Abstract
Herpesviruses have large double-stranded linear DNA genomes that are formed by site-specific cleavage from complex concatemeric intermediates. In this process, only one of the two genomic ends are formed on the concatemer. Although the mechanism underlying this asymmetry is not known, one explanation is that single genomes are cleaved off of concatemer ends in a preferred direction. This implies that cis elements control the direction of packaging. Two highly conserved cis elements named pac1 and pac2 lie near opposite ends of herpesvirus genomes and are important for cleavage and packaging. By comparison of published reports and by analysis of two additional herpesviruses, we found that pac2 elements lie near the ends formed on replicative concatemers of four herpesviruses: herpes simplex virus type 1, equine herpesvirus 1, guinea pig cytomegalovirus, and murine cytomegalovirus. Formation of pac2 ends on concatemers depended on terminal cis sequences, since ectopic cleavage sites engineered into the murine cytomegalovirus genome mediated formation of pac2 ends on concatemers regardless of the orientation of their insertion. These findings are consistent with a model in which pac2 elements at concatemer ends impart a directionality to concatemer packaging by binding proteins that initiate insertion of concatemer ends into empty capsids.
Herpesviruses have large (130 to 235 kb) linear double-stranded DNA genomes that replicate via concatemeric intermediates consisting of head-to-tail linked genomes (1, 3, 17, 22, 23, 31, 32, 41). The concatemers are packaged into preformed capsids and cleaved at precise locations to release unit length genomes within the capsids (31). In previous studies, we analyzed human cytomegalovirus (HCMV) concatemeric DNA for the presence of termini similar to those found on genomic DNA (23). The HCMV genome contains long and short components, or arms, that consist of unique regions flanked by inverted repeats (30). The ends of the genome are therefore referred to as the long arm end and the short arm end. We observed that HCMV concatemers contain terminal restriction fragments from the short arm end of the genome but lack terminal fragments from the long arm end (23). Similar findings of one but not both genomic termini on concatemeric DNA have since been reported for herpes simplex virus type 1 (HSV-1) (22, 32, 41) and equine herpesvirus 1 (EHV-1) (33), suggesting that this may be a characteristic of all herpesviruses. To explain our observation for HCMV, we proposed a model in which short arm termini on concatemer ends are inserted into empty capsids and packaged until full genome lengths have entered and cleavage sites are encountered. Cleavage then releases unit genomes into the capsids, generating long arm termini on the newly formed genomes and short arm termini on the newly formed concatemer ends. The short arm termini on concatemer ends are then free to encounter additional empty capsids and reinitiate the packaging process (23).
The directionality proposed by this model suggests that cis-acting sequences must exist to control the direction of packaging. Two herpesvirus-conserved cis-acting elements, pac1 and pac2, are necessary for the combined process of cleavage and packaging (5, 8, 10–13, 16, 24, 25, 27, 36, 38, 42); however, their specific roles in these processes are not known. We predicted that if pac1 or pac2 functions to control the direction of concatemer packaging, the ends formed on concatemeric DNA from different herpesviruses should consistently contain either pac1 or pac2. Therefore, we sought to determine, with respect to pac1 and pac2, which ends are found on concatemers from different herpesviruses.
As the previous reports detecting one terminus on HSV-1 and EHV-1 concatemers did not specify the corresponding pac element at these ends (22, 32, 33, 41), we examined earlier reports that characterized the sequences present at the genomic termini of these viruses (8, 11) and found that the ends formed on replicative concatemers of both HSV-1 and EHV-1 contain pac2. Because this result could be purely coincidental, we sought to extend the association by examining two additional herpesviruses: murine cytomegalovirus (MCMV) and guinea pig cytomegalovirus (GPCMV).
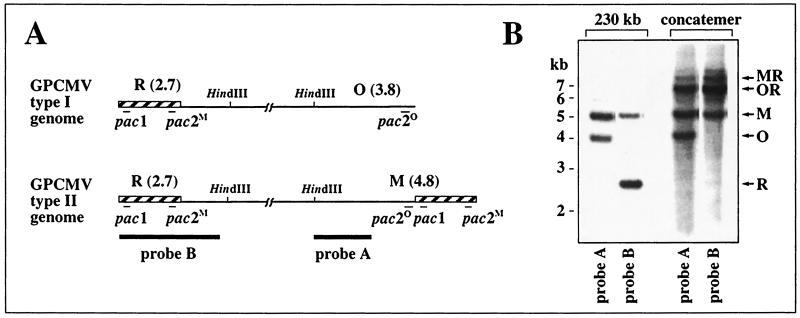
The two genome structures of GPCMV, designated type I and type II, are illustrated in Fig. 1A and differ only in the absence of a 1-kb terminal repeat at the right end of type I genomes (14). Guinea pig embryo fibroblasts (24) were infected with GPCMV strain 22122 (ATCC VR-682) at a multiplicity of infection of 3. After 4 days, infected cells were embedded in agarose blocks and proteinase K treated as described previously (23). The DNAs were then separated on a 1% low-melting-point Seaplaque agarose (FMC Corp.) gel by field-inversion gel electrophoresis (FIGE) as previously described (23). This technique has been shown to retain herpesvirus concatemers in the sample loading blocks while allowing smaller linear forms, such as 230-kb genomes, to migrate into the gel (1, 2, 22–24, 32, 33, 41). After visualizing the DNAs with ethidium bromide and UV light, the sample loading blocks containing concatemeric DNA and sections of the gel containing 230-kb genomic DNA were excised. The blocks were soaked overnight in 10 volumes of 1× HindIII buffer, melted at 68°C, cooled to 37°C, and incubated in a liquid state overnight with 50 U of HindIII. The DNA fragments were then extracted from the agarose by using the Qiaex II kit (Qiagen), digested again with 50 U of HindIII, separated electrophoretically with 0.6% agarose, and transferred to a Nytran nylon membrane (Schleicher and Schuell) as previously described (23). To detect terminal HindIII M and O fragments, the membrane was hybridized with probe A, which consisted of a KpnI-HindIII fragment from the GPCMV HindIII M fragment, cloned in plasmid pGP14 (Fig. 1A) (24). To detect terminal HindIII R fragments, probe A was removed by boiling in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.5% sodium dodecyl sulfate and the membrane was rehybridized with probe B (Fig. 1A), which consisted of GPCMV HindIII R cloned in plasmid pGP48 (24). Because this probe contains the terminal repeat, it also hybridizes with M- but not O-terminal fragments. As expected, the 230-kb intracellular DNA contained terminal fragments M, O, and R (Fig. 1B). Concatemeric DNA contained 6.8-kb OR and 7.8-kb MR junction fragments derived from fusions of genome ends (Fig. 1B) as reported previously (24). Concatemeric DNA also contained significant amounts of M- and O-terminal fragments, but R-terminal fragments were only very faintly detected, even when overexposed relative to probe A (Fig. 1B). As the GPCMV pac1 lies near the R terminus, and the pac2 elements (designated pac2M and pac2O [Fig. 1A]) lie near M and O termini (24), these results indicate that ends bearing pac2 are associated with GPCMV replicative concatemers, whereas ends bearing pac1 are absent.
FIG. 1.
Analysis of GPCMV concatemer termini. (A) The two GPCMV genome structures are illustrated with the terminal repeats (shown as hatched boxes) and the sizes (in kilobases) of terminal HindIII fragments indicated. Thin lines indicate the locations of pac1, pac2M, and the cryptic element pac2O. Thick lines indicate regions used as hybridization probes. (B) Autoradiograph of DNA blot hybridizations to detect GPCMV terminal restriction fragments and concatemer junctions. Concatemeric and 230-kb intracellular DNAs were prepared by FIGE and digested with HindIII, then separated by agarose electrophoresis, transferred to a nylon membrane, and sequentially hybridized with probes A and B.
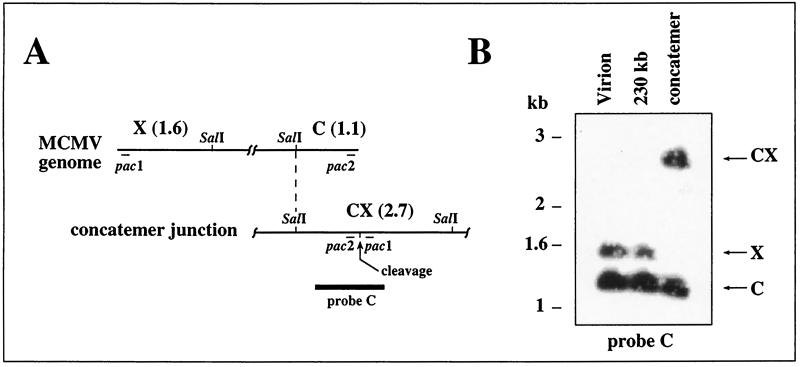
A similar analysis was undertaken for MCMV, whose genome lacks internal repeats and invertible elements but has one copy of a 30-bp repeat at each terminus (21). The ends are named X and C with reference to the EcoRI c and X fragments found at these termini (20) (Fig. 2A). The sequence of the MCMV genome (28) predicts a 1.6-kb SalI fragment from the X terminus and a 1.1-kb SalI fragment from the C terminus, and therefore, a 2.7-kb SalI fragment is predicted from concatemer junctions (Fig. 2A). A β-galactosidase-tagged recombinant MCMV (strain RM461) was used to infect murine NIH 3T3 cells (ATCC CRL1658) at a multiplicity of infection of 3 as previously described (35). DNA was prepared from infected cells 4 days after infection as described above for GPCMV, and in addition, virions were pelleted from culture supernatants by ultracentrifugation, resuspended in agarose, and proteinase K treated as previously described (23). FIGE was then used to isolate concatemeric and 230-kb DNAs from infected cell samples as well as 230-kb DNA from virion samples. FIGE separation, DNA extraction, and restriction enzyme digestion were carried out essentially as described above for GPCMV, except that SalI was used instead of HindIII. The resulting fragments were separated on a 0.6% agarose gel, transferred to a nylon membrane, and hybridized with probe C, which consisted of a 1.9-kb fusion of MCMV terminal sequences cloned in plasmid pON4048 (25) (Fig. 2A). Intracellular 230-kb DNA as well as virion DNA contained the predicted 1.6- and 1.1-kb terminal SalI fragments from each end of the MCMV genome (Fig. 2B). Concatemeric DNA contained a 2.7-kb fragment not found in the genomic DNAs, consistent with a fusion of terminal fragments between adjacent genomes within the concatemer (Fig. 2B). Concatemeric DNA also contained the 1.1-kb C-terminal fragment but completely lacked the 1.6-kb X-terminal fragment (Fig. 2B). As the X terminus contains MCMV pac1 and the C terminus contains MCMV pac2 (21), these data indicate that pac2-containing ends are associated with MCMV replicative concatemers, whereas pac1-containing ends are absent.
FIG. 2.
Analysis of MCMV concatemer termini. (A) The MCMV genome is illustrated, showing terminal SalI C and X fragments. Below it a concatemer junction with the CX junction fragment is illustrated. The sizes of the SalI fragments are indicated (in kilobases). Thin lines indicate the locations of pac1 and pac2, and the thick line indicates the region used as a hybridization probe. (B) Autoradiograph of DNA blot hybridization to detect MCMV terminal restriction fragments and concatemer junctions. Concatemeric, 230-kb intracellular, and 230-kb virion DNAs were prepared by FIGE and digested with SalI, then separated by agarose electrophoresis, transferred to a nylon membrane, and hybridized with probe C.
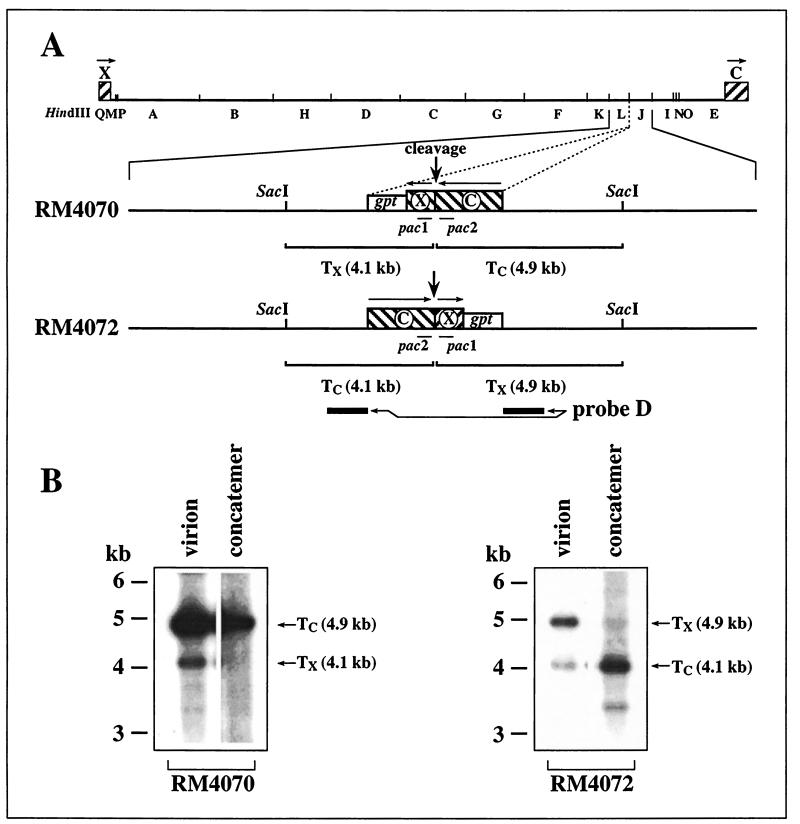
To determine if terminal sequences contain cis elements that define which terminus is formed on the concatemer, we took advantage of two recombinant MCMVs that contain internal cleavage sites engineered in opposite orientations at a location about 40 kb from the right end of the genome (Fig. 3A). In both viruses, 542 bp from the X terminus (nucleotides 1 to 543) were fused with 1,358 bp from the C terminus (nucleotides 228920 to 230278) to create a 1,900-bp cleavage site that was inserted adjacent to an expression cassette for Escherichia coli xanthine-guanine phosphoribosyltransferase (gpt) (used for recombinant virus selection) at nucleotide position 187889 (nucleotide positions are as designated by the published MCMV genomic sequence [28]). Virus RM4072 was constructed by homologous recombination with plasmid pON4072 and selection for gpt expression as previously described (25, 39). Virus RM4070 was constructed from plasmid pON4070 in the same way. Plasmids pON4070 and pON4072 were derived from the same blunt end ligation, as previously described (25), and represent insertion of the gpt/ectopic site-containing fragment into MCMV sequences in opposite orientations. Thus, in virus RM4070, the ectopically inserted terminal sequences form internal inverted repeats of the terminal sequences, whereas in virus RM4072, the ectopic sequences form internal direct repeats (Fig. 3A).
FIG. 3.
Analysis of MCMV ectopic termini. (A) The HindIII restriction map of the entire MCMV genome is illustrated with the C- and X-terminal sequences represented by hatched boxes. Below are expanded the ectopic insertions of RM4070 and RM4072, showing the SacI sites flanking the insertions, the point at which ectopic cleavage occurs (vertical arrow), and the gpt expression cassette (open box). Arrows above the hatched boxes indicate the relative orientations of terminal and ectopic repeated sequences. Shown below each virus insertion are the predicted SacI fragments containing C-terminal (TC) and X-terminal (TX) sequences resulting from cleavage at each ectopic site. Thin lines indicate the locations of pac1 and pac2, and thick lines indicate the regions used as a hybridization probe. (B) Autoradiographs of DNA blot hybridizations to detect ectopic terminal restriction fragments. Concatemeric DNAs and 230-kb virion DNAs from RM4070 and RM4072 were prepared by FIGE and digested with SacI, then separated by agarose electrophoresis, transferred to a nylon membrane, and hybridized with probe D. The positions and molecular sizes of ectopic terminal fragments TC and TX are indicated.
In both RM4070 (unpublished data) and RM4072 (25), the ectopic cleavage sites are cleaved efficiently; therefore, we sought to determine which ectopic termini are formed on concatemeric DNA from these viruses and specifically to determine whether sequences within the ectopic insertions or elsewhere in the genome determine which ends are formed on concatemers. Concatemeric DNA and virion-derived 230-kb DNA were isolated from RM4070- and RM4072-infected cell cultures by FIGE as before, but in this case the DNAs were digested with SacI. The resulting fragments were separated on a 0.6% agarose gel, transferred to a nylon membrane, and hybridized with probe D, which consists of a 1.3-kb BamHI-MluI fragment gel purified from plasmid pON432 (35) and contains sequences from each side of the ectopic insertions but does not contain terminal sequences (Fig. 3A). Virion DNA from both viruses contained the predicted ectopic terminal fragments (Fig. 3B). RM4070 concatemeric DNA contained the 4.9-kb C-terminal SacI fragment but lacked the 4.1-kb X-terminal fragment; similarly, RM4072 concatemeric DNA contained the 4.1-kb C-terminal fragment but lacked the 4.8-kb X-terminal fragment (Fig. 3B). Thus, regardless of the orientation of the insertion relative to sequences elsewhere in the viral genome, cleavage at ectopic sites resulted in pac2-containing C termini on concatemeric DNA, whereas pac1-containing X termini were absent. Therefore, the 1.9-kb fusion of terminal sequences that was inserted at the ectopic sites must contain cis sequences that determine that pac2 ends are formed on concatemers.
Although seemingly obscure, the observation that herpesvirus concatemers contain one but not both genomic termini has far-reaching implications. First, it appears to eliminate from consideration any cleavage/packaging model in which internal cleavage removes genomes from the interior of the concatemer, since this would generate both genomic ends on two smaller but still concatemeric molecules. Second, the observation implies a directionality defined locally by cis elements. This is further supported by evidence from EHV-1 that the cleavage and packaging process is directional with respect to the long unique region of the genome (33). The mechanism used by the large double-stranded DNA bacteriophages satisfies these criteria, since single genomes are cleaved only from concatemer ends, leaving one of the two resulting termini on the concatemer and the other on the newly formed unit genome (4). Directionality is imparted in bacteriophage λ by a cis element on one side of the cleavage site that remains bound to the large subunit of λ terminase after the site is cleaved. The terminase subunit then serves to initiate the next round of packaging by docking the concatemer end to the portal vertex of a phage prohead (6). This docking aspect is recapitulated during packaging of adenovirus DNA, in which a cis sequence element located near one end of the genome initiates packaging of unit genomes into preformed adenovirus capsids (9, 15, 29, 37).
From our results, two conclusions regarding herpesvirus cleavage and packaging can be made. First, there are cis-acting elements located at herpesvirus cleavage sites (and consequently, near genomic termini) that control which ends are formed on replicative concatemers. For MCMV, these elements must lie within the 542 bp of X-terminal or 1,358 bp of C-terminal sequences recapitulated at the ectopic cleavage sites. Second, control is somehow linked to the positions of pac1 and/or pac2. This conclusion is based on the cumulative observations that concatemers from five different herpesviruses (HCMV [23], HSV-1 [11, 22, 32, 41], EHV-1 [8, 33], GPCMV [this study], and MCMV [this study]) lack detectable pac1 ends, yet, with the exception of HCMV (for which pac2 remains undefined), these concatemers contain readily detectable pac2 ends. Within the context of a phage-like mechanism, an attractive model would posit that pac2 on concatemer ends controls the initiation and hence direction of packaging in a λ-like manner by docking concatemer ends with empty capsids.
While well precedented and consistent with the available data, this model should be considered preliminary. Terminal sequences other than pac1 or pac2 could be responsible, and alternative mechanisms are certainly possible. Even within the context of this model, the process of cleavage and packaging is almost certainly more complex. As in bacteriophages (4, 6), because nascent concatemers are probably not formed with authentic genomic termini, it seems likely that a packaging-independent mechanism exists to occasionally cleave concatemers internally in order to create initial ends that can subsequently serve to initiate processive packaging and cleavage. Furthermore, herpesvirus concatemers are almost certainly not simple linear molecules, but are believed to be highly branched as a consequence of frequent recombinations within or between concatemers (1, 2, 22, 23, 32, 34, 41). Resolution of these branches by a viral-encoded alkaline nuclease appears to be important for retention of cleaved DNA within capsids (22). Evidence for frequent inversions of the EHV-1 long unique region within concatemers adds a new dimension of complexity by generating concatemers in which the cleavage frame is disrupted when pac1/pac2 terminal junctions are replaced by pac1/long unique sequence junctions (33). Although these sites lack pac2 elements, they appear to be cleaved, since novel ends terminating with long unique sequences are common on concatemers (33); however, this does not necessarily indicate that cleavage at these sites is efficient, as the ends on concatemers could represent accumulation of rare cleavage events over the course of infection. In most instances, the cleavage/packaging machinery may stall at these sites until an additional inversion event restores the pac2 end of the long unique region to the site. In the context of the model proposed here, concatemer ends lacking pac2 should be unable to serve as substrates for packaging because they lack pac2 elements; indeed, despite their frequent presence on EHV-1 concatemers, ends lacking pac2 are not incorporated on extracellular packaged genomes (33).
Finally, it is a formal possibility that concatemer end formation may not accurately reflect the mechanism of herpesvirus cleavage and packaging. For example, concatemer ends could represent an accumulation of dead-end by-products of the cleavage/packaging process. This latter concern seems improbable, given that formation of HSV-1 concatemer ends occurs early in the replication cycle concomitant with the first indications of genome cleavage (41) and is dependent on the expression of viral genes required for genome cleavage and maturation (19, 40). Even so, direct physical evidence, such as nuclease protection studies to demonstrate that herpesvirus DNA is packaged into capsids in a particular direction or experiments to identify protein-DNA interactions between pac2 and the herpesvirus packaging machinery, is needed to corroborate the directional packaging model.
For HCMV, pac2 elements have not been clearly defined. The inverted repeats of the HCMV short arm are called c sequences and a terminal repeat, or a sequence, is present in one or more copies at the long arm terminus and as a single copy adjacent to the c sequence at the short arm terminus (analogous to GPCMV type II genomes); however, a significant population of short arm termini lack this a sequence and therefore end with a c sequence (36) (analogous to GPCMV type I genomes). A pac1 element is located near one end of the a sequence such that it lies near the long arm end of the genome (26, 36), which is absent from HCMV concatemers (23). A sequence proposed as the HCMV pac2 sequence by Kemble and Mocarski has pac2 sequence characteristics but is located at an unusual position internal to pac1 within the a sequence (18). No sequences with clear pac2-like characteristics are found near the ends found on HCMV concatemers, i.e., near the other end of the a sequence from pac1 or near the terminal end of the c sequence (5, 7, 18, 23, 26). Thus, it is not possible to state that HCMV concatemer ends contain pac2 elements but only that pac1-containing ends are absent.
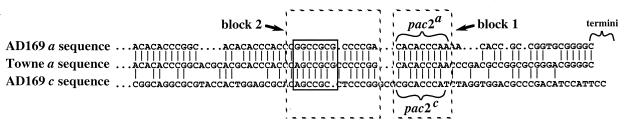
One scenario that reconciles these observations is that HCMV concatemer ends may contain cryptic pac2 elements that function as pac2 elements but lack recognizable pac2 sequence characteristics. Evidence for cryptic pac2 elements in GPCMV was first suggested by alignment of M- and O-terminal sequences, which revealed a conserved region that in M comprised a typical pac2 element (pac2M [Fig. 1A]) but in O was not recognizable as a pac2 element (pac2O [Fig. 1A]) (24). Mutagenic analysis provided clear evidence that pac2O or neighboring sequences function as cryptic cis cleavage signals (24). Since the short arm termini of HCMV similarly exist as ends having or lacking an a sequence, we aligned the short arm terminal regions of a sequences from HCMV strains Towne (26) and AD169 (7) with the terminal region of the AD169 c sequence (7) and identified two blocks of conservation between all three sequences (Fig. 4). The first block, containing sequences provisionally designated pac2a (within the a sequence) and pac2c (within the c sequence), lies a typical distance from the point of cleavage for herpesvirus pac2 elements (Fig. 4). The second block lies distal to the first (relative to the termini) and contains sequences similar to CGCGGCG motifs (Fig. 4), which in some herpesviruses are found distal to pac2 elements (10, 12, 21, 24, 42) and in MCMV play a significant role in cleavage and packaging (25).
FIG. 4.
Alignment of HCMV short arm terminal sequences. HCMV strain AD169 short arm terminal sequences ending in a c sequence are aligned with strain AD169 and Towne short arm terminal sequences ending in an a sequence (7, 26). Vertical lines indicate identical bases, dots indicate gaps added to improve alignments, and brackets indicate the putative cryptic elements pac2a and pac2c. Dashed boxes indicate the two blocks of conservation, and the solid box indicates sequences similar to CGCGGCG motifs (on the complementary strand).
The significance of these conserved regions remains to be determined; however, both are found near HCMV concatemer ends (23) and their locations relative to pac1 are fully consistent with pac1/pac2 arrangements in other herpesviruses. In particular, the arrangement of pac1/pac2a/pac2c in HCMV is highly analogous to the arrangement of pac1/pac2M/pac2O in GPCMV. In the absence of data that functionally define the cis cleavage/packaging elements of HCMV, these putative cryptic pac2 elements are tenable alternatives to the pac2 sequence proposed by Kemble and Mocarski (18) that should be considered when designing experiments to assess in vitro cleavage or DNA binding activities of putative HCMV terminase proteins.
Acknowledgments
This work was supported by Public Health Services grants R21AI43527 (to M.A.M.) and KO8AI01435 (to D.E.N.) from the National Institutes of Health and in part by funds from the A. D. Williams Fund of the Medical College of Virginia, Virginia Commonwealth University, and by grant IN-105V from the American Cancer Society. J.K.H. was supported by a grant from Catholic University, Seoul, Korea.
REFERENCES
- 1.Bataille D, Epstein A. Herpes simplex virus replicative concatemers contain L components in inverted orientation. Virology. 1994;203:384–388. doi: 10.1006/viro.1994.1498. [DOI] [PubMed] [Google Scholar]
- 2.Bataille D, Epstein A L. Equimolar generation of the four possible arrangements of adjacent L components in herpes simplex virus type 1 replicative intermediates. J Virol. 1997;71:7736–7743. doi: 10.1128/jvi.71.10.7736-7743.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ben-Porat T. Replication of herpesvirus DNA. In: Roizman B, editor. The herpesviruses. New York, N.Y: Plenum Press; 1983. pp. 81–86. [Google Scholar]
- 4.Black L W. DNA packaging in dsDNA bacteriophages. Annu Rev Microbiol. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- 5.Broll H, Buhk H J, Zimmermann W, Goltz M. Structure and function of the prDNA and the genomic termini of the gamma2-herpesvirus bovine herpesvirus type 4. J Gen Virol. 1999;80:979–986. doi: 10.1099/0022-1317-80-4-979. [DOI] [PubMed] [Google Scholar]
- 6.Catalano C E, Cue D, Feiss M. Virus DNA packaging: the strategy used by phage lambda. Mol Microbiol. 1995;16:1075–1086. doi: 10.1111/j.1365-2958.1995.tb02333.x. [DOI] [PubMed] [Google Scholar]
- 7.Chee M S, Bankier A T, Beck S, Sohni R, Brown C M, Cerny R, Horsnell T, Hutchinson C A, Kouzarides T, Marignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K, Barrell B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. In: McDougall J K, editor. Cytomegaloviruses. Vol. 154. New York, N.Y: Springer-Verlag; 1990. pp. 125–169. [DOI] [PubMed] [Google Scholar]
- 8.Chowdhury S I, Buhk H J, Ludwig H, Hammerschmidt W. Genomic termini of equine herpesvirus 1. J Virol. 1990;64:873–880. doi: 10.1128/jvi.64.2.873-880.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daniell E. Genome structure of incomplete particles of adenovirus. J Virol. 1976;19:685–708. doi: 10.1128/jvi.19.2.685-708.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davison A J. Structure of the genome termini of varicella-zoster virus. J Gen Virol. 1984;65:1969–1977. doi: 10.1099/0022-1317-65-11-1969. [DOI] [PubMed] [Google Scholar]
- 11.Deiss L P, Chou J, Frenkel N. Functional domains within the a sequence involved in the cleavage-packaging of herpes simplex virus DNA. J Virol. 1986;59:605–618. doi: 10.1128/jvi.59.3.605-618.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deiss L P, Frenkel N. Herpes simplex virus amplicon: cleavage of concatemeric DNA is linked to packaging and involves amplification of the terminally reiterated a sequence. J Virol. 1986;57:933–941. doi: 10.1128/jvi.57.3.933-941.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng H, Dewhurst S. Functional identification and analysis of cis-acting sequences which mediate genome cleavage and packaging in human herpesvirus 6. J Virol. 1998;72:320–329. doi: 10.1128/jvi.72.1.320-329.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao M, Isom H C. Characterization of the guinea pig cytomegalovirus genome by molecular cloning and physical mapping. J Virol. 1984;52:436–447. doi: 10.1128/jvi.52.2.436-447.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammarskjold M L, Winberg G. Encapsidation of adenovirus 16 DNA is directed by a small DNA sequence at the left end of the genome. Cell. 1980;20:787–795. doi: 10.1016/0092-8674(80)90325-6. [DOI] [PubMed] [Google Scholar]
- 16.Hammerschmidt W, Ludwig H, Buhk H J. Specificity of cleavage in replicative-form DNA of bovine herpesvirus 1. J Virol. 1988;62:1355–1363. doi: 10.1128/jvi.62.4.1355-1363.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacob R J, Morse L S, Roizman B. Anatomy of herpes simplex virus DNA. XII. Accumulation of head-to-tail concatemers in nuclei of infected cells and their role in the generation of the four isomeric arrangements of viral DNA. J Virol. 1979;29:448–457. doi: 10.1128/jvi.29.2.448-457.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kemble G W, Mocarski E S. A host cell protein binds to a highly conserved sequence element (pac-2) within the cytomegalovirus a sequence. J Virol. 1989;63:4715–4728. doi: 10.1128/jvi.63.11.4715-4728.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamberti C, Weller S K. The herpes simplex virus type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology. 1996;226:403–407. doi: 10.1006/viro.1996.0668. [DOI] [PubMed] [Google Scholar]
- 20.Marks J R, Spector D H. Fusion of the termini of the murine cytomegalovirus genome after infection. J Virol. 1984;52:24–28. doi: 10.1128/jvi.52.1.24-28.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marks J R, Spector D H. Replication of the murine cytomegalovirus genome: structure and role of the termini in the generation and cleavage of concatenates. Virology. 1988;162:98–107. doi: 10.1016/0042-6822(88)90398-4. [DOI] [PubMed] [Google Scholar]
- 22.Martinez R, Sarisky R T, Weber P C, Weller S K. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J Virol. 1996;70:2075–2085. doi: 10.1128/jvi.70.4.2075-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McVoy M A, Adler S P. Human cytomegalovirus DNA replicates after early circularization by concatemer formation, and inversion occurs within the concatemer. J Virol. 1994;68:1040–1051. doi: 10.1128/jvi.68.2.1040-1051.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McVoy M A, Nixon D E, Adler S P. Circularization and cleavage of guinea pig cytomegalovirus genomes. J Virol. 1997;71:4209–4217. doi: 10.1128/jvi.71.6.4209-4217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McVoy M A, Nixon D E, Adler S P, Mocarski E S. Sequences within the herpesvirus-conserved pac1 and pac2 motifs are required for cleavage and packaging of the murine cytomegalovirus genome. J Virol. 1998;72:48–56. doi: 10.1128/jvi.72.1.48-56.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mocarski E S, Liu A C, Spaete R R. Structure and variability of the a sequence in the genome of human cytomegalovirus (Towne strain) J Gen Virol. 1987;68:2223–2230. doi: 10.1099/0022-1317-68-8-2223. [DOI] [PubMed] [Google Scholar]
- 27.Nasseri M, Mocarski E S. The cleavage recognition signal is contained within sequences surrounding an a-a junction in herpes simplex virus DNA. Virology. 1988;167:25–30. doi: 10.1016/0042-6822(88)90050-5. [DOI] [PubMed] [Google Scholar]
- 28.Rawlinson W D, Farrell H E, Barrell B G. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol. 1996;70:8833–8849. doi: 10.1128/jvi.70.12.8833-8849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson C C, Tibbetts C. Polar encapsidation of adenovirus DNA: evolutionary variants reveal dispensable sequences near the left ends of Ad3 genomes. Virology. 1984;137:276–286. doi: 10.1016/0042-6822(84)90219-8. [DOI] [PubMed] [Google Scholar]
- 30.Roizman B. The Herpesviridae, a brief introduction. In: Roizman B, Whitley R J, Lopez C, editors. The human herpesviruses. New York, N.Y: Raven Press; 1993. pp. 1–9. [Google Scholar]
- 31.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, editors. Fundamental virology. New York, N.Y: Raven Press; 1996. pp. 1048–1066. [Google Scholar]
- 32.Severini A, Morgan A R, Tovell D R, Tyrrell D L. Study of the structure of replicative intermediates of HSV-1 DNA by pulsed-field gel electrophoresis. Virology. 1994;200:428–435. doi: 10.1006/viro.1994.1206. [DOI] [PubMed] [Google Scholar]
- 33.Slobedman B, Simmons A. Concatemeric intermediates of equine herpesvirus type 1 DNA replication contain frequent inversions of adjacent long segments of the viral genome. Virology. 1997;229:415–420. doi: 10.1006/viro.1997.8447. [DOI] [PubMed] [Google Scholar]
- 34.Slobedman B, Zhang X, Simmons A. Herpes simplex virus genome isomerization: origins of adjacent long segments in concatemeric viral DNA. J Virol. 1999;73:810–813. doi: 10.1128/jvi.73.1.810-813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoddart C A, Cardin R D, Boname J M, Manning W C, Abenes G B, Mocarski E S. Peripheral blood mononuclear phagocytes mediate dissemination of murine cytomegalovirus. J Virol. 1994;68:6243–6253. doi: 10.1128/jvi.68.10.6243-6253.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamashiro J C, Filpula D, Friedmann T, Spector D H. Structure of the heterogeneous L-S junction region of human cytomegalovirus strain AD169 DNA. J Virol. 1984;52:541–548. doi: 10.1128/jvi.52.2.541-548.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tibbetts C. Viral DNA sequences from incomplete particles of human adenovirus type 7. Cell. 1977;12:243–249. doi: 10.1016/0092-8674(77)90202-1. [DOI] [PubMed] [Google Scholar]
- 38.Varmuza S L, Smiley J R. Signals for site-specific cleavage of HSV DNA: maturation involves two separate cleavage events at sites distal to the recognition sequences. Cell. 1985;41:793–802. doi: 10.1016/s0092-8674(85)80060-x. [DOI] [PubMed] [Google Scholar]
- 39.Vieira J, Farrell H E, Rawlinson W D, Mocarski E S. Genes in the HindIII J fragment of the murine cytomegalovirus genome are dispensable for growth in cultured cells: insertion mutagenesis with a lacZ/gpt cassette. J Virol. 1994;68:4837–4846. doi: 10.1128/jvi.68.8.4837-4846.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu D, Sheaffer A K, Tenney D J, Weller S K. Characterization of ICP6::lacZ insertion mutants of the UL15 gene of herpes simplex virus type 1 reveals the translation of two proteins. J Virol. 1997;71:2656–2665. doi: 10.1128/jvi.71.4.2656-2665.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X, Efstathiou S, Simmons A. Identification of novel herpes simplex virus replicative intermediates by field inversion gel electrophoresis: implications for viral DNA amplification strategies. Virology. 1994;202:530–539. doi: 10.1006/viro.1994.1375. [DOI] [PubMed] [Google Scholar]
- 42.Zimmermann J, Hammerschmidt W. Structure and role of the terminal repeats of Epstein-Barr virus in processing and packaging of virion DNA. J Virol. 1995;69:3147–3155. doi: 10.1128/jvi.69.5.3147-3155.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]