Abstract
Eukaryotic organisms coevolved with microbes from the environment forming holobiotic meta-genomic units. Members of host-associated microbiomes have commensalic, beneficial/symbiotic, or pathogenic phenotypes. More than 100 years ago, Lorenz Hiltner, pioneer of soil microbiology, introduced the term ‘Rhizosphere’ to characterize the observation that a high density of saprophytic, beneficial, and pathogenic microbes are attracted by root exudates. The balance between these types of microbes decide about the health of the host. Nowadays we know, that for the interaction of microbes with all eukaryotic hosts similar principles and processes of cooperative and competitive functions are in action. Small diffusible molecules like (phyto)hormones, volatiles and quorum sensing signals are examples for mediators of interspecies and cross-kingdom interactions. Quorum sensing of bacteria is mediated by different autoinducible metabolites in a density-dependent manner. In this perspective publication, the role of QS-related activities for the health of hosts will be discussed focussing mostly on N-acyl-homoserine lactones (AHL). It is also considered that in some cases very close phylogenetic relations exist between plant beneficial and opportunistic human pathogenic bacteria. Based on a genome and system-targeted new understanding, sociomicrobiological solutions are possible for the biocontrol of diseases and the health improvement of eukaryotic hosts.
Keywords: control of pathogens, host beneficial microbes, N-acyl-homoserine lactones, One Health concept, opportunistic human pathogens, quorum sensing molecules, systemic induction of tolerance to abiotic stress
Quorum sensing-related activities of beneficial and pathogenic bacteria associated with plant and human have important implications for host health and well-being.
Introduction
As result of the evolution of life on Earth, all higher organisms are living as holobiots characterized by tight interaction with a diversity of microbes. The hologenome concept considers holobionts as units of coevolution and selection of well-adapted pro- and eukaryotic organisms leading to constructive co-operations (Stencel et al. 2018). In the evolution of land plants, soil microbes found roots as most attractive interaction area. More than 100 years ago, the term ‘Rhizosphere’ was defined by Professor Lorenz Hiltner as the plant root-dominated habitat where soil microbes live on the expense of low and high molecular weight root exudates (Hiltner 1904, Hartmann et al. 2008). Symbiotic and plant-beneficial interactions evolved in specific bacteria and fungi contributing to major needs of the plant, such as the supply with essential nutrients like nitrogen from the air and macro- and micronutrients from the soil. Nitrogen-fixing bacteria or mycorrhizal fungi are prominent examples for plant-symbiotic microbes. As already Lorenz Hiltner recognized, not only saprophytic and beneficial but also pathogenic microbes are attracted by root exudates, causing chances and challenges for plant health (Mendes et al. 2013, Abedini et al. 2021). Most recently, rhizosphere phage communities were identified to suppress bacterial plant disease (Yang et al. 2023). Mechanisms have been identified by which plants can form the rhizosphere microbiome in a kind of ‘rhizosphere school’ to support healthy plant development (Berendsen et al. 2012). Biofilms are the natural habitats for microbes, and therefore cooperative and competitive interactions within complex surface microbial biofilms are of key importance during root colonization (Nadell et al. 2016). These interactions decide about success or failure of beneficial microbes to support plant health or of pathogens to develop diseases. In the densely populated biofilms of the rhizosphere, bacteria optimize the expression of their genetic potential using diffusible signal molecules to sense the density of their own and neighbouring population (Fuqua et al. 1994). Quorum sensing (QS) is of central importance for successful adaptation to changing environmental conditions and colonization of and eventual establishment within an eucaryotic host (Mukherjee and Bassler 2019). The rhizosphere also provides a stage for multiple avenues of natural genetic engineering, involving horizontal gene transfer (Van Elsas et al. 2003), transduction of genetic elements with plasmids (Shintani et al. 2020), enhanced mutation rates, and phenotypic variations (Achouak et al. 2004, van de Broek et al. 2005, Li et al. 2012, Lalaouna et al 2011). For example, many bacteria acquired multiple luxI- and/or luxR-QS gene homologues from independent sources via horizontal gene transfer (Lerat and Moran 2004). QS activities are prevalent in commensalic, beneficial/symbiotic, and pathogenic plant-associated bacteria (von Bodman et al. 2003).
The first evidence for the involvement of QS-signals and their perceptions by plant hosts were revealed by Mathesius et al. (2003) analysing the response to AHL-QS-signals by Medicago truncatula and by Schuhegger et al. (2006) studying induction of pathogen resistance after AHL-application to roots of tomato plants. Furthermore, QS-related genes were identified in the endosphere bacterial metagenome of rice by Sessitsch et al. (2012) and in the bacterial microbiome of Populus deltoides (Schaefer et al. 2013). In addition, bacteria harbouring QS-genes were frequently isolated from the rhizosphere and endosphere of plants. The ability to communicate with each other and to trigger essential functions in a timely and optimized manner is an important feature for single cell organisms to efficiently use their genetic potential as an organized community (Hense et al. 2007). Among the QS systems, N-acyl-homoserine lactone (AHL, AI-1) circuits are present in many Gram-negative bacteria (Fuqua et al. 1994). AHLs are small diffusable molecules having different structures consisting of a fatty acid moiety with different chain length (4–20), and l-homoserine lactone (Fig. 1). The fatty acid chain can be modified at the C3-position with a hydroxy- or oxogroup or replaced by a p-coumaroyl- or cinnamon-residue (Schaefer et al. 2008, Ahlgree et al. 2011). AHL-biosynthesis and perception occurs through a two-component LuxI/LuxR system. The LuxI-enzyme catalyses the binding of acylcarrier protein-bound S-adenosylmethionine to the acyl chains resulting in AHLs. The LuxR-receptor binds AHLs and the LuxR-AHL-complex finally activates diverse promotors, including the LuxI-promotor. The special feature of QS-signalling is that small and diffusible molecules are constitutively produced in very low amounts. When AHL-concentrations rise in more dense populations above a critical concentration, called quorum, its biosynthesis is autoinduced. At high signal concentrations, a set of genes (the QS-activated transcriptomes) is induced by the LuxR–AHL complex. This opens up new areas of functions essential, e.g. for efficient colonization of a new host. QS-controlled genes often code for the construction of biofilms, hydrolysis and degradation of nutritional carbon polymers and substances, the induction of virulence, and the synthesis of diverse siderophores and antibiotics (Hense and Schuster 2015). The so-called autoinducer-2 (AI-2), furanosyl borate diester, are produced by some Gram-negative and Gram-positive bacteria (Papenfort and Bassler 2016) (Fig. 1). AI-2 is synthesized by the LuxS-protein, and specific AI-2 receptors (e.g. LuxPQ-receptor protein of Vibrio harveyi). The lack of genomic evidence of AI-2 receptors in some bacteria may suggest a non-QS role for LuxS in these bacteria (Rezzonico and Duffy 2008). In Gram-positive bacteria, like Streptococcus pneumoniae or Staphylococcus aureus, different autoinducing peptides (AI-P) are produced (Fig. 1), which activate e.g. virulence and toxin production during growth in biofilms in a density-dependent manner (Parsek and Greenberg 2005). Further QS-signal molecules are mentioned by Hartmann et al. (2021). Besides QS-signals, root-associated microbes produce a high diversity of signalling compounds, like volatiles (Schultz-Bohm et al. 2017, de Boer et al. 2019), and diverse plant hormones, which influence plant hosts in multiple ways (Gamalero et al. 2023).
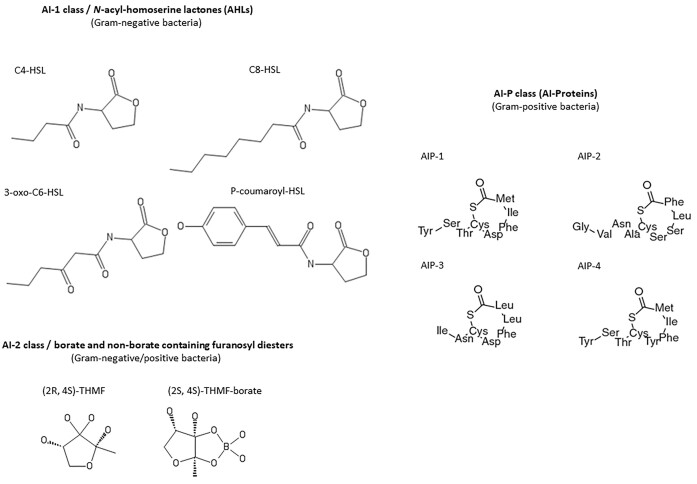
Figure 1.
Structures of QS autoinducers: AI-1 (AHLs), AI-2 (furanosyl borate diester), and AI-P (autoinducer peptide): In AI-1/AHLs, the C3-carbon of the carbonyl fatty acid chain can alternatively carry a hydroxy-residue; the fatty acid chain may also carry a C–C double bond. AI-2 can exist as the boron containing S-THMF-borate (2S, 4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran (QS-active form) or the nonboron form R-THMF (2R, 4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran. The AI-P1, AI-P2, AI-P3, and AI-P4 structures were identified in S. aureus (Schaefer et al. 2008).
Since microbes interact with all eukaryotes in holobiotic-type life forms, comparable signalling mechanisms and principles of cooperation and/or competition are present in all holobionts, which will be discussed in this perspective publication. Likewise, different organs of mammalian hosts, including human, like skin, oral cavity, lung, or gut are colonized with QS-active bacteria of various types, which have beneficial or pathogenic features (Whiteley et al. 2017). It also needs to be considered that in some cases very close phylogenetic relationships exist between plant beneficial and opportunistic human pathogenic bacteria (Berg et al. 2005, 2014, Abreo and Altier 2019, Faoro et al. 2019). In the translation of potentially therapeutic or supportive approaches for sustainable agriculture or human health, the application of metabolites or probiotic microbes with health-supporting signalling capabilities could be of central importance to better control pathogens and support health and well-being of eukaryotic hosts based on natural socio-microbiological mechanisms.
QS-AHL signalling: different AHL functions in plants, LuxR solo receptors, QQ activities, AHL uptake, and signal perception
Many Gram-negative pathogenic bacteria organize their virulence using the AHL-circuit to attack plants (Pollumaa et al. 2012), while symbiotic nitrogen-fixing rhizobia initiate and coordinate root colonization and nodule formation with a diversity of AHL-autoinducers (González and Marketon 2003). The importance and wide distribution of QS-signals in bacteria–plant interaction provide the coevolutionary rational for the sensitive recognition and perception of AHLs by plants leading to an effective stimulation of the innate immune defence system also by beneficial rhizosphere bacteria initiating specific pathogen defence measures or improving abiotic stress tolerance (Schikora et al. 2016). The AHL-based priming of defence is an cost-efficient strategy to fight back pathogens (Shrestha et al. 2020).
LuxR-solo receptors
In addition to the cognate LuxI/LuxR-signal circuit, many Gram-negative bacteria harbour one or several so-called LuxR solos, which are not paired with a classical LuxI-type synthetase, and are thus unable to produce AHLs themselves (González and Venturi 2013). These luxR-solos play a pivotal role in intra- and interspecies, as well as interkingdom, communication. Different classes of LuxR-solo systems can be distinguished, which perceive endogenous and exogenuos AHLs as well as non-AHLs signals (Bez et al. 2023). LuxR regulators are widely distributed bacterial helix-turn-helix transcription factors involved in QS-type mechanisms. They are also found in 50% of the genomes of Gram-positive Actinobacteria for traits at environmental and medical levels in connection with QS and virulence strategies (Santos et al. 2012, Sarveswari and Solomon 2019). As example for non-AHL exogenous signals, host-specific signals of kiwifruit are perceived by PsaR2 LuxR solo in Pseudomonas syringae pv. actinidiae, aimed to induce virulence factors like biofilm formation, motility and endophytic colonization (Cellini et al. 2022). This subclass of LuxR solos is frequently found in plant-associated bacteria, both beneficial/symbiotic or pathogenic bacteria responding to different plant signals (Patel et al. 2013). In some cases the signals are similar to AHLs; e.g. BraR from the stem-nodulating legume symbiont Bradyrhizobium japonicum responds to cinnamoyl-homoserine-HSL, derived from the plant metabolite cinnamon (Ahlgreen et al. 2011). RpaR from Rhodopseudomonas palustris binds to p-coumaroyl-HSL derived from the exogenous p-coumarate (Schaefer et al. 2008). The LuxR-solo QscB receptor and the QscR regulon in the pathogen P. aeruginosa is different from the also present canonical LuxI/LuxR tandem and stimulates virulence activities through increased biosynthesis of the antibiotic phenazine (Fuqua et al. 1994).
QQ activities by AHL hydrolysis, oxidoreductase, and other quenching mechanisms
In the light of the central role of QS for virulence acquisition, the inhibition of QS, including quorum quenching (QQ) by cleaving and inactivating the QS-moieties or by inhibiting the QS-autoinducer action is attracting high attention. Inhibitory mechanisms include blocking the QS-signal synthesis, inhibition of the autoinducer reception, and signal transport (summarized by Hartmann et al. 2021). A large number of especially Gram-positive bacteria and even fungi, but also plant and mammalian hosts, harbour different types of AHL-quenching activities (Grandclément et al. 2016). AHL-signals can undergo different modes of degradation and inactivation in the rhizosphere. Chemical hydrolysis of AHLs occurs at neutral and alkalic pH-values but AHLs are stable at acid pH-values. Efficient enzymatic degradation occurs through AHL-lactonases and AHL-acylases/amidases. In addition, AHL-oxido-reductases were described (Chowdhary et al. 2007). Growth and QQ-activity of Rhodococcus erythropolis R138 was efficiently stimulated by the organic amendment of gammaheptalactone leading to efficient biocontrol of potato (Cirou et al. 2012). Also, a strong inhibition of the QS-regulated pathogen Pectobacterium atrosepticum was shown by gamma-lactone stimulated R. erythropolis resulting in efficient in planta biocontrol (Barbey et al. 2013). Furthermore, a novel type of AHL-acylase of Ochrobactrum sp. A44 was demonstrated to quench the AHL-dependent virulence of P. carotovorum in planta (Czajkowski et al. 2011). In the context of coral reef disease—a case of high global importance—the application of QS-antagonists to white band disease-infected Acropora cervicornis inhibited disease-causing bacteria and stopped coral reef disease development (Certner and Vollmer 2018). Most recently, a novel type IVA secretion system (T4ASS) effector was discovered in Lysobacter enzymogenes OH11 (Liao et al. 2023). This T4ASS is able to deliver a protein (Le1288) into P. fluorescens SPL4, which acts there as a AHL-synthase inhibitor. It was shown that this T4ASS-mechanism is working also to inhibit the human pathogen P. aeruginosa and the plant pathogen Ralstonia solanacearum (Liao et al. 2023). QS-related interventive actions within host-associated microbiomes are of high general relevance also for the balance of beneficial and pathogenic bacteria in mammalian and human health (see below). In Arabidopsis thaliana and some other dicotyledonous plants, especially legumes, fatty acid amid hydrolases cleave AHLs by liberating the fatty acid tail and l-homoserine (HS; Palmer et al. 2014). Interestingly, l-HS is not only able to stimulate root growth but also improve water and nutrient uptake into the plant (Palmer et al. 2014). It was even found that AHL-producing rhizobacteria, like Pseudomonas putida IsoF, itself harbour AHL-degrading activities, leading to reduced intra- and extracellular AHL-concentrations (Fekete et al. 2010). The biosynthesis of C10-HSL in batch as well as contiuous cultures of P. putida IsoF and the simultaneous appearance of the cleavage product l-HS were quantified using high resolving UPLC-measurements and ELISA-technique (Chen et al. 2010, Buddrus-Schiemann et al. 2014). The ecological meaning of this activity could be to limit the AHL-production to a specific culture phase. Alternatively, it could provide plants in another growth phase with the stimulatory l-HS (Palmer et al. 2014).
AHL-uptake
Some plants, like wheat and barley, apparently lack AHL-degrading enzymes and take up unhydrolyzed AHLs. Water soluble C6–C10 AHLs were shown to be taken up into the shoot via active transport (Götz et al. 2007, Sieper et al. 2014). The transport inside the roots occurs in the central cylinder as was shown by autoradiography using 3H-labelled C8- and C10-HSL; the transport was inhibited by orthovanadate, demonstrating that ABC-transporters are involved. The identity of the transported AHLs in the shoots was proven and quantified by AHL-specific monoclonal antibodies, developed by Chen et al. (2010), and AHL-sensor strains (Sieper et al. 2014). In contrast to Hordeum vulgare (cv Barke), the legume Pachyrhizum erosus (L.) did not take up AHL efficiently, as was shown by ultra-performance liquid chromatography (UPLC) and Fourier transform ion cyclotron resonance (FTICR)-mass spectrometry (Götz et al. 2007), due to AHL-degradation in the roots. Further detailed analysis via FTICR-MS and UPLC revealed a metabolism towards C3-hydroxy- and C3-oxo-HSLs in the root compartment especially for C8- and C10-HSL, which may contribute to reduce the transport into the shoot (Götz-Rösch et al. 2015). In addition, a chiral separation of d/l-forms by GC–MS demonstrated that barley selects the l-forms during active transport (Götz et al. 2007, Sieper et al. 2014).
AHL perception as plant growth stimulans and priming agent for pathogen and abiotic stress tolerance
In barley, initial reactions occurs in root cells after treatment with 10 µM C6-, C8-, and C12-HSL (Rankl et al. 2016). Nitric oxide (NO) accumulates in the calyptra and root elongation zone and also the lateral root formation is changed. In addition, increased K+-uptake occurs in root cells, and membrane hyperpolarization is promoted in epidermal root cells especially by C8-HSL (Rankl et al. 2016). Upon application of C6-, C8-, and C10-HSL to the roots, the antioxidative and detoxifying capacities are increased in the shoots of barley (Fig. 2). As compared to control plants, the activity of dehydroascorbate reductase in barley shoots after C10-HSL treatment is greatly increased, whereas superoxide dismutase activity is slightly decreased after application of C6-HSL to the root system (Götz-Rösch et al. 2015). In contrast, the response of antioxidative enzymes in leaves of yam beans was low probably due to reduced uptake of AHLs. In addition, the response of cytosolic glutathion-S-transferase (GST) isoforms in roots and leaves to AHLs were increased or decreased dependent on the iso-form tested in root or shoot compartments in comparison with yam bean (Götz-Rösch et al. 2015). In the light of the observed responses of antioxidant and detoxifying plant activities towards AHLs, these QS-signals may be regarded as strengthening agents or plant antistress boosters.
Figure 2.
AHL interactions with plants (example: Arabidopsis, barley): the perception of AHLs by plants can be divided into interactions with short- and long-carbonyl side chain AHLs. Water-soluble AHLs (like C6- to C10-HSLs) are transported in an active transport process through the central cylinder (Sieper et al. 2014) to the shoot, if AHLs are not degraded by plant lactonases. In the roots, hyperpolarization and K+-uptake occurs and growth and lateral root formation is increased (von Rad et al. 2008, Liu et al. 2012, Rankl et al. 2016). In addition, abiotic and defence gene are activated in the shoots (Götz-Rösch et al. 2015). Lipophilic AHLs like C12- and C14-HSLs are perceived by a membrane protein (ALI1) (Shrestha et al. 2022). In the roots, NO is produced and a systemic signalling cascade to the shoots is activated including salicylic acid and oxylipin (cis-OPA/12-oxo-phytodienolic acid) leading to increased expression of MAP-kinases (MAKs) and defence-related transcription factors WRKY22 and 29 (Shrestha et al. 2019, 2020).
The molecular structure of AHL-autoinducers determines the mode of action towards plant growth stimulation or priming of pathogen resistance (Fig. 2). For example in barley and wheat, which are devoid of AHL-lactonases and -hydrolases, water-soluble AHLs are taken up into the shoots by an energy-dependent transport leading to enzymatic changes in leaves (Sieper et al. 2014, Götz-Rösch et al. 2015). Water-soluble C4-, C6-, and C8 HSLs were also shown to change the phytohormone balance of A. thaliana seedlings, modifying root/shoot growth and metabolism (von Rad et al. 2008). When these AHLs were added to roots (at 10 µM concentration) a multitude of genes, mostly connected with phytohormone regulation, were newly induced in roots and shoots, while others were repressed (von Rad et al. 2008). A special role in this stimulation with C6- and C8-AHLs was identified for the GCR1/GPA1 genes in A. thaliana (Liu et al. 2012). While root growth of GCR1-mutants failed to be stimulated by C6- and C8-HSL, overexpressing mutants showed increased growth responses. In addition, calmodulin receptors were involved in primary root elongation, caused by 3-oxo-C6 HSL (Zhao et al. 2015). Furthermore, AtMYB44 was involved in enhanced elongation of the primary root by increasing cell divisions in the root meristem and enhanced cell elongation in the elongation zone (Zhao et al. 2016); this was accompanied by altering the cytokinin and auxin metabolism in roots. In Arabidopsis and wheat, 3-oxo-C6 HSL was able to enhance salt tolerant growth (Zhao et al. 2020). In 3-oxo-C6-treated salt-stressed plants, the content of chlorophyll as well as the osmolyte proline was increased and the content of malonedialdehyde and the Na+/K+-ratio were decreased (Zhao et al. 2020).
AHLs with long aliphatic chains (C12- and C14-HSL), which cannot be transported into the shoot, are also able to modify plants by conferring systemic resistance towards biotrophic and hemibiotrophic pathogens via altered activation of AtMPK6 (Schikora et al. 2011a). In barley, this response was only present in certain cultivars and therefore has to be regarded as a genetically determined property (Shesthra et al. 2019). In Arabidopsis, as early response specific receptors could be involved to perceive the AHL-signal and to activate a signalling cascade including salicylic acid (SA) and oxylipin 12-oxo-phytodienolic acid (cis-OPDA) (Schenk and Schikora 2015; Shrestha et al. 2020) (Fig. 2). In the presence of flagellin-derived peptide flg22 and C12-HSL, MAP-kinases MPK3, and MPK6 are increasingly stimulated and expressed for a prolonged time along with the upregulation of defence-related transcription factors WRKY22 and WRKY29 (Shrestha et al. 2019) (Fig. 2). Also, glutathione-S-transferase GST6-gene and the heat shock protein Hsp60 are induced. In Arabidosis exposed to mixtures of 3-oxo-C6-, 3-oxo-C8-, 3-oxo-C12-, and 3-oxo-C14-HSLs the response differ as compared to plants treated with single AHLs and jasmonates play an important role (Duan et al. 2023). The fast and stable decreased concentration of COOH-JA-Ile after challenge with flg22 as well as the JA- and SA-affected Arabidopsis mutants strengthened the conclusion that JA-homoeostasis is involved in AHL-priming (Duan et al. 2023). However, a deeper understanding of specific plant factors mediating the response to water-insoluble AHLs, like 3-oxo-C14-HSL, was missing. Most recently, the comparison of wild-type A. thaliana Col-0 and the oxo-C14-HSL insensitive mutant ali1 allowed deeper insights. In Arabidopsis, the gene AtGlcAk2 for glucuronokinase 2 is identical with ali1 (Shrestha et al. 2022) (Fig. 2). Zhao et al. (2013) had described this gene already as a putative kinase from the GHMP kinase family being involved in root and flower development, abscisic acid signalling, and stress response influencing the expression of various ABA-related genes as well as salt stress. MAP-kinase activity measurements, gene expression, and transcriptome analyses as well as pathogenicity assays confirmed a loss of AHL-priming in AtGlcAK2/Ali1 mutants (Shrestha et al. 2022). Furthermore, when fluorescently tagged ALI1-protein was expressed in tobacco leaves, ALI1 colocalized with the plasma membrane, tonoplast, and endoplasmic reticulum in the cell periphery. Thus, the ALI1-protein may be regarded as surface receptor for 3-oxo-C14-HSL and other water-insoluble AHLs. This novel insights may further improve the development of stress resistance of plants, useful for sustainable crop management (Shrestha et al. 2022).
Transgenically modified plants with introduced QS-autoinducer synthesis genes are able to communicate with bacteria in the rhizosphere by altering their QS-controlled activities (Fray et al. 1999). For example, tomato plants harbouring the AHL-biosynthesis genes yenI and lasI from Burkholderia graminis strains altered the activity of these strains in the rhizosphere, leading either to increased or decreased plant growth stimulation or resistance towards salt stress (Barriuso et al. 2008).
QS-/AHL-systems in plant growth promoting and pathogenic bacteria
It has become apparent that within bacterial genera, which are known for efficient rhizosphere and endophytic root colonization, plant beneficial, symbiotic, and even opportunistic human pathogenic bacteria are closely related. Several examples are presented below, which demonstrate that plant beneficial bacteria with probiotic potential exist in the same species with opportunistic pathogens. Therefore, the need for careful evaluation of possible health risks for applications of these bacteria in agricultural or for biotechnological purposes is necessary, if the separation from pathogens are not approved by clear phylogenetic criteria.
A high diversity of AHL-producing, plant beneficial Gram-negative bacteria are now known since several decades to support growth of many crop plants under challenging abiotic and biotic conditions. Gluconacetobacter diazotrophicus PAL5 is an endophytic diazotrophic Gram-negative bacterium, first isolated in 1988 from inside sugar cane stems (Cavalcante and Döbereiner 1988). It can colonize numerous other plant species and confers several beneficial effects including abiotic and biotic stress tolerance and improved plant growth. In addition to the biological nitrogen-fixation activity and the production of several plant hormones, it was shown to harbour an active AHL-QS regulatory system (Bertini et al. 2014) producing eight different QS-signals based on C6-, C8-, C10-, C12-, and C14-HSL (Nieto-Penalver et al. 2012). When G. diazotrophicus PAL5 was inoculated to red rice plants growing under water stress conditions, the expression of the LuxI-gene was strongly stimulated at increasing water deficit conditions. The transcription of the PR1- and PR10-genes along with several antistress defence genes increased, like catalase and superoxide dismutase as well as ascorbate peroxidase (Filgueiras et al. 2020). The induced systemic tolerance to water deficit was accompanied by the accumulation of osmoprotectant solutes and the expression of defence genes against water deficit in plant shoots. Furthermore, mutations in the LuxI/R system of PAL5 resulted in strong reduction in endophytic root colonization of sugar cane seedlings (Hartmann et al. 2019).
Almost 50 years ago, diazotrophic bacteria species of the genus Azospirillum (A. lipoferum and A. brasilense) were isolated and characterized by the group of Johanna Döbereiner in Brazil (Döbereiner and Day 1976). Nowadays, at least 15 different species, mostly of root-associated plant growth-promoting bacteria are officially described within this genus, demonstrating the rich diversity of this efficient rhizosphere bacterium. Phytohormone interactions, based on indole acetic acid (IAA) production and other phytohormones by these bacteria, are prevalent for successful interaction with diverse plants, like wheat, maize, sorghum, causing multiple plant growth-stimulating effects. Although, LuxI-genes and the productions of AHLs were only found in some strains of A. lipoferum (Vial et al. 2006), the addition of C6- and C8-AHLs could stimulate biofilm formation, EPS-production and mobility in strain A. brasilense Ab-V5, a successfully applied A. brasilense inoculant strain (Fukami et al. 2018). It turned out that many Azospirillum spp. strains harbour multiple copies of luxR, so-called LuxR-solo or orphans (Gualpa et al. 2019). This had been documented for other rhizosphere bacteria too (Patel et al. 2013). The most recently characterized diazotrophic plant growth promoting Azospirillum bacterium, A. argentinense type strain Az39 (formerly A. brasilense), was isolated from roots of wheat plants in Argentina (Dos Santos Ferreira et al. 2022). It is a plant growth-promoting rhizobacterium, producing IAA besides other phytohormones and harbours several other plant growth-supporting properties including nitrogen fixation (Cassán and Diaz-Zorita 2016). Surprisingly it hydrolyses AHLs, liberating l-HS (Gualpa et al. 2019). Together with the phytohormone-based interaction, the AHL-degradation activity of Az39 may contribute to its strong plant growth-promotion activity, since l-HS has a plant stimulating ability itslef (Palmer et al. 2014). Interestingly, A. argentinense Az39 like other strains from the A. brasilense cluster (including A. baldaniorum Sp245) harbours up to 25 LuxR-proteins (harbouring LuxR-solo-sequences) providing receptors for AHLs or related QS-signals from other bacteria or even possibly AHL-unrelated signals from the plant for yet unknown interactions. It is a very efficient bioinoculant for gramineae crops in Argentina (Cassán and Diaz-Zorita 2016). The former species Azospirillum amazonense Y1, renamed to Nitrospirillum amazonense, carries a canonical LuxI/R–QS system and has therefore multiple ways of beneficial interactions with plant hosts based on AHL-signalling. Nitrospirillum amazonense strain Y1 is currently used as successful commercial inoculant in sugar cane plantations. Presumably opportunistic human pathogenic bacterial isolates, Roseomonas fauriae and Roseomonas genomospecies 6, were reported (Cohen et al. 2004), which are phylogenetically very closely related to A. brasilense Sp7T. However, DNA–DNA hybridization values of 61.2% and 54.2% place these Roseomonas isolates into a possibly new species within Azospirillum (Hartmann et al. 2019).
The genus Pseudomonas harbours a high number of species many of which are associated with diverse plants, having different plant growth-promoting and protecting properties. For example, P. segetis P6 isolated from Salicornia europaea rhizosphere was characterized to harbour plant growth-promoting activity and QQ-mediated biocontrol (Rodriguez et al. 2020). Seed biopriming of tomato plants with strain P6 resulted in an increase in plant height and weight. Its QQ activity was characterized as an acylase. Thus, strain P6 reduced soft rot symptoms caused by the QS-bacteria Dickeya solani, Pectobacterium atropsepticum, and P. carotovorum on potato and carrot. The QQ-activities of P6 also protected tomato plants against P. syringae pv. tomato, which organizes its virulence through AHL-sensing. Thus, P. segetis P6 may have biotechnological applications. Pseudomonas putida IsoF, isolated from the rhizosphere of tomato, produces two AHLs, 3-oxo-C10, and 3-oxo-C12 from ppuI/ppuR circuit. Interestingly, the Ppu-system controls the expression of a large nonribosomal peptide synthetase, which directs the biosynthesis of two cyclic lipopeptide biosurfactants, putisolvin I and II. Putisolvins inhibit the biofilm formation and also break down existing P. aeruginosa biofilms (Cárcamo-Oyarce et al. 2015). A broad range of AHLs and other QS-active compounds are produced by Pseudomonas strains, which may have plant growth-promoting but also opportunistic human pathogenic potentials (Venturi 2006). For example, a dominant diazotrophic endophytic P. aerugionasa strain PM389 from Pennisetum glaucum (L.) was characterized by Gupta et al. (2013). This diazotrophic bacterium even moves upwards to shoots, and revealed various plant growth-promoting properties including mineral phosphate solubilization, siderophore production, and antagonistic biocontrol properties. Pseudomonas aeruginosa harbours two complete QS-circuits involving AHL signals and a third system using quinolones, which coordinate virulence acquisition and other behaviours (Miranda et al. 2022). The major AHL 3-oxo-C12-HSL regulates virulence gene expression and also induces mammalian cell responses, including apoptosis and immune modulation (see also below).
Rhizobium radiobacter F4 (syn. Agrobacterium tumefaciens) was characterized as an endofungal bacterium of Piriformospora indica (now known as Serendipita indica). Piriformospora indica is known as a plant growth-promoting, mycorrhiza-like fungus, able to stimulate growth and performance of many plants especially under biotic and abiotic stress conditions (Varma et al. 2012). Like P. indica, the free-living bacterium increases plant biomass and enhances resistance against bacterial leaf pathogens and salt stress (Glaeser et al. 2016). Most interestingly, R. radiobacter F4 (RrF4) shows a high degree of similarity to the plant pathogenic R. radiobacter C58, formerly named A. tumefaciens C58. However, it has important differences in the tumour-inducing plasmid (pTi) lacking the T-region (including the ipt-gene) and in the accessory plasmid, because the virH1-gene is truncated (Glaeser et al. 2016). This documents, how a plant beneficial bacterium may have developed from a plant pathogenic one. It could be shown that RrF4 produces a spectrum of QS-mediating AHLs with acyl-chains of C8, C10, and C12 as well as hydroxyl- or oxo-sustitutients at the C3-atom (Alabid et al. 2020), which is quite typical for Rhizobia. In R. radiobacter F4NM13, a lactonase-overexpressing transconjugant of RfF4, the AHLs were missing and also the plant biomass stimulation as well as the systemic resistance was partially compromised in Arabidopsis and wheat. Furthermore, the AHL-deficient transconjugant was lacking cellulose-like fibre scaffolds for efficient root surface attachment. It could not penetrate into the intercellular spaces of the cortex, which is in contrast to the strongly root colonizing endofungal wildtype RrF4 (Alabid et al. 2020). Thus, AHLs contribute to the plant growth stimulation of Rrf4, which may play an important role in the P. indica symbiosis with a high diversity of crop and medicinal plants (Varma et al. 2012).
Acidovorax radicis N35 is an endophytic bacterium with plant growth-promoting activity in barley and wheat (Li et al. 2011); it was characterized to produce only the QS-autoinducer C10-HSL. An araI mutant devoid of C10-HSL production lost the property to efficiently colonize roots of barley. Comparable transcriptome analysis of axenic uninoculated barley seedlings with N35 wild type strain or araI-mutant inoculated plants revealed that the mutant-inoculated barley plants accumulated several flavonoids (Han et al. 2016), which can efficiently inhibit root colonization. It may be concluded from this observation that AHL production of the A. radicis N35 wild type protect it from plant defence responses of flavonoids.
Relationship between plant beneficial and human pathogenic bacteria
In the genus Burkholderia, harbouring saprophytic, beneficial, symbiotic, human pathogenic, or opportunistic pathogenic species, the assessment of the pathogenic potential of each species was for a long time the reason that regulatory authorities banned all environmental release proposals for any Burkholderia strain. The application of whole genome-based comparative software tools together with the assessment of the human pathogenic potential made a clarification possible (Angus et al. 2014). Based on the complete genome sequence data, conserved sequence indels (CSI) were successfully used as molecular marker for the precise identification of three different genera within the Burkholderia cluster: Burkholderia (sensu strictu), with pathogenic and opportunistic pathogenic species, Paraburkholderia, comprising the plant-associated and -beneficial species (Sawana et al. 2014), and Caballeronia, with a group of 12 environmental species (Dobritsa and Samadpour 2016). Several canonical QS-sensing systems mostly based on AHLs and/or also LuxR-solo genes are prevalent in almost all species in the Burkholderia cluster, which are the backbone to efficiently organize either virulence or plant health supporting functions.
It is a matter of fact, that plant pathogens can cross the kingdom border and cause human diseases (as reviewed by Kim et al. 2020). In the other direction, some human pathogens, like Salmonella enterica typhimurium, are known to have affinity to colonize roots of diverse plants like barley, tomato, or Arabidopsis, being even able to colonize the plant hosts endophytically (Kutter et al. 2006, Schikora et al. 2011b, Zarkani et al. 2019). Thus, there are common principles shared by a diversity of bacteria to colonize and enter plant roots as well as mammalian tissues. For example the genus Herbaspirillum harbours well-known efficient plant growth-promoting nitrogen-fixing rhizobacteria, but also clinical isolates in Herbaspirillum species 3 (Baldani et al. 1996). Strains belonging to H. seropedicae and H. frisingense (Kirchhof et al. 2001, Straub et al. 2013) were characterized as plant growth-promoting rhizobacteria, but they are also isolated from human skin wounds, sputum samples of cystis fibrosis patients, or other diseased human organs (Faoro et al. 2019, Oliveira et al. 2021). Herbaspirillum frisingense Mb11, isolated from roots of the energy plant Pennisetum purpureum in Brasil produced AHLs, while the GSF30T, derived from Miscanthus spp. in Freising, Germany, did not synthesize AHLs; both groups of strains efficiently colonized seedlings of Miscanthus and barley endophytically (Rothballer et al. 2008). Within H. hiltneri, isolated from surface disinfected wheat roots, which is phylogenetically apart from the H. seropedica/H. frisingense cluster (Rothballer et al. 2006), nosocomial strains were not yet found. A detailed genomic and proteomic study of clinical and environmental isolates of H. seropedicae revealed that clinical strains have lost the gene sets for biological nitrogen fixation (nif) and the type 3 secretion system (T3SS), which has been described to be essential for the interaction with plants. A different set of accessory genes and genomic islands could be found in the clinical strains, like genes related to lipopolysaccharide (LPS) biosynthesis and neuABC genes, responsible for the biosynthesis of sialic acid. The neuABC-linked LPS was able to increase the bacterial resilience in the mammalian host aiding in the escape from the immune system (Faora et al. 2019). In clinical and environmental isolates of H. frisingense, the genes in the core and accessory genomes were compared and numerous unique clusters could be identified in clinical and rhizosphere strains. Some genomic islands were only found in clinical strains, while others in all strains. Thus, a preadaptation to different hosts was concluded (Oliveira et al. 2021).
In the case of Serratia marcescens, harbouring a wide range of strains from soil, water, and plant surfaces, also opportunistic human pathogens in hospitals and plant growth-promoting bacteria in crops are known. In a pangenome approach, based on available genomic data, whole genome multilocus sequence type schemes (MLSTs) were applied (Abreo and Altier 2019). In most cases, genomes of nosocomial and environmental isolates could be assigned to proposed nosocomial or environmental MLSTs. A minority of nosocomial strains harboured environmental MLSTs, which suggest that these have been recently derived from the environment. One environmental clase had only low numbers of virulence and antibiotic resistance determinants and may represent a group of prospective PGPR strains (Abreo and Altier 2019). In general, it is not entirely clear, whether so-called clinical or nosocomial strains are opportunistic pathogenic bacteria, which cause secondary infections only in immuno-compromised patients and thus worsen conditions in patients. It is a matter of fact that based on general phylogenetic terms, opportunistic pathogenic bacteria can be separated from commensalic and beneficial bacteria only with difficulty. Extensive experimental and genomic assessments of the pathogenic potential of each strain have to be conducted, to allow a case to case decision (Angus et al. 2014; Lee et al. 2014), if the strain should be used as inoculum for plants. A temperature optimum below 37°C is another important criterium for an application in the field. An readdressing of the biosafety level of some already used plant growth promoting rhizobacteria to biosafety level 2 prohibited the continuation of the application in agriculture (Keswani et al. 2019).
Control of the ecological balance of health supporting and threatening bacteria in mammalian habitats
In comparison to the health situation in the rhizosphere, which is strongly influenced by the plant and the soil microbiome (Hartmann et al. 2009, Schreiter et al. 2014), the microbiological and metabolic quality of food has an important role for the establishment of a balanced structure and function as well as a healthy status of human microbiomes in habitats, such as the oral cavity and the gut. From the early life times as baby and infant, edible plant and other food microbiomes together with a nutritionally balanced food with low fat content and a healthy life style are a good basis for a sustainable health state (Berg et al. 2015, Wassermann et al. 2019, Soto-Giron et al. 2021). It was recently shown that a high diversity of potentially health supporting bacteria are associated with fresh fruits and vegetables harbouring functions for an overall healthy and balanced gut microbiome (Wicaksono et al. 2023). In general, a high diversity of human microbiomes is crucial for persistent health. High fat diet causes a clear shift in the gut microbiome, resulting in a decreased ratio of Bacillota/Actinomycetota (formerly named: Firmicutes/Bacteroidetes) (Daniel et al. 2013, Walker et al. 2014) favouring a disbalanced gut microbiome. Specific sulfonolipids as metabolite markers and related bacterial Alistipes and Odoribacter species were found specific for this unhealthy situation in the gut of mice (Walker et al. 2017).
QS-signal production and degradation in the oral cavity
The equilibrium of microbes is maintained through competitive and cooperative interactions. An imbalance of the resident microbiota could be caused by changes in host-dependent habitat conditions or external factors like the quality of food. The development of major oral diseases is usually not dependent on a single oral pathogen, but on the entire microbial community and its activity in the oral cavity (Muras et al. 2020). For example, a reduction of bacterial coaggregation and biofilm formation during dental plaque formation is beneficial for oral and teeth health (Simón-Soro and Mira 2015). Therefore, QS activities, which positively influence biofilm structures, certainly play a major role in caries and peridontal diseases. AI-Ps from Gram-positive bacteria have been identified in different oral streptococci, and AI-2 were frequently found in different Gram-positive and Gram-negative oral pathogens like Streptococcus mutants or Porphyromonas gingivalis (Frias et al. 2001). Furthermore, different isolates of Enterobacter sp., Pseudomonas sp., and Burkholderia sp. from human tongue surface and dental plaque samples were characterized as AHL-producers (Goh et al. 2014). AHLs could also be detected in saliva and sputum samples indicating a possible role in the dental plaque formation (Goh et al 2014). In addition, QQ-activities were detected in many bacterial isolates from healthy and peridontal patients (Muras et al. 2020). These findings were corroborated by the demonstration that the addition of AHL-lactonase Aii20J had an inhibitory effect on the development of oral biofilms. Using confocal laser scanning microscopy, Aii20J inhibited in vitro mixed-species and also saliva biofilms (Muras et al. 2020). Apparently, not only AI-2 inhibitors but also AHL-lactonases or AHL-quenching bacteria are QQ-strategies for the control of oral diseases.
QQ of bacterial virulence by food substances
Many dietary metabolites were demonstrated to interfere with bacterial virulence and inhibition of QS (Dingeo et al. 2020, Fala et al. 2022). For example, pyrogallol competes with bacterial QS signals for receptor binding, polyphenols and lignans sequester bacterial QS molecules, and the flavanon naringenin reduces the production of QS-controlled virulence in P. aeruginosa PAO1. Capsicum frutescens is a spicy chilli pepper, which is rich in bioactive compounds such as capsaicin and luteolin. Capsicum frutescens extract and pure luteolin-inhibited QS in the model bacterium Chromobacterium violaceum and biofilm formation in P. aeruginosa PAO1 (Rivera et al. 2019). Apigenein and luteolin are compounds of Gnapalium hypoleucum DC extracts showing also strong QS-inhibitory activity (Li et al. 2022). In the model bacterium C. violaceum ATCC 12472 vioB, vioC, and vioD genes were strongly downregulated by apigenin and luteolin. The effective treatment of bacterial infections by G. hypoleucum extracts could thus originate from QS inhibition by these natural compounds and provides a potential mechanism for alternative applications of medicinal plants (Li et al. 2022). It was demonstrated that secondary metabolites of medicinal plants, such as terpenoids, flavonoids, and phenolic acids are antibacterial agents. In addition, they exhibit numerous anti-QS mechanisms via the inhibition of autoinducer releases, sequestration of QS-mediated molecules, and deregulation of QS gene expression (Bouyahya et al. 2022).
Microbes in fresh products such as salate or fruits can have a considerable health impact on the gut. Indeed, reconstructed metagenome—assembled genomes from 156 fruits and vegetables revealed that the microbiomes of fresh fruit and vegetables are represented by members of Enterobacteriales, Burkholderiales, and Lactobacillales in the gut microbiome. In these bacterial families diverse QS-signalling activities, but also QQ and quorum-inhibiting microbes and metabolites are represented (Wicaksono et al. 2023).
QS-related microbe–host interactions in the gut
Functional QS-systems are found not only in pathogenic but also in commensalic gut residents or probiotic bacteria (Fujii et al. 2008). In a healthy situation, gut microbiota mutually interacts with coevolved gut epithelial and immune cells in a beneficial reciprocal way (Coquant et al. 2020). QS-signalling of bacteria was shown to have important roles in beneficial bacteria intestinal cross-talk and contribute substantially to establish cross-kingdom symbiotic interactions (Wu and Luo 2021). In the dysbiosis state, like inflammatory bowel disease (IBD), microbe–intestine interactions drive inflammatory responses. The distribution of AHL-compounds detected in the feces of healthy and IBD-subjects correlated with the disease state (Landman et al. 2018). One of the AHL-compounds, 3-oxo-C12:2-HSL, was highly decreased in fecal samples of IBD patients as compared to remission and healthy persons. Thus, the absence of this particular AHL correlated with dysbiosis. Concomitantly, a decreased level of Firmicutes which are indicative for normobiosis indicated dysbiosis. Furthermore, Landman et al. (2018) showed that 3-oxo-C12:2-HSL exerts anti-inflammatory properties on intestinal model cell lines Caco-2/T17. Therefore, AHL-profiles may be considered as noninvasive biomarkers for gut normobiosis. AI-2, which are produced by opportunistic pathogenic bacteria, were successfully demonstrated in vivo to modulate the gut microbiome and cause inflammation (Thompson et al. 2015). ON the other hand, it was demonstrated that mammalian epithelial cells in the gut produce an AI-2 mimicking molecule in response to secreted bacterial factors and tight-junction disruption, which activate QS in bacteria. This was detected by bacterial AI-2 receptors LuxP/LsrB, and by the activation of QS-controlled gene expression of S. typhimurium (Ismail et al. 2016). Thus, members of the gut microbiome could be activated to colonize damaged sites and to repair epithelial tight junctions. In mammalian systems the QS-autoinducer 3-oxo-C12-HSL and similar long acylcarbon-chain AHLs produced by Pseudomonas and Burkholderia spp. are important signalling compounds involved in serious diseases, like COPD. QS-autoinducers are of central importance to coordinate, e.g. biofilm formation and virulence development (Whiteley et al. 2017). While Gram-negative bacteria are known for diverse AHL-production, Gram-positive bacteria, mainly Bacillota and Antinomycetota, are strong players in anti-AHL-QS-quenching systems, possibly acquired by horizontal gene transfer (Rajput and Kumar 2017). QQ-activities of probiotic bacteria were shown to influence the microbial community in the gut as well as immune functions in pigs (Kim et al. 2018). Antagonists of QS-systems of severe pathogens are of great importance, because mechanisms targeting QS have only minor challenge on bacterial cell viability and thus the selection for antibiotic resistant pathogens should be prevented (Zhong and He 2021). In addition, novel screening strategies were developed for QS-inhibitors to combat bacterial infections (Lu et al. 2022).
QS-signalling can organize competing strategies to neighbouring microbes. For example Gram-positive Propionibacteria, like Propionibacterium freudenreichii, have promising probiotic properties (Rabah et al. 2017, Savijoki et al. 2023). Progress in defining such metabolic interactions by in situ screening test was made possible by using biofilm-forming C. violaceum as a QS-reporter and a microscale screening platform. In this way, anti-QS effects of Lactobacillus acidophilus, Lacticaseinbacíllus rhamnosus, P. freudenreichii, and other cheese-associated strains could be identified as potent competitors for virulent pathogens (Savijoki et al. 2023).
Mechanisms of signal perception
Similar to the plant innate immune system, pathogen-associated molecular patterns are recognized by highly sensitive and specific recognition receptors such as the toll-like receptors in mammals. In addition, the adaptive immune system with differentiating dendritic cells (DC) and a guild of T-cells and macrophages are active in a coordinated way building up the most efficient adaptive immune response of mammals. Furthermore, specific receptors are involved for bacterial sensing and perception by the mammalian/human system, including QS-associated molecules and activities (Holm and Vikstrom 2014). In this context, numerous LuxR-solo type orphan genes in the human genome may code for receptors responding to bacterial products, including QS-compounds, but also for unrelated metabolites of the host or other members of complex holobiont in a still unknown way (Yong and Zhong 2013, Uhlig and Hyland 2022). The production of short chain fatty acids (SCFA) produced by commensal bacteria and probiotics in the gastrointestinal tract (GT) contribute to a balanced situation by stimulating e.g. the bacteriocin production of probiotic Lactobacillus strains, which control the virulence status of pathogens (Meng et al. 2021).
G protein-coupled receptors (GPCRs) are potential targets facilitating bacteria–host interactions by microbial-derived molecules including QS-signals. This was shown for the stimulation of root growth by AHLs with C6- and C8-fatty acid chain lengths in A. thaliana (von Rad et al. 2008, Liu et al. 2012) (see above) In general, GPCRs are members of a frequently found gene family in the plant and human genomes, some of which are involved in bacterial sensing (Krasulova and Illes 2021). Other non-GPCR targets were also described in sensing bacterial QS signals.
In mammalian systems, an uptake of 3-oxo-C12-HSL was shown in experimental human lung epithelial cells (Bryan et al. 2010). An microarray of transcriptional responses of lung epithelial cells after exposure to 3-oxo-C12-HSL revealed the expression of several xenobiotic-sensing and drug transport genes. Using radiolabelled autoinducer uptake assays, increased intracellular 3-oxo-C12-HSL levels were found after exposure, which decreased afterwards to background levels. Since this process was inhibited by the ABC transporter ABCA1, it was concluded that mammalian cells detect and take up 3-oxo-C12-HSL, but expel it later after activation of protective transport systems (Bryan et al. 2010).
AHLs were recognized as activators of the cytosolic aryl-hydrocarbon receptors (AhRs), which respond to plant products, xenobiotics, indole molecules, and SCFAs. AhR-activity is differently regulated by distinct QS molecules (Sun et al. 2020), which could constitute a crucial role of AhR in the regulation of host metabolism by pathogenic, commensal, and probiotic bacteria (Karlsson et al. 2012, Natividad et al. 2018). Interestingly, d-tryptophan (d-Trp) was identified as excreted metabolite of several probiotic bacteria, including L. rhamnosus GG, which may interact with the AhR-receptor (Kepert et al. 2017). d-Trp was shown to have stimulatory functions on developing DC-cells, to exert antiallergic responses, and to modify the gut microbiome of mice (Kepert et al. 2017). In addition to known receptor-type sensors, the lipid composition of the cell membrane, and the expression of glycolipids and transmembrane proteins were recently suggested to modulate the perception of QS and other signals and their perception (Uhlig and Hyland 2022).
Bacteria defective in QS signalling are less efficient in colonizing the GT, which was found in commensal, probiotic, and pathogenic strains (Whiteley et al. 2017). For example, Streptococcus gallolyticus subsp. gallolyticus can only colonize the murine intestine when the QS-regulated bacteriocine-like peptides blpA and blpB are produced, since Blp-deficient mutants could not sustain in the intestine. LuxS-signalling of L. rhamnosus GG and Bifidobacterium breve UCC2003 is required for the adhesion to intestinal cells (Jiang et al. 2021) and persistence in the gastro-intestinal (GI) tract (Christiaen et al. 2014).
Interactions with the human immune system
Concerning responses of gut microbiota to host metabolites, Gram-negative bacteria Escherichia coli, Shigella sp., and Salmonella sp. express QseC, a membrane-bound histidine sensor kinase that allows them to react to host-stress signals such as epinephrine and norepinephrine. Qsec inhibitors were developed as antivirulence approach against Gram-negative diseases (Rasko and Sperandio 2010, Curtis and Russell 2014). This interkingdom activation is referred to the autoinducer-3 (AI-3) system leading to the activation of QS gene expression. QseC sensor kinase is involved in GI disease caused by E. coli pathogens in rabbits. The habitat for direct interference of gut microbiota and human host are the inner mucus and the gut epithelial barrier. The structure of the intestinal mucus and epithelial barrier functions are greatly influenced by the host defence status and the QS-activity of the gut microbiome. While probiotics enhance barrier functions in vitro and in animal studies, QS-regulated virulence factors from pathogenic Citrobacterium difficile, E. coli, and S. typhimurium decrease transepithelial resistance by modulation tight barrier junctions. In contrast, QS-associated activities, involving 3-oxo-C12 HSL, were found to negatively impact gut integrity through the activation of inflammatory pathways that impair intestinal barrier function (Adiliaghdam et al. 2019). AHLs, mostly 3-oxo-C12- HSL from P. aeruginosa, stimulate inflammatory responses through the inhibitory interaction with neutrophils, macrophages, and DC, finally causing apoptosis of those immune cells. In this way, 3-oxo-C12-HSL producing bacteria pathogens effectively inhibits attempts oft he immune system to eliminate the aggressor (Coquant et al. 2020). When LPS-stimulated human DC were exposed to 3-oxo-C12-HSL, in vitro flow cell cytometric analyses revealed that important DC surface markers like CD80, CD40, CD184, and HLA-DR were diminished, while 3-oxo-C4-HSL had no effect (Binder T, PhD thesis LMU Munich, 2010). Accordingly, the inflammation inhibitory cytokine IL-10 was decreased and the inflammation stimulatory cytokine IL-8 was increased, leading to a severe damping effect on the immune response. Furthermore, the migration of DC-cells after treatment with LPS and 3-oxo-C12-HSL was decreased as well as their phagocytotic activity. This causes severe health problems of cystic fibrosis patients infected with P. aeruginosa (Cohen et al. 2015). Figure 3 shows a schematic drawing of activities of the autoinducer 3-oxo-C12-HSL of P. aeruginosa towards the human innate and adaptive immune system.
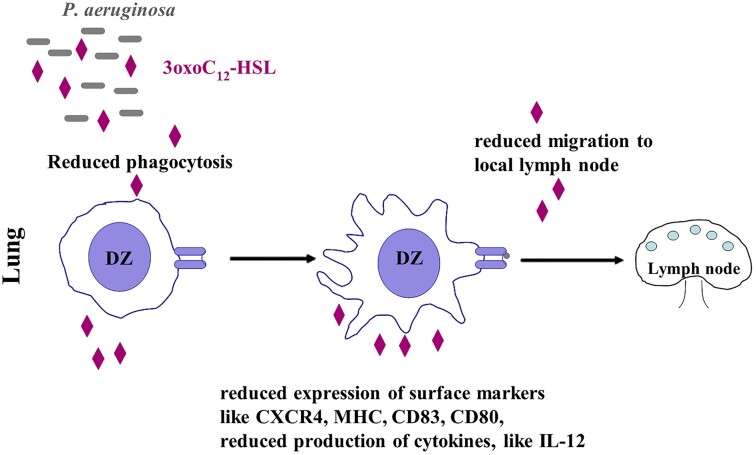
Figure 3.
AHL-interactions with mammalian immune system (example: 3-oxo-C12-HSL of P. aeruginosa and lung) (Binder, PhD thesis, LMU Munich 2010): DC are influenced by 3-oxo-C12-HSL during their ripening process stimulated by lipopolysacharide (LPS), leading to diminished phagocytosis and reduced expression of surface markers, like CXCR4, MHC, CD83, and CD80. The CXCR4-marker is involved in the regulation of migration of DC to lymph nodes, while the reduction of other surface markers lead to the induction of cyclooxygenase (Cox-2) having a modulatory role in inflammation processes; prostaglandines are increased supporting the Th2-response in the lymph node, which support a reduced ripening of DC-cells, meaning reduced inflammation response (Skindersoe et al. 2009). Downregulation of cytokines, like IL-12 and the anti-inflammatory cytokine IL-10, causes a Th1/Th2-imbalance in the infected host (Ritchie et al. 2005).
Conclusions
Microbial health in the rhizosphere is interconnected with ‘One Health’, because the health of each of its components is determined by the omnipresence of microorganisms (Banerjee et al. 2023). Rhizosphere microbiomes are tightly linked to soil and plant microbiomes and their reservoirs for extremely diverse microbioms also influence animal and human health (Hartmann et al. 2009, Schreiter et al. 2014). In the rhizosphere, a rich supply of substrates leaking out of roots guarantees excellent conditions for microbial activities, which also provide the basis for efficient coevolution of environmental and plant microbes with each other in the context of the plant host as master. This includes natural genetic engineering using horizontal gene transfer, plasmid transduction, high mutation rates, and phenotypic switching. In the efficient ‘rhizosphere schools’ of evolution the selection of adapted rhizosphere communities through changing the exudation pattern constantly occurs (Berendsen et al. 2012). Under these selective conditions QS-signalling-based microbe–host interactions towards beneficial/cooperative as well as pathogenic/competitive relations have evolved. The knowledge about responses of plant and mammalian/human hosts to bacterial QS-signalling compounds and their producers are considerable advanced. A large body of data about the interaction of pathogenic, symbiotic/beneficial, and commensalic bacteria accumulated, which are involved in plant or human health (LaSarre and Federle 2013, Mendes et al. 2013, Schikora et al. 2016). In this context, QS research provides new perspectives to better understand the interaction between gut microbiota and the human host (Yang et al. 2022). This may provide an additional key to be able to apply microbiome science for plant and human health (Russ et al. 2023). Nowadays, first experiences about the possibility of translation into practice with recognized specific ‘therapeutic’ microbes and QS-related substances (Uhlig and Hyland 2022), QS-signals (Moshynets et al. 2019), and synthetic microbial communities (syn-coms) (Jiang et al. 2022, Schmitz et al. 2022) are available. Apparently, an effective syn-com may harbour QS-active bacteria, which provide the applied synthetic community a dynamic character to modulate the rhizosphere microbiome and stimulate the plant host to develop abiotic and biotic resistance properties (Andres-Barrao et al. 2017). Further progress in revealing the involvement of QS-regulation in plant as well as in human health is still dependent on further optimization of the sensitivity and specificity of QS-signal analytical approaches (Mellini et al. 2024). Concerning the severe problem of rapid spreading of chronic infections in humans because of the immense dangerous threat by multiresistant pathogens, the application of QS-affecting approaches, which target virulence acquisition processes of pathogens promised to avoid the selection of antibiotic resistances, because growth of pathogens is not directly targeted. This could finally lead to substantial human health improvements in the control of multiresistant pathogens when combined with other strategies (Zhong and He 2021, Naga et al. 2023). However, since QS-systems are important for bacterial fitness, QS-inhibition would unavoidingly affect the targeted pathogens and, may hence impose a selective pressure. Experimental evidences for this problem was published by Maeda et al. (2012) and Imperi et al. (2019). Thus, strategies based on system-level ecologic principles of microbial social behaviour may contribute to successful personalized treatments of patients. Applications of QS-targeted treatments could allow important progress to improve sustainable agriculture and contribute to better control devastating diseases for plant and human health.
Acknowledgements
The generous support by Jürgen Kinder†, Helmholtz Zentrum Munich (former GSF-Research Center for Health and Environment, Neuherberg) for the interdisciplinary research project ‘Molecular interactions in the rhizosphere’ is acknowledged. We greatly appreciate Burkhard Hense†, Michael Schmid†, Peter Schröder, Michael Schloter, Philippe Schmitt-Kopplin, Christian Langebartels, and Susanne Krauss-Etschmann (all GSF-Research Center for Health and Environment) for their commitment to this interdisciplinary project and Adam Schikora (JKI Braunschweig) for his valuable cooperation and support.
Contributor Information
Anton Hartmann, Faculty of Biology, Microbe-Host Interactions, Ludwig-Maximilian-University Munich, Grosshaderner Str. 2, D-82152 Planegg/Martinsried, Germany; Department of Environmental Sciences, Helmholtz Zentrum Munich, German Research Center for Health and Environment, Research Unit Microbe-Plant Interactions, Ingolstädter Landstr. 1, D-85762 Neuherberg, Germany.
Tatiana Binder, Department of Environmental Sciences, Helmholtz Zentrum Munich, German Research Center for Health and Environment, Research Unit Microbe-Plant Interactions, Ingolstädter Landstr. 1, D-85762 Neuherberg, Germany.
Michael Rothballer, Department of Environmental Sciences, Helmholtz Zentrum Munich, German Research Center for Health and Environment, Research Unit Microbe-Plant Interactions, Ingolstädter Landstr. 1, D-85762 Neuherberg, Germany; Helmholtz Zentrum Munich, German Research Center for Health and Environment, Institute of Network Biology, Ingolstädter Landstr. 1 D-85762 Neuherberg, Germany.
Author contributions
Anton Hartmann (Conceptualization, Formal analysis, Funding acquisition, Investigation, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing), Tatiana Binder (Formal analysis, Investigation, Methodology, Visualization), and Michael Rothballer (Conceptualization, Visualization, Writing – review & editing)
Conflict of interest
We declare no conflict of interest.
References
- Abedini D, Jaupite S, Bouwmeester H et al. Metabolic interactions in beneficial microbe recruitment by plants. Curr Opin Biotechnol. 2021;70:241–7. 10.1016/j.copbio.2021.06.015. [DOI] [PubMed] [Google Scholar]
- Abreo E, Altier N. Pangenome of Serratia marcescens strains from nosocomial and environmental origins reveals different populations and the links between them. Sci Rep. 2019;9:46. 10.1038/s41598-018-37118-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achouak W, Conrod S, Cohen V et al. Phenotypic variation of Pseudomonas brassicacearum as a plant-colonization strategy. MPMI. 2004;17:872–9. 10.1094/MPMI.2004.17.8.872. [DOI] [PubMed] [Google Scholar]
- Adiliagham F, Almpani M, Gharedaghi MH et al. Targeting bacterial quorum sensing shows promise in improving intestinal barrier function following burn site infection. Mol Med Report. 2019;19:4057–66. 10.3892/mmr.2019.10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlgren NA, Harwood CS, Schaefer AL et al. Aryl-homoserine lactone quorum sensing in stem-nodulating photosynthetic Bradyrhizobia. Proc Natl Acad Sci USA. 2011;108:7183–8. www.pnas.org/cgi/doi/10.1073/pnas.1103821108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabid I, Hardt M, Imani J et al. The N-acylhomoserine lactone depleted Rhizobium radiobacter mutant RrF4NM13 shows reduced growth-promoting and resistance-inducing activities in mono- and dicotyledonous plants. J Plant Dis Prot. 2020;127:769–81. 10.1007/s41348-020-00360-8. [DOI] [Google Scholar]
- Andres-Barrao C, Lafi FF, Alam I et al. Complete genome sequence analysis of Enterobacter sp. SA187, a plant multi-stress tolerance promoting endopyhtic bacterium. Front Microbiol. 2017;8:2023. 10.3389/fmicb.2017.02023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angus AA, Agapakis CM, Fong S et al. Plant associated symbiotic Burkholderia species lack hallmark strategies required in mammalian pathogenesis. PLoS One. 2014;9:e83779. 10.1371/journal.pone.0083779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldani JI, Pot B, Kirchhof G et al. Emended description of Herbaspirillum; inclusion of Pseudomonas rubrisubalbicans, a mild plant pathogen, as Herbaspirillum rubrisubalbicans comb. nov. and classification of a group of clinical isolates (EF group 1) as Herbaspirillum species 3. Int J Syst Bacteriol. 1996;46:802–10. 10.1099/00207713-46-3-802. [DOI] [PubMed] [Google Scholar]
- Banerjee S, van der Heijden MGA. Soil microbiomes and one health. Nat Rev Micro. 2023;21:6–20. 10.1038/s41579-022-00779-w. [DOI] [PubMed] [Google Scholar]
- Barbey C, Crépin A, Bergeau D et al. In planta biocontrol of Pectobacterium atrosepticum by Rhodococcus erythropolis involves silencing of pathogen communication by the rhodococcal gamma-lactone catabolic pathway. PLoS One. 2013;8:e66642. https://doi.org.10.1371/journal.pone.0066642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barriuso J, Solano BP, Fray RG et al. Transgenic tomato plants alter quorum sensing in plant growth-promoting rhizobacteria. Plant Biotechnol J. 2008;6:442–52. 10.1111/j.1467-7652.2008.00331.x [DOI] [PubMed] [Google Scholar]
- Berendsen RL, Pieterse CMJ, Bakker PAHM. The rhizosphere microbiome and plant health. Trend Plant Sci. 2012;17:478–86. 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- Berg G, Eberl L, Hartmann A. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ Microbiol. 2005;7:1673–85. 10.1111/j.1462-2920.2005.00891.x. [DOI] [PubMed] [Google Scholar]
- Berg G, Erlacher A, Grube M. The edible plant microbiome: importance and health issues. In: Lugtenberg B (ed.), Principles of Plant-Microbe Interactions: Microbes for Sustainable Agriculture. Berlin: Springer, 2015, 419–26. 10.1007/978-3-319-08575-3_44. [DOI] [Google Scholar]
- Berg G, Grube M, Schloter M et al. The plant microbiome and its importance for plant and human health. Front Microbiol. 2014;5:1–2. 10.3389/fmic.2014.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertini EV, Penalver CGN, Leguina AC et al. Gluconacetobacter diazotrophicus PAL5 possesses an active quorum sensing regulatory system. Antonie Van Leeuwenhoek. 2014;106:497–506. 10.1007/s10482-014-0218-0. [DOI] [PubMed] [Google Scholar]
- Bez C, Geller AM, Levy A et al. Cell-cell signaling proteobacterial LuxR solos: a treasure trove of subgroups having different origins, linda, and ecological roles. mSystems. 2023;8:e1039. 10.1128/msystems.01039-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder T. Das N-(3-oxododecanoyl)-L-Homoserinlacton von Pseudomonas aeruginosa Inhibiert Funktionen Humaner Dendritischer Zellen. Ph.D. Thesis, Ludwig-Maximilians-Universität München, 2010. [Google Scholar]
- Bouyahya A, Chamkhi I, Balahbib A et al. Mechanisms, anti-quorum-sensing actions, and clinical trials of medicinal plant bioactive compounds against bacteria: a comprehensive review. Molecules. 2022;27:1484. 10.3390/molecules27051484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan A, Watters C, Koenig L et al. Human transcriptome analysis reveals a potential role for active transport in the metabolism of Pseudomonas aeruginosa autoinducers. Microbes Infect. 2010;12:1042–50. 10.1016/j.micinf.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buddrus-Schiemann K, Rieger M, Mühlbauer M et al. Analysis of N-acyl-homoserine lactone dynamics in continous culture of Pseudomonas putida IsoF using ELISA and UPLC/qTOF-MS related measurements and mathematical models. Anal Bioanal Chem. 2014;406:6373–83. 10.1007/s00216-014-8063-6. [DOI] [PubMed] [Google Scholar]
- Cárcamo-Oyarce G, Lumjiaktase P, Kümmerli R et al. Quorum sensing triggers the stochastic escape of individual cells from Pseudomonas putida biofilms. Nat Commun. 2015;6:5945. 10.1038/ncomms6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassan F, Diaz-Zorita M. Azospirillum spp. in current agriculture: from the laboratory to the field. Soil Biol Biochem. 2016;103:117–30. 10.1016/j.soilbio.2106.08.020. [DOI] [Google Scholar]
- Cavalcante VA, Doebereiner J. A new acid-tolerant nitrogen fixing bacterium associated with sugarcane. Plant Soil. 1988;108:23–31. 10.1007/BF02370096. [DOI] [Google Scholar]
- Cellini A, Buriani G, Correia C et al. Host-specific signal perception by PsaR2 LuxR solo induces Pseudomonas syringae pv. actinidiae virulence traits. Microbiol Res. 2022;260:127048. 10.1016/j.micres.2022.127048. [DOI] [PubMed] [Google Scholar]
- Certner RH, Vollmer SV. Inhibiting bacterial quorum sensing arrests coral disease development and disease-associated microbes. Environ Microbiol. 2018;20:645–57. 10.1111/1462-2920.13991. [DOI] [PubMed] [Google Scholar]
- Chen X, Kremmer E, Gouzy M-F et al. Development and characterization of rat monoclonal antibodies for N-acyl-homoserine lactones. Anal Bioanal Chem. 2010;398:2655–67. 10.1007/s00216-010-4017-9. [DOI] [PubMed] [Google Scholar]
- Chowdhary PK, Keshavan N, Nguyen HQ et al. Bacillus megaterium CYP102A1 oxidation of acyl homoserine lactones and acyl homoserines. Biochemistry. 2007;46:14429–37. 10.1021/bi701945j. [DOI] [PubMed] [Google Scholar]
- Christiaen SE, O'Connell Motherway M, Bottacini F et al. Autoinducer-2 plays a crucial role in gut colonization and probiotic functionality of Bifidobacterium breve UCC2003. PLoS One. 2014;9:e98111. 10.1371/journal.pone.0098111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirou A, Mondy S, An S et al. Efficient biostimulation of native and introduced quorum-quenching Rhodococcus erythropolis populations is revealed by a combination of analytical chemistry, microbiology, and pyrosequencing. Appl Environ Microb. 2012;78:481–92. 10.1128/AEM.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MF, Han XY, Mazzola M. Molecular and physiological comparison of Azospirillum spp. isolated from Rhizoctonia solani mycelia, wheat rhizosphere and human skin wounds. Can J Microbiol. 2004;50:291–7. 10.1139/w04-007 [DOI] [PubMed] [Google Scholar]
- Cohen T, Parker D, Prince A. P. aeruginosa host immune evasion. In: Ramos JL, Goldberg J, Filloux A (eds), Pseudomonas. Dordrecht: Springer, 2015, 3–23. 10.1007/978-94-017-9555-5_1. [DOI] [Google Scholar]
- Coquant G, Grill J-P, Seksik P. Impact of N-acyl-homoserine lactones on gut immunity. Front Immunol. 2020;11:1827. 10.3389/fimmu.2020.01827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MM, Russell R. QseC inhibitors as an antivirulence approach for Gram-negative pathogens. mBio. 2014;5:e02165. 10.1128/mBio.02165-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czajkowski R, Krzyzanowska D, Karczewska J et al. Inactivation of AHLs by Ochrobactrum sp. A44 depends on the activity of a novel class of AHL acylase. Environ Microbio Rep. 2011;3:59–68. 10.1111/j.1758-2229.2010.00188. [DOI] [PubMed] [Google Scholar]
- Daniel H, Desmarchelier C, Hahne H et al. High energy diets alter the composition, proteome and metabolome of the mouse gut microbiota. ISME J. 2013;8:295–308. 10.1038/ismej,2013.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer W, Li X, Meisner A et al. Pathogen suppression by microbial volatile organic compounds in soils. FEMS Microbiol Ecol. 2019;95:fiz105. 10.1093/femsec/fiz105. [DOI] [PubMed] [Google Scholar]
- Dingeo G, Brito A, Samouda H et al. Phytochemicals as modifiers of gut microbial communities. Food Funct. 2020;11:8444–7. 10.1039/s0fo01483d [DOI] [PubMed] [Google Scholar]
- Dobritsa AP, Samadpour M. Transfer of eleven species of the genus Burkholderia to the genus Paraburkholderia and proposal of Caballeronia gen. nov. to accomodate twelve species of the genera Burkholderia and Paraburkholderia. Int J Syst Evol Microbiol. 2016;66:2836–46. 10.1099/ijsem.0.001065. [DOI] [PubMed] [Google Scholar]
- Doebereiner J, Day JM. Associative symbiosis in tropical grasses: characterization of microorganisms and dinitrogen fixing sites. In: Newton WE, Nyman CJN (eds), Proceedings of the First International Symposium on Nitrogen Fixation. Washington: Washington State University Press, 1976, 518–38. [Google Scholar]
- Dos Santos Ferreira N, Coniglio A, Puente M et al. Genome-based reclassification of Azospirillum brasilense Az39 as the type strain of Azospirillum argentinense sp. nov. Int J Syst Evol Microbiol. 2022;72:5475. 10.1099/ijsem.0.005475. [DOI] [PubMed] [Google Scholar]
- Duan Y, Han M, Grimm M et al. Combination of bacterial N-acyl-homoserine lactones primes Arabidopsis defenses via jasmonate metabolism. Plant Physiol. 2023;191:2027–44. 10.1093/plphys/kiad017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falá AK, Alvarez-Ordonez A, Filloux A et al. Quorum sensing in human gut and food microbiomes: significance and potential for therapeutic targeting. Front Microbiol. 2022;13:1002185. 10.3389/fmicb.2022.1002185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faoro H, Oliveira WK, Weiss VA et al. Genome comparison between clinical and environmental strains of Herbaspirillum seropedicae reveals a potential new emerging bacterium adapted to human hosts. BMC Genomics. 2019;20:630. 10.1186/s12864-019-5982-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete A, Kuttler C, Rothballer M et al. Dynamic regulation of AHL-production and degradation in Pseudomonas putida IsoF. FEMS Microbiol Ecol. 2010;72:22–34. 10.1111/j.1574-6941.2009.00828.x. [DOI] [PubMed] [Google Scholar]
- Filgueiras L, Silva R, Almeida I et al. Gluconcetobacter diazotrophicus mitigates drought stress in Oryza sativa L. Plant Soil. 2020;451:57–73. 10.1007/s11104-019-04163-1. [DOI] [Google Scholar]
- Fray RG, Throup JP, Daykin M et al. Plants genetically modified to produce N-acyl-homoserine lactones communicate with bacteria. Nat Biotechnol. 1999;17:1017–20. 10.1038/13717. [DOI] [PubMed] [Google Scholar]
- Frias J, Olle E, Alsina M. Periodontal pathogens produce quorum sensing signal molecules. Infect Immun. 2001;69:3431–4. 10.1128/IAI.69.5.3431-3434.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T, Ingham C, Nakayama J et al. Two homologous Agr-like quorum-sensing systems cooperatively control adherence, cell morphology, and cell viability properties in Lactobacillus plantarum WCFS1. J Bacteriol. 2008;190:7655. 10.1128/JB.01489-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukami J, Abrantes JLF, del Cerro P et al. Revealing strategies of quorum sensing in Azospirillum brasilense strains ab-V5 and Vb-V6. Arch Microbiol. 2018;200:47–56. 10.1007/s00203-017-1422-x. [DOI] [PubMed] [Google Scholar]
- Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–75. 10.1128/jb.176.2.269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamalero E, Lingua G, Glick BR. Ethylene, ACC, and the plant growth promoting enzyme ACC deaminase. Biology. 2023;12:1043. 10.3390/biology12081043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaeser SP, Imani J, Alabid I et al. Non-pathogenic Rhizobium radiobacter RrF4 deploys plant beneficial activity independent of its host Piriformospora indica. ISME J. 2016;10:871–84. 10.1038/ismej.2015.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh SY, Tan W-S, Khan SA et al. Unusual multiple production of N-acyl-homoserine lactones by Burkholderia sp. strain C10B isolated from dentine caries. Sensors. 2014;14:8940–9. 10.3390/s140508940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González JE, Marketon MM. Quorum sensing in nitrogen-fixing rhizobia. Microbiol Mol Biol Rev. 2003;67:574–92. 10.1128/MMBR.67.4.574-592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González JF, Venturi V. A novel widespread inter-kingdom signaling circuit. Trends Plant Sci. 2013;18:167–74. 10.1016/j.tplants.2012.09.007. [DOI] [PubMed] [Google Scholar]
- Götz C, Fekete A, Gebefuegi I et al. Uptake, degradation and chiral discrimination of N-acyl-D/L-homoserine lactones in barley (Hordeum vulgare) and yam bean (Pachyrhizus etosus) plants. Anal Bioanal Chem. 2007;389:1447–57. 10.1007/s00216-007-1579-2. [DOI] [PubMed] [Google Scholar]
- Götz-Rösch C, Sieper T, Fekete A et al. Influence of bacterial N-acyl-homoserine lactones on growth parameters, pigments, antioxidative capacities and the xenobiotic phase II detoxification enzymes in barley and yam bean. Front Plant Sci. 2015;6:205. 10.3389/fpls.2015.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandclément C, Tannières M, Moréra S et al. Quorum quenching: role in nature and applied developments. FEMS Microbiol Rev. 2016;40:86–116. 10.1093/femsre/fuv038. [DOI] [PubMed] [Google Scholar]
- Gualpa J, Lopez G, Nievas S et al. Azospirillum brasilense Az39, a model rhizobacterium with AHL quorum-quenching capacity. J Appl Microbiol. 2019;126:1850–60. 10.1111/jam.14269. [DOI] [PubMed] [Google Scholar]
- Gupta G, Panwar J, Jha PN. Natural occurrence of Pseudomonas aeruginosa, a dominant cultivable diazotrophic endophytic bacterium colonizing Pennisetum glaucum (l.) R. Appl Soil Ecol. 2013;64:252–61. 10.1016/j.apsoil.2012.12.016. [DOI] [Google Scholar]
- Han S, Li D, Trost E et al. Systemic response of barley to the 3-hydroxy-decanoyl-homoserine lactone producing plant beneficial endophytic Acidovorax radicis N35. Front Plant Sci. 2016;7:e1868. 10.3389/fpls.2016.01868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann A, Fischer D, Kinzel L et al. Assessment of the structural and functional diversities of plant microbiota: achievements and challenges—a review. J Adv Res. 2019;19:3–13. 10.1016/j.jare.2019.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann A, Klink S, Rothballer M. Importance of N-acyl-homoserine lactone -based quorum sensing and quorum quenching in pathogen control and plant growth promotion. Pathogens. 2021;10:1561. 10.3390/pathogens10121561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann A, Rothballer M, Schmid M. Lorenz Hiltner: a pioneer in rhizosphere microbial ecology and soil bacteriology research. Plant Soil. 2008;312:7–14. 10.1007/s11104-007-9514-z. [DOI] [Google Scholar]
- Hartmann A, Schmid M, van Tuinen D et al. Plant-driven selection of microbes. Plant Soil. 2009;321:235–57. 10.1007/s11104-008-9814-y. [DOI] [Google Scholar]
- Hense B, Schuster M. Core principles of bacterial autoinducer systems. Microbiol Mol Biol Rev. 2015;79:153–69. 10.1128/MMBR.00024-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hense BA, Kuttler C, Müller J et al. Does efficiency sensing unify diffusion and quorum sensing?. Nat Rev Micro. 2007;5:230–9. 10.1038/nrmicro1600. [DOI] [PubMed] [Google Scholar]
- Hiltner L. Über neuere erfahrungen und probleme auf dem Gebiete der Bodenbakteriologie unter besonderer berücksichtigung der Brache. Arb Dtsch Landwirtsch Gesellsch. 1904;98:59–78. [Google Scholar]
- Holm A, Vikstrom E. Quorum sensing communication between bacteria and human cells: signals, targets, and functions. Front Plant Sci. 2014;5:309. 10.3389/fpls.2014.00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imperi F, Fiscarelli EV, Visaggio D et al. Activity and impact on resistance development of two antivirulence fluoropyrimidine drugs in Pseudomonas aeriginosa. Front Cell Infect Microbiol. 2019;9:49. 10.3389/fcimb.2019.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail AS, Valastyan JS, Bassler BL. A host produced autoinducer-2 mimic activates bacterial quorum sensing. Cell Host Microbe. 2016;19:470–80. 10.1016/j.chom.2016.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G, Zhang Y, Gan G et al. Exploring rhizo-microbiome transplants as a tool for protective plant-microbiome manipulation. ISME Commun. 2022. 10.1038/s43705-022-00094-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Luo Y, Cao X et al. LuxS quorum sensing system mediating Lactobacillus plantarum probiotic characteristics. Arch Microbiol. 2021;2:4141–8. 10.1007/s00203-021-02404-5. [DOI] [PubMed] [Google Scholar]
- Karlsson T, Turkina MV, Vakymenko O et al. The Pseudomonas aeruginosa N-acyl-homoserine lactone quorum sensing molecules target IQGAP1 and modulate epithelial cell migration. PLoS Pathog. 2012;8:e1002953. 10.1371/journal.ppat.1002953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepert I, Fonseca J, Müller C et al. D-tryptophan of probiotic bacteria influences the gut microbiome and allergic airway disease. J Allergy Clin Immunol. 2017;139:1525–35. 10.1016/j.jaci.2016.09.003. [DOI] [PubMed] [Google Scholar]
- Keswani C, Prakash O, Bharti N et al. Re-addressing the biosafety issues of plant growth promoting rhizobacteria. Sci Total Environ. 2019;690:841–52. 10.1016/j.scitotenv.2019.07.046 [DOI] [PubMed] [Google Scholar]
- Kim J-S, Yoon S-J, Park Y-J et al. Crossing the kingdom border: human diseases caused by plant pathogens. Environ Microbiol. 2020;22:2485–95. 10.1111/1462-2920.15028. [DOI] [PubMed] [Google Scholar]
- Kim J, Kim J, Kim Y et al. Influences of quorum quenching probiotic bacteria on the gut microbial community and immune function in weaning pigs. Anim Sci J. 2018;89:412–22. 10.1111/asj.12954. [DOI] [PubMed] [Google Scholar]
- Kirchhof G, Eckert B, Stoffels M et al. Herbaspirillum frisingense sp. nov., a new nitrogen fixing bacterial species occurring in C4-energy plants. Int J Syst Evol Microbiol. 2001;51:157–68. 10.1099/00207713-51-1-157. [DOI] [PubMed] [Google Scholar]
- Krasulova K, Illes P. Intestinal interplay of quorum sensing molecules and human receptors. Biochimie. 2021;189:108–19. 10.1016/j.biochi.2021.06.010. [DOI] [PubMed] [Google Scholar]
- Kutter S, Hartmann A, Schmid M. Colonization of barley (Hordeum vulgare) roots by Salmonella enterica typhimurium and Listeria spp. FEMS Microbiol Ecol. 2006;56:262–71. 10.1111/j.1574-6941.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- Lalaouna D, Fochesato S, Sanchez L et al. Phenotypic switching in Pseudomonas brassicaearum involves GacS- and GacA-dependent small RNAs. Appl Environ Microb. 2012;78:1658–65. 10.1128/AEM.06769-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landman C, Grill JP, Mallet JM et al. Inter-kingdom effect on epithelial cells of the N-acyl homoserine lactone 3-oxo-C12:2 HSL, a major quorum-sensing molecule from gut microbiota. PLoS One. 2018;13:e0202587. 10.1371/journal.pone.0202587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaSarre B, Federle MJ. Exploiting quorum sensing to confuse bacterial pathogens. Microbiol Mol Biol Rev. 2013;77:73–111. 10.1128/MMBR.00046-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M-N, Kim S-K, Li X-H et al. Bacterial virulence analysis using brine shrimp as an infection model in relation to the importance of quorum sensing and proteases. J Gen Appl Microbiol. 2014;60:169–74. 10.2323/jgam.60.169. [DOI] [PubMed] [Google Scholar]
- Lerat E, Moran NA. The evolutionary history of QS systems in bacteria. Mol Biol Evolut. 2004;21:903–13. 10.1093/molbev/msh097. [DOI] [PubMed] [Google Scholar]
- Li D, Rothballer M, Engel M et al. Phenotypic variation in Acidovorax radicis N35 influences plant growth promotion. FEMS Microbiol Ecol. 2012;79:751–62. 10.1111/j.1574-6941.2011.01259.x. [DOI] [PubMed] [Google Scholar]
- Li D, Rothballer M, Schmid M et al. Acidovorax radicis sp. nov., a rhizosphere bacterium isolated from wheat roots. Int J Syst Evol Microbiol. 2011;61:2589–94. 10.1099/ijs.0.025296-0. [DOI] [PubMed] [Google Scholar]
- Li Y-L, Chu Z-Y, Liu G-M et al. The derived components of Gnaphalium hypoleucum DC.reduce quorum sensing of Chromobacterium violaceum. Molecules. 2022;27:4881. 10.3390/molecules27154881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Li Z, Xiong D et al. Quorum quenching by a type IVA secretion system effector. ISME J. 2023;17:1564–77. 10.1038/s41396-023-01457-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Bian Z, Jia Z et al. The GCR1 and GPA1 participate in promotion of A. thaliana primary root elongation induced by N-acyl-homoserine lactones, the bacterial QS-signals. MPMI. 2012;25:677–83. 10.1094/MPMI-10-11-0274. [DOI] [PubMed] [Google Scholar]
- Lu L, Li M, Yi G et al. Screening stragtegies for quorum sensing inhibitors in combating bacterial infections. J Pharm Anal. 2022;12:1–14. 10.1016/j.jpha.2021.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T, Garcia-Contrras R, Pu M et al. Quorum quenching quandary: resistance to antivirulence compounds. ISME J. 2012;6:493–501. 10.1038/ismej.2011.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathesius U, Mulders S, Gao M et al. Extensive and specific responses of an eucaryote to bacterial quorum sensing-signal. Proc Natl Acad Sci USA. 2003;100:1444–9. www.pnas.org/cgi/doi/10.1073/pnas.262672599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellini M, Letizia M, Leoni L et al. Whole-cell biosensors for qualitative and quantitative analysis of quorum sensing signal molecules and the investigation of quorum quenching agents. Methods Mol Biol. 2024;2721:55–67. 10.1007/978-1-0716-3473-8_5. [DOI] [PubMed] [Google Scholar]
- Mendes R, Garbeva P, Raaijmakers J. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev. 2014;38:143–. 10.1111/1574-6976.12068. [DOI] [PubMed] [Google Scholar]
- Meng F, Zhao H, Nie T et al. Acetate activates Lactobacillus bacteriocin synthesis by controlling quorum sensing. Appl Env Microbiol. 2021;87:e0072021. 10.1128/AEM00720-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda SW, Asfahl KL, Dandekar AA et al. Pseudomonas aeruginosa quorum sensing. Exp Med Adv Biol. 2022;1386:95–115. 10.1007/978-3-031-08491-1_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshynets OV, Babenko LM, Rogalsky SP et al. Priming winter wheat seeds with the bacterial quorum sensing signal N-hexanoyl-L-homoserine lactone (C6-HSL) shows potential to improve plant growth and seed yield. PLoS One. 2019;14:e0209460. 10.1371/journal.pone.0209460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Bassler BL. Bacterial quorum sensing in complex and dynamically changing environments. Nat Rev Micro. 2019;17:371–82. https://doil.org/10.1038/S41579-019-0186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muras A, Otero-Casal P, Blanc V et al. Acyl homoserine lactone-mediated quorum sensing in the oral cavity: a paradigm revisited. Sci Rep. 2020;10:9800. 10.1038/s41598-020-66704-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadell CD, Drescher K, Foster KR. Spatial structure, cooperation and competition in biofilms. Nat Rev Micro. 2016;14:589–600. 10.1038/nrmicro.2016.84. [DOI] [PubMed] [Google Scholar]
- Naga NG, El-Badan DE, Ghanem KM et al. It is time for quorum sensing inhibition as alternative strategy of antimicrobial therapy. Cell Commun Signal. 2023;21:133. 10.1186/s12964-023-01154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natividad JM, Agus A, Planchais J et al. Impaired aryl hydrocarbon receptor ligand production by the gut microbiota is a key factor in metabolic syndrome. Cell Metab. 2018;28:737–749.e4. 10.1016/j.cmet.2018.07.001. [DOI] [PubMed] [Google Scholar]
- Nieto-Penalver CG, Bertini EV, de Figueroa LI Identification of N-acyl homoserine lactones produced by Gluconacetobacter diazotrophicus PAL5 cultured in complex and synthetic media. Arch Microbiol. 2012;194:615–22. 10.1007/s00203-012-0794-1. [DOI] [PubMed] [Google Scholar]
- Oliveira WK, Avila HL, Tadra MZ et al. High genomic identity between clinical and environmental strains of Herbasprillum frisingense suggests pre-adaptation to different hosts and intrinsic resistance to multiple drugs. Antibiotics. 2021;10:1409. 10.3390/antibiotics10111409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AG, Senechal AC, Mukherjee A et al. Plant responses to bacterial N-acyl L-homoserine lactones are dependent on enzymatic degradation to L-homoserine. ACS Chem Biol. 2014;9:1834–45. 10.1021/cb500191al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort KP, Bassler BL. Quorum sensing signal response systems in Gram-negative bacteria. Nat Rev Micro. 2016;14:576–88. 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsek MR, Greenberg EP. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 2005;13:27–33. 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Patel HK, Suárez-Moreno ZR, Degrassi G et al. Bacterial LuxR solos have evolved to respond to different molecules including signals from the plant. Front Plant Sci. 2013;4:447. 10.3389/fpls.2013.00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollumaa L, Alamäe T, Mäe A. Quorum sensing and expression of virulence in pectobacteria. Sensors. 2012;12:3327–49. https://doi:10.3390/s121313327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabah H, Rosa do Carmo FL, Jan G. Dairy propionibacteria: versatile probiotics. Microorganisms. 2017;5:24. 10.3390/microorganisms5020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajput A, Kumar M. In silico analyses of conservational, functional and phylogenetic distribution of the LuxI and LuxR homologs in Gram-positive bacteria. Sci Rep. 2017;7:6969. 10.1038/s41598-017-07241-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankl S, Gunsé B, Sieper T et al. Microbial homoserine lactones (AHLs) are effectors of root morphological changes in barley. Plant Sci. 2016;253:130–40. 10.1016/j.plantsci.2016.09014. [DOI] [PubMed] [Google Scholar]
- Rasko DA, Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nat Rev Drug Discov. 2010;9:117–28. 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- Rezzonico F, Duffy B. Lack of genomic evidence of AI-2 receptors suggests a non-quorum sensing role for luxS in most bacteria. BMC Microbiol. 2008;8:154. 10.1186/1471-2180-8-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie AJ, Jansson A, Stallberg J et al. The P. aeruginosa quorum sensing-molecule N-3-oxoC12-homoserine lactone inhibits T-cell differentiation and cytokine production by a mechanism involving an early step in T-cell activation. Infect Immun. 2005;73:1648–55. 10.1128/iai.73.3.1648-1655.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera MLC, Hassimotto NMA, Bueris V et al. Effect of Capsicum frutescens extract, Capsaicin, and luteolin on quorum sensing regulated phenotypes. J Food Sci. 2019;84:1477–86. 10.1111/1750-3841.14648. [DOI] [PubMed] [Google Scholar]
- Rodriguez M, Torres M, Blanco L et al. Plant-growth-promoting and quorum quenching-mediated biocontrol of bacterial phytopathogens by Pseudomonas segetis strain P6. Sci Rep. 2020;10:4110–21. 10.1038//s41598-020-61084-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothballer M, Eckert B, Schmid M et al. Endophytic root colonization of gramineous plants by Herbaspirillum frisingense. FEMS Microbiol Ecol. 2008;66:85–95. 10.1111/j.1574-6941.2008.00582.x. [DOI] [PubMed] [Google Scholar]
- Rothballer M, Schmid M, Klein I et al. Herbaspirillum hiltneri sp. nov., isolated from surface-sterilized roots. Int J Syst Evolut Microbiol. 2006;56:1341–8. 10.1099/ijs.064031-0. [DOI] [PubMed] [Google Scholar]
- Russ D, Fitzpatrick CR, Teixeira PJPL et al. Deep discovery informs difficult deployment in plant microbiome science. Cell. 2023;186:4496. 10.1016/j.cell.2023.08.035 [DOI] [PubMed] [Google Scholar]
- Santos CL, Correia-Neves M, Moradas-Ferreira P et al. A walk into the LuxR regulators of actinobacteria: phylogenomic distribution and functional diversity. PLoS One. 2012;7:e46758. 10.1371/journal.pone.0046758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarveswari HB, Solomon AP. Profile of the intervention potential of the phylum actinobacteria toward quorum sensing and other microbial virulence strategies. Front Microbiol. 2019;10:2073. 10.3389/fmicb.2019.02073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savijoki K, San-Martin-Galindo P, Pitkänen K et al. Food-grade bacteria combat pathogens by blocking AHL-mediated quorum sensing and biofilm formation. Foods. 2023;12:90. 10.3390/foods12010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawana A, Adeolu M, Gupta RS. Molecular signatures and phylogenomic analysis of the genus Burkholderia: proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harbouring environmental species. Front Genet. 2014;5:429. 10.3389/fgene.2014.00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer AL, Greenberg EP, Oliver CM et al. A new class of homoserine lactone quorum sensing signals. Nature. 2008;454:595–9. 10.1038/nature07088. [DOI] [PubMed] [Google Scholar]
- Schaefer AL, Lappala CR, Morlen RP et al. LuxR- and LuxI-type quorum sensing circuits are prevalent in members of the Populus deltoides microbiome. Appl Environ Microb. 2013;79:5745–52. 10.1128/AEM.01417-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk ST, Schikora A. AHL-priming functions via oxylipin and salicylic acid. Front Plant Sci. 2015;5:e784. 10.3389/fpls.2014.00784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schikora A, Schenk ST, Hartmann A. Beneficial effects of bacteria-plant communication based on quorum sensing molecules of the N-acyl-homoserine lactone group. PLoS One. 2016;6:605–12. 10.1007/s11103-016-0457-8. [DOI] [PubMed] [Google Scholar]
- Schikora A, Schenk ST, Stein E et al. N-acyl-homoserine lactone confers resistance towards biotrophic and hemibiotrophic pathogens via altered activation of AtMPK6. Plant Physiol. 2011a;157:1407–18. 10.1104/pp.111.180604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schikora A, Virlogeux-Payant I, Bueso E et al. Conservation of Salmonella infection mechanisms in plants and animals. PLoS One. 2011b;6:e24112. 10.1371/journal.pone.0024112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz L, Yan Z, Schneijderberg M et al. Synthetic bacterial community derived from a desert rhizosphere confers salt stress resilience to tomato in the presence of a soil microbiome. ISME J. 2022;16:1907–20. 10.1038/s41396-022-01238-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiter S, Ding G-C, Heuer H et al. Effect of the soil type on the microbiome in the rhizosphere of field-grown lettuce. Front Microbiol. 2014;5:144. 10.3389/fmicb.2014.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuhegger R, Ihring A, Gantner S et al. Induction of systemic resistance in tomato plants by N-acylhomoserine lactone-producing rhizosphere bacteria. Plant Cell Environ. 2006;29:909–18. 10.1111/j.1365-3040.2005.01471.x [DOI] [PubMed] [Google Scholar]
- Schulz-Bohm K, Martin-Sánchez L, Garbeva P. Microbial volatiles: small molecules with an important role in intra- and inter-kingdom interactions. Front Microbiol. 2017;8:2484. 10.3389/fmicb.2017.02484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessitsch A, Hardoim P, Döring J et al. Functional characteristics of an endophyte community colonizing rice roots as revealed by metagenomic analysis. Mol Plant Microb Interact. 2012;25:28–36. 10.1094/MPMI-08-11-0204. [DOI] [PubMed] [Google Scholar]
- Shintani M, Nour E, Elsayed T et al. Plant-species-dependent increased abundance and diversity of IncP-1 plasmids in the rhizosphere: new insights into their role and ecology. Front Microbiol. 2020;11:e590776. 10.3389/fmicb.2020.590776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha A, Elhady A, Adss S et al. Genetic differences in barley govern the responsiveness to N-acyl homoserine lactone. Phytobiomes J. 2019;3:191–202. 10.1094/PBIOMES-03-19-0015-R. [DOI] [Google Scholar]
- Shrestha A, Grimm M, Ojiro I et al. Impact of quorum sensing molecules on plant growth and immune system. Front Microb. 2020;11:e1545. https://doi.org/10.3389.fmib.2020.01545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha A, Hernández-Reyes C, Grimm M et al. AHL-priming protein 1 mediates N-3-oxo-tetradecanoyl-homoserine lactone priming in Arabidopsis. BMC Biol. 2022;20:268. 10.1186/s12915-022-01464-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieper T, Forczek S, Matucha M et al. N-acyl-homoserine lactone uptake and systemic transport in barley rest upon active parts of the plant. New Phytol. 2014;201:545–55. 10.1111/nph.12519. [DOI] [PubMed] [Google Scholar]
- Simón-Soro A, Mira A. Solving the etiology of dental caries. Trends Microbiol. 2015;23:76–82. 10.1016/j.tim.2014.10.010. [DOI] [PubMed] [Google Scholar]
- Skindersoe ME, Zenthen LH, Brix S et al. P. aeruginosa quorum sensing signal molecule N-3-oxo-C12-homoserine lactone interferes with denditic cell-induced T-cell proliferation. FEMS Immunol Med Microbiol. 2009;55:335–45. 10.1111/j.1574-695X.2008.00533.x. [DOI] [PubMed] [Google Scholar]
- Soto-Giron MJ, Kim J-N, Schott E et al. The edible plant microbiome represents a diverse genetic reservoir with functional potential in the human host. Sci Rep. 2021;11:e24017. 10.1038/s51598-021-03334-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stencel A, Wloch-Salamon DM Some theoretical insights into the hologenome theory of evolution and the role of microbes in speciation. Theory Biosci. 2018;137:197–206. 10.1007/s12064-018-0268-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub D, Rothballer M, Hartmann A et al. The genome of the endophytic bacterium Herbaspirillum frisingense GSF30T identifies diverse strategies in the Herbaspirillum genus to interact with plants. Front Microbiol. 2013;4:168. 10.3389/fmicb.2013.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Ma N, He T et al. Tryptophan (Trp) modulates gut homeostasis via aryl hydrocarbon receptor (AhR). Crit Rev Food Sci Nutr. 2020;60:1760–8. 10.1080/10408398.2019.1598334. [DOI] [PubMed] [Google Scholar]
- Thompson JA, Oliveira RA, Djukovic A et al. Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep. 2015;10:1861–71. 10.1016/j.celpro.2015.02.049. [DOI] [PubMed] [Google Scholar]
- Uhlig F, Hyland NP. Making sense of quorum sensing at the intestinal mucosal interface. Cells. 2022;11:1734. 10.3390/cells11111734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Broek D, Bloemberg GV, Lugtenberg B. The role of phenotypic variation in rhizosphere Pseudomonas bacteria. Environ Microbiol. 2005;7:1686–97. 10.1111/j.1462-2920.2005.00912.x. [DOI] [PubMed] [Google Scholar]
- Van Elsas JD, Turner S, Bailey MJ. Horizontal gene transfer in the phytosphere. New Phytol. 2003;157:525–37. 10.1046/j.1469-8137.2003.00697.x. [DOI] [PubMed] [Google Scholar]
- Varma A, Bakshi M, Lou B et al. Piriformospora indica: a novel plant growth promoting mycorrizal fungus. Agric Res. 2012;1:117–31. 10.1007/s40003-012-0019-5. [DOI] [Google Scholar]
- Venturi V. Regulation of quorum sensing in Pseudomonas. FEMS Microbiol Rev. 2006;30:274–91. 10.1111/j.1574-6976.2005.00012.x. [DOI] [PubMed] [Google Scholar]
- Vial L, Cuny C, Gluchoff-Fiasson K et al. N-acyl-homoserine lactone-mediated quorum-sensing in Azospirillum: an exception rather than a rule. FEMS Microb Ecol. 2006;58:155–68. 10.1111/j.1574-6941.2006.00153.x. [DOI] [PubMed] [Google Scholar]
- von Bodman SB, Bauer WD, Coplin DL. Quorum sensing in plant-pathogenic bacteria. Annu Rev Phytopathol. 2003;41:455–82. 10.1146/annurev.phyto.41.052002.095652. [DOI] [PubMed] [Google Scholar]
- von Rad U, Klein I, Dobrev PI et al. The response of Arabidopsis thaliana to N-hexanoyl-DL-homoserine lactone, a bacterial quorum sensing molecule produced in the rhizosphere. Planta. 2008;229:73–85. 10.1007/s00425-008-0811-4. [DOI] [PubMed] [Google Scholar]
- Walker A, Pfitzner B, Harir M et al. Sulfonolipids as novel metabolite markers of Alistipes and Odoribacter affeced by high-fat diets. Sci Rep. 2017;7:77047. 10.1038/s41598-017-10369-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker A, Pfitzner B, Neschen S et al. Distinct signatures of host-microbial meta-metabolome and gut microbiome in two C57BL/8 mouse strains under high fat diet. ISME J. 2014;8:2380–96. 10.101038/ismej.2014.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassermann B, Müller H, Berg G. An apple a day; which bacteria do we eat with organic and conventional apples?. Front Microbiol. 2019;10:1629. 10.3389/fmib.2019.01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley M, Diggle SP, Greenberg EP. Progress in and promise of bacterial quorum sensing research. Nature. 2017;551:313–20. 10.1038/nature24624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicaksono WA, Cernava T, Wassermann B et al. The edible plant microbiome: evidence for the occurrence of fruit and vegetable bacteria in the human gut. Gut Microbes. 2023;15:2258565. 10.1080/19490976.2023.2258565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Luo Y. Bacterial quorum-sensing systems and their role in intestinal bacteria—host crosstalk. Front Microbiol. 2021;12:611413. 10.3389/fmicb.2021.611413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Wang X, Hou R et al. Rhizosphere phage communities drive soil suppressiveness to bacterial wilt disease. Microbiome. 2023;11:16. 10.1186/s40168-023-01463-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Wang X, Hou R et al. Rhizosphere phage communities drive soil suppressiveness to bacterial wilt disease. Microbiome. 2023;11:16. 10.1186/s40168-023-01463-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Yuan TJ, Wan Y et al. Quorum sensing: a new perspective to reveal the interaction between gut microbiota and host. Bot Stud. 2020;61:293–309. 10.2217/fmb-2021-0217. [DOI] [PubMed] [Google Scholar]
- Yong Y-C, Zhong J-J. Impacts of quorum sensing on microbial metabolism and human health. Adv Biochem Eng Biotech. 2013;131:25–61. 10.1007/10_2012_138. [DOI] [PubMed] [Google Scholar]
- Zarkani AA, Schierstaedt J, Becker M et al. Salmonella adapts to plants and their environment during colonization of tomatoes. FEMS Microbiol Ecol. 2019;95:fiz152. 10.1093/femsec/fiz152. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Li M, Jia Z et al. AtMYB44 positively regulates the enhanced elongation of primary roots induced by N-oxo-hexanoyl-homoserine lactone in Arabidopsis thaliana. MPMI. 2016;29:774–85. 10.1094/mpmi-03-16-0063-r. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Yang X-Y, Li Y et al. N-3-oxo-hexanoyl-homoserine lactone, a bacterial quorum sensing signal, enhances salt tolerance in Arabidopsis and wheat. Bud Stud. 2020;61:8. 10.1186/s40529-020-00283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Yu D, Chang H et al. Regulation and function of Arabidopsis AtGALK2 gene in absciscic acid response signaling. Mol Biol Rep. 2013;40:6605–12. 10.1007/S11033-013-2773-2. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Zhang C, Jia Z et al. Involvement of calmodulin in regulation of primary root elongation by N-3-oxo-hexanoyl-homoserine lactone in Arabidopsis thaliana. Front Plant Sci. 2015;5:807. 10.3389/fpls.2014.00807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, HE S. Quorum sensing inhibition or quenching in Acinetobacter baumannii: the novel therapeutic strategy for new drug development. Front Microbiol. 2021;12:558003. 10.3389/fmicb.2021.558003. [DOI] [PMC free article] [PubMed] [Google Scholar]