Abstract
Background
Plasmodium falciparum erythrocyte membrane protein-1, a variant antigen of the malaria parasite, is potentially a target for the immune response. It would be important to determine whether there are CD4 T cells that recognise conserved regions. However, within the relatively conserved region, there is variation. It is not possible to test T cell responses from small field samples with all possible peptides.
Methods
We have aligned sequences that are relatively conserved between several PfEMP1 molecules, and chosen a representative sequence similar to most of the PfEMP1 variants. Using these peptides as pools representing CIDRα, CIDRβ and DBLβ-δ domains, DBLα domain, and EXON 2 domain of PfEMP1, we measured the CD4 T cell responses of malaria-exposed donors from Benin, West Africa by a FACS based assay.
Results
All the three peptide pools elicited a CD4 T cell response in a proportion of malaria-exposed and non-exposed donors. CD4 T cell proliferation occurs at a relatively higher magnitude to peptide pools from the DBLα and EXON 2 in the malaria-exposed donors living in Benin than in the UK malaria-unexposed donors.
Conclusions
These findings suggest that an immunological recall response to conserved peptides of a variant antigen can be measured. Further testing of individual peptides in a positive pool will allow us to determine those conserved sequences recognised by many individuals. These types of assays may provide information on conserved peptides of PfEMP1 which could be useful for stimulating T cells to provide help to P. falciparum specific B cells.
Background
Plasmodium falciparum infection is a major source of morbidity and mortality in malaria-endemic areas [1]. The manifestation of severe malaria is more common in non-immune children living in, and non-immune visitors to, endemic regions, suggesting that the acquisition of specific immunity is an important factor in preventing morbidity and mortality from malaria infection [1]. However, the immunity to malaria infection is partial, as adults living in malaria hyper-endemic zone can be parasitaemic or indeed develop malarial disease. The immunological mechanisms and the antigens to which acquired immunity is directed, are not fully known, although the erythrocytic stage of the parasite has been reported to be the primary target [2]. The variant antigens, expressed on the surface of P. falciparum infected red blood cells and believed to be a mechanism of immune evasion by the malaria parasite, are important antigens worthy of consideration as targets for malaria immunity. One such antigen used in this study is P. falciparum erythrocyte membrane protein-1 (PfEMP1).
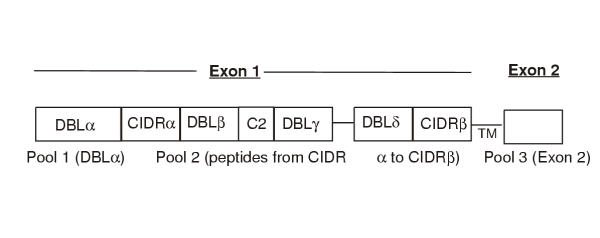
PfEMP1 is a highly variant antigen, with the variable domains within the extra-cellular region coded for by EXON 1 and a relatively conserved intracellular region coded for by EXON 2 (Figure 1). PfEMP1 has been identified as the molecule responsible for intra-vascular sequestration of P. falciparum-infected red blood cells [3,4], a factor which has been associated with the pathogenesis of a severe malaria syndrome (cerebral malaria) [5]. The role of PfEMP1 in cytoadherence, when considered together with the fact, that antibodies to variants of PfEMP1 have been reported to occur in people living in malaria-endemic regions, buttresses the notion that PfEMP1 is indeed an important target for malaria immunity. Furthermore, the number of parasite variants recognised by the sera of these people increases with age and hence, exposure to many parasite variants [2,6]. This indicates that sequential accumulation of antibodies to the different variant molecules present in the local malaria parasite population contributes to the partial immunity that develops to malaria infection. Variant specific antibody responses are likely to require CD4 T cell help. CD4 T cells recognising conserved peptides of a malarial antigen would be potentially more beneficial in malaria immunity, as a memory response from such T cells could provide a rapid help for the variant specific antibody responses. However, it is impractical to determine if a recall response occurs to the conserved portions of a variant antigen by producing recombinant proteins or synthetic peptides of the conserved regions of all known or published sequence of the molecule.
Figure 1.
Three pools of the relatively conserved peptides from PfEMP1. Pool 1 is from the DBLα domain, pool 2 contains two peptides from CIDRα and ten from DBLβ-δ and CIDRβ domains and pool 3 contains conserved peptides from the intracellular EXON 2 domain. The peptides were selected as representative of 15 var gene sequences as described in the Materials and Methods Section. TM stands for transmembrane region.
Hence, we have aligned the sequences of published PfEMP-1 molecules, synthesised a peptide similar to most peptides in a relatively conserved block and pooled the peptides into three groups which correspond to regions of the whole PfEMP1 molecule (Figure 1). The CD4 T cell responses of malaria-exposed donors (from Benin, West Africa) was determined by a flow cytometry method, and compared to the CD4 T cell responses of malaria-unexposed UK donors. Using this method, we have been able to show that CD4 T cells responses to conserved regions of a variant antigen can measured. As the T cell responses to malarial antigens are often low [7] and it has been suggested that cytokines such as IL-10 may inhibit T cell proliferation [8], we have also measured IL-10 production after stimulation with the different peptide pools.
Materials and methods
PfEMP1 sequences, alignment and synthesis
Fifteen PfEMP1 sequences were downloaded from the sequence database maintained at the National Center for Biotechnology Information via PubMed Medline. Gene identification number and names of the sequences are as follows: g1297091, 3D7var1; g6601436, A4 tres var partial; g886375, var-1 Dd2; 1809295, FCR3varT11.1; g914919, MCvar1; g4760402, varCS2 (CSAP) Like; g3845343, PFB 1055c; g3540145, Var IT 4/25/5; g1517814, var ITG 2F6-P21; g914921, var-2 MC132; g886378, var PF 23AFCR3; g2944095, var prot PNG; g5163174, var C2 itG Exon 2; VAR7-like molecule of DD2, (accession number Y11909); and Var MC, (accession number U27338).
The sequences were then aligned using the Megalign program of DNA star computer software. A peptide similar to most peptides in a relatively conserved block was chosen and synthesised by 9-Fluorenylmethoxycarbonyl (Fmoc) chemistry using the multiple peptide synthesiser (Biotech Instrument Limited) as described [9]. The degree of sequence homology of the chosen peptide to the others in a conserved block is shown in Table 1. Three pools of relatively conserved peptides were prepared as shown in Fig. 1 and Table 1. Since previous studies [10-12] show that the peptides recovered from purified MHC class II molecules range from 9 to 30 amino acids or more, we have only chosen peptides from conserved blocks where the amino acid length is not less than 9. Longer peptides were included as they would be processed before presentation by the antigen-presenting cell. Although there are no algorithms or programmes available that can predict MHC class II-binding peptides with probabilities of greater than 50%, screening of the selected peptides using SYFPEITHI database epitope prediction programme [13] predicted that at least 13 out of the peptides used contain one or more epitopes theoretically able to bind one or more HLA-DR molecules.
Table 1.
The degree of sequence homology between chosen peptide and variants within relatively conserved blocks of PfEMP1.
| Peptides in each pool | Length of peptide | Number of variants compared | Identity with variants | Similarity with variants | |
| Pool 1 Peptides from DBLα region | 11 | 10 | 1 | 1aa:11], 2aa:2, 5aa:4, 7aa:1, 8aa:1 | |
| GQCTFNRIKDS | 12 | 10 | 7 | 1aa:3 | |
| 12 | 11 | 0 | 2aa:4, 3aa:2, 4aa:4, 1aa:1 | ||
| GACAPYRRLHLC | 19 | 11 | 1 | 1aa:3, 2aa:7 | |
| HNLLADVCEAAK | 17 | 11 | 1 | 1aa:2, 3aa:3, 1aa:2, 6 aa:2, 10aa:1 | |
| ARSFADIGDIVRGKDLYLG | 19 | 11 | 4 | 1aa:2, 2aa:4, 3aa:1 | |
| LREDWWTANRETVWKAI | 12 | 11 | 1 | 2aa:3, 3aa:1, 4aa:1, 6aa:2, 7aa:3 | |
| VPTYFDYVPQYLRWFEEWA | 16 | 11 | 0 | 2aa:4, 3aa:3, 4aa:2, 5aa:1, 6aa:1 | |
| SRNGYDCTKTIR | 15 | 11 | 0 | 2aa:2, 3aa:2, 4aa:1, 5aa:3, 6aa:1, 7aa:2 | |
| WIDNQKKEFLKQKKKY | |||||
| TFSHTEYREPCPWCG | |||||
| Pool 2 peptides from CIDRα and β and DBLβ-δ regions | KDSIEWRSKLSNCLK | 15 | 11 | 1 | 5aa:2, 6aa:3, 7aa:1, 8aa:3, 9aa:1 |
| CRAMKYSFADLGDII | 15 | 6 | 1 | 1aa:1, 2aa:3, 4aa:1 | |
| ICIPPRRRRLYVGKL | 15 | 10 | 0 | 1aa:1, 2aa:4, 3aa:1, 4aa:2, 6aa:1, 8aa:1 | |
| 11 | 10 | 2 | 1aa:3, 2aa:3, 4aa:1, 8aa:1 | ||
| SAAIETFFLWD | 14 | 9 | 0 | 1aa:1, 2aa:1, 3aa:4, 4aa:2, 5aa:1 | |
| GVIPPDFLRQMFYT | 15 | 10 | 1 | 3aa:5, 4aa:1, 5aa:1, 7aa:1, 10aa:1 | |
| IWNGMICALTYKEKD | 12 | 10 | 4 | 1aa:2, 2aa:1, 3aa:1, 5aa:1, 7aa:1 | |
| RPPYFRYLEEWG | 13 | 6 | 2 | 1aa:2, 2aa:2 | |
| WMTEWAEWFCKEQ | 13 | 10 | 1 | 1aa:2, 5aa:1, 6aa:1 8aa:2, 9aa:1, 10aa:1, | |
| FTYFYLKKKTKSS | 11aa:1 | ||||
| IPTKLSPNRYIPYTSGKYRG- | 27 | 10 | 4 | 1aa:1, 9aa:2, 22aa:1, 23aa:2 | |
| KRYIYLE | 10 | 10 | 4 | 1aa:3, 6aa:2, 9aa:1 | |
| ESEYEEMDIN | 9 | 7 | 5 | 4aa:2 | |
| DHYSDITSS | |||||
| Pool 3 peptides from Exon 2 region | DIYVPGSPKYKTLIEVVLEPS | 21 | 8 | 5 | 1aa:2, 4aa:1 |
| 15 | 10 | 1 | 2aa:4, 3aa:2, 4aa:1, 12 aa:2 | ||
| ITDDEWNQLKKDFIS | 11 | 8 | 2 | 1aa:2, 2aa:1, 6aa:1, 7aa:2 | |
| VDNNTHPTMSR | 10 | 8 | 5 | 1aa:3 | |
| KPFIMSIHDR | 14 | 9 | 3 | 1aa:3, 3aa:1, 7aa:1, 10aa:1 | |
| YSGIDLINDALSGN | 19 | 9 | 0 | 1aa:6, 2aa:2, 14aa:1 | |
| HIDIYDELLKRKENELFGT | 13 | 8 | 3 | 1aa:3, 2aa:2 | |
| NLFHKWLDRHRDM | 11 | 8 | 2 | 1aa:2, 3aa:2, 5aa:1, 6aa:1 | |
| KLKELWENETH | 10 | 7 | 2 | 1aa:2, 2aa:1, 4aa:1, 5aa:1 | |
| MDTILDDLEK |
1] Indicates the number of variant peptides in a conserved block that differs from the peptide used for the T cell assays by the number of amino acids shown as "aa" (for example, 5aa:4 means that 4 variant peptides in a conserved block differ from the peptide used for the T cell assay by 5 amino acids.
Donors
The ages of the Benin malaria exposed donors ranged from 4–45 years (male to female ratio was 4:6). The ages of the UK unexposed control volunteers ranged from 20–45 years (male to female ratio was 3:7). Blood (5 mL from children to 20 mL from adults) was taken from donors living in and around Cotonou, Benin during the dry season of 1992. In this region, malaria is holoendemic with maximal transmission during the rainy season [14]. 28% of the donors were parasitaemic at the time of blood collection, and parasitaemia ranged from 0.0004% to 0.1%. None of the UK volunteers had travelled to a malaria endemic area at any time. Peripheral blood mononuclear cells (PBMC) were separated over Ficoll/paque (Pharmacia Biotech.) using standard procedures. The cells were resuspended in foetal calf serum (FCS) containing 10% DMSO and frozen by controlled-gradient freezing at -70°C for 48h and finally in liquid Nitrogen until use. The Benin samples were transported from Benin to the UK to be analysed simultaneously with the UK samples that were frozen using a similar method. We have shown previously that the CD4 T cell response of PBMC frozen in this manner is as good as that of freshly isolated cells to antigens such as tetanus toxoid [15]. Viability of recovered cells from the Benin samples ranged from 70 to 90% and above 95% for the UK sample. Ethical approval was given by the Vice Dean of Medical School of the University of Benin in accordance with the Helsinki declaration and by the Barnet Health Authority Ethics Committee, UK.
In vitro assay for cell division
PBMC were thawed rapidly at 37°C, washed twice in RPMI 1640, centrifuged at 415 g for 5 min at room temperature and counted. The cells were resuspended at 5 × 106 cells in 250 μL diluent for PKH26 (Sigma) labelling, and stained for 1 min by incubation with PKH26 dye at room temperature as described previously [15,16]. PKH26 dye incorporation into cell membrane was stopped by adding two times the volume of 100% human AB serum. Cells were then washed three times, and staining was assessed by flow cytometry. After labelling and washing the viabililty as assessed by Trypan Blue was greater than 75%. The labelled cells were re-suspended in fresh RPMI 1640 medium supplemented with 2 mM glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin, 10 mM HEPES and 10% heat-inactivated AB serum medium. The cultures were set up in triplicates at 2 × 105 cells/well in a 96 well U-bottomed plate (Nunclon™, Gibco) in a final volume of 200 μL with or without antigen. Test antigens were used at a final concentration of 5 nM for each peptide in a pool. Phytohaemagglutinin (PHA) (Sigma) was used as a control for cell viability at a final concentration of 2.0 μg/mL. Plates were incubated at 37°C, 5% CO2, in a humidified atmosphere for 7 days, after which time supernatants were removed for cytokine measurement and the cells were stained with monoclonal antibodies directed against CD3 and CD4. The relative numbers of dividing CD4+ lymphocytes were determined by flow cytometry.
Antibodies for flow cytometry
The antibody used to determine the phenotype of the responding cells was allophycocyanin (APC) conjugated anti-CD4, 13B8.2 (mouse IgG1) (Pharmingen).
Cultured cells were washed in sterile PBS containing 0.5% w/v bovine serum albumin, 5.0 mM EDTA pH 8.0 and 0.01% w/v sodium azide (Sigma) and incubated with the antibody for 30 mins. Excess antibody was washed off in PBS and the cells fixed in 1% paraformaldehyde in PBS and transferred into FACS tubes for analysis.
Measurement of cell division by flow cytometry
Flow cytometry was performed on a FACscalibur machine using Cell Quest software (Becton Dickinson). Viable cell population was defined by using forward and 90° light scatter. A reduction in the fluorescence intensity of PKH26 labelling was used as an index of cell division [15,16]. The use of APC conjugated antibody allowed the measurement of dividing CD4+ T cells. The flow cytometer was set to acquire events for 2 min. This allows the relative numbers of recovered viable cells within each well (a measure of the magnitude of the response) to be determined. Any cell exhibiting a reduction in PKH26 fluorescence intensity was classified as having divided in response to the antigen. The background number of divided CD4 T cells in the absence of antigen varied widely between donors (30–8152), therefore, a donor was considered to have responded by CD4 T cell division if the mean number of dividing cells in triplicate wells containing an antigen was equal to or above 2 times the mean value of triplicate wells without antigen. The percentage of dividing CD4 population relative to the total population of CD4 positive cells ranged from 1.2% – 10%
Detection of cytokines by ELISA
The amount of IL-10 in the supernatants after 7 days of cultures was measured by ELISA using paired antibody kits (Pharmingen/ Beckton Dickinson, Oxford, UK) and following the manufacturers instructions. The response of a donor was defined as positive when the mean concentration of IL-10 produced in triplicate wells containing antigen was significantly greater (i.e. p < 0.05, Student T test) than the mean concentration of IL-10 production in triplicate wells without antigen of that donor.
The limit of detection for the ELISA assay for IL-10 was 4 pg/mL. The background level of IL-10 production differed widely between individual donors (range: 0 to 149 pg/mL). For this reason, the concentration of IL-10 produced by each donor in response to the peptide antigens was taken as the net concentration after subtraction of the background of that donor without antigen. Standard Errors of the means were usually in the range of 15–20% (not shown).
Statistical analyses
The results were analysed by the Two-tailed Student T test and the Fisher's exact Test.
Results
Prevalence and magnitude of the CD4 T cell proliferation
We have determined the in vitro CD4 T cell division in response to three pools of relatively conserved peptides from three regions of PfEMP1, namely, DBLα (pool 1), CIDRα and CIDRβ + DBL β-δ (pool 2) and EXON 2 (pool 3) (Table 1 and Figure 1). All the three peptide pools elicited a CD4 T cell response in a proportion of malaria-exposed and non-exposed donors (Figure 2). The magnitude of the CD4 T cell division was significantly higher in the Benin malaria-exposed donors when compared to the UK malaria-unexposed donors for peptide pools 1 and 3 (Two tailed Student T test, p = 0.025 and 0.045 for peptide pools 1 and 3, and p = 0.062 for pool 2 peptides) (Table 2). Using a cut off of 2 stimulation indices to define a positive response, the prevalence of positive CD4 T cell response among the malaria-exposed Benin donors to any of the three peptide pools ranged from 21–37% (Figure 2). However, using the Fisher's Exact test, this was not statistically different from the prevalence of the responders among the UK malaria-unexposed donors (4–20%) (Figure 2). The absence of CD4 T cell division in response to the antigens by some donors was not due to loss of cellular viability, as cells from all donors divided in response to PHA, (data not shown).
Figure 2.
CD4 T cell division in response to the three pools of the relatively conserved peptides determined by flow cytometry using PKH26 dye. The results are expressed as stimulation indices; i.e. the mean number of dividing cells in triplicate wells containing an antigen divided by the mean number of dividing cells in triplicate wells without antigen. All responses with stimulation indices greater than or equal to 2 were considered as positive (i.e. those with values above the horizontal line shown on each of the three graphs). The prevalence of responders in each group is given below the graphs.
Table 2.
CD4 T cell proliferation to three pools of relatively conserved peptides of PfEMP-1
| Pools of peptides | Mean CD4 T cell proliferation (Log10) | ||
| Benin donors | UK controls | P value | |
| DBLα (pool 1) | 2.47 ± 0.15 1] (24)2] | 1.94 ± 0.18 (24) | P = 0.025 |
| CIDRα + DBLβ-δ (pool 2) | 2.47 ± 0.14 (23) | 2.02 ± 0.20 (21) | P = 0.062 |
| Exon 2 (pool 3) | 2.44 ± 0.15 (24) | 1.94 ± 0.20 (22) | P = 0.045 |
1] Standard error of the Log10 mean of dividing CD4 T cells in triplicate wells as determined by a flow cytometric assay described in Materials and Methods 2] Total number of donors tested
Prevalence and magnitude of cytokine production
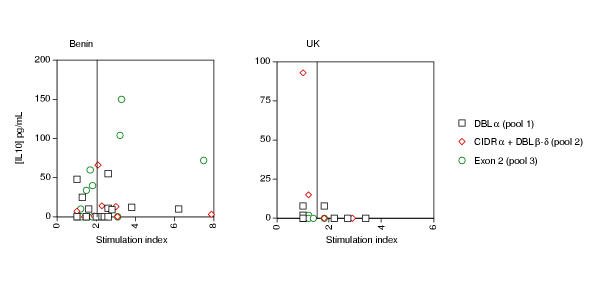
IL-10 was measured in the supernatants after 7 days of culture in the presence of the three PfEMP-1 peptide pools. The prevalence of positive IL-10 responses to any of the peptide pools ranged from 16–21% among the malaria-exposed donors living in Benin and ranged from 0–7% among the malaria-unexposed UK. Because of the small sample size, the differences observed in the prevalence of positive IL-10 responders between the Benin malaria-exposed donors and the UK malaria-unexposed donors was not statistically significant, using the Fisher's Exact test (data not shown). The magnitude of IL-10 production was significantly higher only in the culture supernatants of the Benin malaria-exposed donors stimulated with pool 3 (EXON 2) peptides when compared with the magnitude of IL-10 production in the culture supernatants of the UK non-exposed donors (31.1 ± 10.4 pg/mL and 3.4 ± 1.2 pg/mL respectively; p = 0.03).
Comparison of IL-10 production and CD4 T cell division for each donor indicated an inverse correlation between the 2 responses (Figure 3). UK donors with positive IL-10 production to peptides pools 2 and 3 had no positive CD4 T cell response. Conversely, those UK donors whose CD4 T cells divided in response to the peptide pools did not produce IL-10 (Figure 3). Amongst the Benin donors, only those with the lowest CD4 T cell division produced IL-10 (27% of donors).
Figure 3.
Relationship between IL-10 production and CD4 T cell division (stimulation index) of each donor in response to three pools of relatively conserved peptides of Plasmodium falciparum erythrocyte membrane protein-1. Each symbol represents the net concentration of IL-10 (pg/mL) after substraction of the background level in the absence of antigen, plotted against the stimulation index of CD4 T cell division.
Discussion
In this study we have shown that the magnitude of the CD4 T cell response to peptide pools representing parts of DBLα and Exon 2 domains of PfEMP1 (Table 2) was significantly higher in the Benin malaria-exposed donors when compared to the UK malaria-unexposed donors. These results suggest that, despite the extensive variation of PfEMP1, exposure to P. falciparum may induce a CD4 T cell response to conserved or shared regions of DBLα and EXON 2 of this molecule. A more detailed study with a larger number of PBMC from immune adults would allow us to determine whether individual peptides within these pools were responsible for the enhanced responses, and thus whether conserved peptides might be useful for eliciting CD4 T cells that are able to help variant specific B cell responses.
All CD4 T cell divisions to the peptides were relatively small; only rarely were divisions of greater than 5 stimulation indices observed (Figure 2). These results agree with several earlier studies showing low T cell responses to a variety of malaria antigens [7,16-18] which may in part be caused by selective sequestration of specific cells in lymphoid organs [19,20]. It is also possible in our case that either the peptides selected did not bind the different MHC Class II molecules, or they were not representative of the repertoire of PfEMP1 molecules in the parasite population of this endemic area.
The repertoire of PfEMP1 molecules expressed on parasites in this or any area of P. falciparum transmission is not known. Since the conservation between the fifteen variants sequenced from a variety of parasites from different areas was high, we would expect some of these peptides to be representative of those in the sample population. As tools become available that enable characterisation of the PfEMP1 molecules in different parasites, it will become possible to test T cell responses against peptides from PfEMP1 that are known to be expressed in the parasites infecting the individuals under study.
CD4 T cell responses can be inhibited in vitro and in vivo by cytokines such as TGFβ and IL-10 [21,22]. Therefore we determined whether IL-10 was produced in the culture supernatants of PBMC from Benin and UK donors in response to the three peptide pools. Here we show that greater amounts of IL-10 were produced in the cultures of the Benin malaria-exposed donors stimulated with pool 3 (EXON 2) peptides. Exposed or non-exposed donors, whose cultures produced high levels of IL-10 in response to any of the peptide pools generally had no or low CD4 T cell responses and vice versa. These data, although from a small sample, suggest that strong IL-10 responses are not compatible with CD4 T cell proliferation in vitro. Proliferation assays carried out in the presence of anti-IL-10 antibodies or anti-IL-10 receptor antibodies would clarify the anti-proliferative role of IL-10 in these responses; such studies are underway. The fact that a small fraction of malaria-unexposed UK donors also respond to these peptide pools could indicate that there are cross-reactive or shared T cell epitopes with other antigens among the three peptides pools used in the in vitro stimulation as has been described earlier for some malaria antigen specific T cell clones [23].
Our data suggest that the use of synthetic peptides representing conserved sequences of PfEMP1 might be of value in determining regions of the molecule recognised by T cells of a wide variety of donors exposed to different variants of PfEMP1. Further dissection of the responses to individual peptides may allow us to select regions of PfEMP1 that could be useful for a peptide-based vaccine. It would be very encouraging for vaccine design if CD4 T cell helper responses for variant specific B cells could be directed to common or conserved regions of PfEMP1.
Conclusion
In summary, we have shown that CD4 T cell division to conserved peptides of some regions of PfEMP1 occurs at a relatively higher magnitude in donors living in a malaria endemic zone when compared with malaria-unexposed donors. These findings indicate a secondary response to these peptides and, hence, have important implications in malaria vaccine development, as the relatively conserved peptides inducing the T cell division might be useful for stimulating variant specific B cell help in a multi-component vaccine. At present, we have used peptide mixtures from different regions of PfEMP1 because of limitations in the number of PBMC available from each donor. An important extension of this work is to map out the specific peptides responsible for the induction of the observed CD4 T cell division in the responders and determine whether other variants of the peptides produce a similar response or act as altered peptide ligands. The response of individuals with different HLA class II haplotype to these peptides would also determine their usefulness in vaccination.
Acknowledgments
Acknowledgements
We would like to thank Frank Albano and Ching Li for the critical reading of the manuscript, and the Medical Research Council for funding this study. Part of this work was supported by the EU INCO-DC programme. Latifu Sanni was the recipient of a Wellcome Trust Travelling Research Fellowship (053488).
Contributor Information
Latifu A Sanni, Email: lsanni@nimr.mrc.ac.uk.
Catherine EM Allsopp, Email: cemallsopp@hotmail.com.
Lieke Reubsaet, Email: llreubsaet@hotmail.com.
Ambaliou Sanni, Email: sanni@intnet.bj.
Chris Newbold, Email: CNewbold@hammer.imm.ox.ac.uk.
Virander S Chauhan, Email: virander@icgeb.res.in.
Jean Langhorne, Email: jlangho@nimr.mrc.ac.uk.
References
- Osuntokun BO. Malaria and the nervous system. Afr J Med Med Sci. 1983;12:165–172. [PubMed] [Google Scholar]
- Bull PC, Lowe BS, Kortok M, Molyneux CS, Newbold CI, Marsh K. Parasite antigens on the infected red cell surface are targets for naturally acquired immunity to malaria. Nature Med. 1998;4:358–360. doi: 10.1038/nm0398-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggs BA, Gooze L, Wycherley K, Wilkinson D, Boyd AW, Forsyth KP, Edelman L, Brown GV, Leech JH. Knob-independent cytoadherence of Plasmodium falciparum to the leukocyte differentiation antigen CD36. J Exp Med. 1990;171:1883–1892. doi: 10.1084/jem.171.6.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruch DI, Ma XC, Singh HB, Bi X, Pasloske BL, Howard RJ. Identification of a region of PfEMP1 that mediates adherence of Plasmodium falciparum infected erythrocytes to CD36: conserved function with variant sequence. Blood. 1997;90:3766–3775. [PubMed] [Google Scholar]
- Berendt AR, Turner GDH, Newbold CI. Cerebral malaria: the sequestration hypothesis. Parasitology Today. 1994;10:412–414. doi: 10.1016/0169-4758(94)90238-0. [DOI] [PubMed] [Google Scholar]
- Giha HA, Staalsoe T, Dodoo D, Roper C, Satti GM, Arnot DE, Hviid L, Theander TG. Antibodies to variable Plasmodium falciparum-infected erythrocyte surface antigens are associated with protection from novel malaria infections. Immunol Lett. 2000;71:117–126. doi: 10.1016/S0165-2478(99)00173-X. [DOI] [PubMed] [Google Scholar]
- Riley EM, Jepsen S, Andersson G, Otoo LN, Greenwood BM. Cell-mediated immune responses to Plasmodium falciparum antigens in adult Gambians. Clin Exp Immunol. 1988;71:377–382. [PMC free article] [PubMed] [Google Scholar]
- Plebanski M, Flanagan KL, Lee EA, Reece WH, Hart K, Gelder C, Gillespie G, Pinder M, Hill AV. Interleukin 10-mediated immunosuppression by a variant CD4 T cell epitope of Plasmodium falciparum. Immunity. 1999;10:651–660. doi: 10.1016/s1074-7613(00)80064-3. [DOI] [PubMed] [Google Scholar]
- Fields GB, Noble RL. Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int J Pept Protein Res. 1990;35:161–214. doi: 10.1111/j.1399-3011.1990.tb00939.x. [DOI] [PubMed] [Google Scholar]
- Rudensky A, Preston-Hurlburt P, Hong SC, Barlow A, Janeway CA. Sequence analysis of peptides bound to MHC class II molecules. Nature. 1991;353:622–627. doi: 10.1038/353622a0. [DOI] [PubMed] [Google Scholar]
- Rammensee HG. Chemistry of peptides associated with MHC class I and class II molecules. Curr Opin Immunol. 1995;7:85–96. doi: 10.1016/0952-7915(95)80033-6. [DOI] [PubMed] [Google Scholar]
- Abbas AK, Lichtman AH, Pober JS. Cellular and molecular immunology. Second. Philadelphia: W. B. Saunders Company; 1994. [Google Scholar]
- Rammensee H-G, Bachmann J, Emmerich NN, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–319. doi: 10.1007/s002510050595. http://www.uni-tuebingen.de/uni/kxi/ [DOI] [PubMed] [Google Scholar]
- Goodier M, Krause-Jauer M, Sanni A, Massougbodji A, Sadeler BC, Mitchell GH, Modolell M, Eichmann K, Langhorne J. Gamma delta T cells in the peripheral blood of individuals from an area of holoendemic Plasmodium falciparum transmission. Trans Roy Soc Trop Med Hyg. 1993;87:692–696. doi: 10.1016/0035-9203(93)90299-6. [DOI] [PubMed] [Google Scholar]
- Allsopp CE, Nicholls SJ, Langhorne J. A flow cytometric method to assess antigen-specific proliferative responses of different subpopulations of fresh and cryopreserved human peripheral blood mononuclear cells. J Immunol Methods. 1998;214:175–186. doi: 10.1016/S0022-1759(98)00056-8. [DOI] [PubMed] [Google Scholar]
- Allsopp CEM, Sanni LA, Reubsaet L, Ndungu F, Newbold C, Mwangi T, Marsh K, Langhorne J. CD4 T cell responses to a variant antigen of the malaria parasite, Plasmodium falciparum reythrocyte membrane protein-1, in individuals living in malaria-endemic areas. J Infect Dis. 2002;185:812–819. doi: 10.1086/339521. [DOI] [PubMed] [Google Scholar]
- Doolan DL, Khamboonruang C, Beck HP, Houghten RA, Good MF. Cytotoxic T lymphocyte (CTL) low-responsiveness to the Plasmodium falciparum circumsporozoite protein in naturally-exposed endemic populations: analysis of human CTL response to most known variants. Int Immunol. 1993;5:37–46. doi: 10.1093/intimm/5.1.37. [DOI] [PubMed] [Google Scholar]
- Lalvani A, Hurt N, Aidoo M, Kibatala P, Tanner M, Hill AV. Cytotoxic T lymphocytes to Plasmodium falciparum epitopes in an area of intense and perennial transmission in Tanzania. Eur J Immunol. 1996;26:773–779. doi: 10.1002/eji.1830260408. [DOI] [PubMed] [Google Scholar]
- Hviid L, Theander TG, Abdulhadi NH, Abu-Zeid YA, Bayoumi RA, Jensen JB. Transient depletion of T cells with high LFA-1 expression from peripheral circulation during acute Plasmodium falciparum malaria. Eur J Immunol. 1991;21:1249–1253. doi: 10.1002/eji.1830210523. [DOI] [PubMed] [Google Scholar]
- Hviid L, Kurtzhals JA, Goka BQ, Oliver-Commey JO, Nkrumah FK, Theander TG. Rapid reemergence of T cells into peripheral circulation following treatment of severe and uncomplicated Plasmodium falciparum malaria. Infect Immun. 1997;65:4090–4093. doi: 10.1128/iai.65.10.4090-4093.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti M, Colasante A, Mascetra N, Piantelli M, Musiani P, Aiello FB. IL-10 synergizes with dexamethasone in inhibiting human T cell proliferation. J Pharmacol Exp Therap. 1998;285:915–919. [PubMed] [Google Scholar]
- Fox FE, Ford HC, Douglas R, Cherian S, Nowell PC. Evidence that TGF-beta can inhibit human T-lymphocyte proliferation through paracrine and autocrine mechanisms. Cell Immunol. 1993;150:45–58. doi: 10.1006/cimm.1993.1177. [DOI] [PubMed] [Google Scholar]
- Zevering Y, Amante F, Smillie A, Currier J, Smith G, Houghten RA, Good MF. High frequency of malaria-specific T cells in non-exposed humans. Eur J Immunol. 1992;22:689–696. doi: 10.1002/eji.1830220311. [DOI] [PubMed] [Google Scholar]