Abstract
The type I glycoprotein S of coronavirus, trimers of which constitute the typical viral spikes, is assembled into virions through noncovalent interactions with the M protein. Here we demonstrate that incorporation is mediated by the short carboxy-terminal segment comprising the transmembrane and endodomain. To this aim, we used the virus-like particle (VLP) system that we developed earlier for the mouse hepatitis virus strain A59 (MHV-A59) and which we describe now also for the unrelated coronavirus feline infectious peritonitis virus (FIPV; strain 79-1146). Two chimeric MHV-FIPV S proteins were constructed, consisting of the ectodomain of the one virus and the transmembrane and endodomain of the other. These proteins were tested for their incorporation into VLPs of either species. They were found to assemble only into viral particles of the species from which their carboxy-terminal domain originated. Thus, the 64-terminal-residue sequence suffices to draw the 1308 (MHV)- or 1433 (FIPV)-amino-acid-long mature S protein into VLPs. Both chimeric S proteins appeared to cause cell fusion when expressed individually, suggesting that they were biologically fully active. This was indeed confirmed by incorporating one of the proteins into virions which thereby acquired a new host cell tropism, as will be reported elsewhere.
The first step in virus infection is the binding of the virus particle to a receptor on the target cell. In enveloped viruses, this binding is mediated by one of the viral membrane proteins. Coronaviruses, plus-stranded RNA viruses occurring in various mammalian and avian species including humans, usually carry three proteins in their envelope. Most abundant is the M protein, a triple-spanning membrane glycoprotein the main function of which involves the organization of the viral envelope and the interactions with the nucleocapsid during assembly (for a review, see reference 24). Another component essential in the assembly process is the small E protein. This protein is generally a minor virion constituent (for a review, see reference 29). It is largely embedded within the viral membrane, and only its hydrophilic carboxy terminus protrudes inside the virion (M. J. B. Raamsman, J. Krijnse Locker, A. de Hooghe, A. A. F. de Vries, G. Griffiths, H. Vennema, and P. J. M. Rottier, submitted for publication). The third envelope protein is the spike (S) protein, a type I membrane glycoprotein, trimers of which (8) constitute the characteristic coronavirus spikes. It is this protein that mediates the binding of the virus to the target cell receptor and the subsequent fusion of viral and cellular membranes during entry (for a review, see reference 3).
Coronavirus assembly is not dependent on the S protein. Studies in which the glycosylation and thus the proper folding of the protein were inhibited by treatment of mouse hepatitis virus strain A59 (MHV-A59)-infected cells with tunicamycin revealed that spikeless, noninfectious particles can be formed (12, 23). These observations were confirmed when we (32) and others (1, 2) showed that virus-like particles (VLPs) can be assembled in cells simply from the M and E proteins by the coexpression of their genes; neither the S protein nor a nucleocapsid appeared to be required. These particles, which we found to be morphologically identical to normal virus, did contain spikes if the S gene was also coexpressed.
Incorporation of spikes into coronavirus particles is effected by interactions between the S protein and the M protein. We demonstrated such interactions in MHV-A59-infected cells, in virions, and during coexpression of M and S genes (7, 21, 22). In an extensive mutagenetic analysis of the primary structure requirements of the M protein for M-S interactions, we observed that the amino-terminal domain of M—the domain exposed on the outside of virions—is not involved (7). These observations indicate that the association between the proteins takes place at the level of the membrane, possibly also involving part of the M protein's carboxy-terminal domain. For the S protein, this implies that the interactions would be limited to the small part of the molecule comprising the transmembrane domain and endodomain.
In order to confirm this hypothesis, we have constructed two reciprocal chimeric S proteins composed of the S ectodomain and carboxy-terminal domain of two unrelated coronaviruses. Our aim was to functionally test these proteins by evaluating their assembly into VLPs derived from these viruses. The chimeric spikes were constructed with the S genes of MHV-A59 and of feline infectious peritonitis virus (FIPV; strain 79-1146). These viruses belong to two different groups of coronaviruses which are genetically and serologically very divergent. For the S proteins, the overall amino acid sequence identity is only 27%; maximal identity (44%) occurs in the segment comprising the transmembrane and carboxy-terminal domain. Another distinguishing feature of these S proteins is that the MHV protein is proteolytically cleaved during transport to the cell surface while that of FIPV is not.
Construction of chimeric S genes.
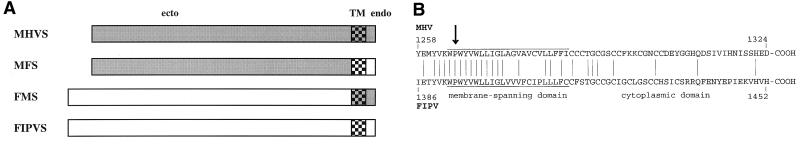
For the construction of the proteins, we have exploited the convenient presence of a StyI restriction site in both S genes located just at the position which encodes the transition between the protein's ectodomain and transmembrane domain, i.e., where the polypeptide enters the lipid membrane. The resulting constructs are depicted in Fig. 1A. One construct (FMS) is composed of the 1,388-amino-acid-long FIPV S ectodomain and the 64-residue transmembrane plus endodomain from MHV-A59 S. The other one (MFS) has the reciprocal structure and consists of 1,260 and 64 amino acids, respectively. The amino acid sequences of the carboxy-terminal regions of the MHV-A59 and FIPV S proteins are compared in Fig. 1B.
FIG. 1.
(A) Spike constructs. MHV-A59 S was expressed from the plasmid pTUMS (32), and the FIPV strain 79-1146 S protein was expressed from pFIPVE2, which was made as follows. A 3′-terminal S fragment was prepared by ligating the XbaI-SalI fragment from pB1 (4) into pUC18, cutting with AccI and SalI, and religating after filling in with Klenow polymerase. From the resulting plasmid p3d, the XbaI-SalI fragment was isolated and used. A middle piece was prepared by isolating the PstI-XbaI fragment from pB1. This fragment and the 3′ XbaI-SalI fragment were ligated into p1A (4), which had been digested with PstI and SalI to give pFIPVE2. Chimeric protein FMS was expressed from pTFMS, which was constructed as follows. Plasmid p3d was digested with HindIII, filled in with Klenow enzyme, and ligated with BglII linkers, resulting in p3dHrB. After the plasmid was cut with StyI and BglII, an MHV S gene fragment was ligated into it; the fragment was prepared by digesting the S gene, obtained as a BamHI fragment from pDGE2 (31), with StyI and taking the small fragment. The resulting p3FM vector was cut with PstI and SalI; into it were ligated the XbaI-SalI fragment from p3d and the PstI-XbaI fragment from pB1. The chimeric gene was finally recloned as a BamHI fragment into pTUG3, resulting in pTFMS. Chimeric protein MFS was expressed from pTMFS, which was prepared starting with p3dHrB. This plasmid was cut with StyI and BamHI, and a BamHI-StyI fragment obtained from the MHV S BamHI gene described above was ligated into it. The chimeric S gene was recloned as a BamHI-SalI fragment into pTUG3 cut with the same enzymes. TM, transmembrane domain; ecto, ectodomain; endo, endodomain. (B) Carboxy-terminal sequences of the MHV-A59 and FIPV spike proteins. The 67 terminal residues of each protein are compared. The arrow indicates the junction point in the chimeric S constructs.
Expression of chimeric S proteins.
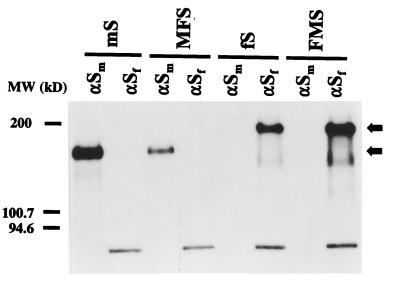
As the gene constructs were placed in plasmids behind a bacteriophage T7 polymerase promoter sequence, they could be tested by expression with the vaccinia virus T7 system (11). Cultures of mouse OST7-1 cells (9) infected with vTF7-3 were transfected in parallel with the plasmids as well as with similar plasmids containing the MHV-A59 and FIPV wild-type S genes. Starting at 5 h postinfection (p.i.), the cells were labeled for 1 h with 35S-amino acids. Cell lysates were then prepared, and immunoprecipitations were carried out on two aliquots of each lysate with the monoclonal antibodies (MAbs) WA3.10 and 23F4.5, known to recognize the ectodomain of the S protein of MHV-A59 (33) and of FIPV (19), respectively. The analysis of the precipitated proteins is shown in Fig. 2. The results demonstrate firstly that the antibodies used are specific and do not cross-react: the wild-type proteins are precipitated only by the proper MAb, not by the other. Secondly, the analysis reveals that the chimeric proteins have the expected properties. The MFS protein comigrates in the gel with the MHV S protein while the mobility of the FMS construct is similar to that of FIPV S.
FIG. 2.
Expression of chimeric spike proteins. Parallel cultures of OST7-1 cells in 35-mm-diameter dishes were infected with vTF7-3 and transfected with plasmids encoding the wild-type and chimeric S proteins described in the legend to Fig. 1. Cells were incubated at 32°C. Starting at 4.5 h p.i., they were starved for 30 min in cysteine- and methionine-free minimal essential medium containing 10 mM HEPES (pH 7.2) without fetal bovine serum. The medium was then replaced by 600 μl of the same containing 100 μCi of 35S in vitro cell labeling mix (Amersham). After a 1-h labeling period, cells were washed with phosphate-buffered saline and solubilized in 1 ml of lysis buffer, TES (20 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1 mM EDTA) containing 1% Triton X-100 and 2 mM phenylmethylsulfonyl fluoride. Nuclei were removed from the cell lysates by centrifugation at 12,000 × g for 10 min at 4°C. For immunoprecipitations, 50-μl aliquots of lysate were diluted with 1 ml of detergent solution (50 mM Tris-HCl [pH 8.0], 62.5 mM EDTA, 0.5% Nonidet P-40, 0.5% Na deoxycholate), and 30 μl of 10% sodium dodecyl sulfate was added. MAbs were then added: 3 μl of hybridoma culture supernatant WA3.10 or 23F4.5, which recognizes the S protein of MHV (αSm) or FIPV (αSf), respectively. Following an overnight incubation at 4°C, immune complexes were adsorbed for 1 h to formalin-fixed Staphylococcus aureus cells (BRL Life Technologies) added as 45 μl of a 10% (wt/vol) suspension. Immune complexes were collected by centrifugation at 12,000 × g and washed three times with radioimmunoprecipitation assay buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.1% sodium dodecyl sulfate, and 1% Na deoxycholate). Pellets were resuspended in 30 μl of Laemmli sample buffer, heated for 2.5 min at 95°C, and analyzed by electrophoresis in a sodium dodecyl sulfate–12.5% polyacrylamide gel followed by fluorography. MW, molecular mass.
Biological activity of chimeric S proteins: cell fusion.
Coronavirus S proteins undergo extensive co- and posttranslational modifications and conformational maturation (for a review, see reference 3). They are extensively glycosylated, become acylated, and undergo formation of multiple intrachain disulfide bonds (20, 21). Most of these events occur during and immediately after synthesis in the endoplasmic reticulum and are critical for the subsequent oligomerization, assembly, and transport processes. In infected cells, the spike complexes are incorporated into viral particles and released with virions from the cell, but a fraction of the complexes is also transported to the plasma membrane where it causes fusion with neighboring cells. Likewise, fusion occurs when the S proteins are expressed individually in the proper cells.
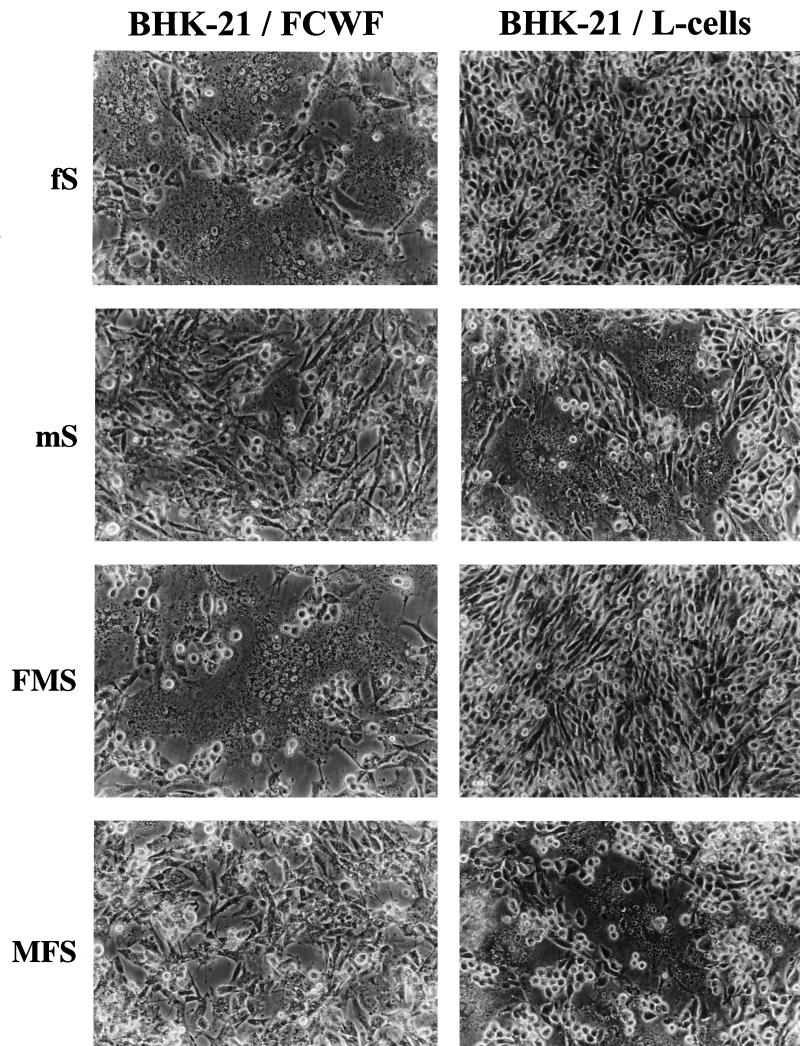
Because the fusion phenotype of an S protein reflects its proper folding and transport to the cell surface as well as a biological property essential for infection, we performed fusion assays with our chimeric constructs. The different S genes were expressed by using the vaccinia virus expression system in BHK-21 cells in which neither of the wild-type S proteins induces fusion by itself. Fusion was evaluated in a coculture assay by overlaying the cell monolayer with mouse L cells or with feline FCWF cells. Pictures of the results are shown in Fig. 3. As predicted, the controls FIPV S (fS) and MHV-A59 S (mS) caused fusion only of the feline and mouse cells, respectively. The chimeric FMS protein induced fusion of the FCWF cells, not of the L cells, consistent with the feline nature of its ectodomain. The reciprocal construct MFS, which derives its ectodomain from MHV S, gave the opposite results, causing fusion only of the mouse cells. The observations demonstrate that the chimeric proteins are processed and transported properly and are biologically active.
FIG. 3.
Fusion properties of the chimeric spike proteins. Subconfluent monolayers of BHK-21 cells grown in 35-mm-diameter dishes were infected with vTF7-3 and transfected with the plasmids encoding MHV-A59 S (mS), FIPV S (fS), and the chimeric S proteins FMS and MFS. At 8 h p.i., the cells were overlaid with either LR7 cells (mouse L cells) or feline FCWF cells. Fusion was followed by light microscopy, and at 24 h p.i., pictures were taken.
Assembly of chimeric S proteins into VLPs.
As the final test to establish whether indeed the incorporation of spikes into coronavirus particles is determined by the carboxy-terminal domain, we analyzed the assembly of the chimeric and wild-type S proteins into both MHV-based and FIPV-based VLPs. Plasmids encoding the M, E, and S proteins were transfected into OST7-1 cells that had been infected with vTF7-3. The proteins were labeled by incubating the cells for 3 h with 35S-amino acids. VLPs secreted into the culture medium were purified by flotation in sucrose gradients. They were subsequently affinity purified with MHV S- and FIPV S-specific MAbs as well as with antisera to other viral structural proteins.
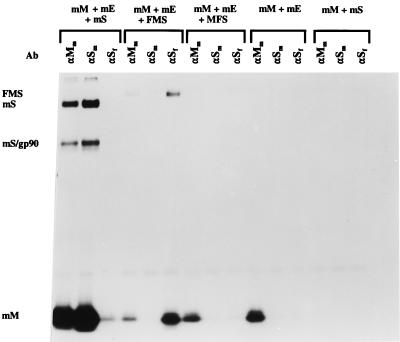
The analyses of the MHV-based VLPs are shown in Fig. 4. Coexpression of all three MHV-A59 wild-type membrane proteins led to the formation of particles that could be affinity isolated as expected by the MAb J1.3 against the MHV M protein ectodomain (6) as well as by the MAb WA3.10 against the MHV S ectodomain (33) but not by the MAb 23F4.5 against the FIPV S ectodomain (19). Control coexpressions of the M and S proteins were not productive, while coexpression of M and E yielded VLPs that could be isolated through their M protein with MAb J1.3 but that were recognized by neither of the anti-S MAbs. Of the chimeric S proteins, only the one with the MHV-derived carboxy-terminal domain (FMS) was incorporated into VLPs. These particles could indeed be collected through their FIPV-specific S ectodomain by using the FIPV S MAb as well as through their M protein with the anti-M MAb and showed the chimeric S protein having a slightly lower electrophoretic mobility than the MHV S protein. When the MFS protein was coexpressed with M and E, VLPs were produced as revealed by the anti-M MAbs, but these could not be isolated by the anti-S MAbs, demonstrating the absence of S protein. As judged from the varying intensities of the M protein bands, the amounts of VLPs produced seemed to differ for the different S proteins coexpressed. This may to some extent be accounted for by differences in the efficiencies with which the different VLPs were affinity isolated by the antibodies. More likely, however, the variations reflect the varying degrees of interference of the different S constructs with the expression of M and E which hampered the reproducible control of VLP production levels. Yet, when we quantitated the radioactivities in the M and S proteins in the VLPs containing MHV S and FMS, calculations revealed that the molar ratios of M and S were rather similar, suggesting that the chimeric S protein is incorporated into viral particles with an efficiency quite similar to that of wild-type S protein.
FIG. 4.
Incorporation of chimeric FMS into MHV-based VLPs. Parallel cultures of OST7-1 cells in 35-mm-diameter dishes were infected with vTF7-3 and transfected with different combinations of plasmids as indicated (mS, mE, and mM represent plasmids encoding the wild-type MHV-A59 S, E, and M proteins, respectively; FMS and MFS refer to plasmids encoding the chimeric S proteins described in the legend to Fig. 1). Cells were incubated at 32°C and labeled from 5 to 8 h p.i. with 35S-amino acids (100 μCi/dish). Culture media (0.8 ml) were harvested, cleared by low-speed centrifugation, mixed with 2.3 ml of 67% sucrose in TM (10 mM Tris-HCl [pH 7.0], 10 mM MgCl2), and transferred into Beckman SW50.1 ultracentrifuge tubes. Each solution was overlaid with 1 ml of 48% sucrose, 0.5 ml of 40% sucrose, and 0.5 ml of 30% sucrose in TM, and the gradients were centrifuged at 36,000 rpm for 43 h. After centrifugation, a fraction consisting of the top 1 ml of each tube was collected. Virus particles were affinity purified from 150 μl of this fraction by addition of 25 μl of MAb J1.3 against the MHV M protein (αMm); 10 μl of MAb WA3.10, which is directed against an epitope in the MHV S ectodomain (αSm); or 3 μl of MAb 23F4.5, which recognizes an epitope in the FIPV S ectodomain (αSf). Samples were processed and analyzed as described for Fig. 2 except that the Staphylococcus aureus immune complexes were washed once with TM instead of three times with radioimmunoprecipitation assay buffer. At the left of the figure, mS and mS/gp90 indicate the positions of the uncleaved and cleaved forms of the S protein, respectively; mM and FMS mark the positions of the M protein and the chimeric S protein, respectively. Ab, antibody.
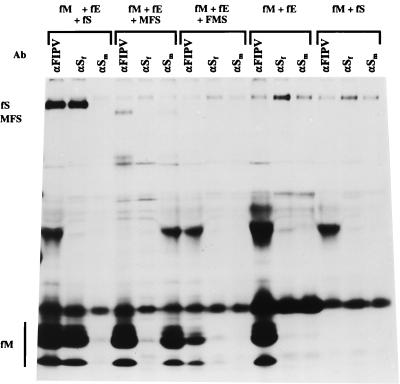
So far, VLPs have been shown only for the coronaviruses MHV (2, 5, 32) and the transmissible gastroenteritis virus of swine (1). In Fig. 5, we show that such particles can similarly be assembled from FIPV envelope proteins. Again, M and E are the minimal requirements, the combination of M and S being unproductive. If wild-type S is coexpressed with M and E, spiked particles which can be affinity purified with anti-FIPV serum and with FIPV S-specific MAbs are formed. Coexpression of the chimeric S proteins shows that now only MFS, the spike protein with the FIPV-derived carboxy terminus, was incorporated, giving rise to particles that could be collected with the MHV S MAb. The reverse construct (FMS) was not incorporated into VLPs. When the radioactivities in the M and S proteins were quantitated for the VLPs produced with wild-type FIPV S and with chimeric MFS, it now appeared that the latter was significantly underrepresented. While this may indicate that this protein is incorporated into particles less efficiently, the result is, at least in part, due to the relatively poor expression that we observed with the MFS construct (data not shown).
FIG. 5.
Incorporation of chimeric MFS into FIPV-based VLPs. Different plasmid combinations were expressed, the proteins were labeled, and the culture media were processed, all as described for Fig. 4. fS, fE, and fM refer to plasmids encoding the wild-type FIPV S, E, and M proteins, respectively; FMS and MFS refer to the chimeric constructs described in the legend to Fig. 1. The αFIPV serum (G73) was from an FIPV-infected cat. Ab, antibody.
The combined data demonstrate that the assembly of spikes into the coronavirus envelope is governed by the S protein's carboxy-terminal domain. Clearly, the 64-residue segment comprising the transmembrane and endodomain is sufficient to interact with the M protein and to draw the 1,308 (MHV)- or 1,433 (FIPV)-residue-long mature (i.e., devoid of its predicted cleaved signal sequence) protein into particles. It will now be interesting to investigate whether this segment is required in its entirety or whether the functional domain can be narrowed down further. In this respect, it is of note that quite substantial homology occurs among transmembrane domains of coronavirus S proteins, particularly on the amino-terminal side of the transmembrane domain where a highly conserved 8-residue sequence (KWPWYVWL) occurs (Fig. 1B). In contrast, besides the generally high cysteine content little similarity exists in the endodomain.
Although for several enveloped viruses a role of the membrane-anchoring and/or cytoplasmic domain has been implicated in the incorporation of membrane proteins, no general conclusions can yet be drawn. Quite inconsistent observations were, for instance, made with well-studied proteins such as the influenza virus hemagglutinin (10, 13, 18) and the rhabdovirus G protein (14–17, 25–28). As an illustration, incorporation of G protein (25) or of heterologous membrane proteins (26, 28) into the vesicular stomatitis virus envelope appeared to occur nonspecifically, i.e., with efficiencies independent of the nature of the transmembrane and cytoplasmic domains, while for the efficient assembly of foreign membrane proteins into the rabies virus envelope the autologous G tail was required (15, 27). A fair comparison with the coronavirus S protein is, however, difficult to make, as for many of these viruses an interaction between these proteins—through their cytoplasmic domain—and the viral core is important (17) or essential (30, 34). For coronaviruses, such interactions are not essential: particle formation is nucleocapsid independent and occurs irrespective of the presence of S protein (12, 23, 32; also the present paper).
The chimeric coronavirus S proteins appeared to be biologically active, causing fusion of reporter cells in a coculture assay (Fig. 3). This indicated that such proteins might mediate infection when incorporated into coronavirions. We have therefore introduced the FMS gene construct into the MHV-A59 genome by targeted RNA recombination, giving rise to a murine coronavirus that, by virtue of its FIPV-derived spike ectodomain, is unable to infect murine cells but has acquired the property to infect and multiply in feline cells (L. Kuo, G.-J. Godeke, M. J. B. Raamsman, P. S. Masters, and P. J. M. Rottier, unpublished data). Not only do these observations confirm the results presented in this paper, the recombinant chimeric MHV also provides a powerful new tool to introduce mutations into the 3′-terminal genomic domain encoding the structural proteins. By using as a recombination partner a donor RNA construct that will restore the wild-type S gene, isolation of mutants can simply be done by selecting for growth on murine cells.
Acknowledgments
We are grateful to Rhône Mérieux (Lyon, France) for providing MAb 23F4.5 and to John Fleming (University of Wisconsin) for the MAb WA3.10.
REFERENCES
- 1.Baudoux P, Carrat C, Besnardeau L, Charley B, Laude H. Coronavirus pseudoparticles formed with recombinant M and E proteins induce alpha interferon synthesis by leukocytes. J Virol. 1998;72:8636–8643. doi: 10.1128/jvi.72.11.8636-8643.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bos E C W, Luytjes W, van der Meulen H, Koerten H K, Spaan W J M. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology. 1996;218:52–60. doi: 10.1006/viro.1996.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cavanagh D. The coronavirus surface glycoprotein. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 73–113. [Google Scholar]
- 4.de Groot R J, van Leen R W, Dalderup M J M, Vennema H, Horzinek M C, Spaan W J M. Stably expressed FIPV peplomer protein induces cell fusion and elicits neutralizing antibodies in mice. Virology. 1989;171:493–502. doi: 10.1016/0042-6822(89)90619-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Haan C A M, Kuo L, Masters P S, Vennema H, Rottier P J M. Coronavirus particle assembly: primary structure requirements of the membrane protein. J Virol. 1998;72:6838–6850. doi: 10.1128/jvi.72.8.6838-6850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Haan C A M, Roestenberg P, de Wit M, de Vries A A F, Nilsson T, Vennema H, Rottier P J M. Structural requirements for O-glycosylation of the mouse hepatitis virus membrane protein. J Biol Chem. 1998;273:29905–29914. doi: 10.1074/jbc.273.45.29905. [DOI] [PubMed] [Google Scholar]
- 7.de Haan C A M, Smeets M, Vernooij F, Vennema H, Rottier P J M. Mapping of the coronavirus membrane protein domains involved in interaction with the spike protein. J Virol. 1999;73:7441–7452. doi: 10.1128/jvi.73.9.7441-7452.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delmas B, Laude H. Assembly of coronavirus spike protein and its role in epitope expression. J Virol. 1990;64:5367–5375. doi: 10.1128/jvi.64.11.5367-5375.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elroy-Stein O, Moss B. Cytoplasmic expression system based on constitutive synthesis of bacteriophage T7 RNA polymerase in mammalian cells. Proc Natl Acad Sci USA. 1990;87:6743–6747. doi: 10.1073/pnas.87.17.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enami M, Enami K. Influenza virus hemagglutinin and neuraminidase glycoproteins stimulate the membrane association of the matrix protein. J Virol. 1996;70:6653–6657. doi: 10.1128/jvi.70.10.6653-6657.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesize bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmes K V, Doller E W, Sturman L S. Tunicamycin resistant glycosylation of coronavirus glycoprotein: demonstration of a novel type of viral glycoprotein. Virology. 1981;115:334–344. doi: 10.1016/0042-6822(81)90115-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin H, Subbarao K, Bagai S, Leser G P, Murphy B R, Lamb R A. Palmitylation of the influenza virus hemagglutinin (H3) is not essential for virus assembly or infectivity. J Virol. 1996;70:1406–1414. doi: 10.1128/jvi.70.3.1406-1414.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mebatsion T, Conzelmann K K. Specific infection of CD4+ target cells by recombinant rabies virus pseudotypes carrying the HIV-1 envelope spike protein. Proc Natl Acad Sci USA. 1996;93:11366–11370. doi: 10.1073/pnas.93.21.11366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mebatsion T, Finke S, Weiland F, Conzelmann K K. A CXCR4/CD4 pseudotype rhabdovirus that selectively infects HIV-1 envelope protein-expressing cells. Cell. 1997;90:841–847. doi: 10.1016/s0092-8674(00)80349-9. [DOI] [PubMed] [Google Scholar]
- 16.Mebatsion T, König M, Conzelmann K K. Budding of rabiesvirus particles in the absence of the spike glycoprotein. Cell. 1996;84:941–951. doi: 10.1016/s0092-8674(00)81072-7. [DOI] [PubMed] [Google Scholar]
- 17.Mebatsion T, Weiland F, Conzelmann K K. Matrix protein of rabies virus is responsible for the assembly and budding of bullet-shaped particles and interacts with the transmembrane spike glycoprotein G. J Virol. 1999;73:242–250. doi: 10.1128/jvi.73.1.242-250.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naim H Y, Roth M G. Basis for selective incorporation of glycoproteins into the influenza virus envelope. J Virol. 1993;67:4831–4841. doi: 10.1128/jvi.67.8.4831-4841.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olsen C W, Corapi W V, Ngichabe C K, Baines J D, Scott F W. Monoclonal antibodies to the spike protein of feline infectious peritonitis virus mediate antibody-dependent enhancement of infection of feline macrophages. J Virol. 1992;66:956–965. doi: 10.1128/jvi.66.2.956-965.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Opstelten D-J E, de Groote P, Horzinek M C, Vennema H, Rottier P J M. Disulfide bonds in folding and transport of the mouse hepatitis virus glycoproteins. J Virol. 1993;67:7394–7401. doi: 10.1128/jvi.67.12.7394-7401.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Opstelten D-J E, de Groote P, Horzinek M C, Rottier P J M. Folding of the mouse hepatitis virus spike protein and its association with the membrane protein. Arch Virol Suppl. 1994;9:319–328. doi: 10.1007/978-3-7091-9326-6_32. [DOI] [PubMed] [Google Scholar]
- 22.Opstelten D-J E, Raamsman M J B, Wolfs K, Horzinek M C, Rottier P J M. Envelope glycoprotein interactions in coronavirus assembly. J Cell Biol. 1995;131:339–349. doi: 10.1083/jcb.131.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rottier P J M, Horzinek M C, van der Zeijst B A M. Viral protein synthesis in mouse hepatitis virus strain A59-infected cells: effect of tunicamycin. J Virol. 1981;40:350–357. doi: 10.1128/jvi.40.2.350-357.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rottier P J M. The coronavirus membrane protein. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 115–139. [Google Scholar]
- 25.Schnell M J, Buonocore L, Botitz E, Ghosh H P, Chernish R, Rose J K. Requirement for a non-specific glycoprotein cytoplasmic domain sequence to drive efficient budding of vesicular stomatitis virus. EMBO J. 1998;17:1289–1296. doi: 10.1093/emboj/17.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schnell M J, Buonocore L, Kretzschmar E, Johnson E, Rose J K. Foreign glycoproteins expressed from recombinant vesicular stomatitis viruses are incorporated efficiently into virus particles. Proc Natl Acad Sci USA. 1996;93:11359–11365. doi: 10.1073/pnas.93.21.11359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schnell M J, Johnson E, Buonocore L, Rose J K. Construction of a novel virus that targets HIV-1-infected cells and controls HIV-1 infection. Cell. 1997;90:849–857. doi: 10.1016/s0092-8674(00)80350-5. [DOI] [PubMed] [Google Scholar]
- 28.Schubert M, Joshi B, Blondel D, Harmison G G. Insertion of the human immunodeficiency virus CD4 receptor into the envelope of vesicular stomatitis virus particles. J Virol. 1992;66:1579–1589. doi: 10.1128/jvi.66.3.1579-1589.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siddell S G. The small-membrane protein. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 181–189. [Google Scholar]
- 30.Suomalainen M, Liljeström P, Garoff H. Spike protein-nucleocapsid interactions drive the budding of alphaviruses. J Virol. 1992;66:4737–4747. doi: 10.1128/jvi.66.8.4737-4747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vennema H, Heijnen L, Zijderveld A, Horzinek M C, Spaan W J M. Intracellular transport of recombinant coronavirus spike proteins: implications for virus assembly. J Virol. 1990;64:339–346. doi: 10.1128/jvi.64.1.339-346.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vennema H, Godeke G-J, Rossen J W A, Voorhout W F, Horzinek M C, Opstelten D-J E, Rottier P J M. Nucleocapsid-independent assembly of coronavirus-like particles by coexpression of viral envelope proteins. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weismiller D G, Sturman L S, Buchmeier M J, Fleming J O, Holmes K V. Monoclonal antibodies to the peplomer glycoprotein of coronavirus mouse hepatitis virus identify two subunits and detect a conformational change in the subunit released under mild alkaline conditions. J Virol. 1990;64:3051–3055. doi: 10.1128/jvi.64.6.3051-3055.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao H, Lindqvist B, Garoff H, von Bonsdorff C H, Liljeström P. A tyrosine-based motif in the cytoplasmic domain of the alphavirus envelope protein is essential for budding. EMBO J. 1994;13:4204–4211. doi: 10.1002/j.1460-2075.1994.tb06740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]