Abstract
Human immunodeficiency virus type 1 Gag protein is cotranslationally myristoylated at the N terminus and targeted to the plasma membrane, where virus particle assembly occurs. Particle assembly requires the ordered multimerization of Gag proteins, yet there is little direct evidence of intermediates of the reaction or of the domains that lead to each stage of the oligomerization process. In this study, following the expression in insect cells of C-terminally truncated Gag proteins and their purification, both the multimeric nature of each Gag protein and the ability to form Gag virus-like particles (VLP) were analyzed. Our results show that (i) the matrix (MA) domain forms a trimer and contributes to a similar level of oligomerization of the assembly-competent Gag; (ii) the p2 domain, located at the capsid/nucleocapsid junction, is essential for a higher order of multimerization (>1,000 kDa); (iii) the latter multimerization is accompanied by a change in Gag assembly morphology from tubes to spheres and results in VLP production; and (iv) N-terminal myristoylation is not required for either of the multimerization stages but plays a key role in conversion of these multimers to Gag VLP. We suggest that the Gag trimer and the >1,000-kDa multimer are intermediates in the assembly reaction and form before Gag targeting to the plasma membrane. Our data identify a minimum of three stages for VLP development and suggest that each stage involves a separate domain, MA, p2, or N-terminal myristoylation, each of which contributes to HIV particle assembly.
Retroviral Gag protein is the main structural component of the virus particle, and a number of expression studies have demonstrated that Gag protein alone is sufficient for the formation of Gag virus-like particles (VLP), the analogue of the authentic immature retroviral particles (7, 16, 23, 45, 48). Gag protein is initially synthesized in the cytosol as a precursor and then follows one of two morphogenetic pathways to the plasma membrane. In the case of type C retroviruses and lentiviruses such as human immunodeficiency virus (HIV), Gag proteins assemble after targeting to the plasma membrane since electron-dense patches of Gag are observed only at the extruded plasma membrane and not in the cytoplasm (15, 38). In contrast, Gag proteins of the B and D types of retrovirus form intracytoplasmic particles which then relocate to the plasma membrane (15, 28). Although these morphogenetic pathways differ, a common basic mechanism of Gag assembly appears likely, as in Mason-Pfizer monkey virus, these pathways can be readily interchanged by subtle mutations (44).
HIV Gag protein consists of four distinct structural domains, the N-terminal matrix domain (MA, p17), the central capsid domain (CA, p24), the nucleocapsid domain (NC, p7), and the C-terminal p6 domain (34). Each is produced by cleavage of the Gag precursor protein by the virion-encoded protease during the maturation process that occurs during or soon after virus particle budding (25, 51). Extensive amino acid deletion, substitution, and complementation experiments have identified which regions of Gag are required for VLP formation. An N-terminal signal combining myristoylation and a region of positive charge is essential for Gag targeting to the plasma membrane, since nonmyristoylated Gag protein obtained by amino acid substitution at the N-terminal glycine, the acceptor site of the myristoyl moiety, failed to produce Gag VLP from the cell surface (4, 16, 18). The N-terminal myristoyl residue is not directly involved in Gag-Gag interaction, however, as nonmyristoylated Gag protein may be rescued into budding Gag VLP by a relatively low level of myristoylated Gag protein (35, 39, 48). A dominant assembly region has been mapped in the C-terminal third of the CA domain that includes the p2 domain which is located at the CA/NC junction (3, 10, 42, 57). The C-terminal region of the CA domain contains the major homology region, where sequence identity between various retrovirus gag genes is the highest (33). Another assembly region has been identified in the central region of the MA domain in a number of studies in which subtle mutations in the region abolished Gag VLP formation (13, 36). In some experimental systems, notably those that express high levels of protein, most of NC and the entire p6 domain have been found to be dispensable for Gag VLP production (16, 23, 24, 49). In other studies, the NC domain has been found to be essential for Gag assembly (6, 9, 26, 53). These apparently conflicting data may be resolved by the NC acting to effectively concentrate Gag protein around an RNA molecule, as it is clear that the determinant for genomic RNA packaging resides in NC (2, 8, 17, 46, 47), although it is also possible that overexpression of Gag protein reveals nonphysiological interactions. A similar set of assembly regions was originally identified as a minimal assembly mechanism for Rous sarcoma virus Gag protein (7, 53).
Despite the mapping of the sequence determinants for Gag VLP formation, their precise roles in the overall process and the stage at which each acts remain uncertain. These regions must be involved in Gag-Gag interactions in the process of Gag assembly. To address these issues, we have purified soluble HIV Gag proteins with a series of C-terminal truncations from expressing cells and analyzed their multimeric state by velocity sedimentation analysis as well as their ability to form VLP. On the basis of our results, we suggest that three discrete events led by each of three separate Gag domains or determinants (MA, p2, and the N-terminal myristoyl residue) sequentially contribute to Gag assembly.
MATERIALS AND METHODS
Construction and expression of a truncated HIV-1 gag gene.
Truncation of the HIV-1 gag gene was carried out by PCR using a forward primer (5′-CGCGGGATCCATGGGTGCGAGAGCGTCAGT-3′) and reverse primers containing an additional six histidine residues at the C termini, 5′-CGCGGAATTCAATGATGATGATGATGATGGTAATTTTGGCTGACCTGACT-3′ (for the MA domain), 5′-CGCGGAATTCAATGATGATGATGATGATGCAAAACTCTTGCCTTATGGCC-3′ (for the MA-CA polyprotein), 5′-CGCGGAATTCAGTGGTGGTGGTGGTGGTGCATTATGGTAGCTGTATTTGTTACT-3′ (for the MA-CA-p2 polyprotein), 5′-CGCGGAATTCAGTGGTGGTGGTGGTGGTGCTTAACCATCTTTCTTTGGTTCC-3′ (for the MA-CA-p2-NCdl1 polyprotein truncated just before the upstream zinc finger motif), 5′-CGCGGAATTCAATGATGATGATGATGATGGCCCTTTTTCCTAGGGGCC-3′ (for the MA-CA-p2-NCdl2 polyprotein truncated just before the downstream zinc finger motif), and 5′-CGCGGAATTCTCAATGATGATGATGATGATGATTAGCCTGTCTCTCAGT-3′ (for the MA-CA-p2-NC polyprotein). For nonmyristoylated Gag constructs, replacement of the N-terminal glycine with alanine was done by PCR using 5′-CGCGGGATCCATGGCTGCGAGAGCGTCAG-3′ as a forward primer. The PCR fragments were cloned into the baculovirus transfer vector pAcCL29-1 (32), and recombinant baculoviruses were obtained by standard procedures.
Virus and cells.
Spodoptera frugiperda (Sf9) cells were propagated at 27°C in TC-100 medium supplemented with 10% fetal bovine serum (FBS). For protein expression, cells were infected with recombinant Autographa californica nuclear polyhedrosis viruses (baculoviruses) containing the truncated HIV type 1 (HIV-1) gag genes at a multiplicity of infection of 2 and cultured for 2 days.
Purification of Gag VLPs and soluble Gag proteins.
Gag VLPs were purified from culture media of Sf9 cells expressing the truncated Gag proteins as described previously (35). Briefly, the culture media were clarified and then centrifuged through 30% (wt/vol) sucrose cushions at 4°C at 24,000 rpm for 2 h. The VLP pellets were resuspended with phosphate-buffered saline and centrifuged on 20 to 60% (wt/vol) sucrose gradients at 4°C at 35,000 rpm overnight. Gag VLPs were obtained by fractionation of the gradients. Soluble Gag proteins were purified from the Sf9 cells as follows. Cells were suspended in binding buffer (20 mM Tris [pH 7.9], 150 mM NaCl, 10 mM imidazole) and disrupted by sonication and the addition of Nonidet P-40 to a final concentration of 0.2%. After centrifugation at 4°C at 15,000 rpm for 30 min, the supernatant was subjected to immobilized metal chelate chromatography (Novagen). Following washes with 25 volumes of binding buffer and 20 volumes of wash buffer (20 mM Tris [pH 7.9], 150 mM NaCl, 60 mM imidazole), bound protein was eluted with 5 volumes of elute buffer (20 mM Tris [pH 7.9], 150 mM NaCl, 1 M imidazole).
Pulse-chase experiment.
Sf9 cells expressing the truncated Gag proteins were metabolically labeled with [35S]methionine-cysteine mixture (50 μCi/ml; New England Nuclear/Du Pont) for 10 min. After the pulse-labeling, the cells were washed with an excess volume of TC-100 medium containing 50-fold-concentrated methionine and cysteine and 10% FBS and then incubated in TC-100 medium containing 10% FBS for the time indicated in the text.
Velocity sedimentation analysis.
Purified Gag proteins were applied onto 15 to 30% (vol/vol) glycerol gradients including 20 mM Tris [pH 7.4]), 100 mM NaCl, 1 mM dithiothreitol, and 0.5 mM EDTA in SW55 tubes and sedimented at 4°C at 48,000 rpm for 40 h (for the MA domain) or 20 h (for the other constructs). To analyze higher orders of Gag multimers, Gag proteins were applied to 20 to 70% (wt/vol) sucrose gradients in phosphate-buffered saline in SW55 tubes and sedimented at 4°C at 35,000 rpm for 3 h. High- and low-molecular-weight calibration kits (Amersham Pharmacia Biotech) were used for sedimentation molecular weight markers.
Protein detection.
Following sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), gels were either stained with Coomassie brilliant blue or subjected to Western blotting (50) using anti-HIV-1 CA (MRC AIDS reagent repository, United Kingdom) and antipolyhistidine monoclonal antibodies (Sigma). For pulse-chase experiment, the gels were subjected to fluorography.
Electron microscopic examination.
Sf9 cells expressing the truncated Gag proteins were fixed in 2.5% glutaraldehyde in 100 mM cacodylate buffer (pH 7.2) and postfixed with 1% osmium tetroxide in 100 mM cacodylate buffer (pH 7.2). The procedure for scanning electron microscopy was described elsewhere (22).
RESULTS
Expression of Gag protein with C-terminal truncation and formation of Gag VLP.
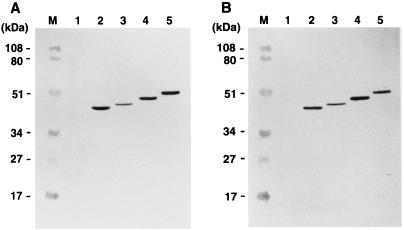
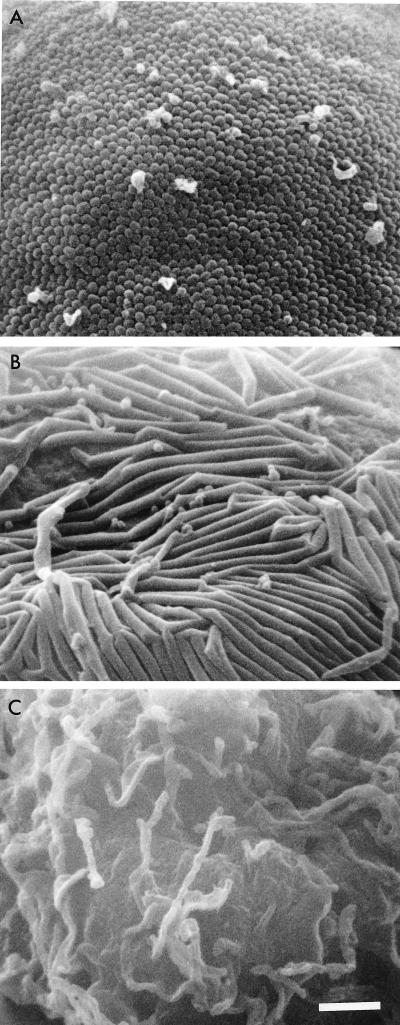
To confirm the minimum C-terminal boundary of the Gag protein necessary for the production of VLP, Gag proteins with a series of C-terminal truncations, each tagged at the C terminus with six histidine residues, were expressed in Sf9 cells by using recombinant baculoviruses. Following expression, Gag VLPs, if present, were harvested from the culture media, and the antigens present were detected by Western blotting using monoclonal antibodies directed to HIV-1 CA (Fig. 1A) and the polyhistidine tag (Fig. 1B). As reported previously (16, 23, 24, 45), any C-terminal truncation that extended beyond the p2 domain abolished Gag VLP production whether detected by anti-HIV-1 CA (Fig. 1A) or antitag (Fig. 1B) antibody. As the six-histidine tags were detected in the Gag VLP fractions and the endpoints for VLP formation were similar to those previously mapped, the presence of the tag clearly had no detrimental effect on Gag VLP production. Other, larger C-terminal extensions to Gag have also been shown not to prevent VLP formation (54). Scanning electron microscopy of the expressed cell surface further confirmed these findings. A number of spherical budding particles were observed at the surface of the cells expressing MA-CA-p2 (Fig. 2A); in contrast, the expression of MA-CA resulted in only long tubular structures protruding from the plasma membrane (Fig. 2B). Thus, the p2 domain at the CA/NC junction is essential for the Gag curvature that gives rise to spherical particles.
FIG. 1.
Detection of Gag VLPs. Gag VLPs were purified from culture media of Sf9 cells expressing Gag with the C-terminal truncations described and subjected to Western blotting using anti-HIV-1 CA (A) and antipolyhistidine (B) monoclonal antibodies. Lanes: M, prestained molecular weight markers (Bio-Rad); 1 to 5, Gag VLP fractions purified from the supernatant of Sf9 cells expressing MA-CA, MA-CA-p2, MA-CA-p2-NCdl1, MA-CA-p2-NCdl2, and MA-CA-p2-NC, respectively.
FIG. 2.
Scanning electron micrographs of Sf9 cells expressing Gag with C-terminal truncations or mock infected. All micrographs are at same magnification; scale bar = 1 μm. (A) Sf9 cells expressing MA-CA-p2; (B) Sf9 cells expressing MA-CA; (C) mock-infected Sf9 cells.
Effect of C-terminal truncations on the multimeric form of Gag protein.
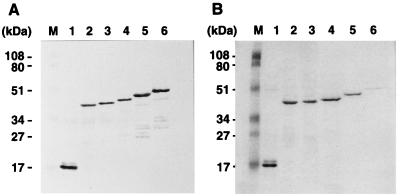
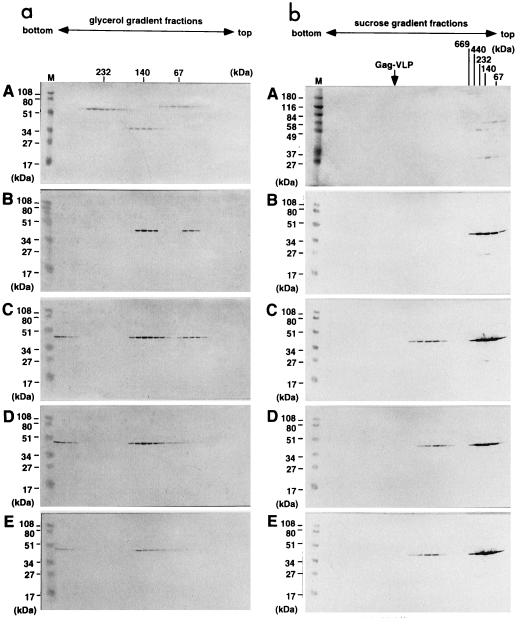
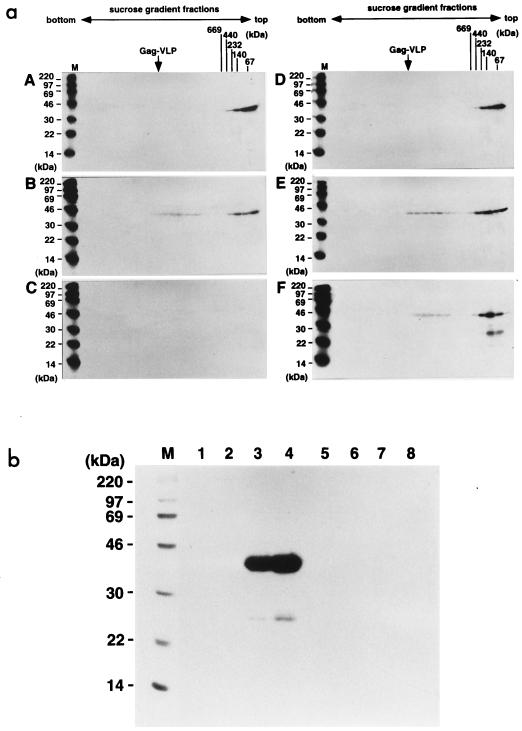
To examine the multimeric nature of the Gag protein expressed by each of the truncated Gag constructs, soluble Gag proteins were purified from the expressed Sf9 cells lysate by use of the polyhistidine tag (Fig. 3). The expression levels of each truncated Gag protein in the cells were broadly equivalent (Fig. 3A) but the yields of purified Gag protein varied (Fig. 3B), most probably due to inaccessibility of the polyhistidine tag to the metal chelate resin. Purified Gag proteins were adjusted to approximately 0.5 mg/ml and subjected to velocity sedimentation analysis. Following centrifugation, fractions containing Gag proteins were detected by Western blotting and compared to molecular mass markers sedimented in parallel. When MA-CA was analyzed on a 15 to 30% glycerol gradient, Gag antigen was detected in fractions corresponding to molecular masses of 40 to 50 and 120 to 150 kDa, equivalent to the monomer and trimer, respectively (Fig. 4a, panel B). The spread of antigen within the gradient did not allow us to rule out the possibility that the higher-molecular-weight form detected was a tetrameric rather than trimeric molecule. In contrast, when the Gag proteins containing the p2 domain (MA-CA-p2, MA-CA-p2-NCdl1, and MA-CA-p2-NCdl2) were analyzed similarly, Gag-reactive antigen was detected in the bottom fractions of the gradients in addition to the antigen in the previously identified fractions (Fig. 4a, panels C to E). The molecular masses of these large Gag multimers were determined by subsequent sedimentation analysis on 20 to 70% sucrose gradients where Gag antigen was present in fractions corresponding to molecular masses greater than 1,000 kDa (Fig. 4b, panels C to E). These results indicate that (i) MA, CA, or a combination of the two allows oligomerization of Gag protein to the level of the Gag trimer (or possibly tetramer) but not to any higher order of multimers detectable by these techniques and (ii) the inclusion of the p2 region at the C terminus of CA allows Gag to multimerize to a state considerably higher than the trimer and is paralleled by the appearance of VLP in the culture supernatant.
FIG. 3.
Expression and purification of soluble Gag proteins from Sf9 cells. (A) Intracellular expression levels of Gag with C-terminal truncations. Sf9 cells expressing each truncated Gag protein were lysed and directly subjected to SDS-PAGE followed by Western blotting using an antipolyhistidine monoclonal antibody (Sigma). (B) Purification yields of Gag with C-terminal truncations. Following disruption of Sf9 cells expressing each truncated Gag protein, soluble Gag proteins were purified from the clarified cell lysates by metal chelate chromatography and subjected to SDS-PAGE followed by staining with Coomassie brilliant blue. Lanes: M, prestained molecular weight markers (Bio-Rad); 1 to 6, MA, MA-CA, MA-CA-p2, MA-CA-p2-NCdl1, MA-CA-p2-NCdl2, and MA-CA-p2-NC, respectively.
FIG. 4.
Sedimentation profiles of soluble Gag proteins on glycerol and sucrose gradients. Following purification, soluble Gag proteins with C-terminal truncations were subjected to velocity sedimentation analysis either on 15 to 30% glycerol gradients to separate low-molecular-weight oligomers (a) or on 20 to 70% sucrose gradients to analyze a higher order of multimer (b). Fractions from the bottom to the top (left to right) were subjected to SDS-PAGE followed by Western blotting using an anti-HIV-1 CA monoclonal antibody. A, high-molecular-weight calibration markers consisting of thyroglobulin (669 kDa = 2 × 330 kDa), ferritin (440 kDa = 2 × 220 kDa), catalase (232 kDa = 4 × 60 kDa), lactate dehydrogenase (140 kDa = 4 × 36 kDa), and albumin (67 kDa) (Amersham Pharmacia Biotech), stained with Coomassie brilliant blue; B, MA-CA; C, MA-CA-p2; D, MA-CA-p2-NCdl1, E, MA-CA-p2-NCdl2, detected by Western blotting. Lanes M show molecular weight markers (Bio-Rad) for SDS-PAGE, and an arrow marks the sedimented position of Gag VLP. Note a higher order of multimer less sedimented than Gag VLP.
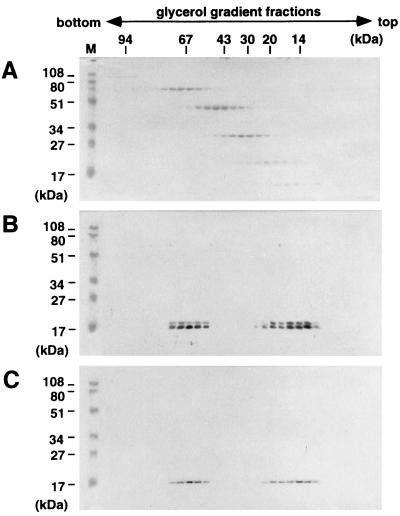
Recently, we and others have shown that MA purified from Escherichia coli cells could be detected as a trimer (21, 37). The traditional view of Gag assembly, however, has been that oligomerization occurs after targeting to the plasma membrane and is dependent on Gag N-terminal myristoylation (4, 16, 18). As Gag protein expressed in E. coli is not myristoylated because of the lack of N-myristoyltransferase activity in E. coli (11), these findings suggest that partial assembly of Gag could occur before membrane localization. To assess whether N-terminal myristoylation plays a role in such partial assembly, wild-type MA and nonmyristoylated MA(G2A), obtained by replacement of the N-terminal glycine with alanine, were expressed by using recombinant baculoviruses, purified from Sf9 cells as before, and analyzed on velocity gradients. Following sedimentation through a 15 to 30% glycerol gradient, both forms of MA were detected at molecular masses of 17 and 51 kDa, equivalent to monomeric and trimeric forms, respectively (Fig. 5). No significant differences between the monomer/trimer ratios of the preparations were observed (compare Fig. 5B and C). These results confirm that the oligomerization of MA, and possibly that of any larger Gag precursor, can proceed to the level of the trimer in the absence of N-terminal myristoylation.
FIG. 5.
Sedimentation profiles of myristoylated and nonmyristoylated MA domains. Purified MA proteins were layered onto 15 to 30% glycerol gradients and centrifuged at 48,000 rpm for 40 h. Fractions from the bottom to the top (left to right) were subjected to SDS-PAGE, and proteins were detected by Western blotting using an antipolyhistidine monoclonal antibody (Sigma). (A) Low-molecular-weight calibration markers for sedimentation (Amersham Pharmacia Biotech) stained with Coomassie brilliant blue; (B) wild-type MA; (C) nonmyristoylated MA(G2A), detected by Western blotting. Lane M shows prestained molecular weight markers (Bio-Rad) for SDS-PAGE.
Role of N-terminal myristoylation in Gag multimerization.
To assess the stage at which N-terminal myristoylation of Gag becomes essential for VLP formation, wild-type MA-CA-p2 (VLP formation competent) and the nonmyristoylated form MA(G2A)-CA-p2 were expressed by using recombinant baculoviruses, and Gag proteins were metabolically pulse-labeled with a [35S]methionine-cysteine mixture for 10 min followed by a chase for 3 or 6 h. Soluble Gag protein was purified from cells at the end of each chase period and analyzed by sedimentation analysis on sucrose gradients. The higher order of Gag multimer (>1,000 kDa) was detected for both forms of Gag protein after a 3-h chase period, suggesting that Gag multimerization to this level occurs within 3 h and does not require N-terminal myristoylation (Fig. 6a, panels B and E). Following a 6-h chase, however, striking differences were observed between the myristoylated and nonmyristoylated forms of Gag protein. No labeled Gag proteins were detected in the sedimentation profile of the myristoylated Gag (Fig. 6a, panel C); in contrast, the nonmyristoylated Gag protein was detected in the same fractions as those observed at the 3-h chase point although some degradation, not present in the 3-h sample, was visible after the 6-h chase (Fig. 6a, panel F). These results suggest that in the case of the myristoylated form of Gag, all labeled molecules had left the >1,000-kDa intracellular pool by 6 h postlabeling and were unavailable for purification from the cytosol by C-terminal affinity chromatography. When the presence of Gag VLPs in the culture media was examined at each chase time, VLPs of the wild-type Gag were detected at the 6-h chase, when no labeled Gag protein was purified from the cells (Fig. 6b, lanes 1 to 4). As expected, no Gag VLPs were released from the cells expressing the nonmyristoylated Gag throughout the chase periods (Fig. 6b, lanes 5 to 8). These findings show that Gag proteins, whether myristoylated or not, can assemble to oligomers and to >1,000-kDa multimers prior to targeting to the plasma membrane. However, N-terminal myristoylation is necessary for the further assembly of Gag to form VLP, the 600S form identified by Royer et al. (45).
FIG. 6.
Time courses of multimerization for myristoylated and nonmyristoylated Gag proteins and incorporation into Gag VLPs. Sf9 cells expressing wild-type MA-CA-p2 and nonmyristoylated MA(G2A)-CA-p2 proteins were pulse-labeled with a [35S]methionine-cysteine mixture for 10 min and then chased for the indicated time with an excess of unlabeled amino acids. (a) Sedimentation profiles of the soluble Gag proteins purified from the cells at each chase period. Following sedimentation, fractions of 20 to 70% sucrose gradients were collected from the bottom to the top (left to right) and subjected to SDS-PAGE followed by fluorography. A to C, wild-type MA-CA-p2 protein; D to F, nonmyristoylated MA(G2A)-CA-p2 protein. A and D, 0-h chase; B and E, 3-h chase; C and F, 6-h chase. Positions of high-molecular-weight calibration markers (described in the legend to Fig. 4) sedimented in parallel and of wild-type Gag VLP (arrow) are indicated. Lanes M show 14C-labeled molecular weight markers for SDS-PAGE (Amersham Pharmacia Biotech). (b) Time course of Gag VLP production. Gag VLPs were purified from the culture media at each chase point and analyzed by SDS-PAGE followed by fluorography. Lanes: M, 14C-labeled molecular weight markers for SDS-PAGE (Amersham Pharmacia Biotech); 1 to 4, Gag VLP fractions of wild-type MA-CA-p2; 5 to 8, Gag VLP fractions of nonmyristoylated MA(G2A)-CA-p2; 1 and 5, 0-h chase; 2 and 6, 3-h chase; 3 and 7, 6-h chase; 4 and 8, 9-h chase.
DISCUSSION
A number of studies of Gag VLP formation using a combination of mutagenesis and expression have shown that mutations in several discrete regions of Gag result in failure to form Gag VLP. Although these studies mapped the regions required for Gag VLP formation, the precise stage in the assembly reaction affected was rarely determined. In this study, we have shown that intermediates are present during Gag assembly and that three discrete domains or determinants, MA, p2, and the N-terminal myristoyl residue, play distinct roles in VLP development.
The trimeric nature of the MA domain was originally observed following crystallographic studies which suggested that the hydrophobic core of the molecule was responsible for trimer formation (21, 41). This was also confirmed by a recent report that both MA and MA-CA were present as a trimer in solution (37). As neither MA or MA-CA alone forms VLP, however, there was some uncertainty as to the role of the trimer in a true VLP assembly reaction. The data presented here, for larger Gag molecules which are VLP competent, suggest, but do not prove, that Gag precursors form trimers and that this oligomer is one of the assembly intermediates in the process of VLP formation. A higher order of Gag multimer with a molecular mass of >1,000 kDa appears to be another assembly intermediate, and multimerization to this level was dependent on the presence of p2 region at the C terminus of CA. No other discrete size classes of intermediate were detected in our studies.
A number of studies have shown that mutations introduced in the C-terminal third of the CA domain or the p2 domain impair Gag VLP production (9, 42, 57). This finding and our data are consistent with earlier studies showing that in Gag ligand affinity assays, the strongest Gag-Gag interactions occur in the p2-to-NC region whereas the C-terminal part of the CA domain (downstream of the major homology region) shows low-affinity Gag-Gag interactions (6, 57). The C-terminal part of CA lacking p2 has been crystallized recently and shown to form a CA dimer (14). The C-terminal domain of CA including the p2 peptide has also been crystallized recently, although the p2 region remained disordered in the determined structure (56). A modeled complete capsid confirms a dimeric structure for CA and highlights the flexibility of the CA domain to allow it to adopt a range of relative orientations (56). We suggest that the p2 domain may influence the flexibility of CA (and probably that of NC) in the context of Gag precursor and act to trigger additional interactions between the Gag molecules, resulting in a higher order of Gag multimerization. An alternative possibility is that the p2 region itself drives multimerization to the >1,000-kDa intermediate in the process of Gag precursor assembly and that the CA dimer interface observed in the crystallographic structure is created only after p2 is removed during the maturation process. In favor of this hypothesis, epitope scanning using quantitative immunoelectron microscopy has shown that the region from p2 to NC is occluded within immature Gag VLP whereas the C-terminal part of the CA domain is relatively exposed (6).
Electron microscopy of cells expressing the various Gag truncations showed that lack of the p2 region result in the production of tubular forms, but no particles were formed (Fig. 2). A deletion of the p2 peptide in the context of complete Gag, in a proviral clone, also led to severe reduction in virus yield and extracellular particles with aberrant morphology that showed tubular and bent electron-dense cores (27). In addition, in vitro assembly studies with purified CA domain alone yielded long tubular structures (19, 20, 52). Interestingly, extension of the CA domain at the N terminus by a short region of MA converted in vitro assembly products from tubes to spheres (19, 52). As the p2 phenotype in our experiments was similar, it is possible that extension of the CA domain at either the N or C terminus affects conformation to allow a spherical assembly phenotype. Consistent with this, CA-p2 obtained by mutation of the CA/p2 cleavage site in a proviral clone gave rise to spherical capsids (1, 55). Although in vitro assembly with CA-p2-NC in the presence of RNA yielded only tubular structures (5, 20), assembly with CA-p2 has not been examined.
Our study showed that neither oligomerization nor assembly to the higher-order (>1,000-kDa) form of Gag required N-terminal myristoylation. Moreover, the ratios of oligomer to monomer and of the higher-order form to oligomer plus monomer were nearly identical for the nonmyristoylated and myristoylated Gag molecules (Fig. 6a). This suggests that both the oligomerization and the higher-order multimerization of Gag are not simply due to the accumulation of nonmyristoylated Gag molecules in the cytoplasm of expressing cells but rather that these multimerizations are in equilibrium and may occur before targeting to the plasma membrane. Similar conclusions regarding the occurrence of Gag multimer formation prior to membrane localization have been made based on the observation of Gag-expressing cells (40) and on the detergent sensitivity of Gag complexes which have suggested a detergent-resistant complex (DRC) of Gag in the cytosol and a detergent-sensitive complex (DSC) at the membrane (30, 31). We speculate that the DRC form equates to the >1,000-kDa multimer observed in this work whereas the DSC form equates to the VLP-competent form. In our experiments, N-terminal myristoylation was required for VLP formation from the Gag assembly intermediates. As the myristoyl group confers upon Gag binding to the plasma membrane, it is likely that the final stages of Gag assembly are facilitated by the concentration of Gag proteins on the plasma membrane as suggested by Nermut et al. (38).
A plausible list of assembly events in authentic VLP development could be Gag oligomerization such as trimerization followed by the higher order (>1,000 kDa) of multimerization in the cytosol and final multimerization to 600S of VLP at the plasma membrane, although the sequential nature of these steps is not proven by our data. Since several large and even entire deletions of the MA domain within the Gag precursor have been shown to have little effect on and even to enhance Gag VLP formation (12, 29, 43), in some situations the step could be bypassed because of other dominant assembly domains within Gag precursor. It is likely that a level of redundancy may be involved in HIV assembly, perhaps ensuring some allowance for virus assembly in a variety of cell types and with moderate sequence variation.
ACKNOWLEDGMENTS
Anti-HIV-1 CA monoclonal antibody was kindly provided by the MRC AIDS reagent repository, United Kingdom.
This work was supported by a grant from Ministry of Health and Welfare, Japan, by a grant from the MRC, and by a grant from Novartis Foundation for the Promotion of Science.
REFERENCES
- 1.Accola M A, Hoglund S, Gottlinger H G. A putative alpha-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J Virol. 1998;72:2072–2078. doi: 10.1128/jvi.72.3.2072-2078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berkowitz R D, Luban J, Goff S P. Specific binding of human immunodeficiency virus type 1 Gag polyprotein and nucleocapsid protein to viral RNAs detected by RNA mobility shift assays. J Virol. 1993;67:7190–7200. doi: 10.1128/jvi.67.12.7190-7200.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borsetti A, Ohagen A, Gottlinger H G. The C-terminal half of the human immunodeficiency virus type 1 Gag precursor is sufficient for efficient particle assembly. J Virol. 1998;72:9313–9317. doi: 10.1128/jvi.72.11.9313-9317.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryant M, Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci USA. 1990;87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell S, Vogt V M. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carriere C, Gay B, Chazal N, Morin N, Boulanger P. Sequence requirements for encapsidation of deletion mutants and chimeras of human immunodeficiency virus type 1 Gag precursor into retrovirus-like particles. J Virol. 1995;69:2366–2377. doi: 10.1128/jvi.69.4.2366-2377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craven R C, Parent L J. Dynamic interactions of the Gag polyprotein. Curr Top Microbiol Immunol. 1996;214:65–94. doi: 10.1007/978-3-642-80145-7_3. [DOI] [PubMed] [Google Scholar]
- 8.Dannull J, Surovoy A, Jung G, Moelling K. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 1994;13:1525–1533. doi: 10.1002/j.1460-2075.1994.tb06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dawson L, Yu X F. The role of nucleocapsid of HIV-1 in virus assembly. Virology. 1998;251:141–157. doi: 10.1006/viro.1998.9374. [DOI] [PubMed] [Google Scholar]
- 10.Dorfman T, Bukovsky A, Ohagen A, Hoglund S, Gottlinger H G. Functional domains of the capsid protein of human immunodeficiency virus type 1. J Virol. 1994;68:8180–8187. doi: 10.1128/jvi.68.12.8180-8187.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duronio R J, Jackson-Machelski E, Heuckeroth R O, Olins P O, Devine C S, Yonemoto W, Slice L W, Taylor S S, Gordon J I. Protein N-myristoylation in Escherichia coli: reconstitution of a eukaryotic protein modification in bacteria. Proc Natl Acad Sci USA. 1990;87:1506–1510. doi: 10.1073/pnas.87.4.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Facke M, Janetzko A, Shoeman R L, Krausslich H G. A large deletion in the matrix domain of the human immunodeficiency virus gag gene redirects virus particle assembly from the plasma membrane to the endoplasmic reticulum. J Virol. 1993;67:4972–4980. doi: 10.1128/jvi.67.8.4972-4980.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freed E O, Orenstein J M, Buckler-White A J, Martin M A. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J Virol. 1994;68:5311–5320. doi: 10.1128/jvi.68.8.5311-5320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gamble T R, Yoo S, Vajdos F F, von Schwedler U K, Worthylake D K, Wang H, McCutcheon J P, Sundquist W I, Hill C P. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 15.Gelderblom H R, Ozel M, Pauli G. Morphogenesis and morphology of HIV. Structure-function relations. Arch Virol. 1989;106:1–13. doi: 10.1007/BF01311033. [DOI] [PubMed] [Google Scholar]
- 16.Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, de Wilde M. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 17.Gorelick R J, Chabot D J, Rein A, Henderson L E, Arthur L O. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J Virol. 1993;67:4027–4036. doi: 10.1128/jvi.67.7.4027-4036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gottlinger H G, Sodroski J G, Haseltine W A. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gross I, Hohenberg H, Huckhagel C, Krausslich H G. N-terminal extension of human immunodeficiency virus capsid protein converts the in vitro assembly phenotype from tubular to spherical particles. J Virol. 1998;72:4798–4810. doi: 10.1128/jvi.72.6.4798-4810.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gross I, Hohenberg H, Krausslich H G. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur J Biochem. 1997;249:592–600. doi: 10.1111/j.1432-1033.1997.t01-1-00592.x. [DOI] [PubMed] [Google Scholar]
- 21.Hill C P, Worthylake D, Bancroft D P, Christensen A M, Sundquist W I. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc Natl Acad Sci USA. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hockley, D. J., M. V. Nermut, C. Grief, J. B. Jowett, and I. M. Jones. Comparative morphology of Gag protein structures produced by mutants of the gag gene of human immunodeficiency virus type 1. J. Gen. Virol. 75:2985–2997. [DOI] [PubMed]
- 23.Hoshikawa N, Kojima A, Yasuda A, Takayashiki E, Masuko S, Chiba J, Sata T, Kurata T. Role of the gag and pol genes of human immunodeficiency virus in the morphogenesis and maturation of retrovirus-like particles expressed by recombinant vaccinia virus: an ultrastructural study. J Gen Virol. 1991;72:2509–2517. doi: 10.1099/0022-1317-72-10-2509. [DOI] [PubMed] [Google Scholar]
- 24.Jowett J B, Hockley D J, Nermut M V, Jones I M. Distinct signals in human immunodeficiency virus type 1 Pr55 necessary for RNA binding and particle formation. J Gen Virol. 1992;73:3079–3086. doi: 10.1099/0022-1317-73-12-3079. [DOI] [PubMed] [Google Scholar]
- 25.Kaplan A H, Manchester M, Swanstrom R. The activity of the protease of human immunodeficiency virus type 1 is initiated at the membrane of infected cells before the release of viral proteins and is required for release to occur with maximum efficiency. J Virol. 1994;68:6782–6786. doi: 10.1128/jvi.68.10.6782-6786.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaye J F, Lever A M. trans-acting proteins involved in RNA encapsidation and viral assembly in human immunodeficiency virus type 1. J Virol. 1996;70:880–886. doi: 10.1128/jvi.70.2.880-886.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krausslich H G, Facke M, Heuser A M, Konvalinka J, Zentgraf H. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J Virol. 1995;69:3407–3419. doi: 10.1128/jvi.69.6.3407-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krausslich H G, Welker R. Intracellular transport of retroviral capsid components. Curr Top Microbiol Immunol. 1996;214:25–63. doi: 10.1007/978-3-642-80145-7_2. [DOI] [PubMed] [Google Scholar]
- 29.Lee P P, Linial M L. Efficient particle formation can occur if the matrix domain of human immunodeficiency virus type 1 Gag is substituted by a myristoylation signal. J Virol. 1994;68:6644–6654. doi: 10.1128/jvi.68.10.6644-6654.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee Y M, Yu X F. Identification and characterization of virus assembly intermediate complexes in HIV-1-infected CD4+ T cells. Virology. 1998;243:78–93. doi: 10.1006/viro.1998.9064. [DOI] [PubMed] [Google Scholar]
- 31.Lee Y M, Liu B, Yu X F. Formation of virus assembly intermediate complexes in the cytoplasm by wild-type and assembly-defective mutant human immunodeficiency virus type 1 and their association with membranes. J Virol. 1999;73:5654–5662. doi: 10.1128/jvi.73.7.5654-5662.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livingstone C, Jones I. Baculovirus expression vectors with single strand capability. Nucleic Acids Res. 1989;17:2366. doi: 10.1093/nar/17.6.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mammano F, Ohagen A, Hoglund S, Göttlinger H G. Role of the major homology region of human immunodeficiency virus type 1 in virion morphogenesis. J Virol. 1994;68:4927–4936. doi: 10.1128/jvi.68.8.4927-4936.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mervis R J, Ahmad N, Lillehoj E P, Raum M G, Salazar F H, Chan H W, Venkatesan S. The gag gene products of human immunodeficiency virus type 1: alignment within the gag open reading frame, identification of posttranslational modifications, and evidence for alternative gag precursors. J Virol. 1988;62:3993–4002. doi: 10.1128/jvi.62.11.3993-4002.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morikawa Y, Hinata S, Tomoda H, Goto T, Nakai M, Aizawa C, Tanaka H, Omura S. Complete inhibition of human immunodeficiency virus Gag myristoylation is necessary for inhibition of particle budding. J Biol Chem. 1996;271:2868–2873. doi: 10.1074/jbc.271.5.2868. [DOI] [PubMed] [Google Scholar]
- 36.Morikawa Y, Kishi T, Zhang W H, Nermut M V, Hockley D J, Jones I M. A molecular determinant of human immunodeficiency virus particle assembly located in matrix antigen p17. J Virol. 1995;69:4519–4523. doi: 10.1128/jvi.69.7.4519-4523.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morikawa Y, Zhang W H, Hockley D J, Nermut M V, Jones I M. Detection of a trimeric human immunodeficiency virus type 1 Gag intermediate is dependent on sequences in the matrix protein, p17. J Virol. 1998;72:7659–7663. doi: 10.1128/jvi.72.9.7659-7663.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nermut M V, Hockley D J, Jowett J B, Jones I M, Garreau M, Thomas D. Fullerene-like organization of HIV gag-protein shell in virus-like particles produced by recombinant baculovirus. Virology. 1994;198:288–296. doi: 10.1006/viro.1994.1032. [DOI] [PubMed] [Google Scholar]
- 39.Park J, Morrow C D. The nonmyristoylated Pr160gag-pol polyprotein of human immunodeficiency virus type 1 interacts with Pr55gag and is incorporated into viruslike particles. J Virol. 1992;66:6304–6313. doi: 10.1128/jvi.66.11.6304-6313.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perrin-Tricaud C, Davoust J, Jones I M. Tagging the human immunodeficiency virus Gag protein with green fluorescent protein. Minimal evidence for colocalisation with actin. Virology. 1999;255:20–25. doi: 10.1006/viro.1998.9573. [DOI] [PubMed] [Google Scholar]
- 41.Rao Z, Belyaev A S, Fry E, Roy P, Jones I M, Stuart D I. Crystal structure of SIV matrix antigen and implications for virus assembly. Nature. 1995;378:743–747. doi: 10.1038/378743a0. [DOI] [PubMed] [Google Scholar]
- 42.Reicin A S, Paik S, Berkowitz R D, Luban J, Lowy I, Goff S P. Linker insertion mutations in the human immunodeficiency virus type 1 gag gene: effects on virion particle assembly, release, and infectivity. J Virol. 1995;69:642–650. doi: 10.1128/jvi.69.2.642-650.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reil H, Bukovsky A A, Gelderblom H R, Gottlinger H G. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 1998;17:2699–2708. doi: 10.1093/emboj/17.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rhee S S, Hunter E. A single amino acid substitution within the matrix protein of a type D retrovirus converts its morphogenesis to that of a type C retrovirus. Cell. 1990;63:77–86. doi: 10.1016/0092-8674(90)90289-q. [DOI] [PubMed] [Google Scholar]
- 45.Royer M, Hong S S, Gay B, Cerutti M, Boulanger P. Expression and extracellular release of human immunodeficiency virus type 1 Gag precursors by recombinant baculovirus-infected cells. J Virol. 1992;66:3230–3235. doi: 10.1128/jvi.66.5.3230-3235.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakaguchi K, Zambrano N, Baldwin E T, Shapiro B A, Erickson J W, Omichinski J G, Clore G M, Gronenborn A M, Appella E. Identification of a binding site for the human immunodeficiency virus type 1 nucleocapsid protein. Proc Natl Acad Sci USA. 1993;90:5219–5223. doi: 10.1073/pnas.90.11.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmalzbauer E, Strack B, Dannull J, Guehmann S, Moelling K. Mutations of basic amino acids of NCp7 of human immunodeficiency virus type 1 affect RNA binding in vitro. J Virol. 1996;70:771–777. doi: 10.1128/jvi.70.2.771-777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith A J, Srinivasakumar N, Hammarskjold M L, Rekosh D. Requirements for incorporation of Pr160gag-pol from human immunodeficiency virus type 1 into virus-like particles. J Virol. 1993;67:2266–2275. doi: 10.1128/jvi.67.4.2266-2275.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spearman P, Wang J J, Vander Heyden N, Ratner L. Identification of human immunodeficiency virus type 1 Gag protein domains essential to membrane binding and particle assembly. J Virol. 1994;68:3232–3242. doi: 10.1128/jvi.68.5.3232-3242.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Towbin H, Staehelin T, Gordon I. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vogt V M. Proteolytic processing and particle maturation. Curr Top Microbiol Immunol. 1996;214:95–131. doi: 10.1007/978-3-642-80145-7_4. [DOI] [PubMed] [Google Scholar]
- 52.von Schwedler U K, Stemmler T L, Klishko V Y, Li S, Albertine K H, Davis D R, Sundquist W I. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 1998;17:1555–1568. doi: 10.1093/emboj/17.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang C T, Lai H Y, Li J J. Analysis of minimal human immunodeficiency virus type 1 gag coding sequences capable of virus-like particle assembly and release. J Virol. 1998;72:7950–7959. doi: 10.1128/jvi.72.10.7950-7959.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang C T, Stegeman-Olsen J, Zhang Y, Barklis E. Assembly of HIV gag-β-galactosidase fusion proteins into virus particles. Virology. 1994;200:524–534. doi: 10.1006/viro.1994.1215. [DOI] [PubMed] [Google Scholar]
- 55.Wiegers K, Rutter G, Kottler H, Tessmer U, Hohenberg H, Krausslich H G. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J Virol. 1998;72:2846–2854. doi: 10.1128/jvi.72.4.2846-2854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Worthylake D K, Wang H, Yoo S, Sundquist W I, Hill C P. Structures of the HIV-1 capsid protein dimerization domain at 2.6 A resolution. Acta Crystallogr Sect D. 1999;55:85–92. doi: 10.1107/S0907444998007689. [DOI] [PubMed] [Google Scholar]
- 57.Zhang W H, Hockley D J, Nermut M V, Morikawa Y, Jones I M. Gag-Gag interactions in the C-terminal domain of human immunodeficiency virus type 1 p24 capsid antigen are essential for Gag particle assembly. J Gen Virol. 1996;77:743–751. doi: 10.1099/0022-1317-77-4-743. [DOI] [PubMed] [Google Scholar]