Abstract
Recently we generated a panel of hepatitis B virus core gene mutants carrying single insertions or deletions which allowed efficient expression of the core protein in bacteria and self-assembly of capsids. Eleven of these mutations were introduced into a eukaryotic core gene expression vector and characterized by trans complementation of a core-negative HBV genome in cotransfected human hepatoma HuH7 cells. Surprisingly, four mutants (two insertions [EFGA downstream of A11 and LDTASALYR downstream of R39] and two deletions [Y38-R39-E40 and L42]) produced no detectable capsids. The other seven mutants supported capsid formation and pregenome packaging/viral minus- and plus-strand-DNA synthesis but to different levels. Four of these seven mutants (two insertions [GA downstream of A11 and EHCSP downstream of P50] and two deletions [S44 and A80]) allowed virion morphogenesis and secretion. The mutant carrying a deletion of A80 at the tip of the spike protruding from the capsid was hepatitis B virus core antigen negative but wild type with respect to virion formation, indicating that this site might not be crucial for capsid-surface protein interactions during morphogenesis. The other three nucleocapsid-forming mutants (one insertion [LS downstream of S141] and two deletions [T12 and P134]) were strongly blocked in virion formation. The corresponding sites are located in the part of the protein forming the body of the capsid and not in the spike. These mutations may alter sites on the particle which contact surface proteins during envelopment, or they may block the appearance of a signal for the transport or the maturation of the capsid which is linked to viral DNA synthesis and required for envelopment.
Hepatitis B virus (HBV) is a major human pathogen causing acute and chronic liver inflammation. It is the prototype of a family of hepatotrophic, enveloped DNA viruses with a very narrow host range, referred to as hepadnaviridae. The spherical virus particle has a diameter of 42 nm and consists of an envelope carrying three surface proteins which surrounds an icosahedral capsid enclosing an open circular, partially double stranded, 3.2-kb DNA as well as the viral DNA polymerase. The capsid has a diameter of 30 nm and is formed by multiple copies of one species of core protein (for a review, see reference 22). This protein comprises 185 amino acids (aa) for genotype A and forms dimers which self-assemble in heterologous expression systems into shells of T=3 and T=4 symmetry (32). The C-terminal 30 aa are very rich in arginine residues and probably bind to the encapsidated viral nucleic acid. The N-terminal 155 aa are sufficient for capsid formation (9) and referred to as the assembly domain. The fold of the assembly domain in core particles has been determined by electron cryomicroscopy (4, 8). The capsid is a very immunogenic antigen (HBV core antigen [HBcAg]), and the corresponding antibody (anti-HBc) mainly binds to a conformation-dependent epitope. An antibody with a different specificity (anti-HBV e antigen [anti-HBe]) which binds also to denatured core protein (24) is formed by only a fraction of infected individuals.
During HBV infection the nucleocapsid is released from the incoming virus into the cytosol, and the viral DNA genome is transported into the nucleus and repaired to give a circular covalently closed episome. This DNA serves as the template for transcription by host factors. A 3.5-kb RNA has two functions: (i) it is the mRNA for translation of core protein and reverse transcriptase/DNA polymerase (P protein), and (ii) it is bound by P protein and packaged by multiple copies of core protein dimers into capsids. The viral DNA genome is then synthesized in the lumen of the capsid by reverse transcription of the 3.5-kb RNA followed by second-strand DNA synthesis. The nucleocapsid can follow two different pathways. It can stay within the cell, in which case its genome contributes to the intracellular amplification of the viral episomes (27). Alternatively, nucleocapsids interact at intracellular membranes of a pre-Golgi compartment with cytosolic domains of viral envelope proteins (5, 23) which are expressed as transmembrane peptides from 2.1- and 2.4-kb mRNAs. This interaction probably initiates and drives budding, resulting in the formation of virions in the lumen of the exocytotic compartment which are released from the cell by secretion.
The destiny of nucleocapsids, either disintegration and release of the genome or envelopment, is regulated. In the duck hepatitis B virus animal model, it was demonstrated that early in infection disintegration and genome amplification prevail, whereas later genome amplification ceases (19) and envelopment of capsids leads to formation of virions. Another step during hepatitis B virion morphogenesis which is unique among enveloped viruses is also regulated: newly formed cytosolic nucleocapsids are not competent for envelopment and are not readily exported. They contain pregenomic RNA as genetic information and not the DNA genome present in virions. Therefore, a maturation step which is linked to the synthesis of the DNA genome by reverse transcription is required to make the nucleocapsid ready for envelopment (10, 29). Apparently, a signal labeling a mature particle as disposed for envelopment is generated on the surface of the capsids during reverse transcription. The nature of this signal, which is probably mediated by the core protein, is unknown.
In this work, we tried to find mutations in the HBV core gene which allow capsid formation and also viral DNA genome synthesis but block nucleocapsid envelopment. Mutations with this phenotype may alter sites on the capsid surface interacting with envelope proteins during envelopment, or they may change the signal generation for, e.g., capsid transport or maturation-dependent envelopment.
MATERIALS AND METHODS
Plasmids.
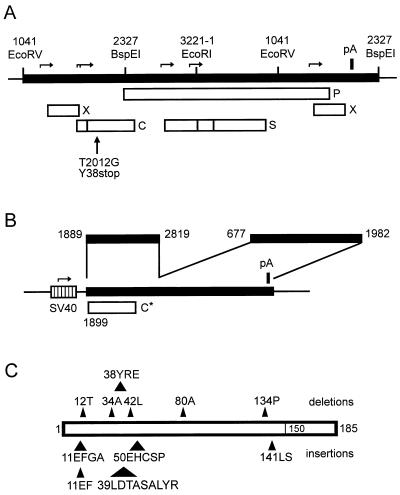
For this study, the HBV genome described by Valenzuela et al. (28) (EMBL accession no. X02763) was used. Numbering of the HBV genome starts at the first deoxycytidine residue of the unique EcoRI site. Plasmid pRVHBV1.5core− is a derivative of plasmid pRVHBV1.5 (6) carrying 1.4 copies of the HBV genome in a head-to-tail configuration inserted into the vector pBluescript KS(+) (Stratagene) (Fig. 1A). Transfection of human hepatoma HuH7 cells with pRVHBV1.5 results in the expression of HBV (5). For construction of pRVHBV1.5core−, the single-point mutation T2012G was introduced into the 5′ region of the HBV DNA inserted in pRVHBV1.5 by site-directed in vitro mutagenesis (18), creating a stop codon at codon 38 of the core gene. A 1,280-bp HindIII-BspEI fragment containing the mutation was recombined back into wild-type (WT) pRVHBV1.5. This part of pRVHBV1.5core− was then sequenced again to ensure that no unintentional mutations were present. HindIII cuts in the polylinker of the vector directly 5′ of the HBV DNA insertion which starts with HBV nucleotide (nt) 1040. BspEI cuts at nt 2327 of the HBV DNA sequence near the 3′ end of the 5′ copy of the core gene in this plasmid.
FIG. 1.
Map of plasmids and mutants. (A) Map of the terminal redundant HBV genome (thick line) carrying a missense mutation at codon 38 of the core gene introduced by changing nt 2012 from T to G (vertical arrow). Boxes indicate open reading frames for the P, X, surface (S), and core (C) proteins. Promoters are indicated by horizontal arrows. The single poly(A) signal (pA) is shown as a thick vertical bar. Thin vertical bars point to restriction enzyme sites; numbers refer to nucleotides of the HBV DNA. (B) Map of the SV40 early promoter (hatched box) expression vector for the HBV core gene. The insertion (thick line) is a fusion of two HBV DNA fragments. The fragment from nt 1889 to 2819 contains a variant of the HBV core gene (C*) carrying some nucleotide changes to introduce unique restriction enzyme sites without altering the coding of the gene (17). The second fragment from nt 677 to 1982 contains the poly(A) signal and a posttranslational regulatory element for the nuclear export of the unspliced mRNA (14). This plasmid was used for trans complemention of the core-negative HBV genome shown in panel A by cotransfection of HuH7 cells, resulting in secretion of virions. (C) Representation of the 11 core gene mutations introduced into the vector shown in panel B and used for trans complementation of the HBV genome shown in panel A. The box represents the core gene. The approximate positions of the six deletions analyzed in this work are indicated by triangles above the box. Numbers indicate the deleted amino acids or the first amino acid residue of the deleted peptide. Positions of the five insertions analyzed are shown below the box. Numbers refer to amino acid residues at which the indicated peptides were C-terminally inserted. The C-terminal arginine-rich domain of the core protein (35 aa) is indicated.
Plasmid pSVcore directs the expression of the HBV core gene under control of the simian virus 40 (SV40) early promoter (Fig. 1B). This plasmid was constructed by first inserting the linker CTAGTGGTCACCA into the unique SpeI site of plasmid pSVBX24H (11) at nt 677 of the HBV sequence in the 3′ part of the S gene. The linker contains a BstEII site. Then this plasmid was cut with BamHI downstream of the SV40 promoter and with BstEII. A BamHI (nt 1889 of the HBV sequence)-BstEII (nt 2819 of the HBV sequence) fragment from plasmid pMK8 (17) containing a derivative of the HBV core gene was inserted (Fig. 1B). The core gene carried several point mutations generating unique restriction sites without changing the amino acid sequence of the core protein (17). Eleven single insertional or deletional mutations in the HBV core gene which have previously been generated and characterized in the context of a bacterial expression vector (17) were separately recombined into the core gene of pSVcore, using unique restriction sites located in the gene. All mutations were confirmed by DNA sequencing.
Nomenclature of mutants.
Six of the 11 mutants have deletions. They are designated by the 2 aa of the WT core protein sequence which were fused by the deletion (e.g., mutant L37-A41 carries a deletion of the three codons corresponding to the core protein peptide Y38-R39-E40). The other five mutants (labeled also by 2 aa X-Y of the WT core protein sequence) carry insertions. The inserted DNA encodes the peptide between and including aa Y and aa X and is located directly 3′ of codon X (e.g., the core protein of mutant A11-E8 has the sequence … K7-E8-F9-G10-A11-E8-F9-G10-A11-T12 … , [the insertion is boldfaced]).
Transfection of HuH7 cells and harvest.
HuH7 cells were seeded in 10-cm-diameter dishes and cultured in Dulbecco's modified essential medium supplemented with 10% (vol/vol) fetal calf serum and with antibiotics/antimycotics at 37°C in a 5% CO2 atmosphere. The cells were subcultured every 3 or 4 days. For transfections, cells were seeded in 10-cm-diameter culture dishes with 10 ml of medium in such a way that they reached approximately 80% confluence after overnight incubation. Five micrograms of pRVHBV1.5core− was mixed with 0.5 ml of 250 mM CaCl2. In case of cotransfections, 5 μg of the WT or mutant plasmid pSVcore was additionally mixed into the CaCl2 solution. The plasmid solution was added slowly while stirring to 0.5 ml of 280 mM NaCl–50 mM HEPES–1.5 mM Na2HPO4 (pH 7.1). After incubation for 30 min at room temperature, the solution was added dropwise to the culture medium of the 10-cm-diameter dish. The culture was placed into the incubator for 9 h. The medium was removed, cells were washed with 10 ml of phosphate-buffered saline (PBS), and 11 ml of fresh medium was added. Four days posttransfection, the culture medium was harvested and spun for 10 min at 4,000 × g. Ten milliliters of the supernatant was taken for further processing. The cells were washed once with 10 ml of PBS and incubated with 1.5 ml of lysis buffer (150 mM NaCl, 50 mM Tris-Cl [pH 7.5], 5 mM MgCl2, 0.2% [vol/vol] Nonidet P-40) for 15 min with agitation. The cell lysate was collected, transferred to a reaction tube, and spun for 5 min at 14,000 rpm in a desktop centrifuge. The supernatant (cleared lysate) was transferred to a fresh tube.
Detection of cytoplasmic capsids by sedimentation.
A cleared lysate from cells of two 10-cm-diameter dishes was prepared in 1 ml of lysis buffer as described above. The cleared lysate (0.8 ml) was layered on top of a sucrose gradient (0.6 ml of 60% [wt/wt], 0.8 ml of 45%, 0.8 ml of 30%, and 0.8 ml of 15% sucrose in PBS) in an SW60 rotor (Beckman Instruments) and spun for 2 h at 38,600 rpm and 10°C. Ten fractions (0.38 ml each) were taken from the top. HBcAg was measured in each fraction by enzyme-linked immunosorbent assay (ELISA) as follows. Immunoglobulin prepared from a human serum with a high anti-HBc and low anti-HBe titer (serum F1451) by ammonium sulfate precipitation was bound to a microtiter plate (Maxisorp U96; Nunc, Roskilde, Denmark) by overnight incubation at 4°C (0.25 μg of immunoglobulin in 0.1 ml of 17.5 mM sodium phosphate buffer [pH 7.6] per well). The wells were washed three times with 0.2 ml of PBS–0.05% (vol/vol) Tween 20, incubated with 0.15 ml of PBS–1% (wt/vol) bovine serum albumin per well for 1 h at 37°C, and washed again as described above. From the undiluted sucrose gradient fractions, 0.1 ml was added; the plate was incubated for 1 h at 37°C and washed as described above. A horseradish peroxidase-labeled anti-HBc antibody (prepared from a sheep immunized with recombinant HBcAg, corresponding to 0.1 μl of serum) was added in 0.1 ml of PBS–1% (wt/vol) bovine serum albumin BSA–20% (vol/vol) fetal calf serum, and the plate was incubated for 1 h at 37°C. After washing as described above, 0.1 ml of 0.1 M citric acid–0.2 M sodium phosphate (pH 7.5)–0.4 mM o-phenylenediamine–0.006% (vol/vol) H2O2 was added, and the dish was incubated for 30 min in the dark. To stop the reaction, 0.1 ml of 2 M H2SO4 was added, and extinction was measured at a wavelength of 492 nm.
Detection of cytoplasmic capsids by agarose gel electrophoresis.
Cleared lysates (1.5 ml) were layered on top of sucrose gradients (1 ml of 40% [wt/wt] and 1.5 ml of 20% sucrose in PBS; SW60 rotor [Beckman Instruments]), and the capsids were sedimented by spinning for 21 h at 50,000 rpm and 20°C. The pellet was resuspended in 50 μl of PBS by sonication (three strokes, 1 s each). Ten microliters of the solution was mixed with 2 μl of sample buffer containing 6× electrophoresis buffer, 0.25% (wt/vol) bromphenol blue, and 30% (vol/vol) glycerol. The sample was loaded onto a 1% (wt/vol) agarose gel prepared in electrophoresis buffer (40 mM Tris-acetate, 1 mM EDTA) as described in reference 25. Electrophoresis was carried out for 90 min at 100 V. The proteins in the gel were blotted overnight onto nitrocellulose membranes (Schleicher & Schuell, Dassel, Germany) by capillary transfer using 10× SSC (1.5 M NaCl, 1.5 M sodium citrate [pH 7.0]) as transfer buffer as described for Southern blots in reference 25. The membrane was blocked by incubation in 30 ml of PBS supplemented with 5% (wt/vol) skim milk powder and 0.1% (vol/vol) Tween 20 for 1 h at room temperature with agitation. The membrane was washed once for 15 min and twice for 5 min with 50 ml of PBS–0.1% (vol/vol) Tween 20 each time and incubated with 10 ml of PBS–5% (wt/vol) skim milk powder–0.1% (vol/vol) Tween 20 and 10 μl of a polyclonal rabbit anti-HBc antibody (DAKO, Hamburg, Germany) for 1 h at room temperature with agitation. The membrane was washed again as described above and incubated in 10 ml of PBS–5% (wt/vol) milk powder–0.1% (vol/vol) Tween 20 and 20 μl of a horseradish peroxidase-labeled second antibody (UnAb-HRP; Eurogentec, Seraing, Belgium) for 1 h at room temperature with agitation. The unbound labeled antibody was removed by washing as described above. HBcAg was detected by incubating the membrane in 6 ml of detection solution (ECL kit; Amersham) for 1 min and exposing the membrane for 1 h to a Kodak Biomax MR film.
Detection of cytoplasmic capsids and secreted virions by the endogenous polymerase reactions.
To control transfection efficiencies, the HBsAg concentrations in a 1:200 dilution of the harvested media from cotransfected cells were measured by ELISA (Sorin Biomedica, Düsseldorf, Germany). A dilution series of a human HBsAg-positive serum with known HBsAg concentration was processed in parallel. The HBsAg concentration in the medium was usually between 80 and 300 ng/ml. Ten milliliters of undiluted harvested medium was used to dissolve 3.8 g of solid CsCl, placed in a Quick-Seal centrifuge tube (Beckman Instruments), overlaid with mineral oil, and spun for 40 h in a Ti70.1 rotor (Beckman Instruments) at 45,000 rpm and 20°C. The tube was cut open at the top, the oil was removed, and 6 ml of the gradient was taken from the top. The density at the surface of the remaining part of the gradient was measured by refractometry and was between 1.30 and 1.31 g/ml, ensuring that virions (density of 1.24 g/ml) were present in the collected part of the gradient, leaving naked nucleocapsids (density of 1.35 g/ml) in the lower part of the gradient, which was discarded. The collected part of the gradient was dialyzed against 1 liter of PBS for 4 h at room temperature. For immunoprecipitation of virions, 30 μl of swollen protein A-Sepharose CL4B beads (Pharmacia) suspended in 0.1 ml of PBS and preincubated overnight at 4°C with 3 μl of goat anti-HBV surface antigen (anti-HBs) (DAKO) was added, and the mixture was incubated for 4 h at room temperature with agitation. The beads were collected by a short spin, transferred to a reaction tube, and washed twice with 1 ml of PBS. After removal of all liquid with a thin glass capillary, 50 μl of 50 mM Tris-Cl (pH 7.5)–75 mM NH4Cl–1 mM EDTA–20 mM MgCl2–0.1% (vol/vol) β-mercaptoethanol–0.5% (vol/vol) Nonidet P-40–0.4 mM dATP–0.4 mM dGTP–0.4 mM dTTP–10 μCi of [α-32P]dCTP (3,000 Ci/mmol) was added for the endogenous polymerase reaction, which was performed at 37°C overnight. Then 50 μl of 1% (wt/vol) sodium dodecyl sulfate–10 mM Tris-Cl (pH 7.5)–10 mM EDTA–0.6 mg of proteinase K per ml–0.8 mg of tRNA per ml was added, followed by incubation at 37°C for 30 min to isolate the labeled viral genome. The mixture was extracted once with 0.1 ml of phenol-chloroform (1:1). Then the DNA was separated from unincorporated radioactive dCTP by precipitation with 25 μl of 10 M ammonium acetate and 250 μl of ethanol, incubation for 15 min at room temperature, and spinning for 15 min in a desktop microcentrifuge. The pellet was dissolved in 0.1 ml of Tris-EDTA buffer, and the DNA was precipitated again as described above. The pellet was dissolved in 10 μl of sample buffer and applied to a 1% (wt/vol) agarose–Tris-acetate-EDTA (TAE) gel. Electrophoresis was done for 90 min at 100 V. The gel was placed on a sheet of Whatman paper, dried by vacuum, and exposed to a Kodak Biomax MR film.
To compare the endogenous polymerase reaction from secreted virions to the reaction from cytoplasmic nucleocapsids, 1 ml of a cleared lysate (taken from 1.5 ml of cleared lysate prepared from a 10-cm-diameter culture dish) was transferred to a fresh tube and stored at −20°C for 2 days. (During this time, the medium was fractionated by CsCl gradient centrifugation so that both samples from medium and cells could be further processed in parallel). After thawing, 10 μl of swollen protein A-Sepharose CL4B beads suspended in 0.1 ml of PBS and prebound to 1 μl of anti-HBc/anti-HBe (DAKO) was added. Immunoprecipitation of capsids and further treatments were done in the same way as for the medium sample, with one exception: exogenous DNA was digested after the endogenous polymerase reaction and prior to the proteinase K treatment by adding 20 μl of 50 mM Tris-Cl (pH 7.5)–75 mM NH4Cl–1 mM EDTA–0.25 mg of DNase I (Boehringer, Mannheim, Germany) per ml and incubating the mixture at 37°C for 30 min.
Southern blotting.
A cleared cell lysate from three 10-cm-diameter culture dishes was prepared, and capsids were immunoprecipitated with 3 μl of anti-HBc/anti-HBe (DAKO) prebound to 30 μl of swollen protein A-Sepharose CL4B beads as described above. Exogenous DNA was digested by adding 50 μl of PBS containing 15 mM MgCl2 and 0.2 mg of DNase I per ml to the beads and incubating the mixture for 30 min at 37°C. Viral DNA was prepared by proteinase K digestion, phenol-chloroform extraction as described above, and ethanol precipitation. Electrophoresis was done in a 1% (wt/vol) agarose–TAE gel. Denaturation and Southern blotting were performed by capillary transfer as described in reference 25, using nylon membranes (Pall, Dreieich, Germany). An EcoRV-linearized unit-length HBV genome isolated by agarose gel electrophoresis was used as a probe. Direct labeling of the probe with alkaline phosphatase, hybridization, and washing were done as described in the protocol for the Amersham AlkPhos direct labeling kit. For comparison, 50 pg of HBV DNA isolated by proteinase K digestion and ethanol precipitation from a human serum was loaded into the gel without or with heat denaturation (5 min at 90°C). In addition, 10 pg of linearized unit-length HBV DNA isolated from EcoRV-treated plasmid pRVHBV1.5 was loaded without or with heat denaturation.
RESULTS
Experimental strategy.
Recently, we generated a panel of HBV core gene mutations which allowed core particle self-assembly by the core protein in the background of a bacterial expression vector (17). However, further phenotypic characterization of the mutants with respect to viral genome synthesis or virion formation is not possible in the prokaryotic system. Therefore, we introduced 11 of the mutations mapped to different regions of the core protein into a eukaryotic expression vector (Fig. 1B). These plasmids were used to complement a core-negative HBV genome (Fig. 1A) in trans in transient cotransfections of the human hepatoma cell line HuH7. This cell line supports replication of the virus when transfected with a terminal redundant linear copy of the viral DNA genome (Fig. 1A). The overlength core-negative HBV genome carries a stop codon at codon 38 of the core gene introduced by site-directed in vitro mutagenesis (Fig. 1A). This mutant HBV genome is blocked in the formation of nucleocapsids and virions but directs the expression of both nucleocapsids and virions (see Fig. 4) when complemented in trans with the expression vector for the WT core protein. The 11 core gene mutants were tested for the ability to form capsids and to allow viral DNA synthesis as well as secretion of virions by this complementation approach.
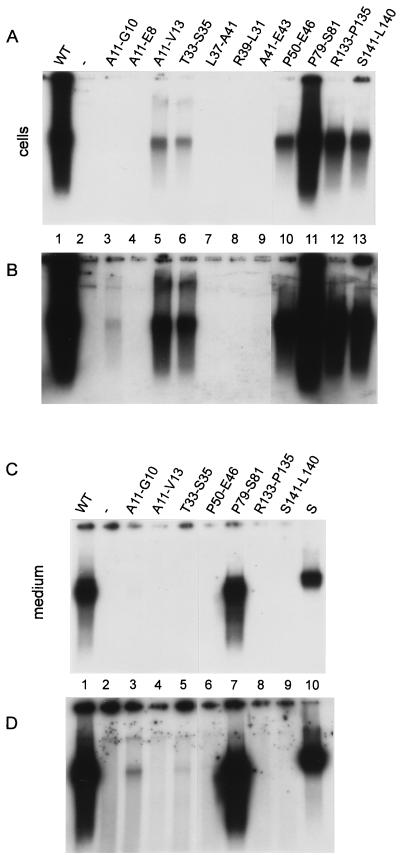
FIG. 4.
Proof of viral particle formation based on endogenous DNA polymerase activity. (A) Capsids from two-thirds of a cleared lysate prepared from cotransfected HuH7 cells from one 10-cm-diameter dish were immunoprecipitated with anti-HBc/anti-HBe. The viral genome was radioactively labeled by the endogenous polymerase activity, isolated, separated in an agarose gel, and visualized by autoradiography. Transfection of the core-negative HBV genome alone (Fig. 1A) gave no signal, as expected (lane 2). Cotransfection with the expression vector for the WT core protein (Fig. 1B) resulted in a strong reaction (lane 1). Results of cotransfections with mutant core genes are shown in lanes 3 to 13. Exposure was for 18 h. (B) Identical to panel A except that the exposure time was 9 days. (C) Virions from the medium of one 10-cm-diameter dish were separated from naked nucleocapsids by CsCl gradient centrifugation, immunoprecipitated with anti-HBs, and detected by the radioactive endogenous polymerase reaction. Only mutant P79-S81 (lane 7) showed a signal comparable to the WT signal (lane 1). In lane S, 2 × 106 labeled virions from a highly viremic human serum were loaded. Exposure was for 18 h. (D) Identical to panel B except that the exposure time was 9 days. Mutants A11-G10 (lane 3), T33-S35 (lane 5), and P50-E46 (lane 7) produced very low but detectable amounts of virions.
Detection of capsids.
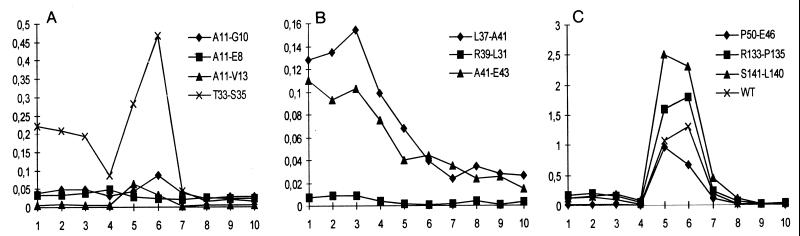
All 11 core mutants formed capsids in Escherichia coli (17). Because it was not certain whether this was also the case in eukaryotic cells, the mutants were tested again in the context of the trans complementation described above. A cleared cell lysate from cotransfected HuH7 cells was fractionated by sucrose gradient centrifugation and tested for HBcAg (Fig. 2). For the WT protein, a single peak appeared in the central fractions (Fig. 2C, fractions 5 and 6) of the gradient, indicative of capsids. Four mutants (T33-S35, P50-E46, R133-P135, and S141-L140) showed a similar profile, suggesting that they also formed capsids. Two further mutants (A11-G10 and A11-V13) generated weak peaks in the central fractions, indicating low amounts of capsids or capsids with reduced HBc antigenicity. Also in bacteria, mutant A11-G10 formed much lower amounts of HBcAg-positive capsids relative to the WT protein. However, mutant A11-V13 was phenotypically WT in E. coli (17). The analysis shows that the capacity of the mutants to assemble capsids in eukaryotic cells cannot be deduced from their phenotype in bacteria. This notion is even more evident from analysis of mutants A11-E8, L37-A41, R39-L31, and A41-E43, which showed no detectable capsid formation, in contrast to their behavior in bacteria (17). In the cases of variants A11-E8 and R39-L31, the whole gradient was HBcAg negative, possibly because the mutant proteins were unstable. In the cases of L37-A41 and A41-E43, the majority of the antigen was found in the upper fractions, probably formed by dimers and possibly higher complexes which smear into the gradient. Clearly, the assembly of these mutants was disturbed in HuH7 cells. Mutant P79-S81 was not tested in this assay. However, this mutant efficiently formed capsids in HuH7 cells, as became obvious from analysis of the endogenous polymerase activity and genome packaged into capsids (see below).
FIG. 2.
Detection of capsids by sucrose gradient sedimentation analysis. Cleared lysates from cotransfected cells (two 10-cm-diameter dishes each) were sedimented through 60 to 15% sucrose gradients. Ten fractions were taken from the top (top fraction is fraction 1) and analyzed for HBcAg. The ordinate indicates optical density of the HBcAg ELISA. The WT protein produced a single peak in fractions 5 and 6 (C), indicative of capsids. Mutant P79-S81 was not tested because it was HBcAg negative.
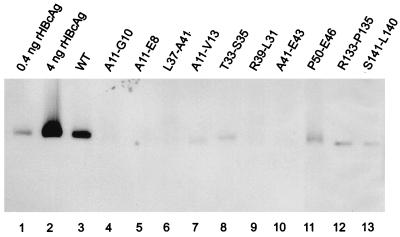
For capsid detection, we used a second assay which has also been used for the characterization of bacterially expressed proteins (17). Capsids from cleared lysates of cotransfected cells were concentrated by pelleting through a sucrose cushion and used for native agarose gel electrophoresis. Core particles running as a band through the gel were blotted onto a nitrocellulose membrane and detected by staining with antibodies (Fig. 3). Recombinant core particles (0.4 ng of core protein) loaded onto the gel resulted in a readily detectable band (Fig. 3, lane 1). The signal from WT-cotransfected HuH7 cells corresponded to approximately 1.5 ng of core protein or approximately 1.9 × 108 to 2.5 × 108 core particles. Mutants A11-G10, A11-E8, L37-A41, R39-L31, and A41-E43 showed no detectable signal, whereas mutants A11-V13, T33-S35, P50-E46, R133-P135, and S141-L140 generated core particles corresponding to approximately 0.2 to 0.4 ng of core protein. The findings are consistent with the analysis by sucrose gradient sedimentation (Fig. 2; Table 1). The only discrepancy (mutant A11-G10) is probably due to different sensitivities of the two assays.
FIG. 3.
Detection of capsids by native agarose gel electrophoresis and Western blotting. Capsids from cell lysates of one 10-cm-diameter dish of cotransfected HuH7 cells were concentrated by pelleting through a sucrose cushion, separated by electrophoresis through a native agarose gel, blotted onto a membrane, and stained with anti-HBc antibodies. For quantification, capsids purified from E. coli corresponding to 0.4 and 4 ng of recombinant core protein (rHBcAg) were loaded in lanes 1 and 2, respectively.
TABLE 1.
Genotypes and phenotypes of HBV core gene mutants
| Name | Mutation | Peptidea | Capsids in E. coli | Capsids in HuH7
|
Viral DNA synthesis | Virions | |
|---|---|---|---|---|---|---|---|
| Sucrose gradient | Agarose gel | ||||||
| WT | + | + | + | + | + | ||
| A11-G10 | Insertion | 11EF | + | +b | − | +b | +b |
| A11-E8 | Insertion | 11EFGA | +b | − | − | − | NDc |
| A11-V13 | Deletion | 12T | + | +b | + | + | − |
| T33-S35 | Deletion | 34A | + | + | + | + | +b |
| L37-A41 | Deletion | 38YRE | + | − | − | − | ND |
| R39-L31 | Insertion | 39LDTASALYR | + | − | − | − | ND |
| A41-E43 | Deletion | 42L | + | − | − | − | ND |
| P50-E46 | Insertion | 50EHCSP | + | + | + | + | +b |
| P79-S81 | Deletion | 80A | + | ND | ND | + | + |
| R133-P135 | Deletion | 134P | + | + | + | + | − |
| S141-L140 | Insertion | 141LS | + | + | + | + | − |
Position of mutation and peptide deleted or inserted (Fig. 1C).
Signal less than 1/10 of WT signal.
ND, not done.
Influence of core mutations on viral DNA synthesis.
To investigate whether the core gene mutations have an impact on pregenome/P protein packaging and DNA genome synthesis, we harvested capsids from cotransfected cells by cell lysis and immunoprecipitation with an anti-HBc/anti-HBe serum. The capsids were subsequently incubated with radioactivity labeled deoxynucleotides. For the WT, the viral DNA polymerase labeled the DNA with these substrates by filling in the single-stranded gap of the viral genome. The labeled genome could be visualized after isolation and agarose gel electrophoresis by autoradiography (Fig. 4). The four mutants showing no detectable particle formation in the former assays (Fig. 2 and 3) were also tested because the endogenous polymerase assay was more sensitive in our hands (compare the WT signal of the endogenous polymerase reaction in Fig. 4B, lane 1, with the WT signal of the Western blot in Fig. 3, lane 3).
All six mutants which allowed capsid formation (A11-G10, A11-V13, T33-S35, P50-E46, R133-P135, and S141-L140 [Fig. 2]) reacted positive in this assay (Fig. 4B, lanes 3, 5, 6, and 10 to 13). The signal was reduced relative to the WT signal (Fig. 4B, lane 1), e.g., for mutant A11-V13 (lane 5) approximately 25-fold (calculated by comparing autoradiographies with different exposure times [data not shown]). For mutant A11-G10, DNA synthesis activity became detectable only on longer exposures (Fig. 4B, lane 3). The signal reductions relative to WT reflected at least in part the reduced amounts of core particles formed by the mutants (compare with Fig. 2 and 3). A seventh mutant (P79-S81) which has not been tested directly for capsid formation produced a WT signal in the endogenous polymerase reaction (Fig. 4A, lane 11). We infer that this mutant also formed capsids because the DNA polymerase activity depends on the capsid structure.
For six of the seven mutants which allowed viral genome synthesis, the HBV DNA was isolated from intracellular capsids immunoprecipitated with anti-HBc/anti-HBe serum and analyzed by Southern blotting (Fig. 5). Results for mutant A11-G10 are not shown because the amount of viral DNA produced by this variant was below the detection limit of our Southern blot technique. The pattern of the viral DNA was similar to the WT pattern in all cases, with predominance of single-stranded DNA and growing strands as well as some partially double stranded DNA. Again, the amount of viral DNA was lower than the WT level and was roughly comparable to the relative levels obtained in the endogenous polymerase reaction (compare Fig. 5 with Fig. 4A). Apparently, the mutations did not induce a detectable block of a specific step in viral DNA synthesis, such as template switches or second-strand DNA synthesis.
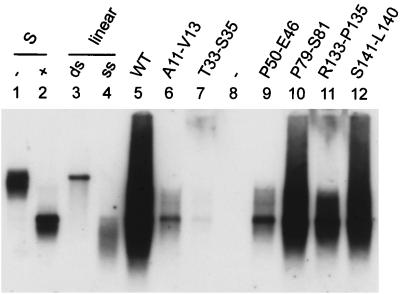
FIG. 5.
Southern blot analysis of viral DNA packaged into cytoplasmic nucleocapsids. Viral DNA was isolated from immunoprecipitated cytoplasmic capsids, using the cleared cell lysate from three 10-cm-diameter dishes of cotransfected HuH7 cells, and analyzed by Southern blotting. A labeled total HBV genome was used as a probe. In lane 1 and 2, 50 pg of HBV-DNA isolated from a viremic human serum was loaded in each lane without (−) or with (+) prior heat denaturation. In lane 3 and 4, 10 pg of linearized HBV DNA isolated from a plasmid was loaded in each lane without (ds) or with (ss) prior heat denaturation. In lane 8, cells were transfected with the core-negative HBV genome alone.
Influence of core mutations on virion formation.
The seven core gene mutants which assembled nucleocapsids were analyzed for the capacity to assemble virions. Transfected HuH7 cells release virions and also naked nucleocapsids into the culture medium. Therefore, virions were first separated from naked nucleocapsids by isopycnic CsCl gradient centrifugation. The gradient was divided into two fractions, using a border density of 1.30 to 1.31 g/ml. The lower, heavier fraction containing naked nucleocapsids (density of 1.35 g/ml) was discarded. Virions (density of 1.24 g/ml) were immunoprecipitated from the upper, lighter fraction after dialysis with an anti-HBs serum and quantified with the help of the endogenous polymerase reaction (Fig. 4C and D). Only mutant P79-S81 (Fig. 4C, lane 7) generated a WT signal (compare with lane 1). Three more mutants (A11-G10, T33-S35, and P50-E46) were able to form very low amounts of virions, as visible on longer exposures (Fig. 4D, lanes 3, 5, and 6). The ratio of the signals from intracellular capsids and extracellular virions was close to the WT ratio for mutant A11-G10 (Fig. 4B and D, lanes 3), drastically reduced for mutant T33-S35 (Fig. 4B [lane 6] and D [lane 5]), and even more reduced for mutant P50-E46 (Fig. 4B [lane 11] and D [lane 7]). Three mutants (A11-V13, R133-P135, and S141-L140), although functional in nucleocapsid morphogenesis as evident from the strong signal in the endogenous polymerase reaction (Fig. 4B, lanes 5, 13, and 14) and Southern blot (Fig. 5, lanes 6, 11, and 12), were blocked in the formation of detectable levels of virions (Fig. 4D, lanes 4, 8, and 9).
DISCUSSION
A genetic study of HBV core protein functions is hampered by the fact that mutations often lead to instability of the protein (31) or block early functions like capsid formation (2). We tried to circumvent this problem by generating and selecting at first a relatively large number of capsid-forming mutants in a prokaryotic system (17). In a second step, 11 variants carrying insertions or deletions in different regions of the protein which were able to assembly capsids in bacteria were randomly chosen. These variants were introduced into a eukaryotic expression vector and characterized in this work by cotransfection of a core-negative HBV genome in eukaryotic cells.
In an initial assay, we tested all mutants again for capsid formation in cotransfected HuH7 cells by using an ELISA technique identical to that employed for the characterization of mutants in bacteria (17). Surprisingly, 4 of the 11 mutants (A11-E8, L37-A41, R39-L31, and A41-E43) were not able to assemble detectable amounts of capsids in this setting, in contrast to their behavior in bacteria. Mutant A11-E8 was expressed in bacteria at levels that may be below the limit of detection in HuH7 cells. Expression of mutant R39-L31, which was close to the WT level in bacteria, was not measurable in HuH7 cells; mutant core proteins L37-A41 and A41-E43 were detectable in eukaryotic cells but blocked in capsid assembly, as evident from the antigen profile in the sucrose gradients (Fig. 2B). We have not determined the reasons for the discrepant results obtained in bacteria and HuH7 cells, but they may differ among various mutants. (i) One conceptual difference is that the core proteins were expressed in the absence and presence of other viral factors in bacteria and in HuH7 cells, respectively. It is known that the pregenome/P protein complex has a profound influence on nucleocapsid assembly (1, 13). (ii) Host factors are involved in capsid assembly and may act differently in prokaryotic and eukaryotic cells; in a cell-free eukaryotic protein expression system, a high-molecular-weight complex of core protein and a 60-kDa protein has been identified as an assembly intermediate (20). Also, a protein kinase not present in bacteria is packaged into nucleocapsids in eukaryotic cells (12, 15, 16). (iii) The local concentrations of core proteins have an influence on particle assembly (26) and are probably higher in the cytosol of bacteria than in HuH7 cells. However, it is difficult to compare local core protein concentrations in the two systems.
Seven mutants were compatible with capsid formation and could therefore be tested for further functions. All seven mutants supported viral DNA synthesis but to different extents. The amount of viral DNA roughly parallels the amount of particulate HBcAg (compare signals in Fig. 2 and 4A) except for mutant A11-V13. The reason for the different reaction of this mutant is unclear. Apparently, all seven mutations allowing particle assembly did not uncouple capsid formation from one of the numerous steps leading to the viral DNA genome, such as packaging of the pregenome/P protein complex, initiation of minus-strand DNA synthesis in the 5′ region of the pregenome, translocation of the DNA polymerase to the 3′ region of the pregenome, elongation of the DNA minus strand, and initiation and elongation of plus-strand synthesis. It is not clear from this limited study whether mutations in the assembly domain of the core protein (M1 to V149) are in principle not able to directly affect one of these steps, which would mean that this domain is not directly involved in viral DNA synthesis. Clearly, mutations in the arginine-rich C-terminal domain, which we have not analyzed, can have a direct impact on genome synthesis (21, 30).
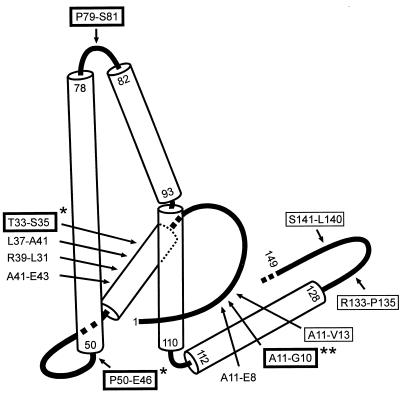
The seven mutants leading to detectable levels of cytoplasmic nucleocapsids were finally tested for virion formation and secretion (Fig. 4C and D). Only mutant P79-S81 produced virions at a level similar to the WT level. The mutation maps to the very tip of the spike protruding from the capsids and formed by a bundle of four alpha helices (7). This mutation alters the structure of the surface in such a way that antibodies of a human anti-HBc-positive, anti-HBe-negative serum did not react with this antigen (17). Apparently, the tip of the spike is not directly involved in the morphogenesis of virions and probably does not establish contacts to cytoplasmic domains of the HBV envelope proteins (5). It is therefore unlikely that peptides which have been found to bind to the tip of the core spikes and to inhibit virion assembly (3) directly interfered with subunit interfaces. It is more likely that these peptides blocked virion assembly by steric hindrance.
Two mutants (T33-S35 and P50-E46) were partially and three mutants (A11-V13, R133-P135, and S141-L140) were strongly blocked in nucleocapsid envelopment and virion secretion (Fig. 4D). The corresponding mutations map to different sites in the core protein fold; at least one site (P50-E46) is located at or near the inner surface of the capsid (Fig. 6) (4, 8). Therefore, it is unlikely that all of these mutations affect a single function of the core protein such as binding to an envelope protein domain. It is possible that some mutations influence the appearance of the envelopment signal which is linked to viral DNA synthesis and required for virion formation (10, 29) or change the intracellular trafficking of capsids. Mutations A11-V13, R133-P135, and S141-L140, strongly blocking virion formation, possibly map in proximity on the outer surface of the particle in the depressions between the spikes (Fig. 6) (4). These sites may be part of a core particle structure which binds to envelope proteins and initiates or drives budding (5).
FIG. 6.
Map of the analyzed mutation in the fold of the core protein. The fold of the core protein assembled into capsids proposed by Böttcher et al. (4) contains loops (lines) and alpha-helical regions (cylinders). The approximate positions of the mutations analyzed in this work are indicated by arrows. Names not framed refer to mutants showing capsid assembly in bacteria but no detectable capsid formation in HuH7 cells; names framed with a thin line indicate mutants allowing nucleocapsid but not virion formation; names framed with a thick line indicate mutants competent for virion morphogenesis. Numbers refer to amino acid positions. The C-terminal arginine-rich domain is not shown. ∗, the amount of secreted virions was less than 1/10 of WT, and the ratio of secreted virions to cytoplasmic nucleocapsids was lower than WT; ∗∗, the amount of cytoplasmic nucleocapsids and virions was less than 1/10 of WT, but the ratio of virions to nucleocapsids was relatively normal.
A striking finding of this study is that minimal alterations of the primary amino acid sequence can have dramatic effects on core protein functions. For example, mutants A11-G10, A11-E8, and A11-V13, carrying insertions of 2 and 4 aa and a deletion of 1 aa, respectively, at identical positions (C terminal of A11), have quite different phenotypes (Table 1): inserting the peptide EFGA lowered capsid formation in bacteria and prevented formation of capsids in eukaryotic cells. Insertion of the peptide EF allowed normal capsid formation in bacteria but formation of fewer capsids in eukaryotic cells, which, however, seemed to be functional in viral DNA genome synthesis and virion formation. And deletion of 1 aa (T12) led to reduced but easily detectable levels of nucleocapsids and blockage of virion formation.
Only 1 of the 11 mutants analyzed (P79-S81) appeared to be WT with respect to replication and particle formation, and even this mutant had an altered phenotype: it did not react with a human anti-HBc positive, anti-HBe-negative serum (17) because the main HBcAg epitope around A80 at the tip of the spike protruding from the capsid was changed. Therefore, in the end, none of the 11 tested mutants was WT with respect to all analyzed parameters. This finding emphasizes the sensitivity of core protein functions to mutations in this protein.
ACKNOWLEDGMENTS
We thank the diagnostic unit of the department for measuring HBsAg concentrations in media of transfected cells.
T.G. was supported by the Deutsche Akademische Austauschdienst. This work was supported by the Deutsche Forschungsgemeinschaft, SFB 402, project C2.
REFERENCES
- 1.Bartenschlager R, Schaller H. Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J. 1992;11:3413–3420. doi: 10.1002/j.1460-2075.1992.tb05420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beames B, Lanford R E. Insertions within the hepatitis B virus capsid protein influence capsid formation and RNA encapsidation. J Virol. 1995;69:6833–6838. doi: 10.1128/jvi.69.11.6833-6838.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Böttcher B, Tsuji N, Takahashi H, Dyson M R, Zhao S, Crowther R A, Murray K. Peptides that block hepatitis B virus assembly: analysis by cryomicroscopy, mutagenesis and transfection. EMBO J. 1998;17:6839–6845. doi: 10.1093/emboj/17.23.6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Böttcher B, Wynne S A, Crowther R A. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 5.Bruss V. A short linear sequence in the pre-S domain of the large hepatitis B virus envelope protein required for virion formation. J Virol. 1997;71:9350–9357. doi: 10.1128/jvi.71.12.9350-9357.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruss V, Ganem D. The role of envelope proteins in hepatitis B virus assembly. Proc Natl Acad Sci USA. 1991;88:1059–1063. doi: 10.1073/pnas.88.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conway J F, Cheng N, Zlotnick A, Stahl S J, Wingfield P T, Belnap D M, Kanngiesser U, Noah M, Steven A C. Hepatitis B virus capsid: localization of the putative immunodominant loop (residues 78 to 83) on the capsid surface, and implications for the distinction between c and e-antigens. J Mol Biol. 1998;279:1111–1121. doi: 10.1006/jmbi.1998.1845. [DOI] [PubMed] [Google Scholar]
- 8.Conway J F, Cheng N, Zlotnick A, Wingfield P T, Stahl S J, Steven A C. Visualization of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature. 1997;386:91–94. doi: 10.1038/386091a0. [DOI] [PubMed] [Google Scholar]
- 9.Gallina A, Bonelli F, Zentilin L, Rindi G, Muttini M, Milanesi G. A recombinant hepatitis B core antigen polypeptide with the protamine-like domain deleted self-assembles into capsid particles but fails to bind nucleic acids. J Virol. 1989;63:4645–4652. doi: 10.1128/jvi.63.11.4645-4652.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerelsaikhan T, Tavis J E, Bruss V. Hepatitis B virus nucleocapsid envelopment does not occur without genomic DNA synthesis. J Virol. 1996;70:4269–4274. doi: 10.1128/jvi.70.7.4269-4274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerhardt E, Bruss V. Phenotypic mixing of rodent but not avian hepadnavirus surface proteins into human hepatitis B virus particles. J Virol. 1995;69:1201–1208. doi: 10.1128/jvi.69.2.1201-1208.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerlich W H, Goldmann U, Muller R, Stibbe W, Wolff W. Specificity and localization of the hepatitis B virus-associated protein kinase. J Virol. 1982;42:761–766. doi: 10.1128/jvi.42.3.761-766.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirsch R C, Lavine J E, Chang L J, Varmus H E, Ganem D. Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as well as for reverse transcription. Nature. 1990;344:552–555. doi: 10.1038/344552a0. [DOI] [PubMed] [Google Scholar]
- 14.Huang Z M, Yen T S. Role of the hepatitis B virus posttranscriptional regulatory element in export of intronless transcripts. Mol Cell Biol. 1995;15:3864–3869. doi: 10.1128/mcb.15.7.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kann M, Thomssen R, Kochel H G, Gerlich W H. Characterization of the endogenous protein kinase activity of the hepatitis B virus. Arch Virol Suppl. 1993;8:53–62. doi: 10.1007/978-3-7091-9312-9_6. [DOI] [PubMed] [Google Scholar]
- 16.Kau J H, Ting L P. Phosphorylation of the core protein of hepatitis B virus by a 46-kilodalton serine kinase. J Virol. 1998;72:3796–3803. doi: 10.1128/jvi.72.5.3796-3803.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koschel M, Thomssen R, Bruss V. Extensive mutagenesis of the hepatitis B virus core gene and mapping of mutations that allow capsid formation. J Virol. 1999;73:2153–2160. doi: 10.1128/jvi.73.3.2153-2160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kunkel T A, Bebenek K, McClary J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 1991;204:125–39. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- 19.Lenhoff R J, Summers J. Coordinate regulation of replication and virus assembly by the large envelope protein of an avian hepadnavirus. J Virol. 1994;68:4565–4571. doi: 10.1128/jvi.68.7.4565-4571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lingappa J R, Martin R L, Wong M L, Ganem D, Welch W J, Lingappa V R. A eukaryotic cytosolic chaperonin is associated with a high molecular weight intermediate in the assembly of hepatitis B virus capsid, a multimeric particle. J Cell Biol. 1994;125:99–111. doi: 10.1083/jcb.125.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nassal M. The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J Virol. 1992;66:4107–4116. doi: 10.1128/jvi.66.7.4107-4116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nassal M. Hepatitis B virus morphogenesis. Curr Top Microbiol Immunol. 1996;214:297–337. doi: 10.1007/978-3-642-80145-7_10. [DOI] [PubMed] [Google Scholar]
- 23.Poisson F, Severac A, Hourioux C, Goudeau A, Roingeard P. Both pre-S1 and S domains of hepatitis B virus envelope proteins interact with the core particle. Virology. 1997;228:115–120. doi: 10.1006/viro.1996.8367. [DOI] [PubMed] [Google Scholar]
- 24.Salfeld J, Pfaff E, Noah M, Schaller H. Antigenic determinants and functional domains in core antigen and e antigen from hepatitis B virus. J Virol. 1989;63:798–808. doi: 10.1128/jvi.63.2.798-808.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook T, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Seifer M, Standring D N. Assembly and antigenicity of hepatitis B virus core particles. Intervirology. 1995;38:47–62. doi: 10.1159/000150414. [DOI] [PubMed] [Google Scholar]
- 27.Tuttleman J S, Pourcel C, Summers J. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell. 1986;47:451–460. doi: 10.1016/0092-8674(86)90602-1. [DOI] [PubMed] [Google Scholar]
- 28.Valenzuela P, Quiroga M, Zaldivar J, Gray R, Rutter W. The nucleotide sequence of the hepatitis B viral genome and the identification of the major viral genes. UCLA Symp Mol Cell Biol. 1980;18:57–70. [Google Scholar]
- 29.Wei Y, Tavis J E, Ganem D. Relationship between viral DNA synthesis and virion envelopment in hepatitis B viruses. J Virol. 1996;70:6455–6458. doi: 10.1128/jvi.70.9.6455-6458.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu M, Summers J. A domain of the hepadnavirus capsid protein is specifically required for DNA maturation and virus assembly. J Virol. 1991;65:2511–2517. doi: 10.1128/jvi.65.5.2511-2517.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan T T, Lin M H, Qiu S M, Shih C. Functional characterization of naturally occurring variants of human hepatitis B virus containing the core internal deletion mutation. J Virol. 1998;72:2168–2176. doi: 10.1128/jvi.72.3.2168-2176.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zlotnick A, Cheng N, Conway J F, Booy F P, Steven A C, Stahl S J, Wingfield P T. Dimorphism of hepatitis B virus capsids is strongly influenced by the C-terminus of the capsid protein. Biochemistry. 1996;35:7412–7421. doi: 10.1021/bi9604800. [DOI] [PubMed] [Google Scholar]