Abstract
The M2 gene of respiratory syncytial virus (RSV) encodes two putative proteins: M2-1 and M2-2; both are believed to be involved in the RNA transcription or replication process. To understand the function of the M2-2 protein in virus replication, we deleted the majority of the M2-2 open reading frame from an infectious cDNA clone derived from the human RSV A2 strain. Transfection of HEp-2 cells with the cDNA clone containing the M2-2 deletion, together with plasmids that encoded the RSV N, P, and L proteins, produced a recombinant RSV that lacked the M2-2 protein (rA2ΔM2-2). Recombinant virus rA2ΔM2-2 was recovered and characterized. The levels of viral mRNA expression for 10 RSV genes examined were unchanged in cells infected with rA2ΔM2-2, except that a shorter M2 mRNA was detected. However, the ratio of viral genomic or antigenomic RNA to mRNA was reduced in rA2ΔM2-2-infected cells. By use of an antibody directed against the bacterially expressed M2-2 protein, the putative M2-2 protein was detected in cells infected with wild-type RSV but not in cells infected with rA2ΔM2-2. rA2ΔM2-2 displayed a small-plaque morphology and grew much more slowly than wild-type RSV in HEp-2 cells. In infected Vero cells, rA2ΔM2-2 exhibited very large syncytium formation compared to that of wild-type recombinant RSV. rA2ΔM2-2 appeared to be a host range mutant, since it replicated poorly in HEp-2, HeLa, and MRC5 cells but replicated efficiently in Vero and LLC-MK2 cells. Replication of rA2ΔM2-2 in the upper and lower respiratory tracts of mice and cotton rats was highly restricted. Despite its attenuated replication in rodents, rA2ΔM2-2 was able to provide protection against challenge with wild-type RSV A2. The genotype and phenotype of the M2-2 deletion mutant were stably maintained after extensive in vitro passages. The attenuated phenotype of rA2ΔM2-2 suggested that rA2ΔM2-2 may be a potential candidate for use as a live attenuated vaccine.
Human respiratory syncytial virus (RSV) has been recognized as a major infectious etiologic agent of pediatric respiratory tract diseases worldwide. RSV is the prototype member of the Pneumovirus genus of the Paramyxoviridae family (23). The RSV genome is a single-stranded negative-sense RNA of 15,222 nucleotides (nt) and encodes 11 proteins: NS1, NS2, N, P, M, SH, G, F, M2-1, M2-2, and L. The nucleoprotein (N protein), the phosphoprotein (P protein), and the major polymerase protein (L protein) are associated with the viral RNA genome in the form of nucleocapsids. The N, P, and L proteins form the viral RNA-dependent RNA polymerase complex for transcription and replication of the RSV genome (13, 33). The G and F proteins are the major integral surface glycoproteins involved in virus entry into cells. The matrix protein (M protein) is a peripheral membrane protein located between viral nucleocapsids and the viral envelope. The small hydrophobic protein (SH protein) is also membrane associated and has counterparts only in the rubulaviruses SV5 (16, 18) and mumps virus (11). Recombinant RSV lacking the SH protein gene replicates very well in tissue cultures, demonstrating that the SH protein is a nonessential protein (3). The NS1, NS2, M2-1, and M2-2 proteins lack known counterparts in other paramyxoviruses. The NS1 and NS2 proteins are nonstructural proteins, and the NS1 protein has been shown to be a potent viral RNA transcription and replication inhibitor (1). Recent work has shown that the NS2 gene is also dispensable for RSV replication in vitro, but small-plaque morphology and reduced replication were observed for the virus lacking the NS2 gene (2, 28).
The RSV M2 gene is located between the genes encoding the F and L proteins and encodes two putative proteins: M2-1 and M2-2. The 22-kDa M2-1 protein is encoded by the 5′-proximal open reading frame of the M2 mRNA, and its open reading frame partially overlaps the second, M2-2, open reading frame by a sequence encoding 10 amino acids (10). The M2-1 protein has been shown to be a transcriptional processivity factor that is involved in RNA transcription elongation (9). The M2-1 protein also decreases RNA transcription termination and facilitates read-through of RNA transcription at each gene junction (14, 15). The predicted M2-2 polypeptide contains 90 amino acids, but the M2-2 protein has not yet been identified intracellularly (10). The M2-2 protein down-regulates RSV RNA transcription and replication in a minigenome model system (9). The significance of this negative effect on RSV RNA transcription and replication in the viral replication cycle is not known.
To examine the function of the M2-2 protein, we generated a recombinant RSV that no longer expresses the M2-2 protein by using a recently developed reverse-genetics system (8, 19). Virus recovery was obtained by cotransfecting the RSV antigenomic cDNA that had the M2-2 open reading frame largely deleted, together with plasmids encoding the N, P, and L proteins, into cells that were infected concomitantly with a recombinant vaccinia virus expressing the T7 RNA polymerase. Viable RSV that lacked M2-2 protein expression was obtained, but it displayed altered growth phenotypes in tissue culture cells and was attenuated in rodent hosts. Our data suggested that the M2-2 protein, although dispensable for virus replication, plays an important role in virus infection and pathogenesis in vivo.
MATERIALS AND METHODS
Cells and viruses.
Monolayer cultures of HEp-2, HeLa, MDBK, LLC-MK2, and Vero cells (obtained from the American Type Culture Collection [ATCC]) were maintained in minimal essential medium containing 10% fetal bovine serum (FBS). MRC5 cells (obtained from the ATCC) were maintained in Dulbecco's modified Eagle medium containing 10% FBS. HEp-2 cells were obtained at passage level 362 and were not used beyond passage level 375. All the other cell lines were used within 20 in vitro passages. Modified vaccinia virus Ankara (MVA-T7) expressing bacteriophage T7 RNA polymerase (26, 32) was provided by Bernard Moss and grown in CEK cells.
Production of polyclonal antibody against the M2-2 protein.
To produce antiserum against the M2-2 protein of RSV, a cDNA fragment encoding the M2-2 open reading frame from nt 8155 to nt 8430 was amplified by PCR and cloned into the pRSETA vector (Invitrogen, Carlsbad, Calif.). The resulting construct, pRSETA/M2-2, was transformed into BL21-Gold(DE3)plysS cells (Strategene, La Jolla, Calif.), and the expression of the His-tagged M2-2 protein was induced by isopropyl-β-d-thiogalactopyranoside (IPTG). The M2-2 fusion protein was purified through HiTrap affinity columns (Amersham Pharmacia Biotech, Piscataway, N.J.) and was used to immunize rabbits. Two weeks after a booster immunization, rabbits were bled and the serum was collected.
Construction of an M2-2 deletion cDNA.
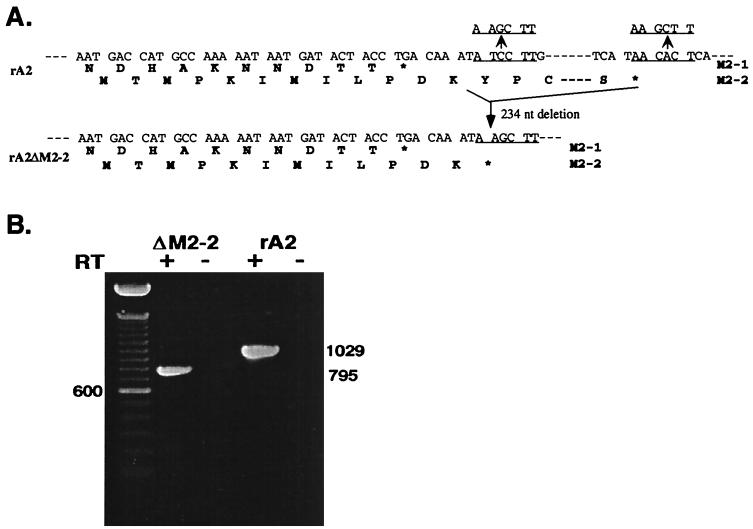
To generate an RSV antigenomic cDNA with an M2-2 deletion (pA2ΔM2-2), a cDNA fragment of 234 nt that contained the majority of the C-terminal part of the M2-2 open reading frame was removed from an antigenomic cDNA clone. The sequence encoding the N-terminal 12 amino acids of the M2-2 open reading frame that mostly overlaps the M2-1 open reading frame was maintained. A two-step cloning procedure was performed to delete the M2-2 open reading frame. Two HindIII restriction enzyme sites were introduced at RSV nt 8197 and nt 8431 in a cDNA subclone (pET-S/B) that contained the RSV SacI (nt 4477)-BamHI (nt 8499) cDNA fragment by use of a Quickchange mutagenesis kit (Strategene). Digestion of this cDNA subclone with the HindIII restriction enzyme removed the 234-nt HindIII cDNA fragment that contained the majority of the M2-2 open reading frame, and the remaining SacI-BamHI fragment with the M2-2 deletion was then cloned into an RSV antigenomic cDNA clone that contained a C to G change at the fourth position of the leader sequence, pRSVC4G (19). The resulting plasmid was designated pA2ΔM2-2 (Fig. 1).
FIG. 1.
Structure of the rA2ΔM2-2 genome and recovery of rA2ΔM2-2. (A) Sequences of the M2 gene in which the M2-1 and M2-2 open reading frames overlap. A total of 234 nt encoding the C-terminal 78 amino acids of the M2-2 protein were deleted through the introduced HindIII sites (underlined). The N-terminal 12 amino acid residues encoded by the M2-2 open reading frame were maintained at the region of overlap with the M2-1 open reading frame. (B) RT-PCR products of rA2ΔM2-2 and rA2 RNAs, obtained with a pair of primers flanking the M2 gene in the presence (+) or absence (−) of reverse transcriptase (RT). The size (in base pairs) of the DNA product derived from rA2 or rA2ΔM2-2 is indicated. The left lane was loaded with a 100-bp DNA size marker.
Recovery of rA2ΔM2-2.
Recombinant RSV was recovered from cDNA as described by Jin et al. (19). Briefly, HEp-2 cells at 80% confluence in a six-well plate were infected with MVA-T7 at a multiplicity of infection (MOI) of 5 PFU/cell for 1 h and then were transfected with plasmids encoding the RSV N, P, and L proteins and pA2ΔM2-2 by use of LipofecTACE (Life Technologies, Gaithersburg, Md.). After 5 h of incubation of the transfected HEp-2 cells at 35°C, the medium was replaced with minimal essential medium containing 2% FBS, and the cells were further incubated at 35°C for 3 days. The rescued virus (rA2ΔM2-2 [recombinant RSV that lacked the M2-2 open reading frame]) recovered from the transfected cells was plaque purified three times and amplified in Vero cells. The virus titer was determined by a plaque assay, and plaques were visualized by immunostaining with polyclonal anti-RSV A2 serum (Biogenesis, Sandown, N.H.).
Growth analysis of recombinant RSV in tissue cultures.
To compare the plaque morphology of rA2ΔM2-2 with that of recombinant RSV A2 (rA2), HEp-2 or Vero cells were infected with each virus and overlaid with semisolid medium composed of 1% methylcellulose and L15 medium (JRH Biosciences, Lenexa, Kans.) with 2% FBS. Five days after infection, infected cells were immunostained with antisera against the RSV A2 strain. Plaque size was determined by measuring plaques from photographed microscopic images. A growth cycle analysis of rA2ΔM2-2 in comparison with rA2 was performed with both HEp-2 and Vero cells. Cells grown in 6-cm dishes were infected with rA2 or rA2ΔM2-2 at an MOI of 0.5. After 1 h of adsorption at room temperature, infected cells were washed three times with phosphate-buffered saline, the medium was replaced with 4 ml of OptiMEM (Life Technologies), and the culture was incubated at 35°C in an incubator containing 5% CO2. At various times postinfection, 200 μl of culture supernatant was collected and stored at −70°C until virus titration. Each aliquot taken was replaced with an equal amount of fresh medium. The virus titer was determined by a plaque assay on Vero cells, using an overlay of 1% methylcellulose and L15 medium containing 2% FBS. To analyze virus replication in different host cells, each cell line grown in six-well plates was infected with rA2ΔM2-2 or rA2 at an MOI of 0.2. Three days postinfection, the culture supernatants were collected, and virus was quantitated by a plaque assay on Vero cells.
RNA extraction, RT-PCR, and Northern blot analysis.
For reverse transcription (RT)-PCR, viral RNA was extracted from rA2ΔM2-2- and rA2-infected cell culture supernatants by use of an RNA extraction kit (RNA STAT-50; Tel-Test, Friendswood, Tex.). Viral RNA was reverse transcribed with reverse transcriptase and a primer complementary to the viral genome from nt 7430 to nt 7449. The cDNA fragment spanning the M2 gene was amplified by PCR with primer V1948 (nt 7486 to nt 7515 for positive sense) and primer V1581 (nt 8544 to nt 8525 for negative sense). The PCR product was analyzed on a 1.2% agarose gel and visualized by ethidium bromide staining.
For Northern blot hybridization analysis, total cellular RNA was extracted from rA2ΔM2-2- or rA2-infected cells by use of an RNA extraction kit (RNA STAT-60; Tel-Test). RNA was electrophoresed on a 1.2% agarose gel containing formaldehyde and transferred to a nylon membrane (Amersham Pharmacia Biotech). The membrane was hybridized with an RSV gene-specific riboprobe labeled with digoxigenin. The hybridized RNA bands were visualized by use of a Dig-Luminescent Detection Kit for Nucleic Acids (Boehringer Mannheim Biochemicals, Indianapolis, Ind.). To detect viral genomic RNA, a 32P-labeled riboprobe specific for the negative-sense F gene or N gene was used in Northern blot hybridization. To detect viral antigenomic RNA and mRNA, a 32P-labeled riboprobe specific for the positive-sense F gene or G gene was used. Hybridization of the membrane with riboprobes was done at 65°C. Membrane washing and signal detection were performed according to standard procedures.
Immunoprecipitation and Western blotting of viral polypeptides.
Virus-specific proteins produced from infected cells were analyzed by immunoprecipitation of the infected-cell extracts or by Western blotting. For immunoprecipitation analysis, Vero cells were infected with virus at an MOI of 1.0 and labeled with 35S-promix (100 μCi each of [35S]Cys and [35S]Met per ml; Amersham, Arlington Heights, Ill.) at 14 to 18 h postinfection. The labeled cell monolayers were lysed with radioimmunoprecipitation assay buffer, and the polypeptides were immunoprecipitated with polyclonal anti-RSV A2 serum (Biogenesis) or anti–M2-2 protein antiserum. Immunoprecipitated polypeptides were electrophoresed on 17.5% polyacrylamide gels containing 0.1% sodium dodecyl sulfate and 4 M urea and detected by autoradiography. For Western blotting analysis, HEp-2 and Vero cells were infected with rA2ΔM2-2 or rA2. At various times postinfection, virus-infected cells were lysed in protein lysis buffer, and the cell lysates were electrophoresed on 17.5% polyacrylamide gels containing 0.1% sodium dodecyl sulfate and 4 M urea. The proteins were transferred to a nylon membrane. Immunoblotting was performed as described in Jin et al. (20) with polyclonal antiserum against M2-1 protein (gift of Jayesh Meanger), NS1 protein, or SH protein (gift of Jose A. Melero).
Virus replication in mice and cotton rats.
Virus replication in vivo was determined with respiratory-tract-pathogen-free 12-week-old BALB/c mice (Simonsen Laboratories, Gilroy, Calif.) and S. hispidus cotton rats (Virion Systems, Rockville, Md.). Mice or cotton rats in groups of six were inoculated intranasally under light methoxyflurane anesthesia with 106 PFU of rA2 or rA2ΔM2-2 in a 0.1-ml inoculum per animal. On day 4 postinoculation, animals were sacrificed by CO2 asphyxiation, and their nasal turbinates and lungs were obtained separately. Tissues were homogenized, and virus titers were determined by a plaque assay on Vero cells. To evaluate immunogenicity and protective efficacy, three groups of mice were inoculated intranasally with rA2, rA2ΔM2-2, or medium only at day 0. Three weeks later, mice were anesthetized, serum samples were collected, and a challenge inoculation of 106 PFU of biologically derived wild-type RSV A2 was administered intranasally. Four days postchallenge, the animals were sacrificed, both nasal turbinates and lungs were harvested, and virus titers were determined by a plaque assay. Serum neutralizing antibodies against RSV A2 were determined by a 60% plaque reduction assay (7) and by immunostaining of RSV-infected cells.
RESULTS
Generation of rA2ΔM2-2.
Previously, we reported the recovery of recombinant RSV from an infectious cDNA clone derived from RSV strain A2 (19). To obtain recombinant RSV in which the expression of the M2-2 open reading frame is ablated, a 234-nt cDNA fragment that encodes the C-terminal 78 amino acids of the M2-2 protein was deleted from the infectious RSV cDNA clone. The N-terminal 12 amino acids that mostly overlapped with the M2-1 open reading frame were maintained, as it was considered likely that these 12 amino acids would not be sufficient to preserve M2-2 protein function (Fig. 1A). The deletion of the M2-2 open reading frame in antigenomic cDNA was confirmed by restriction enzyme digestion and by sequencing across the junction of the deletion. The resulting antigenomic cDNA clone, pA2ΔM2-2, is 14,988 nt long, 234 nt shorter than pRSVC4G.
Since pA2ΔM2-2 was not completely sequenced, two independent clones were obtained and used in the recovery of infectious virus. To recover recombinant RSV with the M2-2 open reading frame largely deleted, pA2ΔM2-2 was transfected, together with plasmids encoding the RSV N, P, and L proteins, under the control of the T7 promoter, into HEp-2 cells which had been infected with a modified vaccinia virus expressing the T7 RNA polymerase (MVA-T7). Culture supernatants from the transfected HEp-2 cells were used to infect fresh HEp-2 or Vero cells to amplify the rescued virus. The recovery of rA2ΔM2-2 was indicated by syncytium formation and confirmed by positive staining of infected cells with polyclonal anti-RSV A2 serum. Recovered rA2ΔM2-2 was plaque purified three times and amplified in Vero cells. To confirm that rA2ΔM2-2 contained the M2-2 deletion, viral RNA was extracted from virus and subjected to RT-PCR with a pair of primers spanning the M2 gene. As shown in Fig. 1B, rA2 yielded a PCR DNA product corresponding to the predicted 1,029-nt fragment, whereas rA2ΔM2-2 yielded a PCR product of 795 nt, 234 nt shorter. Generation of the RT-PCR product was dependent on the RT step, indicating that the product was derived from RNA rather than from DNA contamination. The deletion was also confirmed by sequencing analysis of the 795-nt RT-PCR DNA product derived from rA2ΔM2-2.
Replication of rA2ΔM2-2 in tissue culture cells.
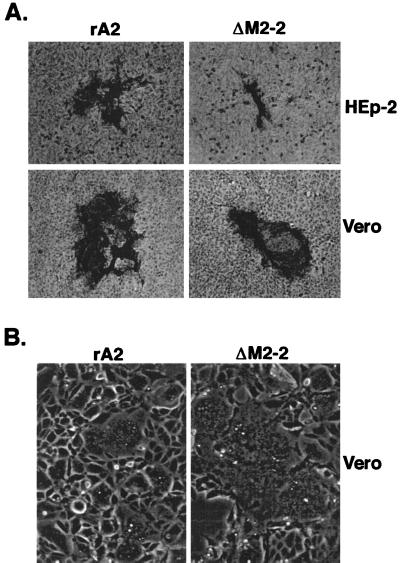
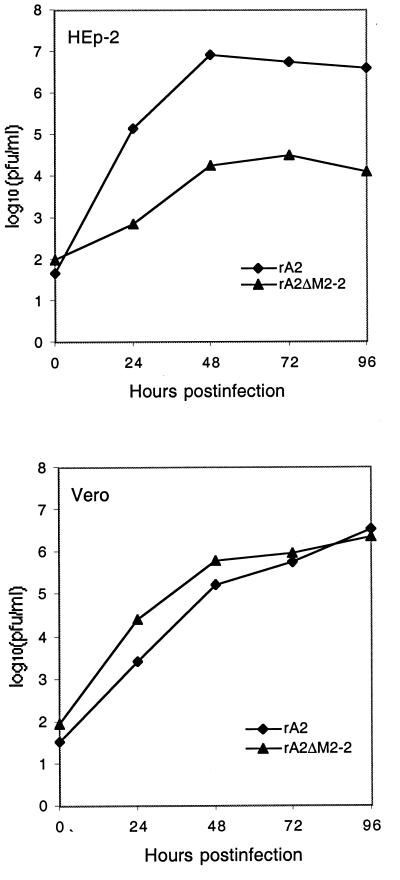
Plaque formation of rA2ΔM2-2 in HEp-2 and Vero cells was compared with that of rA2. As shown in Fig. 2A, rA2ΔM2-2 formed very small plaques in HEp-2 cells, with a reduction in virus plaque size of about fivefold observed for rA2ΔM2-2 compared to rA2. However, only a slight reduction in plaque size (30%) was seen in Vero cells infected with rA2ΔM2-2. In infected Vero cells, rA2ΔM2-2 formed very large syncytia compared to rA2 (Fig. 2B). Increased syncytium formation was not observed in HEp-2 cells. The growth cycle of rA2ΔM2-2 was also compared with that of rA2 in both HEp-2 and Vero cells (Fig. 3). In HEp-2 cells, rA2ΔM2-2 showed very slow growth kinetics, and the peak titer of rA2ΔM2-2 was about 2.0 log units lower than that of rA2. In Vero cells, rA2ΔM2-2 reached a peak titer similar to that of rA2. rA2ΔM2-2 was further examined for its growth properties in various cell lines derived from different hosts with different tissue origins (Table 1). Significantly reduced replication of rA2ΔM2-2, about 2 orders of magnitude less than that of rA2, was observed in infected HEp-2, MRC-5, and HeLa cells, all of human origin. However, the replication of rA2ΔM2-2 was only slightly reduced in MDBK and LLC-MK2 cells, which are derived from bovine and rhesus monkey kidney cells, respectively. It is known that HEp-2 cells from the ATCC contain HeLa cell markers; thus, HEp-2 cells may behave like HeLa cells.
FIG. 2.
Comparison of the abilities of rA2 and rA2ΔM2-2 to form plaques and syncytia. (A) Plaque morphology of rA2ΔM2-2 and rA2. HEp-2 or Vero cells were infected with rA2ΔM2-2 or rA2 under a semisolid overlay composed of 1% methylcellulose and L15 medium containing 2% FBS for 5 days. Virus plaques were visualized by immunostaining with a goat polyclonal anti-RSV antiserum and photographed under a microscope. (B) Comparison of syncytium formation. Vero cells were infected with rA2 and rA2ΔM2-2 at an MOI of 0.5 and incubated in liquid medium (OptiMEM) at 35°C for 40 h. The infected cell monolayers were photographed without any treatment.
FIG. 3.
Growth curves of rA2ΔM2-2 in HEp-2 and Vero cells. Vero cells or HEp-2 cells were infected with rA2ΔM2-2 or rA2 at an MOI of 0.5, and aliquots of medium were harvested at 24-h intervals. The virus titers were determined by a plaque assay on Vero cells. The virus titer at each time point is an average from two experiments with two independent isolates for both viruses.
TABLE 1.
Levels of replication of rA2ΔM2-2 and rA2 in various cell linesa
| Cell line | Host | Tissue origin | Titer (log10 Pfu/ml)
|
|
|---|---|---|---|---|
| rA2 | rA2ΔM2-2 | |||
| Vero | Monkey | Kidney | 6.1 | 6.1 |
| HEp-2 | Human | Larynx | 6.2 | 4.3 |
| MDBK | Bovine | Kidney | 6.1 | 5.5 |
| MRC-5 | Human | Lung | 5.5 | 3.0 |
| HeLa | Human | Cervix | 6.6 | 4.5 |
| LLC-MK2 | Monkey | Kidney | 6.7 | 6.1 |
Cells were infected with rA2 or rA2ΔM2-2 at an MOI of 0.2; at 72 h postinfection, the culture supernatants were harvested, and the levels of virus replication were determined by a plaque assay on Vero cells. Each virus titer was averaged from two experiments with two independent isolates for both viruses.
rA2ΔM2-2 mRNA synthesis.
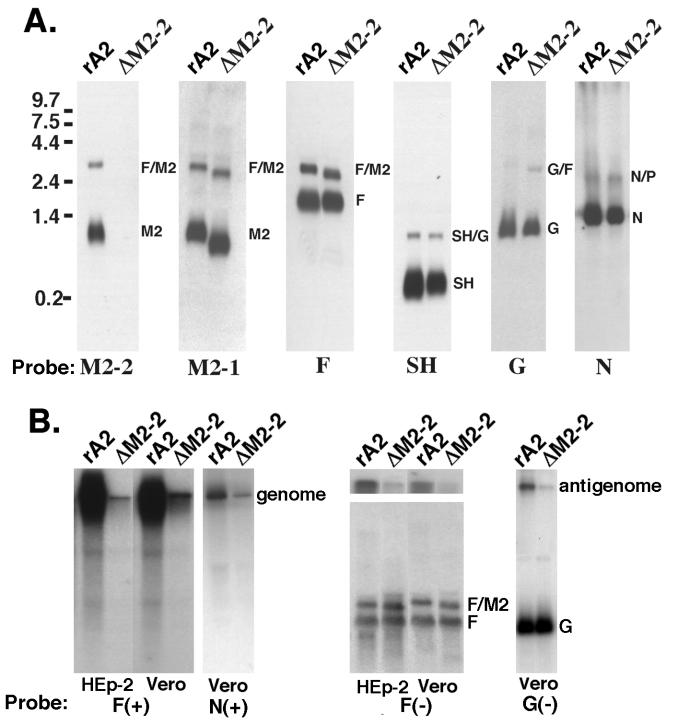
To examine mRNA synthesis from rA2ΔM2-2 and rA2, the accumulation of M2 mRNA and the other viral mRNA products in infected Vero cells was analyzed by Northern blot hybridization. Hybridization of the blot with a riboprobe specific for the M2-2 open reading frame did not reveal any signal in rA2ΔM2-2-infected cells. Instead, a short M2 mRNA was detected in rA2ΔM2-2-infected cells by a riboprobe specific for the M2-1 open reading frame (Fig. 4A). These observations confirmed that the M2-2 open reading frame was deleted from rA2ΔM2-2. The accumulation of the other nine RSV mRNA transcripts was also examined, and the amounts of each mRNA were found to be comparable between rA2ΔM2-2- and rA2-infected cells. Examples of Northern blots probed with riboprobes specific for the N, SH, G, or F genes are also shown in Fig. 4A. Slightly faster migration of F-M2 bicistronic mRNA was also discernible due to the deletion of the M2-2 open reading frame.
FIG. 4.
Viral RNA expression by rA2ΔM2-2 and rA2. (A) Total RNA was extracted from rA2- or rA2ΔM2-2-infected Vero cells (MOI, 1.0) at 48 h postinfection, separated by electrophoresis on 1.2% agarose–2.2 M formaldehyde gels, and transferred to nylon membranes. Each blot was hybridized with a digoxigenin-labeled riboprobe specific for the M2-2, M2-1, F, SH, G, or N gene. The sizes of the RNA markers are indicated on the left. (B) HEp-2 and Vero cells were infected with rA2 or rA2ΔM2-2 for 24 h, and total cellular RNA was extracted. An RNA Northern blot was hybridized with a 32P-labeled riboprobe specific for the negative-sense F gene to detect viral genomic RNA in both HEp-2 and Vero cells or an N gene probe to detect viral genomic RNA in Vero cells only. A 32P-labeled riboprobe specific for the positive-sense F gene was used to detect viral antigenomic RNA and F mRNA in HEp-2 and Vero cells, and a G gene probe was used to detect antigenomic RNA and G mRNA in Vero cells only. The top panel of the Northern blot on the right (F gene probe) was taken from the top portion of the gel shown in the lower panel and was exposed for 1 week to show the antigenome. The lower panel of that Northern blot was exposed for 3 h to show the F mRNA. The genome, antigenome, F mRNA, and dicistronic F-M2 RNA are indicated.
The M2-2 protein was previously reported to be a potent transcriptional negative regulator in a minigenome replication assay. However, the lack of M2-2 protein expression did not appear to affect viral mRNA production in infected cells. To determine if the levels of viral antigenomic and genomic RNAs of rA2ΔM2-2 were affected by the M2-2 deletion, we examined the amounts of viral genomic and antigenomic RNAs produced in infected Vero and HEp-2 cells by Northern hybridization. Hybridization of the infected total cellular RNA with a 32P-labeled F or N gene riboprobe specific for the negative-sense genomic RNA indicated that much less genomic RNA was produced in cells infected with rA2ΔM2-2 than in cells infected with rA2 (Fig. 4B). A duplicate membrane was hybridized with a 32P-labeled F or G gene riboprobe specific for the positive-sense RNA. Very little antigenomic RNA was detected in cells infected with rA2ΔM2-2; the amount of the F or G mRNA in rA2ΔM2-2-infected cells was comparable to that in rA2-infected cells. It is very striking that the levels of both genomic and antigenomic RNAs in rA2ΔM2-2-infected cells were significantly reduced. Quantitation of the ratio of genomic and antigenomic RNA amounts to the viral mRNA amount indicated that at least a 10-fold reduction in antigenomic and genomic RNA amounts was observed in rA2ΔM2-2-infected cells. Therefore, it appears that RSV genome and antigenome syntheses were down-regulated due to the M2-2 deletion. This down-regulation was seen in both Vero and HEp-2 cells and thus was not cell type dependent. This phenomenon has been observed with different riboprobes and two different rA2ΔM2-2 isolates (Fig. 4B).
rA2ΔM2-2 protein synthesis.
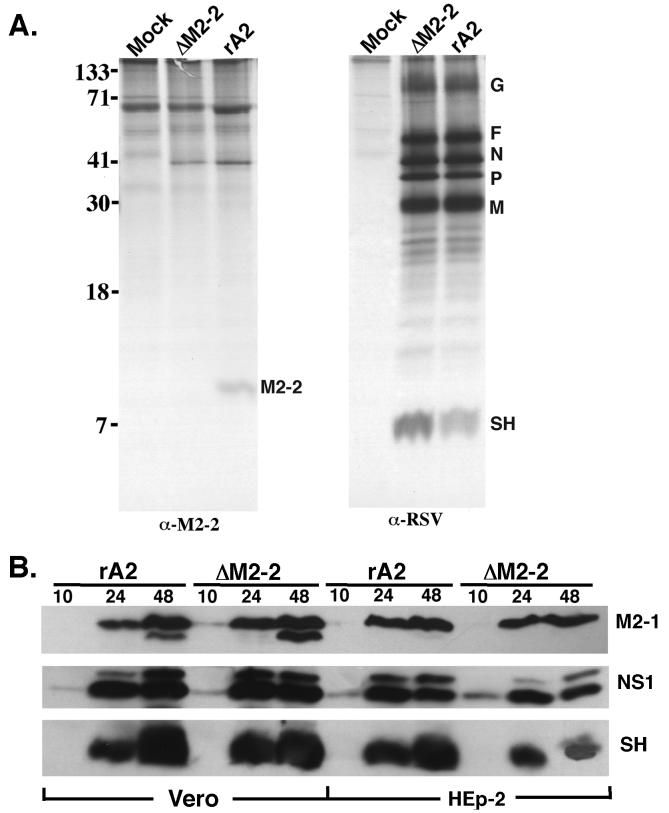
Since the putative M2-2 protein has not been identified in RSV-infected cells previously, it was necessary to demonstrate that the M2-2 protein is indeed encoded by RSV and produced in infected cells. We produced a polyclonal antiserum against the M2-2 fusion protein expressed in a bacterial expression system. Immunoprecipitation of rA2-infected Vero cell lysates with anti–M2-2 protein antibody produced a protein band of approximately 10 kDa, which is the predicted size for the M2-2 polypeptide. This polypeptide was not detected in rA2ΔM2-2-infected cells (Fig. 5A), confirming that the M2-2 protein is a product produced by RSV and that its expression was ablated from rA2ΔM2-2. The overall polypeptide pattern of rA2ΔM2-2 was indistinguishable from that of rA2. However, it was noted by immunoprecipitation that slightly higher levels of the P and SH proteins were produced in rA2ΔM2-2-infected Vero cells. Nevertheless, as noted by Western blotting analysis, comparable amounts of the SH protein were produced in cells infected with rA2ΔM2-2 or rA2 (Fig. 5B).
FIG. 5.
Viral protein expression in rA2ΔM2-2- and rA2-infected cells. (A). Mock-infected or rA2ΔM2-2- and rA2-infected Vero cells (MOI, 1.0) were metabolically labeled with 35S-promix (100 μCi/ml) between 14 and 18 h postinfection. Cell lysates were prepared for immunoprecipitation with goat polyclonal anti-RSV or rabbit polyclonal anti–M2-2 antiserum. Immunoprecipitated polypeptides were separated on a 17.5% polyacrylamide gel containing 4 M urea and processed for autoradiography. The position of each viral protein is indicated on the right, and the molecular weight size markers (in thousands) are indicated on the left. (B) Time course of RSV protein expression by rA2 and rA2ΔM2-2. HEp-2 and Vero cells were infected with rA2 or rA2ΔM2-2 at an MOI of 1.0. At 10, 24, or 48 h postinfection, total infected cellular polypeptides were separated on a 17.5% polyacrylamide gel containing 4 M urea. Proteins were transferred to a nylon membrane, and the blot was probed with polyclonal antisera against the M2-1, NS1, or SH protein.
Western blotting was used to determine the rate and accumulation of protein synthesis by rA2ΔM2-2 in both Vero and HEp-2 cell lines. HEp-2 or Vero cells were infected with rA2ΔM2-2 or rA2; at various times postinfection, the infected cells were harvested, and the polypeptides were separated on a 17.5% polyacrylamide gel containing 4 M urea. The proteins were transferred to a nylon membrane and probed with polyclonal antisera against the three accessory proteins: M2-1, NS1 and SH. Protein expression levels for all three viral proteins were very similar for rA2ΔM2-2 and rA2 in both HEp-2 and Vero cells (Fig. 5B). Synthesis of the NS1 protein was detected at 10 h postinfection, slightly earlier than that of the M2-1 and SH proteins because the NS1 protein is the most abundant protein in infected cells due to its 3′ proximal location. Similar protein synthesis rates and levels were also observed for both rA2ΔM2-2 and rA2 when the membrane was probed with a polyclonal antiserum against RSV (data not shown). Comparable levels of the M2-1 protein were detected for both viruses, indicating that deletion of the M2-2 open reading frame did not affect the level of the M2-1 protein, which is translated by the same M2 mRNA.
Replication of rA2ΔM2-2 in mice and cotton rats.
To evaluate the levels of attenuation and immunogenicity of rA2ΔM2-2, the replication of rA2ΔM2-2 in the upper and lower respiratory tracts of mice and cotton rats was examined. Mice in groups of six were inoculated with 106 PFU of rA2ΔM2-2 or rA2 intranasally. Animals were sacrificed at 4 days postinoculation; their nasal turbinates and lung tissues were harvested and homogenized, and levels of virus replication in these tissues were determined by a plaque assay. Geometric mean titers of virus replication and standard errors obtained from two experiments are shown in Table 2. rA2ΔM2-2 exhibited at least a 2.0-log unit reduction of replication in both nasal turbinates and lungs of infected mice. rA2ΔM2-2 replication was detected only in 1 or 2 of 12 infected mice. The replication was limited; only a few plaques were observed at a 10−1 dilution of the tissue homogenates. A high level of rA2 replication was detected in both the upper and the lower respiratory tracts of mice. A similar degree of attenuation of rA2ΔM2-2 was also observed in cotton rats. Despite its restricted replication in mice, rA2ΔM2-2 induced significant resistance to challenge with wild-type RSV A2 (Table 2). When mice previously inoculated with rA2ΔM2-2 or rA2 were inoculated intranasally with 106 PFU of wild-type A2 virus, no wild-type A2 virus replication was detected in the upper and lower respiratory tracts. Therefore, rA2ΔM2-2 was fully protective against wild-type A2 virus challenge.
TABLE 2.
Replication of rA2ΔM2-2 and rA2 in mice and protection against wild-type RSV A2 challenge
| Immunizing virus | Virus replicationa (mean log10 PFU/g of tissue ± SE) in:
|
RSV A2 replication after challengeb (mean log10 PFU/g of tissue ± SE) in:
|
||
|---|---|---|---|---|
| Nasal turbinates | Lungs | Nasal turbinates | Lungs | |
| rA2 | 3.72 ± 0.33 | 4.0 ± 0.13 | <1.4 | <1.4 |
| rA2ΔM2-2 | <1.4 | <1.4 | <1.4 | <1.4 |
| None (control) | <1.4 | <1.4 | 3.53 ± 0.17 | 4.10 ± 0.13 |
Groups of 12 BALB/c mice were immunized intranasally with 106 PFU of the indicated virus on day 0. The level of infected virus in the indicated tissues was determined by a plaque assay at day 4, and the mean log10 titer ± standard error per gram of tissue was determined.
Groups of 6 BALB/c mice were challenged intranasally with 106 PFU of RSV A2 on day 21 and sacrificed 4 days later. The replication of wild-type RSV A2 in tissues was determined by a plaque assay, and the mean log10 titer ± standard error per gram of tissue was determined.
The immunogenicity of rA2ΔM2-2 was also examined. Mice in groups of six were infected with rA2ΔM2-2 or rA2; 3 weeks later, serum samples were collected. The serum neutralization titer was determined by a 60% plaque reduction assay. The neutralization titer obtained from rA2ΔM2-2-infected mice was comparable to that of rA2; mice infected with both viruses had a 60% plaque reduction titer at mean dilutions of 1:32 to 1:64, whereas the prebleed serum had an RSV neutralization titer of 1:4. Since the detectable neutralization titer was low, as was also reported previously (17), we thus tested the RSV-specific antibody by immunostaining of RSV-infected cells. The serum obtained from rA2ΔM2-2-infected mice immunostained RSV plaques at dilutions similar to that of rA2, confirming that the RSV-specific antibody was produced in rA2ΔM2-2-infected mice at a level similar to that of rA2.
DISCUSSION
In this study, we reported the construction of a recombinant RSV that lacked the majority of the M2-2 open reading frame (rA2ΔM2-2). The recovery of rA2ΔM2-2 was confirmed by RT-PCR, sequencing, Northern blot hybridization, and immunoprecipitation analyses. A recombinant RSV that lacked M2-2 protein expression was viable in cell cultures, but it replicated poorly in several host cell lines and rodent hosts. The cell-type-dependent replication of rA2ΔM2-2 suggested that a host factor(s) might be involved in RSV replication in the absence of the M2-2 protein.
RSV encodes five unique proteins: NS1, NS2, SH, M2-1, and M2-2. It has been reported that the NS2 and SH genes are dispensable for RSV replication in vitro (3, 28). Our experiments demonstrated that the M2-2 gene is also not essential for RSV replication in cell cultures. Therefore, the M2-2 gene is the third gene that has been reported to be dispensable for RSV replication. It is very interesting that RSV has evolved to encode at least three nonessential genes in its genome. These accessory genes must therefore provide certain auxiliary functions for virus replication in hosts. Deletion of the RSV SH gene did not appear to affect virus replication in vitro (3), a result very similar to what was reported for recombinant SV5 lacking the SH gene (16). On the contrary, slightly better replication of the SH knockout RSV (rA2ΔSH) in certain cell lines has been observed. Although it is not attenuated in the lower respiratory tracts of mice (3), rA2ΔSH is attenuated in the lower respiratory tracts of chimpanzees (31). Removal of the NS2 gene impairs virus growth for both human RSV and bovine RSV (2, 28). The NS2 knockout RSV (rA2ΔNS2), in which tandem stop codons were introduced, reverted rapidly to restore NS2 protein expression (28). When inoculated into chimpanzees intranasally and intratracheally, rA2ΔNS2 is slightly attenuated in the upper respiratory tract but highly attenuated in the lower respiratory tract (31). These results indicate that the NS2 protein plays an important role in full virus replication capacity. The data presented in this study indicated that the M2-2 protein, although not essential for RSV viability, is an accessory factor that is able to substantially support virus growth in vitro and in vivo. Further studies are needed to understand the mechanisms by which the M2-2 protein facilitates virus growth in different cell types and in animal hosts.
The M2-2 protein has been identified as a strong inhibitor to RSV minigenome replication in vitro (9). The effect of inhibition of RNA synthesis by the M2-2 protein is reminiscent of the inhibitory transcription function of the M protein of vesicular stomatitis virus (5, 24). If this inhibitory effect is exerted late in infection, decreased RNA synthesis may be beneficial for the virus in restricting excess cytopathogenicity that may abort further progeny production and in rendering nucleocapsids quiescent prior to budding. Indeed, we have observed that cells infected with rA2ΔM2-2 exhibited a cytopathogenic effect earlier and had larger syncytia than cells infected with wild-type virus. However, down-regulation instead of up-regulation of RNA genome replication was observed in cells infected with rA2ΔM2-2. Much reduced amounts of antigenomic and genomic RNAs were detected in rA2ΔM2-2-infected cells. Northern blot hybridization data indicated that the amount of mRNA production in rA2ΔM2-2-infected cells was comparable to that in cells infected with wild-type RSV. Viral proteins expressed by rA2ΔM2-2 were also comparable to those synthesized by wild-type RSV. The data obtained in experiments reported here did not provide any evidence that the M2-2 protein inhibited RSV RNA transcription and replication. The possibility that the M2-2 protein could affect mRNA stability in infected cells has not been examined. It is possible that the M2-2 protein is involved in the switch from RNA transcription to replication and thus results in reduced antigenomic and genomic RNA synthesis in cells infected with rA2ΔM2-2.
The HEp-2 cell line is fully permissive to wild-type RSV replication. However, replication of rA2ΔM2-2 in this cell line was reduced; rA2ΔM2-2 formed very small plaques and replicated to a low titer in HEp-2 cells. To delineate the M2-2 protein function that is required for efficient RSV replication in HEp-2 cells, we compared RNA and protein syntheses of rA2ΔM2-2 and rA2 in both Vero and HEp-2 cells. Surprisingly, the amounts of RNAs and proteins expressed from rA2ΔM2-2-infected HEp-2 cells were comparable to those expressed from rA2-infected cells. The genomic and antigenomic RNA synthesis of rA2ΔM2-2 in HEp-2 cells is similar to that in Vero cells. Previously, it was reported that the addition of a small amount of the M2-2 protein increased the packaging of the minigenome in vitro (29). It is likely that the poor replication of rA2ΔM2-2 in HEp-2 cells is due to a defect at a later stage of virus replication, possibly during the virus assembly process. rA2ΔM2-2 formed very large syncytia in infected Vero cells. Preliminary data indicated that rA2ΔM2-2 is more fusogenic (data not shown) in a cytoplasmic content mixing experiment (16). How the lack of the M2-2 protein affected RSV fusion activity remains to be investigated.
Impaired virus replication and reduced virus pathogenicity due to the loss of virus accessory proteins have been reported for Sendai virus and measles virus. The C protein of Sendai virus inhibits viral mRNA synthesis and amplification of the Sendai virus minigenome in a promoter-specific manner (4, 27). However, up-regulation instead of down-regulation of transcription, translation, and genome replication was seen in Sendai virus that lacked the C protein. The virus lacking the C protein is highly attenuated in the natural murine host (22). The C protein of measles virus is dispensable for virus replication in Vero cells (25) but is required for efficient measles virus replication in human peripheral blood cells (12). The V protein of both Sendai virus and measles virus is also associated with virus pathogenicity (21, 30). As was observed when accessory genes were deleted from these other paramyxoviruses, we found that deletion of the M2-2 open reading frame rendered RSV attenuated in the upper and lower respiratory tracts of mice and cotton rats. Since other RSV proteins are targets of immunity (6), the absence of the M2-2 protein in a vaccine virus would not compromise the immunogenicity of RSV. rA2ΔM2-2 resembled rA2 in its ability to induce RSV-specific antibodies with neutralizing function and to protect mice against the replication of wild-type challenge virus in both the upper and lower respiratory tracts. Virus with M2-2 gene deletion is easily distinguishable from wild-type virus and genetically stable, making rA2ΔM2-2 a potential candidate vaccine for human use.
ACKNOWLEDGMENTS
We thank Yang He and Linda Zhu of the Aviron tissue culture facility for supplying tissue culture cells; Roderick Tang, Robert Brazas, and Tai-An Cha for suggestions and help; and Ann Arvin and Richard Spaete for critical review of the manuscript. We are grateful to Jayesh Meanger for providing antiserum against the M2-1 protein and Jose A. Melero for providing antiserum against the SH protein.
REFERENCES
- 1.Atreya P L, Peeples M E, Collins P L. The NS1 protein of human respiratory syncytial virus is a potent inhibitor of minigenome transcription and RNA replication. J Virol. 1998;72:1452–1461. doi: 10.1128/jvi.72.2.1452-1461.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchholz U J, Finke S, Conzelmann K K. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol. 1999;73:251–259. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bukreyev A, Whitehead S S, Murphy B R, Collins P L. Recombinant respiratory syncytial virus from which the entire SH gene has been deleted grows efficiently in cell culture and exhibits site-specific attenuation in the respiratory tract of the mouse. J Virol. 1997;71:8973–8982. doi: 10.1128/jvi.71.12.8973-8982.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cadd T, Garcin D, Tapparel C, Itoh M, Homma M, Roux L, Curran J, Kolakofsky D. The Sendai paramyxovirus accessory C proteins inhibit viral genome amplification in a promoter-specific fashion. J Virol. 1996;70:5067–5074. doi: 10.1128/jvi.70.8.5067-5074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll A R, Wagner R R. Role of the membrane (M) protein in endogenous inhibition of in vitro transcription by vesicular stomatitis virus. J Virol. 1979;29:134–142. doi: 10.1128/jvi.29.1.134-142.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cherrie A H, Anderson K, Wertz G W, Openshaw P J. Human cytotoxic T cells stimulated by antigen on dendritic cells recognize the N, SH, F, M, 22K, and 1b proteins of respiratory syncytial virus. J Virol. 1992;66:2102–2110. doi: 10.1128/jvi.66.4.2102-2110.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coates H V, Alling D W, Chanock R M. An antigenic analysis of respiratory syncytial virus isolates by a plaque reduction neutralization test. Am J Epidemiol. 1965;83:299–313. doi: 10.1093/oxfordjournals.aje.a120586. [DOI] [PubMed] [Google Scholar]
- 8.Collins P L, Hill M G, Camargo E, Grosfeld H, Chanock R M, Murphy B R. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc Natl Acad Sci USA. 1995;92:11563–11567. doi: 10.1073/pnas.92.25.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins P L, Hill M G, Cristina J, Grosfeld H. Transcription elongation factor of respiratory syncytial virus, a nonsegmented negative-strand RNA virus. Proc Natl Acad Sci USA. 1996;93:81–85. doi: 10.1073/pnas.93.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins P L, Wertz G W. The envelope-associated 22K protein of human respiratory syncytial virus: nucleotide sequence of the mRNA and a related polytranscript. J Virol. 1985;54:65–71. doi: 10.1128/jvi.54.1.65-71.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elango N, Kovamees J, Varsanyi T M, Norrby E. mRNA sequence and deduced amino acid sequence of the mumps virus small hydrophobic protein gene. J Virol. 1989;63:1413–1415. doi: 10.1128/jvi.63.3.1413-1415.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Escoffier C, Manie S, Vincent S, Muller C P, Billeter M, Gerlier D. Nonstructural C protein is required for efficient measles virus replication in human peripheral blood cells. J Virol. 1999;73:1695–1698. doi: 10.1128/jvi.73.2.1695-1698.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grosfeld H, Hill M G, Collins P L. RNA replication by respiratory syncytial virus (RSV) is directed by the N, P, and L proteins; transcription also occurs under these conditions but requires RSV superinfection for efficient synthesis of full-length mRNA. J Virol. 1995;69:5677–5686. doi: 10.1128/jvi.69.9.5677-5686.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardy R W, Harmon S B, Wertz G W. Diverse gene junctions of respiratory syncytial virus modulate the efficiency of transcription termination and respond differently to M2-mediated antitermination. J Virol. 1999;73:170–176. doi: 10.1128/jvi.73.1.170-176.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardy R W, Wertz G W. The product of the respiratory syncytial virus M2 gene ORF1 enhances readthrough of intergenic junctions during viral transcription. J Virol. 1998;72:520–526. doi: 10.1128/jvi.72.1.520-526.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He B, Leser G P, Paterson R G, Lamb R A. The paramyxovirus SV5 small hydrophobic (SH) protein is not essential for virus growth in tissue culture cells. Virology. 1998;250:30–40. doi: 10.1006/viro.1998.9354. [DOI] [PubMed] [Google Scholar]
- 17.Herlocher M L, Ewasyshyn M, Sambhara S, Gharaee-Hermani M, Cho D, Lai J, Klein M, Maassab H F. Immunological properties of plaque purified strains of live attenuated respiratory syncytial virus (RSV) for human vaccine. Vaccine. 1999;17:172–181. doi: 10.1016/s0264-410x(98)00155-8. [DOI] [PubMed] [Google Scholar]
- 18.Hiebert S W, Paterson R G, Lamb R A. Identification and predicted sequence of a previously unrecognized small hydrophobic protein, SH, of the paramyxovirus simian virus 5. J Virol. 1985;55:744–751. doi: 10.1128/jvi.55.3.744-751.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin H, Clarke D, Zhou Z-Y H, Cheng X, Coelingh K, Bryant M, Li S. Recombinant human respiratory syncytial virus (RSV) from cDNA and construction of subgroup A and B chimeric RSV. Virology. 1998;251:206–214. doi: 10.1006/viro.1998.9414. [DOI] [PubMed] [Google Scholar]
- 20.Jin H, Leser G P, Zhang J, Lamb R A. Influenza virus hemagglutinin and neuraminidase cytoplasmic tails control particle shape. EMBO J. 1997;16:1236–1247. doi: 10.1093/emboj/16.6.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato A, Kiyotani K, Sakai Y, Yoshida T, Shioda T, Nagai Y. Importance of the cysteine-rich carboxyl-terminal half of V protein for Sendai virus pathogenesis. J Virol. 1997;71:7266–7272. doi: 10.1128/jvi.71.10.7266-7272.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurotani A, Kiyotani K, Kato A, Shioda T, Sakai Y, Mizumoto K, Yoshida T, Nagai Y. Sendai virus C proteins are categorically nonessential gene products but silencing their expression severely impairs viral replication and pathogenesis. Genes Cells. 1998;3:111–124. doi: 10.1046/j.1365-2443.1998.00170.x. [DOI] [PubMed] [Google Scholar]
- 23.Lamb R A, Kolakofsky D. Paramyxoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1177–1204. [Google Scholar]
- 24.Peeples M E. Paramyxovirus M proteins: pulling it all together and taking it on the road. In: Kingsbury D W, editor. The paramyxoviruses. New York, N.Y: Plenum Publishing Corp.; 1991. pp. 427–456. [Google Scholar]
- 25.Radecke F, Billeter M A. The nonstructural C protein is not essential for multiplication of Edmonston B strain measles virus in cultured cells. Virology. 1996;217:418–421. doi: 10.1006/viro.1996.0134. [DOI] [PubMed] [Google Scholar]
- 26.Sutter G, Ohlmann M, Erfle V. Non-replicating vaccinia vector efficiently expresses bacteriophage T7 RNA polymerase. FEBS Lett. 1995;371:9–12. doi: 10.1016/0014-5793(95)00843-x. [DOI] [PubMed] [Google Scholar]
- 27.Tapparel C, Hausmann S, Pelet T, Curran J, Kolakofsky D, Roux L. Inhibition of Sendai virus genome replication due to promoter-increased selectivity: a possible role for the accessory C proteins. J Virol. 1997;71:9588–9599. doi: 10.1128/jvi.71.12.9588-9599.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teng M N, Collins P L. Altered growth characteristics of recombinant respiratory syncytial viruses which do not produce NS2 protein. J Virol. 1999;73:466–473. doi: 10.1128/jvi.73.1.466-473.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teng M N, Collins P L. Identification of the respiratory syncytial virus proteins required for formation and passage of helper-dependent infectious particles. J Virol. 1998;72:5707–5716. doi: 10.1128/jvi.72.7.5707-5716.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tober C, Seufert M, Schneider H, Billeter M A, Johnston I C, Niewiesk S, ter Meulen V, Schneider-Schaulies S. Expression of measles virus V protein is associated with pathogenicity and control of viral RNA synthesis. J Virol. 1998;72:8124–8132. doi: 10.1128/jvi.72.10.8124-8132.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitehead S S, Bukreyev A, Teng M N, Firestone C Y, St. Claire M, Elkins W R, Collins P L, Murphy B R. Recombinant respiratory syncytial virus bearing a deletion of either the NS2 or SH gene is attenuated in chimpanzees. J Virol. 1999;73:3438–3442. doi: 10.1128/jvi.73.4.3438-3442.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wyatt L S, Moss B, Rozenblatt S. Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology. 1995;210:202–205. doi: 10.1006/viro.1995.1332. [DOI] [PubMed] [Google Scholar]
- 33.Yu Q, Hardy R W, Wertz G W. Functional cDNA clones of the human respiratory syncytial (RS) virus N, P, and L proteins support replication of RS virus genomic RNA analogs and define minimal trans-acting requirements for RNA replication. J Virol. 1995;69:2412–2419. doi: 10.1128/jvi.69.4.2412-2419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]