Abstract
Cellular pathways that detect DNA damage are useful for identifying genes that suppress DNA damage, which can cause genome instability and cancer predisposition syndromes when mutated. We identified 199 high-confidence and 530 low-confidence DNA damage-suppressing (DDS) genes in Saccharomyces cerevisiae through a whole-genome screen for mutations inducing Hug1 expression, a focused screen for mutations inducing Ddc2 foci, and data from previous screens for mutations causing Rad52 foci accumulation and Rnr3 induction. We also identified 286 high-confidence and 394 low-confidence diverse genome instability-suppressing (DGIS) genes through a whole-genome screen for mutations resulting in increased gross chromosomal rearrangements and data from previous screens for mutations causing increased genome instability as assessed in a diversity of genome instability assays. Genes that suppress both pathways (DDS+ DGIS+) prevent or repair DNA replication damage and likely include genes preventing collisions between the replication and transcription machineries. DDS+ DGIS− genes, including many transcription-related genes, likely suppress damage that is normally repaired properly or prevent inappropriate signaling, whereas DDS− DGIS+ genes, like PIF1, do not suppress damage but likely promote its proper, nonmutagenic repair. Thus, induction of DNA damage markers is not a reliable indicator of increased genome instability, and the DDS and DGIS categories define mechanistically distinct groups of genes.
Keywords: DNA damage signaling, DNA repair, genome instability
Introduction
Cells have sensitive mechanisms to identify and signal the presence of DNA damage. Mutations affecting these pathways have been linked to cancer predisposition syndromes in humans and increased genome instability in both model organisms and human cells and tumors (Vogelstein and Kinzler 2004; Friedberg et al. 2013; Carbone et al. 2020; Sharma et al. 2020). Markers for the presence of DNA damage have been widely exploited as in vivo sensors for increased levels of DNA damage (Sun et al. 1996; Maser et al. 1997; Nelms et al. 1998; Zhong et al. 1999; Downs et al. 2000; Paull et al. 2000; Melo et al. 2001; Fernandez-Capetillo et al. 2002; Wang et al. 2002; Lou et al. 2003; Stewart et al. 2003).
One important class of DNA damage response (DDR) markers includes targets of the DNA damage checkpoint. In Saccharomyces cerevisiae, the DNA damage checkpoint is triggered by a protein kinase cascade comprised of Tel1ATM or Mec1ATR-Ddc2ATRIP, Rad53CHEK2, and Dun1 (Supplementary Fig. 1; Putnam, Jaehnig, et al. 2009; Ciccia and Elledge 2010; Lanz et al. 2019). Markers of checkpoint activation include (1) the accumulation of phosphorylated forms of histone H2A (the S. cerevisiae equivalent of γ-H2AX), Rad53CHEK2, RPA, Rad54, and Rad55 (Sun et al. 1996; Bashkirov et al. 2000, 2006; Downs et al. 2000; Herzberg et al. 2006; Smolka et al. 2007; Chen et al. 2010; Lanz et al. 2021) and (2) changes in the levels of ribonucleotide reductase (RNR)-related proteins, including Rnr1–4, Sml1, and Hug1 (Supplementary Fig. 1; Huang et al. 1998; Zhao et al. 2001). Together, these responses trigger DNA damage checkpoint activation, which can arrest cells at the G2/M cell cycle transition and increase the deoxyribonucleotide (dNTP) levels to promote DNA replication and DNA repair. Induction of dNTP levels is the essential function of the Mec1ATR-Ddc2ATRIP and Rad53CHEK2 proteins in S. cerevisiae (Desany et al. 1998; Huang et al. 1998; Zhao et al. 1998; Basrai et al. 1999; Lee and Elledge 2006; Zhang et al. 2006; Lee et al. 2008; Wu and Huang 2008; Meurisse et al. 2014).
A second important class of the DDR markers includes proteins that form discrete nuclear foci in response to DNA damage. Nuclear foci of DNA repair proteins that form and colocalize with double-strand breaks (DSBs) have been identified, including the Mre11-Rad52-Xrs2 complex, Rad52, Rad51, Rad54, Rad55, Rad59, Rdh54, and RPA (Maser et al. 1997; Nelms et al. 1998; Melo et al. 2001; Lisby et al. 2003, 2004; Lisby and Rothstein 2009). These foci have been used to elucidate an ordered assembly pathway for DSB repair proteins in S. cerevisiae (Lisby et al. 2004). Foci have also been observed for other DNA repair proteins as well as components of the DNA damage checkpoint, such as Ddc2ATRIP and Ddc1RAD9A/RAD9B (Melo et al. 2001; Makhnevych et al. 2009; Hombauer et al. 2011; Tkach et al. 2012; Yimit et al. 2015).
Despite their utility, the consistency between these different DDR markers and their correlation with the results of genome instability assays has not been systematically investigated. Here, we have performed genome-wide screens for mutations in S. cerevisiae that induce Hug1 expression and induce the accumulation of gross chromosomal rearrangements (GCRs) detected using the duplication-mediated GCR assay (Putnam, Hayes, et al. 2009) as well as a screen for mutations causing increased Ddc2 foci using a focused set of query mutations. We have combined these data with data from 3 previous genome-wide DDR marker screens (Alvaro et al. 2007; Hendry et al. 2015; Styles et al. 2016), the results of 14 different studies identifying mutations that cause increased genome instability as assessed in a range of different genome instability assays (Chen and Kolodner 1999; Yuen et al. 2007; Andersen et al. 2008; Putnam, Hayes, et al. 2009; Chan and Kolodner 2011; Stirling et al. 2011; Zhang et al. 2012, 2013; Putnam et al. 2016; Nene et al. 2018; Srivatsan et al. 2019; Novarina et al. 2020), and systematic screens for mutations that alter the cell cycle distribution in log-phase cells (Koren et al. 2010; Hoose et al. 2012; Soifer and Barkai 2014). We used these data to define sets of 199 high-confidence and 530 low-confidence genes that suppress DNA damage levels and 286 high-confidence and 394 low-confidence genes that suppress genome instability during unperturbed cell growth; high-confidence genes are those that were identified in multiple assays. Genes that act in DNA damage suppression and genome instability suppression (DDS+ DGIS+) are strongly linked to preventing or repairing endogenous damage arising during DNA replication. Remarkably, we also identified many high-confidence genes that only suppress DNA damage (DDS+ DGIS−) or only suppress genome instability (DDS− DGIS+). The DDS+ DGIS− genes may suppress DNA damage that is only rarely misrepaired to cause genome instability or may suppress inappropriate signaling, whereas the DDS− DGIS+ genes do not suppress damage but likely promote the correct repair of damage when it occurs. Together, our results provide mechanistic insights into the prevention and processing of spontaneous DNA damage in unperturbed cells.
Materials and methods
Bait strain construction
The bait strain containing the HUG1-EGFP marker was created by integrating a EGFP-hphNT1 cassette amplified by PCR onto the 3′ end of the HUG1 coding sequencing in the strain RDKY7635 (Putnam et al. 2016) to create RDKY8174 (MATα his3Δ200 hom3-10 leu2Δ0 trp1Δ63 ura3Δ0 lyp1::TRP1 cyh2-Q38K iYFR016C::PMFA1-LEU2 can1::PLEU2-NAT yel072w::CAN1-URA3 HUG1-EGFP.hphNT1); note that this strain contains the dGCR assay markers. The bait strain containing the DDC2-EGFP marker was created by a multistep process (see Supplementary Methods S1) to introduce markers required for systematic mating, the NUP49-mCherry fluorescent marker, and the Ddc2-EGFP fluorescent marker to create RDKY8934 (MATα his3Δ200 leu2Δ0 trp1Δ63 ura3Δ0 can1 cyh2-Q38K iYFR016C:PMFA1-URA3 NUP49-mCherry.hphNT1 DDC2-EGFP.HIS3MX6).
Systematic genetic crosses
The HUG1-EGFP containing bait strain RDKY8174 was crossed twice to a selected set of the BY4741 MATa strains in S. cerevisiae deletion collection (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), which was enriched for genes predicted to be involved in genome stability (Putnam et al. 2012), and once to the entire collection of BY4741 MATa strains using a RoToR pinning robot (Singer Instruments). The DDC2-EGFP.HIS3MX6 containing bait strain was crossed to a set of 468 BY4741 MATa deletion strains containing mutations selected as causing increased Hug1-EGFP expression or increased GCR rates or the leu2Δ::kanMX4 control mutation. Systematic mating followed a modified synthetic genetic array (SGA) protocol (Tong and Boone 2006) involving multiple steps: diploid selection, sporulation, diploid killing, and haploid selection (see Supplementary Methods S1). Due to the different markers in the two bait strains, different selective media were used at some steps (see Supplementary Methods S1).
DNA content analysis
Cell cycle analysis was conducted as previously described (Vanoli et al. 2010). In brief, 1 × 107 cells were collected by centrifugation and resuspended in 70% ethanol for 16 h. Cells were then washed in 0.25 M Tris-HCl (pH 7.5), resuspended in the same buffer containing 2 mg/mL of RNaseA (Sigma Aldrich), incubated at 37°C for at least 1 h, and then treated overnight with proteinase K (1 mg/mL; Sigma Aldrich) at 37°C. Cells were collected by centrifugation, resuspended in 200 mM Tris-HCl (pH 7.5) buffer containing 200 mM NaCl and 80 mM MgCl2, and stained in the same buffer containing 1 µM SYTOX green (Invitrogen). Samples were then diluted 10-fold in 50 mM Tris-HCl (pH 7.8), sonicated in a Cole-Palmer 8891 Water Bath Sonicator, and analyzed using a Becton Dickinson FACScan instrument. This FACS analysis verified that all the strains used for analysis in experiments from this study were haploids.
Measurement of Hug1-EGFP levels
To measure Hug1-EGFP expression, logarithmic-phase cultures were grown in YPD medium in 96 well plates and centrifuged, and the cells were resuspended in 100-µL sterile water and sonicated in a Cole-Palmer 8891 Water Bath Sonicator. The cells were then analyzed using a BD LSR Fortessa analytical cytometer with a High-Throughput Sampler. Excitation was at 488 nm, and the fluorescence signal was collected through a 505-nm long-pass filter and a HQ510/20 band-pass filter (Chroma Technology Corp). For each sample, 30,000 events were recorded. The mean value of the GFP fluorescence in each sample was calculated using FlowJo software and normalized to the mean GFP value of wild-type cells.
Measurement of Ddc2-GFP foci
Cells were grown in a complete synthetic medium (Sigma Aldrich) to log phase and examined by live imaging using an Olympus BX43 fluorescence microscope with a 60× 1.42 PlanApo N Olympus oil-immersion objective. GFP fluorescence was detected using a Chroma FITC filter set and captured with a Qimaging QIClick CCD camera. Nuclear boundaries were identified by Nup49-mCherry fluorescence, and only Ddc2 foci contained within nuclei were scored. Images were analyzed using Meta Morph Advanced 7.7 imaging software, keeping processing parameters constant within each experiment.
Cell cycle distribution data analysis
The cell cycle distributions of log-phase cells for the MATa BY4741 haploid deletion collection and the homozygous diploid BY4743 deletion collection were previously published by others (Koren et al. 2010; Hoose et al. 2012; Soifer and Barkai 2014). For the diploid deletion collection data, the published supplement containing cell cycle parameters from the Dean–Jett–Fox model (Fox 1980) as implemented in FlowJo was used (Hoose et al. 2012). For the haploid deletion collection data, FACS data were downloaded from the FlowRepository (Spidlen et al. 2012) and were analyzed using the Dean–Jett–Fox model in FlowJo version 10 (FlowJo 2019). Analyses of these data to remove aggregates, merge multiple observations, and project onto ternary plots are described in Supplementary Methods S1.
Genome-wide GCR screen
Haploid cells resulting from the cross of RDKY8174, which contains the dGCR assay markers, with the BY4741 deletion collection were grown in YPD in 96-well plates and patched in triplicate on YPD plates using a Singer RoToR robot. Patched YPD plates were then replica plated onto GCR selection medium plates [6.7-g yeast nitrogen base (Fisher Scientific), 2-g CSM-Arg-Ura dropout mix (US Biological), 60-mg uracil (Sigma Aldrich), 1-g 5-fluoroorotic acid (US Biological), 60-mg L-canavanine sulfate (Sigma Aldrich), 20-g glucose (Fisher Scientific), and 24-g bacto-agar (Fisher Scientific) per liter]. After 3–4 days of growth, the GCR selection medium plates were photographed and the number of papillae in each patch was counted. For each set of 3 plates, the distribution of papillae per patch was determined and fitted to a normal distribution. The P-values for each patch were then calculated and then a combined P-value for each mutant was determined using Fisher's sumlog procedure as implemented in R. Using a false discovery rate (FDR) of 0.05, 197 mutations of 193 genes were identified as causing increased GCRs. These hits were then rescreened by manual repatching in triplicate onto YPD plates and rescreening by replica plating onto GCR selective medium plates and counting papillae. A cutoff ratio, 1.8, for the fold increase relative to the average number of papillae per patch was determined by maximizing the Youden index using a set of 94 true positive mutations and 921 true negative mutations from previous studies. This cutoff identified 52 significant hits, 24 of which were previously identified as causing increased GCR rates in quantitative assays and 6 of which were previously identified as not causing increased GCR rates in quantitative assays. Selected mutations were then reconstructed in RDKY7635 (systematic dGCR assay; MATα hom3–10 ura3Δ0 leu2Δ0 trp1Δ63 his3Δ200 lyp1::TRP1 cyh2-Q38K iYFR016C::PMFA1-LEU2 can1::PLEU2-NAT yel072w::CAN1-URA3), and GCR rates were measured using published methods (Srivatsan et al. 2018).
Quantification and statistical analysis
All statistical analyses were performed with R version 4.1.1. Significance for the Hug1-EGFP expression data was performed by fitting to a Gaussian distribution as described in the main text, Fig. 1, and Supplementary Fig. 1. Significance for the Ddc2-GFP foci data was performed using 95% cutoffs of the wild-type data as described in the main text and Fig. 2. Significance for the cell cycle distribution was performed as described in the Supplemental Methods based on the two-independent Gaussian error model.
Fig. 1.
Distribution of Hug1-EGFP levels. a) Histograms of Hug1-EGFP levels in individual cells from log-phase cultures of the control (leu2Δ) strain and 3 mutant strains with increased Hug1-EGFP levels. b) Distribution of the fold-increased Hug1-EGFP levels for all mutants analyzed. The dashed line is the center of the fitted normal distribution, and solid lines are FDR < 0.05 significance cutoffs (Supplementary Fig. 3c and d).
Fig. 2.
Induction of Ddc2-EGFP foci in mutant strains. a) Representative images of Ddc2-EGFP foci (triangles) for the control (leu2Δ) and mre11Δ strains. b) Distribution of the fraction of cells with Ddc2-EGFP foci. The dashed horizontal line is the average of the leu2Δ control strain measurements, and the solid horizontal lines correspond to the 95 and 5% percentiles of the control strain measurements.
Results
Identification of genes suppressing Hug1-EGFP induction
Fluorescence-activated cell sorting (FACS) determines fluorescence intensity on a per-cell basis, making it suitable to monitor fluorescence responses even in strains with a slow growth phenotype. To identify mutations inducing DNA damage signaling, we first constructed a strain containing the HUG1-EGFP marker. We found that the HUG1-EGFP marker gave the largest change in expression upon hydroxyurea treatment in preliminary FACS experiments when compared to 3 other potential markers (Supplementary Fig. 2), and previous studies demonstrated that the HUG1 promoter is responsive to a wide variety of DNA-damaging agents, but not other types of cellular stress (Benton et al. 2007; Ainsworth et al. 2012). We crossed this strain to the S. cerevisiae BY4741 deletion collection using a SGA protocol (Supplementary Fig. 3a; Tong and Boone 2006; Putnam et al. 2016). For downstream analyses, we added a BY4741 leu2Δ::kanMX4 control strain to the deletion collection by generating a strain in which the BY4741 leu2Δ0 deletion was replaced with a leu2Δ::kanMX4 deletion. Unperturbed log-phase cultures of the resulting MATa haploid strains that contained HUG1-EGFP and the gene deletion of interest were analyzed by high-throughput FACS for both DNA content and EGFP fluorescence. We used the DNA content measurements to identify isolates with diploid DNA content or with a mixture of cells with haploid and diploid DNA content; data from these problematic isolates were excluded from our analyses. Fold Hug1-EGFP induction levels were calculated by normalizing the average EGFP fluorescence for each strain to the fluorescence of control strains from the same experiment (Fig. 1a; Supplementary Table 1). Because subsets of the deletion collection were independently mated and measured up to 3 times, 4,791 deletion mutants derived from 5,781 independently generated strains were analyzed; repeated measurements from independent biological isolates were highly correlated (Supplementary Fig. 3b).
We combined the Hug1-EGFP expression data for the 4,791 deletion mutants determined here with our previous data for 394 strains with hypomorphic mutations in essential genes (Fig. 1b; Srivatsan et al. 2019). The resulting fold Hug1-EGFP values were normally distributed with a mean of 1.06 and a standard deviation of 0.13, which indicates that most mutations do not substantially alter Hug1 expression (Supplementary Fig. 3c). More strains in the experimental distribution had increased Hug1 expression than were predicted by a fitted normal distribution, indicating the presence of mutations that induce Hug1 expression during unperturbed growth (Supplementary Fig. 3c and d). Using a FDR of 0.05, 479 mutations affecting 416 genes were identified as causing increased Hug1-EGFP expression (1.39- to 35.8-fold induction; Supplementary Table 1). Details of these genes are described below.
Induction of Ddc2 foci imperfectly correlates with Hug1 induction
Hug1 induction is a downstream readout of the Mec1 DNA damage checkpoint (Supplementary Fig. 1; Huang et al. 1998; Basrai et al. 1999). We therefore examined an upstream marker of Mec1 activation, the accumulation of Ddc2 foci (also called Lcd1 foci; Melo et al. 2001; Waterman et al. 2019). We constructed a MATα bait strain expressing a functional Ddc2-EGFP fusion protein (Supplementary Fig. 4a) and the Nup49-mCherry nuclear pore fusion protein, which allows for scoring of only nuclear Ddc2 foci. We crossed this bait strain using SGA (Supplementary Fig. 4b) to a focused set of 469 BY4741 MATa strains enriched for deletion mutations that induced Hug1 expression. The resulting 444 recovered haploid strains were grown to log phase and analyzed by fluorescence microscopy to determine the fraction of cells containing nuclear Ddc2 foci (Fig. 2a; Supplementary Table 2). Because this focused screen was biased for mutations inducing DNA damage, we assessed significance thresholds using the 5th and 95th percentile of the leu2Δ control strain observations. Only 4 of the strains had significantly reduced levels of Ddc2 foci (fraction of cells with Ddc2 foci < 0.015): npl3Δ, ydjlΔ, dus1Δ, and mec1Δ sml1Δ. In contrast, 153 strains had significantly increased levels of Ddc2 foci (fraction of cells with Ddc2 foci > 0.048; Fig. 2b), and many, but not all, of these mutant strains were also identified in the Hug1 induction screen as having increased levels of Hug1 expression (see below).
DDR screens implicate DNA replication errors as a major source of spontaneous DNA damage
To better understand the mutations causing induced Hug1 expression and increased Ddc2 foci levels, we compared the data generated here with data from previous screens for mutations affecting DNA damage responses. These screens included (1) induction of Rnr3 expression (Hendry et al. 2015); (2) induction of Rad52 foci in diploid strains, which are intermediates during homologous recombination of DSBs and replication fork damage (Lisby et al. 2001) (Rad52D screen; Alvaro et al. 2007); and (3) induction of Rad52 foci in haploid strains (Rad52H screen; Styles et al. 2016). Together, these screens evaluated 5,976 mutations and identified 729 DNA damage-suppressing (DDS) genes, including 199 high-confidence DDS genes identified in multiple screens and 530 low-confidence DDS genes identified in only 1 screen (Fig. 3a–c; Supplementary Table 3). In general, mutations in genes identified by multiple screens tended to cause larger induction responses (Fig. 3d), consistent with the idea that mutations causing high levels of DNA damage are robustly identified in multiple screens using different DNA damage response readouts. Because only some of the screens analyzed alleles of essential genes and because some strains may not have been recovered from crosses, some low-confidence genes are likely to be bona fide DDS genes as these factors reduce the possibility that a gene could be scored as a high-confidence DDS gene.
Fig. 3.
Comparison of DDR marker screens. a) Histogram of the number of screens genes were identified in. High-confidence genes were identified in multiple screens. Low-confidence genes were identified in one screen. b) Total number of genes identified in each screen. c) Percentage of high-confidence DDR genes identified in each screen, which was calculated by dividing the number of high-confidence genes identified in each screen b) by the total count of 199 high-confidence DDR genes. d) Mutations of genes identified in more screens cause stronger effects. Mutations are grouped by the number of screens the affected gene was identified in, and the screen results are rank ordered by the magnitude of the signal in the quantitative assays (100% = strongest signal and 0% = weakest signal). The fractional median rank is plotted for each group in each screen (red = Hug1, green = Rnr3, blue = Ddc2, and purple = Rad52D). e) Plot of the Rnr3 expression Z-score vs the fold increase in Hug1-EGFP expression. Lines are significance thresholds; the Z-score threshold of 2 was used in the original study (Hendry et al. 2015). f) The fraction of cells with Ddc2-EGFP foci vs the fold Hug1-EGFP expression plotted as for d). g) The fraction of diploid cells with Rad52 foci vs the fold Hug1-EGFP expression plotted as for d). The threshold of 0.15 was one used in the original study (Alvaro et al. 2007). h) A subset of high-confidence DDS genes, including all genes identified at least 3 times, grouped by function.
We directly compared the quantitative data from the Hug1 screen with the data from the Rnr3 (Fig. 3e), Ddc2 foci (Fig. 3f), and Rad52D foci screens (Fig. 3g). Consistent with the similarities of HUG1 and RNR3 regulation (Supplementary Fig. 1; Huang et al. 1998; Basrai et al. 1999), we observed a modest correlation between the data from the Hug1 and Rnr3 screens (Fig. 3e); 4,466 mutations were tested in both assays, and 73 mutations induced both markers, whereas 316 mutations only induced Hug1 and 61 mutations only induced Rnr3. This modest correlation may reflect the fact that induction of Rnr3 was a less sensitive DNA damage sensor than induction of Hug1 (Supplementary Fig. 1). Comparison with the Ddc2 screen results distinguished 2 groups of Hug1-inducing mutations among the 434 mutations tested in both assays. For mutations identified in both the Hug1 and Ddc2 screens (Fig. 3f, upper right quadrant; 110 mutations), the damage marker levels were correlated, suggesting that the mutations cause both increased DNA damage and increased Mec1-Ddc2-Hug1 signaling. For the uncorrelated mutations that only induced Hug1 (Fig. 3f, lower right quadrant; 144 mutations), these mutations may induce Hug1 through mechanisms independent of increased DNA damage (Basrai et al. 1999). The results from the Rad52D foci screen had relatively poor correlation with the results from the Hug1 screen; of the 4,541 mutations measured in both assays, only 36 mutations caused significant induction of both Hug1 expression and Rad52 foci (Fig. 3g). This poor correlation may result from the fact that increased Rad52 foci can be caused by defects resulting in (1) increased DNA damage, (2) increased processing of normal levels of endogenous DNA damage by HR, and (3) delays in the turnover of HR intermediates, the latter two of which may not be recognized as increased DNA damage. Other foci markers used to monitor the DNA damage response, including BRCA1, BRCA2, RAD51, MDC1, 53BP1, and RPA (Zhong et al. 1999; Paull et al. 2000; Wang et al. 2002; Fernandez-Capetillo et al. 2002; Lou et al. 2003; Stewart et al. 2003), may also be affected by these types of indirect effects. The indirect influences on Rad52 foci accumulation are echoed by the diverse roles of some of the genes identified (Alvaro et al. 2007), including transcription initiation (IRC1), clathrin-mediated vesicle trafficking (IRC6), degradation of cysteine (IRC7), and proteasome assembly (IRC25).
Analysis of the high-confidence DDS genes provides mechanistic insights into processes that generate and/or suppress the accumulation of DNA damage in unperturbed cells (Fig. 3h). High-confidence DDS genes include genes involved in DNA replication (CSM3, CTF4, DBF4, DNA2, DPB4, MCM10, MRC1, ORC1, ORC2, POL1, POL12, POL2, POL3, POL31, POL32, PRI1, PRI2, RAD27, RTT105, RFA2, and TOF1), DNA replication-associated histone disassembly and assembly (ASF1, CAC2, HST3, POB3, RLF2, and RTT109), sister chromatid cohesion (CTF18, CTF8, CHL1, DCC1, and RAD61), telomere homeostasis (EST1, EST2, EST3, and STN1), cell cycle control (CLB5, CLN3, DBF2, SIC1, and SWI6), and sumoylation, ubiquitination, and proteolysis (CUZ1, DOA1, PRE4, RPN11, RPN4, RPT4, RPT6, SEM1, SLX5, SLX8, SMT3, ULP1, and UMP1). In addition, genes involved in pathways known to repair and restart DNA replication forks were also identified, including homologous recombination (MRE11, RAD50, XRS2, SAE2, RAD51, RAD52, RAD54, RAD55, RAD57, RAD59, SGS1, RMI1, TOP3, MMS4, and MUS81), the Rtt101 cullin complex (MMS1, MMS22, RTT101, and RTT107), postreplication repair (MMS2, RAD18, RAD5, RAD6, and UBC13), and genes implicated in other repair pathways (APN1, DIA2, ELG1, ESC2, IRC3, MLH1, MSH2, NSE1, RAD1, RNH202, RNH203, RRM3, SLX5, SLX8, and WSS1). Genes encoding 2 components of the Rad17-Ddc1-Mec3 complex were also identified (RAD17 and DDC1; Fig. 3h), indicating a role for the Mec1 DNA damage checkpoint in avoiding the accumulation of spontaneous DNA damage. Remarkably, the RAD17 and DDC1 genes were not identified in the Hug1 and Rnr3 expression screens; this is consistent with their roles in triggering the checkpoint required to derepress HUG1 and RNR3. Mutations disrupting HUG1 and RNR3 repression (Basrai et al. 1999) were also identified in both the Hug1 and Rnr3 expression assays, including rfx1Δ/crt1Δ, isw2Δ, itc1Δ, sin4Δ, hda1Δ, and hda3Δ, but were generally not identified in the other DDR screens, consistent with these genes functioning in transcription repression and not functioning directly in the DDR. The high-confidence list also includes genes involved in transcription, including genes encoding subunits of the mediator (SIN4 and SRB5), SAGA (ADA2, NGG1, and SGF73), Swr1 (SWC4, VPS72), COMPASS (SWD1), and Rpd3S/L (RPD3) complexes, and genes involved in transcriptional elongation (DST1, SGV1, and SPT4) and transcriptional silencing (ESC1, ESC2, and SPT21). The identification of transcriptional genes in both the high- and low-confidence lists is consistent with the possibility that transcriptional defects cause DNA damage, potentially by increasing collisions between the replication fork and the transcription machinery (Brambati et al. 2015). Taken together, these results indicate that DNA replication is a major source of spontaneous DNA damage in S. cerevisiae cells.
Cell cycle analysis of DDR-inducing mutations is consistent with damage caused by DNA replication errors
A major function of DNA damage signaling is to trigger cell cycle checkpoints to prevent cell division in the presence of DNA damage (Blackford and Jackson 2017). We, therefore, reasoned that DDS gene mutations ought to perturb the timing of the cell cycle. To test this, we analyzed published FACS data for the S. cerevisiae haploid (Koren et al. 2010; Soifer and Barkai 2014) and diploid (Hoose et al. 2012) deletion collections to identify mutations that altered the percentage of cells in the G1, S, and G2 phases of the cell cycle during unperturbed growth (Supplementary Table 4). We first fitted each observation (Fig. 4a) so that the percentages of cells in the G1, S, and G2 phases summed to 100% (Fig. 4b). We then plotted each observation as a point on a ternary plot so that multiple observations could be combined and that cell cycle perturbations could be measured (Fig. 4c). The percentage of G1, S, and G2 cells was 21.5%, 25.1%, and 53.4% for the haploid control (n = 292) and 22.9%, 24.1%, and 53.0% for the diploid control (n = 219; Supplementary Fig. 5). Using these data, strains with cell cycle progression defects were identified (Fig. 4d and e; Supplementary Table 4).
Fig. 4.
Cell cycle distributions for mutant strains with and without increased levels of DNA damage markers. a) Example FACS analyses of log-phase cultures of a haploid S. cerevisiae control strain and haploid strains with enriched G1 (hem14Δ), S (met7Δ), and G2 (ste50Δ) populations. FACS data (Koren et al. 2010) were plotted using FlowJo (FlowJo 2019) with cell count vs the measured SYBR-green area for each cell. Peaks for these haploid strains are at 1n and 2n DNA content. b) Stacked bar plots for the cell cycle distributions for the observations in panel a derived using FlowJo with the Dean–Jett–Fox model (Fox 1980) and fitted to account for cell–cell aggregation (see ‘Materials and methods’). c) Ternary plot of the cell cycle distributions of the observations in a). Samples only having G1, S, or G2 cells would be at the top, left, or right vertex, respectively. d) Ternary plot diagram for the cell cycle distribution of haploid strains colored by the DDR category (black = all, blue = low-confidence, and red = high-confidence). The cross is the position of the wild-type control strain. Strains outside the circle have significantly altered cell cycle distributions (P < 0.01). Inset shows the average deviation of all genes (black), high-confidence DDR genes (red), and low-confidence DDR genes (blue) relative to the geometric median of the control strain. e) Ternary plot diagram for diploid strains displayed as in d). f) The cumulative distribution of the percent of cells in the G1, S, and G2 phases for all mutations (black) and for the high- (red) and low- (blue) confidence DDR mutations. In almost all cases, the all-mutation and low-confidence-mutation curves overlap. Vertical lines indicate the percentage found in the control samples. P-values calculated by the Kolmogorov–Smirnov test with the distribution of all mutations as the null hypothesis; n.s., not significant.
DDS gene mutations as a group tended to perturb the cell cycle distribution of log-phase cultures. Comparison of the effects of all of the mutations tested with the effects of the mutations in high-confidence DDS genes revealed that the DDS mutations caused a general decrease in the percentage of cells in G1 phase (P-values of <2 × 10−16 and 0.002 for the haploid and diploid data, respectively; Kolmogorov–Smirnov test; Fig. 4f) and an increase in the percentage of cells in G2 phase (P-values of 4 × 10−9 and 2 × 10−5; Fig. 4f). This shift, along with a very minor or no effect on the percentage of S-phase cells, suggests that DDS gene defects cause an accumulation of cells with or very near to a 2N DNA content, which could be due to defects in the completion of DNA replication and/or triggering of the G2/M DNA damage checkpoint. In contrast, the distribution of the low-confidence DDS mutations was generally not different than the distribution of all mutations, most of which (88%) were not identified in any DDS screen (Fig. 4f). The results of this analysis are consistent with the idea that the accumulation of damage in many DDS gene mutants occurs during S phase and is consistent with the identification of high-confidence DDS genes that are associated with DNA replication.
Genome-wide screen for deletion mutations causing increased GCR rates
In previous studies, we tested mutations in individual S. cerevisiae genes of interest and conducted a partial screen of the S. cerevisiae deletion collection (Chen and Kolodner 1999; Putnam, Hayes, et al. 2009; Chan and Kolodner 2011; Putnam et al. 2016; Nene et al. 2018; Srivatsan et al. 2019) and a screen of a set of mutations in an extensive subset of S. cerevisiae essential genes to identify mutations that cause increased GCR rates and identify genome instability-suppressing (GIS) genes. To facilitate comparisons between DDS genes and GIS genes, we performed a screen of the entire S. cerevisiae deletion collection for mutations causing increased GCR rates to expand our previous analyses to cover all nonessential genes. The strains recovered from the Hug1-EGFP cross also contained the duplication-mediated (dGCR) assay, which we included because increased dGCR rates can be detected using patch tests (Putnam, Hayes, et al. 2009). Haploid strains containing the dGCR assay and deletion mutations from the S. cerevisiae deletion collection were grown in 96-well plates, patched in triplicate using a Singer RoToR robot on YPD plates, and then replica plated onto GCR selection medium plates (Supplementary Fig. 6a). Our initial screen of patches identified 197 mutations affecting 193 genes as causing significantly increased number of papillae per patch (FDR < 0.05; Supplementary Fig. 6b). As expected, known mutations causing higher dGCR rates were more likely to be identified than those causing lower dGCR rates (Supplementary Fig. 6c). We then rescreened 620 strains, including the 197 strains with an FDR < 0.05, which resulted in identifying 52 candidate mutations that caused the largest increased patch scores, 24 of which were previously known to cause increased GCR rates (Supplementary Fig. 6d and e); this analysis defined 28 new candidate GIS genes that were incorporated into the analyses performed in this study (Supplementary Table 5). We performed a limited validation study in which mutant strains were reconstructed followed by measurement of GCR rates and found that 2 previously unidentified mutations causing the greatest number of papillae in the patch tests (lip2Δ and mrm1Δ) caused increased dGCR rates in reconstructed strains (Supplementary Table 6). It is unclear how these 2 defects cause increased GCR rates, as LIP2 encodes an enzyme that adds lipoic acids to mitochondrial proteins, and MRM1 encodes a mitochondrial RNA methylase. Combined with the results of previous studies, the number of known GCR-causing mutations from this and previous analyses is 419 (of 5,255 mutations tested), corresponding to 334 GCR-suppressing genes (Supplementary Table 5), many but not all of which have been extensively validated.
Genome instability assays identify both general and assay-specific mutations
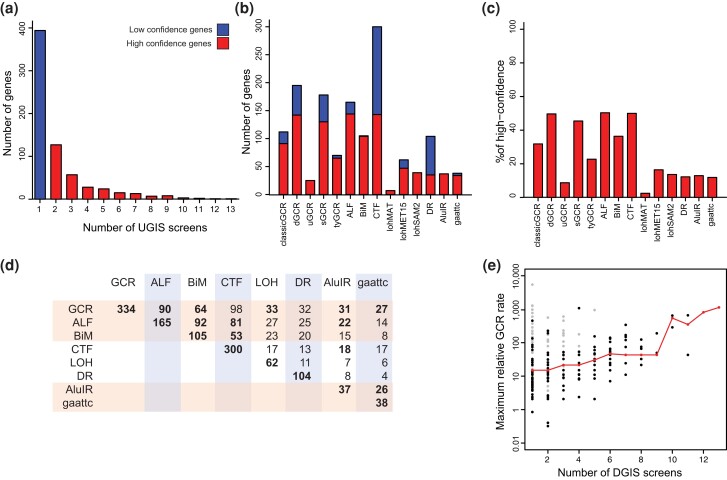
We combined data from studies using 14 genome instability assays to more comprehensively analyze genes thought to act to suppress genome instability. The assays used in these studies included (1) multiple GCR assays (Supplementary Table 5; Chen and Kolodner 1999; Putnam, Hayes, et al. 2009; Chan and Kolodner 2011; Putnam et al. 2016; Nene et al. 2018; Srivatsan et al. 2019), (2) an inverted Alu repeat instability assay (Zhang et al. 2013), (3) a GAA/TTC repeat instability assay (Zhang et al. 2012), (4) a direct repeat stability assay (Novarina et al. 2020), (5) chromosomal stability screens including the A-like faker (ALF) assay, the biallelic mating (BiM) assay, and the chromosome transmission failure (CTF) assay (Yuen et al. 2007; Stirling et al. 2011), and (6) screens for the loss of heterozygosity at the MAT, MET15, and SAM2 loci (lohMAT, lohMET15, and lohSAM2; Andersen et al. 2008). Together, these screens evaluated 7,313 mutations and identified 286 high-confidence genes identified by multiple assays and 394 low-confidence genes identified by 1 assay (Fig. 5a; Supplementary Table 7). Remarkably, the overlap between the results of these screens is worse than that observed for the DDR screens (Fig. 5b–d), as no single screen identified more than 50% of the high-confidence genes. Commonly identified mutations, such as defects in DNA replication and homologous recombination, likely induce of damage that is misrepaired. Mutations restricted to one or a few assays likely reflect aspects of the different genetic readouts, such as the differences between assays identifying increased chromosomal rearrangements (GCR, DR, AluIR, and GAA/TTC stability assays), defects in chromosome transmission (ALF, BiM, and CTF assays), and increased sister chromosome recombination (ALF, BiM, and loh assays). Despite this, there is a trend in which mutations identified in more screens tend to cause greater increases in rates in GCR assays (Fig. 5e); note that some mutations, particularly hypomorphic mutations of essential genes, have been tested in a smaller number of assays and hence are more likely to be scored as low-confidence genes. Because of a broad range of genetic readouts in these assays, we call the identified genes the DGIS gene set.
Fig. 5.
Comparison of DGIS screens. a) Histogram of the number of screens genes were identified in. High-confidence genes were identified in multiple screens. Low-confidence genes were identified in one screen. b) Total number of genes identified in each screen. c). Percentage of high-confidence DGIS genes identified in each screen, which was calculated by dividing the number of high-confidence genes identified in each screen b) by the total count of 286 high-confidence DGIS genes. d) Number of shared genes between assays. GCR, combination of the classicGCR, dGCR, uGCR, sGCR, and tyGCR assays. LOH, combination of the lohMAT, lohMET15, and lohSAM2 assays. e) Comparison of the maximum fold increased GCR rates of deletion mutants (black points) and hypomorphic mutants (gray points) with the number of screens that those alleles have been identified in. The line indicates the position of the median fold increased GCR rate for the alleles for each number of DGIS screens.
Mutations causing increased genome instability are not restricted to those causing increased DNA damage signaling
Functional categorization of the high-confidence DDS and DGIS genes reveals that many are involved in common pathways, particularly DNA replication and the DNA damage response; however, the high-confidence DGIS genes also contain genes associated with cell cycle control, chromosome cohesion, and chromosome segregation (Fig. 6a). Direct comparison of the high- and low-confidence DDS and DGIS genes reveals that there are many genes identified in both types of assays (DDS+ DGIS+ genes), as well as genes identified in only one of the 2 types of assays (DDS+ DGIS− and DDS− DGIS+ genes; Fig. 6b; Supplementary Table 8). Consistent with the role of the DDR in arresting the cell cycle, the mutations with the strongest effects on the cell cycle (reducing the percentage of G1 cells and increasing the percentage of G2 cells) were mostly restricted to high-confidence DDS genes as was previously observed (Fig. 4f), whereas mutations in DDS− DGIS+ genes did not have a significant effect on cell cycle distributions (Fig. 6c).
Fig. 6.
DDS and DGIS genes only partially overlap. a) Breakdown of the high-confidence DDS and DGIS genes into different functional categories shows that most categories are shared, with the exception that cell cycle control, chromosome cohesion, and chromosome segregation contain more genes in the high-confidence DGIS gene group than the high-confidence DDS gene group. b) Venn diagram of the high-confidence and low-confidence DDS and DGIS genes. c) Significant changes in the haploid log-phase cell cycle distribution for genes in different parts of the Venn diagram calculated as for Fig. 4f. d) Model for and examples of different categories of mutations.
DDS+ DGIS+ genes might be expected to suppress DNA damage and subsequent DNA damage signaling and genomic rearrangements (Fig. 6d). The genes/pathways identified by the DDS+ DGIS+ genes include the replication fork protection complex (CSM3, MRC1, and TOF1; Baretic et al. 2020), the Rtt101 cullin complex that regulates the stability of and repair events at stalled replication forks (MMS1, MMS22, RTT101, and RTT107; Duro et al. 2008; Vaisica et al. 2011; Buser et al. 2016), individual enzymes implicated in replication fork progression and repair events in the vicinity of the replication fork (RRM3 and WSS1; Lopez-Mosqueda et al. 2016; Munoz-Galvan et al. 2017; Maddi et al. 2020), and pathways that promote replication fork reversal, including an Mph1-dependent and Smc5/6 regulated pathway (MPH1 and genes encoding the Smc5-6 complex) and postreplication repair (MMS2, RAD5, RAD6, RAD18, and UBC13; Blastyak et al. 2007; Zheng et al. 2011; Xue et al. 2015). We also robustly identified genes involved in homologous recombination and DSB repair (RAD50, MRE11, XRS2, RAD1, RAD10, RAD52, RAD51, RAD59, RAD54, RAD55, RAD57, TOP3, SGS1, RMI1, MMS4, and MUS81) and genes involved in ribonucleotide excision repair (RNH201, RNH202, and RNH203). Several DNA repair-associated pathways were identified among the DDS+ DGIS+ pathways, including components of the DNA damage checkpoint (RAD24, RAD17, DDC1, RAD9, TEL1, and RAD53), protein sumoylation and sumo-targeted ubiquitination (SMT3, ULP1, SLX5, SLX8, SIZ1, and ESC2), and the nuclear pore (NUP133, NUP60, NUP120, NUP145, NUP84, and NUP85), which is known to play a role in HR.
The DDS+ DGIS− genes could either correspond to genes identified by mutations that result in increased damage where the damage is correctly repaired in most cases or to genes that prevent the DNA damage signaling components from recognizing normal DNA structures such as replication forks that do not correspond to DNA damage (Fig. 6d). These genes tend not to include DNA replication or repair genes but rather include genes involved in transcription (ADA2, GAL11, GCR2, NGG1, RPD3, RPN4, SGF73, SIN4, SRB5, SUB1, SUM1, SWD1, and SWI4), some aspects of ubiquitination (CUZ1, DOA1, and UBC4), genes associated with cell polarity, the actin cytoskeleton, and later steps during mitosis (AIM44, BEM1, BEM2, BEM4, BUD3, DBF2, ECM33, ELM1, NUM1, SKT5, SLA1, and VRP1), and the NatB N-terminal acetyltransferase (MDM20 and NAT3). One intriguing possibility raised by this is that collisions between DNA replication forks and transcriptional machinery induce DNA damage that is efficiently repaired in most cases without generating genome rearrangements consistent with the weak effect that most R-loop causing mutations show in GCR assays (Gomez-Gonzalez et al. 2009; Wahba et al. 2011).
The DDS− DGIS+ genes likely promote the correct repair of DNA damage but do not normally suppress the formation of DNA damage. DDS− DGIS+ mutations include (1) pif1 that causes defects in suppression of de novo telomere addition reactions at DNA damage; (2) different checkpoint mutations that disrupt the DDR and cell cycle delay/arrest in response to DNA damage (chk1, ddc2, dun1, mec1, mec3, and mrc1-aq) allowing aberrant repair to occur; (3) translesion synthesis, which may prevent the accumulation of DNA breaks during replication caused by an inability of the replicative DNA polymerases to extend past lesions or misincorporated bases (rad30, rev1, rev3, rev7, and srs2); and (4) mutations like exo1 that by altering DSB resection, but not the overall efficiency of DNA repair, could change the balance between HR and pathways leading to misrepair.
Discussion
Here, we investigated the relationship between the effect of mutations on inducing various DDR markers and inducing genome instability during unperturbed cell growth. Since these measurements have been made during unperturbed growth, the effects of the mutations observed are due to the accumulation and processing of endogenously generated DNA damage, which are most likely generated during DNA replication based on the analyses performed here. The results of 6 screens for mutations inducing DNA damage response markers, which employed 5 different types of assays, and from 14 screens for mutations that cause increased genome instability, which employed 6 different types of genome instability assays, identified 199 high-confidence DDS genes that were found in multiple screens, and similarly identified 286 high-confidence DGIS genes. Low-confidence genes may be due to (1) screening of a mutation in only a few assays, particularly for essential genes not present in the deletion collection, (2) false positive mutations, particularly for mutations that have not been extensively validated, and (3) mutations reflecting key differences between assay readouts, as some mutations are known to increase GCR formation in some GCR assays and not others and mutations involved primarily in chromosome transmission defects are unlikely to be identified in GCR assays.
It was our initial expectation that we would primarily identify genes in which mutations would both induce DNA damage response markers and genome instability because increased DNA damage would be likely to drive increased genome instability. Many such DDS+ DGIS+ genes were identified, and, based on their functional properties, these DDS+ DGIS+ genes are likely to act to (1) suppress DNA damage during replication, (2) promote repair of this DNA damage, and/or (3) prevent this DNA damage from being misrepaired. The DDS+ DGIS+ DNA repair pathways are consistent with at least 2 nonexclusive mechanisms by which cells process DNA replication defects. First, stalled replication forks may undergo fork reversal, which converts the 3-way replication fork junction to a 4-way junction in which the nascent strands are paired. This regressed fork contains a new double-stranded DNA end that can be extended by DNA polymerases, isomerized to rebuild a functional fork, or subject to homologous recombination with the template strand to rebuild a functional fork. Second, stalled or regressed replication forks may be subject to enzymatic cleavage to generate a double-stranded DNA break on 1 daughter duplex that can undergo homologous recombination with the sister chromatid, which is in proximity due to the cohesion of sister chromatids. In both mechanisms, replication fork-mediated and cohesin-mediated coordination of the damaged and undamaged duplexes would be expected to both promote repair and help channel the damage away from misrepair events (for example nonallelic homologous recombination or targeting of the double-stranded end in the regressed fork by the nonhomologous end joining or de novo telomere addition pathways). These misrepair events would give rise to genome rearrangements observable by one or more of the genome instability assays analyzed here. Repair-associated pathways, such as the DNA damage checkpoint, sumoylation by Mms21, SUMO-targeted ubiquitin ligation by the nuclear pore-associated Slx5-Slx8 complex, and protein degradation, have been implicated in the regulation of components of the fork repair processes outlined above (Argunhan et al. 2021; Cappadocia et al. 2021; Dhingra and Zhao 2021).
The identification of additional categories of genes suppressing DNA damage and genome instability (e.g. DDS− DGIS+ and DDS+ DGIS−) besides the DDS+ DGIS+ category was not initially expected but has important biological implications. An important difference between these types of assays is that cytological assays for DDR markers measure responses in the bulk of the cells (e.g. the “averaged” response), whereas genome instability assays detect increased levels of rare events, which are generally not observable in the absence of genetic selection. The DDS− DGIS+ genes could reflect genes in which mutations result in the misrepair of normal levels of spontaneous DNA damage that is not detected as induction of DNA damage or result in the inappropriate recognition of normal DNA as damage. The DDS+ DGIS− genes likely reflect genes in which mutations cause increased DNA damage where the DNA damage is correctly repaired such that genome instability does not occur. Consistent with the idea that the DDS and DGIS gene categories define mechanistically distinct groups of genes, DDS− DGIS+ and DDS+ DGIS+ mutations are functionally distinct in mutation accumulation experiments, where multiple cultures are grown in parallel and analyzed after many generations of growth. In these experiments, mre11Δ and rad27Δ strains (DDS+ DGIS+) accumulated large numbers of rearrangements, whereas pif1Δ strains (DDS− DGIS+) did not (Serero et al. 2014), despite having similar GCR rates (Chen and Kolodner 1999). These differences not only affect how they can be studied in model organisms but also have implications as to how these defects can be therapeutically targeted in human cancers. An important implication of the existence of different classes of DDS and DGIS genes and the imperfect overlap of DDR markers with genome instability suggests caution in inferring the presence or lack of increased genome instability based on the presence or lack of markers for increased DNA damage.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. John Petrini (Sloan Kettering Institute) for helpful comments on the manuscript.
Contributor Information
Bin-Zhong Li, Ludwig Institute for Cancer Research, San Diego Branch, La Jolla, CA 92093-0669, USA.
Richard D Kolodner, Ludwig Institute for Cancer Research, San Diego Branch, La Jolla, CA 92093-0669, USA; Department of Cellular and Molecular Medicine, University of California San Diego, La Jolla, CA 92093-0669, USA; Moores-UCSD Cancer Center, University of California San Diego, La Jolla, CA 92093-0669, USA; Institute of Genomic Medicine, University of California San Diego, La Jolla, CA 92093-0669, USA.
Christopher D Putnam, Ludwig Institute for Cancer Research, San Diego Branch, La Jolla, CA 92093-0669, USA; Department of Medicine, University of California San Diego, La Jolla, CA 92093-0669, USA.
Data availability
All data, yeast strains, and plasmids underlying this article will be shared on request to the corresponding author. Supplementary Tables 1–3 contain the Hug1-EGFP measurements, the Ddc2-EGFP foci measurements, and the compiled information for the high- and low-confidence DDS genes. Supplementary Table 4 contains the cell cycle distribution data for the mutant strains. Supplementary Tables 5–7 contain the list of GCR-suppressing genes, GCR rates, and the high- and low-confidence genome instability-suppressing genes. Supplementary Table 8 contains the merged information for all mutations.
Supplemental material available at G3 online.
Funding
This work was funded by National Institutes of Health R01 GM26017 to RDK and the Ludwig Institute for Cancer Research to RDK and CDP.
Literature cited
- Ainsworth WB, Rome CM, Hjortso MA, Benton MG. 2012. Construction of a cytosolic firefly luciferase reporter cassette for use in PCR-mediated gene deletion and fusion in Saccharomyces cerevisiae. Yeast. 29(12):505–517. doi: 10.1002/yea.2931. [DOI] [PubMed] [Google Scholar]
- Alvaro D, Lisby M, Rothstein R. 2007. Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS Genet. 3(12):e228. doi: 10.1371/journal.pgen.0030228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen MP, Nelson ZW, Hetrick ED, Gottschling DE. 2008. A genetic screen for increased loss of heterozygosity in Saccharomyces cerevisiae. Genetics. 179(3):1179–1195. doi: 10.1534/genetics.108.089250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argunhan B, Iwasaki H, Tsubouchi H. 2021. Post-translational modification of factors involved in homologous recombination. DNA Repair (Amst). 104:103114. doi: 10.1016/j.dnarep.2021.103114. [DOI] [PubMed] [Google Scholar]
- Baretic D, Jenkyn-Bedford M, Aria V, Cannone G, Skehel M, Yeeles JTP. 2020. Cryo-EM structure of the fork protection complex bound to CMG at a replication fork. Mol Cell. 78(5):926–940.e13. doi: 10.1016/j.molcel.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashkirov VI, Herzberg K, Haghnazari E, Vlasenko AS, Heyer WD. 2006. DNA damage-induced phosphorylation of Rad55 protein as a sentinel for DNA damage checkpoint activation in S. cerevisiae. Methods Enzymol. 409:166–182. doi: 10.1016/S0076-6879(05)09010-5. [DOI] [PubMed] [Google Scholar]
- Bashkirov VI, King JS, Bashkirova EV, Schmuckli-Maurer J, Heyer WD. 2000. DNA repair protein Rad55 is a terminal substrate of the DNA damage checkpoints. Mol Cell Biol. 20(12):4393–4404. doi: 10.1128/MCB.20.12.4393-4404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basrai MA, Velculescu VE, Kinzler KW, Hieter P. 1999. NORF5/HUG1 is a component of the MEC1-mediated checkpoint response to DNA damage and replication arrest in Saccharomyces cerevisiae. Mol Cell Biol. 19(10):7041–7049. doi: 10.1128/MCB.19.10.7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton MG, Glasser NR, Palecek SP. 2007. The utilization of a Saccharomyces cerevisiae HUG1P-GFP promoter-reporter construct for the selective detection of DNA damage. Mutat Res. 633(1):21–34. doi: 10.1016/j.mrgentox.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Blackford AN, Jackson SP. 2017. ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol Cell. 66(6):801–817. doi: 10.1016/j.molcel.2017.05.015. [DOI] [PubMed] [Google Scholar]
- Blastyak A, Pinter L, Unk I, Prakash L, Prakash S, Haracska L. 2007. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol Cell. 28(1):167–175. doi: 10.1016/j.molcel.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambati A, Colosio A, Zardoni L, Galanti L, Liberi G. 2015. Replication and transcription on a collision course: eukaryotic regulation mechanisms and implications for DNA stability. Front Genet. 6:166. doi: 10.3389/fgene.2015.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buser R, Kellner V, Melnik A, Wilson-Zbinden C, Schellhaas R, Kastner L, Piwko W, Dees M, Picotti P, Maric M et al. 2016. The replisome-coupled E3 ubiquitin ligase Rtt101Mms22 counteracts Mrc1 function to tolerate genotoxic stress. PLoS Genet. 12(2):e1005843. doi: 10.1371/journal.pgen.1005843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappadocia L, Kochanczyk T, Lima CD. 2021. DNA asymmetry promotes SUMO modification of the single-stranded DNA-binding protein RPA. EMBO J. 40(22):e103787. doi: 10.15252/embj.2019103787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone M, Arron ST, Beutler B, Bononi A, Cavenee W, Cleaver JE, Croce CM, D'Andrea A, Foulkes WD, Gaudino G, et al. 2020. Tumour predisposition and cancer syndromes as models to study gene-environment interactions. Nat Rev Cancer. 20(9):533–549. doi: 10.1038/s41568-020-0265-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JE, Kolodner RD. 2011. A genetic and structural study of genome rearrangements mediated by high copy repeat Ty1 elements. PLoS Genet. 7(5):e1002089. doi: 10.1371/journal.pgen.1002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Kolodner RD. 1999. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat Genet. 23(1):81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]
- Chen SH, Albuquerque CP, Liang J, Suhandynata RT, Zhou H. 2010. A proteome-wide analysis of kinase-substrate network in the DNA damage response. J Biol Chem. 285(17):12803–12812. doi: 10.1074/jbc.M110.106989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. 2010. The DNA damage response: making it safe to play with knives. Mol Cell. 40(2):179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desany BA, Alcasabas AA, Bachant JB, Elledge SJ. 1998. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 12(18):2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhingra N, Zhao X. 2021. Advances in sumo-based regulation of homologous recombination. Curr Opin Genet Dev. 71:114–119. doi: 10.1016/j.gde.2021.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs JA, Lowndes NF, Jackson SP. 2000. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature. 408(6815):1001–1004. doi: 10.1038/35050000. [DOI] [PubMed] [Google Scholar]
- Duro E, Vaisica JA, Brown GW, Rouse J. 2008. Budding yeast Mms22 and Mms1 regulate homologous recombination induced by replisome blockage. DNA Repair (Amst). 7(5):811–818. doi: 10.1016/j.dnarep.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Chen HT, Celeste A, Ward I, Romanienko PJ, Morales JC, Naka K, Xia Z, Camerini-Otero RD, Motoyama N et al. 2002. DNA damage-induced G2-M checkpoint activation by histone H2AX and 53BP1. Nat Cell Biol. 4(12):993–997. doi: 10.1038/ncb884. [DOI] [PubMed] [Google Scholar]
- FlowJo . 2019. Flowjo Software. Ashland (OR): Becton, Dickinson and Company. [Google Scholar]
- Fox MH. 1980. A model for the computer analysis of synchronous DNA distributions obtained by flow cytometry. Cytometry. 1(1):71–77. doi: 10.1002/cyto.990010114. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Elledge SJ, Lehmann AR, Lindahl T, Muzi-Falconi M. 2013. DNA repair, mutagenesis, and other responses to DNA damage. Cold Sprint Harbor Laboratory Press. [Google Scholar]
- Gomez-Gonzalez B, Felipe-Abrio I, Aguilera A. 2009. The S-phase checkpoint is required to respond to R-loops accumulated in Tho mutants. Mol Cell Biol. 29(19):5203–5213. doi: 10.1128/MCB.00402-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendry JA, Tan G, Ou J, Boone C, Brown GW. 2015. Leveraging DNA damage response signaling to identify yeast genes controlling genome stability. G3 (Bethesda). 5(5):997–1006. doi: 10.1534/g3.115.016576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzberg K, Bashkirov VI, Rolfsmeier M, Haghnazari E, McDonald WH, Anderson S, Bashkirova EV, Yates JR, 3rd, Heyer WD. 2006. Phosphorylation of Rad55 on serines 2, 8, and 14 is required for efficient homologous recombination in the recovery of stalled replication forks. Mol Cell Biol. 26(22):8396–8409. doi: 10.1128/MCB.01317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombauer H, Campbell CS, Smith CE, Desai A, Kolodner RD. 2011. Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair intermediates. Cell. 147(5):1040–1053. doi: 10.1016/j.cell.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoose SA, Rawlings JA, Kelly MM, Leitch MC, Ababneh QO, Robles JP, Taylor D, Hoover EM, Hailu B, McEnery KA et al. 2012. A systematic analysis of cell cycle regulators in yeast reveals that most factors act independently of cell size to control initiation of division. PLoS Genet. 8(3):e1002590. doi: 10.1371/journal.pgen.1002590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Zhou Z, Elledge SJ. 1998. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell. 94(5):595–605. doi: 10.1016/S0092-8674(00)81601-3. [DOI] [PubMed] [Google Scholar]
- Koren A, Soifer I, Barkai N. 2010. Mrc1-dependent scaling of the budding yeast DNA replication timing program. Genome Res. 20(6):781–790. doi: 10.1101/gr.102764.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz MC, Dibitetto D, Smolka MB. 2019. DNA damage kinase signaling: checkpoint and repair at 30 years. EMBO J. 38(18):e101801. doi: 10.15252/embj.2019101801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz MC, Yugandhar K, Gupta S, Sanford EJ, Faca VM, Vega S, Joiner AMN, Fromme JC, Yu H, Smolka MB. 2021. In-depth and 3-dimensional exploration of the budding yeast phosphoproteome. EMBO Rep. 22(2):e51121. doi: 10.15252/embr.202051121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YD, Elledge SJ. 2006. Control of ribonucleotide reductase localization through an anchoring mechanism involving Wtm1. Genes Dev. 20(3):334–344. doi: 10.1101/gad.1380506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YD, Wang J, Stubbe J, Elledge SJ. 2008. Dif1 is a DNA-damage-regulated facilitator of nuclear import for ribonucleotide reductase. Mol Cell. 32(1):70–80. doi: 10.1016/j.molcel.2008.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R. 2004. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 118(6):699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Lisby M, Mortensen UH, Rothstein R. 2003. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat Cell Biol. 5(6):572–577. doi: 10.1038/ncb997. [DOI] [PubMed] [Google Scholar]
- Lisby M, Rothstein R. 2009. Choreography of recombination proteins during the DNA damage response. DNA Repair (Amst). 8(9):1068–1076. doi: 10.1016/j.dnarep.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Rothstein R, Mortensen UH. 2001. Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci U S A. 98(15):8276–8282. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Mosqueda J, Maddi K, Prgomet S, Kalayil S, Marinovic-Terzic I, Terzic J, Dikic I. 2016. SPRTN is a mammalian DNA-binding metalloprotease that resolves DNA-protein crosslinks. eLife. 5:e21491. doi: 10.7554/eLife.21491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Z, Minter-Dykhouse K, Wu X, Chen J. 2003. MDC1 is coupled to activated CHK2 in mammalian DNA damage response pathways. Nature. 421(6926):957–961. doi: 10.1038/nature01447. [DOI] [PubMed] [Google Scholar]
- Maddi K, Sam DK, Bonn F, Prgomet S, Tulowetzke E, Akutsu M, Lopez-Mosqueda J, Dikic I. 2020. Wss1 promotes replication stress tolerance by degrading histones. Cell Rep. 30(9):3117–3126.e3114. doi: 10.1016/j.celrep.2020.02.018. [DOI] [PubMed] [Google Scholar]
- Makhnevych T, Sydorskyy Y, Xin X, Srikumar T, Vizeacoumar FJ, Jeram SM, Li Z, Bahr S, Andrews BJ, Boone C et al. 2009. Global map of sumo function revealed by protein–protein interaction and genetic networks. Mol Cell. 33(1):124–135. doi: 10.1016/j.molcel.2008.12.025. [DOI] [PubMed] [Google Scholar]
- Maser RS, Monsen KJ, Nelms BE, Petrini JH. 1997. Hmre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol Cell Biol. 17(10):6087–6096. doi: 10.1128/MCB.17.10.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo JA, Cohen J, Toczyski DP. 2001. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 15(21):2809–2821. doi: 10.1101/gad.903501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurisse J, Bacquin A, Richet N, Charbonnier JB, Ochsenbein F, Peyroche A. 2014. Hug1 is an intrinsically disordered protein that inhibits ribonucleotide reductase activity by directly binding Rnr2 subunit. Nucleic Acids Res. 42(21):13174–13185. doi: 10.1093/nar/gku1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Galvan S, Garcia-Rubio M, Ortega P, Ruiz JF, Jimeno S, Pardo B, Gomez-Gonzalez B, Aguilera A. 2017. A new role for Rrm3 in repair of replication-born DNA breakage by sister chromatid recombination. PLoS Genet. 13(5):e1006781. doi: 10.1371/journal.pgen.1006781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelms BE, Maser RS, MacKay JF, Lagally MG, Petrini JH. 1998. In situ visualization of DNA double-strand break repair in human fibroblasts. Science. 280(5363):590–592. doi: 10.1126/science.280.5363.590. [DOI] [PubMed] [Google Scholar]
- Nene RV, Putnam CD, Li BZ, Nguyen KG, Srivatsan A, Campbell CS, Desai A, Kolodner RD. 2018. Cdc73 suppresses genome instability by mediating telomere homeostasis. PLoS Genet. 14(1):e1007170. doi: 10.1371/journal.pgen.1007170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novarina D, Desai R, Vaisica JA, Ou J, Bellaoui M, Brown GW, Chang M. 2020. A genome-wide screen for genes affecting spontaneous direct-repeat recombination in Saccharomyces cerevisiae. G3 (Bethesda). 10(6):1853–1867. doi: 10.1534/g3.120.401137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 10(15):886–895. doi: 10.1016/S0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- Putnam CD, Allen-Soltero SR, Martinez SL, Chan JE, Hayes TK, Kolodner RD. 2012. Bioinformatic identification of genes suppressing genome instability. Proc Natl Acad Sci U S A. 109(47):E3251–E3259. doi: 10.1073/pnas.1216733109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam CD, Hayes TK, Kolodner RD. 2009. Specific pathways prevent duplication-mediated genome rearrangements. Nature. 460(7258):984–989. doi: 10.1038/nature08217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam CD, Jaehnig EJ, Kolodner RD. 2009. Perspectives on the DNA damage and replication checkpoint responses in Saccharomyces cerevisiae. DNA Repair (Amst). 8(9):974–982. doi: 10.1016/j.dnarep.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam CD, Srivatsan A, Nene RV, Martinez SL, Clotfelter SP, Bell SN, Somach SB, de Souza JE, Fonseca AF, de Souza SJ, et al. 2016. A genetic network that suppresses genome rearrangements in Saccharomyces cerevisiae and contains defects in cancers. Nat Commun. 7(1):11256. doi: 10.1038/ncomms11256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serero A, Jubin C, Loeillet S, Legoix-Ne P, Nicolas AG. 2014. Mutational landscape of yeast mutator strains. Proc Natl Acad Sci U S A. 111(5):1897–1902. doi: 10.1073/pnas.1314423111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R, Lewis S, Wlodarski MW. 2020. DNA repair syndromes and cancer: insights into genetics and phenotype patterns. Front Pediatr. 8:570084. doi: 10.3389/fped.2020.570084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolka MB, Albuquerque CP, Chen SH, Zhou H. 2007. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci U S A. 104(25):10364–10369. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soifer I, Barkai N. 2014. Systematic identification of cell size regulators in budding yeast. Mol Syst Biol. 10(11):761. doi: 10.15252/msb.20145345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spidlen J, Breuer K, Rosenberg C, Kotecha N, Brinkman RR. 2012. Flowrepository: a resource of annotated flow cytometry datasets associated with peer-reviewed publications. Cytometry A. 81(9):727–731. doi: 10.1002/cyto.a.22106. [DOI] [PubMed] [Google Scholar]
- Srivatsan A, Li B, Sanchez DN, Somach SB, da Silva VL, de Souza SJ, Putnam CD, Kolodner RD. 2019. Essential Saccharomyces cerevisiae genome instability suppressing genes identify potential human tumor suppressors. Proc Natl Acad Sci U S A. 116(35):17377–17382. doi: 10.1073/pnas.1906921116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivatsan A, Putnam CD, Kolodner RD. 2018. Analyzing genome rearrangements in Saccharomyces cerevisiae. Methods Mol Biol. 1672:43–61. doi: 10.1007/978-1-4939-7306-4_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. 2003. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 421(6926):961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- Stirling PC, Bloom MS, Solanki-Patil T, Smith S, Sipahimalani P, Li Z, Kofoed M, Ben-Aroya S, Myung K, Hieter P. 2011. The complete spectrum of yeast chromosome instability genes identifies candidate CIN cancer genes and functional roles for ASTRA complex components. PLoS Genet. 7(4):e1002057. doi: 10.1371/journal.pgen.1002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styles EB, Founk KJ, Zamparo LA, Sing TL, Altintas D, Ribeyre C, Ribaud V, Rougemont J, Mayhew D, Costanzo M et al. 2016. Exploring quantitative yeast phenomics with single-cell analysis of DNA damage foci. Cell Syst. 3(3):264–277.e210. doi: 10.1016/j.cels.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Fay DS, Marini F, Foiani M, Stern DF. 1996. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 10(4):395–406. doi: 10.1101/gad.10.4.395. [DOI] [PubMed] [Google Scholar]
- Tkach JM, Yimit A, Lee AY, Riffle M, Costanzo M, Jaschob D, Hendry JA, Ou J, Moffat J, Boone C et al. 2012. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat Cell Biol. 14(9):966–976. doi: 10.1038/ncb2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, Boone C. 2006. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol Biol. 313:171–192. doi: 10.1385/1-59259-958-3:171. [DOI] [PubMed] [Google Scholar]
- Vaisica JA, Baryshnikova A, Costanzo M, Boone C, Brown GW. 2011. Mms1 and Mms22 stabilize the replisome during replication stress. Mol Biol Cell. 22(13):2396–2408. doi: 10.1091/mbc.e10-10-0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoli F, Fumasoni M, Szakal B, Maloisel L, Branzei D. 2010. Replication and recombination factors contributing to recombination-dependent bypass of DNA lesions by template switch. PLoS Genet. 6(11):e1001205. doi: 10.1371/journal.pgen.1001205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW. 2004. Cancer genes and the pathways they control. Nat Med. 10(8):789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- Wahba L, Amon JD, Koshland D, Vuica-Ross M. 2011. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol Cell. 44(6):978–988. doi: 10.1016/j.molcel.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Matsuoka S, Carpenter PB, Elledge SJ. 2002. 53BP1, a mediator of the DNA damage checkpoint. Science. 298(5597):1435–1438. doi: 10.1126/science.1076182. [DOI] [PubMed] [Google Scholar]
- Waterman DP, Zhou F, Li K, Lee CS, Tsabar M, Eapen VV, Mazzella A, Haber JE. 2019. Live cell monitoring of double strand breaks in S. cerevisiae. PLoS Genet. 15(3):e1008001. doi: 10.1371/journal.pgen.1008001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Huang M. 2008. Dif1 controls subcellular localization of ribonucleotide reductase by mediating nuclear import of the R2 subunit. Mol Cell Biol. 28(23):7156–7167. doi: 10.1128/MCB.01388-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X, Sung P, Zhao X. 2015. Functions and regulation of the multitasking FANCM family of DNA motor proteins. Genes Dev. 29(17):1777–1788. doi: 10.1101/gad.266593.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yimit A, Riffle M, Brown GW. 2015. Genetic regulation of DNA2 localization during the DNA damage response. G3 (Bethesda). 5(9):1937–1944. doi: 10.1534/g3.115.019208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen KW, Warren CD, Chen O, Kwok T, Hieter P, Spencer FA. 2007. Systematic genome instability screens in yeast and their potential relevance to cancer. Proc Natl Acad Sci U S A. 104(10):3925–3930. doi: 10.1073/pnas.0610642104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Saini N, Sheng Z, Lobachev KS. 2013. Genome-wide screen reveals replication pathway for quasi-palindrome fragility dependent on homologous recombination. PLoS Genet. 9(12):e1003979. doi: 10.1371/journal.pgen.1003979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Shishkin AA, Nishida Y, Marcinkowski-Desmond D, Saini N, Volkov KV, Mirkin SM, Lobachev KS. 2012. Genome-wide screen identifies pathways that govern GAA/TTC repeat fragility and expansions in dividing and nondividing yeast cells. Mol Cell. 48(2):254–265. doi: 10.1016/j.molcel.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, An X, Yang K, Perlstein DL, Hicks L, Kelleher N, Stubbe J, Huang M. 2006. Nuclear localization of the Saccharomyces cerevisiae ribonucleotide reductase small subunit requires a karyopherin and a WD40 repeat protein. Proc Natl Acad Sci U S A. 103(5):1422–1427. doi: 10.1073/pnas.0510516103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Chabes A, Domkin V, Thelander L, Rothstein R. 2001. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J. 20(13):3544–3553. doi: 10.1093/emboj/20.13.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Muller EG, Rothstein R. 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 2(3):329–340. doi: 10.1016/S1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- Zheng XF, Prakash R, Saro D, Longerich S, Niu H, Sung P. 2011. Processing of DNA structures via DNA unwinding and branch migration by the S. cerevisiae Mph1 protein. DNA Repair (Amst). 10(10):1034–1043. doi: 10.1016/j.dnarep.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q, Chen CF, Li S, Chen Y, Wang CC, Xiao J, Chen PL, Sharp ZD, Lee WH. 1999. Association of BRCA1 with the hRad50-hMre11-p95 complex and the DNA damage response. Science. 285(5428):747–750. doi: 10.1126/science.285.5428.747. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data, yeast strains, and plasmids underlying this article will be shared on request to the corresponding author. Supplementary Tables 1–3 contain the Hug1-EGFP measurements, the Ddc2-EGFP foci measurements, and the compiled information for the high- and low-confidence DDS genes. Supplementary Table 4 contains the cell cycle distribution data for the mutant strains. Supplementary Tables 5–7 contain the list of GCR-suppressing genes, GCR rates, and the high- and low-confidence genome instability-suppressing genes. Supplementary Table 8 contains the merged information for all mutations.
Supplemental material available at G3 online.