Abstract
Herpes simplex virus type 1 (HSV-1) gene UL14 is located between divergently transcribed genes UL13 and UL15 and overlaps the promoters for both of these genes. UL14 also exhibits a substantial overlap of its coding region with that of UL13. It is one of the few HSV-1 genes for which a phenotype and protein product have not been described. Using mass spectrometric and immunological approaches, we demonstrated that the UL14 protein is a minor component of the virion tegument of 32 kDa which is expressed late in infection. In infected cells, the UL14 protein was detected in the nucleus at discrete sites within electron-dense nuclear bodies and in the cytoplasm initially in a diffuse distribution and then at discrete sites. Some of the UL14 protein was phosphorylated. A mutant with a 4-bp deletion in the central region of UL14 failed to produce the UL14 protein and generated small plaques. The mutant exhibited an extended growth cycle at low multiplicity of infection and appeared to be compromised in efficient transit of virus particles from the infected cell. In mice injected intracranially, the 50% lethal dose of the mutant was reduced more than 30,000-fold. Recovery of the mutant from the latently infected sacral ganglia of mice injected peripherally was significantly less than that of wild-type virus, suggesting a marked defect in the establishment of, or reactivation from, latent infection.
The herpes simplex virus type 1 (HSV-1) genome contains 74 different known protein-coding genes (18, 20). The proteins encoded by the majority of these genes have been detected, and mutant phenotypes of most genes have been determined in cell culture. In addition, the phenotypes of some mutants have been investigated in animal models. UL14 is one of the few remaining HSV-1 genes for which no phenotypic data are available. The probable reason for this is that the UL14 coding region is relatively small (219 codons), contains the promoters of two flanking genes (UL13 and UL15), and overlaps the UL13 coding region for over a third of its length (Fig. 1A). These features make design of a UL14 mutant nontrivial. The commonest approach to constructing HSV-1 mutants, deletion of a sizeable portion of the coding region accompanied by insertion of a marker gene, is unlikely to leave expression of UL13 and UL15 unaffected. A mutant containing a minimal change in the UL14 coding sequence has most chance of leaving the flanking genes functionally intact.
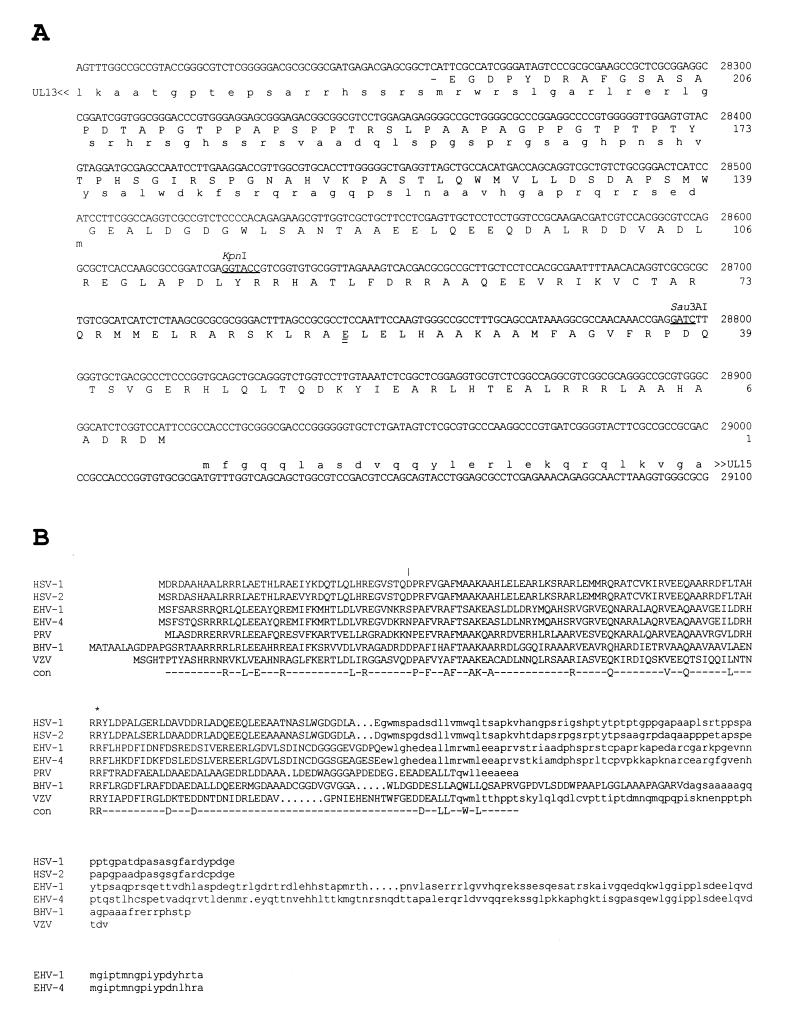
FIG. 1.
(A) Location of HSV-1 UL14 with respect to flanking genes. The predicted amino acid sequence of the UL14 protein (oriented leftward) is aligned in uppercase with the appropriate part of the HSV-1 genome sequence. The N-terminal portions of the UL13 (oriented leftward) and UL15 proteins (oriented rightward) are shown in lowercase. The KpnI and Sau3AI sites at which mutations were introduced are singly underlined. The first residue of the C-terminal portion of the UL14 protein that was present in the UL14-GST fusion protein is doubly underlined. (B) Alignment of the predicted amino acid sequence of HSV-1 UL14 with counterparts in other alphaherpesviruses. Residues affected by the overlapping UL13 coding region are in lowercase. The last UL14 wild-type residue coded by UL14D is indicated by an asterisk, and the last wild-type residue coded by UL14I is shown by a vertical line. Sequences are from HSV-1 (reference 20, as corrected in reference 18), HSV-2 (18), equine herpesvirus 1 (EHV-1) (35), EHV-4 (36), pseudorabies virus (PRV) (17), bovine herpesvirus 1 (BHV-1) (37), and varicella-zoster virus (VZV) (16).
UL14 is conserved in the alphaherpesviruses, and the coding region overlaps that of UL13 (Fig. 1B). Most conserved residues are located in the nonoverlapping region, and the overlapping region seems to encode a variable-length C-terminal domain that is poorly conserved in sequence. Positional counterparts of UL14 are present in betaherpesviruses (such as gene UL96 in human cytomegalovirus and gene 68 in human herpesvirus 6) and gammaherpesviruses (such as gene BGLF3.5 in Epstein-Barr virus and gene 35 in herpesvirus saimiri), but these are not convincingly related to the alphaherpesvirus proteins at the amino acid sequence level.
In this report, the HSV-1 UL14 protein is identified as a minor component of the virion tegument. The growth properties of a viable mutant with a 4-bp deletion in the coding region indicate that UL14 functions at some stage of transit of virus particles from infected cells and is required for complete neurovirulence and efficient establishment of, or reactivation from, latency.
MATERIALS AND METHODS
Cells and viruses.
Rabbit skin (RS), human fetal lung (HFL), and human melanoma (MeWo) cells were grown in Dulbecco's modified Eagle medium supplemented with 8% (vol/vol) fetal calf serum, 2 mM glutamine, nonessential amino acids, penicillin (100 U/ml), and streptomycin (100 μg/ml). Vero cells and HEp-2 cells were maintained in Dulbecco's medium supplemented with 10% newborn calf serum, penicillin (100 U/ml), and streptomycin (100 μg/ml).
Wild-type HSV-1 strain 17 syn+ was obtained as a standard viral stock (8) or reconstructed by cosmid recombination (12). A plaque-purified stock of the former with stable high virulence (1) was used for in vivo analyses.
Low-multiplicity growth curves were obtained by infecting cell monolayers in 35-mm petri dishes at a multiplicity of infection (MOI) of 0.001 PFU/cell at 37°C. Virus was harvested at various times after infection by scraping the cells into the medium and sonicating and was then titrated in duplicate on RS cells. High-multiplicity growth curves were obtained similarly except that RS monolayers were infected at an MOI of 5 PFU/cell. In some experiments, the proportion of virus released from cells was determined by separating intact infected cells from the medium by centrifugation, sonicating the cells in fresh medium, and titrating the infected cell medium and sonicated cells separately. Viral stocks used as input were titrated before and after experiments were conducted.
Construction of UL14 mutants.
A mutant (UL14D) containing a 4-bp deletion in the UL14 coding region was constructed by cosmid recombination (12). The coordinates given below relate to the updated HSV-1 DNA sequence (18). Deletion mutants in cos6 (141221 to 29733; a fused form of the right and left genome ends) were produced by linearization with KpnI, treatment with T4 DNA polymerase in the presence of four deoxynucleoside triphosphates, religation, and transfection into Escherichia coli DH5α. This had the effect of removing 4 bp at the cleaved KpnI site. A series of cosmids lacking individual KpnI sites was characterized by restriction endonuclease digestion. One cosmid lacking the KpnI site at 28624 within UL14 was digested with PacI to release the insert and cotransfected into RS cells with four other PacI-cleaved cosmids representing the rest of the genome.
Since UL14 is located within an area of overlap between cos6 and cos28 (24697 to 64405), a proportion of the progeny virus was wild type. A population of very small plaques was observed against this wild-type background, however, and several were selected. One plaque was subjected to an additional round of plaque purification, and a virus stock was prepared. The absence of the KpnI site in UL14 (Fig. 1A) was confirmed by Southern blot hybridization of infected cell DNA with an appropriate probe (12). The presence of an origin of replication, which is located in the region of overlap between two cosmids (cos28 and cos14) and may be deleted from cosmid-reconstituted viruses (12), was confirmed similarly. The deletion in UL14 was verified by sequencing a PCR product derived from infected cell DNA.
An additional mutant with an insertion in UL14 was constructed by using cosmid recombination, and its structure was confirmed as described above. This mutant (UL14I) had a 16-bp insertion containing stop codons in all three frames at the Sau3AI site at 28795 (Fig. 1A), converting GATC to GATCTAATCTAGATTAGATC.
UL14D was restored to wild type by infecting RS monolayers with the mutant at 0.01 PFU/cell and transfecting cells at 1 h postinfection with a cloned HSV-1 BglII fragment (25378 to 30676) containing the entire UL14 gene. Infected cells were harvested at 5 days postinfection and sonicated, and virus was plated at 10-fold dilutions on fresh monolayers. A large plaque was selected at 4 days postinfection and plaque purified once more, and a virus stock was prepared. Restoration of the deleted base pairs in the rescued mutant (UL14R) was verified by sequencing a PCR product derived from infected cell DNA.
Generation of UL14 antibody.
A PCR product of the 3′ portion of the UL14 coding region was generated by using the primers GCCCACTTGGAATTCGAGGCGCGGCTA and TGAGACGAGCGGCTCATTCGCCATC. The underlined C was altered from the G residue in the HSV-1 sequence to produce an EcoRI site in codon 57 in the UL14 coding region (Fig. 1A). The fragment (approximately 450 bp) was gel purified, treated with T4 DNA polymerase in the presence of the four deoxynucleoside triphosphates, and digested with EcoRI. The DNA fragment was cloned into pGEX 4T-1 (Pharmacia) between the EcoRI and SmaI sites, such that UL14 sequences (codons 58 to 219) were maintained in frame with, and downstream from, the sequence encoding glutathione S-transferase (GST). The junction between the two coding regions was confirmed by DNA sequencing.
Expression of the GST-UL14 fusion protein was induced in cultures of E. coli BL21 by the addition of 0.3 mM isopropyl-β-d-thiogalactopyranoside at 30°C for 1 h. The protein was purified by affinity chromatography on glutathione-Sepharose beads according to the directions of the manufacturer (Pharmacia). A female New Zealand White rabbit was immunized three times with purified protein (50 μg per injection) emulsified in complete and incomplete Freund's adjuvant as described previously (3).
Purification and fractionation of virions.
Wild-type virions and L particles (virions lacking capsids) were purified from infected MeWo cell medium by centrifugation on 5 to 15% (wt/vol) Ficoll gradients (34). Virions were treated with 1% Nonidet P-40 to fractionate soluble envelope proteins from insoluble capsid-tegument material (2). L particles were fractionated similarly into envelope and tegument proteins.
Radioisotopic labeling of infected cells.
Extracts from cells infected at 5 PFU/cell and labeled with 25 μCi of l-[35S]methionine per ml from 6 to 10 h postinfection were prepared as described previously (23). Extracts from cells labeled with 100 μCi of [32P]orthophosphate per ml in phosphate-free medium were prepared similarly.
Electron microscopy.
For conventional electron microscopy, HEp-2 cells infected with UL14D or UL14R were fixed in 2.5% (wt/vol) glutaraldehyde in 0.1 M sodium cacodylate (pH 7.4) at 15 h postinfection. Fixed samples were washed in 0.1 M sodium cacodylate (pH 7.4), treated with 2% osmium tetroxide, washed again, and treated with 2% (wt/vol) aqueous uranyl acetate. Samples were dehydrated in ethanol and then acetone and were infiltrated with Epon-Araldite. Thin sections were cut and stained with 2% (wt/vol) aqueous uranyl acetate and then 2% (wt/vol) lead citrate. Samples were viewed with a Zeiss 902 electron microscope.
Immunological detection of the UL14 protein.
Immunoprecipitation and immunoblotting were carried out by using standard protocols.
For immunofluorescence experiments, six-well plates containing sterile glass coverslips were seeded with 4 × 104 HEp-2 cells per 10-cm2 well. The cells were allowed to attach overnight and then were infected at 5 PFU/cell with UL14D or UL14R. Cells were fixed in ice-cold methanol for 10 min at 6, 9, or 16 h postinfection and then rinsed in phosphate-buffered saline (PBS). Fixed cells were incubated for 1 h at 37°C in PBS containing 1% (wt/vol) bovine serum albumin and 10% (vol/vol) human serum (Sigma S2145) to block nonspecific binding of rabbit immunoglobulin by the HSV-1-encoded Fc receptor. After extensive washing in PBS, cells were reacted for 1 h at 37°C with the UL14 antiserum (1:1,000) diluted in PBS containing 1% bovine serum albumin. The cells were again washed in PBS and reacted with Texas red-conjugated donkey anti-rabbit immunoglobulin (Jackson Immunoresearch Laboratories, Inc.) for 1 h at 37°C. Samples were washed in PBS, rinsed once in distilled water, and mounted in Vectashield mounting medium (Vector Laboratories, Inc.). Confocal microscopy was performed as described previously (25) except that samples were scanned only for Texas red fluorescence. Digital images were recorded with COMOS software (Bio-Rad).
For immunogold labeling, HEp-2 cells infected at 2 PFU/cell with UL14D or UL14R were fixed at 15 h postinfection in 2.5% (wt/vol) glutaraldehyde–4% (wt/vol) paraformaldehyde in 0.1 M sodium phosphate (pH 7.4). Fixed samples were washed at 25°C in 0.1 M sodium phosphate (pH 7.4), dehydrated at 25°C and then −20°C in ethanol, and infiltrated at −20°C with LR White. UV irradiation was used to polymerize the plastic, samples were sectioned, and sections were blocked in PBS containing 0.1% (vol/vol) Triton X-100 and 1% (wt/vol) fish gelatin. Samples were then incubated for 3 h with the UL14 antiserum (1:500). After extensive washing in PBS containing 0.1% (vol/vol) Triton X-100 and 1% (wt/vol) fish gelatin, samples were incubated for 1 h with 12-nm gold bead-conjugated donkey anti-rabbit immunoglobulin (1:100). Samples were washed extensively, stained in 2% (wt/vol) aqueous uranyl acetate followed by 2% (wt/vol) lead citrate, and viewed with a Philips 201 electron microscope.
Gel electrophoresis.
Protein samples were reduced and denatured and then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Gels were fixed and stained with Coomassie blue, or fixed, dried, and autoradiographed or exposed to screens for analysis in a Bio-Rad Fluor-S MultiImager, or electroblotted for mass spectrometric analysis or immunoblotting.
Mass spectrometry.
Proteins separated by SDS-PAGE were electroblotted to polyvinylidene difluoride membranes, stained with sulforhodamine, and subjected to trypsin digestion as described previously (10, 28). The masses of resulting peptides were determined with a Finnigan Lasermat laser desorption mass spectrometer, and parent proteins were identified by using the Massmap program to search proteins with predicted masses of 20 to 40 kDa in release 14.0 of the National Center for Biotechnology Information Entrez database (15, 27).
Neurovirulence assays.
Pathogenicity of wild-type virus (as a standard viral stock or reconstructed from cosmids), UL14D, and UL14R was determined following intracranial inoculation into mice. Three-week-old female BALB/c mice were injected in the left cerebral hemisphere with 25 μl of appropriate 10-fold serial dilutions of each virus (22). At the time of inoculation, the virus was titrated on RS cells to ensure that the correct dose had been administered. Animals were monitored on a daily basis, deaths were scored up to day 21 postinjection, and 50% lethal dose (LD50) values were calculated according to the Spearman-Karber formula (39).
Reactivation assays.
Four-week-old female BALB/c mice were injected in the left rear footpad with 25 μl of appropriate 10-fold serial dilutions of each virus (32). At the time of inoculation, the virus was titrated on RS cells to ensure that the correct dose had been administered. Surviving animals were maintained for at least 6 weeks postinjection to allow the virus to establish a latent infection. To determine the amount of latent virus established in the dorsal root ganglia (DRG) and the spread of virus in the peripheral nervous system, animals were sacrificed and the 10 ipsilateral DRG supplying the footpad were individually explanted and monitored for 21 days for shedding of infectious virus by transferring the supernatants every 2 days to indicator RS cells and scoring for cytopathic effect (32).
RESULTS
Initial identification of the UL14 protein.
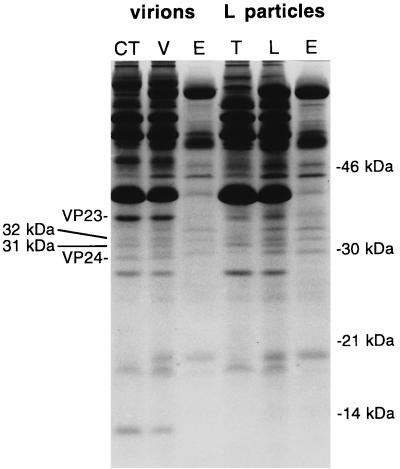
Figure 2 shows a Coomassie blue-stained gel of purified HSV-1 virion and L particles. Proteins that fractionate with the capsid-tegument (from virions) or tegument (L particles) and envelope are also shown. The lanes were heavily loaded to allow detection of minor proteins. Two minor virion protein bands migrating at 32 and 31 kDa fractionated with the capsid-tegument. These two bands were also present in L particles and fractionated with the tegument.
FIG. 2.
Coomassie blue-stained 15% polyacrylamide gel of proteins present in HSV-1 virions (V) and L particles (L) and fractions derived therefrom (CT, capsid-tegument; T, tegument; E, envelope). The 32- and 31-kDa bands and capsid proteins VP23 and VP24 are indicated on the left; positions of marker proteins are indicated on the right.
To identify the genes encoding the 32- and 31-kDa proteins, samples from the capsid-tegument were isolated on membranes and digested with trypsin, and the resulting peptides were subjected to mass spectrometry. Table 1 shows the masses of individual peptides determined in one mass spectrometry experiment. The data indicated the presence of a protein common to both bands plus at least one protein unique to each. Of the five peptides (column 1, not in bold type) common to both bands, four (column 2, in bold type) matched peptides (column 4) predicted from the UL51 gene product (allowing for mass errors of ±3.0 Da), causing the UL51 protein to score with the Massmap program as the most likely protein in the database to produce these peptides. The UL51 protein is predicted to contain 244 residues and have a mass of 25,470 Da.
TABLE 1.
Comparison of experimental tryptic peptide masses obtained from the 31- and 32-kDa protein bands with those predicted from the UL14, UL51, and UL7 proteinsa
| Exptl masses (Da)
|
Hypothetical mass (Da) of corresponding peptide
|
|||
|---|---|---|---|---|
| 32-kDa protein | 31-kDa protein | UL14 | UL51 | UL7 |
| 840.6 | 837.9 | |||
| 864.1 | ||||
| 882.4 | 881.0 | |||
| 921.3 | ||||
| 1,014.5 | 1,016.2 | |||
| 1,036.2 | 1,034.2 | |||
| 1,045.3 | 1,042.3 | |||
| 1,047.2 | ||||
| 1,120.6 | 1,121.3 | 1,124.3 | ||
| 1,169.0 | 1,164.2 | |||
| 1,275.5 | 1,278.3 | 1,277.5 | ||
| 1,421.0 | ||||
| 1,609.7 | ||||
| 1,731.9 | 1,730.0 | |||
| 1,889.7 | ||||
| 2,151.7 | 2,151.4 | |||
| 2,186.1 | 2,183.4 | |||
| 2,343.4 | 2,344.7 | |||
| 2,823.0 | 2,819.9 | 2,818.2 | ||
| 3,005.5 | 3,005.5 | 3,006.5 | ||
Boldface and italicized values are explained in the text.
When eight peptides (Table 1, column 1, in bold type) specific to the 32-kDa band were analyzed, five matched peptides (column 3) predicted from the UL14 protein (allowing for mass errors of ±2.7 Da), causing the UL14 protein to score as the most probable parental protein. The UL14 protein is predicted to contain 219 residues and have a mass of 23,933 Da.
Identification of the protein specific to the 31-kDa band was problematic. When six peptides (Table 1, column 2, in italic type) absent from the 32-kDa band were analyzed, four (column 5) matched predicted products of the UL7 protein (allowing for mass errors of ±3.1 Da). With Massmap, the UL7 protein scored as the third-most-probable parental protein; the higher-scoring proteins originated from vaccinia virus and yeast and were thus likely to be irrelevant. The UL7 protein is predicted to contain 296 residues and have a mass of 33,059 Da.
The conclusions drawn tentatively from these data are that the 32-kDa band consists of a mixture of the UL14 and UL51 proteins, and the 31-kDa band contains the UL51 protein in addition to another component, possibly the UL7 protein. The low abundance of these proteins and their presence as mixtures precluded determination of the number of molecules present per virion. Comparison with reported values (26) for the capsid proteins indicated in Fig. 2, VP23 (estimated at 572 ± 67 molecules, geometrically computed at 640 copies) and VP24 (estimated at 147 ± 67 molecules), suggests that the 31-kDa band represents about 150 molecules per virion and the 32-kDa band represents fewer than 100 molecules per virion. Given that the 32-kDa band consists of two protein species and that sufficient amounts of each were present for peptides to be detected, the UL14 protein is probably present at no more than a few dozen molecules per virion.
Construction of UL14 mutants.
In the initial stages of this work, a mutant (UL14I) containing a 16-bp insert at a Sau3AI site in UL14 was constructed (Fig. 1). In principle, the mutated gene could specify a protein consisting of the N-terminal 40 residues of the UL14 protein fused to two other residues coded in an alternative reading frame. This mutant exhibited a small-plaque phenotype but suffered from instability in that it readily generated large wild-type plaques after passage. This presumably occurred via loss of the inserted sequence by recombination, in this case between flanking Sau3AI sites, as observed previously for another insertion mutant (12). Consequently, UL14I was not analyzed further.
The mutant used in subsequent studies (UL14D) lacked 4 bp at the KpnI site (Fig. 1) and could potentially generate a protein consisting of the N-terminal 96 residues of the UL14 protein fused to 42 other residues coded in an alternative reading frame. This mutant exhibited a plaque phenotype similar to that of UL14I. In one experiment, mean plaque diameters ± standard deviations (SD) at 3 days postinfection on RS cells were 1.4 ± 0.3 mm for wild-type virus, 0.3 ± 0.06 mm for UL14D, and 0.28 ± 0.05 mm for UL14I. The use of this deletion mutant precluded reversion to wild type by recombination. Phenotypic reversion by mutation at a second site could occur in principle, but in no study did UL14D generate large plaques characteristic of such pseudorevertants. Therefore, we consider that the mutation in UL14D is stable.
Immunological detection of the UL14 protein.
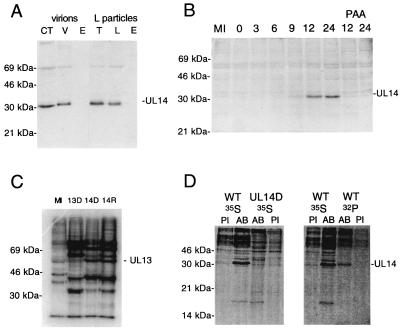
Figure 3A shows results for fractionated virions immunoblotted with the UL14 antiserum. The antiserum identified a 32-kDa protein strongly and a 69-kDa protein weakly in virions and L particles. These proteins fractionated with the capsid-tegument from virions and the tegument from L particles. Preimmune serum did not react with the 32-kDa protein but did react with the 69-kDa protein (data not shown). These results confirm that the UL14 protein is a 32-kDa species present in the tegument, as indicated by the mass spectrometric data.
FIG. 3.
Immunological identification of the UL14 protein and assessment of UL13 expression. (A) HSV-1 virions (V) or L particles (L) and fractions derived therefrom (CT, capsid-tegument; T, tegument; E, envelope) were separated on a 10% polyacrylamide gel, transferred to a Hybond ECL membrane, and immunoblotted with the UL14 antiserum. Positions of the UL14 protein and marker proteins are indicated. (B) HSV-1-infected cells harvested at various times postinfection were separated on a 10% polyacrylamide gel, transferred to a Hybond ECL membrane, and immunoblotted with the UL14 antiserum. Positions of the UL14 protein and marker proteins are indicated. The mock-infected (MI) sample was harvested at 24 h. Two samples were maintained in the presence of phosphonoacetic acid and harvested at 12 and 24 h postinfection. (C) In vitro-phosphorylated nuclear extracts from mock-infected cells (MI) or cells infected with a UL13 null mutant (13D [11]), UL14D (14D), or UL14R (14R) were separated on a 10% polyacrylamide gel and autoradiographed. Positions of the labeled UL13 protein and marker proteins are indicated. (D) 35S- or 32P-labeled proteins from cells infected with wild-type HSV-1 (WT) or UL14D were immunoprecipitated with preimmune serum (PI) or the UL14 antiserum (AB), separated on 15% polyacrylamide gels, and phosphorimaged. Positions of the UL14 protein and marker proteins are indicated.
Other experiments on occasion demonstrated the presence of the UL14 protein in purified capsids (data not shown). However, the presence of the protein in variable amounts and at levels very much lower than that in virions was interpreted as being most likely due to contamination rather than to the specific presence of the UL14 protein in capsids.
Expression and phosphorylation of the UL14 protein.
Figure 3B shows that the UL14 protein is expressed late in infection, first being detected at 9 h postinfection by immunoblotting and in increasing amounts at least until 24 h postinfection. Lack of expression at 12 or 24 h postinfection in the presence of phosphonoacetic acid indicates that the UL14 protein is in the late kinetic class. Reactivity with the UL14 protein was not detected when preimmune serum was used (data not shown). Immunoblotting experiments demonstrated that two late HSV-1 genes, US11 and UL38, are expressed with similar kinetics in cells infected with wild-type virus or UL14D (data not shown). This indicates that the phenotype of UL14D is not due to general retardation of gene expression.
In principle, it is possible that even a 4-bp deletion within the UL14 coding sequences could compromise expression of UL13 and UL15. Expression of these genes in UL14D was therefore assessed. The viability of UL14D indicates a priori that at least some UL15 protein was produced, since UL15 is essential for growth in cell culture (30). Moreover, the small-plaque phenotype of UL14D was unaltered by growth on an RS-derived cell line that expresses functional UL15 protein (4) (data not shown). UL13 is not required for growth of HSV-1 in cell culture, and a mutant unable to express UL13 exhibited a plaque diameter about half that of wild-type virus and showed a slight impairment in yield at low MOI (11). UL14D and UL14R expressed the UL13 protein in similar amounts, as assayed by in vitro phosphorylation of nuclear extracts prepared at 23 h postinfection (13) (Fig. 3C). Thus, it appears unlikely that UL14D is defective in expression of UL15 or UL13. As expected, the mutant formed small plaques on a UL12-expressing cell line, S22 (9).
Figure 3D shows that the UL14 antiserum was able to immunoprecipitate a 32-kDa protein from extracts of cells infected with wild-type virus, again confirming the identification of the UL14 protein. The 32-kDa protein was not precipitated from UL14D-infected cell extracts by the UL14 antiserum or from lysates of cells infected with wild-type virus or UL14D by preimmune serum. Figure 3D shows that the UL14 antiserum also precipitated a 32P-labeled protein with a mobility marginally less than that of the bulk of the 35S-labeled UL14 protein. Again, this protein was not recognized by preimmune serum in wild type- or UL14D-infected cell extracts or by the UL14 antiserum in UL14D-infected cell extracts (data not shown). These results indicate that a proportion of the UL14 protein in infected cells is phosphorylated; the slightly lower mobility of these molecules would be consistent with the presence of phosphate groups.
Intracellular localization of the UL14 protein.
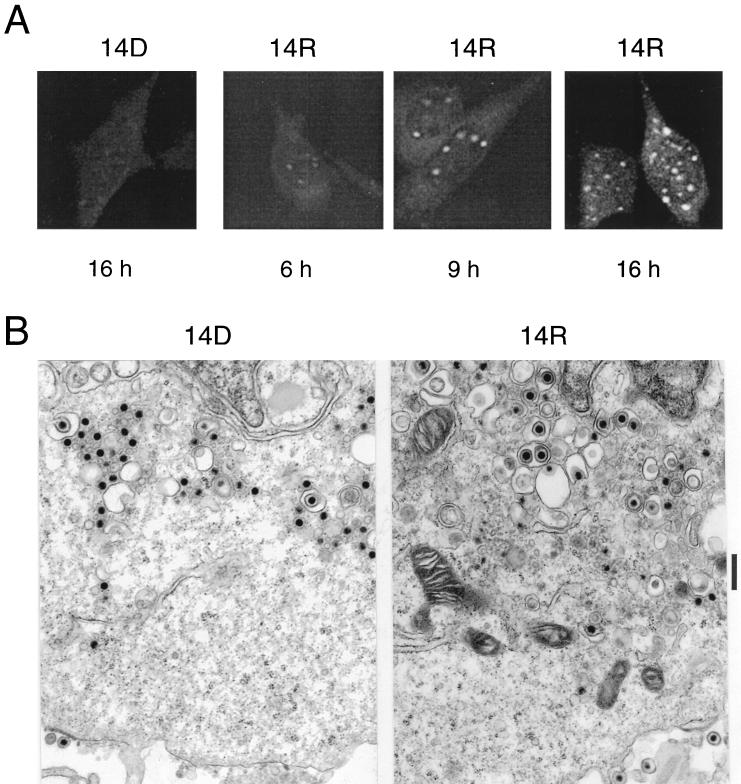
To test the specificity of the UL14 antiserum in indirect immunofluorescence assays, HEp-2 cells were infected with UL14D, fixed and permeabilized at 16 h postinfection, and reacted with the antiserum. Little fluorescence was detected (Fig. 4A, 14D), indicating that the antiserum does not cross-react with host or other viral proteins in immunofluorescence assays.
FIG. 4.
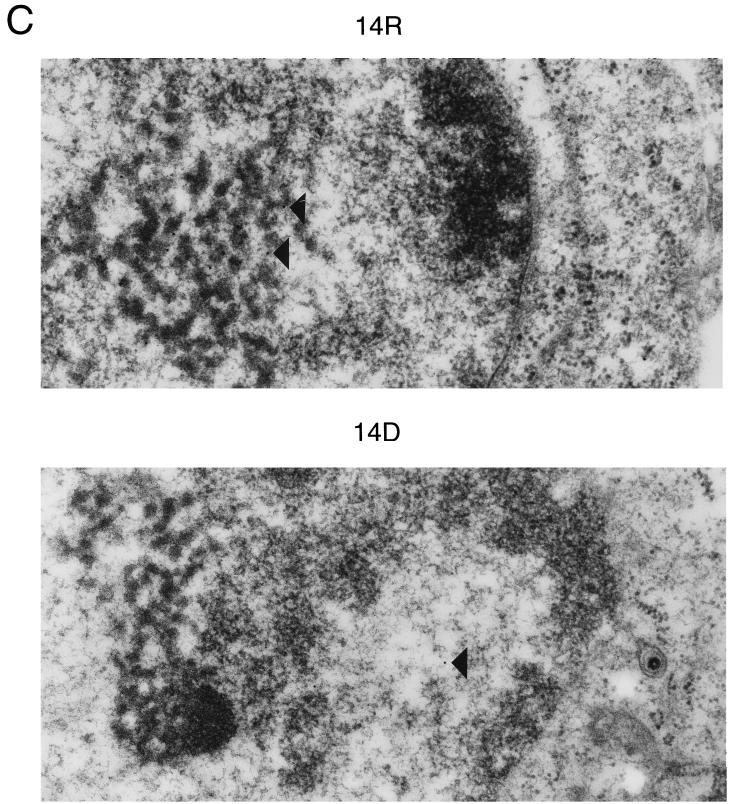
Localization of the UL14 protein in infected cells. (A) Confocal images of HEp-2 cells infected with UL14R (14R) or UL14D (14D), fixed at 6, 9, or 16 h postinfection, and reacted with the UL14 antiserum. (B) Electron photomicrographs at 15 h postinfection of cells infected with UL14D or UL14R. The nucleus is at the top in each panel. The vertical bar indicates 1 μm. (C) Electron photomicrographs of cells infected with UL14R or UL14D and labeled with immunogold particles at 15 h postinfection. Representative gold beads are indicated by arrowheads. The nucleus is to the left. The vertical bar indicates 1 μm.
To examine the intracellular distribution of the UL14 protein, HEp-2 cells were infected with UL14R, fixed and permeabilized at various times postinfection, and reacted with the UL14 antiserum (Fig. 4A, 14R). At 6 h postinfection, faint diffuse cytoplasmic staining, as well as faintly fluorescent, intranuclear aggregates, was visible. By 9 h postinfection, the intranuclear fluorescence had intensified and was clearly visible at the nuclear periphery. The diffuse cytoplasmic fluorescence had also intensifed. At 16 h postinfection, the cytoplasmic staining was somewhat greater than that at 9 h postinfection, with foci of intense fluorescence visible. Intranuclear staining did not appear to change markedly between 9 and 16 h postinfection; 10 to 40 intensely staining foci were present within each infected cell nucleus.
Electron microscopic examination indicated that in the perinuclear region of UL14D-infected HEp-2 cells at 15 h postinfection, the great majority of particles were nonenveloped or in the process of becoming enveloped or de-enveloped (Fig. 4B). The perinuclear region of UL14R-infected cells also contained nonenveloped particles, but many intact virions were present in vesicles. No differences were discernible between UL14D and UL14R capsids in negatively stained samples. These experiments also indicated that extracellular virions were approximately sevenfold less abundant in the UL14D sections (data not shown).
Using immunogold electron microscopy, the distribution of the UL14 protein was examined in thin sections of HEp-2 cells infected with UL14D or UL14R (Fig. 4C). At 15 h postinfection, only a few randomly distributed gold beads (four to five beads per field at ×20,000 magnification) were visible in the nuclei and cytoplasm of cells infected with UL14D. In UL14R-infected cells, gold beads (10 to 50 per field) were concentrated in areas containing electron-dense material within nuclei. These areas ranged from loosely packed intranuclear regions, such as that visible toward the left of the UL14R panel in Fig. 4C, to very dense, compact areas (not shown).
Growth properties of UL14D in vitro.
Although UL14D is viable in cell culture, it was apparent that it is compromised in its growth properties. In one experiment, measurements of mean plaque diameters ± SD at 3 days postinfection on RS cells yielded 1.5 ± 0.15 mm for wild-type virus, 1.5 ± 0.19 mm for UL14R, and 0.42 ± 0.08 mm for UL14D. A plaque size defect of similar magnitude was noted on HFL, Vero, and MeWo cells.
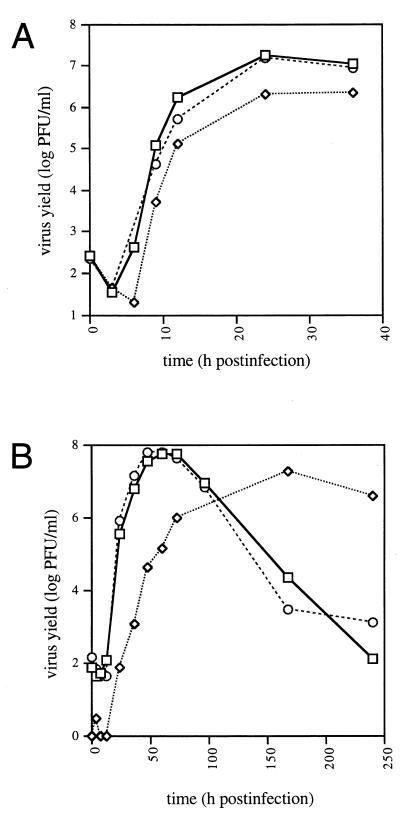
Figure 5A shows that when RS cells were infected at high MOI, the growth kinetics of UL14D were slightly delayed in comparison with the wild type and UL14R, and the final yield of virus was seven- to eightfold less. Figure 5B shows that when cells were infected at low MOI, the maximum yield of UL14D was three- to fourfold less than the yield of the wild type or UL14R but took about three times longer to attain. The decrease in titer of the wild type and UL14R at later times was a consequence of prolonged incubation after virus production had ceased. The lower titer of UL14D at early stages of the growth curve was observed repeatedly. The growth kinetics of UL14D at low MOI in HFL cells were similar to those in RS cells (data not shown). Measurements of the ratio of viral particles to PFU were carried out on virus harvested at peak titers after infection of RS cells at low MOI. The results revealed no significant difference between UL14D and wild-type virus or UL14R (data not shown).
FIG. 5.
Growth curves of wild-type virus (□), UL14R (○), and UL14D (◊) in RS cells infected at high MOI (5 PFU/cell) (A) and low MOI (0.001 PFU/cell) (B).
The observation that the final yield of UL14D was depressed but was largely independent of MOI, whereas the time taken to reach maximum yield was dependent on MOI, suggested that the mutant may have a defect in spread of virus from cell to cell. To investigate this, we infected RS cells at high MOI and determined the proportion of the total virus yield released from cells at 24 h postinfection. From three independent experiments, UL14D released an average of 19.1% (±10.3% SD) and 17.1% (±8.2% SD) of the proportion of virus released into the medium by the wild type and UL14R, respectively. This result indicates that UL14D is released inefficiently from infected cells.
Growth properties of UL14D in vivo.
After intracranial inoculation into mice, standard wild-type virus or wild-type virus reconstructed from cosmids had similar LD50s of 50 and 100 PFU per animal, respectively. In contrast, the LD50 of UL14D was greater than 3 × 106 PFU per animal. There were no deaths in the group of animals administered the highest dose (106 PFU per animal). UL14R exhibited an LD50 of 70, closely comparable to that of wild-type virus.
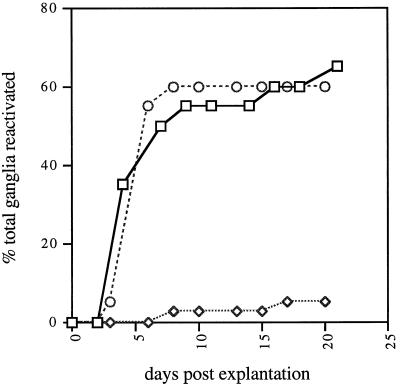
Figure 6 shows that wild-type virus reconstructed from cosmids and UL14R reactivated from mouse DRG at similar rates and to similar levels. In contrast, UL14D reactivated to a much lower level even at a 10-fold-higher dose (no reactivation was detected at lower doses). Virus reactivated from mice infected with UL14D exhibited the characteristic small plaque size of UL14D and was shown to retain the mutation by sequencing a PCR product derived from infected cell DNA.
FIG. 6.
Reactivation of wild-type virus (□; 106 PFU/animal), UL14R (○; 106 PFU/animal), and UL14D (◊; 107 PFU/animal) from explanted mouse DRG.
DISCUSSION
The work described in this report shows that the product of HSV-1 UL14 gene is a 32-kDa protein that is present in the virion as a minor component of the tegument. The size of the UL14 protein estimated by SDS-PAGE is thus larger than that predicted from the primary amino acid sequence (24 kDa). The UL14 protein is expressed with late kinetics, and some of the protein seems to be phosphorylated in infected cells. It is not known whether the protein present in the tegument is phosphorylated.
Immunofluorescence experiments indicated that the UL14 protein is present at earlier stages of infection in a diffuse distribution in the cytoplasm and at discrete sites in the nucleus, and then at discrete sites in the cytoplasm at later times. We view it as likely that the structures identified by immunogold labeling correspond to the intranuclear foci apparent from immunofluorescence. Although the fixation protocol used in this technique makes elucidation of ultrastructural detail less than optimal, the structures with which the UL14 protein was associated are reminiscent of the previously described intranuclear mixed-density bodies that are readily detectable in HSV-1-infected cells (5, 7, 31). Gold beads were also present in the cytoplasm of UL14R-infected cells and, although they might be expected to be associated with virions, did not appear to be associated with obvious ultramicroscopic structures.
As anticipated, UL14D failed to produce the UL14 protein. It is possible in principle for UL14D to produce a C-terminally truncated form of the UL14 protein containing the N-terminal 96 residues. This protein was not detected in any immunological experiments (for example, Fig. 3D). However, the epitopes in the UL14 protein recognized by the antibody were not identified, and thus it is still possible that UL14D produces a truncated protein that retains a degree of function. However, UL14I is capable of producing a protein that contains only the N-terminal 40 residues of the UL14 protein and is thus less likely to retain UL14 function. This mutant gave plaques the same size as those of UL14D. UL14I was not used in subsequent experiments owing to its propensity to revert to wild type.
The HSV-1 tegument consists of a complex mixture of viral proteins. Many of these proteins, regardless of abundance in the virion, are not essential for viral growth in cell culture, but most are required for optimal growth. Moreover, the genes encoding many tegument proteins, including three of the most abundant (the UL47, UL48, and UL49 proteins), are confined to the alphaherpesviruses and thus are relatively recent in evolutionary terms. In contrast, a number of minor tegument proteins are encoded by genes that appear to have counterparts in beta- and gammaherpesviruses. With the caveat that evidence for conservation is in some cases (such as UL14) limited to positional data only, these genes probably originated during a more ancient era of herpesvirus existence. UL14 has now been identified as a member of this class, along with such genes as UL36 (24), UL37 (21), UL13 (13), UL51 (14), and possibly UL16 (25).
In addition to giving an initial identification of the UL14 protein, mass spectrometry indicated tentatively that the UL51 protein is also a tegument protein. This is in accord with previously published results (14) which showed that the UL51 protein is present in the tegument as three forms with apparent mobilities of 27, 29, and 31 kDa. It is not known whether our 31- and 32-kDa forms correspond to two of these species or whether they represent minor forms with lower mobility.
It is intriguing that a null mutant in UL51 behaves similarly to UL14D in producing small plaques, yielding less virus, and being compromised in spread in confluent cell monolayers (6). Also, like the UL14 protein, the UL51 protein is phosphorylated in infected cells (14). Mass spectrometry indicated that the UL7 protein, along with the UL51 protein, may also be a component of the 31-kDa tegument band. This preliminary conclusion must await immunological confirmation. The UL7 counterpart in bovine herpesvirus 1 was not detected as a major constituent of virions, but a mutant exhibited delayed kinetics of virus production, a reduced yield, and an impairment in release of virions from infected cells (33). UL7 and UL51, like UL14, have apparent counterparts in beta- and gammaherpesviruses.
The functions of most tegument proteins remain unknown. Roles could be envisaged in entry, egress, and stability of virus particles, in addition to providing a means of introducing factors at the start of infection that modulate cellular processes (such as the UL41 protein, which stimulates shutoff of host protein synthesis) or promote initiation of the viral replicative cycle (such as the UL48 protein, which stimulates transcription of immediate-early genes). The growth properties of UL14D indicate a defect in spread of virus from cell to cell and thus a role for the UL14 protein in some aspect of egress. Electron microscopy showed that the perinuclear region of UL14D-infected cells contains more nonenveloped particles than UL14R. The interpretation of this observation depends on whether addition of tegument and envelope are nuclear or cytoplasmic processes or some combination of the two. It has been reported that at least three tegument proteins (the UL48, US11, and UL49 proteins) are added at nuclear sites (29, 38). Nuclear localization indicates that the UL14 protein could be incorporated into particles in this compartment. However, the reported confinement of at least one tegument protein (the UL49 protein) to the cytoplasmic compartment implies that addition of part of the tegument and the virion envelope occur in the cytoplasm (19). Thus, the UL14 protein may be required for efficient envelopment as particles bud into cytoplasmic vesicles. Alternatively, it may prevent premature fusion of enveloped particles with vesicle membranes or may have a role in subsequent transport of virions, with delay increasing the likelihood of fusion.
The effects of the absence of UL14 are particularly striking in vivo. UL14D is highly attenuated in its ability to replicate in the mouse brain and in its ability to establish or reactivate from latent infection. The latter observation may reflect the compromised growth properties of the mutant rather than an effect on the mechanisms for establishing or reactivating from latency per se. The observed phenotype of UL14D in vitro and in vivo serves to reinforce the view that the tegument, as a complex association of viral proteins, contributes in several different ways to successful viral growth and thus to the sophistication of the herpesvirus life cycle.
ACKNOWLEDGMENTS
We are grateful to our colleagues Bernard Roizman, for making available the RS cell line, Sandy Weller, for providing the S22 cell line, and Duncan McGeoch, for critical reading of the manuscript. Electron microscopy was carried out at the Integrated Microscopy Center, Cornell University.
The studies at Cornell University were supported by Public Health Service grants R01-GM50740 to J.D.B. and F32-GM18164 to N.S.T.
REFERENCES
- 1.Adams R. Identification and characterisation of the protein encoded by gene UL49A of herpes simplex virus type 1, and investigation of its role in the virus lifecycle. Ph.D. thesis. Glasgow, United Kingdom: University of Glasgow; 1999. [Google Scholar]
- 2.Adams R, Cunningham C, Davison M D, MacLean C A, Davison A J. Characterization of the protein encoded by gene UL49A of herpes simplex virus type 1. J Gen Virol. 1998;79:813–823. doi: 10.1099/0022-1317-79-4-813. [DOI] [PubMed] [Google Scholar]
- 3.Baines J D, Roizman B. The UL10 gene of herpes simplex virus 1 encodes a novel glycoprotein, gM, which is present in the virion and in the plasma membrane of infected cells. J Virol. 1993;67:1441–1452. doi: 10.1128/jvi.67.3.1441-1452.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baines J D, Cunningham C, Nalwanga D, Davison A J. The UL15 gene of herpes simplex virus type 1 contains within its second exon a novel open reading frame that is translated in frame with the UL15 gene product. J Virol. 1997;71:2666–2673. doi: 10.1128/jvi.71.4.2666-2673.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baines J D, Jacob R J, Simmerman L, Roizman B. The herpes simplex virus 1 UL11 proteins are associated with cytoplasmic and nuclear membranes and with nuclear bodies of infected cells. J Virol. 1995;69:825–833. doi: 10.1128/jvi.69.2.825-833.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barker D E, Roizman B. Identification of three genes nonessential for growth in cell culture near the right terminus of unique sequences of long component of herpes simplex virus 1. Virology. 1990;177:684–691. doi: 10.1016/0042-6822(90)90534-x. [DOI] [PubMed] [Google Scholar]
- 7.Brasch K, Ochs R L. Nuclear bodies (NBs): a newly “rediscovered” organelle. Exp Cell Res. 1992;202:211–223. doi: 10.1016/0014-4827(92)90068-j. [DOI] [PubMed] [Google Scholar]
- 8.Brown S M, Ritchie D A, Subak-Sharpe J H. Genetic studies with herpes simplex type 1. The isolation of temperature-sensitive mutants, their arrangement into complementation groups and recombination analysis leading to a linkage map. J Gen Virol. 1973;18:329–346. doi: 10.1099/0022-1317-18-3-329. [DOI] [PubMed] [Google Scholar]
- 9.Carmichael E P, Kosovsky M J, Weller S K. Isolation and characterization of herpes simplex virus type 1 host range mutants defective in viral DNA synthesis. J Virol. 1988;62:91–99. doi: 10.1128/jvi.62.1.91-99.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coull J M, Pappin D J C. A rapid fluorescent staining procedure for proteins electroblotted onto PVDF membranes. J Protein Chem. 1990;9:259–260. [Google Scholar]
- 11.Coulter L J, Moss H W M, Lang J, McGeoch D J. A mutant of herpes simplex virus type 1 in which the UL13 protein kinase gene is disrupted. J Gen Virol. 1993;74:387–395. doi: 10.1099/0022-1317-74-3-387. [DOI] [PubMed] [Google Scholar]
- 12.Cunningham C, Davison A J. A cosmid-based system for constructing mutants of herpes simplex virus type 1. Virology. 1993;197:116–124. doi: 10.1006/viro.1993.1572. [DOI] [PubMed] [Google Scholar]
- 13.Cunningham C, Davison A J, Dolan A, Frame M C, McGeoch D J, Meredith D M, Moss H W M, Orr A C. The UL13 virion protein of herpes simplex virus type 1 is phosphorylated by a novel virus-induced protein kinase. J Gen Virol. 1992;73:303–311. doi: 10.1099/0022-1317-73-2-303. [DOI] [PubMed] [Google Scholar]
- 14.Daikoku T, Ikenoya K, Yamada H, Goshima F, Nishiyama Y. Identification and characterization of the herpes simplex virus type 1 UL51 gene product. J Gen Virol. 1998;79:3027–3031. doi: 10.1099/0022-1317-79-12-3027. [DOI] [PubMed] [Google Scholar]
- 15.Davison A J, Davison M D. Identification of structural proteins of channel catfish virus by mass spectrometry. Virology. 1995;206:1035–1043. doi: 10.1006/viro.1995.1026. [DOI] [PubMed] [Google Scholar]
- 16.Davison A J, Scott J E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986;67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 17.de Wind N, Domen J, Berns A. Herpesviruses encode an unusual protein-serine/threonine kinase which is nonessential for growth in cultured cells. J Virol. 1992;66:5200–5209. doi: 10.1128/jvi.66.9.5200-5209.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dolan A, Jamieson F E, Cunningham C, Barnett B C, McGeoch D J. The genome sequence of herpes simplex virus type 2. J Virol. 1998;72:2010–2021. doi: 10.1128/jvi.72.3.2010-2021.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elliott G, O'Hare P. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J Virol. 1999;73:4110–4119. doi: 10.1128/jvi.73.5.4110-4119.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 21.McLauchlan J, Liefkens K, Stow N D. The herpes simplex virus type 1 UL37 gene product is a component of virus particles. J Gen Virol. 1994;75:2047–2052. doi: 10.1099/0022-1317-75-8-2047. [DOI] [PubMed] [Google Scholar]
- 22.MacLean A R, Ul-Fareed M, Robertson L, Harland J, Brown S M. Herpes simplex virus type 1 deletion variants 1714 and 1716 pinpoint neurovirulence-related sequences in Glasgow strain 17+ between immediate early gene 1 and the 'a‘ sequence. J Gen Virol. 1991;72:631–639. doi: 10.1099/0022-1317-72-3-631. [DOI] [PubMed] [Google Scholar]
- 23.MacLean C A, Robertson L M, Jamieson F E. Characterization of the UL10 gene product of herpes simplex virus type 1 and investigation of its role in vivo. J Gen Virol. 1993;74:975–983. doi: 10.1099/0022-1317-74-6-975. [DOI] [PubMed] [Google Scholar]
- 24.McNabb D S, Courtney R J. Analysis of the UL36 open reading frame encoding the large tegument protein (ICP1/2) of herpes simplex virus type 1. J Virol. 1992;66:7581–7584. doi: 10.1128/jvi.66.12.7581-7584.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nalwanga D, Rempel S, Roizman B, Baines J D. The UL16 gene product of herpes simplex virus 1 is a virion protein that colocalizes with intranuclear capsid proteins. Virology. 1996;226:236–242. doi: 10.1006/viro.1996.0651. [DOI] [PubMed] [Google Scholar]
- 26.Newcomb W W, Trus B L, Booy F P, Steven A C, Wall J S, Brown J C. Structure of the herpes simplex virus capsid: molecular composition of the pentons and the triplexes. J Mol Biol. 1993;232:499–511. doi: 10.1006/jmbi.1993.1406. [DOI] [PubMed] [Google Scholar]
- 27.Pappin D J C, Hojrup P, Bleasby A J. Rapid identification of proteins by peptide-mass fingerprinting. Curr Biol. 1993;3:327–332. doi: 10.1016/0960-9822(93)90195-t. [DOI] [PubMed] [Google Scholar]
- 28.Pappin D J C, Rahman D, Hansen H F, Jeffrey W, Sutton C W. Peptide-mass fingerprinting as a tool for the rapid identification and mapping of cellular proteins. In: Atassi M, Appella E, editors. Methods in protein structural analysis. New York, N.Y: Plenum Press; 1995. pp. 161–173. [Google Scholar]
- 29.Pomeranz L E, Blaho J A. Modified VP22 localizes to the cell nucleus during synchronized herpes simplex virus type 1 infection. J Virol. 1999;73:6769–6781. doi: 10.1128/jvi.73.8.6769-6781.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poon A P W, Roizman B. Characterization of a temperature-sensitive mutant of the UL15 open reading frame of herpes simplex virus 1. J Virol. 1993;67:4497–4503. doi: 10.1128/jvi.67.8.4497-4503.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puvion-Dutilleul F, Pichard E. Viral alkaline nuclease in intranuclear dense bodies induced by herpes simplex infection. Biol Cell. 1986;58:15–22. doi: 10.1111/j.1768-322x.1986.tb00485.x. [DOI] [PubMed] [Google Scholar]
- 32.Robertson L M, MacLean A R, Brown S M. Peripheral replication and latency reactivation kinetics of the non-neurovirulent herpes simplex virus type 1 variant 1716. J Gen Virol. 1992;73:967–970. doi: 10.1099/0022-1317-73-4-967. [DOI] [PubMed] [Google Scholar]
- 33.Schmitt J, Keil G M. Identification and characterization of the bovine herpesvirus 1 UL7 gene and gene product which are not essential for virus replication in cell culture. J Virol. 1996;70:1091–1099. doi: 10.1128/jvi.70.2.1091-1099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szilágyi J F, Cunningham C. Identification and characterization of a novel non-infectious herpes simplex virus-related particle. J Gen Virol. 1991;72:661–668. doi: 10.1099/0022-1317-72-3-661. [DOI] [PubMed] [Google Scholar]
- 35.Telford E A R, Watson M S, McBride K, Davison A J. The DNA sequence of equine herpesvirus-1. Virology. 1992;189:304–316. doi: 10.1016/0042-6822(92)90706-u. [DOI] [PubMed] [Google Scholar]
- 36.Telford E A R, Watson M S, Perry J, Cullinane A A, Davison A J. The DNA sequence of equine herpesvirus 4. J Gen Virol. 1998;79:1197–1203. doi: 10.1099/0022-1317-79-5-1197. [DOI] [PubMed] [Google Scholar]
- 37.Vlcek C, Benes V, Lu Z, Kutish G F, Paces V, Rock D, Letchworth G J, Schwyzer M. Nucleotide sequence analysis of a 30-kb region of the bovine herpesvirus 1 genome which exhibits a colinear gene arrangement with the UL21 to UL4 genes of herpes simplex virus. Virology. 1995;210:100–108. doi: 10.1006/viro.1995.1321. [DOI] [PubMed] [Google Scholar]
- 38.Ward P L, Ogle W O, Roizman B. Assemblons: nuclear structures defined by aggregation of immature capsids and some tegument proteins of herpes simplex virus 1. J Virol. 1996;70:4623–4631. doi: 10.1128/jvi.70.7.4623-4631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wardlaw A C. Practical statistics for experimental biologists. New York, N.Y: Wiley-Interscience Publications; 1987. [Google Scholar]