Abstract
The phosphoprotein (P) gene of rabies virus (CVS strain) was cloned and expressed in bacteria. The purified protein was used as the substrate for phosphorylation by the protein kinase(s) present in cell extract prepared from rat brain. Two distinct types of protein kinases, staurosporin sensitive and heparin sensitive, were found to phosphorylate the P protein in vitro by the cell extract. Interestingly, the heparin-sensitive kinase was not the ubiquitous casein kinase II present in a variety of cell types. Further purification of the cell fractions revealed that the protein kinase C (PKC) isomers constitute the staurosporin-sensitive kinases α, β, γ, and ζ, with the PKCγ isomer being the most effective in phosphorylating the P protein. A unique heparin-sensitive kinase was characterized as a 71-kDa protein with biochemical properties not demonstrated by any known protein kinases stored in the protein data bank. This protein kinase, designated RVPK (rabies virus protein kinase), phosphorylates P protein (36 kDa) and alters its mobility in gel to migrate at 40 kDa. In contrast, the PKC isoforms do not change the mobility of unphosphorylated P protein. RVPK appears to be packaged in the purified virions, to display biochemical characteristics similar to those of the cell-purified RVPK, and to similarly alter the mobility of endogenous P protein upon phosphorylation. By site-directed mutagenesis, the sites of phosphorylation of RVPK were mapped at S63 and S64, whereas PKC isomers phosphorylated at S162, S210, and S271. Involvement of a unique protein kinase in phosphorylating rabies virus P protein indicates its important role in the structure and function of the protein and consequently in the life cycle of the virus.
Like viruses belonging to the rhabdovirus and paramyxovirus family, Rabies virus (RV), a member of Lyssavirus genus, contains a linear nonsegmented RNA genome of negative polarity. The ribonucleoprotein (RNP) complex contains the genome RNA enwrapped by the nucleocapsid protein (N) and the RNA polymerase, which contains a large protein (L) and the phosphoprotein (P) (1). Both L and P proteins of RV, like the corresponding proteins in rhabdoviruses and paramyxoviruses, are involved in the transcription and replication of the genome RNA (1, 46). Although the L proteins of this class of viruses show significant similarity in amino acid sequence, the P proteins appear to be highly divergent and nonhomologous. However, all P proteins are structurally similar in being highly acidic and phosphorylated and in playing a common vital role as transcription factors for the function of the corresponding L proteins. The P proteins, in addition to providing the transcription function of the L protein, appear to play an important role in the replicative process as well (19, 22, 28, 34, 36). In this role, the P protein forms a complex intracellularly with the N protein to impart an undefined replication-competent form to the latter which enables it to encapsidate nascent RNA chains during the replicative reaction (12, 28, 35). Interestingly, phosphorylation of the P protein appears not to be required for formation of this complex (19, 42, 43), suggesting a possible involvement of phosphorylation/dephosphorylation of P protein in the so-called switch of the RNA polymerase from transcriptive to replicative mode. Thus, the structures and functions of the P proteins of these viruses have been the subject of studies in several laboratories.
In the recent past, a systematic effort has been made to characterize the cellular protein kinases that phosphorylate the P proteins of these viruses and to establish the role, if any, of this posttranslational modification in P-protein function. Interestingly, a diverse group of cellular kinases have been found to phosphorylate different P proteins of this class of viruses (13, 15). For example, casein kinase II (CKII) and an L-protein-associated protein kinase (LAK) are specifically used for phosphorylation of the P protein of vesicular stomatitis virus (VSV) (2, 3), whereas the protein kinase Cζ (PKCζ) isoform phosphorylates the P proteins of human parainfluenza virus type 3 (HPIV3) and Sendai virus (14, 24, 25), although a unique proline-directed protein kinase has also been shown to be involved in Sendai virus P-protein phosphorylation (5). In contrast, both CKII and PKCζ are used by measles virus and canine distemper virus (11, 31) P proteins, and the PKCɛ isomer is used for Borna virus P-protein phosphorylation (39). Involvement of specific cellular kinases for the phosphorylation of different P proteins raises the important question as to their precise roles in the structures and functions of these proteins. Recently, by biochemical and reverse genetics approach, an obligate role of phosphorylation in the P-protein function of VSV (34) and HPIV3 (14, 25) has been established. For other viral P proteins studied so far, the functional role of phosphorylation remains to be firmly established.
In this work, we have studied phosphorylation of the RV P protein by cellular kinases, using rat brain tissue homogenate as the source of protein kinase and bacterially expressed P protein as the substrate. By extensive purification of the cell extracts, we have purified a unique heparin-sensitive non-CKII protein kinase that specifically phosphorylates the RV P protein. The precise identity of this kinase remains unknown. We also demonstrate that several isomers of PKC phosphorylate P protein of RV. Among them, PKCγ seems to be used preferentially by the P protein. Both the unique protein kinase and PKC isomers phosphorylate at specific sites on the P protein, resulting in the formation of distinct phosphorylated forms of P protein distinguishable by polyacrylamide gel electrophoresis (PAGE) analyses. Interestingly, the unique protein kinase seems to be preferentially packaged within the purified virions. The functional role of these protein kinases in RV replication remains to be determined.
MATERIALS AND METHODS
Cell cultures and virus.
The CVS (challenge virus standard) strain of RV was grown on BSR cells (a clone of BHK-21 cells) and purified by centrifugation in a 10 to 40% sucrose gradient as described elsewhere (40). For 35S labeling of virus, cell culture medium was replaced 20 h after infection by methionine-free minimal essential medium supplemented with 50 μCi of [35S]methionine and [35S]cysteine (specific activity, >1,000 Ci/mmol; Amersham) per ml for 24 h.
Expression of RV P protein in Escherichia coli.
The cDNA clone of RV (CVS strain) P gene was subcloned into the bacterial expression vector pET-3a as described earlier (2). For expression, the positive recombinant plasmid was introduced into E. coli BL21/DE3, and the expressed recombinant P protein was then purified through phosphocellulose and DE52 ion-exchange chromatography. All mutant P genes were similarly cloned, expressed, and purified.
Purification of cellular kinases from rat brain.
Rat brain tissue homogenate was prepared, and an S100 fraction was made (27). The S100 fraction was passed through a DEAE column equilibrated with buffer A (25 mM Tris [pH 8.0], 7.5% sucrose, 1 mM dithiothreitol [DTT], 0.5 mM EDTA) containing 0.12 M NaCl. Both the unbound (DE-UB) and bound (DE-B) fractions (eluted with a 0.12 to 0.5 M NaCl gradient in buffer A) were collected. The DE-UB fractions were dialyzed against 25 mM potassium phosphate buffer (pH 7.5) containing 1 mM DTT and 5% glycerol and were loaded onto a hydroxylapatite column equilibrated in the same phosphate buffer. Different isoforms of conventional PKC were step eluted with 75, 100, and 150 mM potassium phosphate buffer as described elsewhere (33). The peak fraction from each step eluate was dialyzed in 25 mM potassium phosphate buffer, rechromatographed through a hydroxylapatite column, and processed similarly. The peak fractions containing kinase activity were stored at −80°C. The DE-B fractions having the P-protein phosphorylating activity were combined and passed through a phosphocellulose column equilibrated with buffer A containing 0.2 M NaCl. P-protein phosphorylating activity was collected in unbound (PC-UB) and bound (PC-B) fractions (eluted at 0.6 M NaCl). The PC-B fractions were then loaded onto a heparin-Sepharose column at 50 mM NaCl salt concentration. The active fractions, eluted at 0.8 M NaCl, were combined (HS-B1) and loaded onto a hydroxylapatite column equilibrated with 50 mM potassium phosphate buffer (pH 7.5). P-protein phosphorylating kinase binds to the column which can be eluted at 500 mM potassium phosphate buffer, pH 7.5 (HAP-B). The HAP-B fractions were then rechromatographed through a heparin-Sepharose column. The active fractions eluted at 0.8 M NaCl were then combined (to form what we have designated RVPK [RV protein kinase]) and stored at −80°C.
Protein kinase assay.
In vitro phosphorylation of P protein was carried out in a 20-μl reaction containing 0.2 to 0.5 μg of recombinant P protein, 5 μCi of [γ32P]ATP, 50 μM ATP, 50 mM Tris-HCl (pH 8.0), 2 mM MgCl2, 1 mM DTT, and the indicated amount of kinase. To check the PKC activity, 200 μM CaCl2, 100 μg of phosphatidylserine per ml, and 100 μg of diacylglycerol per ml were also added in the reaction buffer (33). The reaction mixture was incubated at 30°C for 30 min. The reaction was stopped by adding sodium dodecyl sulfate (SDS) sample loading buffer and electrophoresed on a 10% polyacrylamide gel containing SDS. The gels were then subjected to autoradiography.
For assay of virion-associated protein kinase (RNPK) activity, purified virions (2.0 μg) were disrupted with 0.1% Triton X-100 and incubated with kinase reaction buffer (20 mM Tris-HCl (pH 8), 10 mM MgCl, 2 mM DTT, 10 μCi of [γ-32P]ATP, 50 μM unlabeled ATP) and incubated at 30°C for 30 min. Reactions were stopped by adding SDS sample loading buffer and analyzed by electrophoresis through an SDS–12% polyacrylamide gel followed by autoradiography. Quantification of radioactivity was performed with a PhosphorImager (Molecular Dynamics). 35S-labeled virus or unlabeled virus was incubated as previously described except that unlabeled ATP was added in the reaction.
Peptide mapping of the phosphorylated P protein.
Either the wild-type or mutant P protein was phosphorylated with RVPK (HAP-B fraction) in the presence of [γ-32P]ATP. The radiolabeled P protein was electrophoresed through an SDS–10% acrylamide gel and were visualized by brief autoradiography. The 32P-labeled bands were excised, electroeluted, and concentrated. The 32P-labeled proteins were digested with LysC or chymotrypsin essentially as described elsewhere (7). The digests were then analyzed by electrophoresis in a 20% polyacrylamide gel and autoradiographed (7).
RESULTS
Phosphorylation of E. coli-expressed P protein of RV by fractionated tissue extract.
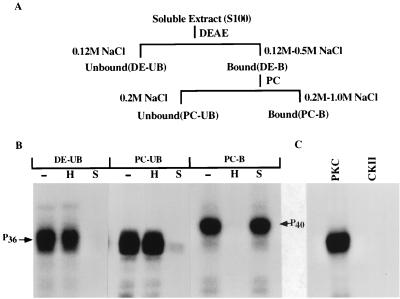
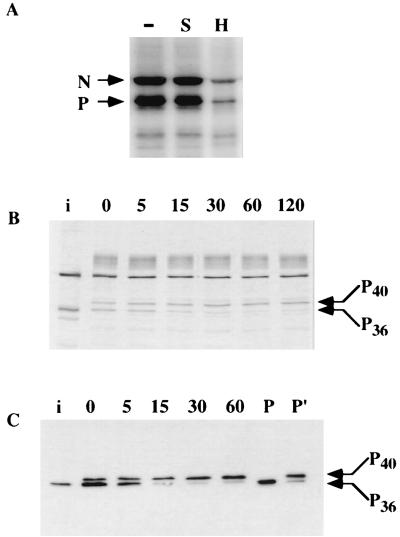
A full-length cDNA of RV (CVS strain) P protein was expressed in E. coli by using an inducible expression vector, pET3a, as described earlier (2). The recombinant unphosphorylated P protein was purified by fractionation on DEAE and phosphocellulose column chromatography and used in subsequent reactions as the substrate for phosphorylation by the cellular protein kinases purified from cell extract. Initially an S100 soluble extract of rat brain was prepared and subjected to fractionation by DEAE-cellulose and phosphocellulose chromatography as shown in Fig. 1A. Various bound and unbound fractions from both column fractions were used to phosphorylate recombinant P protein in the presence of [γ-32P]ATP in vitro. In each phosphorylation reaction, either heparin (an inhibitor of CKII) or staurosporin (an inhibitor of PKC) was added to characterize the protein kinase(s). As shown in Fig. 1B, although all fractions efficiently phosphorylated RV Protein the protein kinases in fractions DE-UB and PC-UB were sensitive to staurosporin, whereas fraction PC-B was sensitive to heparin. These results strongly suggested that fractions DE-UB and PC-UB contained PKC subtypes and fraction PC-B contained CKII. Surprisingly, as shown in Fig. 1C, the P protein was not phosphorylated by authentic CKII whereas commercial PKC effectively phosphorylated it. We concluded from these results that the protein kinase present in fraction PC-B is not CKII, although it is heparin sensitive, and may represent a unique class of protein kinase. In addition, recombinant unphosphorylated P protein of 36K kDa (P36) migrated slower (as P40) in PAGE upon phosphorylation by fraction PC-B (Fig. 1). The migration rate remained unchanged when recombinant P protein was phosphorylated by fractions DE-UB and PC-UB. Thus, it seems that different phosphorylated forms of RV P protein are produced following phosphorylation by the different protein kinases present in DE-UB, PC-UB, and PC-B fractions.
FIG. 1.
Phosphorylation of recombinant P protein of RV by rat brain extract. (A) Schematic presentation of the purification of soluble extract (S100) by DEAE-cellulose and phosphocellulose (PC) column chromatography rat brain tissue homogenate. The unbound (UB) and bound (B) fraction from each column are noted in parentheses. (B) Bacterially expressed recombinant P protein of RV was incubated with [γ-32P]ATP and different fractions, as indicated above the lanes, in the absence (−) or presence of heparin (H) or staurosporine (S). The positions of migration of the phosphorylated P proteins are indicated. (C) Recombinant P protein was incubated with commercially available PKC or recombinant CKII.
Characterization of PKC isoforms in fractions DE-UB and PC-UB.
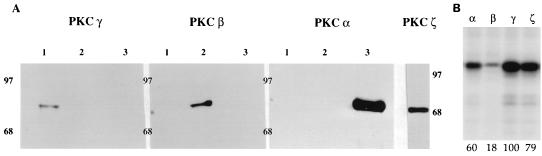
First, we fractionated further both DE-UB and PC-UB fractions by chromatography on a hydroxylapatite column, and different isomers of PKC (α, β, and γ) were eluted by a sodium phosphate step gradient. Each fraction (1, 2, and 3) was tested for different isomers of PKC by Western blot analyses (Fig. 2A). Similarly, the PKCζ normally present in fraction PC-UB (13) was purified and analyzed by Western blotting with authentic PKCζ. As shown in Fig. 2A, fractions 1, 2, and 3 cross-reacted with PKC isoforms γ, β, and α, respectively, and PKCζ antibodies reacted specifically with the PC-UB fraction. The separation of these three isomers was absolute since no fraction cross-reacted with another in Western blot analyses (Fig. 2A). The purified PKC isoforms were then tested for phosphorylation activity on P protein. As shown in Fig. 2B, all four isoforms of PKC phosphorylated P protein but to different degrees. PKCγ consistently phosphorylated the P protein with the highest efficiency, followed by ζ, α, and β. In each assay, equal unit activity of each PKC isoform, equalized on the basis of peptide phosphorylation by PKC isomers, was used with an equivalent amount of P as substrate. These results suggest that at least four isoforms of PKC are able to phosphorylate the P protein, with PKCγ being the most effective.
FIG. 2.
Analysis of the PKC isoforms purified from rat brain extract by Western blotting and by phosphorylating activity. (A) Each conventional isoform of PKC purified from the DE-UB fraction as described in Materials and Methods was assayed for the presence of conventional PKC isoforms (α, β, and γ) by Western blot analysis. The antibody used for each blot is indicated at the top. Lanes 1, 2, and 3 represent fractions eluted with 75, 100, and 150 mM potassium phosphate buffer, respectively. PKCζ, a novel isoform normally present in the PC-UB fraction, was assayed by anti-ζ antibody. Positions of migration of molecular weight markers are shown in kilodaltons on each side. (B) Recombinant P protein was incubated with [γ-32P]ATP and identical unit activity of different isoforms of PKC, marked above each lane. Numbers at the bottom denote percent phosphorylation, taking the highest level of phosphorylation by PKCγ as 100%.
Purification and characterization of the heparin-sensitive protein kinase.
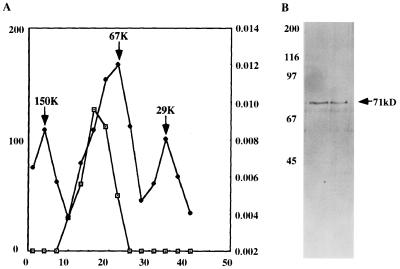
Fraction PC-B (Fig. 1A) which contained the unique heparin-sensitive non-CKII protein kinase, was subjected to a series of chromatographic purification steps as described in Materials and Methods. The final eluted fraction from the heparin-Sepharose column was nearly homogeneous and efficiently phosphorylated the P protein in vitro (not shown). It was then subjected to glycerol gradient centrifugation followed by PAGE of the peak fraction. As shown in Fig. 3A, the protein kinase sedimented at a position slightly higher than that of the control, bovine serum albumin (67 kDa). This is consistent with the PAGE analysis (Fig. 3B), where the protein migrated slower than the 67-kDa marker, with a molecular weight of approximately 71,000. From these analyses it seems that a heparin-sensitive protein kinase (designated RVPK) with a molecular weight of 71,000 is involved in the phosphorylation of P protein of RV, in addition to specific PKC isoforms.
FIG. 3.
Determination of molecular weight of RVPK by glycerol gradient centrifugation. (A) Purified RVPK obtained after the second heparin-Sepharose column step, as detailed in Materials and Methods, was loaded on a 5 to 25% glycerol gradient and run in an SW40 rotor at 36,000 rpm for 42 h at 4°C. Fractions (3 drops/fraction) were collected from the bottom. Alternative fractions were assayed for the ability to phosphorylate the P protein and plotted (squares); a parallel gradient was run with the known molecular weight protein mixture, and the optical densities at 280 nm of the fractions were plotted (circles). The peak position of migration of each known molecular weight protein is marked by an arrow. (B) Two fractions showing highest protein kinase activity were analyzed on an SDS–10% acrylamide gel and silver stained. Numbers on the left indicate positions of migration (in kilodaltons) of molecular weight markers; the number on the right indicates the deduced molecular mass of the stained protein.
Different phosphorylated forms of RV P protein following phosphorylation by kinases.
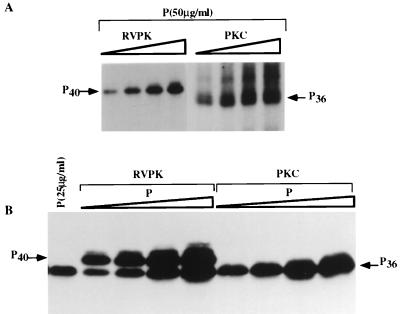
Having purified the PKC isoforms and the heparin-sensitive RVPK from tissue homogenate, we next studied the effect of phosphorylation on the electrophoretic mobility of the recombinant P protein, which would indicate different phosphorylated forms, if any, of the P protein. As shown in Fig. 4A, with increasing concentration of RVPK, the P protein originally migrating as P36 shifted its mobility and migrated slower, as P40. In contrast, with phosphorylation by any one of the PKC isoforms the migration position of the P protein remained unchanged; i.e., it migrated as P36. These results strongly suggest that phosphorylation by RVPK and PKC isoforms led to the formation of different phosphorylated forms of P protein in vitro. This effect of phosphorylation on the mobility of P was more pronounced when different concentrations of P protein were phosphorylated by a fixed amount of RVPK or PKC isoforms in the presence of unlabeled ATP followed by Western blot analyses with 25E6, a monoclonal antibody for P protein (38). As shown in Fig. 4B, the unphosphorylated P migrating as P36 migrated slower with increasing phosphorylation by RVPK, whereas the migration position of P remained virtually unchanged upon phosphorylation by PKC isoforms. It is noteworthy that a slower-migrating minor band appears slightly above either P36 or P40, depending on the phosphorylation of P by PKC or RVPK (Fig. 4). However, the slower-migrating band which appears upon phosphorylation by PKC (Fig. 4A) does not cross-react with the anti-P antibody (Fig. 4B). Again the slower-migrating band that appears after phosphorylation by RVPK in the Western blot (Fig. 4B) is not detected by 32P assay (Fig. 4A). At this point, the precise identities of these bands are unknown. Thus, it seems that two distinct phosphorylated forms of P protein are formed upon phosphorylation by RVPK and PKC isomers.
FIG. 4.
Mobility of P protein following phosphorylation by different kinases. (A) P protein (50 μg/ml) was incubated with increasing concentrations of RVPK (HAP-B) fraction or PKC (DE-UB) fraction (0.025, 0.05, 0.1, and 0.2 μg/μl) in the presence of [γ-32P]ATP. (B) Different concentration (0.1 μg/μl) of either RVPK or PKC in the presence of unlabeled ATP. The reaction mixture was then electrophoresed on an SDS–10% acrylamide gel followed by Western blot analysis against P antibody, using ECL reagent. Positions of migration of radiolabeled P protein are marked by P36 and P40.
Characterization of RNPK in RV.
Protein kinases have been found to be associated with the RNP of the purified virions of many enveloped viruses (9, 18, 21, 26, 30, 32). It is generally believed to be a phenomenon of random packaging of cellular kinases, possibly during virus maturation. However, in the recent past, this contention has changed particularly for VSV and HPIV3, where a selective packaging of CKII and the PKCζ isoform, respectively, the protein kinases that are involved in phosphorylation of the corresponding P proteins, was observed (14, 20). These findings raised the possibility that RV also packages the heparin-sensitive kinase and all or some of the PKC isoforms involved in P phosphorylation. Accordingly, purified RV was disrupted by Triton X-100 and incubated directly with [γ-32P]ATP. As shown in Fig. 5A, efficient phosphorylation of RNP-associated P protein as well as N protein was observed; the latter protein has previously been shown to be phosphorylated both in vitro and in vivo (47). Interestingly, the RNPK was highly sensitive to heparin but not to staurosporin. We did, however, observe a low level of heparin-resistant activity, which may represent a low level of PKC activity within the virion. Moreover, disrupted RV did not phosphorylate casein in vitro (not shown), indicating that the RNPK is probably a unique protein kinase similar to RVPK purified from the cell extract. To test whether the RNPK changes the mobility of the P protein as observed for the RVPK purified from cell extract, a kinetic analyses of the phosphorylation of endogenous virion P protein was carried out in vitro. When we incubated 35S-labeled virus with unlabeled ATP (Fig. 5B) or carried out Western blot analyses of unlabeled virus with unlabeled ATP (C), we detected a time-dependent shift of P protein mobility from P36 to P40 similar to that observed in Fig. 4. These results indicated that RV selectively packaged the heparin-sensitive protein kinase during maturation and remained associated with the RNP complex within the virion. Western blotting of the RNPK with antibodies against PKC isomers detected a small amount of PKCζ subtype only (data not shown).
FIG. 5.
Phosphorylation of viral P protein with RNPK. (A) Purified RV (25 μg/ml) was disrupted by Triton X-100 and incubated with [γ-32P]ATP in absence (−) or presence of heparin (25 μg/ml; H) or staurosporine (200 nM; S). Positions of migration of N and P proteins are indicated. (B) [35S]Met-labeled purified virus was similarly incubated with unlabeled ATP for different time periods, indicated (in minutes) above the lanes. i, [35S]Met-labeled infected cell RNP; P36 and P40, positions of slower- and faster-migrating P protein, respectively. (C) Purified RV (25 μg/ml) was incubated with unlabeled ATP for different time periods as indicated (in minutes) above the lanes. The samples were then run on an SDS–10% polyacrylamide gel followed by Western blot analysis with anti-P antibody. i, [35S]Met-labeled infected cell RNP; P and P′, bacterially expressed recombinant P protein phosphorylated with PKC (DE-UB) and RVPK (HAP-B), respectively. Positions of migration of the phosphorylated P proteins are indicated by P36 and P40.
Next, we carried out a detailed biochemical characterization of the RVPK for comparison with properties of the RNPK. As detailed in Table 1, virtually all biochemical parameters tested were identical for the two enzymes. We conclude from these results that the unique protein kinase is responsible for the phosphorylation of P protein both in vitro and probably in vivo.
TABLE 1.
Comparison of biochemical properties of RVPK and RNPKa
| Enzyme | Effect of:
|
Requirement for divalent cation:
|
Effect of ATP concn at 2 mM Mg2+ (% relative activity)
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Heparin (25 μg/ml | Staurosporine (200 nM) | Rottlerin (50 μM) | GF109203X (10 μM) | PKA-peptide (5 μg/ml) | Sphingosine (2 mM) | Phosphatidylserine (800 μg/ml) | Arachidonic acid (40 μM) | Ca2+ | Mg2+ | Mn2+ | Co2+ | Zm2+ | 2 mM | 4 mM | 6 mM | |
| RVPK | 100% inhibition | No inhibition | No inhibition | No inhibition | No inhibition | 90% inhibition | No effect | No effect | No | Yes | No | No | No | 100 | 50 | 5 |
| RNPK | 80% inhibition | No inhibition | 15% inhibition | No inhibition | No inhibition | 83% inhibition | No effect | No effect | No | Yes | No | No | No | 100 | 49 | 7 |
All reactions were carried out in a 25-μl reaction volume containing either 20 μg of recombinant P and purified RVPK per ml or 25 μg of purified virus per ml. For requirement for divalent cations, “no” or “yes” represents the absolute need for activity.
Determination of the phosphorylation sites on RVP.
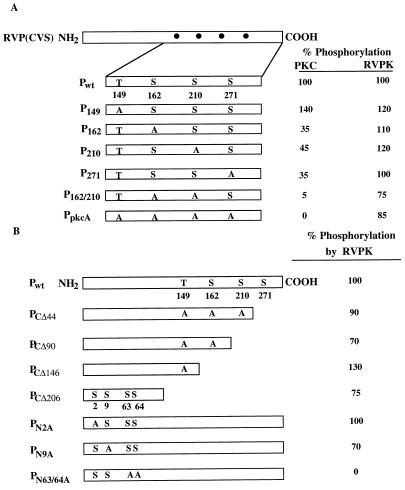
We next wanted to determine the site(s) of phosphorylation by RVPK and the PKC isoforms on the P protein. From the amino acid sequence of P protein of the CVS strain of RV, we first located the S and the T residues that are present within the consensus motif of the PKC phosphorylation site, i.e., S/T-X-K/R. This exercise led us to select four sites (T149, S162, S210, and S271) which are located within the PKC consensus motif. We carried out systematic site-directed mutagenesis at those sites, altering S or T to A, expressed the mutant proteins in E. coli, and then purified and tested them for phosphorylation by RVPK and PKC. As shown in Fig. 6, altering S162 and S210 to A resulted in a drastic reduction in the ability of PKC to phosphorylate P protein (95%), whereas phosphorylation by RVPK was reduced by only 25%. These results suggest that PKC phosphorylation sites are probably S162, S210, and S271, which are presumably different from the RVPK sites which may be located at a different domain in the P protein.
FIG. 6.
Determination of P phosphorylation sites for RVPK. (A) A diagram of the entire P protein is shown at the left hand; the dots represent computer-predicted PKC sites. An enlargement of that region shows the positions of one threonine (T) and three serine (S) residues. The mutant P proteins at these sites are shown. PpkcA is the mutant P protein in which all four sites were changed to alanine. Phosphorylation of each mutant as a percentage of the wild-type level is shown at the right. PKC (DE-UB) and RVPK (HS-B) were used for analyses. (B) The entire P protein is drawn schematically along with the computer-predicted PKC sites. Subscript numbers in C-terminal mutant designations represent the number of amino acids deleted from the C terminus. The mutated serines and threonine in the PKC motif are shown. The number in the subscript of each N-terminal mutant designation denotes the position of serine altered to alanine, keeping the PKC motifs unaltered. Percent phosphorylation of each mutant by purified RVPK (HAP-B) is shown at the right.
We then carried out a systematic deletion mapping of the RVPK phosphorylation site(s) by progressive deletion from the C-terminal end of the P protein. As shown in Fig. 6B, the mutant PCΔ206, which lacked all of the PKC sites, could still be phosphorylated efficiently (75%) by RVPK. Examination of the remaining N-terminal fragment revealed the presence of four S residues located at positions 2, 9, 63, and 64 which may the potential sites for RVPK. Since no phosphorylated threonine residue was detected by phosphoamino acid analysis, (data not shown), we did not include the threonine residues present in the 91-amino-acid long N-terminal fragment for mutational analysis. By site-directed mutagenesis of the above serine residues to alanine, it was found that both S63 and S64 are the probable sites of phosphorylation by RVPK (Fig. 6B). Thus, the phosphorylation sites of RVPK and PKC are located at the N- and C-terminal regions of the P protein, respectively.
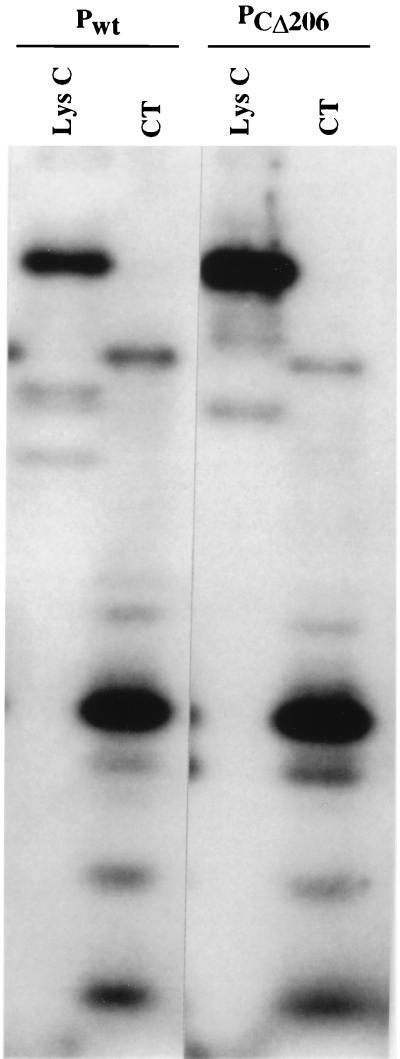
Since both wild-type P (Pwt) and the deletion mutant PCΔ206 can be phosphorylated almost equally by RVPK, RVPK may phosphorylate Pwt at sites different from those in PCΔ206; the latter sites may have been exposed due to deletion of 206 amino acids from the carboxy terminus. To test this, we phosphorylated both Pwt and PCΔ206 by RVPK; then the eluted labeled proteins were digested with chymotrypsin or LysC and subjected to gel electrophoresis. As shown in Fig. 7, virtually all peptides migrated identically, demonstrating that both Pwt and PCΔ206 are phosphorylated by RVPK at identical sites, i.e., S63 and S64.
FIG. 7.
Peptide mapping of the phosphorylated P protein. 32P-labeled Pwt and PCΔ206 phosphorylated by RVPK (HAP-B) were gel purified as described in Materials and Methods. Each labeled protein was then digested with either LysC or chymotrypsin (CT), as shown above each lane.
DISCUSSION
In this study, we identified and characterized the cellular protein kinases involved in the phosphorylation of recombinant rabies virus (CVS strain) P protein, the transcription factor subunit of the RNA polymerase complex. Similar studies on bacterially expressed P protein of nonsegmented negative-strand RNA viruses revealed the involvement of a diverse class of protein kinases responsible for phosphorylating the transcription factors. These enzymes ranged from the ubiquitous cellular protein kinase CKII to rare PKC isoforms ζ and ɛ (4, 14, 24, 31, 39). Our present study demonstrated that in addition to several isomers of PKC, the RV P protein utilizes for its phosphorylation a unique heparin-sensitive non-CKII protein kinase which appears to be selectively packaged within the matured virions. The unique protein kinase (designed RVPK) is a 71-kDa polypeptide that is sensitive to heparin and sphingosine, requires specifically Mg2+, and is optimally active at an ATP concentration of 2 mM. We have so far been unable to identify this protein kinase by comparing its biochemical properties with those of other known cellular protein kinases in the protein data bank. Clearly, determination of the amino acid sequence of this kinase is essential to find a match with a protein which may or may not turn out to be a bona fide protein kinase.
Our interesting finding is that RVPK seems to be directly involved in altering the secondary structure of P protein such that it migrates slower than the unphosphorylated or PKC isomer-phosphorylated P protein (Fig. 4). In RV virions as well as in infected cells, two phosphorylated forms of RV P proteins have been found: a so-called hyperphosphorylated form (P40) and a hypophosphorylated form (P36). Also observed are several minor forms, P35, P31, and P30, which appear to be smaller N-terminally truncated products apparently resulting from translation initiation at alternate AUG codons located in frame within the P protein (8). The P36 form is found as the principal product both in the virion and in infected cells (45). The P40 form appears to be present transiently during normal infection but accumulates when cells are treated with okadaic acid, an inhibitor of protein phosphatases (44). Thus, it seems that P40 and P36 phosphorylated forms are interconvertible, and our data suggest that the two forms are produced upon phosphorylation by RVPK and PKC isomers, respectively. In this respect, the RV P protein is similar to the VSV P protein studied previously (7), where the phosphorylated P1 and P2 forms are produced upon phosphorylation by CKII and LAK, respectively (2, 7). In earlier studies, expression of RV P protein in insect cells by infection of recombinant baculovirus yielded P protein which was not phosphorylated (37). This may have been due to dephosphorylation of P protein in vivo by active cellular phosphatases present in the insect cells or due to the absence of a specific kinase. It is important to note that although we purified RVPK from rat brain tissue homogenate, it is present in BHK cells in which RV is grown (data not shown). Thus, selective utilization of this kinase strongly suggests its role in the structure and function of the P protein.
The involvement of PKC isomers in RV P phosphorylation, on the other hand, is also quite interesting. Although four isomers (α, β, γ, and ζ) appear to be involved in the phosphorylation process, the γ isomer consistently appears to be used in preference to the other three. Since PKCγ activity is found predominantly in the brain and the ζ and α isomers are present in all cell types, involvement of PKCγ may suggest the possible neurotropism routinely observed during RV infection (6, 16, 41, 46). Another possibility could be that because RV is neurotropic for another reason, its P protein is predominantly phosphorylated by PKCγ.
Another interesting finding is the specific packaging of RVPK within the mature RV virions. It is apparent from Table 1 that RNPK demonstrated biochemical properties identical to those of the RVPK purified from brain tissue homogenate. Moreover, it phosphorylated in vitro the endogenous P protein to generate two phosphorylated forms, P36 and P40, similar to the purified RVPK (Fig. 3). In addition to RVPK activity, a small amount of PKCζ was found in the virions of the CVS strain by Western blot analysis (data not shown). These findings are similar to those for VSV, HPIV3, and Sendai virus, which also package the protein kinases involved in P-protein phosphorylation. Thus, it seems that the specific kinases interact with the viral P proteins of nonsegmented negative-strand viruses during replication and remain associated with the virions during morphogenesis. It is important to point out that these studies were carried out with the CVS strain of RV grown in BHK cells. It is quite possible that different strains of RV grown in different cell lines may package different protein kinases involved in that particular virus-host system.
Finally, it is apparent from our site-directed mutagenesis studies that sites of phosphorylation of RVPK and PKC isomers are at opposite ends of the P protein, N and C terminal, respectively. Phosphorylation at the N-terminal end must lead to some conformational change in the P protein leading to slower migration in PAGE (Figs. 4 and 5). In contrast, C-terminal phosphorylation does not cause any such change in the electrophoretic mobility of P protein. This situation is highly analogous to that for the VSV P protein, where CKII phosphorylation produces only the P1 form and LAK-mediated phosphorylation yields the P2 form (2, 7, 23). Thus, it may be incorrect to consider the P40 form of RV the hyperphosphorylated from since phosphorylation of only two sites in the P protein resulted in the formation of P40. It is possible that, as for VSV, phosphorylation of the RV P protein causes a distinct change in the structural property of the protein which may lead to its proper association with the L protein. In fact, for VSV P (New Jersey serotype), biophysical measurements confirmed that phosphorylation altered the secondary structure of the protein (10). In addition, phosphorylation of VSV P (both Indiana and New Jersey serotypes) results in oligomerization of the protein, leading eventually to its activation (10, 17). Similar studies with RV P protein would certainly provide some insight into the structure of the protein as it relates to its function. It is important to mention here that unlike P of the CVS strain, P of the HEP-Flurry strain of RV does not contain an S residue at position 63. Since a heparin-sensitive protein kinase has recently been implicated in phosphorylating the P protein of this strain (44), it is likely that S64 is the principal phosphorylation site for RVPK. This predication can be confirmed by mutational analyses. Interestingly, P proteins of the PV, PM, and ERA strains of RV all have only S63, although the ERA strain has T at position 64 (4, 29). Involvement of RVPK in these strains is yet to be established. Notwithstanding, active phosphorylation of S63 and S64 of the P protein of the CVS strain coupled with the fact that RVPK is predominately packaged within the virion strongly suggests that this unique protein kinase plays a role in the life cycle of RV. Further studies along this line would clearly be informative.
ACKNOWLEDGMENTS
This work was supported by an NIH grant to A.K.B. and by CNRS UPR 9053.
We are grateful to Anne Flamand for helpful discussions.
REFERENCES
- 1.Banerjee A K, Barik S, De B P. Gene expression of nonsegmented negative strand RNA viruses. Pharmacol Ther. 1991;51:47–70. doi: 10.1016/0163-7258(91)90041-j. [DOI] [PubMed] [Google Scholar]
- 2.Barik S, Banerjee A K. Sequential phosphorylation of the phosphoprotein of vesicular stomatitis virus by cellular and viral protein kinases is essential for transcription activation. J Virol. 1992;66:1109–1118. doi: 10.1128/jvi.66.2.1109-1118.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barik S, Banerjee A K. Phosphorylation by cellular casein kinase II is essential for transcriptional activity of vesicular stomatitis virus phosphoprotein. Proc Natl Acad Sci USA. 1992;89:6570–6574. doi: 10.1073/pnas.89.14.6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byrappa S, Pan Y B, Gupta K C. Sendai virus P protein is constitutively phosphorylated at serine 249: high phosphorylation potential of the P protein. Virology. 1996;216:228–234. doi: 10.1006/viro.1996.0052. [DOI] [PubMed] [Google Scholar]
- 5.Charlton K M, Casey G A. Experimental rabies in skunks. Immunofluorescence light and electron microscope studies. Lab Investig. 1979;41:36–44. [PubMed] [Google Scholar]
- 6.Chen J L, Das T, Banerjee A K. Phosphorylated status of vesicular stomatitis virus P protein in vitro and in vivo. Virology. 1997;228:200–212. doi: 10.1006/viro.1996.8401. [DOI] [PubMed] [Google Scholar]
- 7.Chenik M, Chebli K, Blondel D. Translation initiation at alternate in-frame AUG codons in the rabies virus phosphoprotein mRNA is mediated by a ribosomal leaky scanning mechanism. J Virol. 1995;69:707–712. doi: 10.1128/jvi.69.2.707-712.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinton G M, Burge B W, Huang A S. Phosphoproteins of VSV: identity and interconversion of phosphorylated forms. Virology. 1979;99:84–94. doi: 10.1016/0042-6822(79)90039-4. [DOI] [PubMed] [Google Scholar]
- 9.Clinton G M, Guerina N G, Guo H, Huang A S. Host-dependent phosphorylation and kinase activity associated with vesicular stomatitis virus. J Virol. 1982;257:3313–3319. [PubMed] [Google Scholar]
- 10.Das T, Gupta A K, Sims P W, Gelfand C A, Jentoft J E, Banerjee A K. Role of cellular casein kinase II in the function of the phosphoprotein (P) subunit of RNA olymerase of vesicular stomatitis virus. J Biol Chem. 1995;270:24100–24107. doi: 10.1074/jbc.270.41.24100. [DOI] [PubMed] [Google Scholar]
- 11.Das T, Shuster A, Schneider-Schaulies S, Banerjee A K. Involvement of cellular casein kinase II in the phosphorylation of measles virus P protein: identification of phosphorylation sites. Virology. 1995;211:218–226. doi: 10.1006/viro.1995.1394. [DOI] [PubMed] [Google Scholar]
- 12.Davis N L, Arnheiter H, Wertz G W. Vesicular stomatitis virus N and NS proteins from multiple complexes. J Virol. 1986;59:751–754. doi: 10.1128/jvi.59.3.751-754.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De B P, Banerjee A K. Role of host proteins in gene expression of negative strand RNA viruses. Adv Virus Res. 1997;48:169–204. doi: 10.1016/s0065-3527(08)60288-2. [DOI] [PubMed] [Google Scholar]
- 14.De B P, Gupta S, Gupta S, Banerjee A K. Cellular protein kinase C isoform ζ regulates human parainfluenza virus type 3 replication. Proc Natl Acad Sci USA. 1995;92:5204–5208. doi: 10.1073/pnas.92.11.5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De B P, Das T, Banerjee A K. Role of cellular kinases in the gene expression of negative strand RNA viruses. Biol Chem. 1997;378:489–493. [PubMed] [Google Scholar]
- 16.Dietzschold B, Wiktor T J, Trojanowski J Q, Macfarlan R I, Wunner W H, Torres-Anjel M J, Koprowski H. Differences in cell-to-cell spread of pathogenic and apathogenic rabies virus in vivo and in vitro. J Virol. 1985;56:12–18. doi: 10.1128/jvi.56.1.12-18.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao Y, Lenard J. Multimerization and transcriptional activation of the phosphoprotein (P) of vesicular stomatitis virus by casein kinase II. EMBO J. 1995;14:1240–1247. doi: 10.1002/j.1460-2075.1995.tb07107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grubman M J, Baxt B, La Torre J L, Bachrach H L. Identification of a protein kinase activity in purified foot-and-mouth disease virus. J Virol. 1981;39:455–462. doi: 10.1128/jvi.39.2.455-462.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta A K, Banerjee A K. Expression and purification of vesicular stomatitis virus N-P complex from Escherichia coli: role in genome RNA transcription and replication in vitro. J Virol. 1997;71:4264–4271. doi: 10.1128/jvi.71.6.4264-4271.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta A K, Das T, Banerjee A K. Casein kinase II is the P protein phosphorylating cellular kinase associated with the ribonucleoprotein complex of purified vesicular stomatitis virus. J Gen Virol. 1995;76:365–372. doi: 10.1099/0022-1317-76-2-365. [DOI] [PubMed] [Google Scholar]
- 21.Hatanaka M, Twiddy E, Gilden R V. Protein kinase associated with RNA tumor viruses and other budding RNA viruses. Virology. 1972;47:536–538. doi: 10.1016/0042-6822(72)90297-8. [DOI] [PubMed] [Google Scholar]
- 22.Howard M, Davis N, Patton J, Wertz G. Roles of vesicular stomatitis virus N and NS protein in viral RNA replication. In: Mahy B W J, Kolakofsky D, editors. The biology of negative strand viruses. Amsterdam, The Netherlands: Elsevier; 1987. pp. 134–140. [Google Scholar]
- 23.Hsu C H, Kingsbury D W. NS phosphoprotein of vesicular stomatitis virus: subspecies separated by electrophoresis and isoelectric focusing. J Virol. 1982;42:342–345. doi: 10.1128/jvi.42.1.342-345.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huntley C C, De B P, Banerjee A K. Phosphorylation of Sendai virus phosphoprotein by cellular protein kinase C ζ. J Biol Chem. 1997;272:16578–16584. doi: 10.1074/jbc.272.26.16578. [DOI] [PubMed] [Google Scholar]
- 25.Huntley C C, De B P, Banerjee A K. Human parainfluenza virus type 3 phosphoprotein: identification of serine 333 as the major site for PKCζ phosphorylation. Virology. 1995;211:561–567. doi: 10.1006/viro.1995.1438. [DOI] [PubMed] [Google Scholar]
- 26.Imblum R L, Wagner R R. Protein kinase and phosphorylation of vesicular stomatitis virus. J Virol. 1974;13:113–124. doi: 10.1128/jvi.13.1.113-124.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kikkawa U, Takai Y, Minakuchi R, Inohara S, Nishizuka Y. Calcium activated phospholipid dependent protein kinase from rat brain. J Biol Chem. 1982;257:13341–13348. [PubMed] [Google Scholar]
- 28.La Ferla F M, Peluso R W. The 1:1 N-NS complex of vesicular stomatitis virus is essential for efficient genome replication. J Virol. 1989;63:3852–3857. doi: 10.1128/jvi.63.9.3852-3857.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larson J K, Wunner W H. Nucleotides ans deduced amino-acid sequences of the nominal nonstructural phosphoprotein of the ERA, PM and CVS-11 strains of rabies virus. Nucleic Acids Res. 1990;18:7172. doi: 10.1093/nar/18.23.7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemaster S, Roizman B. Herpes simplex virus phosphoproteins. II. Characterization of the virion protein kinase and of the polypeptides phosphorylated in the virus. J Virol. 1980;35:798–811. doi: 10.1128/jvi.35.3.798-811.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Z, Huntley C C, De B P, Das T, Banerjee A K, Oglesbee M J. Phosphorylation of canine distemper virus P protein by protein kinase C-ζ and casein kinase II. Virology. 1997;232:198–206. doi: 10.1006/viro.1997.8548. [DOI] [PubMed] [Google Scholar]
- 32.Moyer S A, Summers D F. Phosphorylation of vesicular stomatitis virus in vivo and in vitro. J Virol. 1974;13:455–465. doi: 10.1128/jvi.13.2.455-465.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogita K, Ono Y, Kikkawa U, Nishizuka Y. Expression separation and assay of protein kinase C subspecies. Methods Enzymol. 1991;200:228–234. doi: 10.1016/0076-6879(91)00142-j. [DOI] [PubMed] [Google Scholar]
- 34.Pattnaik A K, Hwang L, Li T, England N, Mathur M, Das T, Banerjee A K. Phosphorylation within the amino-terminal acidic domain I of the phosphoprotein of vesicular stomatitis virus is required for transcription but not for replication. J Virol. 1997;71:8167–8175. doi: 10.1128/jvi.71.11.8167-8175.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peluso R W. Kinetic, quantitative, and functional analysis of multiple forms of vesicular stomatitis virus nucleocapsid protein in infected cells. J Virol. 1988;62:2799–2807. doi: 10.1128/jvi.62.8.2799-2807.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peluso R W, Moyer S A. Viral proteins required for the in vitro replication of vesicular stomatitis virus defective interfering particle genome RNA. Virology. 1988;162:369–376. doi: 10.1016/0042-6822(88)90477-1. [DOI] [PubMed] [Google Scholar]
- 37.Prehaud C, Nel K, Bishop D L. Baculovirus expressed rabies virus M1 protein is not phosphorylated: it forms multiple complexes with expressed rabies N protein. Virology. 1992;189:766–770. doi: 10.1016/0042-6822(92)90602-l. [DOI] [PubMed] [Google Scholar]
- 38.Raux H, Iseni F, Lafay F, Blondel D. Mapping of monoclonal antibody epitopes of the rabies virus P protein. J Gen Virol. 1997;78:119–124. doi: 10.1099/0022-1317-78-1-119. [DOI] [PubMed] [Google Scholar]
- 39.Schwemmle M, De B P, Shi L, Banerjee A K, Lipkin I. Borna disease virus P protein is phosphorylated by a calcium independent protein kinase C and casein kinase II. J Biol Chem. 1997;272:21818–21823. doi: 10.1074/jbc.272.35.21818. [DOI] [PubMed] [Google Scholar]
- 40.Seif I, Coulon P, Rollin E, Flamand A. Rabies virulence: effect on pathogenicity and sequence characterization of rabies virus mutations affecting antigenic site III of the glycoproteins. J Virol. 1985;53:926–934. doi: 10.1128/jvi.53.3.926-934.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shankar V, Dietzschold B, Koprowski H. Direct entry of rabies virus into the central nervous system without prior local replication. J Virol. 1991;65:2736–2738. doi: 10.1128/jvi.65.5.2736-2738.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takacs M A, Das T, Banerjee A K. Mapping of interacting domains between the nucleocapsid protein and the phosphoprotein of vesicular stomatitis virus by using a two hybrid system. Proc Natl Acad Sci USA. 1993;90:10375–10379. doi: 10.1073/pnas.90.21.10375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takacs M A, Banerjee A K. Efficient interaction of the vesicular stomatitis virus P protein with the L protein and the N protein in cells expressing the recombinant proteins. Virology. 1995;208:821–826. doi: 10.1006/viro.1995.1219. [DOI] [PubMed] [Google Scholar]
- 44.Takamatsu F, Asakawa N, Morimoto K, Eriguchi Y, Toriumi H, Kawai A. Possible relationship between the two forms of the non-catalytic subunit (P protein) Microbiol Immunol. 1998;42:761–771. doi: 10.1111/j.1348-0421.1998.tb02350.x. [DOI] [PubMed] [Google Scholar]
- 45.Tuffereau C, Fischer S, Flammand A. Phosphorylation of the N and M1 proteins of rabies virus. J Gen Virol. 1985;66:2285–2289. doi: 10.1099/0022-1317-66-10-2285. [DOI] [PubMed] [Google Scholar]
- 46.Wagner R R. Rhabdoviridae and their replication. In: Knipe D M, Fields B N, editors. Fields virology. New York, N.Y: Raven Press Ltd.; 1990. pp. 867–881. [Google Scholar]
- 47.Yang J, Koprowski H, Dietzschold B, Fu Z F. Phosphorylation of rabies virus nucleoprotein regulates viral RNA transcription and replication by modulating leader RNA encapsidation. J Virol. 1999;73:1661–1664. doi: 10.1128/jvi.73.2.1661-1664.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]