Abstract
The 3′ termini of the genomic and antigenomic RNAs of human respiratory syncytial virus (RSV) are identical at 10 of the first 11 nucleotide positions and 21 of the first 26 positions. These conserved 3′-terminal sequences are thought to contain the genomic and antigenomic promoters. Furthermore, the complement of each conserved sequence (i.e., the 5′ end of the RNA it encodes) might contain an encapsidation signal. Using an RSV minigenome system, we individually mutated each of the last seven nucleotides in the 5′ trailer region of the genome. We analyzed effects of these mutations on encapsidation of the T7 polymerase-transcribed negative-sense genome, its ability to function as a template for RSV-driven synthesis of positive-sense antigenome and mRNA, and the ability of this antigenome to be encapsidated and to function as template for the synthesis of more genome. As a technical complication, mutations in the last five nucleotides of the trailer region were found to affect the efficiency of the adjoining T7 promoter over more than a 10-fold range, even though three nonviral G residues had been included between the core promoter and the trailer to maximize the efficiency of promoter activity. This was controlled in all experiments by monitoring the levels of total and encapsidated genome. The efficiency of encapsidation of the T7 polymerase-transcribed genome was not affected by any of the trailer mutations. Furthermore, neither the efficiency of positive-sense RNA synthesis from the genome nor the efficiency of encapsidation of the encoded antigenome was affected by the mutations. However, nucleotide substitution at positions 2, 3, 6, or 7 relative to the 5′ end of the trailer blocked the production of progeny genome, whereas substitution at positions 1 and 5 allowed a low level of genome production and substitutions at position 4 were tolerated. Position 4 is the only one of the seven positions examined that is not conserved between the 3′ ends of genomic and antigenomic RNA. The mutations that blocked the synthesis of progeny genome thus limited RNA replication to one step, namely, the synthesis and encapsidation of antigenome. Restoration of terminal complementarity for one of the trailer mutants by making a compensatory mutation in the leader region did not restore synthesis of genomic RNA, confirming that its loss was not due to reduced terminal complementarity. Interestingly, this leader mutation appeared to prevent antigenome synthesis with only a slight effect on mRNA synthesis, apparently providing a dissociation between these two synthetic activities. Genomes in which the terminal 24 or 325 nucleotides of the trailer have been deleted were competent for encapsidation and the synthesis of mRNA and antigenomic RNA, further confirming that terminal complementarity was not required for these functions.
Respiratory syncytial virus (RSV) is a nonsegmented negative-strand RNA virus in the Pneumovirus genus of the Paramyxoviridae family (6, 18). RSV is a major cause of serious respiratory disease in infants and adults and is a major target for vaccine and antiviral drug development. As is typical for the nonsegmented negative-strand RNA viruses, RSV genomic RNA is associated tightly with the nucleocapsid N protein to form an RNase-resistant helical nucleocapsid. This encapsidated genomic RNA is the template used by the viral polymerase to synthesize the positive-sense RNAs, namely, the 10 subgenomic viral mRNAs and the antigenomic RNA. The antigenome is a complete positive-sense copy of the genome. It is an intermediate in RNA replication and, like the genome, is encapsidated with N protein. In addition to the N protein, the nucleocapsid-associated proteins include the large L protein and the phosphoprotein P. The L protein contains conserved polymerase motifs and likely directs RNA synthetic functions as well as capping and methylation. The P protein appears to serve both as a polymerase cofactor and as a chaperone that keeps free N protein soluble and available for assembly with nascent genomic or antigenomic RNA (16). In contrast to other nonsegmented negative-strand viruses, RSV mRNA synthesis involves an additional viral protein, the M2-1 protein, which confers transcriptional processivity and increases the frequency of polymerase readthrough across intergenic junctions (7, 8, 10, 14).
The polymerase can engage in transcription, producing subgenomic mRNAs, or in RNA replication, a two-step process producing in turn antigenomic RNA and progeny genomic RNA (6, 18). To initiate either of these processes, the polymerase is presumed to bind at a genomic promoter at the 3′ end of the genomic template. Transcription involves a stop-start mechanism guided by conserved signals at the gene boundaries. Specifically, each gene begins with a 10-nucleotide gene start (GS) signal, which directs transcriptional initiation, and ends with a 12- to 13-nucleotide gene end (GE) signal, which directs polyadenylation and release of the completed transcript (16, 17). The polymerase proceeds down the genome, transcribing each gene in turn. The same template, and ostensibly the same promoter, is used for the synthesis of the antigenome, the first step in RNA replication, but the GS and GE signals are ignored. In addition, it is thought that nascent replication products are encapsidated cosynthetically. The factors which determine whether the genomic template engages in transcription versus replication are not well understood. One proposal for nonsegmented negative-strand RNA viruses in general is that there is a balance between replication and transcription which is mediated by encapsidation of the nascent antigenomic RNA (18). However, at least in the case of RSV, the proposed switch to replication at the expense of transcription could not be reproduced in a minigenome system by overexpression of the N and P proteins (11).
There is a high degree of sequence identity between the 3′ ends of genomic RNA and antigenomic RNA: specifically, 10 of the first 11 nucleotides (nt) and 21 of the first 26 nt are identical, after which the amount of sequence identity is insignificant (7, 20). It seems likely that this high degree of sequence identity is due to a conserved 3′ promoter structure. It might also reflect the presence of a conserved encapsidation signal at the 5′ end of the encoded antigenomic or genomic RNA. As a consequence of these conserved sequences, genomic and antigenomic RNA each has complementarity between its 3′ and 5′ ends, and thus each has the potential to form a panhandle structure. It has been suggested that base pairing between the 3′ and 5′ ends of the genome of the rhabdovirus vesicular stomatitis virus (VSV) is important in RNA replication (27), although other studies with the paramyxovirus Sendai virus appear to rule out a role for terminal complementarity (26).
In the present study, we used an RSV minigenome system to study the roles of the conserved terminal sequences in encapsidation, RNA replication, and transcription. We focused on the seven terminal nucleotides of the 5′ trailer region, which are complementary with the leader except at position 4. This sequence might be part of a possible encapsidation signal in the genomic RNA, and its complement at the 3′ end of the antigenome is likely to be part of the antigenomic promoter. We examined mutations at each of the seven positions to determine their importance in encapsidation and RNA synthesis. We also examined deletions of part or all of the trailer region. None of the point mutations affected encapsidation of the genome produced by T7 polymerase or its function as a template for producing antigenome and mRNA, but mutations at four of these positions blocked the accumulation of progeny genome, while mutations at three other positions had an intermediate effect or no inhibitory effect. The trailer deletions did not affect encapsidation or the template function of genome produced by T7 polymerase, providing evidence that neither transcription nor replication involves terminal complementarity. We also describe the importance of carefully controlling for effects on the magnitude of T7-mediated transcription of the genome plasmid.
MATERIALS AND METHODS
Cells and transfection.
HEp-2 cells were maintained in Opti-MEM (Life Technologies, Inc., Gaithersburg, Md.) containing 2% fetal bovine serum (FBS) and tested every 2 months to confirm the absence of mycoplasma. Transfections were performed in 35-mm wells of HEp-2 cells with a mixture of pTM1-N (0.4 μg), pTM1-P (0.2 μg), pTM1-L (0.1 μg), and minigenome plasmid (0.2 μg), as well as 12 μl of LipofectACE (Life Technologies, Inc.), unless otherwise stated (13). This mix (0.2 ml) was incubated in the absence of serum for 45 min at room temperature before the addition of 0.8 ml of Opti-MEM–2% FBS that included vTF7-3, a recombinant vaccinia virus that expresses T7 polymerase (12). Cells were transfected overnight in the absence of antibiotics, and medium was replaced the next morning with Opti-MEM–2% FBS. Transfected cells were harvested at 44 h and RNA extracted with Trizol (Life Technologies, Inc.). To analyze encapsidated RNA, cells were resuspended in 10 mM Tris (pH 7.5)–10 mM NaCl–1 mM CaCl2–1% Triton X-100–0.5% sodium deoxycholate containing 1 μg of micrococcal nuclease (S7 nuclease; Boehringer Mannheim GmbH, Mannheim, Germany) per ml and gently vortexed, incubated at 30°C for 30 min (1, 21), and extracted with Trizol.
Northern blotting.
RNA was electrophoresed on 1.5% agarose gels containing 0.44 M formaldehyde, transferred to Nytran Plus by using a Turbo Blotter (Schleicher & Schuell, Keene, N.H.) and 8 mM NaOH as buffer, and fixed by UV cross-linking (Stratalinker; Stratagene). 32P-labeled RNA probes were synthesized in vitro by T7 RNA polymerase (Boehringer Mannheim) in the presence of [32P]CTP (Amersham Life Science, Inc., Arlington Heights, Ill.) and incubated with the blots, as previously described (12). Positive-sense probe was synthesized from antigenome C4 plasmid which was linearized at an NcoI site located in the downstream region of the chloramphenicol acetyltransferase (CAT) gene, resulting in a transcript of approximately 600 nt which included the upstream (leader) end of the antigenome and most of the CAT open reading frame (ORF) but lacked the downstream (trailer) end (12). Negative-sense probe was synthesized from minigenome C2 plasmid which was linearized at the XbaI site marking the upstream end of the CAT gene, resulting in a transcript of approximately 852 nt which included the complete CAT ORF and the downstream (trailer) end of the genome (12). The blots were exposed to film (BioMax Film; Kodak, Rochester, N.Y.) with an enhancing screen at −70°C and to phosphor screens for quantitation in a STORM phosphorimager (Molecular Dynamics, Sunnyvale, Calif.).
Plasmid mutagenesis and isolation.
Genome trailer mutants 3A, 4G, 4C, 5A, and 7C were prepared by PCR amplification of a PstI-HindIII fragment which begins at the end of the CAT gene in the minigenome C2 cDNA and spans the trailer region and T7 promoter. Amplification was done with a positive-sense primer which hybridized within the CAT gene and a mutagenic negative-sense primer which hybridized to the T7 promoter and the end of the trailer region and contained the desired mutation. The PCR product was cloned into the PstI-HindIII window of minigenome plasmid C2. Mutants 1C, 2G, and 6C were prepared by the method of Byrappa et al. (3). Briefly, two primers were used, one of which contained the desired mutation, abutting each other but facing in the opposite direction on the genome plasmid, C2. These primers were used to PCR amplify the plasmid with Vent polymerase (New England Biolabs, Inc.) for 18 cycles. The tube was immediately removed from the thermocycler, and EDTA was added to stop the reaction. The product was isolated by agarose gel electrophoresis, ligated, and used to transform competent Escherichia coli DH10B cells. All mutations were confirmed by nucleotide sequence analysis.
RESULTS
Effect of trailer mutations on T7 transcription.
We examined the effects of nucleotide substitutions in the last seven positions of the 5′-terminal trailer region of RSV. This was done with a reconstituted transcription-replication system based on the negative-sense RSV-CAT minigenome C2 (13), as outlined in Fig. 1. The cDNA is bordered on the 5′ end relative to the encoded genome by the core promoter for T7 RNA polymerase. There are three nonviral G residues between the trailer and the core promoter in order to increase promoter efficiency. The cDNA is bordered on the 3′ end by a hammerhead ribozyme and a T7 terminator. Ribozyme-mediated cleavage would generate the 3′ end of the genome. T7 RNA polymerase is provided by infection with the recombinant vaccinia virus, vTF7-3 (12).
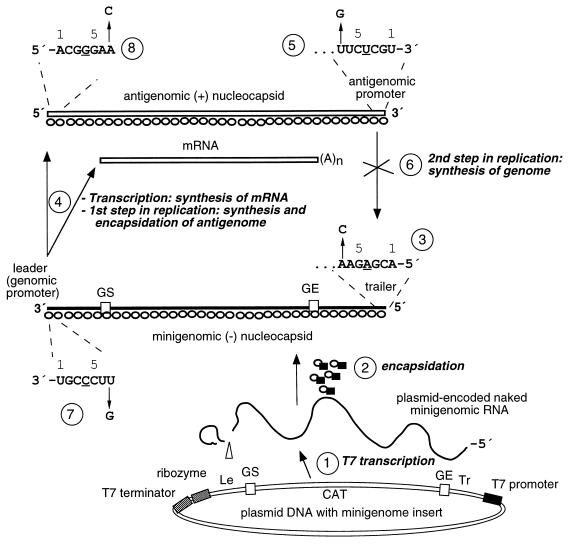
FIG. 1.
Steps involved in reconstituted encapsidation, RNA replication, and transcription of the wt RSV-CAT C2 minigenome and derivatives containing nucleotide substitutions. The cDNA-encoded genome is 934 nt long and contains, in 3′-to-5′ order, the 44-nt RSV leader region, the 10-nt NS1 GS signal, the upstream 32 nt of the nontranslated region of the NS1 gene, a 669-nt negative-sense copy of the CAT ORF, the last 12 nucleotides of the nontranslated region of the L gene, the 12-nucleotide L GE signal, and the 155-nucleotide trailer region. (Step 1) Naked minigenomic RNA is synthesized from transfected plasmid by T7 RNA polymerase supplied by the coinfecting vaccinia virus recombinant, vTF7-3. Self-cleavage by the ribozyme is indicated with an open triangle. Other cotransfected plasmids bearing individual RSV ORFs (not shown) are transcribed by T7 polymerase and then translated into support proteins. (Step 2) Soluble N and P support proteins (open circles and filled squares, respectively) encapsidate the naked RNA, rendering it resistant to digestion with micrococcal nuclease. This step is very inefficient, probably because it is not associated with the RSV polymerase (see Discussion). (Step 3) This step illustrates the 7C mutation in the genome trailer, in which position 7 relative to the 5′ end of the genome is changed from A to C (negative sense). The three nonviral G residues present at the 5′ end of the T7 transcript are not shown and, in any event, do not interfere with replication either of minigenomes or of complete recombinant virus (7, 24). (Step 4) The encapsidated plasmid-supplied genome (parental or mutant) serves as template for the synthesis of antigenome (RNA replication) and CAT mRNA (transcription) by the reconstituted RSV polymerase. The antigenome is encapsidated efficiently, probably concurrent with synthesis. (Step 5) The 7C trailer mutation at the 5′ end of the trailer, from step 3 above, results in a complementary substitution in the 3′ end of the encoded antigenome. (Step 6) The second step of RNA replication, the synthesis of genome, is blocked in the 7C mutant (Results). (Step 7) This illustrates a second point mutation, at leader position 7, which was introduced into the 7C trailer mutant in order to restore terminal complementarity at position 7. (Step 8) The 7G leader mutation results in a complementary mutation in the 5′ end of the encoded antigenome.
In these experiments, the wild-type or mutant RSV genome was expressed alone or was complemented by cotransfection of plasmids encoding RSV support proteins. The scheme of plasmid-based, reconstituted RSV encapsidation, transcription and RNA replication is shown in Fig. 1. The substitutions in the trailer region are shown in Fig. 2. At 44 h posttransfection, the cells were harvested and lysates were prepared. The cells from one well were processed directly for RNA purification, while the cells from a duplicate well were detergent lysed and digested with micrococcal nuclease to destroy naked RNA before being processed for RNA purification. RNA was analyzed by Northern blot hybridization with strand-specific riboprobes to detect total and micrococcal nuclease-resistant negative-sense RNA, as well as total positive-sense RNA (Fig. 3A to C, respectively).
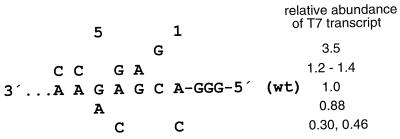
FIG. 2.
Mutations introduced into the 5′ trailer region of the wt C2 minigenome and effect on the intracellular accumulation of T7 transcript. The 5′-terminal 7 nt of the C2 genome are shown as a negative-sense sequence, 3′ to 5′. The various single point mutations which were introduced are shown. Those listed above the wt sequence are ones which either did not affect or increased the level of intracellular T7 transcript. Mutations listed below are ones which reduced the level of T7 transcript.
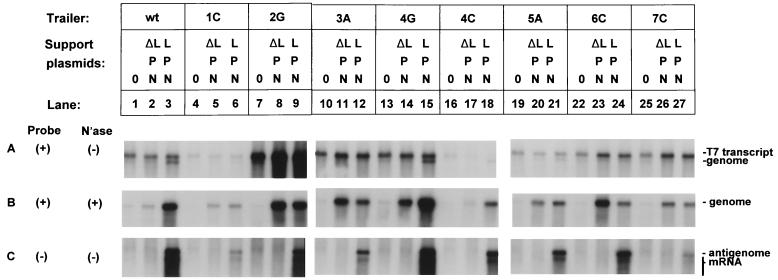
FIG. 3.
Effects of individual point mutations in the minigenome trailer region on T7 transcription, genome encapsidation, synthesis of positive-sense mRNA and antigenome, and synthesis of negative-sense progeny genome. Duplicate wells of HEp-2 cells were infected with vTF7-3 to provide T7 RNA polymerase and transfected (13) with wt minigenome plasmid C2 (lanes 1 to 3) or mutant genome plasmid 1C (lanes 4 to 6), 2G (lanes 7 to 9), 3A (lanes 10 to 12), 4G (lanes 13 to 15), 4C (lanes 16 to 18), 5A (lanes 19 to 21), 6C (lanes 22 to 24), or 7C (lanes 25 to 27). Lanes marked “0” received no additional plasmids (lanes 1, 4, 7, 10, 13, 16, 19, 22, and 25). Lanes marked “NPΔL” received N, P, and ΔL support plasmids, the latter encoding a nonfunctional L protein (lanes 2, 5, 8, 11, 14, 17, 20, 23, and 26). Lanes marked “NPL” received N, P, and functional L plasmids (lanes 3, 6, 9, 12, 15, 18, 21, 24, and 27). RNAs were harvested at 44 h, and lysates were prepared. One lysate of each pair was processed directly for RNA purification, and the resulting total intracellular RNA was subjected to electrophoresis on formaldehyde gels and analyzed by Northern blot hybridization with positive-sense (A) or negative-sense (C) riboprobe. The other lysate of each pair was subjected to digestion with micrococcal nuclease (N'ase), and the remaining RNA was purified and analyzed by Northern blot hybridization with positive-sense riboprobe (B). Film exposure for panels B and C was 12 h, and for panel A it was 1.5 h. Quantitation of the hybridization patterns shown in this figure are presented in Table 1.
When the wild-type genome C2 plasmid was transfected into vTF7-3-infected cells in the absence of support plasmids, a large amount of plasmid-derived negative-sense RNA accumulated, the product of T7 RNA polymerase (Fig. 1, step 1, and Fig. 3A, lane 1). With this particular genome, the most abundant negative-sense species typically is genome linked to uncleaved ribozyme (designated “T7 transcript” in Fig. 3A), which migrates slightly more slowly than correctly sized genomic RNA, as described previously (13). We speculate that this species accumulates because it is stabilized by the attached uncleaved ribozyme, perhaps due to its folded structure. Correctly sized genome also was detected, although it typically was much less abundant, probably reflecting extensive degradation due to its nonpolyadenylated, uncapped nature.
We then examined the accumulation of negative-sense RNA synthesized from plasmids encoding genomes with single point mutations in the trailer region (Fig. 2 and 3). In preliminary studies, we had found that single point mutations in the three nucleotide positions immediately adjacent to the T7 core promoter (i.e., the promoter not including transcribed sequence) had strong effects on the efficiency of transcription by T7 RNA polymerase (13; J. Cristina and P. L. Collins, unpublished data). The introduction of a pyrimidine or, to a lesser extent, a G-to-A change reduced the efficiency of T7 transcription in vitro, whereas converse changes had the opposite effect. This is consistent with published studies (for example, reference 19). To avoid this problem, the C2 genome cDNA had been designed to contain three additional nonviral G residues between the trailer and the T7 core promoter to enhance transcription and preclude effects of sequence mutation or domain swaps adjacent to the promoter (13). The insertion of these three Gs was sufficient to avoid the effects of point mutations in the trailer on T7 transcription in vitro (data not shown).
Surprisingly, however, in the present study we found that the magnitude of T7 transcription in transfected cells remained sensitive to substitutions in the region adjoining the promoter, the three additional G residues notwithstanding, and the length of the sensitive region was longer than expected. This can be seen by comparing the amount of T7 transcript which accumulated intracellularly from genome plasmids transfected in the absence of support plasmid (lanes marked “0” in Fig. 3A). These differences are quantified for this representative experiment in Table 1 (column marked “T7 transcript”). Specifically, mutant 1C, in which the terminal nucleotide of the trailer region was changed from the purine A to the pyrimidine C (negative sense), exhibited a substantial reduction in the accumulation of negative-sense RNA compared to wild type (wt) (Fig. 3A, compare lanes 1 and 4). Conversely, a change of pyrimidine C to purine G at the second nucleotide from the end in mutant 2G resulted in a >3-fold increase in accumulation compared to wt (compare lanes 1 and 7). Mutants 3A (G to A, lane 10) and 4G (A to G, lane 13), which are purine to purine changes, were comparable to wt, while a purine-to-pyrimidine change in 4C (A to C, lane 16) was much lower, and a G-to-A change in 5A (lane 19) was slightly lower. Purine-to-pyrimidine changes beyond the first five trailer nucleotides, 6C (A to C, lane 22) and 7C (A to C, lane 25), had no detrimental effect. Since the constructs also contain three nonviral G residues adjacent to the promoter, a total of 8 nt adjacent to the T7 promoter affect the accumulation of T7 transcripts intracellularly.
TABLE 1.
Quantitation of RNA species produced in transfected cells programmed with plasmid encoding a minigenome bearing one of various trailer mutationsa
| Trailer mutant | T7 transcriptb | Encapsidated genome (protected from nuclease)c
|
Total positive-sense RNA, NPL | Relative genome template activityd | ||
|---|---|---|---|---|---|---|
| 0e | NPΔLf | NPLg | ||||
| wt | 37,700h (1.0)i, 100j | 1,288,h3.4j | 3,757,h10j | 19,257,h51j (5.1)k | 76,451h (1.0)l | 3.97 (1.0)m |
| 1C | 17,600 (0.46), 100 | 0, 0 | 4,076, 23 | 5,252, 30 (1.3) | 13,760 (0.18) | 2.62 (0.66) |
| 2G | 132,400 (3.5), 100 | 576, 0.44 | 22,118, 17 | 18,071 14 (0.82) | 57,569 (0.75) | 3.19 (0.80) |
| 3A | 45,900 (1.3), 100 | 2,006, 4.4 | 18,263, 40 | 13,579, 30 (0.74) | 24,818 (0.32) | 1.83 (0.47) |
| 4G | 44,000 (1.2), 100 | 4,017, 9.1 | 17,776, 40 | 30,217, 69 (1.7) | 100,394 (1.3) | 3.32 (0.84) |
| 4C | 11,400 (0.30), 100 | 1,242, 11 | 3,577, 31 | 11,715, 103 (3.3) | 44,409 (0.58) | 3.79 (0.95) |
| 5A | 33,000 (0.88), 100 | 1,564, 4.7 | 9,223, 28 | 11,711, 35 (1.3) | 37,714 (0.49) | 3.22 (0.81) |
| 6C | 49,800 (1.3), 100 | 1,415, 2.8 | 20,368, 41 | 11,120, 22 (0.55) | 45,214 (0.59) | 4.07 (1.03) |
| 7C | 51,900 (1.4), 100 | 1,411, 2.7 | 9,608, 19 | 8,444, 16 (0.88) | 16,290 (0.21) | 1.93 (0.49) |
Data from the representative experiment shown in Fig. 3.
Amount of plasmid-derived T7 transcript with uncleaved ribozyme, averaged from the three support conditions 0, NPΔL, and NPL (e.g., for wt, Fig. 3A, lanes 1, 2, and 3). This is taken as a surrogate measure of the amount of total T7-synthesized minigenome (see the text).
Amount of minigenome remaining after micrococcal nuclease treatment under the indicated support conditions: 0, NPΔL, and NPL (e.g., for wt, Fig. 3B, lanes 1, 2, and 3).
The relative activity of each minigenome as template for the synthesis of positive-sense RNA was determined by dividing the total amount of positive-sense RNA by the total amount of encapsidated negative-sense RNA (NPL) for that minigenome. Since the values for positive sense versus negative sense are from separate blots, the quotient is an arbitrary unit. Compared between minigenomes, it gives an indication of relative template activity.
Residual undigested minigenome, reflecting a background due to incomplete and variable nuclease digestion in the crude cell lysates.
Reflects encapsidation by N and P in the absence of active RSV polymerase.
Reflects genome synthesized and encapsidated in the presence of active RSV polymerase.
Phosphorimager signal units.
Values in parentheses are amount of T7 transcript relative to wt as 1.0.
The numbers in boldface are values normalized to the T7 transcript value for that minigenome as 100.
Encapsidation value relative to NPΔL for that minigenome as 1.0 is given in parentheses. Values of >1 indicate genome amplification by the RSV polymerase. Values of <1.5 are considered marginal.
Values in parentheses are relative to wt as 1.0.
Values in parentheses are relative to wt as 1.0.
Effect of trailer mutations on the encapsidation of the T7 genome transcript.
We then investigated whether any of the nucleotide substitutions in the plasmid-derived mutant genomes affected encapsidation, namely, their assembly into nucleocapsids (Fig. 1, step 2). This was done in the experiment shown in Fig. 3 by measuring the amount of micrococcal nuclease-resistant genome synthesized in cells receiving genome plasmid alone or receiving in addition the N, P, and ΔL support plasmids, the latter encoding a nonfunctional L protein. This mix supplied the proteins (N and P) necessary for encapsidation without reconstituting the viral polymerase. When no support plasmids were provided, genomic RNA would not be encapsidated and should be sensitive to degradation by micrococcal nuclease (Fig. 3, lanes “0” in panel B versus panel A, and note that the exposure time for panel B was eight times that of panel A; also, see Table 1), although as noted below there sometimes was a background of residual RNA due to incomplete nuclease digestion. When the N and P support plasmids were provided, a band of protected genome was evident for the wt genome and for each mutant (Fig. 3B, lanes marked NPΔL). This species was quite abundant for each genome except for the 4C mutant (Fig. 3B, lane 17). The reduced accumulation of protected genome in that case reflects the reduced efficiency of T7-expressed, plasmid-derived genome (Fig. 3A, lane 16), as described above. Thus, each genome retained the capacity to be assembled into a nucleocapsid by N and P proteins supplied in trans (Fig. 3B, lanes NPΔL). The proportion of T7 RNA polymerase-produced genome that was encapsidated relative to the amount of T7 transcript containing the ribozyme varied from 10 to 41% (Table 1, column NPΔL). It should be noted that these values do not exactly reflect the efficiency of encapsidation of the plasmid-derived RNA. This is because the T7 transcript containing uncleaved ribozyme represents only a fraction of the total plasmid-derived material, much of which was likely degraded. Hence, it serves as a surrogate marker. In addition, the apparent differences in encapsidation efficiency between certain minigenomes were not consistent between experiments and thus represent experimental variability rather than significant differences.
Effect of trailer mutations on the production of progeny antigenome.
If plasmid-derived genomic RNA is encapsidated properly, the genomic promoter at its 3′ end (Fig. 1, step 3) should be fully functional, since this end was not subjected to mutagenesis. However, there is the possibility that RNA synthesis is influenced by interaction between the genomic termini, as suggested for VSV (27). To investigate the ability of each genome to form a functional template for the synthesis of positive-sense RNA (Fig. 1, step 4), the total intracellular RNA from cells infected and transfected as described above was analyzed by Northern blot by using a negative-sense riboprobe (Fig. 3C, note that the length of film exposure for panel C was the same as for panel B and eight times longer than for panel A). As would be expected, positive-sense RNAs were not synthesized either in the absence of support plasmids or when N and P were provided together with nonfunctional L (for example, Fig. 3C, lanes 1 and 2). When N, P, and functional L were provided, the genomic template was copied into a discrete band of positive-sense antigenomic RNA and a smear of subgenomic mRNA (Fig. 3C, lane 3). The mRNA appears as a smear because it contains a high proportion of prematurely terminated species due to the absence of the M2-1 antitermination factor (8, 10). The M2-1 protein was specifically omitted here because otherwise the intense band of full-length mRNA obscured the antigenomic RNA. Analysis of positive-sense RNAs from lysates treated with micrococcal nuclease confirmed that the antigenome was completely protected, whereas most of the mRNA was degraded (not shown).
Both antigenomic RNA and mRNA were synthesized by each of the mutants and in the same relative proportions. This indicates that functional genomic nucleocapsids had been formed. In all cases, the antigenome was protected from micrococcal nuclease digestion (not shown). The total amount of positive-sense RNA produced by different genomes varied. This depended on the amount of encapsidated genomic RNA available as template, which in turn had two sources: (i) T7 transcription from transfected plasmid, as described above, and (ii) amplification by RNA replication mediated by the reconstituted RSV polymerase, as described in the next section.
Effect of the trailer mutations on the function of the antigenomic promoter.
Once antigenomic RNA is synthesized and encapsidated, it should be able to direct the production of progeny genome by the RSV polymerase (Fig. 1, step 6). This is clearly evident for the wt genome by examination of total negative-strand RNA (Fig. 3A, compare lane 2 to lane 3). The correctly sized genome migrates more rapidly than the T7 transcript genome-ribozyme, and an increase in its amount in the presence of the complete polymerase is indicative of RNA replication. The amount of genome-sized RNA produced by T7 polymerase (Fig. 3A, lane 2) was barely detectable, while the amount of genome that accumulated in the presence of the L protein was as prominent as the T7 transcript (lane 3). A similar pattern of amplification was observed with mutants 4G and 4C (lanes 15 and 18), though in the case of 4C the much-lower level of amplified genome reflected the much-lower level of T7 transcript, as described above. Mutants 1C and 5A exhibited some genome amplification, though the amount produced was less than the T7 transcript (lanes 6 and 21), indicating a decrease in the efficiency of replication.
Genome amplification was not evident in any of the other mutants, indicating that their antigenome was not functional as a template. Thus, the single nucleotide substitutions at positions 2, 3, 6, and 7 inactivated the antigenome template, and the mutations at positions 1 and 5 decreased its efficiency. In other work, mutational analysis of the leader region showed that changing position 4 from its natural assignment of G to C (negative sense) resulted in a large increase in RNA synthesis (13), which we have found to be due to enhanced RNA replication (unpublished data). The finding that a comparable increase was not observed in the position 4 trailer mutants is indicative of a difference between the leader and trailer regions.
To confirm these conclusions and to test the amplified genomes for encapsidation, we examined the negative-sense RNA resistant to micrococcal nuclease when treated prior to RNA purification. This is shown in Fig. 3B and Table 1, which compares the amount of genome protected by N and P in the absence or presence of functional L (compare the NPΔL and NPL lanes). Substantially more genome was encapsidated in the presence of L than in its absence for the wt, 4G, and 4C genomes, and slightly more for the 1C and 5A genomes, demonstrating the synthesis and encapsidation of progeny genome driven by the reconstituted RSV polymerase. None of the other mutants displayed an increase in encapsidated genome, indicating that their antigenomes were inactive as templates. In the particular experiment shown in Fig. 3, the intensity of encapsidated genome 6C in the presence of L (Fig. 3B, lane 24) was reduced due to a loading error; in other experiments, it was equivalent to the intensity of the corresponding band in lane 23.
Comparison of genome template activities.
We estimated the relative efficiency of each genome template for the synthesis of positive-sense RNA. This was done by dividing the total amount of positive-sense RNA product for each genome (Fig. 3C, lanes 3, 6, 9, 12, 15, 18, 21, 24, and 27; Table 1, column “total positive-sense RNA”) by the corresponding amount of encapsidated genome available as template (Fig. 3B, lanes 3, 6, 9, 12, 15, 18, 21, 24, and 27; Table 1, column “NPL”). The values for positive- and negative-sense RNA came from different blots and cannot be compared directly, but the quotient obtained for each genome provided an estimate in arbitrary units of relative template activity that can be compared between genomes (Table 1, column “Relative genome template activity”). The values were very similar, varying by 2.2-fold or less, and thus the wt and various mutant genomes were very similar with regard to the ability to serve as template for the synthesis of positive-sense RNA.
Effects of double and triple trailer mutants.
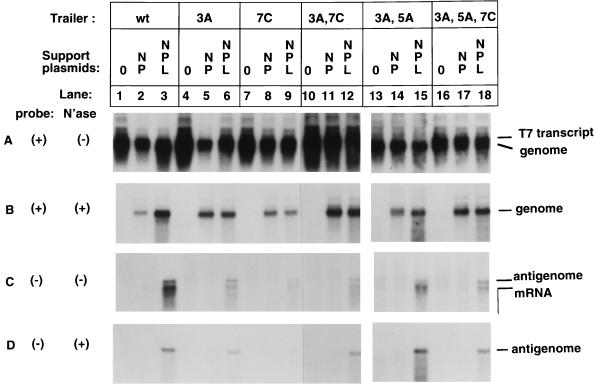
We also prepared mutant genomes that combined the 3A, 5A, and 7C mutations in pairs or altogether (Fig. 4). The various mutant genomic RNAs produced by T7 polymerase were encapsidated (Fig. 4B, compare lanes 4 and 5, 7 and 8, 10 and 11, 13 and 14, and 16 and 17; note that the exposure time was identical for all panels). The encapsidated mutant genomes functioned as template for the synthesis of both antigenome and mRNA (Fig. 4C, lanes 6, 9, 12, 15, and 18), and the antigenome was encapsidated (Fig. 4D, lanes 6, 9, 12, 15, and 18). In addition, none of the mutant genomes was amplified detectably in the presence of the replicase, as judged by comparing the amount of encapsidated genome present in the presence of N and P versus N, P, and L (Fig. 4B, compare lanes 5 and 6, 8 and 9, 11 and 12, 14 and 15, and 17 and 18). In the particular experiment shown in Fig. 4, the amount of encapsidated genome which accumulated in the presence of N, P and L for the double mutant 3A 5A (panel B, lane 15) was marginally greater than the amount which accumulated in the presence of N and P (lane 14), but this was not observed in repeat experiments and thus represents experimental variability, which is not unexpected in experiments involving multiple samples and nuclease treatments. In summary, the double and triple trailer mutants behaved like the individual mutants. In particular, the presence of the various combinations of two or three trailer mutations did not affect the encapsidation and template function of genomic RNA.
FIG. 4.
Effects of multiple point mutations in the minigenome trailer region on T7 transcription, genome encapsidation, synthesis of positive-sense mRNA and antigenome, and synthesis of negative-sense progeny genome. The trailer mutations 3A, 5A, and 7C were inserted individually (3A and 7C), in pairs (3A, 7C and 3A, 5A), and as a triplet (3A, 5A, 7C) into the C2 minigenome. The experiment was performed basically as described in the legend for Fig. 3 except that the nonfunctional ΔL plasmid was not included in the N and P plasmid condition (lanes 2, 5, 8, 11, and 14). Results from the individual mutations and from the combined mutant are presented.
Effect of restoring terminal complementarity on RNA replication.
The point mutations at positions 1 to 3 and 5 to 7 reduce the extent of terminal complementarity of both genomic and antigenomic RNA. Previously, terminal complementarity was shown to enhance the replication of VSV genomes (27). Thus, disruption of complementarity by the point mutants described here might be a factor in the inhibition of genome synthesis. Since all of the genomes synthesized antigenomic RNA, and in the same proportion to mRNA as wt (Fig. 3C), the disruption of terminal complementarity apparently did not affect the ability of the genome to function as template. However, it was possible that complementarity is more important for the function of the antigenome template.
Therefore, we restored complementarity to one of these trailer mutants, 7C. This was accomplished by changing the seventh nucleotide in the leader of this construct from U to G (negative sense, Fig. 1, step 7). Because leader position 7 is part of the sequence that the hammerhead ribozyme uses to align itself for cleavage of the T7 transcript (12), it was possible that this change might affect the efficiency of ribozyme cleavage. To avoid this variable, we replaced the hammerhead ribozyme with that of hepatitis delta virus (23), which does not involve complementarity within the adjacent transcript. This was done for the wt C2 genome, resulting in genome C41, and for genomes containing the trailer 7C mutation alone, the leader 7G mutation alone, and both mutations together.
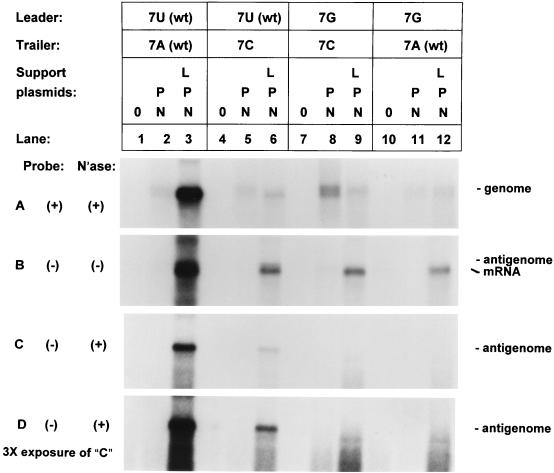
In the presence of the N, P, and L proteins, the wt genome was amplified by RNA replication as described above, resulting in a much greater accumulation of encapsidated genome than in the absence of L protein. This was evident from examination of nuclease-protected negative-sense RNA (Fig. 5A, compare lanes 2 and 3). As described above, the trailer 7C mutant was encapsidated (panel A, compare lanes 4 and 5) and was not amplified (panel A, compare lanes 5 and 6) but was functional as template for the synthesis of positive-sense RNA (panel B, lane 6). The genome containing the 7G mutation in the leader region also was encapsidated (panel A, compare lanes 10 and 11) and served as template for the synthesis of positive-sense RNA (panel B, lane 12). Unexpectedly, however, the only positive-sense RNA detected was mRNA. Antigenomic RNA was greatly reduced or absent in both the total positive-sense RNA (panel B, lane 12) and that which was resistant to micrococcal nuclease (panels C and D, lane 12; note that panel D is a threefold-longer exposure of panel C). In comparison, the wt genome and the 7C trailer mutant produced a clear band of encapsidated antigenomic RNA (panels C and D, lanes 3 and 6, respectively). The genome containing the 7G leader mutation also was not amplified by RNA replication (panel A, compare lanes 11 and 12), as would be expected given the lack of synthesis of the antigenome intermediate.
FIG. 5.
Restoration of complementarity between the genome ends at the position 7 point mutation. Trailer mutant 7C (lanes 4 to 6) is compared with a mutant containing both the trailer 7C mutation and a compensatory mutation in the leader, 7G (lanes 7 to 9). This second mutation restores the complementarity of the ends to the wt level. A third mutant with only the leader 7G mutation was also tested (lanes 10 to 12). Each of these mutants and the wt minigenome, C41, contains a delta ribozyme instead of the hammerhead ribozyme used in genomes described in the previous experiments. Transfections were performed in HEp-2 cells as described in Fig. 3. Blots in panels A, B, and C were exposed to film for 2 h. Panel D is the same blot as panel C, but it was exposed to film for 6 h to confirm the absence of antigenome in lanes 9 and 12.
The 7C 7G double mutant was encapsidated (Fig. 5, panel A, compare lanes 7 and 8) and served as the template for the synthesis of positive-sense RNA (panel B, lane 9). However, as was the case with the 7G leader mutant, the double mutant produced mRNA but not antigenomic RNA (panels B, C, and D, lane 9). Thus, the 7G leader mutation has the effect of ablating the accumulation of antigenome without preventing the synthesis of mRNA, and this effect was not relieved when terminal complementarity was restored by including the 7C trailer mutation. Like the 7C trailer mutant and 7G leader mutant, the double mutant was not amplified by the RSV polymerase (panel A, compare lanes 8 and 9). Thus, substitution at position 7 of either the genomic or antigenomic 3′ terminus destroys its ability to participate in RNA replication, and restoring complementarity by a compensatory mutation at the other end of the template does not relieve this effect. This indicates that the effects of the position 7 mutations are not due to loss of terminal complementarity but rather are direct effects on a local cis-acting signal, probably the genomic (leader 7G mutant) and antigenomic (trailer 7C mutant) promoters.
Effect of large trailer deletions on genome encapsidation and template function.
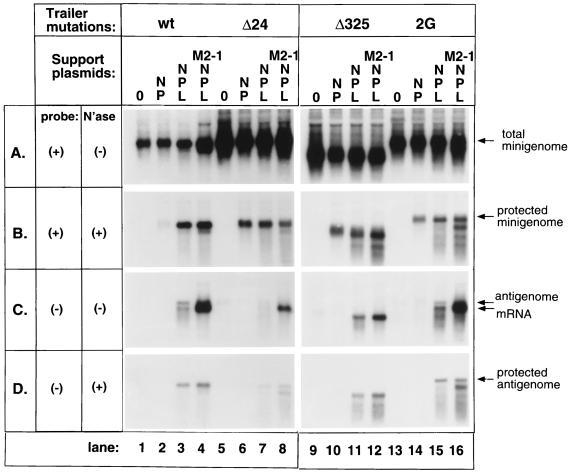
As described above, single substitutions in the last 7 nt of genomic RNA, or double or triple mutations at positions 3, 5 and 7, had no discernible effect on its encapsidation or template function. We therefore examined the effect of two large terminal deletions. In mutant Δ24, the last 24 nt of the 155-nt trailer region were deleted, whereas mutant Δ325 sustained deletion of the complete trailer region and the adjacent GE signal of the CAT transcription cassette. The sequence (negative sense) at the 5′ terminus of each deletion genome is 3′-UGCUCUAUAA-5′ for Δ24 and 3′-AAAGUGGUAC-5′ for Δ325 compared to 3′-AAAAAGAGCA-5′ for the wt genome (mutant nucleotide assignments which are identical to wt are underlined). Both of these genomes were cleaved with the hepatitis delta virus ribozyme. The two mutants were examined in parallel with wt genome C41 and a version of trailer mutant 2G, both of which also employ the hepatitis delta virus ribozyme.
As described above, the wt and 2G genomes were encapsidated, and the wt genome was amplified, whereas the 2G mutant was not (Fig. 6B, compare lanes 2 and 3 and lanes 14 and 15). Interestingly, in this particular experiment, the addition of functional L seemed to render the nucleocapsid somewhat sensitive to micrococcal nuclease (panel B, compare lane 14 with lane 15), and this was further augmented by the addition of M2-1 (panel B, lane 16). The significance of this is unknown; it might mean that active polymerase exposes sites for nuclease attack in the nucleocapsid, but this is not considered further here. The wt and 2G genomes were both active as template for the synthesis of positive-sense RNA by N, P, and functional L (panel C, lanes 3 and 15), as described above. Also, as might be expected, both templates were sensitive to the antitermination activity of the M2-1 protein, which enhanced the synthesis of complete mRNA (panel C, lanes 4 and 16). Finally, antigenomic RNA synthesized from the wt and 2G templates was fully protected from micrococcal nuclease (compare panel C to panel D, lanes 3 and 15).
FIG. 6.
Effects of deletion at the 5′ genome trailer region on T7 transcription, genome encapsidation, synthesis of positive-sense mRNA and antigenome, and synthesis of negative-sense progeny genome. HEp-2 cells were infected with vTF7-3 and transfected with wt minigenome plasmid C41 (wt, lanes 1 to 4) or mutant minigenome plasmids Δ24 (lanes 5 to 8), Δ325 (lanes 9 to 12), or trailer 2G (lanes 13 to 16). Lanes marked “0” received no additional plasmids (lanes 1, 5, 9, and 13). Lanes marked “NP” received N and P support plasmids (lanes 2, 6, 10, and 14). Lanes marked “NPL” received N, P, and L plasmids (lanes 3, 7, 11, and 15). Lanes marked “NPLM2-1” received N, P, L, and M2-1 plasmids (lanes 4, 8, 12, and 16). RNAs were harvested and processed with (B and D) or without (A and C) micrococcal nuclease treatment, as indicated, and were analyzed by Northern blot hybridization with a negative (C and D)- or positive (A and B)-sense riboprobe, as indicated.
In comparison, both the Δ24 and the Δ325 genomes were encapsidated (Fig. 6B, compare lanes 5 and 6 and lanes 9 and 10). As would be expected, neither genome was amplified by the RSV polymerase (panels A and B, compare lanes 6 and 7 and lanes 10 and 11). Both functioned as a template for the synthesis of positive-sense RNAs (panel C, lanes 7, 8, 11, and 12), although the activity of the Δ24 genome was somewhat lower than might have been expected based on the amount of encapsidated genome (panel B, lanes 7 and 8 and lanes 11 and 12). Whereas the Δ24 genome was clearly sensitive to the antitermination activity of M2-1, as evidenced by the increased synthesis of full-length mRNA (panel C, lane 8), little mRNA accumulated for the Δ325 genome (panel C, lane 12). This was not surprising since the deletion removed the GE signal and thus would result in the synthesis of nonpolyadenylated mRNA which likely would be unstable (17). The ability of the antigenomic RNA synthesized by each deletion mutant to be encapsidated was confirmed by analysis of micrococcal nuclease-resistant positive-sense RNA (panel D, lanes 7, 8, 11, and 12). Thus, genomes which had sustained large deletions in their trailer regions and lacked terminal complementarity were encapsidated and functioned as template for the synthesis of antigenome and mRNA, and the antigenome was encapsidated.
DISCUSSION
The trailer region is thought to have two roles during RNA replication: it encodes the antigenomic promoter, and it might contain a sequence that initiates genome encapsidation. We tested the effects of substitution at each of the seven terminal trailer nucleotide positions. None of the mutations affected encapsidation or template activity of the T7-transcribed genome. However, most of the substitutions prevented the encoded antigenome from serving as a template to synthesize progeny genome. As a result, RNA synthesis by the reconstituted RSV polymerase was restricted to that of positive-sense molecules, namely, the synthesis of mRNA and the synthesis and encapsidation of antigenomic RNA. The genome template was not amplified and instead was restricted to that supplied from the plasmid. Thus, the synthesis of mRNA, and the synthesis and encapsidation of antigenomic RNA, can be analyzed under conditions where the abundance of the template is controlled.
An important control in this work was that we examined the abundance of T7-transcribed genome, the pool for nucleocapsid formation. We also directly monitored the abundance of initial encapsidated genome and of genome after amplification by replication. These controls typically have not been part of published studies, but it is clear that they are critical in any situation where mutations can affect the synthesis or stability of the T7-transcribed replicon. It was well known that the sequence immediately downstream of the T7 core promoter can affect its activity and that purines yield higher activity than pyrimidines and G more than A. However, the length of sensitive sequence was not appreciated. For example, Milligan et al. (19) found strong effects in vitro only for the first two positions. We had attempted to preclude such effects by including three nonviral G residues as the first nucleotides of the transcript, which mimic the first three transcribed nucleotides of the authentic promoter. Indeed, this was sufficient to eliminate effects in vitro (unpublished data). Surprisingly, however, we found that substitutions involving the first 5 nt of the trailer region were associated with strong effects on transcription intracellularly. Together with the three nonviral G residues, this indicates that the first eight transcribed nucleotides influence the efficiency of T7 transcription, and the first seven strongly so, varying over a range of 10-fold or more. Since these experiments were performed in transfected cells rather than in the test tube, we cannot rule out effects on transcript stability due to the point mutations. However, the simpler and more likely explanation is that these substitutions affect promoter activity. Furthermore, identical transfection experiments were performed in 293 cells, in which naked RNA is considerably more stable. These experiments showed the same pattern of T7 transcript accumulation (data not shown), supporting the idea that the differences observed here were not due to effects on transcript stability. Consistent with the higher activities observed here with purine residues, the five strongest promoters in bacteriophage T7 initiate transcription with six purines, GGGAGA, whereas all but one of the 10 weaker promoters have pyrimidine substitutions within these 6 nt (9). Furthermore, the footprint of T7 polymerase has been shown to extend 5 nt beyond the core promoter (5). We suspect that the effect of these initial nucleotides was not evident in in vitro transcription reactions because the large excess of added purified polymerase compensated for reductions in the efficiency of initiation.
Not surprisingly, the amount of genome which became encapsidated was influenced by the level of T7 transcript, and in turn the level of RSV-mediated RNA synthesis was strongly influenced by the level of encapsidated template. If we had not controlled for this variable, it would have led to misinterpretation of the effects of certain mutations on replication. For example, the amount of antigenome and mRNA which accumulated in cells in the presence of N, P, and L with the 4C trailer mutant was substantially lower than for the wt, 2G, and 4G mutants and was similar to that of the 5A and 6C genomes (Fig. 3C and Table 1). Taken by itself, that observation would have suggested that the 4C genome is defective in RNA synthesis, but such a conclusion would have been incorrect. On the contrary, examination of the controls indicated that 4C was one of only two mutants (the other being 4G) in which RSV-directed RNA synthesis appeared to be relatively unaffected. Instead, the low level of positive-sense RNA synthesis reflected the low accumulation of initial T7 transcript. Conversely, the 2G mutant resembled the wt genome in the total amount of positive-sense RNA synthesized intracellularly. However, the obvious interpretation that this mutation did not greatly affect RNA synthesis would have been incorrect since, on the contrary, the genome was completely defective in amplification, and the high level of encapsidated genome observed was due solely to the enhanced synthesis of initial T7 transcripts.
None of the trailer mutations affected encapsidation of T7-supplied genome. For both wt and mutant genomes, this process was inefficient, as has been noted previously in a VSV minigenome system (22), although values for the efficiency of encapsidation could not be determined because much of the plasmid-derived transcript appeared to be degraded. It may be that the RSV genome is not preferentially encapsidated under these conditions. Indeed, in some of our experiments a small fraction of mRNA also was encapsidated, as has been described in a Sendai virus minigenome system (4). In contrast, essentially all of the antigenomic RNA which accumulated due to the reconstituted RSV polymerase was encapsidated, and thus this process appeared to be much more efficient, although we cannot exclude the possibility that additional unencapsidated antigenome was made and quickly degraded. Also, in the case of genomes which were competent for genome amplification, such as the wt and the 4G and 4C mutants, essentially all of the amplified genome also was encapsidated. Thus, replicon RNA which was synthesized by T7 polymerase was encapsidated inefficiently, whereas that produced by the reconstituted RSV polymerase was encapsidated efficiently.
It is generally thought that encapsidation of the genomic and antigenomic RNAs of nonsegmented negative-strand RNA viruses initiates at a cis-acting signal near the 5′ end of each RNA and occurs concurrent with chain elongation (18). There is evidence for such a signal in the case of VSV (2, 21, 25) and of rabies virus (29). If there is such a signal for RSV, it was not evident in the highly inefficient encapsidation of genome which was synthesized by T7 RNA polymerase. In these experiments, the synthesis of replicon RNA by T7 RNA polymerase or by the RSV polymerase occurred at the same time and in the same transfected cells, and thus ostensibly under the same conditions except for the difference in template (plasmid versus nucleocapsid) and polymerase (T7 versus RSV). In particular, the requirement that the nascent transcript be available for cosynthetic encapsidation should have been fulfilled in each case. However, there may be some unknown aspect of T7-mediated RNA synthesis which specifically interferes with cosynthetic encapsidation. In these experiments, none of the trailer point mutations, either alone or in combination, and neither of the two trailer deletions affected the efficiency of encapsidation of the T7-transcribed genome. Thus, from these experiments, there is no evidence of a cis-acting signal for RSV. This will be investigated further by examining smaller RNAs that contain just the 5′ end of the genome or antigenome versus heterologous negative controls. An alternative explanation is that a specific RNA initiation signal does not exist for RSV and that the replicase complex (N, P, and L) directs encapsidation, whereas the transcriptase (N, P, L, and M2-1) does not. We are in the process of mapping leader positions important in the synthesis of encapsidated antigenome, and it may be that this will shed further light on whether there is an RNA encapsidation signal.
While none of the mutations affected encapsidation of the T7 produced genome, nucleotides 2, 3, 6, and 7 appeared to be absolutely critical for the function of the antigenomic template, and positions 1 and 5 also appeared to be important. We have not yet tested the effect in this system of substituting other nucleotides at these positions or of making substitutions at additional sequence positions. It is interesting that of the seven positions tested, only position 4 was flexible, in that A, C, and G are all capable of functioning. This nucleotide position is also the only one of the first 11 nt of the antigenomic promoter that is not conserved in the genomic promoter. The leader region (genomic promoter) of wt RSV strain A2 contains G at this position (negative sense) (20). Certain attenuated strains contain C at this position, although this substitution did not appear to be part of the attenuation phenotype (28). In other work, we found that this 4C leader substitution results in a substantial enhancement of genome RNA synthesis, which we have now determined to be an effect on enhancing the production of encapsidated antigenome (unpublished data). In contrast, in the trailer region, the substitution of the wt A assignment at trailer position 4 with C or G did not have much effect on RNA synthesis. This indicates a difference between the genomic and antigenomic promoters.
The point mutations which blocked the synthesis of progeny genome, namely, at trailer positions 2, 3, 6, and 7, might directly inactivate the antigenomic promoter. This is the explanation we favor. Alternatively, they might act by preventing encapsidation of the nascent genomic RNA. This explanation seems less likely since these mutations did not prevent encapsidation of genomic RNA synthesized from plasmid. In addition, the possibility existed that the inhibitory effect of the trailer mutations was due to reduced terminal complementarity, as has been described for VSV (27). However, restoration of terminal complementarity by the further introduction of a compensatory mutation in the genome leader did not restore RNA replication. Furthermore, genomes which had sustained large deletions at the end of trailer, and thus lacked terminal complementarity altogether, retained the capacity to function as templates for the synthesis of antigenomic RNA. This indicated that terminal complementarity of the RSV genome is not important for its function as template. Thus, we conclude that the mutations in the trailer region which inhibited replication did so by direct effects on the primary structure of a cis-acting element, probably the antigenomic promoter.
It is interesting that the one leader mutation which was examined here, the 7G mutation, inhibited accumulation of antigenomic RNA, a replication product, but did not inhibit mRNA synthesis. One possibility is that this substitution specifically affected promoter function for replication but not transcription. Alternatively, perhaps antigenomic RNA was produced but not encapsidated and therefore was degraded. These possibilities will be further investigated.
ACKNOWLEDGMENTS
We thank Kailash Gupta for helpful discussions on mutagenesis, Myron Hill and Ena Camargo for technical assistance, Juan Cristina for preliminary results based on an RNA transfection assay, Rachel Fearns for helpful discussions, and Brian Murphy, Robert Chanock, and Rachel Fearns for review of the manuscript.
REFERENCES
- 1.Baker S C, Moyer S A. Encapsidation of Sendai virus genome RNAs by purified NP protein during in vitro replication. J Virol. 1988;62:834–838. doi: 10.1128/jvi.62.3.834-838.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blumberg B M, Kolakofsky D. N protein of vesicular stomatitis virus selectively encapsidates leader RNA in vitro. Cell. 1983;32:559–567. doi: 10.1016/0092-8674(83)90475-0. [DOI] [PubMed] [Google Scholar]
- 3.Byrappa S, Gavin D K, Gupta K C. A highly efficient procedure for site-specific mutagenesis of full-length plasmids using Vent DNA polymerase. Genome Res. 1995;5:404–407. doi: 10.1101/gr.5.4.404. [DOI] [PubMed] [Google Scholar]
- 4.Cadd T, Garcin D, Tapparel C, Itoh M, Homma M, Roux L, Curran J, Kolakofsky D. The Sendai paramyxovirus accessory C proteins inhibit viral genome amplification in a promoter-specific fashion. J Virol. 1996;70:5067–5074. doi: 10.1128/jvi.70.8.5067-5074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapman K A, Gunderson S I, Anello M, Wells R D, Burgess R R. Bacteriophage T7 late promoters with point mutations: quantitative footprinting and in vivo expression. Nucleic Acids Res. 1988;16:4511–4524. doi: 10.1093/nar/16.10.4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins P, McIntosh K, Chanock R. Respiratory syncytial virus. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven; 1996. [Google Scholar]
- 7.Collins P L, Hill M G, Camargo E, Grosfeld H, Chanock R M, Murphy B R. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc Natl Acad Sci USA. 1995;92:11563–11567. doi: 10.1073/pnas.92.25.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins P L, Hill M G, Cristina J, Grosfeld H. Transcription elongation factor of respiratory syncytial virus, a nonsegmented negative-strand RNA virus. Proc Natl Acad Sci USA. 1996;93:81–85. doi: 10.1073/pnas.93.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunn J J, Studier F W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983;166:477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- 10.Fearns R, Collins P L. Role of the M2-1 transcription antitermination protein of respiratory syncytial virus in sequential transcription. J Virol. 1999;73:5852–5864. doi: 10.1128/jvi.73.7.5852-5864.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fearns R, Peeples M E, Collins P L. Increased expression of the N protein of respiratory syncytial virus stimulates minigenome replication but does not alter the balance between the synthesis of mRNA and antigenome. Virology. 1997;236:188–201. doi: 10.1006/viro.1997.8734. [DOI] [PubMed] [Google Scholar]
- 12.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grosfeld H, Hill M G, Collins P L. RNA replication by respiratory syncytial virus (RSV) is directed by the N, P, and L proteins; transcription also occurs under these conditions but requires RSV superinfection for efficient synthesis of full-length mRNA. J Virol. 1995;69:5677–5686. doi: 10.1128/jvi.69.9.5677-5686.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardy R W, Wertz G W. The product of the respiratory syncytial virus M2 gene ORF1 enhances readthrough of intergenic junctions during viral transcription. J Virol. 1998;72:520–526. doi: 10.1128/jvi.72.1.520-526.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horikami S M, Curran J, Kolakofsky D, Moyer S A. Complexes of Sendai virus NP-P and P-L proteins are required for defective interfering particle genome replication in vitro. J Virol. 1992;66:4901–4908. doi: 10.1128/jvi.66.8.4901-4908.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuo L, Fearns R, Collins P L. Analysis of the gene start and gene end signals of human respiratory syncytial virus: quasi-templated initiation at position 1 of the encoded mRNA. J Virol. 1997;71:4944–4953. doi: 10.1128/jvi.71.7.4944-4953.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuo L, Grosfeld H, Cristina J, Hill M G, Collins P L. Effects of mutations in the gene-start and gene-end sequence motifs on transcription of monocistronic and dicistronic minigenomes of respiratory syncytial virus. J Virol. 1996;70:6892–6901. doi: 10.1128/jvi.70.10.6892-6901.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamb R, Kolakofsky D. Paramyxoviridae: the viruses and their replication. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven; 1996. [Google Scholar]
- 19.Milligan J F, Groebe D R, Witherell G W, Uhlenbeck O C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mink M A, Stec D S, Collins P L. Nucleotide sequences of the 3′ leader and 5′ trailer regions of human respiratory syncytial virus genomic RNA. Virology. 1991;185:615–624. doi: 10.1016/0042-6822(91)90532-g. [DOI] [PubMed] [Google Scholar]
- 21.Moyer S A, Baker S C, Lessard J L. Tubulin: a factor necessary for the synthesis of both Sendai virus and vesicular stomatitis virus RNAs. Proc Natl Acad Sci USA. 1986;83:5405–5409. doi: 10.1073/pnas.83.15.5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pattnaik A K, Ball L A, LeGrone A W, Wertz G W. Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell. 1992;69:1011–1020. doi: 10.1016/0092-8674(92)90619-n. [DOI] [PubMed] [Google Scholar]
- 23.Perrotta A T, Been M D. A pseudoknot-like structure required for efficient self-cleavage of hepatitis delta virus RNA. Nature. 1991;350:434–436. doi: 10.1038/350434a0. [DOI] [PubMed] [Google Scholar]
- 24.Samal S K, Collins P L. RNA replication by a respiratory syncytial virus RNA analog does not obey the rule of six and retains a nonviral trinucleotide extension at the leader end. J Virol. 1996;70:5075–5082. doi: 10.1128/jvi.70.8.5075-5082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smallwood S, Moyer S A. Promoter analysis of the vesicular stomatitis virus RNA polymerase. Virology. 1993;192:254–263. doi: 10.1006/viro.1993.1028. [DOI] [PubMed] [Google Scholar]
- 26.Tapparel C, Roux L. The efficiency of Sendai virus genome replication: the importance of the RNA primary sequence independent of terminal complementarity. Virology. 1996;225:163–171. doi: 10.1006/viro.1996.0584. [DOI] [PubMed] [Google Scholar]
- 27.Wertz G W, Whelan S, LeGrone A, Ball L A. Extent of terminal complementarity modulates the balance between transcription and replication of vesicular stomatitis virus RNA. Proc Natl Acad Sci USA. 1994;91:8587–8591. doi: 10.1073/pnas.91.18.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitehead S S, Juhasz K, Firestone C-Y, Collins P L, Murphy B R. Recombinant respiratory syncytial virus (RSV) bearing a set of mutations from cold-passaged RSV is attenuated in chimpanzees. J Virol. 1998;72:4467–4471. doi: 10.1128/jvi.72.5.4467-4471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J, Koprowski H, Dietzschold B, Fu Z F. Phosphorylation of rabies virus nucleoprotein regulates viral RNA transcription and replication by modulating leader RNA encapsidation. J Virol. 1999;73:1661–1664. doi: 10.1128/jvi.73.2.1661-1664.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]