives. The LSM 980 confocal light path has two multialkali photomultiplier tubes (PMTs) and two GaAsP PMTs for four track imaging. Confocal images were acquired using a 63×/1.4 NA Plan-Apochromat Oil DIC M27 objective. For some experiments utilizing the LSM 980 confocal, a linear Wiener filter deconvolution step (Zeiss LSM Plus) was used at the end of image acquisition with 1.2× Nyquist sampling (Figs. 3B,C, 7C–E). Z-Stack images were acquired and analyzed off-line in ImageJ/Fiji (Schindelin et al., 2012). Images within each experiment were acquired using the same microscope settings. Brightness and contrast were adjusted identically across samples in Fiji to improve visibility of images for publication. Figures were composed in Adobe Illustrator 2023 (Adobe Systems), with graphs assembled in R (version 4.2.3).
Figure 3.
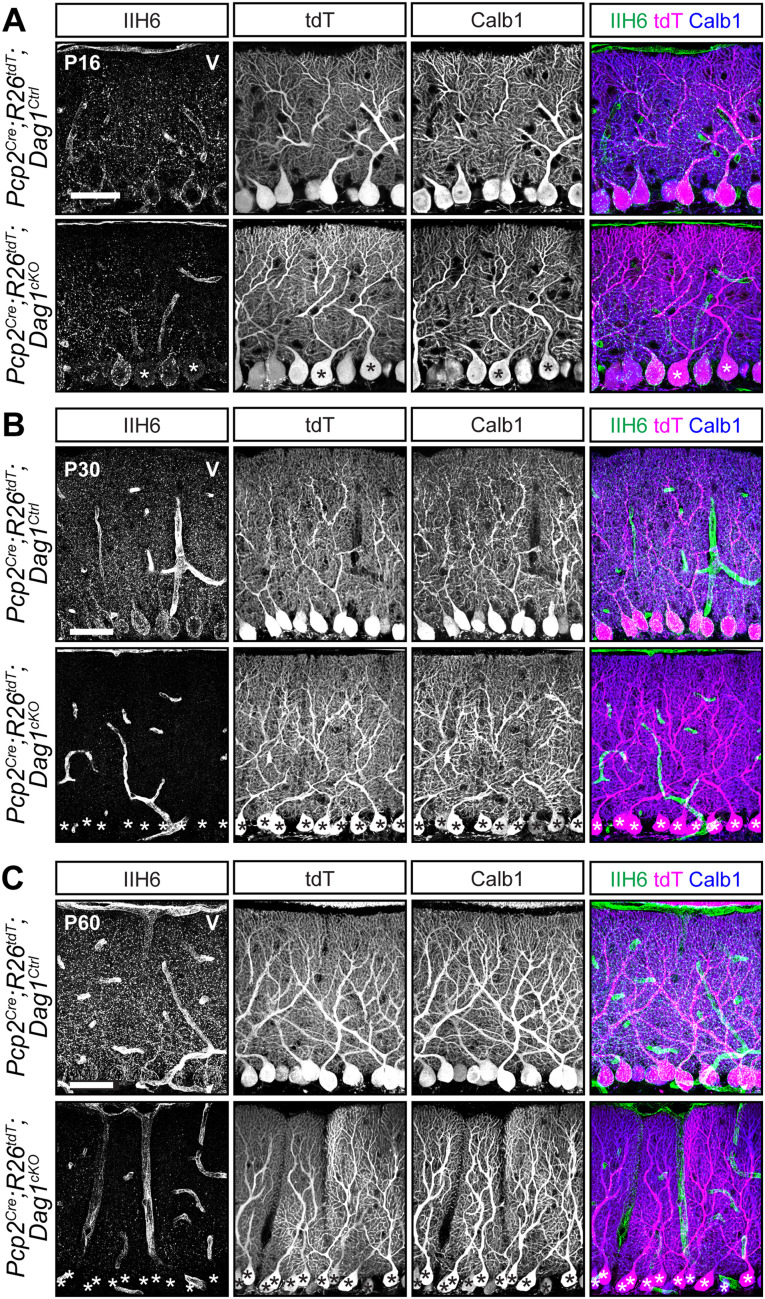
Dystroglycan protein loss lags behind Pcp2Cre recombination of fluorescent tdTomato reporter. A–C, Lobule 5 PCs of Pcp2Cre;ROSA26LSL-tdTomato;Dag1cKOs and littermate controls immunostained for α-Dystroglycan (IIH6, green) to visualize Dag1 protein and Calbindin (Calb1, blue) to label PCs. Cre recombination was assessed by native fluorescence of the tdTomato reporter (tdT, magenta). Expression was evaluated at (A) P16, (B) P30, and (C) P60. Asterisks denote PCs with no detectable IIH6 signal. Scale bars, 50 µm.
Figure 7.
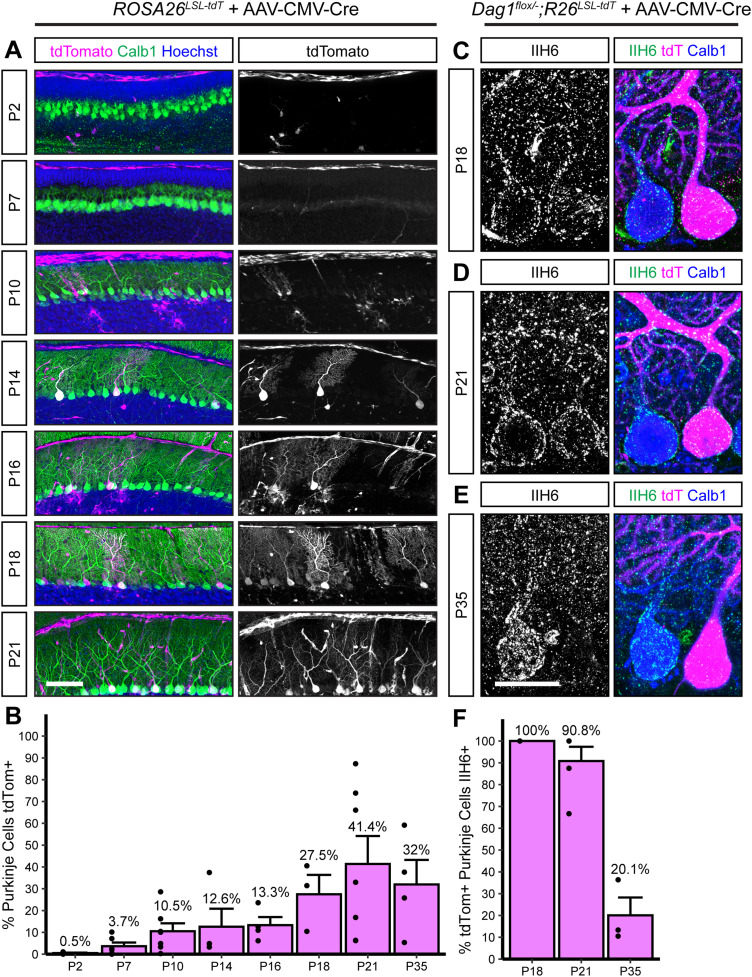
Viral delivery of Cre under the ubiquitous CMV promoter takes several weeks to recombine the Dag1 floxed locus in PCs. A, Dilute AAV8-CMV-Cre (1.00 × 1012) was injected into the lateral ventricles of P0 ROSA26LSL-tdTomato reporter mice. Cerebella were collected at timepoints from P2 to P35 to observe tdTomato expression. Sagittal sections were immunolabeled with Calbindin (Calb1, green) to label PCs and Hoechst (blue) to label nuclei. Cre recombination was assessed by native fluorescence of the tdTomato reporter (magenta). Scale bar, 100 µm. B, Quantification of the percentage of PCs that express tdTomato in A. (P2N = 4; P7N = 6; P10N = 7; P14N = 4; P16N = 4; P18N = 3; P21N = 7; P35N = 4 mice.) C–E, Dag1flox/−;ROSA26LSL-tdTomato mice were injected with dilute AAV8-CMV-Cre at P0, and brains were collected at (C) P18, (D) P21, and (E) P35. Cerebellar sections were counterstained with Calbindin (Calb1, blue) to label PCs and IIH6 (green) to label Dag1 protein. Cre-expressing PCs were visualized with native tdTomato expression (magenta). Scale bar, 25 µm. F, Quantification of the percentage of tdTomato-expressing PCs that are IIH6 immunoreactive in C–E. (P18N = 3; P21N = 3; P35N = 3 mice.)
TdTomato time course image analysis
Images of sagittal cerebellar vermis sections stained for chicken anti-Calbindin and goat anti-tdTomato were taken on a Zeiss Axio Observer Z1 inverted microscope using either a 10×/0.3 NA or 20×/0.8 NA objective, using stitched tiles when necessary to encompass PCs in Lobules 5 and 6. Images were centered where the folia of Lobules 5 and 6 meets. Every PC in the image was manually counted for Calbindin and tdTomato expression. The exact number of PCs counted varied between timepoints as the cerebellum continues to expand rapidly across the timepoints examined (P2 through P35). The percentage of Calbindin+ PCs that were also tdTomato+ was the reported proxy of Cre recombination. The percentage of tdTomato expression from multiple tissue sections was averaged together for a single mouse, and 3–7 mice were analyzed at each timepoint.
Results
Pcp2Cre drives gradual Cre-mediated recombination in PCs
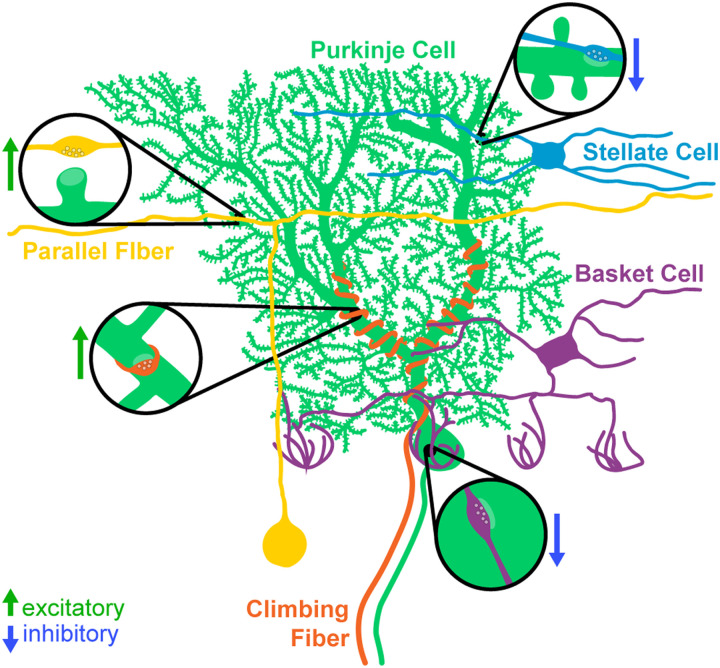
The simple, well-defined, and stereotyped circuitry of the cerebellum makes it an ideal model system for studying synapse development and maintenance (Fig. 1). At the center of the circuit are PCs, which are the primary output neurons of the cerebellum and project their axons to the deep cerebellar nuclei and the vestibular nuclei in the brainstem. PCs receive excitatory inputs from two different sources. Parallel fibers originate from cerebellar granule cells, the most numerous neuron type in the brain, and provide a large number of weak excitatory inputs to the dendrites of PCs (Itō, 1984; Palay and Chan-Palay, 2012). Climbing fibers originate from excitatory neurons in the inferior olive, and their axons wrap around the primary dendritic branches of PCs, forming strong excitatory contacts (Itō, 1984; Palay and Chan-Palay, 2012). In mouse, PCs initially receive inputs from multiple climbing fibers, which undergo activity-dependent pruning during the first 3 weeks of postnatal development until a 1:1 ratio is achieved (Crepel et al., 1976; Bosman et al., 2008; Bosman and Konnerth, 2009, but see Busch and Hansel, 2023). These inputs represent one of the best-studied examples of synaptic competition in the central nervous system (CNS). PCs receive the majority of their inhibitory inputs from two types of molecular layer interneurons (MLIs): basket cells (BCs) and stellate cells (SCs; Itō, 1984; Palay and Chan-Palay, 2012). BCs form inhibitory contacts on the soma and proximal dendrites of PCs, whereas SCs innervate the distal dendrites. Each BC/SC contacts multiple PCs in the same sagittal plane. There are also recurrent inhibitory connections between PCs (Altman, 1972; Bernard and Axelrad, 1993; Witter et al., 2016). PCs in the mouse are born between Embryonic Days (E) 11–13 and elaborate their dendrites over the first 4 postnatal weeks (Miale and Sidman, 1961; Altman, 1972; Yuasa et al., 1991). They begin to receive excitatory and inhibitory inputs after the first postnatal week, and the development of the circuit is largely complete by P28 (Altman, 1972; Kapfhammer, 2004).
Figure 1.
Major synaptic inputs onto PCs in the cerebellar cortex. PCs (green) receive excitatory inputs from two populations: parallel fibers originating from granule cells (yellow) and climbing fibers originating from the inferior olive (orange). The MLIs (BCs, purple; SCs, blue) provide inhibitory input onto PCs.
The most commonly used Cre lines for targeting PCs express Cre under the control of Pcp2 (Purkinje cell protein 2, previously referred to as L7), a gene exclusively expressed by PCs in the brain. There are three Pcp2Cre lines deposited at The Jackson Laboratory: (1) B6.Cg-Tg(Pcp2-cre)3555Jdhu/J (RRID: IMSR_JAX:010536), commonly referred to as BAC-Pcp2-IRES-Cre (Zhang et al., 2004); (2) B6.129-Tg(Pcp2-cre)2Mpin/J (RRID: IMSR_JAX:004146), commonly referred to as L7Cre-2 (Barski et al., 2000); and (3) Tg(Pcp2-cre)1Amc (RRID: IMSR_JAX:006207), commonly referred to as L7-Cre (Lewis et al., 2004). All three transgenic lines were generated with similar bacterial artificial chromosome (BAC) constructs. The L7Cre and L7Cre-2 lines used the same BAC, whereas the sequence used for the generation of the BAC-Pcp2-IRES-Cre line contained more flanking sequence of Pcp2 into which the IRES-Cre sequence was inserted (Smeyne et al., 1995; Barski et al., 2000; Lewis et al., 2004; Zhang et al., 2004).
As all three available Pcp2Cre lines are BAC transgenics, differences in insertion site can result in different patterns of expression. The L7-Cre line exhibits Cre expression in PCs as early as E17, though the Cre expression pattern varies between parasagittal planes early in development, expanding to all PCs by adulthood (Lewis et al., 2004). Although the developmentally early expression of Cre is advantageous for the excision of genes prior to synaptogenesis, this mouse line is only available by cryorecovery. A key limitation of the BAC-Pcp2-IRES-Cre and L7Cre-2 lines is that Cre expression turns on gradually during the postnatal period of PC development. We chose to analyze the BAC-Pcp2-IRES-Cre line (hereafter referred to as Pcp2Cre) as it exhibits expression restricted to PCs, whereas the L7Cre-2 line (hereafter referred to as L7Cre) exhibits “off-target” Cre expression in other cellular populations both within and outside of the cerebellum (Barski et al., 2000; Zhang et al., 2004; Witter et al., 2016).
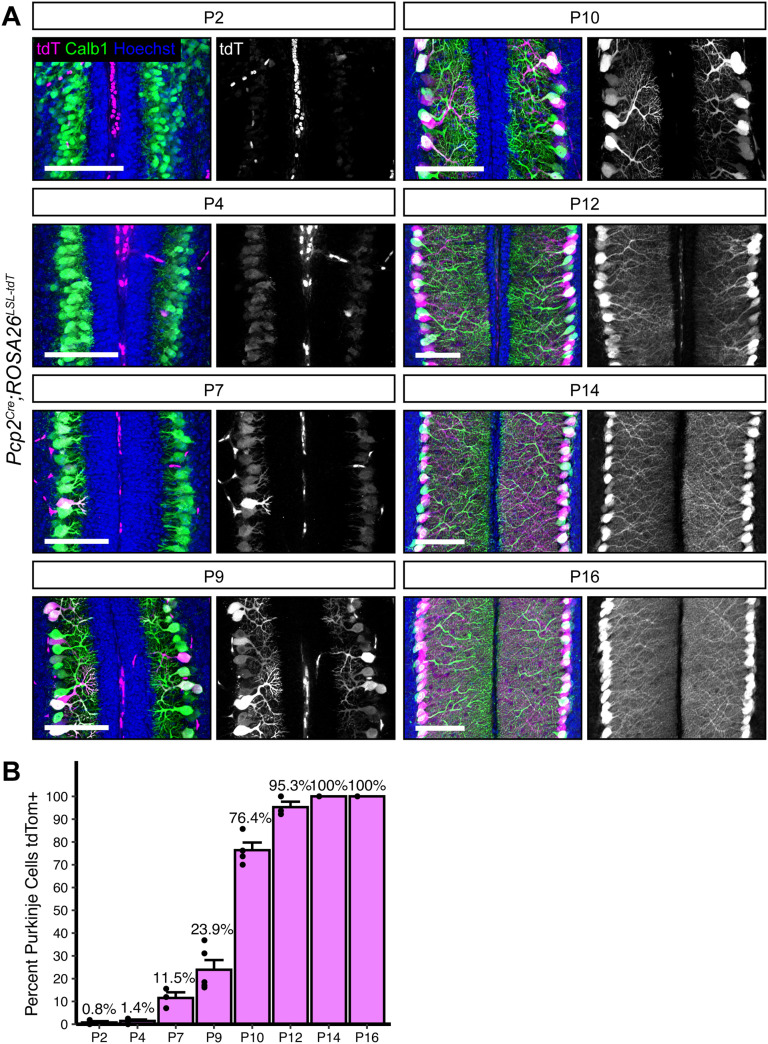
To rigorously and precisely define the period of Cre-mediated recombination in PCs, we analyzed cerebellar Lobule 5/6 from Pcp2Cre;Ai14 brains, which carry a Cre-dependent tdTomato reporter knocked into the ROSA26 locus (Pcp2Cre;ROSA26LSL-tdTomato). The first tdTomato+ PCs were visible at P7 (11.5% of all PCs), and this increased to 23.9% positive at P9. There was an appreciable increase at P10, with 76.4% of PCs labeled by tdTomato, and recombination of tdTomato was complete by P14 (Fig. 2A,B). As previously reported, this line shows high specificity for PCs, as we saw no other tdTomato+ neuron types in the cerebellum. There was no evidence of variation in tdTomato expression between parasagittal planes, suggesting that Pcp2Cre expression does not correlate with the presence of zebrin stripes. However, the identification of parasagittal variations early in development (<P10) would be difficult due to the low number of Cre-expressing PCs at these timepoints.
Figure 2.
Pcp2Cre drives Cre recombination gradually in the second postnatal week. A, Pcp2Cre was crossed to a tdTomato reporter to observe the time course of Cre recombination. Cerebella were analyzed at timepoints from P2 to P16. Cre recombined cells are labeled with tdTomato (magenta). PCs are visualized with Calbindin immunostaining (green). Nuclei (blue) are most evident in granule cells. Scale bars, 100 µm. B, The percentage of all PCs that were tdTomato+ was quantified at each timepoint. (P2N = 3; P4N = 4; P7N = 3; P9N = 5; P10N = 3; P12N = 3; P14N = 3; P16N = 4 mice).
A key consideration when using Cre lines is the timing of protein loss following deletion of floxed alleles. The presence of fluorescent reporters like tdTomato is frequently used as a proxy for Cre-mediated recombination, particularly in situations where antibodies for the targeted proteins are lacking. However, this does not account for the timing of mRNA and protein turnover following deletion of the targeted allele, which can vary widely. This is exemplified by Dystroglycan (Dag1), a cell adhesion molecule present postsynaptically at PC→MLI synapses. Dag1 is a stable protein, with a half-life of ∼25 d in the skeletal muscle (Novak et al., 2021). Recent work has shown that Dag1 is required for PC→MLI synapse maintenance (Briatore et al., 2020). However, this study was unable to examine its role in synapse formation due to the gradual recombination driven by the L7Cre line. We generated Pcp2Cre;Dag1flox/− mice (hereafter referred to as Pcp2Cre;Dag1cKO) to see if starting with one allele of Dag1 already deleted would result in more rapid protein loss. To assess loss of Dag1 by immunohistochemistry, we used the antibody IIH6, which detects the mature matriglycan chains that are specific to Dag1. At P16, a timepoint at which all PCs are tdTomato+ in Pcp2Cre mice (Fig. 2), we still observed punctate Dag1 staining consistent with synaptic localization in the majority of Pcp2Cre;Dag1cKO PCs (Fig. 3A). Analysis at P30, a period after developmental synaptogenesis is complete, showed a loss of Dag1 protein in Pcp2Cre;Dag1cKO PCs by immunohistochemistry, which appeared identical at P60 (Fig. 3B). The incomplete loss of Dag1 protein in Pcp2Cre;Dag1cKO PCs at P16 highlights a limitation of using a Cre line that turns on after gene expression initiates when the protein is particularly stable, as many synaptic molecules are.
Widespread NestinCre recombination does not extend to PCs
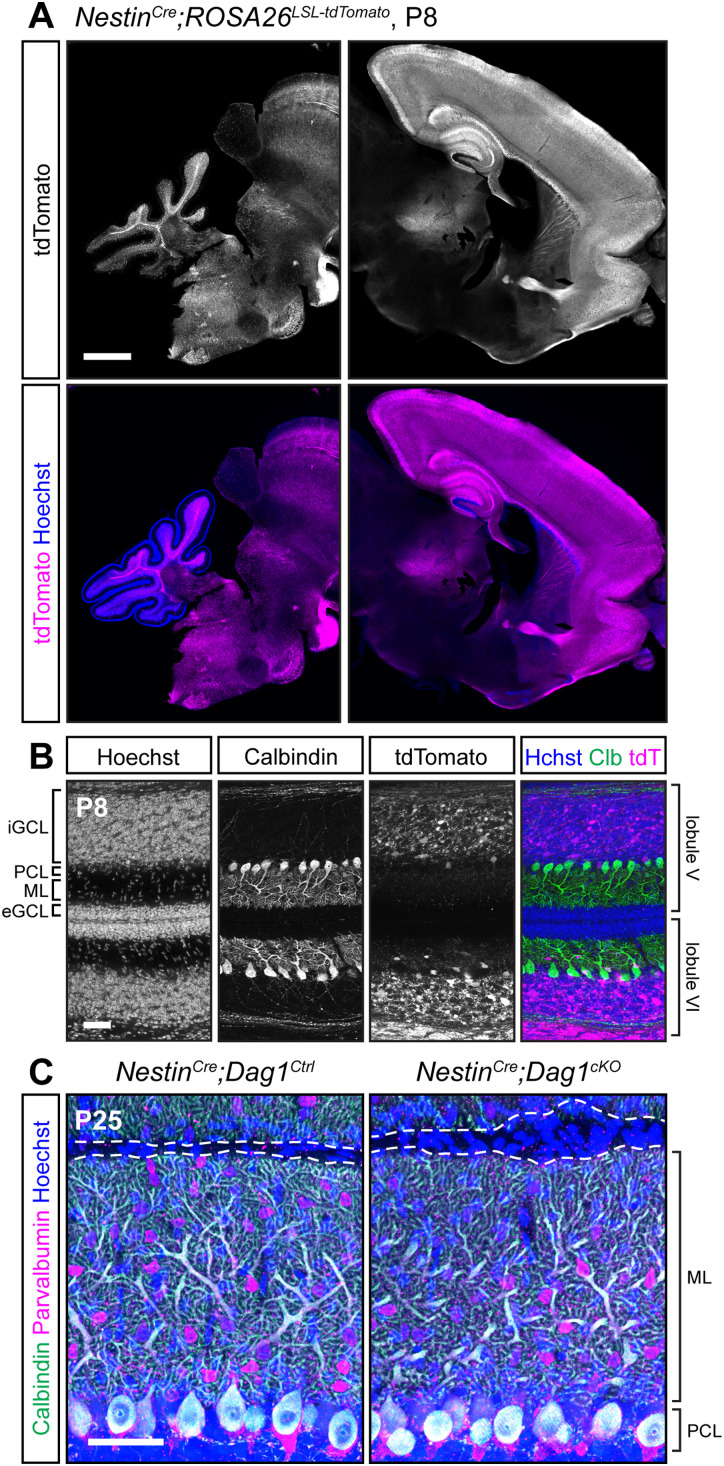
We sought to identify a Cre line that would express in PCs prior to synaptogenesis. We first examined NestinCre, which has widespread expression in the CNS beginning at E10.5 (Tronche et al., 1999; Graus-Porta et al., 2001). Examination of brains from NestinCre;ROSA26LSL-tdTomato mice at P8 showed extensive recombination in the forebrain but minimal recombination in the cerebellum (Fig. 4A). Higher magnification images confirmed that there was no recombination in PCs, MLIs, or cerebellar granule neurons at P8 (Fig. 4B). Consistent with previous reports, there were mild perturbations in cerebellar granule neuron migration in NestinCre;Dag1cKO mice due to Dystroglycan's function in the Bergmann glia (Fig. 4C; Nguyen et al., 2014). It is important to note the particular NestinCre line that we used, B6.Cg-Tg(Nes-cre)1Kln/J (Tronche et al., 1999; RRID, IMSR_JAX:003771), as there are a number of different NestinCre lines that have been generated, some of which have been used to delete genes from PCs with success (Sun et al., 2014; Takeo et al., 2021).
Figure 4.
NestinCre does not drive Cre recombination in cerebellar PCs. A, Fluorescent tdTomato reporter (magenta) in a sagittal brain slice of a P8 NestinCre;ROSALSL-tdTomato mouse. Nuclei are stained in blue. Right and left panels are of the same image but adjusted differently as fluorescence in the forebrain is much brighter than the rest of the brain. Scale bar, 1,000 µm. B, Higher magnification view of tdTomato expression pattern in cerebellar Lobules 5 and 6 at P8 (magenta). PCs are immunolabeled with anti-Calbindin (green). Scale bar, 50 µm. (eGCL, external granule cell layer; iGCL, internal granule cell layer; PCL, Purkinje cell layer; ML, molecular layer.) C, P25 NestinCre;Dag1cKO and littermate control PCs in cerebellar Lobule 5. Calbindin (green) labels PCs; PV (magenta) labels PCs and MLIs; Hoechst (blue) labels nuclei. Dashed line shows the edge of PC dendrites at the pial surface. Scale bar, 50 µm.
Embryonic expression of Ptf1aCre in GABAergic precursors results in stochastic deletion of Dag1 from PCs despite ubiquitous reporter expression
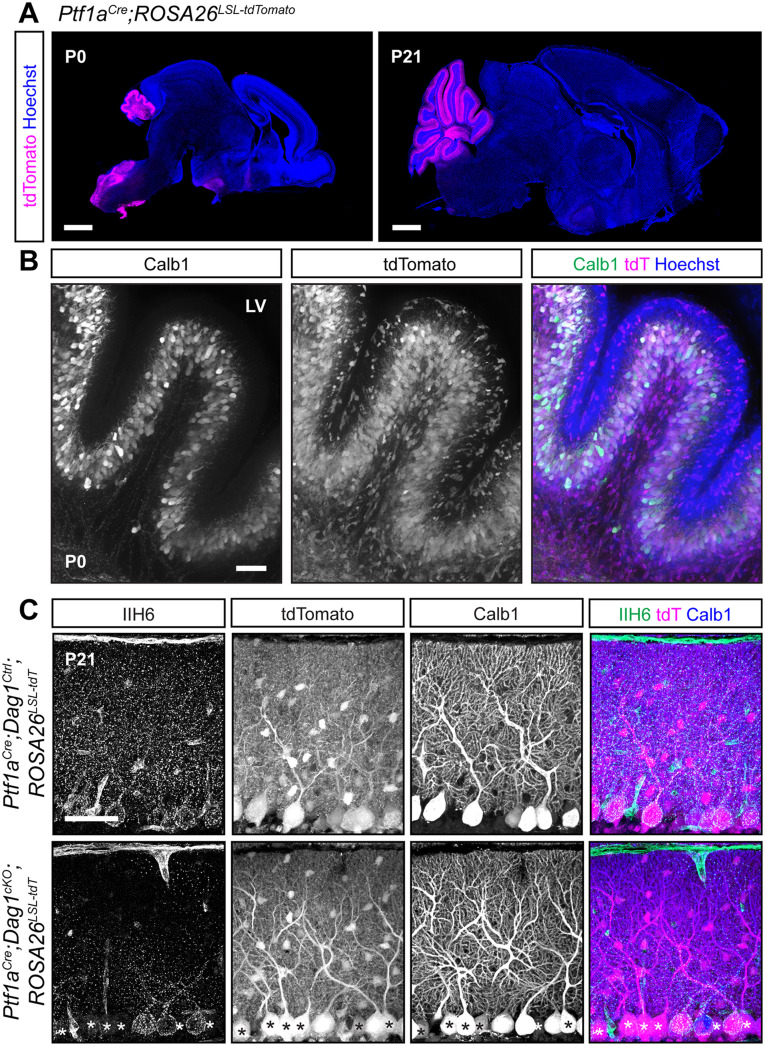
We next tested a Ptf1aCre line, which expresses Cre from the endogenous Ptf1a locus. Ptf1a is a transcription factor that is required for GABAergic neuronal fate in the cerebellum (Hoshino et al., 2005). Examination of Ptf1aCre;ROSA26LSL-tdTomato brains at P0 showed strong tdTomato fluorescence in the nascent cerebellum which colocalized with Calbindin, a marker for PCs (Fig. 5A,B). Examination of adult (P21) brains showed tdTomato in all PCs and MLIs (Fig. 5A,C). The early expression of tdTomato suggested that this line may be useful for developmental deletion of synaptogenic genes in PCs, although with reduced cellular specificity. However, when we examined Dag1 protein localization at P21, we found that Ptf1aCre;Dag1cKO PCs showed mosaic loss of Dag1 protein, despite all PCs being tdTomato+ (Fig. 5C). This result was surprising, as we anticipated that the early expression of Cre in PCs would delete the Dag1 allele before it is expressed at appreciable levels. This difference between robust tdTomato expression and mosaic Dag1 deletion may be due to the transient expression of Ptf1a in GABAergic precursors in conjunction with the known sensitivity of the particular ROSA26LSL-tdTomato floxed locus in the Ai14 line to Cre-mediated recombination (Jin and Xiang, 2019; Luo et al., 2020). In this situation, the transient Cre activity can potentially drive recombination of the reporter allele without driving recombination of the Dag1 floxed allele. These results suggest that the Ptf1aCre line is of limited utility for studying deletion of proteins in GABAergic cerebellar neurons, as the presence of tdTomato is not predictive of loss of the targeted allele.
Figure 5.
Early Cre recombination in developing PCs and MLIs with Ptf1aCre results in a mosaic loss of Dystroglycan protein despite uniform reporter expression. A, Ptf1aCre;ROSA26LSL-tdTomato shows that Cre recombination, as reported by tdTomato (magenta), is evident at P0 and restricted to the cerebellum. Scale bars, 1,000 µm. B, A higher magnification view of cerebellar Lobule 5 at P0. PCs are labeled with Calbindin (Calb1, green), and Cre recombination is reported by native fluorescence of tdTomato (tdT, magenta). Nuclei are visualized with Hoechst (blue). Scale bar, 50 µm. C, Immunostaining in Lobule 5 of P21 cerebella from Ptf1aCre;ROSA26LSL-tdTomato;Dag1cKO and littermate controls for α-Dystroglycan (IIH6, green) to visualize Dag1 protein and Calbindin (Calb1, blue) to label PCs. Cre recombination was assessed by native fluorescence of the tdTomato reporter (tdT, magenta). Asterisks denote PCs with no detectable IIH6 signal. Scale bar, 50 µm.
Calb1Cre drives stable Cre expression and Dag1 deletion in all PCs prior to synaptogenesis
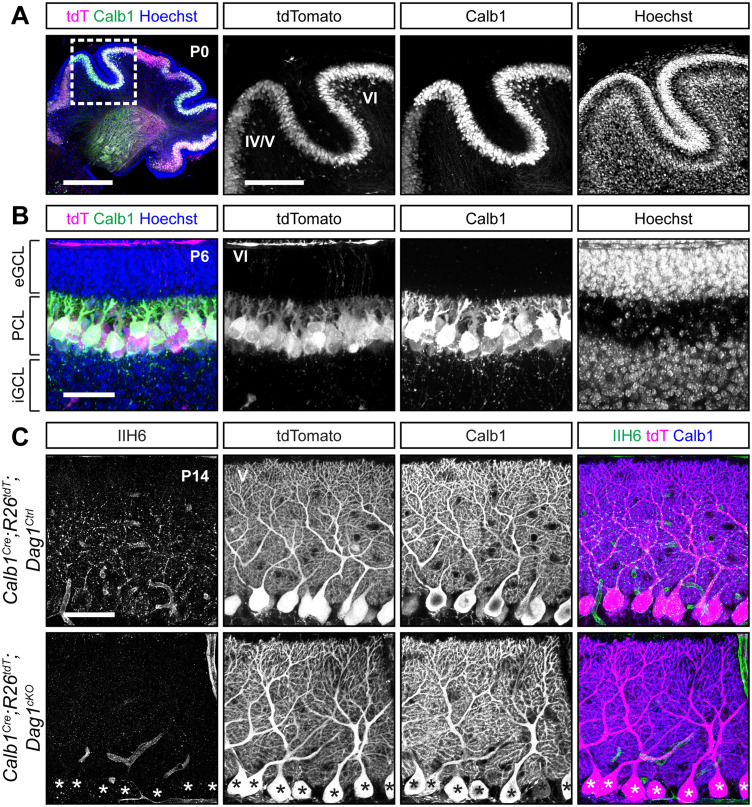
In the course of characterizing the Ptf1aCre line, we observed abundant Calbindin staining in PCs at P0 (Fig. 5), and previous work has shown Calb1 expression in the cerebellum as early as E14.5 (Morales and Hatten, 2006). While Calbindin is widely expressed in multiple neuronal subtypes in the majority of the CNS, within the cerebellum its expression is highly selective for PCs. Calb1-IRES-Cre-D mice (hereafter referred to as Calb1Cre) express Cre from the endogenous Calb1 locus while retaining Calb1 expression (Daigle et al., 2018). Analysis of Calb1Cre;ROSA26LSL-tdTomato brains showed tdTomato signal in all PCs at P0 (Fig. 6A). Further analysis of staining at P6 and P14 continued to show that recombination is specific to PCs (Fig. 6B,C). We next examined Dag1 deletion in Calb1Cre;Dag1cKO mice at P14 during the period of active synaptogenesis in PCs. In contrast to the Pcp2Cre;Dag1cKO mice, which retained Dag1 at P16, we saw a complete loss of punctate Dag1 signal in PCs at P14 in Calb1Cre;Dag1cKO cerebella. These results highlight the utility of the Calb1Cre line to study the effect of gene knock-out in PCs early in development.
Figure 6.
Calb1Cre drives Cre recombination early in development resulting in complete loss of synaptic PC Dystroglycan protein. A, B, The cerebellum of (A) P0 (insets show Lobules 4/5 and 6) and (B) P6 (Lobule 6) Calb1Cre;ROSA26LSL-tdTomato mouse immunolabeled with Calbindin (Calb1, green) to label PCs and Hoechst (blue) to label nuclei. Cre recombination is reported by native tdTomato fluorescence (tdT, magenta). (eGCL, external granule cell layer; iGCL, internal granule cell layer; PCL, Purkinje cell layer.) Scale bars: A, 500 µm, 250 µm (inset); B, 50 µm. C, Immunostaining in Lobule 5 of P14 cerebella from Calb1Cre;ROSA26LSL-tdTomato;Dag1cKO and littermate controls for α-Dystroglycan (IIH6, green) to visualize Dag1 protein and Calbindin (Calb1, blue) to label PCs. Cre recombination was assessed by native fluorescence of the tdTomato reporter (tdT, magenta). Asterisks denote PCs with no detectable IIH6 signal. Scale bar, 50 µm.
AAV delivery of Cre to PCs results in gradual Cre expression over the course of 3 weeks
In addition to transgenic Cre lines, viral-mediated delivery of Cre is commonly used to delete synaptogenic genes throughout the brain. One advantage of viral-mediated deletion is the ability to test cell-autonomous deletion of target alleles in a mosaic fashion by titrating the amount of virus. However, this approach is limited by the timing of viral transduction, subsequent Cre expression, and recombination. To better define this time course in PCs, we examined recombination of tdTomato in ROSA26LSL-tdTomato mice injected in the lateral cerebral ventricles at P0 with dilute AAV8-CMV-Cre. The first tdTomato+ PCs were observed ∼P7–P10, with the number of positive cells increasing to maximal levels by P18 (Fig. 7A,B). This timing is similar to the recombination driven by Pcp2Cre, limiting its utility to examining synapse maintenance. Examination of Dag1 protein in Dagflox/−;ROSA26LSL-tdTomato mice injected with AAV8-CMV-Cre at P0 showed that loss of Dag1 lagged behind tdTomato expression, similar to what was observed in the Pcp2Cre;Dag1cKO PCs (Fig. 7C–F). All tdTomato-expressing PCs were immunoreactive for Dag1 protein at P18, and most (90.8 ± 6.5%) remained Dag1 immunoreactive at P21 (Fig. 7C,D,F). By P35 the majority of tdTomato-expressing PCs lacked Dag1 immunoreactivity (Fig. 7E,F). This lag between tdTomato expression and loss of Dag1 protein implies that it can take 2 or more weeks for Dag1 synaptic protein to turn over.
Discussion
The Cre-lox system has proven to be an invaluable tool for studying the role of genes in a population-specific manner as conditional gene deletion is often required to avoid lethality of constitutive deletion. However, our results highlight the need for rigorous validation to ensure that a transgenic Cre line drives recombination and protein loss in expected cell types and timepoints. While the timing of Cre expression in a given line should be consistent across different conditional lines, the recombination efficiency of the floxed allele and the stability of existing protein that must be turned over before a cell can be deemed a “knock-out” can vary. For these reasons, it is important to use a Cre-dependent reporter to evaluate specificity, along with a method for evaluating loss of protein (or mRNA) to validate deletion.
Each Cre-dependent reporter line may have different advantages and limitations. The ROSA26 and TIGRE loci are popular sites for insertion of Cre-dependent fluorescent reporters due to their genomic accessibility. However, even reporters within the same loci (i.e., ROSA26) can show differences in recombination efficiency, which could lead to confusion about patterns of Cre expression (Madisen et al., 2010; Daigle et al., 2018). There have also been cases reported in which a Cre line may become lethal or cause health complications when crossed to certain fluorescent reporter lines but not others (Daigle et al., 2018). Here we used the popular Ai14 line which expresses LSL-tdTomato in the ROSA26 locus. The Ai14 line is particularly sensitive to recombination, likely due to the proximity of the two loxP sites to one another. This could lead to off-target reporter expression and requires especially rigorous validation of any knock-out when paired with inducible Cre lines which require administration of tamoxifen to activate the Cre protein (Álvarez-Aznar et al., 2020; Luo et al., 2020).
Inducible Cre lines (e.g., CreERT2) are often used to either control the timing of Cre activity or to restrict Cre activity to a subset of a cellular population. However, because of the transient nature, inducible Cre lines require especially careful validation. CreERT2 consists of Cre fused to the hormone-binding domain of the estrogen receptor (Feil et al., 1997; Feil et al., 2009). Under baseline conditions, CreERT2 localization is excluded from the nucleus. Once tamoxifen is administered, it is metabolized into 4-hydroxytamoxifen, a synthetic ligand of the estrogen receptor that binds to CreERT2 and permits localization to the nucleus. After tamoxifen has been cleared from the system, CreERT2 is once again excluded from the nucleus, restricting CreERT2 activity to roughly 24 h after tamoxifen administration (Feil et al., 1997). Since the effect of tamoxifen is both transient and dose-dependent, it is prudent that a Cre-dependent reporter is used to identify which cells experienced Cre-mediated recombination. However, due to differences in sensitivity to recombination between floxed alleles, reporter expression cannot be used alone to predict loss of protein (Feil et al., 2009).
Based on expression of tdTomato driven by Pcp2Cre, we observed that Pcp2Cre drives Cre expression selectively but gradually in PCs beginning at P7 and reaching all PCs by P14 (Fig. 2). This time course is similar to what has been reported for the related L7Cre line (Barski et al., 2000; Lewis et al., 2004). However, when L7Cre was used to delete Dag1 from PCs, loss of protein wasn't complete until P90 (Briatore et al., 2020), whereas we observed loss of Dag1 protein by P30 (Fig. 3). This difference in the loss of protein is likely due to differing breeding strategies. Briatore and colleagues report crossing L7Cre;Dag1flox/flox mice with Dag1flox/flox mice to generate L7Cre;Dag1flox/flox mutants and Dag1flox/flox littermate controls. While this is an efficient breeding strategy allowing for the use of all progeny in a litter, the use of flox/flox mutants may not be ideal for studying Dag1 specifically. Dag1 is a stable protein, and in this case, Cre must recombine both alleles of Dag1 before protein loss can begin. We crossed Pcp2Cre;Dag1+/− mice to Dag1flox/flox mice to generate Pcp2Cre;Dag1flox/− mutants and Pcp2Cre;Dag1flox/+ littermate controls. Not only does this approach control for potential off-target effects of Cre expression by ensuring that both controls and mutants are Pcp2Cre expressing; Cre only needs to recombine one floxed allele of Dag1. This alone could account for the faster loss of protein. However, this approach requires ensuring that heterozygous control mice lack a phenotype. While L7Cre and Pcp2Cre remain useful tools for studying the effect of losing protein later in development, an inducible Cre such as Pcp2CreERT2 with tamoxifen administration at or after P14 might be more useful for standardizing the timing of gene deletion across PCs, avoiding gradual Cre expression seen in the Pcp2Cre line. However, as described above, tamoxifen administration represents a new variable with additional optimization and validation required.
The time course of Cre expression in PCs with intracerebroventricular delivery of AAV8-CMV-Cre at P0 was similar to that observed with Pcp2Cre (Figs. 2, 7). One advantage of viral delivery is the ability to achieve sparse or mosaic Cre expression. This can also be achieved using inducible CreERT2 lines with low-dose tamoxifen; however, virally expressed Cre remains expressed in the cell after transduction, whereas Cre activity with CreERT2 is temporally limited to ∼24 h after tamoxifen administration (Feil et al., 1997; Kaspar et al., 2002; Ahmed et al., 2004). This increased stability from virally expressed Cre is therefore less likely to result in expression of a Cre-dependent reporter without recombination of target floxed alleles, which can occur following transient CreERT2 activity induced by low doses of tamoxifen (Fernández-Chacón et al., 2019; Tian and Zhou, 2021). The timing of AAV8 viral transduction limits the utility of this approach to later developmental processes. The use of AAVs for gene transduction has become standard in the field, and new viral capsids to expand the utility of the approach are under rapid development. Different capsids can exhibit differences in tropism, efficiency, and transduction timeline (Haery et al., 2019). While the most widely used capsids show a delay in transduction of several days to a couple weeks, newer developments can speed up that timeline considerably. A recent study identified an AAV-SCH9 serotype that can transduce neurons in 24–48 h, making it useful for studying developmental processes (Zheng et al., 2024).
Development of genetic tools for targeting inhibitory populations of neurons has lagged behind those available for targeting excitatory populations. Within the cerebellum, PCs (the primary output of the cerebellum) and MLIs (local interneurons) are both GABAergic, making it challenging to separate the two based on common markers for inhibitory populations (e.g., VGATCre, GAD2Cre). Both populations also express PV, meaning PVCre will cause recombination in both PCs and MLIs. Dlx5/6Cre is becoming increasingly popular to target forebrain interneuron populations; however, Dlx5/6 is not expressed in the cerebellum. Here we have described a suite of tools for targeting PCs for either genetic labeling or gene deletion at various developmental stages: Calb1Cre for embryonic deletion (Fig. 6), Pcp2Cre for early postnatal deletion (Figs. 2, 3), and AAV-Cre for deletion during or after postnatal development (Fig. 7). We also highlight important considerations when identifying and validating new tools for conditional genetic deletion applicable to any population of cells.
Synthesis
Reviewing Editor: Niraj Desai, National Institute of Neurological Disorders and Stroke
Decisions are customarily a result of the Reviewing Editor and the peer reviewers coming together and discussing their recommendations until a consensus is reached. When revisions are invited, a fact-based synthesis statement explaining their decision and outlining what is needed to prepare a revision will be listed below. The following reviewer(s) agreed to reveal their identity: Jason Pugh.
Both reviewers and I were impressed by your study and think it offers useful tools for the neuroscience community. The reviewers' detailed comments are appended below.
The one I would particularly draw your attention to is Reviewer 2's third point on Pcp2-cre)1Amc/J. After our consultation, the reviewers and I agreed that new experiments using this mouse line were not strictly necessary, but that some discussion or clarification was. See, in this regard, Reviewer 1's comment on the subject, which is appended at the very end.
The other comments would not require additional experiments.
REVIEWER 1
The is an excellent technical paper that merits publication after a few very minor revisions. In the Materials and Methods subsection detailing the mouse strains authors should cite the original paper that described the use of female Sox2 Cre mice to derive a hemizygous null; Vincent and Robertson 2003 Genesis 37: 54-56. In Figure 2B the number of cells counted from which the graph was plotted should be stated. In the Discussion the last sentence of paragraph 4 needs to be corrected. In paragraph 5 of the Discussion, a reference is needed to support the statement "This increased stability from virally-expressed Cre is therefore less likely to result in expression of a Cre-dependent reporter without recombination of target floxed alleles, which can occur following transient CreERT2 activity induced by low doses of tamoxifen". Rather than using IIH6, the name of the Dag1 mAb, in the figures it might be better to use the name of the protein.
REVIEWER 2
Major comments:
1. Jackson labs and previous papers have used the nomenclature of PCP2-Cre/Jdhu/J (JAX strain 010536) and PCP2-Cre/2Mpin/J (JAX strain 004146) to describe the "PCP2-cre" and "L7-cre" mouse lines. It would be helpful to clarify that these different terminologies refer to the same mouse lines.
2. A previous paper (Witter et al., 2016; PMID: 27346533) showed the PCP2/Jdhu/J line is specific to Purkinje cells, but the PCP2/Mpin/J line produces significant recombination in MLIs, including this information in the results or discussion regarding the L7/Mpin/J line would be helpful to those looking for the best tools for genetic manipulation of Purkinje cells.
3. The manuscript would be improved by including similar experiments with the (Pcp2-cre)1Amc/J (JAX Strain 006207). Previous work has shown that this line can produce Cre expression in Purkinje cells as early as E17, with all Purkinje cells showing expression by birth (Lewis et al., 2004, PMID: 15183722; Zhou et al., 2020, PMID: 15183722). This line is only available by cryo-recovery at Jackson labs, and so may be beyond the scope of a revision (but may be available from other labs who have used it recently), however, given the primary objective to conditionally manipulate gene expression in early development in Purkinje cells, experimental validation of the PCP2/amc/J line seems fitting.
4. On a related note, Lewis et al., 2004 (PMID: 15183722) report Cre expression following zebrin stripes in early development in the (Pcp2-cre)1Amc/J strain which later expanded to expression in all Purkinje cells. Did the authors here observe any differences in tdTomato or dystroglycan KO across zebrin positive or negative stripes at any stage?
5. The paragraph on inducible Cre lines (CreERt2) in the discussion feels a little out of place as the results do not provide any data on these types of lines.
Minor comments:
1. Page 2 of intro, 4th line from the bottom, delete "provide".
2. Nestin-Cre results section, "It IS important..."
3. Last line of Prf1a-Cre section, "as the presence OF tdTomato..."
4. Last line of 4th discussion paragraph, delete "Jahna"
REVIEWER 1 additional comment on Pcp2-cre)1Amc/J
There appear to be three different Pcp2Cre strains available form Jackson Labs: Tg(Pcp2-cre)3555Jdhu (JAX #010536 donated by Jian-dong Huang), which the authors used; Tg(Pcp2-cre)2Mpin (JAX #004146 donated by Michael Meyer), and Tg(Pcp2-cre)1Amc (JAX #006207 donated by Andrew McMahon). I agree with [Reviewer 1] that this should be clarified. The expression in Purkinje cells of beta galactosidase in Rosa26lacZ mice crossed with the Tg(Pcp2-cre)1Amc strain does not appear to be in all cells of the pcl (Lewis at al Figure 4), but still could be mentioned in the Discussion. It appears that Tg(Pcp2-cre)1Amc and Tg(Pcp2-cre)2Mpin strains were generated form the same plasmid (Smeyne et al. (Mol Cell Neuroscience 6:230) that contains ~1kb of the promoter and a mutated AUG, whereas Tg(Pcp2-cre)3555Jdhu was made using a BAC encompassing more flanking sequence in which an IREScre gene is inserted in the last exon of Pcp2 (exon 4). Since these are conventional transgenics, copy number and site of insertion may be account for differences in expression. The choice of Tg(Pcp2-cre)3555Jdhu by the authors seems to be the most appropriate.
Author Response
We thank the reviewers and editors for their positive assessment of our submission "Tools for Cre-mediated conditional deletion of floxed alleles from developing cerebellar Purkinje cells". We agree with the assessment that this study "offers useful tools for the neuroscience community". We appreciate the reviewers' thoroughness, and for bringing to our attention some points that we missed in the original submission. We have attempted to address all of the reviewers' concerns, as described point-by-point below:
REVIEWER 1:
The is an excellent technical paper that merits publication after a few very minor revisions.
1. In the Materials and Methods subsection detailing the mouse strains authors should cite the original paper that described the use of female Sox2 Cre mice to derive a hemizygous null; Vincent and Robertson 2003 Genesis 37: 54-56.
This was an oversight on our part and this citation is now included in the Mouse Strains and Genotyping section of Materials and Methods.
2. In Figure 2B the number of cells counted from which the graph was plotted should be stated.
We have clarified the analysis approach in the methods section. Since all of the cells in an imaging window were counted for the quantification, the exact number varied from as many as ~500 Purkinje cells at P2 to as few as ~50 Purkinje cells at later timepoints. We have also added animal "Ns" to the figure legends.
3. In the Discussion the last sentence of paragraph 4 needs to be corrected.
Done 4. In paragraph 5 of the Discussion, a reference is needed to support the statement "This increased stability from virally-expressed Cre is therefore less likely to result in expression of a Cre-dependent reporter without recombination of target floxed alleles, which can occur following transient CreERT2 activity induced by low doses of tamoxifen".
We have included two citations to support this statement (Fernández-Chacón et al., 2019; Tian &Zhou, 2021).
5. Rather than using IIH6, the name of the Dag1 mAb, in the figures it might be better to use the name of the protein.
We prefer to use the name of the antibody in this context, because IIH6 does not recognize the Dag1 core protein, but rather the posttranslational matriglycan chains that are unique to Dag1. We have added a sentence clarifying this in the results section in an effort tobe transparent with the functionality of the antibody.
REVIEWER 2 Major comments:
1. Jackson labs and previous papers have used the nomenclature of PCP2-Cre/Jdhu/J (JAX strain 010536) and PCP2-Cre/2Mpin/J (JAX strain 004146) to describe the "PCP2-cre" and "L7-cre" mouse lines. It would be helpful to clarify that these different terminologies refer to the same mouse lines.
Thank you for this comment, we agree that the names/terminologies for these different lines can lead to some confusion. We have rewritten the first portion of the results section to introduce all three Pcp2-Cre lines that are available at Jackson Labs using their nomenclature and including the RRID numbers. We subsequently clarifying how we refer to the two "live repository" lines as Pcp2-Cre and L7-Cre.
2. A previous paper (Witter et al., 2016; PMID: 27346533) showed the PCP2/Jdhu/J line is specific to Purkinje cells, but the PCP2/Mpin/J line produces significant recombination in MLIs, including this information in the results or discussion regarding the L7/Mpin/J line would be helpful to those looking for the best tools for genetic manipulation of Purkinje cells.
Thank you for bringing this to our attention. We have now Included the Witter 2016 citation in the sentence where we mention off target recombination with the L7/Mpin/J line.
3. The manuscript would be improved by including similar experiments with the (Pcp2-cre)1Amc/J (JAX Strain 006207). Previous work has shown that this line can produce Cre expression in Purkinje cells as early as E17, with all Purkinje cells showing expression by birth (Lewis et al., 2004, PMID: 15183722; Zhou et al., 2020, PMID: 15183722). This line is only available by cryo-recovery at Jackson labs, and so may be beyond the scope of a revision (but may be available from other labs who have used it recently), however, given the primary objective to conditionally manipulate gene expression in early development in Purkinje cells, experimental validation of the PCP2/amc/J line seems fitting.
While we agree that it would be useful and informative to have a comparison with the (Pcp2-cre)1Amc/J line, as the reviewer notes, this line is only available through "cryo-recovery" at Jackson Labs. Since this would require several months to obtain, we agree that this is beyond the scope of a revision. However, we now include a discussion of this line alongside the Pcp2/Jdhu and L7/Mpin lines in the results section.
4. On a related note, Lewis et al., 2004 (PMID: 15183722) report Cre expression following zebrin stripes in early development in the (Pcp2-cre)1Amc/J strain which later expanded to expression in all Purkinje cells. Did the authors here observe any differences in tdTomato or dystroglycan KO across zebrin positive or negative stripes at any stage? Thank you for bringing this to our attention. While we did not specifically examine whether recombination followed zebrin stripes, we did not see any evidence of parasagittal stripes for any of the lines we examined. We did not see a striped pattern when whole cerebella were examined under a fluorescent dissection microscope. In addition, serial sagittal sections were cut on a vibratome, which presumably would have contained sections that were negative for tdTomato if recombination followed zebrin stripes, but we did not observe this. Therefore, this pattern of recombination appears unique to the (Pcp2-cre)1Amc/J line.
5. The paragraph on inducible Cre lines (CreERt2) in the discussion feels a little out of place as the results do not provide any data on these types of lines.
While it is true that we didn't characterize any CreER lines in this study, we thought this point was worth mentioning. Specifically, because we are arguing for essential validation approaches with conditional knockouts, we thought it was worthwhile highlighting that CreER lines can present a unique situation that needs to be specifically addressed/examined. We would prefer to leave this point in the discussion.
Minor comments:
1. Page 2 of intro, 4th line from the bottom, delete "provide".
Done 2. Nestin-Cre results section, "It IS important..." Done 3. Last line of Prf1a-Cre section, "as the presence OF tdTomato..." Done 4. Last line of 4th discussion paragraph, delete "Jahna" Done
References
- Ahmed BY, et al. (2004) Efficient delivery of Cre-recombinase to neurons in vivo and stable transduction of neurons using adeno-associated and lentiviral vectors. BMC Neurosci 5:4. 10.1186/1471-2202-5-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman J (1972) Postnatal development of the cerebellar cortex in the rat. II. Phases in the maturation of Purkinje cells and of the molecular layer. J Comp Neurol 145:399–463. 10.1002/cne.901450402 [DOI] [PubMed] [Google Scholar]
- Álvarez-Aznar A, Martínez-Corral I, Daubel N, Betsholtz C, Mäkinen T, Gaengel K (2020) Tamoxifen-independent recombination of reporter genes limits lineage tracing and mosaic analysis using CreERT2 lines. Transgenic Res 29:53–68. 10.1007/s11248-019-00177-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastassiadis K, Fu J, Patsch C, Hu S, Weidlich S, Duerschke K, Buchholz F, Edenhofer F, Stewart AF (2009) Dre recombinase, like Cre, is a highly efficient site-specific recombinase in E. coli, mammalian cells and mice. Dis Model Mech 2:508–515. 10.1242/dmm.003087 [DOI] [PubMed] [Google Scholar]
- Barski JJ, Dethleffsen K, Meyer M (2000) Cre recombinase expression in cerebellar Purkinje cells. Genesis 28:93–98. [DOI] [PubMed] [Google Scholar]
- Bernard C, Axelrad H (1993) Effects of recurrent collateral inhibition on Purkinje cell activity in the immature rat cerebellar cortex—an in vivo electrophysiological study. Brain Res 626:234–258. 10.1016/0006-8993(93)90584-A [DOI] [PubMed] [Google Scholar]
- Bosman LWJ, Konnerth A (2009) Activity-dependent plasticity of developing climbing fiber–Purkinje cell synapses. Neuroscience 162:612–623. 10.1016/j.neuroscience.2009.01.032 [DOI] [PubMed] [Google Scholar]
- Bosman LWJ, Takechi H, Hartmann J, Eilers J, Konnerth A (2008) Homosynaptic long-term synaptic potentiation of the “winner” climbing fiber synapse in developing Purkinje cells. J Neurosci 28:798–807. 10.1523/JNEUROSCI.4074-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briatore F, Pregno G, Di Angelantonio S, Frola E, De Stefano ME, Vaillend C, Sassoè-Pognetto M, Patrizi A (2020) Dystroglycan mediates clustering of essential GABAergic components in cerebellar Purkinje cells. Front Mol Neurosci 13:164. 10.3389/fnmol.2020.00164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch SE, Hansel C (2023) Climbing fiber multi-innervation of mouse Purkinje dendrites with arborization common to human. Science 381:420–427. 10.1126/science.adi1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn RD, et al. (2002) Disruption of Dag1 in differentiated skeletal muscle reveals a role for dystroglycan in muscle regeneration. Cell 110:639–648. 10.1016/S0092-8674(02)00907-8 [DOI] [PubMed] [Google Scholar]
- Crepel F, Mariani J, Delhaye-Bouchaud N (1976) Evidence for a multiple innervation of Purkinje cells by climbing fibers in the immature rat cerebellum. J Neurobiol 7:567–578. 10.1002/neu.480070609 [DOI] [PubMed] [Google Scholar]
- Daigle TL, et al. (2018) A suite of transgenic driver and reporter mouse lines with enhanced brain-cell-type targeting and functionality. Cell 174:465–480.e22. 10.1016/j.cell.2018.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lecea L, del Río J, Soriano E (1995) Developmental expression of parvalbumin mRNA in the cerebral cortex and hippocampus of the rat. Mol Brain Res 32:1–13. 10.1016/0169-328X(95)00056-X [DOI] [PubMed] [Google Scholar]
- Doischer D, Hosp JA, Yanagawa Y, Obata K, Jonas P, Vida I, Bartos M (2008) Postnatal differentiation of basket cells from slow to fast signaling devices. J Neurosci 28:12956–12968. 10.1523/JNEUROSCI.2890-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymecki SM, Tomasiewicz H (1998) Using flp-recombinase to characterize expansion ofWnt1-expressing neural progenitors in the mouse. Dev Biol 201:57–65. 10.1006/dbio.1998.8971 [DOI] [PubMed] [Google Scholar]
- Feil S, Valtcheva N, Feil R (2009) Inducible cre mice. In: Gene knockout protocols, Ed 2. (Wurst W, Kühn R, eds.), pp 343–363. New York, NY: Humana Press. 10.1007/978-1-59745-471-1_18 [DOI] [PubMed] [Google Scholar]
- Feil R, Wagner J, Metzger D, Chambon P (1997) Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun 237:752–757. 10.1006/bbrc.1997.7124 [DOI] [PubMed] [Google Scholar]
- Fernández-Chacón M, Casquero-García V, Luo W, Francesca Lunella F, Ferreira Rocha S, Del Olmo-Cabrera S, Benedito R (2019) iSuRe-Cre is a genetic tool to reliably induce and report Cre-dependent genetic modifications. Nat Commun 10:2262. 10.1038/s41467-019-10239-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graus-Porta D, Blaess S, Senften M, Littlewood-Evans A, Damsky C, Huang Z, Orban P, Klein R, Schittny JC, Müller U (2001) Β1-class integrins regulate the development of laminae and folia in the cerebral and cerebellar cortex. Neuron 31:367–379. 10.1016/S0896-6273(01)00374-9 [DOI] [PubMed] [Google Scholar]
- Gu H, Marth JD, Orban PC, Mossmann H, Rajewsky K (1994) Deletion of a DNA polymerase β gene segment in T cells using cell type-specific gene targeting. Science 265:103–106. 10.1126/science.8016642 [DOI] [PubMed] [Google Scholar]
- Haery L, et al. (2019) Adeno-associated virus technologies and methods for targeted neuronal manipulation. Front Neuroanat 13:Article 93. https://www.frontiersin.org/articles/10.3389/fnana.2019.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Lewis P, Pevny L, McMahon AP (2002) Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech Dev 119:S97–S101. 10.1016/S0925-4773(03)00099-6 [DOI] [PubMed] [Google Scholar]
- Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, Arber S (2005) A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLOS Biol 3:e159. 10.1371/journal.pbio.0030159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino M, et al. (2005) Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron 47:201–213. 10.1016/j.neuron.2005.06.007 [DOI] [PubMed] [Google Scholar]
- Itō M (1984) The cerebellum and neural control. UK: Raven Press. [Google Scholar]
- Jin K, Xiang M (2019) Transcription factor Ptf1a in development, diseases and reprogramming. Cell Mol Life Sci 76:921–940. 10.1007/s00018-018-2972-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner AL, Skarnes WC, Rossant J (1989) Production of a mutation in mouse En-2 gene by homologous recombination in embryonic stem cells. Nature 338:Article 6211. 10.1038/338153a0 [DOI] [PubMed] [Google Scholar]
- Kapfhammer JP (2004) Cellular and molecular control of dendritic growth and development of cerebellar Purkinje cells. Prog Histochem Cytochem 39:131–182. 10.1016/j.proghi.2004.07.002 [DOI] [PubMed] [Google Scholar]
- Kaspar BK, et al. (2002) Adeno-associated virus effectively mediates conditional gene modification in the brain. Proc Natl Acad Sci U S A 99:2320–2325. 10.1073/pnas.042678699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller BH, Hagemann LJ, Doetschman T, Hagaman JR, Huang S, Williams PJ, First NL, Maeda N, Smithies O (1989) Germ-line transmission of a planned alteration made in a hypoxanthine phosphoribosyltransferase gene by homologous recombination in embryonic stem cells. Proc Natl Acad Sci U S A 86:8927–8931. 10.1073/pnas.86.22.8927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PM, Gritli-Linde A, Smeyne R, Kottmann A, McMahon AP (2004) Sonic hedgehog signaling is required for expansion of granule neuron precursors and patterning of the mouse cerebellum. Dev Biol 270:393–410. 10.1016/j.ydbio.2004.03.007 [DOI] [PubMed] [Google Scholar]
- Li Y, Erzurumlu RS, Chen C, Jhaveri S, Tonegawa S (1994) Whisker-related neuronal patterns fail to develop in the trigeminal brainstem nuclei of NMDAR1 knockout mice. Cell 76:427–437. 10.1016/0092-8674(94)90108-2 [DOI] [PubMed] [Google Scholar]
- Luo L, et al. (2020) Optimizing nervous system-specific gene targeting with Cre driver lines: prevalence of germline recombination and influencing factors. Neuron 106:37–65.e5. 10.1016/j.neuron.2020.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, et al. (2010) A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13:Article 1. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miale IL, Sidman RL (1961) An autoradiographic analysis of histogenesis in the mouse cerebellum. Exp Neurol 4:277–296. 10.1016/0014-4886(61)90055-3 [DOI] [PubMed] [Google Scholar]
- Morales D, Hatten ME (2006) Molecular markers of neuronal progenitors in the embryonic cerebellar anlage. J Neurosci 26:12226–12236. 10.1523/JNEUROSCI.3493-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakhai H, Sel S, Favor J, Mendoza-Torres L, Paulsen F, Duncker GIW, Schmid RM (2007) Ptf1a is essential for the differentiation of GABAergic and glycinergic amacrine cells and horizontal cells in the mouse retina. Development 134:1151–1160. 10.1242/dev.02781 [DOI] [PubMed] [Google Scholar]
- Navabpour S, Kwapis JL, Jarome TJ (2020) A neuroscientist’s guide to transgenic mice and other genetic tools. Neurosci Biobehav Rev 108:732–748. 10.1016/j.neubiorev.2019.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H, Ostendorf AP, Satz JS, Westra S, Ross-Barta SE, Campbell KP, Moore SA (2014) Glial scaffold required for cerebellar granule cell migration is dependent on dystroglycan function as a receptor for basement membrane proteins. Acta Neuropathol Commun 2:Article 58. 10.1186/2051-5960-1-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak JS, et al. (2021) Interrogation of dystrophin and dystroglycan complex protein turnover after exon skipping therapy. J Neuromuscul Dis 8:S383–S402. 10.3233/JND-210696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palay SL, Chan-Palay V (2012) Cerebellar Cortex: Cytology and Organization. Germany: Springer Berlin Heidelberg. [Google Scholar]
- Park Y-N, Masison D, Eisenberg E, Greene LE (2011) Application of the FLP/FRT system for conditional gene deletion in yeast Saccharomyces cerevisiae. Yeast 28:673–681. 10.1002/yea.1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter GB, Petryniak MA, Shevchenko E, McKinsey GL, Ekker M, Rubenstein JLR (2009) Generation of Cre-transgenic mice using Dlx1/Dlx2 enhancers and their characterization in GABAergic interneurons. Mol Cell Neurosci 40:167–186. 10.1016/j.mcn.2008.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9:Article 7. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzberg PL, Goff SP, Robertson EJ (1989) Germ-line transmission of a c-abl mutation produced by targeted gene disruption in ES cells. Science 246:799–803. 10.1126/science.2554496 [DOI] [PubMed] [Google Scholar]
- Smeyne RJ, Chu T, Lewin A, Bian F, Crisman SS, Kunsch C, Lira SA, Oberdick J (1995) Local control of granule cell generation by cerebellar Purkinje cells. Mol Cell Neurosci 6:230–251. 10.1006/mcne.1995.1019 [DOI] [PubMed] [Google Scholar]
- Sun M-Y, Yetman MJ, Lee T-C, Chen Y, Jankowsky JL (2014) Specificity and efficiency of reporter expression in adult neural progenitors vary substantially among nestin-CreERT2 lines. J Comp Neurol 522:1191–1208. 10.1002/cne.23497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeo YH, et al. (2021) GluD2- and Cbln1-mediated competitive interactions shape the dendritic arbors of cerebellar Purkinje cells. Neuron 109:629–644.e8. 10.1016/j.neuron.2020.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H, et al. (2011) A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71(6):995–1013. 10.1016/j.neuron.2011.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Zhou B (2021) Strategies for site-specific recombination with high efficiency and precise spatiotemporal resolution. J Biol Chem 296:100509. 10.1016/j.jbc.2021.100509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schütz G (1999) Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet 23:Article 1. 10.1038/12703 [DOI] [PubMed] [Google Scholar]
- Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML (2000) Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and Tris (HotSHOT). BioTechniques 29:52–54. 10.2144/00291bm09 [DOI] [PubMed] [Google Scholar]
- Tsien JZ, Chen DF, Gerber D, Tom C, Mercer EH, Anderson DJ, Mayford M, Kandel ER, Tonegawa S (1996) Subregion- and cell type–restricted gene knockout in mouse brain. Cell 87:1317–1326. 10.1016/S0092-8674(00)81826-7 [DOI] [PubMed] [Google Scholar]
- Tsien JZ, Huerta PT, Tonegawa S (1996) The essential role of hippocampal CA1 NMDA receptor–dependent synaptic plasticity in spatial memory. Cell 87:1327–1338. 10.1016/S0092-8674(00)81827-9 [DOI] [PubMed] [Google Scholar]
- Vincent SD, Robertson EJ (2003) Highly efficient transgene-independent recombination directed by a maternally derived SOX2CRE transgene. Genesis 37:54–56. 10.1002/gene.10226 [DOI] [PubMed] [Google Scholar]
- Witter L, Rudolph S, Pressler RT, Lahlaf SI, Regehr WG (2016) Purkinje cell collaterals enable output signals from the cerebellar cortex to feed back to Purkinje cells and interneurons. Neuron 91:312–319. 10.1016/j.neuron.2016.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa S, Kawamura K, Ono K, Yamakuni T, Takahashi Y (1991) Development and migration of Purkinje cells in the mouse cerebellar primordium. Anat Embryol 184:195–212. 10.1007/BF01673256 [DOI] [PubMed] [Google Scholar]
- Zhang XM, Ng AHL, Tanner JA, Wu WT, Copeland NG, Jenkins NA, Huang JD (2004) Highly restricted expression of Cre recombinase in cerebellar Purkinje cells. Genesis 40:45–51. 10.1002/gene.20062 [DOI] [PubMed] [Google Scholar]
- Zheng X, et al. (2024) Massively parallel in vivo Perturb-seq reveals cell-type-specific transcriptional networks in cortical development. Cell 187:1–13. 10.1016/j.cell.2024.04.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijlstra M, Li E, Sajjadi F, Subramani S, Jaenisch R (1989) Germ-line transmission of a disrupted β2microglobulin gene produced by homologous recombination in embryonic stem cells. Nature 342:Article 6248. 10.1038/342435a0 [DOI] [PubMed] [Google Scholar]