Abstract
Human immunodeficiency virus type 1 (HIV-1) infects the central nervous system (CNS) and plays a direct role in the pathogenesis of AIDS dementia. However, mechanisms underlying HIV-1 gene expression in the CNS are poorly understood. The importance of CCAAT/enhancer binding proteins (C/EBP) for HIV-1 expression in cells of the immune system has been recently reported. In this study, we have examined the role and the molecular mechanisms by which proteins of the C/EBP family regulate HIV-1 gene transcription in human brain cells. We found that NF-IL6 acts as a potent activator of the long terminal repeat (LTR)-driven transcription in microglial and oligodendroglioma cells. In contrast, C/EBPγ inhibits NF-IL6-induced activation. Consistent with previous data, our transient expression results show cell-type-specific NF-IL6-mediated transactivation. In glial cells, full activation needs the presence of the C/EBP binding sites; however, NF-IL6 is still able to function via the minimal −40/+80 region. In microglial cells, C/EBP sites are not essential, since NF-IL6 acts through the −68/+80 LTR region, containing two binding sites for the transcription factor Sp1. Moreover, we show that functional interactions between NF-IL6 and Sp1 lead to synergistic transcriptional activation of the LTR in oligodendroglioma and to mutual repression in microglial cells. We further demonstrate that NF-IL6 physically interacts with the nuclear receptor chicken ovalbumin upstream promoter transcription factor (COUP-TF), via its DNA binding domain, in vitro and in cells, which results in mutual transcriptional repression. These findings reveal how the interplay of NF-IL6 and C/EBPγ, together with Sp1 and COUP-TF, regulates HIV-1 gene transcription in brain cells.
Human immunodeficiency virus type 1 (HIV-1) infects the central nervous system (CNS) and causes a multitude of clinical complications such as AIDS dementia (12, 44). Microglial cells, the CNS resident macrophages (42), are the most productively infected cells in the brain (43, 45, 60, 62). Recent studies have proposed that astrocytes, oligodendrocytes and more rarely neuronal cells, which harbor a restricted infection with HIV-1, also contribute to the development of CNS disease (16, 39). Infected microglia was recently shown to transmit the virus to oligodendrocytes (2). However, the cellular and molecular mechanisms responsible for the neurological damage are not yet resolved.
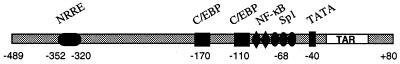
Once HIV-1 is integrated in the host chromosome, transcription of the viral genome is governed by interactions of viral and cellular transcription factors with the long terminal repeat (LTR) (for reviews, see references 13 and 27). Only few studies have examined the nature and the role of transcription factors which control HIV-1 gene expression in brain cells. The transcription factor NF-κB was shown to activate transcription via κB regulatory sequences of the LTR, both in neurons (25, 46) and in astrocytes (55). Our recent data have revealed the importance of the orphan nuclear receptor COUP-TFI/Ear3 (36, 58, 59), which activates HIV-1 gene expression in brain cells. We have described that in human oligodendroglioma TC-620 cells, COUP-TF functions as a potent transcriptional activator by acting on the −352/−320 DNA target site, the nuclear receptor response element (NRRE) (50) (Fig. 1); in contrast, in microglial cells, COUP-TF mediates transcriptional activation by acting on the −68/+29 proximal promoter region, via direct physical and functional interactions with the Sp1 transcription factor (47).
FIG. 1.
Localization of some binding sites in the LTR of HIV-1. NRRE, C/EBP, NF-κB, Sp1, TATA elements, and the TAR region are indicated.
NF-IL6, a member of the C/EBP family of transcription factors, was discovered to play a central role in the control of HIV-1 gene expression in cells of the immune system. The C/EBP family belongs to a class of basic region-leucine zipper (bZIP) proteins, which include C/EBPα (4), C/EBPβ (1, 6, 11, 63), C/EBPγ (Ig/EBP [9, 48]), and C/EBPδ (6) (for a review, see reference 61). Various groups independently reported the cloning of C/EBPβ cDNAs from human (NF-IL6 [1]), rat (LAP [11]), and mouse (C/EBPβ [6]; CRP2 [63]). These proteins contain a leucine zipper domain and a DNA binding basic region located in the C-terminal half of the protein. NF-IL6 was shown to activate transcription from the HIV-1 LTR in Jurkat and HepG2 cells (57) and in the promonocytic cell line U937 (19, 56). The formation of C/EBP–NF-κB heterodimers (53) was shown to synergistically activate transcription of the HIV-1 genome in teratocarcinoma cells via the NF-κB sequences (49). Recent studies have demonstrated that C/EBP proteins and their binding sites are required for HIV-1 replication in promonocytic U937 cells (18) and in primary macrophages but not in CD4+ T lymphocytes (17). In glioblastoma U318 and neuroblastoma SHSY5Y cells, mouse C/EBP proteins were unable to activate HIV gene transcription and downregulated the HIV-1 promoter activity (38).
In this study, we have investigated the molecular mechanisms by which C/EBP proteins regulate HIV-1 gene transcription in human microglial and glial cells of the brain. We first show that two members of the C/EBP family, NF-IL6 and C/EBPγ, are expressed in human brain cells. Our results show that NF-IL6 stimulates LTR-driven transcription via cell-type-specific mechanisms. Moreover, our data reveal the existence of novel functional interactions between NF-IL6 and the transcription factors Sp1 and COUP-TF which lead to cell-type-specific synergistic or inhibitory transcriptional effects. These data describe complex interactions between the transcriptional activators NF-IL6, Sp1, COUP-TF, and the transdominant negative inhibitor C/EBPγ in the regulation of HIV-1 gene transcription in CNS cells.
MATERIALS AND METHODS
Plasmids constructs.
The LTR (JR-FL)-chloramphenicol acetyltransferase (CAT) and LTR (LAI)-CAT vectors were described previously (50, 51). Construction of the glutathione S-transferase (GST)–COUP-1 and GST–COUP-3 vectors of Sp1 in pBluescript KS− was described previously (47). The cDNAs coding for NF-IL6 and NF-IL6ΔSp1 were excised from pEF-NF-IL6 and pEF-NF-IL6ΔSpl (gift from S. Akira, Osaka, Japan) (34) with EcoRI and religated into the EcoRI site of pcDNA3 containing the T7 promoter. To construct CMV (cytomegalovirus)-C/EBPβ, the cDNA coding for C/EBPβ was excised from pMSV-C/EBPβ (gift of A. Henderson, New York, N.Y.) with EcoRI and XhoI and religated into the EcoRI and XhoI sites of pcDNA3 containing the CMV promoter. The HIV-1(HXB2) LTRΔGC vector (gift of B. Sawaya, Philadelphia, Pa.) contains a −76/−40 deletion.
Cell culture, transfections, and CAT assays.
Human microglial (21) and astrocytoma U373-MG cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and 10 mM HEPES in the presence of penicillin-streptomycin (100 U/ml). Human oligodendroglioma TC-620 cells (35) were grown in Iscove medium containing 10% non-heat-inactivated fetal calf serum and 1% gentamicin. When indicated, cells were treated with interleukin-1 (IL-1; 100 U/ml), IL-6 (400 U/ml), or tumor necrosis factor alpha (TNF-α; 100 U/ml) (Genzyme) over a period of 24 h.
Cells (106) were either transfected by the calcium phosphate precipitation method as described previously (47) with 1 pmol of plasmid reporter DNA or cotransfected with reporter DNA (1 pmol) and one of the following expression vectors: MSV (Moloney sarcoma virus)-C/EBPβ or CMV-C/EBPβ (0.5 pmol), CMV-C/EBPγ (0.5 pmol; gift of A. Henderson, New York, N.Y.), pEF-NF-IL6 (0.5 pmol; gift of S. Akira, Hyogo, Japan), RSV-COUP-TF (0.2 pmol; gift of M. J. Tsai, Houston, Tex.), or CMV-Sp1 (0.5 pmol; gift of R. Tjian, Berkeley, Calif.). Each transfection was done in duplicate and repeated a minimum of three separate times with at least two different plasmid preparations. When indicated, cells were treated with phorbol myristate acetate (PMA; 10 ng/ml) 24 h after transfection and incubated for another 24 h before harvesting. Cell extracts were prepared 48 h after transfection. CAT assays were performed as described previously (50). Reaction mixtures containing 15 μg of protein were incubated at 37°C for 2 h.
Western blot analysis.
Nuclear proteins (10 μg) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10 or 15% polyacrylamide gel) and transferred to nitrocellulose paper. Membranes were preincubated with 3% bovine serum albumin in phosphate-buffered saline (PBS) overnight at 4°C and were probed with polyclonal antibodies (1:2,000 dilution) directed against C/EBPα (sc-61X; Santa Cruz Biotechnology), C/EBPβ (sc-150X), C/EBPδ (sc-636X), or C/EBPγ (sc-7658X) for 90 min at room temperature in PBS–0.1% Tween 20 (PBST). After three washes with PBST, membranes were incubated with peroxidase-labeled anti-rabbit antibody (1:7,500 dilution; Santa Cruz Biotechnology) for 40 min and washed four times for 30 min in PBST. The signal was visualized by enhanced chemiluminescence (ECL kit; Amersham).
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were performed with nuclear proteins as described previously (50). Mixtures were incubated for 15 min at 4°C, and protein-DNA complexes were analyzed by electrophoresis on a 6% polyacrylamide gel in 0.25× Tris-borate-EDTA. For supershift assays, antibodies directed against C/EBPα, C/EBPβ, C/EBPδ (Santa Cruz Biotechnology), C/EBPγ (gift from A. Henderson, Columbia University, New York, N.Y.), DBP (gift from P. Fonjallaz, Geneva, Switzerland), USF (Santa Cruz Biotechnology), or normal rabbit serum were mixed with nuclear proteins for 4 h at 4°C prior to addition of the probe. The sequence of the synthetic PRII oligonucleotide is 5′-GGAGAGGGGCGATTGGGCAACCCGG-3′ (50).
GST fusion protein interaction assay.
GST and GST fusion proteins were expressed in Escherichia coli BL-21(DE3). Overnight cultures of bacteria that were newly transformed with the plasmids were diluted with 20 volumes of medium, cultured for several hours to an optical density at 600 nm of 0.6, and induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside at 37°C for 3 h. Bacteria from 125 ml of culture were harvested and resuspended in 1.5 ml of NETN (20 mM Tris [pH 8], 100 mM NaCl, 1 mM EDTA, 0.5% NP-40, 10 μg of leupeptin/ml, 10 μg of pepstatin/ml, 10 μg of aprotinin/ml, 1 mM phenylmethylsulfonyl fluoride). The lysates were sonicated, and after centrifugation, the supernatants were mixed with glutathione-Sepharose 4B beads (40 μl; Pharmacia) at 4°C overnight in NETN buffer. The 35S-labeled input protein was prepared by in vitro translation using the TNT T7 system (Promega) according to the manufacturer's suggestions. The coated beads (40 μl) were washed with NETN and further incubated for 2 h at 4°C with 15 μl of the total in vitro-translated protein reaction mixture in a final volume of 300 μl of binding buffer (50 mM Tris-Cl [pH 7.6], 50 mM NaCl, 0.02% Tween 20, 0.02% bovine serum albumin) containing antiproteases as in NETN. After extensive washing with washing buffer (50 mM Tris-Cl [pH 7.6], 150 mM NaCl, 0.02% Tween 20) containing the antiproteases, the bound proteins were dissociated by boiling for 5 min in Laemmli sample buffer and subjected to SDS-PAGE.
Immunoprecipitations.
Nuclear proteins were resuspended in 400 μl of TNE (50 mM Tris [pH 8.0], 1% NP-40, 2 mM EDTA, cocktail of protease inhibitors), mixed with protein A-agarose beads (20 μl), and gently shaken for 1 h at 4°C. The suspension was briefly centrifuged, and the supernatant was mixed with 3 μl of anti-Sp1 or anti-C/EBPβ antibody or preimmune serum. After overnight incubation at 4°C, protein A-agarose (30 μl) was added and mixed for 2 h. After extensive washing of the beads with TNE, 15 μl of beads was processed for SDS-PAGE and Western blotting as described previously (50). To detect Sp1, the bound proteins were dissociated by boiling for 5 min in Laemmli sample buffer containing β-mercaptoethanol (10%) and subjected to SDS-PAGE. To detect COUP-TF, beads were resuspended in Laemmli buffer without β-mercaptoethanol and were subjected to SDS-PAGE without previous boiling; the blot was probed with COUP-TF antiserum (gift from S. K. Karathanasis, Philadelphia, Pa.).
RESULTS
Pattern of C/EBP expression in human brain cells.
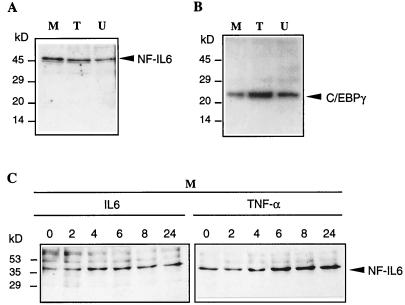
To determine the expression pattern of endogenous proteins of the C/EBP family in different human brain cells, we performed a Western blot analysis with nuclear protein extracts from human oligodendroglioma TC-620, astrocytoma U373-MG, and microglial cells, using a set of specific polyclonal antibodies. Only NF-IL6 (Fig. 2A) and C/EBPγ (Fig. 2B) were detected in glial and microglial cells. C/EBPγ with a predicted molecular mass 16.4 kDa, known to be ubiquitously expressed (48), was detected at higher molecular mass than expected (Fig. 2B). Although the expression of C/EBPδ was found in rat cortical astrocytes (64), we were unable to detect the expression of C/EBPδ in human microglial and glial cells (results not shown).
FIG. 2.
Western blot analysis of nuclear proteins from human brain cells. (A and B) Nuclear protein extracts were prepared from oligodendroglioma TC-620 (lanes T), astrocytoma U373-MG (lanes U), and microglial (lanes M) cells. Proteins (10 μg) were subjected to SDS-PAGE (15% gel), transferred to a nitrocellulose membrane, and probed with antibodies of the C/EBP family. The same gel was cut in two and probed with anti-C/EBPβ (reactive with C/EBPβ and NF-IL6; Santa Cruz Biotechnology) (A) and anti-C/EBPγ (B). The signal was visualized with the Amersham ECL system. (C) Microglial cells were stimulated with IL-6 and TNF-α over a 24-h period. Nuclear proteins were subjected to SDS-PAGE (10% gel), transferred to a nitrocellulose membrane, and probed as in panel A. Positions of prestained molecular weight markers (Bio-Rad) are indicated on the left.
Inflammatory cytokines such as IL-1, IL-6, and TNF-α are elevated in patients with AIDS (5, 28). Therefore, we analyzed the time course of NF-IL6 expression in cells treated with various cytokines over a 24-h period. While IL-1 was unable to significantly alter the level of expression (results not shown), IL-6 and in particular TNF-α led to a significant increase in the level of NF-IL6 expression (Fig. 2C).
NF-IL6 stimulates LTR-driven HIV-1 gene transcription in microglial and glial cells via cell-type-specific mechanisms.
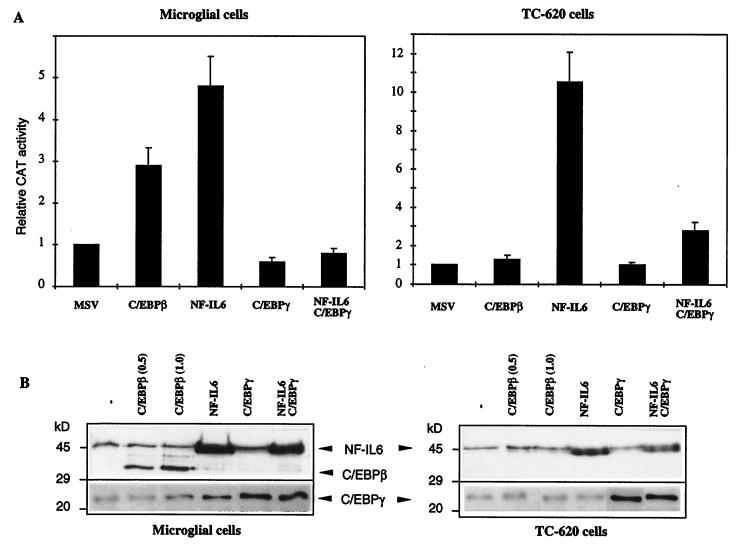
To study the effect of C/EBPβ/NF-IL6 on HIV-1 gene transcription in brain cells, transfection experiments were performed with an LTR-CAT reporter vector containing the CAT gene under the control of the HIV-1(LAI) LTR region and vectors expressing either the mouse C/EBPβ or the human NF-IL6 protein. The human NF-IL6 protein was expressed from the pEF-NF-IL6 vector (0.5 pmol), under the control of the elongation factor 1α promoter (1). The mouse C/EBPβ protein was expressed from the MSV-C/EBPβ vector (0.5 pmol), under the control of the MSV LTR (6). The results in Fig. 3A show that in microglial cells, C/EBPβ acted as a weak transcriptional activator of the HIV-1 genome, since LTR-driven CAT expression was stimulated threefold. C/EBPβ was unable to affect HIV-1 gene transcription in oligodendroglioma TC-620 cells, as already reported for glioblastoma U138MG cells (38). Interestingly, NF-IL6 functioned as a potent activator of LTR-driven transcription, since CAT activity was stimulated 4.8- and 10.5-fold in microglial and TC-620 cells, respectively (Fig. 3A). In astrocytoma U373-MG cells, results were similar with those obtained for TC-620 cells (results not shown).
FIG. 3.
Functional effects of C/EBPβ, C/EBPγ, and NF-IL6 on HIV-1 LTR-driven transcription in human microglial and glial cells. (A) Histogram showing relative CAT activities in microglial and oligodendroglioma TC-620 cells transfected with the LTR-CAT reporter, either alone or in the presence of the indicated expression vectors. The control parental MSV vector contains no cDNA insert. Vectors expressing the mouse C/EBPβ and the human NF-IL6 protein were used. (B) Western blot analysis of nuclear proteins (10 μg) extracted from cells transfected for 48 h with the indicated expression vectors (0.5 pmol). Two concentrations (0.5 and 1.0 pmol) were used for C/EBPβ, as indicated. Blots were probed with the mouse, rat, and human protein-reactive anti-C/EBPβ antibodies (sc-150X; Santa Cruz Biotechnology). Arrows show the endogenous and overexpressed human NF-IL6 protein, the overexpressed mouse C/EBPβ protein, and the endogenous and overexpressed C/EBPγ protein.
Since these two proteins were expressed from different promoters, we examined their level of overexpression in the different cell types. We performed a Western blot analysis with nuclear proteins extracted from cells transfected for 48 h with each vector. We used 0.5 pmol of pEF-NF-IL6 and two concentrations (0.5 and 1.0 pmol) of MSV-C/EBPβ DNA. The blots were probed with anti-C/EBPβ antibodies reactive with the mouse, rat, and human proteins. Figure 3B shows that in addition to the endogenous human NF-IL6 protein, the exogenous NF-IL6 protein was overexpressed in both cell types but at a higher level in microglial cells. In contrast, the mouse C/EBPβ protein, well expressed in microglial cells, was not expressed in TC-620 cells, suggesting that the MSV promoter does not function in TC-620 cells. As seen in Fig. 3B, NF-IL6 and C/EBPβ have distinct molecular masses, since the corresponding human and mouse genes encode proteins with predicted molecular masses of 36 and 31 kDa, respectively (1, 6). These Western blotting results correlate perfectly with the effect and absence of effect of C/EBPβ on LTR-directed transcription in microglial and TC-620 cells, respectively. In microglial cells, where both proteins function as transcriptional activators, it is interesting that the level of CAT stimulation can be correlated with the level of protein overexpression. We further constructed a pcDNA3-C/EBPβ vector in which C/EBPβ is under the control of the CMV promoter, known to function well in microglial and TC-620 cells. Western blots performed with extracts from cells transfected with this vector showed a high level of C/EBPβ expression in microglial cells but no protein expression in TC-620 cells, similar to the results found with the pMSV-C/EBPβ vector, suggesting a control of C/EBPβ expression or degradation in a cell-type-specific manner (results not shown).
The C/EBPγ protein contains the basic and leucine zipper domains but lacks the transcriptional activation domain (9). C/EBPγ was expressed from the CMV-C/EBPγ vector, under the control of the CMV promoter. The CMV promoter was previously shown to function well in microglial and TC-620 cells (47), since it is able to overexpress the Sp1 protein, which functions as a strong transcriptional activator in these cells (see Fig. 6). As expected, overexpression of C/EBPγ (Fig. 3B) did not significantly alter the basal level of transcription; transfection of both C/EBPγ and NF-IL6 vectors in equimolar amounts led to an inhibition of NF-IL6-induced stimulation (Fig. 3A). As a control, the pcDNA3 control plasmid containing only the CMV promoter without any cDNA insert did not affect NF-IL6-mediated CAT stimulation, showing that inhibition of NF-IL6-mediated transcriptional stimulation results from C/EBPγ protein expression (result not shown). As seen in Fig. 3B, cotransfection of both NF-IL6 and C/EBPγ led to a level of overexpression of NF-IL6 similar to that obtained with NF-IL6 alone. This result again demonstrates the previously described transdominant negative effect of C/EBPγ (9). Taken together, these results indicate that HIV-1 gene transcription is controlled by the relative amounts of NF-IL6 and C/EBPγ, thus highlighting the importance of physiological stimuli such as cytokine stimulation which increase the ratio of NF-IL6 to C/EBPγ.
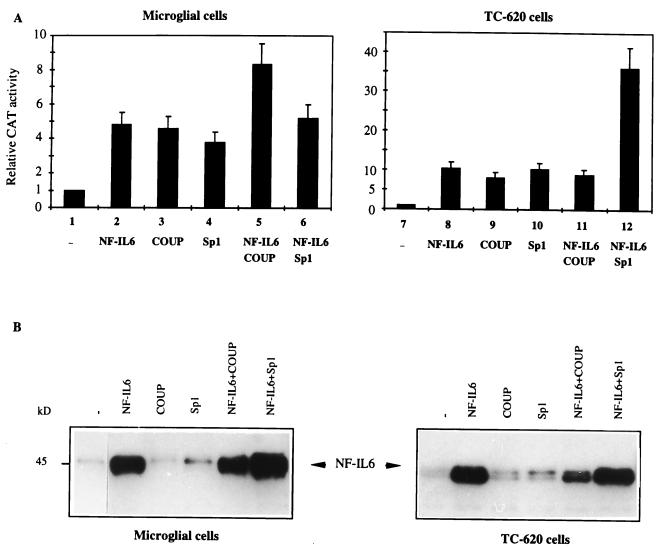
FIG. 6.
Regulation of HIV-1 gene transcription by NF-IL6, COUP-TF, and Sp1 in human microglial and glial cells. (A) Transient expression experiments were performed by cotransfecting HIV-1 LTR-CAT (1 pmol) with vectors expressing NF-IL6 (0.5 pmol), COUP-TF (0.2 pmol), and Sp1 (0.5 pmol) as indicated. Histograms show CAT activities expressed relative to the value obtained with the LTR-CAT reporter vector. Values correspond to an average of at least three independent experiments done in duplicate. (B) Western blot analysis of nuclear proteins (10 μg) extracted from cells transfected for 48 h with the indicated expression vectors (0.5 pmol). Blots were probed with the mouse, rat, and human protein-reactive anti-C/EBPβ antibodies (sc-150X; Santa Cruz Biotechnology), which recognizes the endogenous and overexpressed human NF-IL6 protein.
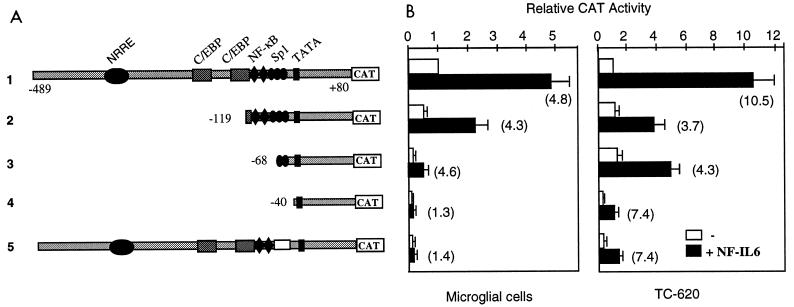
To identify the LTR region that mediates the NF-IL6 response in brain cells, we performed transfection experiments using LTR-CAT vectors containing 5′ deletions of the LTR region. The results (Fig. 4) show that the mechanisms of NF-IL6 action are dependent on the cell type. In microglial cells, deletion of the C/EBP sites up to position −119 (construct 2) and of the NF-κB sites up to position −68 (construct 3) did not abolish the 4.8-fold transcriptional stimulation. With −68/+80 LTR-CAT, NF-IL6 was still able to exert a 4.6-fold transcriptional stimulation. The activation was abolished only by the removal of the two Sp1 binding sites between positions −68 and −40 (construct 4), which indicates that the action of NF-IL6 is mediated via the minimal −68/+80 LTR region. To further confirm that NF-IL6 needs the presence of the two Sp1 elements located within the −68/−40 GC-rich region, we used LTR-CAT containing a deletion of the GC-rich region (construct 5). As expected, NF-IL6 was unable to activate transcription of this deleted LTR in microglial cells.
FIG. 4.
Analysis of the NF-IL6-responsive element in the HIV-1 LTR in human microglial and glial cells. (A) LTR-CAT constructs used in transient expression assays. The LTR region of HIV-1(LAI) was 5′-end deleted. The LTR of HIV-1(HXB2) contains a deletion within the Sp1 region. (B) Relative CAT activities in brain cells transfected with the LTR-CAT constructs, either alone or in the presence of the NF-IL6 expression vector. CAT activities are expressed relative to that of the LTR-CAT construct, taken as 1. Numbers in parentheses indicate the fold stimulation induced by NF-IL6. Values correspond to an average of at least three independent experiments done in duplicate.
In contrast, in TC-620 cells, the C/EBP sites contribute to mediate the NF-IL6 action, since their deletion (constructs 2 and 3) reduced CAT activity to about half. In these cells, the Sp1 sites were not essential for NF-IL6 responsiveness (construct 4); this was confirmed with construct 5: in the absence of Sp1 sites, NF-IL6 was still able to stimulate transcription. Moreover, although the basal level of transcription was strongly reduced with −40/+80 LTR-CAT, the basal sequences containing only the TATA region were still able to mediate part of the NF-IL6 response. These results show that in TC-620 cells, the action of NF-IL6 is mediated by the C/EBP sites and the TATA region. Similar results (not shown) were obtained for astrocytoma U373-MG cells.
Analysis of nuclear proteins present in microglial and glial cells which bind to the C/EBP sites in the LTR of HIV-1.
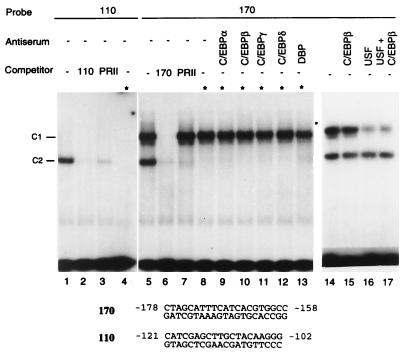
Two high-affinity C/EBP sites centered around positions −170 and −110 of the LTR were shown to be necessary for HIV-1 replication in macrophages but not in T cells (17). To investigate whether C/EBP proteins present in human brain cells interact with these sites, we performed EMSAs with nuclear proteins isolated from microglial and glial cells (Fig. 5). Two complexes, C1 and C2, were formed with oligonucleotide probe 170 (lane 5), while only one complex, C2, was formed with probe 110 (lane 1). The formation of complex C2 was abolished in the presence of an excess of unlabeled homologous or heterologous competitor 110 or PRII, known as a binding site for C/EBP in the promoter of the human transferrin gene (52) (lanes 2, 3, 6, and 7). The formation of complex C1 was specific, since it was abolished only by an excess of homologous competitor (lane 6). To identify which complex contained C/EBP, we first used the property of C/EBP to be heat resistant; after incubation of nuclear extracts at 80°C for 5 min, the formation of complex C2 was abolished (lanes 4 and 8), indicating that the protein binding to oligonucleotide 110 does not belong to the C/EBP family. In contrast, the formation of complex C1 was unaltered (lane 8). When supershift experiments were performed in the presence of C/EBP-α, -β, -δ, or -γ or DBP antibodies, both complexes C1 and C2 were unaffected (lanes 9 to 13), indicating that no protein of the C/EBP family present in nuclear extracts is able to interact with the 170 site. However, by gel shift assays performed with the 170 probe and in vitro-translated NF-IL6, we confirmed previous reports that NF-IL6 is able to bind to the 170 site (49); interestingly, in vitro-translated C/EBPβ was unable to bind to the 170 sequence (results not shown).
FIG. 5.
Binding of nuclear proteins present in microglial cells to the C/EBP sites of the HIV-1 LTR. Nuclear protein extracts from microglial cells (5 μg) were assayed by gel retardation assay with 1 ng of 32P-end-labeled oligonucleotide probe 110 or 170. The DNA-protein complexes C1 and C2 are indicated. For supershift assays, 1 μl of the indicated antiserum was mixed with nuclear proteins and incubated 4 h prior addition of probe 170. The sequence of the synthetic oligonucleotides present in the LTR is indicated. The PRII sequence is a C/EBP binding site in the promoter of the transferrin gene (52). Asterisks indicate nuclear extracts that were heated for 5 min at 80°C.
It has been reported that the basic loop helix leucine zipper proteins TFE3 and USF bind to the E box that overlaps the −170 bp region (10, 57) and compete with NF-IL6 for binding to the 170 sequence (19). Our supershift experiments using anti-USF antibodies confirmed that USF forms the majority of complex C1 (Fig. 5A, lanes 15 to 17). These findings indicate that although NF-IL6 is able to interact with the 170 site, in the cellular context where NF-IL6 is not overexpressed, USF rather than NF-IL6 binds to this site.
Regulation of HIV-1 LTR-directed gene transcription by NF-IL6, Sp1, and COUP-TF in unstimulated and PMA-stimulated brain cells.
We have recently demonstrated that in microglial cells, the −68/−40 GC-rich region is the binding site for transcription factors Sp1 and Sp3 (47). While Sp1 activates transcription, Sp3 functions as an inhibitor of LTR-driven transcription (47). This prompted us to investigate how NF-IL6 modulates Sp1-mediated transcriptional stimulation. Combinations of the expression vectors for NF-IL6 and Sp1 were cotransfected with the LTR-CAT reporter vector (Fig. 6A).
In microglial cells, overexpression of both NF-IL6 and Sp1 proteins (lane 6) did not alter CAT activity obtained with each individual protein (lanes 2 and 4), suggesting a mutual repression of both proteins on the full-length LTR. In contrast, in TC-620 cells, a synergistic stimulation of transcription was observed: while overexpression of either NF-IL6 or Sp1 led to 10.5- or 10.2-fold stimulation of CAT activity, respectively (lanes 8 and 10), their combined action led to a 36-fold transcriptional increase (lane 12). Similarly, with the −68/+80 LTR-CAT vector, the combination of NF-IL6 and Sp1 led to a synergistic 39-fold stimulation (results not shown). These results demonstrate the cell type specificity of the transcriptional effects mediated by NF-IL6 and Sp1 and further suggest that NF-IL6 interacts with Sp1, directly or indirectly. To test whether these interactions are direct, we performed coimmunoprecipitation experiments with extracts from microglial cells transfected with NF-IL6 and Sp1 expression vectors. Results did not reveal any direct interaction between the two proteins.
We next investigated how NF-IL6 modulates HIV-1 gene transcription in the presence of the nuclear receptor COUP-TF, since we recently demonstrated physical and functional interactions between Sp1 and COUP-TF, leading to a synergistic transcriptional stimulation via the −68/+29 LTR region in microglial cells (47). In these cells, overexpression of NF-IL6 and COUP-TF led to an additive effect with LTR-CAT (lanes 2, 3, and 5). In contrast, in TC-620 cells, CAT activity was slightly altered by the combination of both proteins (lane 11) compared with each protein alone (lanes 8 and 9), suggesting a mutual inhibition through cross-coupling interactions. These results indicate cell-type-specific functional interactions between NF-IL6 and COUP-TF.
To examine whether overexpression of COUP-TF or Sp1 altered the expression level of the NF-IL6 protein, we performed Western blotting experiments with nuclear extracts from microglial and TC-620 cells transfected with either NF-IL6, NF-IL6 and COUP-TF, or NF-IL6 and Sp1 expression vectors (Fig. 6B). Results show that overexpression of COUP-TF did not modify the level of NF-IL6 expression in microglial cells but led to an additive transcriptional effect (Fig. 6A, lane 5); in TC-620 cells, Sp1 appeared to slightly decrease the overexpressed level NF-IL6 compared with NF-IL6 alone; however, this overexpressed level was still sufficient for a transcriptional stimulation similar to that of NF-IL6 (Fig. 6A, lane 11). In microglial cells, overexpression of Sp1 appeared to increase the level of NF-IL6 but did not lead to any additive or synergistic transcriptional effect, suggesting that the level of NF-IL6 alone was already saturating (Fig. 6A, lane 6); interestingly, in TC-620 cells, where a synergistic transcriptional effect was detected (Fig. 6A, lane 12), overexpression of Sp1 did not alter the level of overexpressed NF-IL6. These results support the idea of direct or indirect cell-type-specific interactions of these proteins in the regulation of HIV-1 gene transcription.
These data concern the immediate-early phase, where viral transcription proceeds through dependence on solely cellular transcription factors present in the nucleus. Recent reports showed that the formation of NF-κB–C/EBP heterodimers results in a potent activation of HIV-1 LTR in NTera cells (49). It was therefore of interest to examine how NF-IL6 regulates HIV-1 gene transcription in conditions where stimuli such as PMA induce NF-κB nuclear translocation. Cells were cotransfected with the LTR-CAT reporter and various combinations of NF-IL6, Sp1, and COUP-TF expression vectors and were treated with PMA for 24 h. The presence of NF-κB in nuclear extracts was controlled by gel shift experiments (results not shown). Comparison of CAT activities measured with untreated and PMA-stimulated cells revealed a dramatic effect in TC-620 cells, where PMA treatment stimulated four- to fivefold the basal activity as well as the Sp1-, COUP-TF-, and NF-IL6-mediated transcriptional activities. In microglial cells, PMA treatment led to a twofold stimulation of transcription (Table 1).
TABLE 1.
Fold induction following treatment with PMAa
| Expression vector(s) | Relative CAT activityb
|
|
|---|---|---|
| Microglial cells | TC-620 cells | |
| None | 1.6 | 4.2 |
| NF-IL6 | 1.9 | 4.5 |
| COUP | 1.9 | 4.1 |
| Sp1 | 1.9 | 3.6 |
| NF-IL6 + COUP | 1.9 | 4.2 |
| NF-IL6 + Sp1 | 2.2 | 2.3 |
Cells were cotransfected with the LTR-CAT reporter construct and the indicated expression vectors and then cultured in the absence or the presence of PMA (10 ng/ml) for 24 h. CAT activities were determined 48 h after transfection.
Mean of three independent experiments done in duplicate. Standard deviation did not exceed 20%.
NF-IL6 and COUP-TF interact in vitro and in cells.
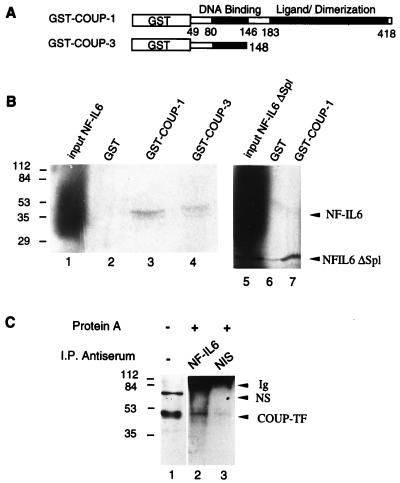
Our transient expression data suggest functional interactions between NF-IL6 and COUP-TF. We therefore investigated whether these proteins are able to interact physically. To examine in vitro interactions, we analyzed the ability of in vitro-translated NF-IL6 in the presence of [35S]methionine to interact with the GST–COUP-1 fusion protein (Fig. 7A). SDS-PAGE analysis of proteins retained by glutathionine-Sepharose shows that 35S-labeled NF-IL6 associates with GST-COUP-1 (Fig. 7B, lane 3). This association is specific, since NF-IL6 was bound to GST–COUP-1 but not to GST alone (lane 2).
FIG. 7.
NF-IL6 interacts with COUP-TF in vitro and in vivo. (A) Schematic representation of GST-COUP-TF constructs. (B) Visualization of the in vitro interaction between NF-IL6 and COUP-TF. 35S-labeled full-length NF-IL6 (lane 1) and truncated NF-IL6ΔSp1 (lane 5) translated in wheat germ lysate (Promega) were incubated with bacterially expressed GST, GST–COUP-1, or GST–COUP-3 immobilized on glutathione-Sepharose beads. After extensive washing, the bound proteins were eluted and analyzed by SDS-PAGE followed by autoradiography (lanes 2 to 4, 6 and 7). (C) Coimmunoprecipitation of NF-IL6 and COUP-TF. Nuclear protein extracts from microglial cells (50 μg) were immunoprecipitated (I.P.) in the presence of anti-NF-IL6 antibodies (lane 2) or nonimmune serum (NIS; lane 3), followed by Western blotting with COUP-TF antibodies. Lane 1 corresponds to nonimmunoprecipitated nuclear proteins. Positions of size standards are indicated in kilodaltons. Ig, immunoglobulin; NS, nonspecific band.
To localize the domain of COUP-TF that mediates interaction with NF-IL6, we used the GST–COUP-3 expression vector encoding residues 49 to 148 of COUP-TF (Fig. 7A). Results from GST pull-down experiments show that GST–COUP-3 was still able to mediate association with NF-IL6 (Fig. 7B, lane 4), confirming that the N-terminal part of COUP-TF containing the DNA binding domain is sufficient for the interaction with NF-IL6 in vitro.
To localize the domain of NF-IL6 involved in the interaction with COUP-TF, we constructed the NF-IL6ΔSpl vector, expressing the C-terminal DNA binding domain under the control of the T7 promoter in pcDNA3. The results of GST pull-down assays showed that 35S-labeled NF-IL6ΔSpl was still able to associate with GST-COUP (lane 7) but not with the control GST (lane 6). Taken together, these results indicate that NF-IL6 and COUP-TF associate via their DNA binding domains.
These in vitro results were confirmed in vivo by immunoprecipitation experiments with extracts from microglial cells that had been transfected with NF-IL6 and COUP-TF vectors (Fig. 7C). Antibodies directed against NF-IL6 (lane 2), but not nonimmune serum (lane 3), were able to immunoprecipitate endogenous COUP-TF species as visualized by Western blotting with COUP-TF antibodies.
DISCUSSION
In this study, we have investigated the regulation of HIV-1 gene transcription in human microglial cells, which are the primary target of HIV-1 infection in the CNS, compared with oligodendroglioma cells. We show here that NF-IL6 and C/EBPγ proteins are present in microglial and glial cells. While NF-IL6 acts as a potent activator of LTR-driven transcription, its transcriptional ability is repressed by C/EBPγ, previously described as a transdominant negative regulator (9). Therefore, the levels of these two proteins, as well as the levels of cytokines such as IL-1 and TNF-α, known to be increased during HIV-1 infection, appear critical in the transcriptional regulation of the HIV-1 genome.
A number of studies have shown that transactivation of the LTR by NF-IL6 includes pathways that can bypass a requirement for direct interaction of NF-IL6 with its binding site, either through a direct cooperation between NF-IL6 and NF-κB (49) or through the basal transcriptional machinery (56). Distinct mechanisms of transcriptional regulation were detected depending on cell type. In monocytic U937 cells, the basal LTR sites retained half of NF-IL6 responsiveness, while in hepatoma HepG2 cells, response to NF-IL6 was achieved with the Sp1 element (56). These authors suggested that activation of the LTR by NF-IL6 involves the basal transcription machinery as well as cooperation with other transcription factors. Our transient expression results for brain cells support these data and show that NF-IL6 can activate HIV-1 LTR-driven transcription, independently of the C/EBP binding sequence, in a cell-type-specific manner. In microglial cells, the minimal −68/+80 LTR region containing two Sp1 binding sites is sufficient for NF-IL6-mediated stimulation. In oligodendroglioma cells, the C/EBP sites contribute to full NF-IL6 responsiveness; moreover, in these glial cells, like in U937 monocytic cells (56), the basal −40/+80 region containing only the TATA site retains a significant fraction of NF-IL6 responsiveness. Moreover, our in vitro data indicate that in the normal cellular context of brain cells, no C/EBP protein binds to the C/EBP sites present in the LTR. It is the USF protein rather than NF-IL6 which binds to the 170 C/EBP site. Since NF-IL6 transactivates the HIV-1 LTR via the −68/+80 or the −40/+80 region, our data indicate that the C/EBP sites are not essential for HIV-1 gene transcription in microglial and glial cells.
Our cotransfection data provide evidence for functional interactions between NF-IL6 and Sp1. In TC-620 cells, a functional cooperation between NF-IL6 and Sp1 leads to a synergistic transcriptional stimulation, further enhanced in the presence of PMA, resulting in a dramatic 90-fold increase in HIV-1 gene transcription. In contrast, in microglial cells, HIV-1 gene transcription is regulated by a mutual inhibition between NF-IL6 and Sp1.
The essential role of the Sp1 transcription factor (24) in the regulation of basal transcription and in Tat-mediated transactivation of the HIV-1 LTR has been well established (15, 23, 54). A direct interaction between Sp1 and the viral protein Tat during transactivation has been described (22, 26). A cooperative interaction between Sp1 and NF-κB, bound to the two adjacent binding sites, is required for optimal HIV-1 enhancer activation and inducible HIV-1 gene expression (31, 33, 41). A physical interaction between Sp1 and the p53 tumor suppressor gene has been found in the TNF-induced transcriptional activation of the HIV-1 LTR (14). Our recent data have demonstrated a physical and functional interaction between Sp1 and the nuclear receptor COUP-TF in microglial cells (47). Although previous reports have shown that NF-IL6 and Sp1 can work in conjunction to activate the CYP2D5 gene (30) and the CD11c integrin gene (32), similar to our coimmunoprecipitation data, no evidence for a direct association between these two transcription factors could be demonstrated. Therefore, interactions of NF-IL6 with Sp1 may involve transcriptional coactivators such as p300, recently described to interact directly with C/EBPβ and NF-IL6 (37). An indirect interaction between Sp1 and p300 via the progesterone receptor (40), another member of the nuclear hormone receptor superfamily, has also been recently described.
We have previously described that in microglial cells, a direct interaction of the N-terminal DNA binding domain of the nuclear receptor COUP-TF with the Sp1 protein results in a synergistic transcriptional activation of the HIV-1 genome (47). In contrast, in oligodendrocytes, COUP-TF stimulates HIV-1 gene transcription by direct interactions with its DNA target site (50). Two members of the COUP-TF family, COUP-TFI/Ear3 and COUP-TFII/ARP-1 are also able to directly target components of the basal transcription machinery, such as the basal transcription factor TFIIB, via the activation domain of COUP-TF (20). Here our findings reveal that the DNA binding domain of COUP-TF is able to associate directly with the C-terminal DNA binding domain of NF-IL6. This interaction, demonstrated both in vitro and in cells, results in a relative inhibition of the transcriptional stimulation mediated by each factor.
The NF-IL6 protein, like all members of the C/EBP family, consists of three structural components: a C-terminal leucine zipper, a basic DNA binding region, and an N-terminal transactivating region (61). Previous studies have demonstrated an association of the C-terminal bZIP region of C/EBP with the Rel homology domain of NF-κB (29, 34, 53). A physical interaction occurs between C/EBPs and the retinoblastoma protein Rb during terminal adipocyte differentiation (8), leading to an activation of NF-IL6. In this case, two distinct regions of NF-IL6 are involved in the binding to the hypophosphorylated form of Rb in vitro and in cells (7). In addition, the viral protein Tat was also shown to physically interact with NF-IL6 in vitro and in vivo (3).
The cell-type-specific mode of action of COUP-TF may help explain the functional data obtained in the presence of Sp1 and NF-IL6. In microglial cells, where COUP-TF functions by direct interaction with the Sp1 protein, NF-IL6 competes with Sp1 for binding to the N-terminal part of COUP-TF, which could result in the observed mutual inhibition. In contrast, in TC-620 cells, where COUP-TF binds to the NRRE site, NF-IL6 is free to interact with proteins bound to Sp1 and thus to synergistically stimulate transcription.
Our findings establish the role of NF-IL6 as a potent transcriptional activator of LTR-directed HIV-1 gene expression in brain cells. They highlight novel mechanisms of HIV-1 gene regulation involving functional interactions between NF-IL6 and the transcription factors Sp1 and COUP-TF, which lead to cell-type-specific regulations in various brain cells. C/EBP proteins were recently shown to be required for provirus induction in monocytic cell lines (18) and in primary macrophages but not in T cells (17). It is therefore essential in our following experiments to further investigate the role of NF-IL6 in HIV-1 replication and induction of latent HIV-1 provirus in different brain cells.
ACKNOWLEDGMENTS
We thank N. Israël and J. Clements for providing the vectors containing the LAI and JR-FL LTRs, respectively. We are grateful to S. Akira for providing the NF-IL6 vectors, to A. Henderson for providing the C/EBPγ vector and antibodies, to R. Tjian for providing the Sp1 expression vector, and to S. K. Karathanasis for providing the COUP-TF antibodies.
This work was supported by the Institut National de la Santé et de la Recherche Médicale (INSERM), the Agence Nationale des Recherches sur le SIDA, the Fondation pour la Recherche Medicale, and the association Le Cercle d'Emeraude. C.S. received a fellowship from INSERM.
REFERENCES
- 1.Akira S, Isshiki H, Sugita T, Tanabe O, Kinoshita S, Nishio Y, Nakajima T, Hirano T, Kishimoto T. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 1990;9:1897–1906. doi: 10.1002/j.1460-2075.1990.tb08316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albright A V, Strizki J, Harouse J M, Lavi E, O'Connor M, Gonzalez S F. HIV-1 infection of cultured human adult oligodendrocytes. Virology. 1996;217:211–219. doi: 10.1006/viro.1996.0108. [DOI] [PubMed] [Google Scholar]
- 3.Ambrosino C, Ruocco M R, Chen X, Mallardo M, Baudi F, Trematerra S, Quinto I, Venuta S, Scala G. HIV-1 Tat induces the expression of the interleukin-6 (IL6) gene by binding to the IL6 leader RNA and by interacting with CAAT enhancer-binding protein β (NF-IL6) transcription factors. J Biol Chem. 1997;272:14883–14892. doi: 10.1074/jbc.272.23.14883. [DOI] [PubMed] [Google Scholar]
- 4.Birkenmeier E H, Gwynn B, Howard S, Jerry J, Gordon J I, Landschulz W H, McKnight S L. Tissue-specific expression, developmental regulation, and genetic mapping of the gene encoding CCAAT/enhancer binding protein. Genes Dev. 1989;3:1146–1156. doi: 10.1101/gad.3.8.1146. [DOI] [PubMed] [Google Scholar]
- 5.Breen E C, Rezai A R, Nakajima K, Beall G N, Mitsuyasu R T, Hirano T, Kishimoto T, Martinez M O. Infection with HIV is associated with elevated IL-6 levels and production. J Immunol. 1990;144:480–484. [PubMed] [Google Scholar]
- 6.Cao Z, Umek R M, McKnight S L. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 1991;5:1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- 7.Chen P L, Riley D J, Chen K S, Lee W H. Retinoblastoma protein directly interacts with and activates the transcription factor NF-IL6. Proc Natl Acad Sci USA. 1996;93:465–469. doi: 10.1073/pnas.93.1.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen P L, Riley D J, Chen Y, Lee W H. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev. 1996;10:2794–2804. doi: 10.1101/gad.10.21.2794. [DOI] [PubMed] [Google Scholar]
- 9.Cooper C, Henderson A, Artandi S, Avitahl N, Calame K. Ig/EBP (C/EBP gamma) is a transdominant negative inhibitor of C/EBP family transcriptional activators. Nucleic Acids Res. 1995;23:4371–4377. doi: 10.1093/nar/23.21.4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Adda di Fagagna F, Marzio G, Gutierrez M I, Kang L Y, Falaschi A, Giacca M. Molecular and functional interactions of transcription factor USF with the long terminal repeat of human immunodeficiency virus type 1. J Virol. 1995;69:2765–2775. doi: 10.1128/jvi.69.5.2765-2775.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Descombes, P., M. Chojkier, S. Lichtsteiner, E. Falvey, and U. Schibler. LAP, a novel member of the C/EBP gene family, encodes a liver-enriched transcriptional activator protein. Genes Dev. 4:1541–1551. [DOI] [PubMed]
- 12.Fauci A S. The human immunodeficiency virus: infectivity and mechanisms of pathogenesis. Science. 1988;239:617–622. doi: 10.1126/science.3277274. [DOI] [PubMed] [Google Scholar]
- 13.Garcia J A, Gaynor R B. The human immunodeficiency virus type-1 long terminal repeat and its role in gene expression. Prog Nucleic Acid Res Mol Biol. 1994;49:157–196. doi: 10.1016/s0079-6603(08)60050-1. [DOI] [PubMed] [Google Scholar]
- 14.Gualberto A, Baldwin A S. p53 and Sp1 interact and cooperate in the tumor necrosis factor-induced transcriptional activation of the HIV-1 long terminal repeat. J Biol Chem. 1995;270:19680–19683. doi: 10.1074/jbc.270.34.19680. [DOI] [PubMed] [Google Scholar]
- 15.Harrich D, Garcia J, Wu F, Mitsuyasu R, Gonazalez J, Gaynor R. Role of Sp1-binding domains in in vivo transcriptional regulation of the human immunodeficiency virus type 1 long terminal repeat. J Virol. 1989;63:2585–2591. doi: 10.1128/jvi.63.6.2585-2591.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatch W C, Pousada E, Losev L, Rashbaum W K, Lyman W D. Neural cell targets of human immunodeficiency virus type 1 in human fetal organotypic cultures. AIDS Res Hum Retroviruses. 1994;10:1597–1607. doi: 10.1089/aid.1994.10.1597. [DOI] [PubMed] [Google Scholar]
- 17.Henderson A J, Calame K L. CCAAT/enhancer binding protein (C/EBP) sites are required for HIV-1 replication in primary macrophages but not CD4(+) T cells. Proc Natl Acad Sci USA. 1997;94:8714–8719. doi: 10.1073/pnas.94.16.8714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henderson A J, Connor R I, Calame K L. C/EBP activators are required for HIV-1 replication and proviral induction in monocytic cell lines. Immunity. 1996;5:91–101. doi: 10.1016/s1074-7613(00)80313-1. [DOI] [PubMed] [Google Scholar]
- 19.Henderson A J, Zou X, Calame K L. C/EBP proteins activate transcription from the human immunodeficiency virus type 1 long terminal repeat in macrophages/monocytes. J Virol. 1995;69:5337–5344. doi: 10.1128/jvi.69.9.5337-5344.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ing N H, Beekman J M, Tsai S Y, Tsai M J, O'Malley B W. Members of the steroid hormone receptor superfamily interact with TFIIB (S300-II) J Biol Chem. 1992;267:17617–17623. [PubMed] [Google Scholar]
- 21.Janabi N, Peudenier S, Heron B, Ng K H, Tardieu M. Establishment of human microglial cell lines after transfection of primary cultures of embryonic microglial cells with the SV40 large T antigen. Neurosci Lett. 1995;195:105–108. doi: 10.1016/0304-3940(94)11792-h. [DOI] [PubMed] [Google Scholar]
- 22.Jeang K T, Chun R, Lin N H, Gatignol A, Glabe C G, Fan H. In vitro and in vivo binding of human immunodeficiency virus type 1 Tat protein and Sp1 transcription factor. J Virol. 1993;67:6224–6233. doi: 10.1128/jvi.67.10.6224-6233.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones K A, Kadonaga J T, Luciw P A, Tjian R. Activation of the AIDS retrovirus promoter by the cellular transcription factor, Sp1. Science. 1986;232:755–759. doi: 10.1126/science.3008338. [DOI] [PubMed] [Google Scholar]
- 24.Kadonaga J T, Carner K R, Masiarz F R, Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987;51:1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- 25.Kaltschmidt C, Kaltschmidt B, Neumann H, Wekerle H, Baeuerle P A. Constitutive NF-κB activity in neurons. Mol Cell Biol. 1994;14:3981–3992. doi: 10.1128/mcb.14.6.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamine J, Chinnadurai G. Synergistic activation of the human immunodeficiency virus type 1 promoter by the viral Tat protein and cellular transcription factor Sp1. J Virol. 1992;66:3932–3936. doi: 10.1128/jvi.66.6.3932-3936.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kingsman S M, Kingsman A J. The regulation of human immunodeficiency virus type-1 gene expression. Eur J Biochem. 1996;240:491–507. doi: 10.1111/j.1432-1033.1996.0491h.x. [DOI] [PubMed] [Google Scholar]
- 28.Lahdevirta J, Maury C P, Teppo A M, Repo H. Elevated levels of circulating cachectin/tumor necrosis factor in patients with acquired immunodeficiency syndrome. Am J Med. 1988;85:289–291. doi: 10.1016/0002-9343(88)90576-1. [DOI] [PubMed] [Google Scholar]
- 29.LeClair K P, Blanar M A, Sharp P A. The p50 subunit of NF-κB associates with the NF-IL6 transcription factor. Proc Natl Acad Sci USA. 1992;89:8145–8149. doi: 10.1073/pnas.89.17.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee Y H, Williams S C, Baer M, Sterneck E, Gonzalez F J, Johnson P F. The ability of C/EBPβ but not C/EBPα to synergize with an Sp1 protein is specified by the leucine zipper and activation domain. Mol Cell Biol. 1997;17:2038–2047. doi: 10.1128/mcb.17.4.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y C, Mak G, Franza B R. In vitro study of functional involvement of Sp1, NF-κB/Rel, and AP1 in phorbol 12-myristate 13-acetate-mediated HIV-1 long terminal repeat activation. J Biol Chem. 1994;269:30616–30619. [PubMed] [Google Scholar]
- 32.Lopez-Rodriguez C, Botella L, Corbi A L. CCAAT-enhancer-binding proteins (C/EBP) regulate the tissue specific activity of the CD11c integrin gene promoter through functional interactions with Sp1 proteins. J Biol Chem. 1997;272:29120–29126. doi: 10.1074/jbc.272.46.29120. [DOI] [PubMed] [Google Scholar]
- 33.Majello B, Deluca P, Hagen G, Suske G, Lania L. Different members of the Sp1 multigene family exert opposite transcriptional regulation of the long terminal repeat of HIV-1. Nucleic Acids Res. 1994;22:4914–4921. doi: 10.1093/nar/22.23.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsusaka T, Fujikawa K, Nishio Y, Mukaida N, Matsushima K, Kishimoto T, Akira S. Transcription factors NF-IL6 and NF-κB synergistically activate transcription of the inflammatory cytokines, interleukin 6 and interleukin 8. Proc Natl Acad Sci USA. 1993;90:10193–10197. doi: 10.1073/pnas.90.21.10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merrill J E, Matsushima K. Production of and response to interleukin 1 by cloned human oligodendroglioma cell lines. J Biol Regul Homeost Agents. 1988;2:77–86. [PubMed] [Google Scholar]
- 36.Miyajima N, Kadowaki Y, Fukushige S, Shimizu S, Semba K, Yamanashi Y, Matsubara K, Toyoshima K, Yamamoto T. Identification of two novel members of erbA superfamily by molecular cloning: the gene products of the two are highly related to each other. Nucleic Acids Res. 1988;16:11057–11074. doi: 10.1093/nar/16.23.11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mink S, Haenig B, Klempnauer K-H. Interaction and functional collaboration of p300 and C/EBPβ. Mol Cell Biol. 1997;17:6609–6617. doi: 10.1128/mcb.17.11.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mondal D, Alam J, Prakash O. NF-κB site-mediated negative regulation of the HIV-1 promoter by CCAAT/enhancer binding proteins in brain-derived cells. J Mol Neurosci. 1994;5:241–258. doi: 10.1007/BF02736725. [DOI] [PubMed] [Google Scholar]
- 39.Nuovo G J, Gallery F, MacConnell P, Braun A. In situ detection of polymerase chain reaction-amplified HIV-1 nucleic acids and tumor necrosis factor-α RNA in the central nervous system. Am J Pathol. 1994;144:659–666. [PMC free article] [PubMed] [Google Scholar]
- 40.Owen G I, Richer J K, Tung L, Takimoto G, Horwitz K B. Progesterone regulates transcription of the p21(WAF1) cyclin-dependent kinase inhibitor gene through Sp1 and CBP/p300. J Biol Chem. 1998;273:10696–10701. doi: 10.1074/jbc.273.17.10696. [DOI] [PubMed] [Google Scholar]
- 41.Perkins N D, Edwards N L, Duckett C S, Agranoff A B, Schmid R M, Nabel G J. A cooperative interaction between NF-κB and Sp1 is required for HIV-1 enhancer activation. EMBO J. 1993;12:3551–3558. doi: 10.1002/j.1460-2075.1993.tb06029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perry V H, Lawson L J, Reid D M. Biology of the mononuclear phagocyte system of the central nervous system and HIV infection. J Leukoc Biol. 1994;56:399–406. doi: 10.1002/jlb.56.3.399. [DOI] [PubMed] [Google Scholar]
- 43.Peudenier S, Hery C, Montagnier L, Tardieu M. Human microglial cells: characterization in cerebral tissue and in primary culture, and study of their susceptibility to HIV-1 infection. Ann Neurol. 1991;29:152–161. doi: 10.1002/ana.410290207. [DOI] [PubMed] [Google Scholar]
- 44.Portegies P, Brew B J. Update on HIV-related neurological illness. AIDS. 1991;5:S211–217. doi: 10.1097/00002030-199101001-00029. [DOI] [PubMed] [Google Scholar]
- 45.Price R W, Brew B, Sidtis J, Rosenblum M, Scheck A C, Cleary P. The brain in AIDS: central nervous system HIV-1 infection and AIDS dementia complex. Science. 1988;239:586–592. doi: 10.1126/science.3277272. [DOI] [PubMed] [Google Scholar]
- 46.Rattner A, Korner M, Walker M D, Citri Y. NF-κB activates the HIV promoter in neurons. EMBO J. 1993;12:4261–4267. doi: 10.1002/j.1460-2075.1993.tb06110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rohr O, Aunis D, Schaeffer E. COUP-TF and Sp1 interact and cooperate in the transcriptional activation of the human immunodeficiency virus type 1 long terminal repeat in human microglial cells. J Biol Chem. 1997;272:31149–31155. doi: 10.1074/jbc.272.49.31149. [DOI] [PubMed] [Google Scholar]
- 48.Roman C, Platero J S, Shuman J, Calame K. Ig/EBP-1: a ubiquitously expressed immunoglobulin enhancer binding protein that is similar to C/EBP and heterodimerizes with C/EBP. Genes Dev. 1990;4:1404–1415. doi: 10.1101/gad.4.8.1404. [DOI] [PubMed] [Google Scholar]
- 49.Ruocco M R, Chen X E, Ambrosino C, Dragonetti E, Liu W M, Mallardo M, Defalco G, Palmieri C, Franzoso G, Quinto I, Venuta S, Scala G. Regulation of HIV-1 long terminal repeats by interaction of C/EBP(NF-IL6) and NF-κB/Rel transcription factors. J Biol Chem. 1996;271:22479–22486. doi: 10.1074/jbc.271.37.22479. [DOI] [PubMed] [Google Scholar]
- 50.Sawaya B E, Rohr O, Aunis D, Schaeffer E. Chicken ovalbumin upstream promoter transcription factor, a transcriptional activator of HIV-1 gene expression in human brain cells. J Biol Chem. 1996;271:23572–23576. doi: 10.1074/jbc.271.38.23572. [DOI] [PubMed] [Google Scholar]
- 51.Sawaya B E, Rohr O, Aunis D, Schaeffer E. Regulation of human immunodeficiency virus type 1 gene transcription by nuclear receptors in human brain cells. J Biol Chem. 1996;271:22895–22900. doi: 10.1074/jbc.271.37.22895. [DOI] [PubMed] [Google Scholar]
- 52.Sawaya B E, Schaeffer E. Transcription of the human transferrin gene in neuronal cells. Nucleic Acids Res. 1995;23:2206–2211. doi: 10.1093/nar/23.12.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stein B, Cogswell P C, Baldwin A J. Functional and physical associations between NF-κB and C/EBP family members: a Rel domain-bZIP interaction. Mol Cell Biol. 1993;13:3964–3974. doi: 10.1128/mcb.13.7.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sune C, Garcia-Blanco M A. Sp1 transcription factor is required for in vitro basal and Tat-activated transcription from the human immunodeficiency virus type 1 long terminal repeat. J Virol. 1995;69:6572–6576. doi: 10.1128/jvi.69.10.6572-6576.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taylor J P, Khalili K. Activation of HIV-1 transcription by Tat in cells derived from the CNS: evidence for the participation of NF-κB—a review. Adv Neuroimmunol. 1994;4:291–303. doi: 10.1016/s0960-5428(06)80270-6. [DOI] [PubMed] [Google Scholar]
- 56.Tesmer V M, Bina M. Regulation of HIV-1 gene expression by NF-IL6. J Mol Biol. 1996;262:327–335. doi: 10.1006/jmbi.1996.0516. [DOI] [PubMed] [Google Scholar]
- 57.Tesmer V M, Rajadhyaksha A, Babin J, Bina M. NF-IL6-mediated transcriptional activation of the long terminal repeat of the human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1993;90:7298–7302. doi: 10.1073/pnas.90.15.7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang L H, Tsai S Y, Cook R G, Beattie W G, Tsai M J, O'Malley B W. COUP transcription factor is a member of the steroid receptor superfamily. Nature. 1989;340:163–166. doi: 10.1038/340163a0. [DOI] [PubMed] [Google Scholar]
- 59.Wang L H, Tsai S Y, Sagami I, Tsai M J, O'Malley B W. Purification and characterization of chicken ovalbumin upstream promoter transcription factor from HeLa cells. J Biol Chem. 1987;262:16080–16086. [PubMed] [Google Scholar]
- 60.Watkins B A, Dorn H H, Kelly W B, Armstrong R C, Potts B J, Michaels F, Kufta C V, Dubois D M. Specific tropism of HIV-1 for microglial cells in primary human brain cultures. Science. 1990;249:549–553. doi: 10.1126/science.2200125. [DOI] [PubMed] [Google Scholar]
- 61.Wedel A, Ziegler H H. The C/EBP family of transcription factors. Immunobiology. 1995;193:171–185. doi: 10.1016/s0171-2985(11)80541-3. [DOI] [PubMed] [Google Scholar]
- 62.Wiley C A, Schrier R D, Nelson J A, Lampert P W, Oldstone M B. Cellular localization of human immunodeficiency virus infection within the brains of acquired immune deficiency syndrome patients. Proc Natl Acad Sci USA. 1986;83:7089–7093. doi: 10.1073/pnas.83.18.7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williams S C, Cantwell C A, Johnson P F. A family of C/EBP-related proteins capable of forming covalently linked leucine zipper dimers in vitro. Genes Dev 1991. 1991;5:1553–1567. doi: 10.1101/gad.5.9.1553. [DOI] [PubMed] [Google Scholar]
- 64.Yano S, Fukunaga K, Takiguchi M, Ushio Y, Mori M, Miyamoto E. Regulation of CCAAT/enhancer-binding protein family members by stimulation of glutamate receptors in cultured rat cortical astrocytes J. Biol Chem. 1996;271:23520–23527. doi: 10.1074/jbc.271.38.23520. [DOI] [PubMed] [Google Scholar]