Abstract
Machine learning has made great progress in the field of medicine, especially in oncology research showing significant potential. In this paper, the application of machine learning in the study of cholangiocarcinoma was discussed. By developing a novel intra-tumor heterogeneity feature, the study successfully achieved accurate prediction of prognosis and immunotherapy effect in patients with cholangiocarcinoma. This study not only provides strong support for personalized treatment, but also provides key information for clinicians to develop more effective treatment strategies. This breakthrough marks the continuous evolution of machine learning in cancer research and brings new hope for the future development of the medical field. Our study lays a solid foundation for deepening the understanding of the biological characteristics of cholangiocarcinoma and improving the therapeutic effect, and provides a useful reference for more extensive cancer research.
Keywords: Machine learning, Intratumor heterogeneity signature, Survival prognosis, Cholangiocarcinoma, Immune microenvironment, Commentary
Through the analysis of a large number of literature, we found that the prevalence of cholangiocarcinoma was increasing year by year, especially in specific age groups and geographical regions, which provided strong support for further research on the etiology of cholangiocarcinoma. The latest research [1] shows that the increase in morbidity and mortality of cholangiocarcinoma is mainly attributed to intrahepatic cholangiocarcinoma, while extrahepatic cholangiocarcinoma tends to stabilize or decrease.
Intrahepatic cholangiocarcinoma and extrahepatic cholangiocarcinoma differ greatly in tumor heterogeneity and prognosis, but there is no distinction in the original study article. Therefore, it is very necessary to conduct graded studies on intrahepatic cholangiocarcinoma and extrahepatic cholangiocarcinoma. Feng Y, et al. [2] show that there is significant heterogeneity in the prognosis of patients with cholangiocarcinoma, which is closely related to the heterogeneity within the tumor. Golino JL, et al. [3] found that some patients respond well to traditional treatments, while others show varying degrees of resistance. Kamp EJCA, et al. [4] suggests an urgent need for individualized treatment strategies to improve patient survival and quality of life. Kitagawa A, et al. [5] showed that immunotherapy achieved significant results in a subset of patients, but a subset of patients did not benefit. Through in-depth analysis of the response mechanism of immunotherapy and trying to find out the differences between different patients, we can provide a more accurate basis for individual treatment.
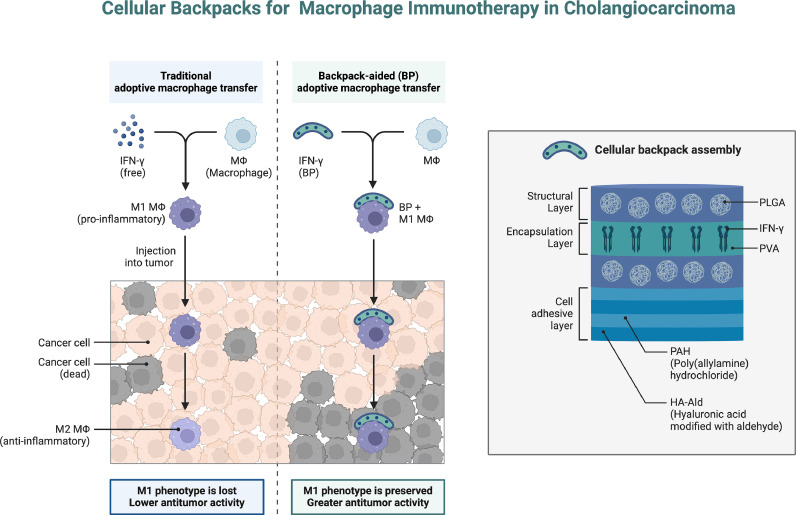
Relevant studies [[3], [4], [5]] have highlighted the association between cholangiocarcinoma and macrophage immunotherapy and revealed its extended mechanism of action. Through in-depth analysis, it was found that macrophages play a key role in the cholangiocarcinoma microenvironment, regulating the immune response and influencing the immunotherapy effect of the tumor. Lin Y, et al. [6] found that activation of macrophages can promote antigen presentation and T cell activation, improving the immune response. In addition, macrophages play a balancing role in regulating inflammatory responses and immunosuppressive factors in the tumor microenvironment, creating more favorable conditions for immunotherapy This diagram describes how the cell backpack is assembled in 4 layers: 1) a PLGA layer as structural support; 2) a PVA-encapsulated reservoir loaded with IFN-γ to maintain the M1 macrophage phenotype; 3) a second PLGA layer for system integrity; and 4) a cell adhesion layer created by PAH and HA-Ald to keep the backpack attached to the macrophage. Injecting M1 macrophages with cell packs into a mouse model of cholangiocarcinoma killed the tumor (Fig. 1).
Fig. 1.
Macrophage immunotherapy in cholangiocarcinoma.
In order to better understand and predict the complexity of cholangiocarcinoma, the relevant researchers introduced machine learning algorithms to extract the characteristics of intra-tumor heterogeneity through the learning and analysis of large-scale data. Ma L, et al. [7] compared with traditional statistical methods, machine learning algorithms have more powerful pattern recognition abilities and can reveal the differences among individual patients with cholangiocarcinoma more comprehensively and accurately. Providing a new means for predicting the prognosis of cholangiocarcinoma and the effect of immunotherapy and promoting the development of cholangiocarcinoma treatment has positive guiding significance.
I was very interested to read the paper by Chen et al. [8] on the successful construction of a prognostic model predicting the effect of immunotherapy for cholangiocarcinoma by applying machine learning algorithms and further revealing the intra-tumor heterogeneity of cholangiocarcinoma. In this study, the authors used a variety of statistical methods for integrated machine learning to construct a prognostic model of ITH-related features (IRS) for cholangiocarcinoma. In addition, the authors further applied bioinformatics techniques to single-cell analysis to clarify the association between immune cell subtypes and validate the biological function of the hub gene through in vitro cell experiments.
The results of this study [8] found that the AUC of the 2-year, 3-year, and 4-year ROC curves in the TCGA cohort were 0.955, 0.950, and 1.000, respectively. The lower the IRS score, the lower the tumor immune dysfunction and rejection score, the lower the tumor microsatellite instability score, the lower the immune escape score, the lower the MATH score, and the higher the mutation burden score in patients with cholangiocarcinoma. This is consistent with previous findings [[8], [9], [10]] reported in related studies. Single-cell sequencing and in vitro cell assays were used to validate the large database cholangiocarcinoma prognostic model. The authors highlighted specific ligand receptor pairs between fibroblasts, macrophages, and epithelial cells, including COL4A1-(ITGAV+ITGB8) and COL1A2-(ITGAV+ITGB8). There is a strong association, and down-regulation of BET1L can inhibit the proliferation, migration, and invasion of bile duct cancer cells and promote their apoptosis.
Chen et al. [8] cleverly applied machine learning to predict prognosis and immunotherapy outcomes in cholangiocarcinoma, successfully exploiting intratumoral heterogeneity. Its advantage is that, through comprehensive and in-depth data learning, it improves the interpretation accuracy of individual patient differences and lays the foundation for individualized treatment. The findings highlight the unique advantages of machine learning in complex cancer research, providing an innovative path for the practice of precision medicine in the future. However, the research still has to face the challenges of data quality and sample size, as well as the lack of explanatory power of the model. Nevertheless, this study has had a positive and far-reaching impact on the in-depth exploration of intra-tumor heterogeneity, the development of individualized therapy, and the wide application of machine learning in the medical field, which is worthy of further attention and exploration by the medical community.
CRediT authorship contribution statement
Liusheng Wu: Writing – original draft. Xiaoqiang Li: Writing – review & editing. Jun Yan: Writing – review & editing.
Declaration of competing interest
None.
Acknowledgments
None.
References
- 1.Greten T.F., Schwabe R., Bardeesy N., Ma L., Goyal L., Kelley R.K., Wang X.W. Immunology and immunotherapy of cholangiocarcinoma. Nat. Rev. Gastroenterol. Hepatol. 2023 Jun;20(6):349–365. doi: 10.1038/s41575-022-00741-4. Epub 2023 Jan 25. PMID: 36697706. [DOI] [PubMed] [Google Scholar]
- 2.Feng Y., Zhao M., Wang L., Li L., Lei J.H., Zhou J., Chen J., Wu Y., Miao K., Deng C.X. The heterogeneity of signaling pathways and drug responses in intrahepatic cholangiocarcinoma with distinct genetic mutations. Cell Death. Dis. 2024 Jan 11;15(1):34. doi: 10.1038/s41419-023-06406-7. PMID: 38212325; PMCID: PMC10784283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Golino J.L., Wang X., Maeng H.M., Xie C. Revealing the heterogeneity of the tumor ecosystem of cholangiocarcinoma through single-cell transcriptomics. Cells. 2023 Mar 10;12(6):862. doi: 10.3390/cells12060862. PMID: 36980203; PMCID: PMC10047686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamp E.J.C.A., Peppelenbosch M.P., Doukas M., Verheij J., Ponsioen C.Y., van Marion R., Bruno M.J., Groot Koerkamp B., Dinjens W.N.M., de Vries A.C. Primary sclerosing cholangitis-associated cholangiocarcinoma demonstrates high intertumor and intratumor heterogeneity. Clin. Transl. Gastroenterol. 2021 Oct 5;12(10):e00410. doi: 10.14309/ctg.0000000000000410. PMID: 34608877; PMCID: PMC8500610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kitagawa A., Osawa T., Noda M., Kobayashi Y., Aki S., Nakano Y., Saito T., Shimizu D., Komatsu H., Sugaya M., Takahashi J., Kosai K., Takao S., Motomura Y., Sato K., Hu Q., Fujii A., Wakiyama H., Tobo T., Uchida H., Sugimachi K., Shibata K., Utsunomiya T., Kobayashi S., Ishii H., Hasegawa T., Masuda T., Matsui Y., Niida A., Soga T., Suzuki Y., Miyano S., Aburatani H., Doki Y., Eguchi H., Mori M., Nakayama K.I., Shimamura T., Shibata T., Mimori K. Convergent genomic diversity and novel BCAA metabolism in intrahepatic cholangiocarcinoma. Br. J. Cancer. 2023 Jun;128(12):2206–2217. doi: 10.1038/s41416-023-02256-4. Epub 2023 Apr 19. PMID: 37076565; PMCID: PMC10241955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin Y., Peng L., Dong L., Liu D., Ma J., Lin J., Chen X., Lin P., Song G., Zhang M., Liu Y., Rao J., Wei C., Lu Y., Zhang S., Ding G., Peng Z., Lu H., Wang X., Zhou J., Fan J., Wu K., Gao Q. Geospatial immune heterogeneity reflects the diverse tumor-immune interactions in intrahepatic Cholangiocarcinoma. Cancer Discov. 2022 Oct 5;12(10):2350–2371. doi: 10.1158/2159-8290.CD-21-1640. PMID: 35853232. [DOI] [PubMed] [Google Scholar]
- 7.Ma L., Wang L., Khatib S.A., Chang C.W., Heinrich S., Dominguez D.A., Forgues M., Candia J., Hernandez M.O., Kelly M., Zhao Y., Tran B., Hernandez J.M., Davis J.L., Kleiner D.E., Wood B.J., Greten T.F., Wang X.W. Single-cell atlas of tumor cell evolution in response to therapy in hepatocellular carcinoma and intrahepatic cholangiocarcinoma. J. Hepatol. 2021 Dec;75(6):1397–1408. doi: 10.1016/j.jhep.2021.06.028. Epub 2021 Jun 30. PMID: 34216724; PMCID: PMC8604764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X., Sun B., Chen Y., Xiao Y., Song Y., Liu S., Peng C. Machine learning developed an intratumor heterogeneity signature for predicting prognosis and immunotherapy benefits in cholangiocarcinoma. Transl. Oncol. 2024 May;43 doi: 10.1016/j.tranon.2024.101905. Epub 2024 Feb 22. PMID: 38387388; PMCID: PMC10899030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiang X., Liu Z., Zhang C., Li Z., Gao J., Zhang C., Cao Q., Cheng J., Liu H., Chen D., Cheng Q., Zhang N., Xue R., Bai F., Zhu J. IDH mutation subgroup status associates with intratumor heterogeneity and the tumor microenvironment in intrahepatic Cholangiocarcinoma. Adv. Sci. (Weinh) 2021 Sep;8(17) doi: 10.1002/advs.202101230. Epub 2021 Jul 11. PMID: 34250753; PMCID: PMC8425914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cantallops Vilà P., Ravichandra A., Agirre Lizaso A., Perugorria M.J., Affò S. Heterogeneity, crosstalk, and targeting of cancer-associated fibroblasts in cholangiocarcinoma. Hepatology. 2024 Apr 1;79(4):941–958. doi: 10.1097/HEP.0000000000000206. Epub 2023 Jan 3. PMID: 37018128. [DOI] [PubMed] [Google Scholar]