Abstract
Primate gamma-2 herpesviruses (rhadinoviruses) have so far been found in humans (Kaposi's sarcoma-associated herpesvirus [KSHV], also called human herpesvirus 8), macaques (Macaca spp.) (rhesus rhadinovirus [RRV] and retroperitoneal fibromatosis herpesvirus [RFHV]), squirrel monkeys (Saimiri sciureus) (herpesvirus saimiri), and spider monkeys (Ateles spp.) (herpesvirus ateles). Using serological screening and degenerate consensus primer PCR for the viral DNA polymerase gene, we have detected sequences from two distinct gamma-2 herpesviruses, termed Chlorocebus rhadinovirus 1 (ChRV1) and ChRV2, in African green monkeys. ChRV1 is more closely related to KSHV and RFHV, whereas ChRV2 is closest to RRV. Our findings suggest the existence of two distinct rhadinovirus lineages, represented by the KSHV/RFHV/ChRV1 group and the RRV/ChRV2 group, respectively, in at least two Old World monkey species. Antibodies to members of the RRV/ChRV2 lineage may cross-react in an immunofluorescence assay for early and late KSHV antigens.
Kaposi's sarcoma-associated herpesvirus (KSHV), or human herpesvirus 8 (HHV8) (9), is found in all clinical forms of Kaposi's sarcoma, in primary effusion lymphomas (7, 8, 9), and in some cases of multicentric Castleman's disease (14, 27). KSHV is currently classified as a member of the rhadinovirus subgroup of gammaherpesviruses (9, 23). Rhadinoviruses have been found in many species, including cattle, mice, and both Old World and New World primates (1, 2, 3, 9, 11, 12, 17, 21, 22, 25). Viruses that infect New World monkeys include herpesvirus saimiri (HVS), which infects the squirrel monkey, and herpesvirus ateles (HVA), which infects the spider monkey (1, 2, 3). Other than humans, the macaques of Asia are the only Old World primate species documented thus far to harbor rhadinoviruses. Rhesus rhadinovirus (RRV) is widespread among rhesus macaques (Macaca mulatta), grows well in tissue culture (11), and has been fully sequenced (25). Its genome organization is very similar to that of KSHV, with homologues of several potentially pathogenic KSHV genes, including those for K1, vIL-6, vIRFs, a D-type cyclin, vFLIP, G-protein-coupled receptor, and the membrane protein K15 (16, 25). However, of those KSHV genes that have been tentatively linked to pathogenicity, RRV lacks a K12 homologue and has only one vMIP, in contrast to the three in KSHV (25). Because of this extensive similarity between RRV and KSHV, RRV has been proposed as the macaque equivalent of KSHV, and macaques infected with RRV are potentially a suitable animal model for KSHV infection. Recently, coinfection of rhesus monkeys with RRV and simian immunodeficiency virus (SIVmac239) was implicated in the induction of multicentric lymphoproliferative disorder, reminiscent of multicentric Castleman's disease (31). However, degenerate consensus primer PCR for the herpesvirus DNA polymerase and glycoprotein B genes has identified other rhadinovirus sequences in macaques suffering from retroperitoneal fibromatosis (RF) (21). RF is a mesenchymal neoplasm with vascular components and is also associated with simian retrovirus 2, which causes immunosuppression and an AIDS-like disease in macaques (6, 21, 28). Because of these features, RF in macaques had previously been suggested as an animal model for KS (28). Both the rhesus macaque and the pigtailed macaque (Macaca nemestrina) harbor RF herpesviruses (RFHVMm and RFHVMn) which appear to be more closely related to KSHV than KSHV is to RRV (5, 21). In view of reports that some currently circulating KSHV strains may have undergone a recombination event with a related rhadinovirus (15, 18, 19, 32), and the high prevalence of KSHV in sub-Saharan Africa (reviewed in reference 24), we decided to investigate the existence of KSHV-related viruses in African primates.
To determine the seroprevalence of KSHV-related viruses, we studied sera from 78 captive African green monkeys (AGMs), housed at the Paul-Ehrlich-Institut, Langen, Germany, including three subspecies of Chlorocebus aethiops: C. aethiops aethiops (grivet), C. aethiops pygerythrus (vervet), and C. aethiops sabaeus (sabaeus). Some animals were naturally infected with SIVAGM and simian T-lymphotrophic virus type 1. None of the animals had been housed with species other than AGMs at the institute. The sera were tested for cross-reactivity with KSHV orf65/VP19 by enzyme-linked immunosorbent assay (26) and for unspecified early and late antigens by immunofluorescent antibody assay (IFA) (Advanced Biotechnology Inc., Columbia, Md.) (10). Sera from six animals (7.8%) reacted against orf65/VP19 alone, and sera from 37 (47.4%) reacted in the lytic IFA. Sera from only two (2.5%) animals reacted in both assays. Fifty sera were also tested for antibodies to the latency-associated nuclear antigen (LANA) by IFA (13, 26), but none were positive.
In a separate study carried out in the Department of Human Retrovirology at the University of Amsterdam (N. Renwick et al., unpublished data), 201 plasma or serum samples from Cercopithecus, Chlorocebus, Miopithecus, Cercocebus, Macaca, Mandrillus, Colobus, Presbytis, Pan, Pongo, Callithrix, Cebus, Lagothrix, Saguinus and Saimiri species were screened for antibodies to orf73 (LANA) or orf65/VP19 as previously described (20). Three were found to have antibodies to recombinant orf65/VP19, seven had antibodies against recombinant orf73 (LANA), and three had antibodies in both assays. Positive animals included Chlorocebus aethiops, Chlorocebus aethiops pygerythrus, Cercopithecus albogularis, Cercopithecus ascanius (two), Cercopithecus cephus, Cercopithecus mona, Cercopithecus nictitans, Cercopithecus pogonias, Colobus guereza, Macaca nemestrina, Macaca sylvanus, and Papio cynocephalus anubis.
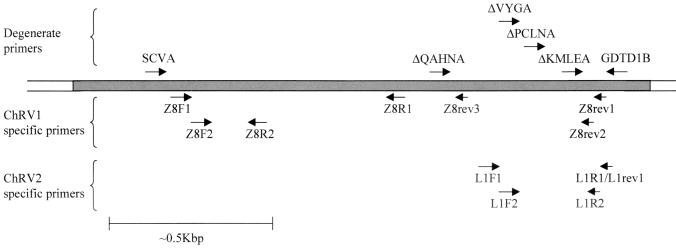
We initially chose five animals (one vervet, three grivets, and one unknown subspecies) from the Paul-Ehrlich-Institut AGMs with cross-reactive antibodies against orf65/VP19 and attempted to amplify herpesvirus DNA polymerase sequences from peripheral blood mononuclear cell (PBMC) DNA (extracted with the Qiagen whole-blood kit) by consensus PCR based on the method previously described (21). We modified the reported primers after including sequence from RFHVMm, RFHVMn, and RRV (11, 21, 25) and designed an additional primer (SVCA) to allow amplification of a longer DNA polymerase sequence fragment (Fig. 1; Table 1). PCR cycling conditions were 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, for 30 cycles. After three rounds of heminested PCR (GDTD1B and ΔVYGA in the initial round, followed by GDTD1B and ΔPCLNA, followed by GDTD1B and ΔKMLEA [Fig. 1; Table 1]), a fragment of 88 bp (excluding primer sequence) with 48.9% nucleotide identity with KSHV was obtained from animal Z8, a vervet monkey. Specific reverse primers (Z8rev1 and Z8rev2 [Fig. 1; Table 1]) were designed from this sequence and used in nested PCRs with the primer ΔQAHNA to obtain 454 bp of viral sequence (excluding primers). Another heminested PCR, using Z8rev2, a new specific primer Z8rev3, and SVCA (Fig. 1; Table 1) then yielded a further 1,348 bp of viral sequence. After all sequences were assembled, 1,802 bp (excluding primers) were obtained for this virus, termed Chlorocebus rhadinovirus 1 (ChRV1). A similar screen by consensus PCR of another 21 PBMC DNA samples from animals with cross-reactive antibodies in the IFA and 7 with no evidence of cross-reactivity yielded a markedly different herpesvirus DNA polymerase sequence from a seronegative monkey, L1. The virus was termed ChRV2. By using a PCR strategy similar to that used for ChRV1, but with ChRV2-specific primers (L1rev1 and L1R2 [Fig. 1; Table 1]), 454 bp of sequence (excluding primers) was determined for ChRV2.
FIG. 1.
Positions of primers for consensus and virus-specific (ChRV1 and ChRV2) PCR for DNA polymerase.
TABLE 1.
Sequences of primers used for consensus and virus (ChRV1 and ChRV2)-specific PCR for DNA polymerase
| Primer | Orientation | 5′→3′ sequencea | Position in KSHV genomeb
|
|
|---|---|---|---|---|
| Start | End | |||
| Degenerate | ||||
| GDTD1Bc | − | CGGCATGCGACAAACACGGAGTCNGTRTCNCCRTA | 13643 | 13610 |
| ΔQAHNAd | + | CCCAGTATCATNCARGCRCACAA | 13133 | 13154 |
| ΔVYGAd | + | ACMTGTAACGCGGTKTACGGSTTYACVGG | 13409 | 13437 |
| ΔPCLNAd | + | GTCGCCTCTGGCATCCTVCCDTGCMTNAA | 13439 | 13467 |
| ΔKMLEAd | + | CAGGGCAGGAARATGCTGGARACRTCNMAGGC | 13490 | 13521 |
| SVCA | + | CCAGYGTNTGYGTKAACG | 11784 | 11801 |
| ChRV1 specific | ||||
| Z8F1 | + | ACGATGTTATTTCTACACCCGCG | 11812 | 11834 |
| Z8F2 | + | GAATCTAACGTCGACGCGACCAG | 12050 | 12072 |
| Z8R1 | − | CTCATGGACCTCAAACACGGATC | 12607 | 12584 |
| Z8R2 | − | GCTGGAGGTCTTCCCATCCGC | 12200 | 12180 |
| Z8rev1 | − | TGAAGTGGGCTTCTGTCC | 13602 | 13584 |
| Z8rev2 | − | TCTGTCTTTGCAATAGGTGGC | 13573 | 13562 |
| Z8rev3 | − | AGTGTGGAGTAACAGAGG | 13171 | 13155 |
| ChRV2 specific | ||||
| L1F1 | + | CCGGACGGGACCTGCACCTG | 13178 | 13197 |
| L1F2 | + | TACGAAACGTTCGCGCTCAGCG | 13221 | 13242 |
| L1R1 | − | ACGCGGAATCTCGCGCCGGG | 13605 | 13585 |
| L1R2 | − | GCAGCTCGTCCGGGGTCAGG | 13552 | 13533 |
| L1rev1 | − | ACGACGCGGAATCTCGCGCCGGG | 13602 | 13585 |
a N indicates A, G, C, or T; R indicates A or G; M indicates A or C; K indicates G or T; S indicates G or C; Y indicates C or T; V indicates C, G, or A.
Nucleotide numbers as in reference 23. Positions for virus-specific primers correspond to equivalent residues in the KSHV genome.
Described in reference 21.
Modified from primers described in reference 21.
A comparison of nucleotide and amino acid identities among primate rhadinoviruses (Table 2) indicates that ChRV1 is most closely related to RFHVMn, RFHVMm, and KSHV (72.2, 68.5, and 70.9% nucleotide identity, respectively). It appears to be least closely related to the two New World rhadinoviruses HVS and HVA (60.6 and 61.3% nucleotide identity), with an intermediate degree of relatedness to RRV and ChRV2 (63.5 and 65.4% identity) (Table 2). When ChRV2 is compared with all these viruses, the same three groupings are apparent, with ChRV2 being closest to RRV (84.1% nucleotide identity), most distant from HVS and HVA (55.7 and 52.9% nucleotide identity), and the KSHV/RFHV/ChRV1 group in an intermediate position (60.6 to 65.9% nucleotide identities). The same three groupings are also seen when the GC content is tabulated and, to some extent, also for the CpG ratios of individual viruses (Table 3).
TABLE 2.
Nucleotide and amino acid identities between ChRV1, ChRV2, and other gammaherpesviruses
| Virus | % Identity witha
|
|||
|---|---|---|---|---|
| ChRV1
|
ChRV2
|
|||
| Nucleotide | Protein | Nucleotide | Protein | |
| KSHV | 70.9 (66.6) | 79.5 (78.3) | 65.9 | 76.2 |
| ChRV1 | 60.6 | 70.9 | ||
| ChRV2 | 65.4 (ND) | 70.9 (ND) | ||
| RFHVMm | 68.5 (ND) | 81.5 (ND) | 60.8 | 70.2 |
| RFHVMn | 72.2 (ND) | 82.8 (ND) | 63.0 | 72.2 |
| RRV | 63.5 (59.9) | 68.2 (67.7) | 84.1 | 88.7 |
| HVS | 60.6 (50.6) | 64.2 (61.9) | 55.7 | 70.9 |
| HVA | 61.3 (49.5) | 64.9 (61.6) | 52.9 | 68.2 |
| EBV | 59.1 (50.1) | 57.6 (67.6) | 60.1 | 57.6 |
Numbers refer to values obtained in a comparison of the 454-bp fragment, which is available for all viruses. Numbers in parentheses indicate identity for the 1,802-bp fragment, which is not available for ChRV2, RFHVMm, and RFHVMn. ND, sequence not determined.
TABLE 3.
GC content and CpG dinucleotide frequencies in DNA polymerase fragments from gammaherpesviruses
| Virus | GC content (%)
|
CpG ratioa
|
||
|---|---|---|---|---|
| 1,802-bp equivalent | 454-bp equivalent | 1,802-bp equivalent | 454-bp equivalent | |
| KSHV | 53.9 | 54.3 | 0.80 | 0.91 |
| ChRV1 | 54.1 | 54.5 | 0.96 | 0.93 |
| ChRV2 | NDb | 63.8 | ND | 1.15 |
| RFHVMm | ND | 51.0 | ND | 1.13 |
| RFHVMn | ND | 55.9 | ND | 0.96 |
| RRV | 59.0 | 59.8 | 1.18 | 1.25 |
| HVA | 36.2 | 38.9 | 0.45 | 0.29 |
| HVS | 35.2 | 40.0 | 0.29 | 0.28 |
| EBV | 62.6 | 64.9 | 0.70 | 0.73 |
Observed frequency/expected frequency of CpG dinucleotides for the mononucleotide composition. (Expected frequency = proportion C × proportion G × [sequence length − 1].)
ND, sequence not determined.
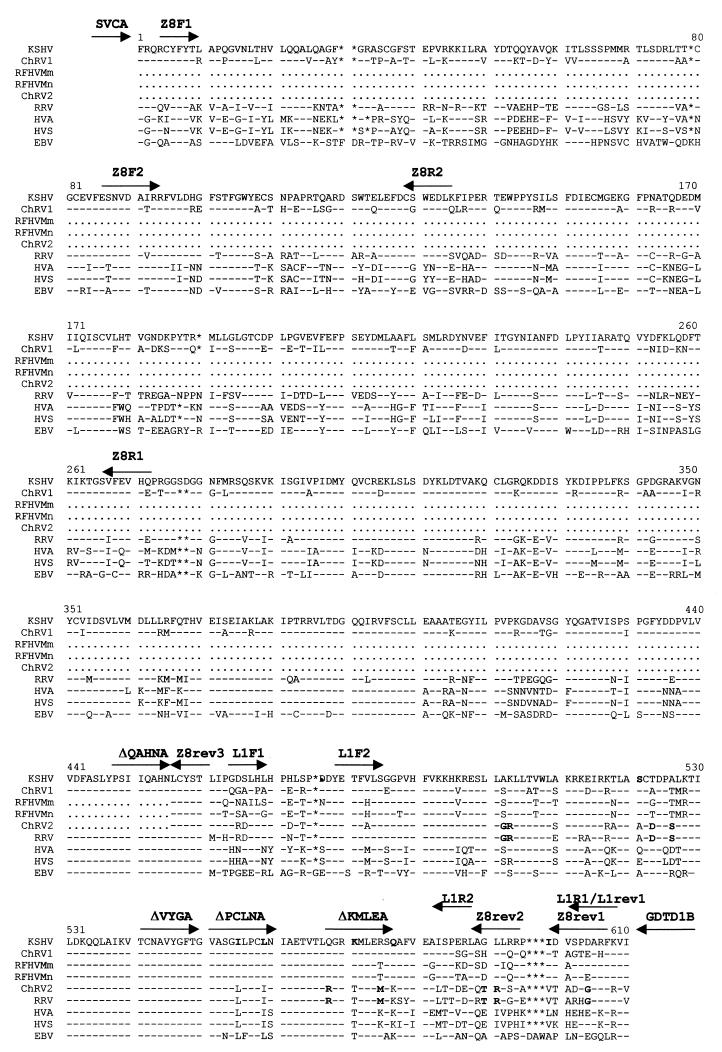
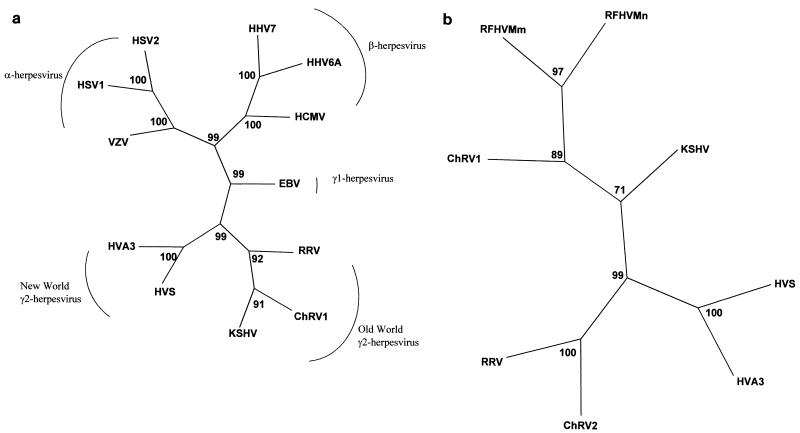
An alignment of the amino acid sequence corresponding to the 1,802-bp fragment is shown in Fig. 2 and reveals sequence motifs shared by ChRV2 and RRV or the KSHV/RFHV/ChRV1 group. By maximum likelihood analysis of the 1,802-bp DNA sequence (which is not available for ChRV2 and RFHVMm/RFHVMn) (Fig. 3a), ChRV1 appears most closely related to KSHV, followed by RRV and, in a separate branch, by the New World rhadinoviruses HVS and HVA. The same branching order was obtained with other phylogenetic methods, including neighbor joining analysis and parsimony (not shown), and supported by high bootstrap values (Fig. 3a). Similar analyses were carried out on the 454-bp DNA polymerase fragment available for other rhadinoviruses, including ChRV2 and RFHVMm/RFHVMn. A neighbor joining tree of the corresponding 151-amino-acid protein sequence (Fig. 3b) illustrates the existence of three separate lineages among rhadinoviruses, one for the New World viruses HVS and HVA and two for the Old World viruses. In spite of being derived from different species living, respectively, in Asia and Africa, RRV and ChRV2 cluster together, as do KSHV, ChRV1, and RFHVMm/RFHVMn. Furthermore, viruses from the same species (ChRV1 and ChRV2 from AGMs and RRV and RFHVMm from rhesus macaques) group separately from each other. This strongly suggests that at least two Old World primate species harbor two fairly distinct rhadinovirus lineages. In humans, one of these lineages is represented by KSHV, whereas the other lineage currently lacks a known representative.
FIG. 2.
Alignment of a 599- to 608-amino-acid fragment (primers SCVA and GDTD1B), using CLUSTALX (EMBL, Heidelberg, Germany), of ChRV1 with published rhadinovirus and Epstein-Barr virus sequences. The shorter sequences of ChRV2, RFHVMm, and RFHVMn are also shown (primers ΔQAHNA and GDTD1B). Gaps for optimal alignment are indicated by asterisks. Amino acid residues identical to the sequence of KSHV are indicated by dashes. Undetermined sequence is indicated by dots. Residues that are conserved within the KSHV/RFHV/ChRV1 lineage and within the RRV/ChRV2 lineage are shown in bold. In addition, ChRV1 and KSHV both have a deletion relative to the other sequences at position 190, where the sequence for the RFHVs is undetermined. Positions of degenerate and virus-specific primers are shown. The origin of sequences is as described in the legend to Fig. 3a.
FIG. 3.
(a) DNA maximum likelihood tree for the 1,802-bp fragment (primers SCVA and GDTD1B) of DNA polymerase. Sequences were aligned by using CLUSTALX and analyzed by using the DNAML program (PHYLIP version 3.5c; J. Felsenstein and the University of Washington). One hundred replica samplings were subjected to bootstrap analysis (SEQBOOT). The tree is unrooted. Other sequences included and their EMBL accession numbers are as follows: HHV1/HSV1 (X04771), HHV2/HSV2 (M16321), HHV3/VZV (X04370), HHV4/Epstein-Barr virus (V01555), HHV5/HCMV (AF133589), HHV6A (X83413), HHV7 (U43400), HHV8/KSHV (U75698), HVS (X64346), HVA (HVA3) (AF083424), RFHVMm (AF005479), RFHVMn (AF005478), RRV (AF029302). (b) Neighbor-joining protein distance tree for the 151 amino acid residues encoded by the 454-bp fragment (primers ΔQAHNA and GDTD1B) of DNA polymerase. Sequences were aligned by using CLUSTALX and analyzed by using the PROTDIST and NEIGHBOR programs in PHYLIP. One hundred replica samplings were analyzed. The tree is unrooted.
Further specific primers were designed (Z8F1, Z8F2, Z8R1, and Z8R2 for ChRV1 and L1F1, L1F2, and L1R1 [with L1R2] for ChRV2 [Fig. 1; Table 1]) and used to screen DNA from PBMCs of 68 AGMs for the presence of either virus. PCRs were repeated to ensure the veracity of results, in some cases on new aliquots of PBMCs. Six other monkeys were found to be infected with an ChRV1-like virus; an animal from the Amsterdam panel (a red-tailed monkey, Cercopithecus ascanius) and five AGMs from the Paul-Ehrlich-Institut (two grivets and three unknown subspecies). Hence, ChRV1-like viruses appear to be infrequent among captive African monkeys, or they are difficult to detect because of low viral load. In this, they resemble the other members of this lineage, KSHV and RFHVMm/RFHVMn; KSHV is detected in only approximately 10 to 20% of serologically reactive individuals (4, 24, 30), although concurrent human immunodeficiency virus type 1 infection, or iatrogenic immunosuppression, markedly increases the risk of developing Kaposi's sarcoma in KSHV-infected individuals (4, 26, 30), and RFHVMm-RFHVMn is detected only in animals with RF or after immune suppression (6). Sequence analysis of the PCR products obtained from these animals with the ChRV1 diagnostic primers (Z8F2 and Z8R2; 107 bp of sequence, excluding primers) indicated that the sequences obtained from the six Chlorocebus aethiops animals, Z8 and five other positive animals, were identical. PCRs were repeated for these animals, with the same sequence being determined. The sequence from the more distantly related red-tailed monkey (Cercopithecus ascanius) (29) differed from ChRV1 by 2 nucleotides (98.1% nucleotide identity), with the substitutions A-G (KSHV nucleotide 12093 [23]), and G-A (KSHV nucleotide 12147), leading to Glu-Gly and Gly-Asp substitutions, respectively. While more extensive sequence comparisons among different cercopithecine species are required, this could indicate a higher degree of sequence conservation among ChRV1-like viruses than among RFHVs in different macaque species (21). However, we cannot currently exclude transmission of ChRV1 among cercopithecine monkeys in captivity. As determined with the ChRV2 diagnostic primers (L1F1, L1F2, L1R1, and L1R2 [Fig. 1; Table 1]), 22 AGMs (33.3%) were PCR positive for the ChRV2-type virus. This is again reminiscent of RRV, the other member of the second lineage (Fig. 3), which is easily isolated from different macaques (11, 25); serological studies, using purified RRV as the antigen, found evidence for a high prevalence of the virus (∼50% positivity rate) (11). We have also recently detected ChRV2-like sequences in 3 of 11 wild-caught Cercopithecus mona animals, which are quite closely related to C. aethiops and Cercopithecus ascanius species (29; Greensill et al., unpublished data).
We investigated whether detection, by PCR, of ChRV1 or ChRV2 correlated with the presence of cross-reacting antibodies to KSHV, as determined by orf65/vp19 ELISA or lytic IFA. Among 66 AGMs (18 grivets, 4 vervets, 2 sabaeus, and 42 unknown) also tested by PCR, reactivity in lytic IFA was widespread (37 of 66; 56%) but particularly high in those in which ChRV2 could be detected by PCR (16 of 22; 73.7%), compared to those in which it was not (21 of 44; 47.8%; P = 0.054). No such correlation was found between reactivity in orf65/vp19 ELISA and ChRV2 detection (0 of 22 ChRV2 PCR-positive animals had antibodies to orf65/VP19, whereas 5 of the remaining ChRV2 PCR-negative animals had orf65/VP19 antibodies). ChRV1 was detected in only seven animals in our study, so any correlation between antibody reactivity and infection with the virus is difficult to resolve. However, of the seven ChRV1 PCR positive animals, two had antibodies to orf65/VP19, three had antibodies detected in the lytic IFA, and two were nonseroreactive. Of the remaining 73 ChRV1 PCR-negative animals, 10 had antibodies to orf65/VP19.
The existence of two rhadinovirus lineages in at least two Old World monkey species could suggest the existence of a similar situation in the great apes, and perhaps in humans. The correlation of PCR-detectable ChRV2 with reactivity in lytic IFA is in accord with the concept that this assay, in particular when carried out at low serum dilutions, may detect antibodies against a related virus. Whether this explains some cases of lytic IFA reactivity in humans where KSHV infection could not be confirmed by other, more specific assays remains to be seen. Evidence for recombination of the right-hand end of the KSHV genome with a related rhadinovirus has recently been reported (15, 18, 19, 32). It is conceivable that this could have occurred as the result of a coinfection in humans with two related viruses. In this study we have found two examples, with an AGM from Paul-Ehrlich-Institut colony and the Amsterdam red-tailed monkey, of coinfection with representatives of the ChRV1 and ChRV2 groups.
If the existence of two separate lineages of gamma-2 herpesviruses in different primate species is confirmed by more extensive sequence analysis it may be necessary to derive a nomenclature that distinguishes between these two branches.
Nucleotide sequence accession numbers.
The 1,802-bp sequence of ChRV1 and the 454-bp sequence of ChRV2 have been deposited in GenBank under accession no. AJ251573 and AJ251574, respectively.
Acknowledgments
We are grateful to Helen Williams for technical assistance and to D. Ablashi of Advanced Biotechnologies Inc. for providing the lytic IFA kits.
This study was supported by the Oxenhale Trust and the Northwest Cancer Research Fund.
REFERENCES
- 1.Albrecht J-C. Primary structure of the Herpesvirus ateles genome. J Virol. 2000;74:1033–1037. doi: 10.1128/jvi.74.2.1033-1037.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albrecht J-C, Nicholas J, Biller D, Cameron K R, Biesinger B, Newman C, Wittmann S, Craxton M A, Coleman H, Fleckenstein B, Honess R W. Primary structure of the herpesvirus saimiri genome. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albrecht J-C, Friedrich U, Kardinal C, Koehn J, Fleckenstein B, Feller S M, Biesinger B. Herpesvirus ateles gene product Tio interacts with nonreceptor protein tyrosine kinases. J Virol. 1999;73:4631–4639. doi: 10.1128/jvi.73.6.4631-4639.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ariyoshi K, Schim van der Loeff M, Corrah T, Cham F, Cook P M, Whitby D, Weiss R A, Schulz T F. Kaposi's sarcoma and human herpesvirus 8 (HHV8) in HIV-1 and HIV-2 infection in The Gambia. J Hum Virol. 1998;1:193–199. [PubMed] [Google Scholar]
- 5.Bosch M L, Strand K B, Rose T L. Gammaherpesvirus sequence comparisons. J Virol. 1998;72:8458–8459. doi: 10.1128/jvi.72.10.8458-8459.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosch M L, Harper E, Schmidt A, Strand K B, Thormahlen S, Thouless M E, Wang Y. Activation in vivo of retroperitoneal fibromatosis-associated herpesvirus, a simian homologue of human herpesvirus-8. J Gen Virol. 1999;80:467–475. doi: 10.1099/0022-1317-80-2-467. [DOI] [PubMed] [Google Scholar]
- 7.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences are present in AIDS-related body cavity based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 8.Cesarman E, Moore P S, Rao P H, Inghirami G, Knowles D M, Chang Y. In vitro establishment and characterization of two AIDS-related lymphoma cell-lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995;86:2708–2714. [PubMed] [Google Scholar]
- 9.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;265:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 10.Chatlynne L G, Lapps W, Handy M, Huang Y Q, Masood R, Hamilton A S, Said J W, Koeffler H P, Kaplan M H, Friedman-Kien A, Gill P S, Whitman J E, Ablashi D V. Detection and titration of human herpesvirus-8-specific antibodies in sera from blood donors, acquired immunodeficiency syndrome patients, and Kaposi's sarcoma patients using a whole virus enzyme-linked immunosorbent assay. Blood. 1998;92:53–58. [PubMed] [Google Scholar]
- 11.Desrosiers R C, Sasseville V G, Czajak S C, Zhang X, Mansfield K G, Kaur A, Johnson R P, Lackner A A, Jung J U. A herpesvirus of rhesus monkeys related to the human Kaposi's sarcoma-associated herpesvirus. J Virol. 1997;71:9764–9769. doi: 10.1128/jvi.71.12.9764-9769.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Efstathiou S, Ho Y M, Hall S, Styles C J, Scott S D, Gompels U A. Murine herpesvirus 68 is genetically related to the gammaherpesviruses Epstein-Barr virus and herpesvirus saimiri. J Gen Virol. 1990;71:1365–1372. doi: 10.1099/0022-1317-71-6-1365. [DOI] [PubMed] [Google Scholar]
- 13.Gao S-J, Kingsley L, Li M, Zheng W, Parravicini C, Ziegler J, Newton R, Rinaldo C R, Saah A, Phair J, Detels R, Chang Y, Moore P S. KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi's sarcoma. Nat Med. 1996;2:925–928. doi: 10.1038/nm0896-925. [DOI] [PubMed] [Google Scholar]
- 14.Gessain A, Sudaka A, Briere J, Fouchard N, Nicola M-A, Rio B, Arborio M, Troussard X, Audouin J, Diebold J, de The G. Kaposi's sarcoma associated herpes-like virus (human herpesvirus type 8) DNA sequences in multicentric Castleman's disease: is there any relevant association in non-human immunodeficiency virus-infected patients? Blood. 1996;87:414–416. [PubMed] [Google Scholar]
- 15.Glenn M, Rainbow L, Aurade F, Davison A, Schulz T F. Identification of a spliced gene from Kaposi's sarcoma-associated herpesvirus encoding a protein with similarities to latent membrane proteins 1 and 2a of Epstein-Barr virus. J Virol. 1999;73:6953–6963. doi: 10.1128/jvi.73.8.6953-6963.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaleeba J A R, Bergquam E P, Wong S W. A rhesus macaque rhadinovirus related to Kaposi's sarcoma-associated herpesvirus 8 encodes a functional homologue of interleukin-6. J Virol. 1999;73:6177–6181. doi: 10.1128/jvi.73.7.6177-6181.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore P S, Gao S-J, Dominguez G, Cesarman E, Lungu O, Knowles D M, Garber R, McGeoch D J, Pellett P, Chang Y. Primary characterization of a herpesvirus-like agent associated with Kaposi's sarcoma. J Virol. 1996;70:549–558. doi: 10.1128/jvi.70.1.549-558.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicholas J, Zong J C, Alcendor D J, Ciufo D M, Poole L J, Sarisky R T, Chiou C J, Zhang X, Wan X, Guo H G, Reitz M S, Hayward G S. Novel organizational features, captured cellular genes, and strain variability within the genome of KSHV/HHV8. J Natl Cancer Inst Monogr. 1998;23:79–88. doi: 10.1093/oxfordjournals.jncimonographs.a024179. [DOI] [PubMed] [Google Scholar]
- 19.Poole L J, Zong J-C, Ciufo D M, Alcendor D J, Cannon J S, Ambinder R, Orenstein J M, Reitz M S, Hayward G S. Comparison of genetic variability at multiple loci across the genomes of the major subtypes of Kaposi's sarcoma-associated herpesvirus reveals evidence for recombination and for two distinct types of open reading frame K15 alleles at the right-hand end. J Virol. 1999;73:6646–6660. doi: 10.1128/jvi.73.8.6646-6660.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Renwick N, Halaby T, Weverling G J, Dukers N H, Simpson G R, Coutinho R A, Lange J M, Schulz T F, Goudsmit J. Seroconversion for human herpesvirus 8 during HIV infection is highly predictive of Kaposi's sarcoma. AIDS. 1998;12:2481–2488. doi: 10.1097/00002030-199818000-00018. [DOI] [PubMed] [Google Scholar]
- 21.Rose T M, Strand K B, Schultz E R, Schaefer G, Rankin G W, Jr, Thouless M E, Tsai C-C, Bosch M L. Identification of two homologs of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in retroperitoneal fibromatosis of different macaque species. J Virol. 1997;71:4138–4144. doi: 10.1128/jvi.71.5.4138-4144.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rovnak J, Quackenbush S L, Reyes R A, Baines J D, Parrish C R, Casey J W. Detection of a novel bovine lymphotropic herpesvirus. J Virol. 1998;72:4237–4242. doi: 10.1128/jvi.72.5.4237-4242.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russo J J, Bohenzky R A, Chien M-C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi's sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schulz T F. Epidemiology of Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. Adv Cancer Res. 1999;76:121–160. doi: 10.1016/s0065-230x(08)60775-7. [DOI] [PubMed] [Google Scholar]
- 25.Searles R P, Bergquam E P, Axthelm M K, Wong S W. Sequence and genomic analysis of a rhesus monkey rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J Virol. 1999;73:3040–3053. doi: 10.1128/jvi.73.4.3040-3053.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simpson G R, Schulz T F, Whitby D, Cook P M, Boshoff C, Rainbow L, Howard M R, Gao S-J, Bohenzky R A, Simmonds P, Lee C, de Ruiter A, Hatzakis A, Tedder R S, Weller I V D, Weiss R A, Moore P S. Prevalence of Kaposi's sarcoma associated herpesvirus infection measured by antibodies to recombinant capsid protein and latent immunofluorescence antigen. Lancet. 1996;349:1133–1138. doi: 10.1016/S0140-6736(96)07560-5. [DOI] [PubMed] [Google Scholar]
- 27.Soulier J, Grollet L, Oskenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay M-F, Clauvel J-P, Raphael M, Degos L, Sigaux F. Kaposi's sarcoma associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 28.Tsai C C, Tsai C-C, Roodman S T, Woon M-D. Mesochymo-proliferative disorders (MPD) in simian AIDS associated with SRV-2 infection. J Med Primatol. 1990;19:189–202. [PubMed] [Google Scholar]
- 29.van der Kuyl A C, Kuikun C L, Dekker J T, Goudsmit J. Phylogeny of African monkeys based upon mitochondrial 12S rRNA sequences. J Mol Evol. 1995;40:173–180. doi: 10.1007/BF00167111. [DOI] [PubMed] [Google Scholar]
- 30.Whitby D, Howard M R, Tenant-Flowers M, Brink N S, Copas A, Boshoff C, Hatziouannou T, Suggett F E A, Aldam D M, Denton A S, Miller R F, Weller I V D, Weiss R A, Tedder R S, Schulz T F. Detection of Kaposi's sarcoma-associated herpesvirus (KSHV) in peripheral blood of HIV-infected individuals predicts progression to Kaposi's sarcoma. Lancet. 1995;364:799–802. doi: 10.1016/s0140-6736(95)91619-9. [DOI] [PubMed] [Google Scholar]
- 31.Wong S W, Bergquam E P, Swanson R M, Lee F W, Shiigi S M, Avery N A, Fanton J W, Axthelm M K. Induction of B cell hyperplasia in simian immunodeficiency virus-infected rhesus macaques with the simian homologue of Kaposi's sarcoma-associated herpesvirus. J Exp Med. 1999;190:827–840. doi: 10.1084/jem.190.6.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zong J C, Ciufo D M, Alcendor D J, Wan X, Nicholas J, Browning P J, Rady P L, Tyring S K, Orenstein J M, Rabkin C S, Su I J, Powell K F, Croxson M, Foreman K E, Nickoloff B J, Alkan S, Hayward G S. High-level variability in the ORF-K1 membrane protein gene at the left end of the Kaposi's sarcoma-associated herpesvirus genome defines four major virus subtypes and multiple variants or clades in different human populations. J Virol. 1999;73:4156–4170. doi: 10.1128/jvi.73.5.4156-4170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]