Abstract
Objective: To determine if host genetics may be a risk factor for severe blastomycosis.
Design: A cohort of patients who had contracted blastomycosis underwent targeted SNP (single nucleotide polymorphism) genotyping. The genetics of these patients were compared to a set of age and gender-matched controls and between patients with severe versus mild to moderate blastomycosis.
Setting: The Marshfield Clinic Health System in central and northern Wisconsin
Participants: Patients with a diagnosis of blastomycosis prior to 2017 were contacted for enrollment in this study. A phone hotline was also set up to allow interested participants from outside the Marshfield Clinic Health System to request enrollment.
Methods: SNP frequency was assessed for significant differences between the patient cohort and controls and between patients with severe versus mild to moderate blastomycosis. We also tested the effect of Blastomyces species identified in clinical isolates on disease symptoms and severity.
Results: No significant differences were found in SNP frequency between cases and controls or between those with severe or mild to moderate blastomycosis. We did detect significant differences in symptom frequency and disease severity by Blastomyces species.
Conclusions: Our study did not identify any genetic risk factors for blastomycosis. Instead, the species of Blastomyces causing the infection had a significant effect on disease severity.
Keywords: Blastomycosis, Fungal infection, Host genetics
Dimorphic fungi are a major cause of severe disease, estimated to cause over 300 million severe infections per year globally.1 One of these diseases, blastomycosis, is endemic to the Mississippi, Ohio, and St. Lawrence River valleys, as well as the Great Lakes region, in the United States. Blastomycosis is most often caused by two species, Blastomyces dermatitidis and Blastomyces gilchristii, which have differing clinical presentations.2,3 B. gilchristii typically infects younger people and remains a pulmonary infection, while B. dermatitidis is more likely to cause disease in people with co-morbidities and to cause a disseminated infection.3 Two additional species, Blastomyces helicus and Blastomyces percusus, have also been identified as less common disease-causing agents, while Blastomyces emzantsi was recently described in South Africa.4–6 For all species, infection occurs through the inhalation of spores found in moist, organic-rich soils.7,8 Exposure often occurs along waterways and is linked to activities that disturb the soil, such as gardening. Once exposed, there is an incubation period of 3-15 weeks before symptoms develop.9 An unknown percentage of people exposed never develop symptoms, revealed through incidental findings of Blastomyces and positive histoplasmin skin tests following a point-source outbreak in individuals not diagnosed with blastomycosis.10,11 However, many patients with blastomycosis require hospitalization and intensive care, with 4.3-6.3% of cases resulting in death.9
Related dimorphic fungi, such as Coccidiomyces, are known to disproportionately affect specific populations.12,13 There is also evidence to suggest that the genetics of the person infected may play a role in Blastomyces infection and that genetic variants associated with higher risk of infection with dimorphic fungi are more likely to be found in certain populations. In 2009-2010, an outbreak of blastomycosis occurred in Wisconsin that disproportionately affected persons of Hmong ethnicity.14 Genome-wide sequencing of these patients revealed variants potentially involved with immune response, specifically the gene encoding interleukin-6 (IL-6), which regulates interleukin-17 response.15 People with these genetic variants may be more susceptible to infection with Blastomyces, more likely to develop symptoms, or progress to severe, life-threatening disease.
The reasons underlying the wide range of clinical outcomes from blastomycosis are largely unknown. We hypothesized that host genetics play a significant role in disease severity during infection with Blastomyces. Therefore, we aimed to test for genetic variants linked to more severe disease using a cohort of genotyped patients with blastomycosis from the Marshfield Clinic Health System.
Methods
Setting and Participants
Blastomycosis is highly endemic to Wisconsin. While globally a rare infection, blastomycosis has a particularly high incidence (10-40 per 100,000 population) in northern Wisconsin.7,16 Many of these patients seek treatment through the Marshfield Clinic Health System, which spans central and northern Wisconsin.
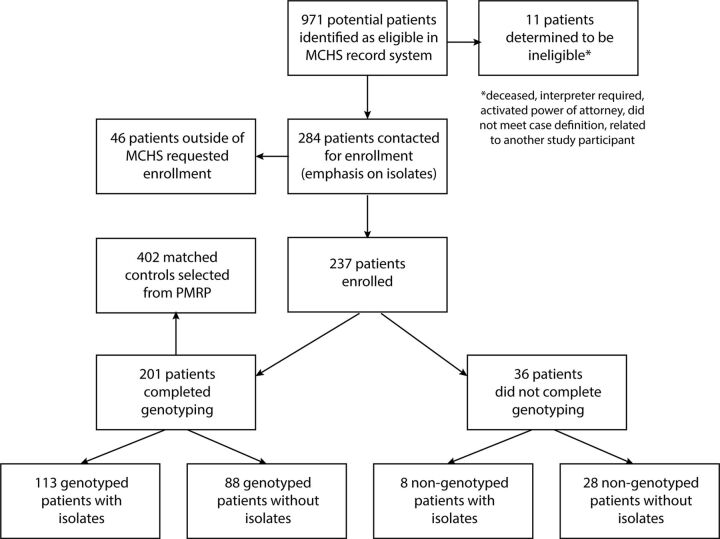
Patients with a diagnosis of blastomycosis were identified in clinical records of the Marshfield Clinic Health System under a prior Institutional Review Board and considered eligible for this study (Figure 1). Many of these patients had previously participated in clinical research, and chart reviews had already been completed. We identified 971 eligible patients and had another 46 patients from other health systems call in to a hotline to request enrollment. Of this group, 284 eligible participants were contacted via letter to offer enrollment in this study, with a focus on those with previously bio-banked isolates from their Blastomyces infection. Participants were asked to consent to a medical records request and to provide a blood sample for genotype analysis. If no response was received to the letter, patients were then contacted via phone or email. Patients who were deceased, required an interpreter, had an activated power of attorney, were related to another study participant, or had incomplete contact information were not contacted.
Figure 1.
Flowchart of recruitment, enrollment, and study completion. Eligible patients were identified based on chart review in the Marshfield Clinic Health System, in addition to setting up a phone hotline for interested participants from other health systems. After exclusions and searching specifically for patients with clinical isolates available, 284 patients were contacted for enrollment. Of those contacted, 83% enrolled in our cohort. Within the cohort, 85% consented to SNP genotyping, and 51% had genotyped clinical isolates of Blastomyces.
We enrolled 237 patients, a response rate of 83% (Figure 1). Data were collected via chart abstraction on patient demographics and co-morbidities, clinical disease characteristics, and treatments. Clinical isolates of Blastomyces were available for 120 patients in our study. Of this cohort, 201 patients were genotyped at 550,601 key SNP locations.
Age-matched controls, for whom the same genotype data were available, were selected from the Personalized Medicine Research Project (PMRP) biobank, which had enrolled Marshfield Clinic Health System adult patients as participants.17 Two controls were matched to each patient with exclusion criteria of any previous diagnosis of blastomycosis. Since the PMRP database does not contain pediatric patients, cases under the age of 18 were matched to 18 year old controls.
Ethics Statement
This study and its protocol were approved by the institutional review board at the Marshfield Clinic Health System. Written informed consent was obtained from each enrolled or parent of each enrolled child. The authors attest that the study was performed in accordance with the protocol.
Outcome Measures and Exposure Variables
We assessed genotypic variation between patients diagnosed with blastomycosis and those without this diagnosis and patients with mild to moderate versus severe blastomycosis. For this study, a severe case of blastomycosis was defined as a case requiring hospitalization. Mild to moderate disease was defined as managed in an outpatient setting.
We investigated two potential exposure variables in this study. The first was host genotype, i.e., the genetics of the person with blastomycosis. We tested associations between genotypes and a blastomycosis diagnosis and between mild to moderate and severe blastomycosis. The second variable was the species of Blastomyces identified in the clinical isolate. We tested the association between species and disease severity for this exposure variable.
Genotyping Methods
For human genotyping, patients in this study consented to either a blood draw or a buccal swab. For blood draws, 6 mL of blood was collected. DNA was extracted using the Puregene EP DNA Purification Kit and quantitated using a Qubit Fluorometer (ThermoFisher Scientific, Grand Island, NY). DNA samples were sent to the Center for Inherited Disease Research (CIDR) at Johns Hopkins University for genotyping using the HumanCore BeadChip (Illumina), targeted towards a set of 550,601 SNPs known to have variation. Data are available on dbGaP under study number “phs003426”.
Clinical isolates of Blastomyces from patients in this cohort were obtained from Marshfield Labs as part of a previously established biorepository and stored at −20C until extraction. Isolates were extracted and genotyped as described in Meece, et al., 2013.3 Briefly, DNA was extracted using the QIAamp DNA mini kit and tissue protocol (QIAGEN) with minor modifications.18 Isolates were genotyped using PCR.19 This method allows us to distinguish B. dermatitidis from B. gilchristii.
Data Analysis and Significance
Data analysis was performed in R (version 4.1.0) using RStudio and the packages “tidyverse” (version 1.3.1) and “glmnet” (version 4.1.4) (Friedman et al., 2010).20–23 To determine whether case characteristics differed according to pathogen species, patient demographics and symptoms were first compared between B. gilchristii and B. dermatitidis using Pearson’s chi-squared test for count data and the Wilcoxon sum rank test for continuous data. Confidence intervals for count data were calculated using equality of proportions hypothesis testing. Variables found to be significant at P < 0.05 in these univariate statistics were next assessed via relaxed fit, cross-validated lasso regression in glmnet to determine their predictive value. This included age at symptom onset, presence of a fever, presence of skin lesions, hospitalization, and observation of consolidation, infiltrate, or opacity via x-ray of the chest.
Genotype data was compared between groups using PLINK (version 1.07).24 This toolset performs genome-wide association analysis to determine if any SNPs are significantly outside of their expected equilibrium values. We compared genotypes between our blastomycosis cases and our age-matched controls, as well as between those in our cohort with severe blastomycosis (requiring hospitalization) to those with mild to moderate blastomycosis using case/control association analysis. Of the 550,601 SNPs sequenced, there were 220,187 in our dataset that had no variation, so 330,414 were retained for analysis. Due to the large number of SNPs tested, we implemented a Bonferonni correction on the resulting P values.
Because the large number of SNPs sequenced reduced our ability to detect significance, we next targeted our dataset to only SNPs in genes previously linked to increased risk or severity of blastomycosis, histoplasmosis, and coccidioidomycosis. These genes included IFN(g)R1, IL12R(b)1, STAT3, STAT1, GATA2, MyD88, IL-1R1, and IL-6.25 Of our sequenced SNPs, 61 were within these genes, and a Bonferroni correction was still applied.
Results
Patient Demographics
Cases in this dataset were predominantly male [138 (58%)], consistent with previous research suggesting that males are more likely to be exposed to Blastomyces (Table 1).26,27 The majority of patients were white [216 (91%)], reflecting the demographics of central and northern Wisconsin. The mean age of patients at the age of symptom onset was 44-years-old, with a median of 46-years and a standard deviation 17. Many patients in this dataset had co-morbidities, with 89 (38%) reporting a cardiac condition, 27 (11%) reporting a pulmonary condition, and 11 (5%) reporting a neurological condition. Of the patients, 35 (15%) had diabetes or hypoglycemia, and 38 (16%) were immunocompromised. In this cohort, 118 (50%) patients had never smoked, 65 (27%) had smoked in the past, and 50 (21%) were current smokers.
Table 1.
Differences in patient demographics, underlying medical conditions, and clinical presentation associated with Blastomyces species. we identified the species of Blastomyces as a significant variable explaining differences in clinical progression between patients diagnosed with blastomycosis. Demographic variables and co-morbidities were not associated with Blastomyces species, but several symptoms were significantly associated with species.
| Variable category | Variable tested | B. gilchristii (80 patients) | B. dermatitidis (40 patients) | P value | 95% Confidence Interval |
|---|---|---|---|---|---|
| Demographics | |||||
| Gender | 59% male | 63% male | 0.84 | −24 – 17% | |
| Smoking (past or current) | 38% | 58% | 0.35 | −11 – 34% | |
| Underlying Conditions | |||||
| Cardiac | 29% | 45% | 0.12 | −36 – 4% | |
| Pulmonary | 9% | 18% | 0.27 | −24 – 6% | |
| Neurological | 4% | 10% | 0.34 | 4 – 10% | |
| Diabetes | 11% | 13% | 1.00 | −15 – 12% | |
| Immunocompromised | 14% | 18% | 0.79 | −20 – 12% | |
| Obesity | 51% | 38% | 0.36 | −7 – 35% | |
| Symptoms | |||||
| Fever | 79% | 35% | < 0.01 | −63 – −25% | |
| Fatigue | 60% | 33% | 0.01 | −47 – −8% | |
| Cough | 94% | 78% | 0.02 | −32 – −0.4% | |
| Chest pain | 54% | 33% | 0.05 | −41 – −1% | |
| Shortness of breath | 65% | 21% | 0.03 | −43 – −2% | |
| Skin lesion | 6% | 35% | < 0.01 | 12 – 46% | |
An age- and gender-matched control group of patients with no prior diagnosis of blastomycosis was selected from the PMRP database, which contains 18,239 patients enrolled in 2002–2005 in Marshfield, Wisconsin and its surrounding zip codes.17 In the full PMRP cohort, 41% of patients were diagnosed with a cardiovascular condition, 46% had asthma or chronic obstructive pulmonary disease, and 10% had diabetes.
Association of Patient Genetics with Blastomycosis
We genotyped SNPs for patients in this cohort to determine if there was a genetic association with more or less severe disease. We defined severe disease as requiring hospitalization. After controlling for Blastomyces genotype and correcting for multiple testing, no SNPs were significantly associated with either contracting blastomycosis as compared to our control group or disease severity within our case group. Because the large number of SNPs in our dataset may reduce our ability to detect significant differences, we repeated these analyses on SNPs found in genes previously associated with fungal disease.28 No significant differences were found in this reduced dataset in either the cases versus control or severe versus mild to moderate disease groups.
Clinical Presentation
The most common symptom of blastomycosis was a cough, with 206 (87%) patients reporting this symptom. Roughly half of patients experienced the following symptoms: fever [152 (64%)], fatigue [137 (56%)], and chills [103 (43%)]. Skin lesions were observed in 35 (15%) patients. A computed tomography (CT) scan and radiograph of the chest was obtained for all patients in this cohort. Via CT scan, consolidation or opacity of the chest was seen in 117 (50%) patients, and 70 (30%) had nodules, 51 (22%) had masses, and 53 (22%) had adenopathy. Results differed when obtained via radiograph, with 199 (84%) patients presenting with consolidation or opacity, 45 (19%) with nodules, 29 (12%) with masses, and 9 (4%) with adenopathy. Pulmonary only infections were present in 188 (79%) patients, while 46 (19%) infections were disseminated to other locations. In this cohort, 135 (57%) patients in this cohort were hospitalized. The mean length of hospital stay was 12 days, with a median of 9 and a standard deviation of 10. In this study, 37 (16%) patients received intensive care, and 19 (8%) patients were intubated.
Blastomyces Genotypes
Clinical isolates were obtained from 120 patients in this cohort and genotyped. Of these samples, 40 (33%) were identified as B. dermatitidis, and 80 (66%) were identified as B. gilchristii. B. dermatitidis was much more likely to cause a disseminated infection than B. gilchristii (P < 0.001, CI = 30 – 74%), with 18 (45%) of B. dermatitidis infections becoming disseminated compared to 6 (8%) of B. gilchristii infections. The most common location of dissemination for both species was the skin [5 (6%) for B. gilchristii and 15 (35%) for B. dermatitidis, P < 0.001, CI = 26 – 74%]. The mean age of diagnosis also differed by species (P = 0.02, CI = 2 – 16 years). The mean age of symptom onset for B. gilchristii was 40-years-old, while the mean age for B. dermatitidis was 49-years. No co-morbidities were significantly associated with either species. Patients with B. gilchristii were more likely to be hospitalized than those with B. dermatitidis [48 (60%) versus 11 (28%)] (P < 0.01, CI = 13 - 48%).
Of the symptoms reported, B. gilchristii was more likely to cause a fever than B. dermatitidis (P < 0.001, CI = 25 - 63%), with 63 (79%) patients with B. gilchristii reporting this symptom compared to 14 (35%) patients with B. dermatitidis. The only other symptom that differed significantly between Blastomyces species was the presence of skin lesions, indicating dissemination to the skin as previously described. For diagnosis, consolidation, infiltrate, or opacity of the lungs observed via chest radiograph was more often found in patients with B. gilchristii [73 (91%)] than B. dermatitidis [27 (68%)] (P < 0.01, CI = 12 – 64%). This difference was not significant when consolidation was observed via CT scan [B. gilchristii: 40 (50%), B. dermatitidis: 18 (45%), P = 0.75, CI = 16 – 26%].
We next used lasso regression to assess combinations of variables and their ability to predict Blastomyces genotype. Of the variables found to be significant in univariate analysis, only presence of a fever and skin lesions were predictive of Blastomyces species in the lasso regression model (fever: P < 0.05, skin lesions: P < 0.05). Hospitalization was not significant at P = 0.06.
Discussion
Symptom severity in blastomycosis can span anywhere from asymptomatic to fatal disease, but the underlying cause of this range of clinical outcomes has not yet been described. We genotyped a set of 550,601 variable SNPs from a cohort of patients with blastomycosis in the Marshfield Clinic Health System to test the hypothesis that the genotype of the human host would be associated with disease severity in blastomycosis. We did not find variability in any genetic factors correlated more or less severe blastomycosis. However, we did find the genotype of Blastomyces could predict the clinical progression of the disease. B. gilchristii tends to infect younger people, lead to hospitalization, and is less likely to disseminate than B. dermatitidis. Skin lesions are more commonly found with B. dermatitidis, while a fever is more indicative of B. gilchristii.
While previous studies have identified genetic factors associated with susceptibility to infection with dimorphic fungi, including Blastomyces, Histoplasma, and Coccidioidomyces, our study did not find any SNPs associated with increased risk of hospitalization or risk of contracting blastomycosis. Our analysis of genetic differences between our cohort and our control group found no significant SNPs associated with susceptibility to blastomycosis. We note that many patients in the control group were not residing in areas where blastomycosis is endemic and, therefore, were highly unlikely to have been exposed to Blastomyces. Even in Wisconsin, exposure to this organism is suspected to be so rare that most people will not have encountered Blastomyces in the environment, making studying susceptibility difficult. There is currently no reliable test to determine previous exposure to Blastomyces.
Another reason why we did not detect any SNPs significantly associated with disease severity could be the homogeneity of patient demographics in this study. In our cohort, 91% of patients were white; this is equivalent to the 2020 United States census data for Wood County, where the Marshfield Clinic Healthcare System is headquartered. However, this does not provide opportunity to investigate blastomycosis across diverse populations and may not have allowed us to detect the trends observed with other dimorphic fungi. For example, a previous study of Hmong patients diagnosed with blastomycosis in Wisconsin identified genetic variants of interest located in the genes encoding IL-6; our study likely did not include enough non-white participants or SNPs within IL-6 to replicate this result.15 Additionally, this study used whole-genome sequencing, while we tested a pre-determined set of SNPs. Another cohort study of blastomycosis in the Marshfield Clinic Health system found that Hispanic white, American Indian/Alaskan Native, and Asian patients were younger and healthier than average in the cohort, but were more likely to be hospitalized for blastomycosis.29 We also note that our dataset was predominantly male (58%). This is commonly observed in blastomycosis datasets,30,31 but as females have a higher rate of mortality from blastomycosis,30 may have skewed our results.
Instead, we found that etiologic Blastomyces species was a better predictor of disease severity. Previous research has already described differing clinical progressions between B. gilchristii and B. dermatitidis, and this study confirms those findings.3 We also further identified the signs and symptoms associated with each species. A case of blastomycosis with a fever is highly likely to be caused by B. gilchristii and less likely to lead to infection outside the lungs, but has a high chance of requiring hospitalization. The presence of a skin lesion suggests B. dermatitidis and is associated with disseminated infection in multiple organ systems, but care can more likely be managed in an outpatient setting. Treatment for both species is the same. However, knowledge that there are two distinct progressions of blastomycosis may guide clinicians in diagnosing this disease. Rapid, PCR-based genotyping of Blastomyces samples is now possible and could allow clinicians to better predict which patients may require hospitalization and which patients should undergo additional testing for disseminated infection.31
This study has several important limitations, including that a sample size of 201 may not be sufficient to detect more subtle differences in allele variation. We also targeted specific SNPs rather than performing whole genome sequencing; while economical, this method covers only a very small portion of the human genome. Finally, due to the nature of study design and the approach to patient recruitment, our dataset does not include the most extreme outcomes of blastomycosis. Those who did not survive blastomycosis could not consent to genotyping, while those with asymptomatic blastomycosis would not have sought treatment. Therefore, we cannot eliminate the possibility of genetic susceptibility to blastomycosis, even though we did not detect any such associations in our study. It is also possible that social constructs such as delayed access to medical care may also contribute to severe blastomycosis.
This study also has several strengths, including its sample size. Because blastomycosis is so rare, 201 patients represents the largest blastomycosis cohort to date. Targeted-SNP sequencing allowed us to effectively genotype these patients, as well as obtaining matched data from controls sequenced as part of the PMRP. The extensive chart review completed for this research was used to further investigate the impacts of demographics, underlying conditions, and Blastomyces species on clinical progression of this life-threatening and endemic disease.
Conclusions
In this cohort study of patients with blastomycosis, we were able to confirm and identify differing clinical presentations of B. dermatitidis and B. gilchristii. We did not identify any host genetic variation associated with contracting blastomycosis or with being a more severe case of blastomycosis. However, we did not assess the exact SNPs reported in the literature that were previously associated with susceptibility to infection with dimorphic fungi or severity of that infection. A future research study including a wider range of disease severity (fatal cases and asymptomatic cases) and patient diversity using whole genome sequencing instead of SNP genotyping may be able to detect genetic variation that our study did not. Investigation of social factors associated with disease severity may also reveal additional effects. However, additional evidence supporting disparately different clinical presentations of blastomycosis may assist clinicians in diagnosis and management of this serious and endemic fungal disease.
Acknowledgments
We thank Dr. Tonia Carter for advice in planning the genotype analysis and for critical review of this manuscript.
References
- 1.Gnat S, Łagowski D, Nowakiewicz A, Dyląg M.. A global view on fungal infections in humans and animals: infections caused by dimorphic fungi and dermatophytoses. J Appl Microbiol. 2021;131(6):2688-2704. doi: 10.1111/jam.15084. [DOI] [PubMed] [Google Scholar]
- 2.Brown EM, McTaggart LR, Zhang SX, Low DE, Stevens DA, Richardson SE.. Phylogenetic analysis reveals a cryptic species Blastomyces gilchristii, sp. nov. within the human pathogenic fungus Blastomyces dermatitidis [published correction appears in PLoS One. 2016 Dec 9;11(12):e0168018]. PLoS One. 2013;8(3):e59237. doi: 10.1371/journal.pone.0059237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meece JK, Anderson JL, Gruszka S, Sloss BL, Sullivan B, Reed KD.. Variation in clinical phenotype of human infection among genetic groups of Blastomyces dermatitidis. J Infect Dis. 2013;207(5):814-822. doi: 10.1093/infdis/jis756. [DOI] [PubMed] [Google Scholar]
- 4.Dukik K, Muñoz JF, Jiang Y, et al. Novel taxa of thermally dimorphic systemic pathogens in the Ajellomycetaceae (Onygenales). Mycoses. 2017;60(5):296-309. doi: 10.1111/myc.12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maphanga TG, Birkhead M, Muñoz JF, et al. Human Blastomycosis in South Africa Caused by Blastomyces percursus and Blastomyces emzantsi sp. nov., 1967 to 2014. J Clin Microbiol. 2020;58(3):e01661-19. doi: 10.1128/JCM.01661-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwartz IS, Wiederhold NP, Hanson KE, Patterson TF, Sigler L.. Blastomyces helicus, a New Dimorphic Fungus Causing Fatal Pulmonary and Systemic Disease in Humans and Animals in Western Canada and the United States. Clin Infect Dis. 2019;68(2):188-195. doi: 10.1093/cid/ciy483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benedict K, Roy M, Chiller T, Davis JP.. Epidemiologic and ecologic features of blastomycosis: a review. Curr Fungal Infect Rep. 2012;6(4):327-335. doi: 10.1007/s12281-012-0110-1. [DOI] [Google Scholar]
- 8.Reed KD, Meece JK, Archer JR, Peterson AT.. Ecologic niche modeling of Blastomyces dermatitidis in Wisconsin. PLoS One. 2008;3(4):e2034. doi: 10.1371/journal.pone.0002034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McBride JA, Gauthier GM, Klein BS.. Clinical manifestations and treatment of blastomycosis. Clin Chest Med. 2017;38(3):435-449. doi: 10.1016/j.ccm.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson JL, Hall MC, Meece JK.. Incidental findings of blastomycosis lung nodules in five asymptomatic patients. Med Mycol Case Rep. 2018;21:63-65. doi: 10.1016/j.mmcr.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein BS, Bradsher RW, Vergeront JM, Davis JP.. Development of long-term specific cellular immunity after acute Blastomyces dermatitidis infection: assessments following a large point-source outbreak in Wisconsin. J Infect Dis. 1990;161(1):97-101. doi: 10.1093/infdis/161.1.97. [DOI] [PubMed] [Google Scholar]
- 12.Benedict K, McCotter OZ, Brady S, et al. Surveillance for Coccidioidomycosis - United States, 2011-2017. MMWR Surveill Summ. 2019;68(7):1-15. Published 2019 Sep 20. doi: 10.15585/mmwr.ss6807a1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Odio CD, Marciano BE, Galgiani JN, Holland SM.. Risk factors for disseminated coccidioidomycosis, United States. Emerg Infect Dis. 2017;23(2):308-311. doi: 10.3201/eid2302.160505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roy M, Benedict K, Deak E, et al. A large community outbreak of blastomycosis in Wisconsin with geographic and ethnic clustering. Clin Infect Dis. 2013;57(5):655-662. doi: 10.1093/cid/cit366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merkhofer RM Jr, O’Neill MB, Xiong D, et al. Investigation of Genetic Susceptibility to Blastomycosis Reveals Interleukin-6 as a Potential Susceptibility Locus. mBio. 2019;10(3):e01224-19. Published 2019 Jun 18. doi: 10.1128/mBio.01224-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baumgardner DJ, Buggy BP, Mattson BJ, Burdick JS, Ludwig D.. Epidemiology of blastomycosis in a region of high endemicity in north central Wisconsin. Clin Infect Dis. 1992;15(4):629-635. doi: 10.1093/clind/15.4.629. [DOI] [PubMed] [Google Scholar]
- 17.McCarty CA, Mukesh BN, Giampietro PF, Wilke RA.. Healthy People 2010 disease prevalence in the Marshfield Clinic Personalized Medicine Research Project cohort: opportunities for public health genomic research. Per Med. 2007;4(2):183-190. doi: 10.2217/17410541.4.2.183. [DOI] [PubMed] [Google Scholar]
- 18.Meece JK, Anderson JL, Klein BS, et al. Genetic diversity in Blastomyces dermatitidis : implications for PCR detection in clinical and environmental samples. Med Mycol. 2010;48(2):285-290. doi: 10.3109/13693780903103952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meece JK, Anderson JL, Fisher MC, Henk DA, Sloss BL, Reed KD.. Population genetic structure of clinical and environmental isolates of Blastomyces dermatitidis, based on 27 polymorphic microsatellite markers. Appl Environ Microbiol. 2011;77(15):5123-5131. doi: 10.1128/AEM.00258-11. Published online August 1, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2020. https://www.R-project.org/ [Google Scholar]
- 21.RStudio Team . RStudio: Integrated Development for R. RStudio, Inc.; 2019. http://www.rstudio.com/ [Google Scholar]
- 22.Wickham H, Averick M, Bryan J, et al. Welcome to the Tidyverse. Journal of Open Source Software. 2019;4(43):1686. doi: 10.21105/joss.01686. [DOI] [Google Scholar]
- 23.Friedman J, Hastie T, Tibshirani R.. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw. 2010;33(1):1-22. doi: 10.18637/jss.v033.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559-575. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merkhofer RM, Klein BS.. Advances in understanding human genetic variations that influence innate immunity to fungi. Front Cell Infect Microbiol. 2020;10:69. doi: 10.3389/fcimb.2020.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baumgardner DJ, Halsmer SE, Egan G.. Symptoms of pulmonary blastomycosis: northern Wisconsin, United States. Wilderness Environ Med. 2004;15(4):250-256. doi: 10.1580/1080-6032(2004)015[0250:SOPBNW]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 27.Kralt D, Light B, Cheang M, et al. Clinical characteristics and outcomes in patients with pulmonary blastomycosis. Mycopathologia. 2009;167(3):115-124. doi: 10.1007/s11046-008-9163-7. [DOI] [PubMed] [Google Scholar]
- 28.Merkhofer RM, Klein BS.. Advances in Understanding Human Genetic Variations That Influence Innate Immunity to Fungi. Front Cell Infect Microbiol. 2020;10:69. Published 2020 Feb 28. doi: 10.3389/fcimb.2020.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson JL, Frost HM, King JP, Meece JK.. Racial Differences in Clinical Phenotype and Hospitalization of Blastomycosis Patients. Open Forum Infect Dis. 2019;6(11):ofz438. doi: 10.1093/ofid/ofz438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rush B, Lother S, Paunovic B, Mooney O, Kumar A.. Outcomes With Severe Blastomycosis and Respiratory Failure in the United States. Clin Infect Dis. 2021;72(9):1603-1607. doi: 10.1093/cid/ciaa294. [DOI] [PubMed] [Google Scholar]
- 31.Fritsche TR, Anderson JL, Bassi D, Hall MC, Boyle TR, Meece JK.. Direct Tissue PCR and Genotyping for Species Identification in a Case of Laryngeal Blastomycosis. Ear Nose Throat J. 2023;102(4):NP157-NP160. doi: 10.1177/0145561321991342 [DOI] [PubMed] [Google Scholar]