Abstract
The skin of vertebrates is the outermost organ of the body and serves as the first line of defense against external aggressions. In contrast to mammalian skin, that of teleost fish lacks keratinization and has evolved to operate as a mucosal surface containing a skin-associated lymphoid tissue (SALT). Thus far, IgT representing the prevalent Ig in SALT have only been reported upon infection with a parasite. However, very little is known about the types of B cells and Igs responding to bacterial infection in the teleost skin mucosa, as well as the inductive or effector role of the SALT in such responses. To address these questions, in this study, we analyzed the immune response of trout skin upon infection with one of the most widespread fish skin bacterial pathogens, Flavobacterium columnare. This pathogen induced strong skin innate immune and inflammatory responses at the initial phases of infection. More critically, we found that the skin mucus of fish having survived the infection contained significant IgT- but not IgM- or IgD-specific titers against the bacteria. Moreover, we demonstrate the local proliferation and production of IgT+ B cells and specific IgT titers, respectively, within the SALT upon bacterial infection. Thus, our findings represent the first demonstration that IgT is the main Ig isotype induced by the skin mucosa upon bacterial infection and that, because of the large surface of the skin, its SALT probably represents a prominent IgT-inductive site in fish.
In contrast to five major Ig classes (IgM, IgG, IgA, IgD, and IgE) in mammals, only three Ig isotypes are present in teleosts, including IgM, IgT/IgZ, and IgD (1). Teleost IgM is the most abundant Ig class that is involved in systemic immunity upon infection or vaccination (2–5). Secreted IgD has been identified and found coating a significant portion of the fish microbiota (6–8); however, its immune function against pathogens remains largely unknown. Teleost IgT has been demonstrated to play a specialized role in mucosal immunity akin to that of mammalian secretory IgA (sIgA) (6–10). Similar to sIgA, secreted IgT (sIgT) plays a key function both in protecting fish mucosal sites against pathogens (7–12) as well as in promoting microbiota homeostasis at mucosal surfaces (13). With regards to its role in pathogen control, we and others have shown that sIgT is the main Ig induced in several fish mucosal-associated lymphoid tissues (MALTs) upon pathogen insult (7–12). Thus far, pathogen-specific IgT responses have been documented in the gut, skin, gill, nasal, buccal, and pharyngeal mucosal areas (7–12). However, in-depth studies assessing the local induction of IgT have only been performed in some of the fish MALTs, including those of the gills, nose, buccal, and pharyngeal tissues (7, 8, 11, 12). Moreover, such local responses have been assessed with the use of only one pathogen type, the parasite Ichthyophthirius multifiliis, a pathogen that is responsible for important losses to both wild and farmed populations of salmonid species.
At this point, nothing is known about the local induction of IgT responses in the skin of fish, by far the largest mucosal surface in these species. Moreover, pathogen-mediated IgT responses in that surface are restricted to one study that used the parasite I. multifiliis as the pathogen model (9), although it is unknown at this point whether bacterial pathogens are capable of inducing similar responses. Anatomically, the skin presents a similar structure in all vertebrates, which is generally composed of two layers (epidermis and dermis) (9). However, it should be noted that vertebrates of different taxonomic statuses have faced unique evolutionary pressures that have continuously shaped their own skin structures and components. These histological and componential differences in the skin of vertebrates are reflected also in their specific immune strategies (14). It is worth noting that the term skin-associated lymphoid tissue (SALT) was first used in reference to mammals (15), and that in these species, the main cellular constituents of the skin epidermis are specialized epithelial cells known as keratinocytes, Langerhans cells, and T cells (16). Moreover, the skin dermis of mammals is home to a diverse array of specialized immune cells, including Ag-presenting dermal dendritic cells, T cells, B cells, and NK cells, as well as mast cells, monocytes, and macrophages (17), indicating that many immunological processes take place in the dermal layer of the mammalian SALT (9). Interestingly, teleost fish have also been shown to contain a SALT (2). However, in contrast to mammalian skin, that of teleosts behaves as a mucosal surface because it lacks keratinization, harbors abundant mucus, and its living epithelial cells are in direct contact with the water environment where they live. Although the fish dermis is mainly composed of dense connective tissue with a large amount of collagen fibers and is devoid for the most part of immune cells, its epidermis has been shown thus far to contain B cells, T cells, macrophages, and granulocytes as the main immune cells (9). Moreover, it is in the epidermis where significant immune responses have also been reported (18, 19). To our knowledge, IgT responses in the skin were first described by us and we showed that IgT-specific responses against a parasite were chiefly detected in the skin mucus, whereas IgM-specific titers were for the most part observed in the serum (9). Whether IgT and IgT+ B cell responses are locally induced in the skin is an important question that remains to be addressed. In that regard, it is important to note that in mammals not all IgA responses are induced locally in all existing mucosal sites (20). For example, IgA found in some mucosal areas (e.g., female genital tract) and secretory glands (e.g., salivary glands and lacrimal glands) is derived from B cells that are locally induced in other mucosal sites (e.g., nasopharynx-associated lymphoid tissue and gut-associated lymphoid tissue) (21–23).
As stated above, only one report has described the induction of skin IgT-specific titers against a pathogen I. multifiliis, and local IgT responses have only been assessed, with the use of the same pathogen, only in some fish mucosal sites but not the skin. Prior to the discovery of the role of IgT in mucosal immunity, several studies in teleosts showed the induction of modest IgM-specific titers against several bacterial pathogens. More recently, the study was unable to detect IgM-specific titers in the skin mucus of rainbow trout exposed to Flavobacterium psychrophilum, which prompted the authors to suggest that IgT might play a role against this bacterial pathogen (24). Thus, in this study, we aimed to evaluate the specific contributions of the different fish Igs to skin bacterial pathogen in a representative teleost and to assess whether such responses could be induced locally in the SALT. In this study, we choose Flavobacterium columnare, one of the most widespread fish skin bacterial pathogens. F. columnare is a long Gram-negative rod in the family Flavobacteriaceae, one of the main phyletic lines within the Bacteroidetes group from the domain bacteria (25), which can survival in both fresh and brackish water throughout the world (26).This pathogen can cause columnaris disease that affects a variety of freshwater fishes, leading significant losses both in wild and farmed fish populations (27–33). Columnaris disease is characterized by a pronounced erosion and necrosis of mucosal tissues, including the fin, skin, and gills, and it can cause particularly high mortalities. Secreted enzymes, such as proteases and chondroitin sulfate lyases, have been potential F. columnare virulence factors, contributing to the branchial and cutaneous necrosis (34, 35). Although we have shown in the past that F. columnare induces dominant IgT- and IgM-specific responses in the gill mucosa and serum, respectively, of rainbow trout, thus far, little is known with regards to the pathogenesis of columnaris in the fish skin as well as the innate and adaptive immune mechanisms induced by this pathogen in that surface upon infection.
To assess the mucosal immune responses induced in the skin by F. columnare, we infected rainbow trout with a newly constructed fluorescent version of this pathogen following a bath exposure strategy. Our findings show that trout skin elicited strong innate and adaptive immune responses to F. columnare. More critically, we show that F. columnare–specific Ig responses in the skin mucus were mainly represented by the IgT isotype and that such responses were induced locally, as shown by the local proliferation and accumulation of IgT+ B cells. Moreover, when cultured in vitro, skin explants produced F. columnare–specific IgT titers. These data show for the first time, to our knowledge, that the use of a fluorescent version of F. columnare was critical to further understand the pathogenesis of this important fish pathogen. More importantly, we demonstrate the previously undiscovered capacity of the fish skin to induce potent mucosal IgT-specific responses against a bacterial pathogen and that such responses are induced locally upon mucosal infection.
Materials and Methods
Ethics statement
All experimental protocols were performed in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Ministry of Science and Technology of China. They were approved by the Scientific Committee of Huazhong Agricultural University (permit HZAUFI-2017–013). The procedures done to minimize suffering of fish followed the Chinese Association for Laboratory Animal Sciences.
Fish
Rainbow trout (~5 g) were obtained from a fish farm in Shiyan (Hubei, China). Fish were only purchased when no signs of disease or abnormal behavior were observed (i.e., absence of damage in skin, fin, and gill tissues, normal breathing and swimming behavior). These fish were transported to Huazhong Agriculture University and maintained in aerated aquarium tanks with a water recirculation system, including internal biofilters and thermostatic temperature control. Fish were acclimatized for at least 2 wk at 16°C and fed daily with commercial trout pellets at a rate of 1% biomass during the whole experiment periods. During this time, water conditions were examined on a daily basis, including pH, conductivity, nitrite/nitrate/ammonia content, and oxygen levels. Fish did not show any signs of disease or abnormal behavior during the acclimation period. The feeding was terminated 2 d prior to sacrifice.
F. columnare culture and construction of GFP-expressing F. columnare G4 strain
The strain G4 of F. columnare used in this study was provided by Prof. Nie (36) and conserved at −80°C. F. columnare strains were grown at 28°C in Shieh medium or on Shieh agar. The GFP-expressing F. columnare (GFP–F. columnare) G4 strain was constructed by using the method previously described by Li et al. (37). Briefly, a fused fragment carrying PompA of Flavobacterium johnsoniae) and gfp was amplified by PCR using the primers pr44-Bam (5′-CGCGGATCCGGCAGCGCATACCAAAGAACACTTAG-3′) and pr38-Pst (5′-GCTAGCTGCAGCAGATCTATTTGTATAGTTCATCCA-3′) with pAS43 as a template. The PCR product was digested with BamHI and PstI and cloned into the corresponding sites of pCP23 to generate plasmid pCP-gfp. This plasmid was transferred into Escherichia coli S17–1λpir and then was introduce into G4 by conjugation. The GFP-labeled strain was screened on Shieh agar containing 1 μg/ml tobramycin and 10 μg/ml tetracycline.
Growth curve determination
To compare the growth rates of the GFP–F. columnare and wild-type F. columnare G4 strain in vitro, both strains were streaked on Shieh agar at 28°C for 24 h. The colonies were then inoculated into Shieh broth and incubated at 28°C with shaking at 150 rpm. The OD was measured at 600 nm with a 2-ml cuvette using NanoPhotometer NP80 (Implen, Munich, Germany). Until the OD600 reached 0.4, 500 μl of such early exponential phase were inoculated in 25 ml Shieh broth and incubated at 28°C with shaking at 150 rpm. The OD600 of the culture liquid from each group was measured every 1 h for 13 h.
Rainbow trout challenges
The method used for F. columnare infection followed that described previously (7, 38, 39), with slight modification. Fish (~5 g per fish) were randomly chosen and exposed by immersion with wild-type or GFP–F. columnare at a final concentration of 1 × 106 CFU/ml for 4 h at 16°C and then migrated into the aquarium containing new aquatic water. To detect the localization of F. columnare in trout, 30 fish were infected with GFP–F. columnare, and tissue samples, including skin, gill, and fin were collected at 1, 2, 4, 7, and 14 d postinfection (dpi). For the study of immune responses in trout, there were two types of challenges performed. In the first group (infected fish), 60 fish were challenged by wild-type G4 strain. Tissue samples, including skin, head kidney, and spleen, were collected at 1, 2, 4, 7, 14, and 28 dpi. Skin mucus and serum were taken 4 wk later as the infected group (fish survived 28 d after one exposure). In the second group, another 60 fish were monthly exposed to wild-type F. columnare for 75 d, with the final concentration of 1 × 106 CFU/ml for 4 h at 16°C. Fish samples mentioned above were taken 2 wk after the last challenge as the survivor group (fish survived 75 d after three exposures). Both experiments were performed at least three independent times. As a control group (mock infected), the same number of fish were maintained in similar tanks and were exposed to the same culture medium without pathogens. In all experiments, the anterior dorsal skin was sampled for histology study, the posterior dorsal skin was sampled for extracting DNA or RNA, the ventral skin was sampled for coating plates, and the whole unilateral skin was sampled for collecting mucus or cells.
The detection of F. columnare in trout
To detect the F. columnare in rainbow trout after challenge, different tissue samples, including skin, gills, and fins were collected, weighed and homogenized in tissue lysis buffer using steel beads and shaking (60 HZ for 1 min) by TissueLyser II (Jingxin Technology). The DNA of different tissue samples was then extracted and amplified by PCR using the 16S rRNA-specific primers F1 (5′-GCCCAGAGAAATTTGGAT-3′) and R1 (5′-TGCGATTACTAGCGAATCC-′ of F. columnare (40). The PCR products were extracted from a 2% agarose gel, and images were acquired using a Gel Doc XR+ (Bio-Rad Laboratories). Moreover, skin tissue samples were collected from both control and infected fish and homogenized in 1 ml of PBS (filtered with 0.22 μm) for coating plates, and the colonies were observed using a fluorescence microscope.
RNA isolation and quantitative real-time PCR analysis
Total RNAwas isolated by the TRIzol (Invitrogen) method as we previously described (38). Approximately 2 μl of RNA (1000 ng in total) was reverse transcribed into cDNA with Hifair III First-Strand cDNA Synthesis SuperMix for qPCR (YEASEN) following the manufacturer’s instructions. The synthesized cDNA was diluted four times and then used as a template for quantitative real-time PCR (qRT-PCR) analysis. The qRT-PCRs were performed in triplicate and each contained 1 μl of a diluted cDNA template, 5 μl of SYBR Green qPCR Mix (2 ×; Monad) and 150 nM forward and reverse primers in a 10-μl reaction volume. The amplification profile was performed as an initial denaturation step at 95°C for 5 min, then for 40 cycles of 95°C for 10 s and 58°C for 30 s. A dissociation protocol was carried out by reading fluorescence at every degree between 72 and 95°C after thermos cycling to confirm that only a single band of the correct size was amplified. We calculated the relative fold changes by using the 2−∆∆Ct method, and elongation factor 1a (EF-1α) was used as an internal control. The specific primers used for this experiment are listed in Supplemental Table I.
RNA-sequencing libraries and differential expression analysis
Nine samples (control, 2 dpi, and 14 dpi groups contain three samples each, with five fish per sample) from rainbow trout were used for the RNA-sequencing (RNA-seq) library construction and analyzed as we previous described (41). Briefly, polyadenylated RNA fragments were purified by a Dynabeads mRNA Purification Kit and fragmented by RNA fragmentation buffer. Random hexamer primer and Moloney murine leukemia virus Reverse Transcriptase (RNase H) were used for first-strand cDNA synthesis. Second-strand cDNA synthesis was subsequently performed using DNA polymerase I and RNase H. These dscDNA fragments were end repaired, and a single “A” was added at the 3′ ends before ligating to Illumina paired-end sequencing adaptors. PCR was amplified using Phusion High-Fidelity PCR Master Mix and Illumina primers with the condition of 98°C for 60 s, then 15 cycles of 98°C for 10 s and 65°C for 75 s, concluding with 65°C for 5 min to create the final cDNA library. All RNA-seq data were generated by Illumina paired-end sequencing with a read length of 150 bp. Reads were mapped to the Oncorhynchus mykiss genome using STAR (42) with default parameters. The mapped reads were analyzed by featureCounts (43). Differential expression was estimated by the edgeR package (44). We excluded the genes with low expression (counter-per-million <1 in three or more samples) from downstream analysis. The resulting genes were considered as differentially expressed genes (DEGs) if the false discovery rate (FDR) ≤0.05 and |log2 (fold change)| ≥1. For further understanding of the DEGs, we carried out a gene ontology enrichment using KOBAS (45) to determine the biological processes that were significantly altered following bacterial infection.
Collection of serum and skin mucus
MS-222 was used to anesthetize the trout, and after that, the blood was obtained from the caudal vein by syringe and transferred into an Eppendorf tube, then serum was collected by centrifugation for 15 min at 4°C, 5000 × g (10). As for the skin mucus, we used the method described by Xu et al. (9). Briefly, we transferred the scraping from the surface of skin into an Eppendorf tube and then blew it 30 times repeatedly through a 1-ml syringe (Shanghai Jinta Medical Co., Shanghai, China). Subsequently, the skin mucus was collected by centrifuge. To remove the skin bacteria, we centrifuged the previously treated skin mucus by 10,000 × g ultracentrifugation, then the skin mucus supernatant was obtained.
Western blot
Skin mucus and serum under nonreducing condition were separated by 4–15% SDS-PAGE Ready Gel (Bio-Rad Laboratories) according to previous research (7). For Western blot analysis, the gels were transferred onto PVDF membranes (Bio-Rad Laboratories). Thereafter, the membranes were blocked with 8% skim milk and incubated with primary anti-trout IgT (polyclonal Ab [pAb] rabbit IgG) (10), anti-trout IgM (1.14, mAb mouse IgG1) (46) or biotinylated anti-trout IgD (mAb mouse IgG1) (47), followed by incubation with peroxidase-conjugated Anti-Rabbit, Anti-Mouse IgG (Invitrogen) or streptavidin (Invitrogen). Immunoreactive bands were visualized using the enhanced chemiluminescent reagent (Advansta) and scanned by a GE Amersham Imager 600 imaging system (GE Healthcare). For quantitative analysis of Igs in serum and skin mucus, the signal strength of each band was analyzed by using ImageQuant TL software (GE Healthcare). Thereafter, the concentration of IgT, IgM, and IgD were determined as previously reported by us (7, 9) by plotting the obtained signal strength values on a standard curve generated for each blot using known amounts of purified trout IgT, IgM, or IgD.
Histology, light microscopy, and immunofluorescence microscopy studies
The paraffin sections of tissues were obtained for histology or immunofluorescence study as previously reported (41). To detect the histological structure and mucus cells in skin, the sections were stained with H&E and Alcian blue (AB). To observe the distribution of GFP–F. columnare in skin, gills, and fins, the sections were stained with DAPI (1 μg/ml; Invitrogen). In the immunofluorescence study, anti-trout IgT (pAb rabbit IgG, 0.5 μg/ml), anti-trout IgM (1.14, mAb mouse IgG1, 1 μg/ml), or anti-trout, poly-Ig receptor (pIgR) Ab (pAb rabbit IgG, 0.5 μg/ml) (10) were used to assess the IgT+ and IgM+ B cells or pIgR+ cells in skin sections as in a previous study (9). As controls, the rabbit IgG prebleed and the mouse IgG1 isotype Abs were used at the same concentrations. All images were gained by Olympus BX53 fluorescence microscope (Olympus) and processed by iVision–Mac scientific imaging processing software (Olympus).
Proliferation of B cells in the skin of trout
The proliferation of B cells in skin and head kidney were assessed in control and survivor fish by injection with 5-ethynyl-20-deoxyuridine (EdU; Invitrogen) as previously reported (7). Briefly, control and survivor fish (~15 g) were anesthetized with MS-222 and i.v. injected with 200 μg EdU (Invitrogen). Twenty-four hours later, the leukocytes from trout skin and head kidney were isolated for flow cytometry study using the existing methodology as described previously (7–9). To detect IgT+ or IgM+ B cells, ~5 × 106 cells were incubated with anti-trout IgT (41.8, mAb mouse IgG2b, 1 μg/ml) (10) or anti-trout IgM (1.14, mAb mouse IgG1, 1 μg/ml) (46), and Alexa Fluor 488–goat anti-mouse IgG (Invitrogen) was used as a secondary Ab to be incubated subsequently. The EdU+ cells were then stained according to the manufacturer’s instructions (Click-iT EdU Alexa Fluor 647 Flow Cytometry Assay Kit; Invitrogen). Using a CytoFLEX flow cytometer (Beckman Coulter) and FlowJo software (Tree Star), the percentages of EdU+ cells within the IgM+ or IgT+ B cells populations were presented.
Tissue explants culture
First, fish were treated with MS-222 for conveniently removing blood from the caudal vein. The tissues of head kidney, spleen, and skin (~50 mg) were then taken from each fish and soaked with 75% ethanol for 15 s to kill the surface bacteria. Thereafter, the tissues were submerged twice in PBS and moved to 400 μl of prepared DMEM (Invitrogen) supplemented with 10% FBS, 100 μg/ml streptomycin, 200 μg/ml amphotericin B, and 250 μg/ml gentamicin sulfate. Last, tissues in the medium were cultured (5% CO2, 17°C) for 7 d, and the tissue culture fluid was collected and stored until use.
Binding of trout Igs to F. columnare
In this study, we measured the titers of F. columnare–specific Igs in fluid samples (skin mucus, serum, and tissue explant medium) by pull-down assay as previously described (7). Briefly, 40 μl of bacteria suspension (4 × 108 CFU) were incubated with diluted fluid samples for 4 h at 4°C with continuous shaking in a 300-μl volume. Dilutions were made with PBS containing 1% BSA (pH 7.2). Thereafter, the bacteria were washed three times with PBS, and bound proteins were eluted with 2 × Laemmli sample buffer (Bio-Rad Laboratories), then analyzed by Western blot as clarified above.
Statistical analysis
An unpaired Student t test (Excel version 11.0; Microsoft) and one-way ANOVA with Bonferroni correction (Prism version 6.0; GraphPad) were used for analysis of differences between two groups. The p values ≤0.05 were considered statistically significant.
Results
Pathological changes in trout skin after bacterial infection
To evaluate morphological changes in trout skin, we used a bath challenge model with F. columnare that results in the infection of fish mucosal tissues and elicits strong immune responses (38, 39). However, because of a lack of specific Ab to this bacterial strain, a recombinant F. columnare strain containing a plasmid expressing GFP was constructed to gain insight into the pathogenesis and progression of the bacterial infection in fish skin (Supplemental Fig. 1A). Under the same culture conditions, the GFP-expressing and wild-type F. columnare strains grew at similar rate in Shieh medium (Supplemental Fig. 1B). Critically, both the GFP-expressing and wild-type F. columnare strains elicited very similar mortality rates when used at the same concentration (Supplemental Fig. 1C). Together, these results indicated that the growth and virulence of the GFP–F. columnare were not influenced by GFP or the plasmid. At 4 dpi with GFP–F. columnare, we observed that the classical symptoms of skin lesions: fin rot and gill necrosis appeared in the trout (Fig. 1A). The different locations of skin samples were marked in Supplemental Fig. 1D. Importantly, high expression of F. columnare–specific 16S rRNA accumulated in the skin, gills, and fins of trout at 4 dpi (Fig. 1B). Notably, by using a fluorescence microscope, a large number of F. columnare with green fluorescence was observed in the fin, skin, and gill tissues from 4-d-infected fish when compared with that of control fish (Fig. 1C). In addition, the tissue homogenates of trout skin from control fish and 4-d-infected fish were cultured on Shieh agar, and the yellow-green color bacterial colonies showed characteristic rhizoid shapes that were detected only from homogenate samples of infected trout skin (red arrows). Moreover, the single colonies isolated from the plate were grown in pure culture and displayed characteristic elongated rod-shaped F. columnare bacteria with green fluorescence (Fig. 1D). By fluorescence microscopy, a time-series observation of GFP–F. columnare in skin tissue showed that much of the bacteria was found in the skin epithelium and that bacteria accumulated in trout skin as early as 1 dpi (Fig. 1E). Moreover, similar to these results, qRT-PCR analysis showed that the F. columnare load in trout skin increased significantly from 1 dpi and peaked at day 4 (Fig. 1F), which further confirmed that the bacteria succeeded in invading the skin mucosal tissue.
FIGURE 1.
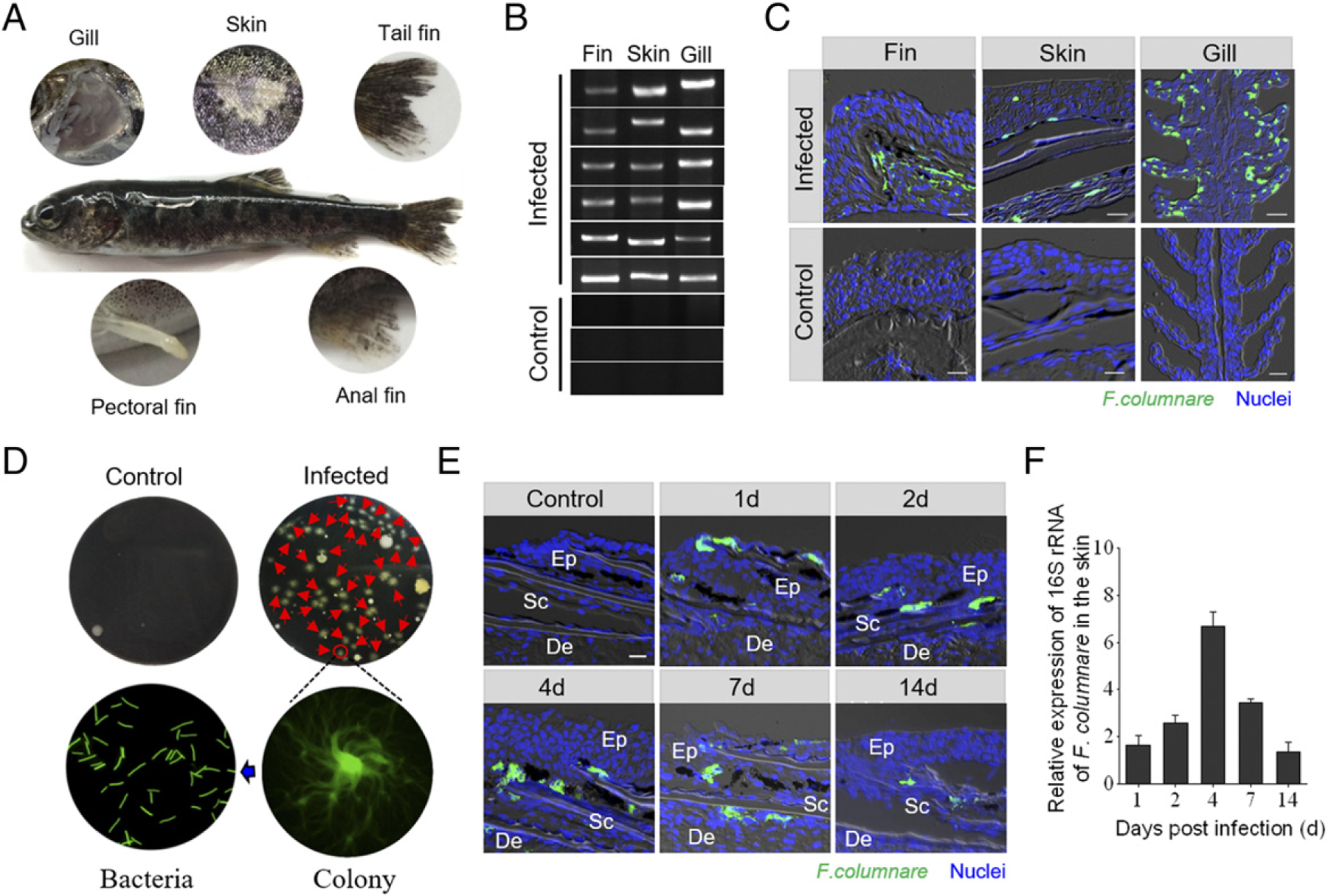
. Invasion of F. columnare to the skin of rainbow trout. (A) The severe phenotype of rainbow trout was observed at 4 dpi with GFP–F. columnare. (B) The PCR products of F. columnare 16S rRNA gene of the fins, skin, and gill tissues from three control and six infected fish at 4 dpi were electrophoresed in 2% agarose, and the bands were recorded using the gel imaging system. (C) Histological examination of GFP–F. columnare by fluorescence in trout fins, skin, and gill paraffinic sections from infected fish at 4 dpi. Differential interference contrast images show merged staining with F. columnare (green) and nuclei (blue). Scale bar, 20 μm. (D) The culture plates from trout skin of control fish (upper left) and 4-d-infected fish (upper right), respectively. Red triangles indicate single colony of GFP–F. columnare. The colony image is a magnified view of a circled colony from infected fish by fluorescence microscope (lower right). The bacteria image is an observation of bacterial solution obtained by circled colony expansion (lower left). (E) Localization of GFP–F. columnare in trout skin of control fish and infected fish at 1, 2, 4, 7, and 14 dpi. Scale bar, 20 μm. (F) The relative expression of 16S rRNA of F. columnare in infected fish versus control fish measured at days 1, 2, 4, 7, and 14 dpi in skin of rainbow trout (n = 6). Data are representative of three independent experiments (mean ± SEM). De, dermis; Ep, epidermis; Sc, scale.
In combination with paraffin sections and staining (H&E or AB), morphological changes were observed in the skin epidermis. We observed a conspicuous decrease in both the thickness of the skin epidermis and the number of mucus cells in the infected trout skin (Fig. 2A). Moreover, the thickness of the trout skin epidermis reduced significantly at 2 dpi, and it remained relatively stable for up to 14 d (Fig. 2B). In addition, the significantly reduced number of mucus cells in the skin epidermis occurred at 1, 2, and 7 dpi (Fig. 2C).
FIGURE 2.
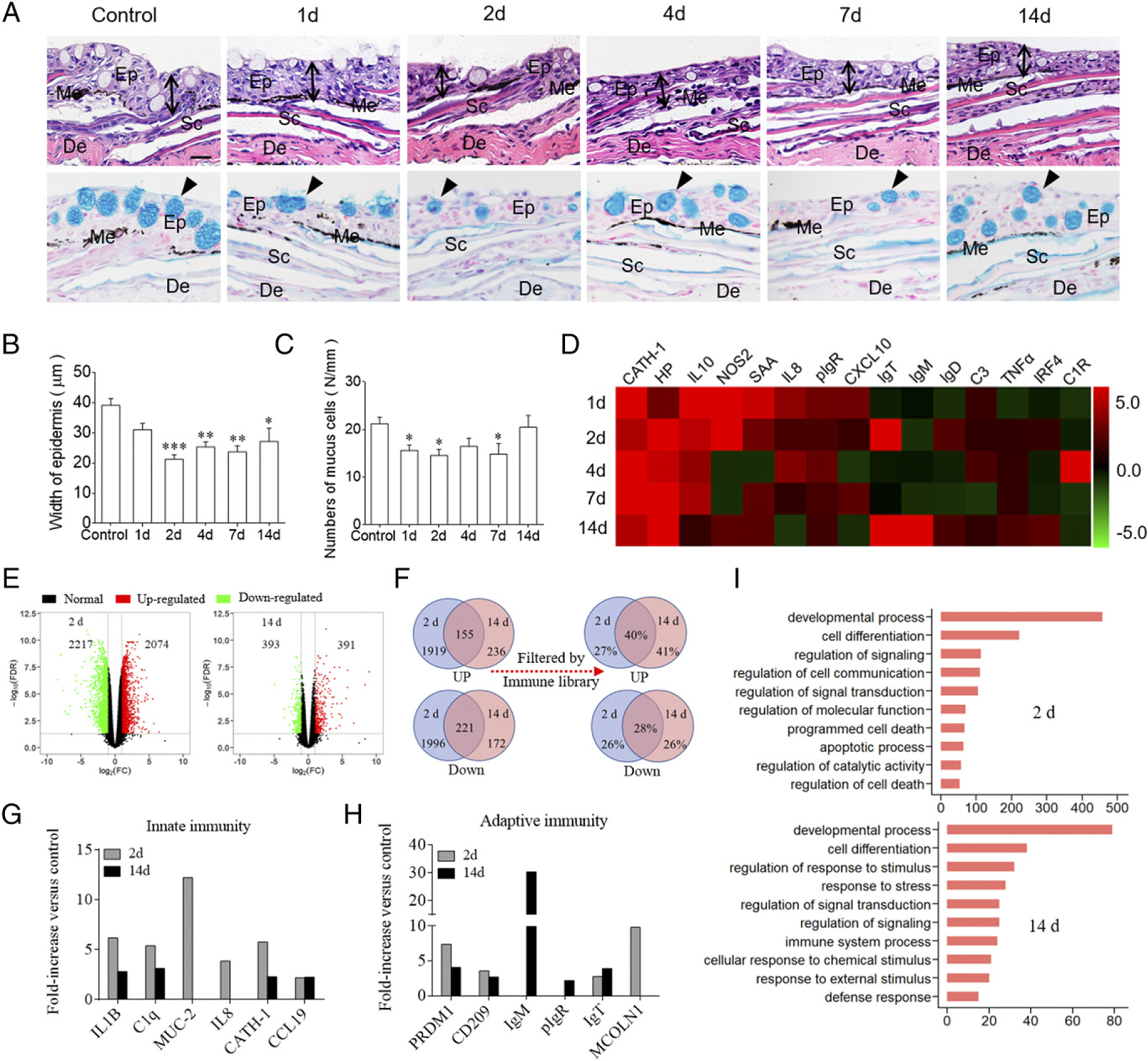
Pathological changes and immune responses in the skin of rainbow trout following F. columnare infection. (A) Histological examination by H&E and AB staining of skin from trout infected with F. columnare at days 1, 2, 4, 7, and 14 dpi, as well as uninfected control fish (n = 6). The black lines with double arrows and black triangle indicate the width of skin epidermis and the mucus-secreting cells, respectively. Scale bar, 20 mm. (B) The width of skin epidermis of control and bacteria-infected fish counted from (A) (upper) (n = 6). (C) The number of mucus cells per millimeter in the epidermis of trout skin from control and bacteria-infected fish counted from (A) (lower) (n = 6). (D) Heat map illustrates results from qRT-PCR of mRNA expression levels for selected immune markers in F. columnare–infected fish versus control fish measured at 1, 2, 4, 7, and 14 dpi in the skin (n = 6). The color value represents fold change. (E) Volcano plot showing the overlap of genes upregulated or downregulated in the skin of rainbow trout at days 2 (left) and 14 (right) after infection with F. columnare versus control fish. Red spots are expression of fold change >2 and FDR <0.05; green spots are expression of fold change <2 and FDR >0.05; black spots mean no difference in expression. (F) Venn diagrams showed the statistical results of DEGs, including upregulated and downregulated genes and the percentage of immune genes after the DEGs filtered by rainbow trout immune genes libraries in the trout skin at days 2 and 14 after infection with F. columnare versus control fish. (G and H) Representative innate (G) and adaptive (H) immune genes modulated by F. columnare infection at 2 and 14 dpi (n = 9). (I) Biological processes that were significantly altered in rainbow trout at days 2 and 14 after infection with F. columnare versus control fish revealed by RNA-seq studies. Fold change differences between control and bacteria-infected samples were calculated using cutoff of 2-fold. Data are representative of three independent experiments (mean ± SEM). *p < 0.05, **p < 0.01, ***p < 0.001, unpaired Student t test. De, dermis; Ep, epidermis; Me, melanophores; Sc, scale.
Bacterial infection elicits strong immune responses in trout skin
Using qRT-PCR, we detected the mRNA expression levels of 15 immune-related genes and cell markers, including cytokines (IL-8, IL-10, TNF-α, and IFN regulatory factor 4, [IRF4]), cathelicidin, complement 3 (C3), pIgR, and IgH genes (IgT, IgM, and IgD) (Fig. 2D; primers were shown in Supplemental Table I). These studies indicated a strong immune response occurred in trout skin (Fig. 2D), spleen (Supplemental Fig. 2A), and head kidney (Supplemental Fig. 2B) after challenge with F. columnare. Interestingly, significantly upregulated immune-related genes (e.g., IL-10, inducible NO synthase [NOS2], and serum amyloid A [SAA]) were detected in trout skin as early as 1 d, whereas in trout head kidney (Supplemental Fig. 2A) and spleen (Supplemental Fig. 2B), immune responses were delayed upon challenge with F. columnare. It is worth noting that day 2 and day 14 were most correlated in terms of the strength of the skin immune responses. Given the time point of expression of the immune-related genes and significant histological responses from 2 dpi to 14 dpi, we chose these two time points for subsequent high-throughput transcriptome sequencing analysis. RNA-Seq libraries were constructed from nine samples that separately represented three groups (control, 2, and 14 d) which were sequenced on an Illumina platform. The National Center for Biotechnology Information Sequence Read Archive accession number for the RNA-seq dataset used in this study is https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA674968&o=acc_s%3Aa. Upon F. columnare challenge, we found significantly modified mRNA expression of 4291 genes (2074 genes upregulated and 2217 genes downregulated) at 2 dpi, and 784 genes (391 genes upregulated and 393 genes downregulated) at 14 dpi, respectively (Fig. 2E). More than 25% of DEGs indicated immune-related genes after filtering by the immune gene library of O. mykiss (Fig. 2F). Importantly, using RNA-seq, we found significantly modified mRNA expression of innate immune genes (Fig. 2G; e.g., ILs, chemokines, antibacterial peptides, and mucins) and adaptive immune genes (Fig. 2H; e.g., genes associated with Ag processing and presentation, and Igs) at both days 2 and 14 among the immune-related genes. Gene ontology analysis showed that these DEGs were enriched for genes involved in developmental processes, cell differentiation, signaling transduction, programmed cell death, and immune response (Fig. 2I).These data indicated that F. columnare invasion could induce intense and long-term innate as well as adaptive immune responses. To validate the DEGs identified by RNA-seq, we chose 12 upregulated and 12 downregulated genes for qRT-PCR confirmation. The result showed a significant correlation of the expression values determined by RNA-seq and qRT-PCR at each time point (Supplemental Fig. 2C–F), suggesting that the RNA-seq results performed with the same accuracy as that of qRT-PCR in the determination of gene expression in vivo.
Pathogen load decreased in fish skin after reinfection
The fish from the infected group (i.e., infected by F. columnare for 28 d) and the survivor group (infected monthly with F. columnare [i.e., three rounds of infection] until 75 d after first infection) (Fig. 3A), as well as the uninfected control group (control) were challenged by bath with a high dose (5 × 106 CFU/ml) of GFP–F. columnare for 4 h at 16°C (Fig. 3B). Approximately 50% of the control fish died at 1–4 d postchallenge, whereas all the fish from infected and survivor groups survived the challenged fish, as they presumably induced protective immunity because of their prior exposures to F. columnare. All fish were euthanized at 14 d postchallenge to evaluate bacterial load in the skin tissues of all three challenged groups (i.e., control-challenge, infected-challenge, and survivor-challenge). Histopathology of the skin of both infected and survivor fish showed no significant changes in the epidermis compared with the control fish (Fig. 3C, 3D), and the number of mucus-secreting cells also recovered to normal levels (Fig. 3C, 3E). Using fluorescence microscopy and qRT-PCR, the infected-challenge and survivor-challenge groups had markedly limited green fluorescence signal and lower 16S rRNA expression of F. columnare compared with those of the uninfected control-challenged fish (Fig. 3F, 3G). These data strongly suggest that pre-exposure to F. columnare protects trout skin from the bacterial reinfection, indicating that mucosal adaptive immunity had been induced in trout skin to fight pathogen invasion.
FIGURE 3.
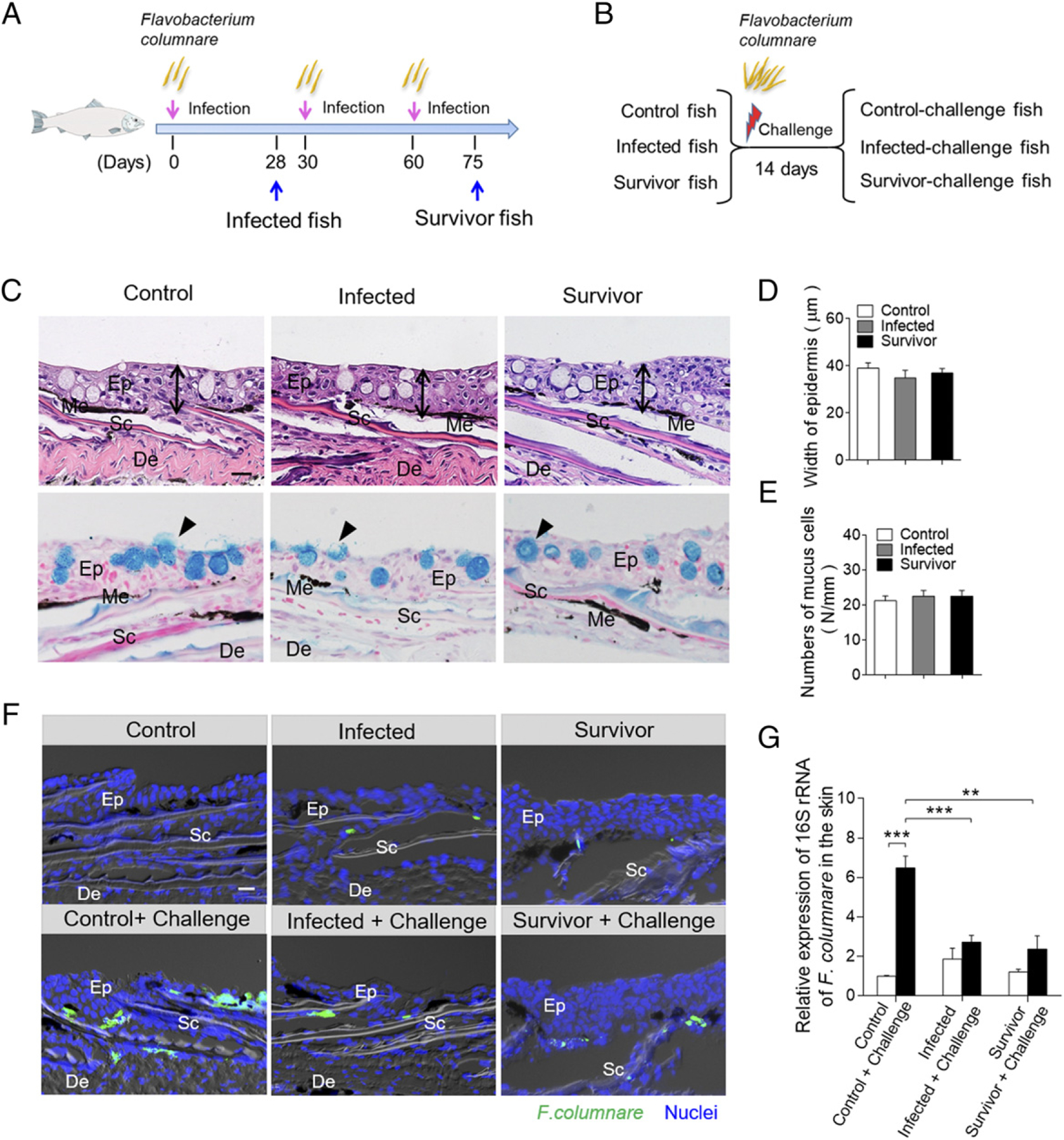
Bacterial load of the skin from survivor fish upon F. columnare reinfection. (A) Scheme of the strategy of infection with F. columnare to obtain infected and survivor group fish. (B) The fish from control (previously uninfected), infected, and survivor fish were challenged with a high dose of F. columnare. (C) Histological examination by H&E and AB staining of skin from fish infected for 28 d and survivor fish (n = 6). The black lines with double arrows and black triangle indicate the width of skin epidermis and the mucus-secreting cells, respectively. Scale bar, 20 μm. (D) The width of skin epidermis of control fish, infected fish, and survivor fish counted from (A) (upper) (n = 6). (E) The number of mucus cells per millimeter in the epidermis of trout skin from control, infected, and survivor fish counted from (A) (lower) (n = 6). (F) Differential interference contrast images of fluorescence analysis showed the GFP–F. columnare location and load in the skin from control and control-challenge fish (left), 28-d-infected and 28-d–infected-challenge fish (middle), and survivor fish and survivor-challenge fish (right). These challenge fish were sampled at 4 d after reinfection with GFP–F. columnare (n = 6). Scale bar, 20 μm. (G) The relative expression of 16S RNA of F. columnare in the trout skin tissue of control, control-challenge, infected, infected-challenge, survivor, and survivor-challenge group were measured by qRT-PCR (n = 6 fish per group). Data are representative of at least three independent experiments (mean ± SEM). **p < 0.01, ***p < 0.001, unpaired Student t test. De, dermis; Ep, epidermis; Me, melanophores; Sc, scale.
B cell responses and local proliferation in trout skin after bacterial infection
Our previous studies have demonstrated that IgT play a specialized role in skin mucosal immunity against an aquatic parasite (25). The decreases of F. columnare load in infected and survivor trout skin after rechallenge with a higher dose of the same pathogen suggest whether IgT plays an important role in mucosal adaptive immune responses against this bacterial pathogen. To evaluate the role of skin mucosal Igs in response to bacterial challenge, we detected Ig and B cell responses in trout skin infected with F. columnare by bath challenge. Using immunofluorescence analysis, we found very few IgT+ B cells as well as IgM+ B cells in the skin of control fish (Fig. 4A; isotype control Ab staining is shown in Supplemental Fig. 3A). However, in fish infected after 28 d with F. columnare, a moderate (~2-fold) but significant increase in the number of IgT+ B cells was detected in the skin (Fig. 4A, 4B). Importantly, when compared with control fish, we observed a notable accumulation of IgT+ B cells in the skin of survivor fish. In stark contrast to this, there was no change in the number of IgM+ B cells, whether in infected or survivor fish groups, when compared with that of control fish (Fig. 4A, 4B).
FIGURE 4.

Accumulation and proliferation of IgT+ B cells in the skin of rainbow trout following F. columnare infection. (A) Differential interference contrast images of immunofluorescence staining on trout skin paraffinic sections from control fish, infected, and survivor fish stained for IgT (green) and IgM (red); nuclei are stained with DAPI (blue). The top and bottom lines are different repetitions. Scale bar, 20 μm. (B) The number of IgT+ and IgM+ B cells in skin paraffin sections of control, infected, and survivor fish infected with F. columnare are counted (n = 12; original magnification ×20). *p < 0.05, ***p < 0.001, one-way ANOVA with Bonferroni correction. (C) Representative flow cytometry dot plot showing proliferation of IgT+ B cells and IgM+ B cells in skin leukocytes of control and survivor fish (n = 12). (D) Percentage of EdU+ cells from the total skin IgT+ or IgM+ B cell populations in control and survivor fish (n = 12). (E) Representative flow cytometry dot plot showing proliferation of IgT+ B cells and IgM+ B cells in head kidney leukocytes of control and survivor fish. (F) Percentage of EdU+ cells from the total head kidney IgT+ or IgM+ B cell populations in control or survivor fish (n = 12). Data are representative of at least three independent experiments (mean ± SEM). **p < 0.01, ***p < 0.001 by unpaired Student t test in (D) and (F). De, dermis; Ep, epidermis; Sc, scale.
To confirm whether the substantial increase of IgT+ B cells in the survivor fish skin was due to proliferative IgT+ B cell in local skin mucosa or to the migration of these cells from other immune tissues, such as systemic lymphoid organs and other MALT, the in vivo proliferation of B cells in control and survivor fish was detected by caudal vein injection of these fish with fluorescent EdU. Using flow analysis, the percentage of proliferating IgT+ B cells (EdU+ IgT+ B cells) showed a significant increase in the skin of survivor fish (~7.20%) when compared with control fish (~3.45%), whereas there was no change in the percentage of IgM+ B cells (EdU+ IgM+ B cells) between control and survivor fish (Fig. 4C, 4D). Interestingly, we found a notable rise of EdU+ IgM+ B cells in the head kidney of survivor fish (~6.23%) compared with control fish (~2.89%), whereas no differences in the proliferation of IgT+ B cells were detected (Fig. 4E, 4F).
Bacteria-specific Ig responses in trout skin
To further study the skin Ig responses in trout skin after F. columnare infection, we performed SDS-PAGE and Western blot analysis. Along with the significant increase of IgT+ B cells observed in the skin of infected and survivor fish, the IgT protein level in the skin mucus of the both groups were ~8- and ~15-fold higher, respectively, when compared with that of control fish, whereas the levels of IgM and IgD did not differ (Fig. 5A). In serum, ~2-fold and ~5-fold increases of IgT concentration were detected in infected and survivor fish, respectively. Conversely, the concentration of IgM in serum was much higher than that of IgT and had ~4- and 3-fold increases in infected and survivor fish, respectively, when compared with those of control fish (Fig. 5E). In contrast, the IgD protein concentration did not change significantly in either the skin mucus or serum of the same fish groups (Fig. 5A, 5E).
FIGURE 5.

Ig responses in the skin mucus and serum from infected and surviving fish. (A) Concentration of IgT, IgM, and IgD in skin mucus from control, infected, and survivor fish (n = 12). (B) Western blot analysis of IgT-, IgM- and IgD-specific binding to F. columnare in skin mucus (dilution 1:2) from infected and survivor fish. (C and D) IgT-, IgM- and IgD-specific binding to F. columnare in dilutions of skin mucus from infected (C) and survivor (D) fish evaluated by densitometric analysis of immunoblots and presented as relative values to those of control fish (n = 12–18). (E) Concentration of IgT, IgM, and IgD in serum from control, infected, and survivor fish (n = 12). (F) Western blot analysis of IgT-, IgM- and IgD-specific binding to F. columnare in serum (dilution 1:10) from infected and survivor fish. (G and H) IgT-, IgM- and IgD-specific binding to F. columnare in dilutions of serum from infected (G) and survivor (H) fish evaluated by densitometric analysis of immunoblots and presented as relative values to those of control fish (n = 12–18). Data are representative of at least three independent experiments (mean ± SEM). *p < 0.05, **p < 0.01, ***p < 0.001 by unpaired Student t test.
The results of large increases in IgT+ B cells and IgT protein levels in the skin of infected and survivor fish suggested the generation of bacteria-specific IgT immune responses. To verify this hypothesis, using a pull-down assay, we measured the capacity of skin mucus Igs binding to F. columnare. Compared with control fish, we detected significantly higher (~5-fold) bacteria-specific IgT binding up to 1:40 diluted skin mucus of infected and survivor fish, respectively (Fig. 5B–D), although bacteria-specific IgT binding was detected in serum with only in the 1:10 dilution of the survivor group (Fig. 5F, 5H). In contrast, ~3-fold and ~2.4-fold higher bacteria-specific IgM binding was observed in up to 1:100 serum dilutions both from infected and survivor fish, respectively (Fig. 5G, 5H). However, bacteria-specific IgD binding is hardly detected in either the skin mucus or the serum from infected and survivor fish (Fig. 5A–H).
To further test the local production of pathogen-specific Igs in trout mucosal and systemic tissues, respectively, we analyzed bacteria-specific Ig titers of culture medium derived from cultured skin, head kidney, and spleen explants of control and survivor fish (Fig. 6A–F). We detected bacteria-specific IgT binding up to the 1:100 dilution of medium of cultured skin explants from survivor fish, whereas very low bacteria-specific IgM responses (Fig. 6A, 6D) were detected (only in the 1:2 dilution in the same medium). In contrast, dominant bacteria-specific IgM binding (up to the 1:10 dilution) was observed in the medium of survivor head kidney and spleen explants, whereas similar (up to the 1:10 dilution) and lower (up to the 1:2 dilution) bacteria-specific IgT responses were detected in the medium of survivor head kidney and spleen explants, respectively (Fig. 6B, 6C, 6E, 6F). Interestingly, we could not detect any bacteria-specific IgD binding in either the mucosal (skin) or systemic (head kidney and spleen) tissue explants from survivor fish (Fig. 6A–F).
FIGURE 6.
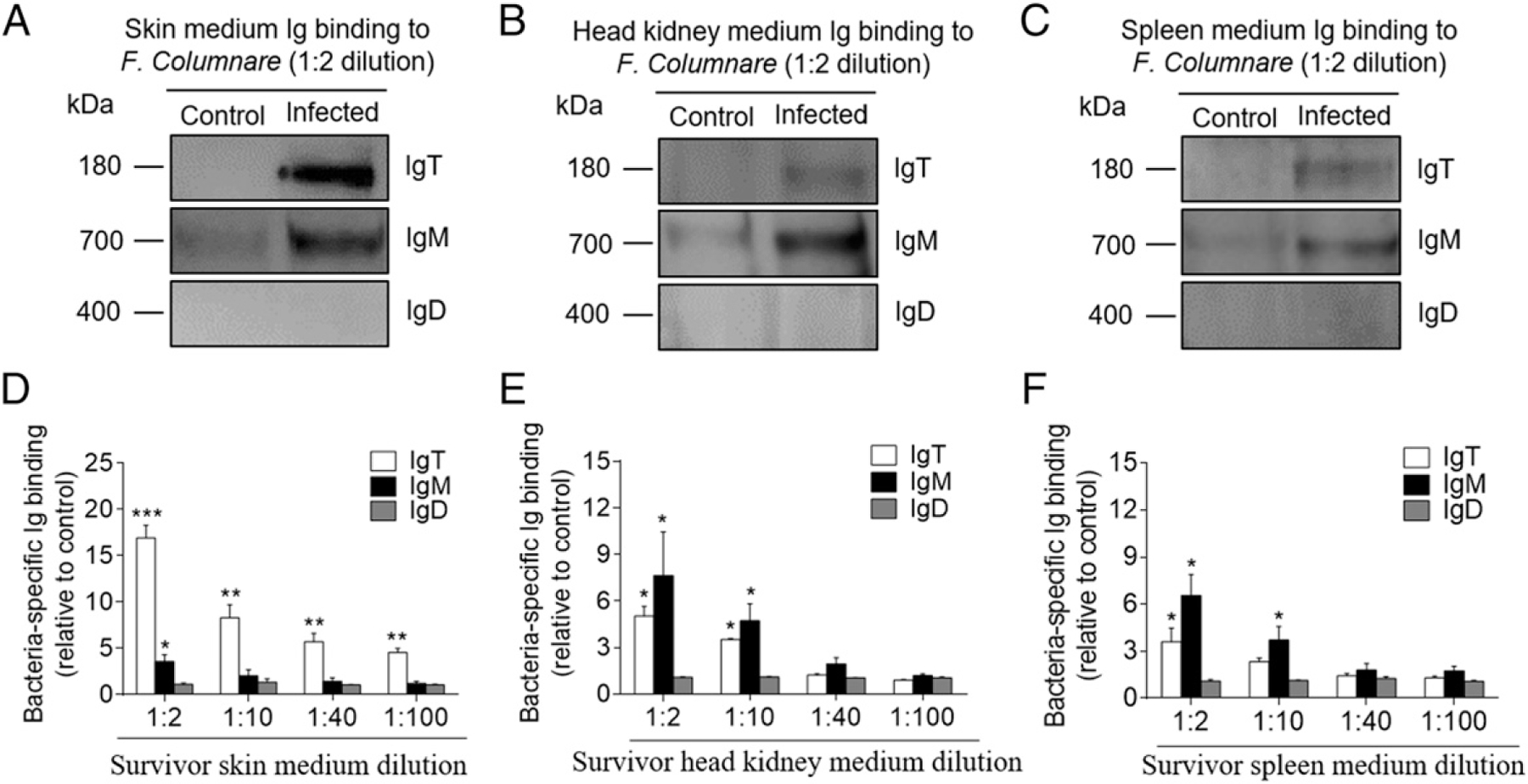
Local IgT-, IgM- and IgD-specific responses in skin explants of survivor fish. (A–C) Skin, head kidney, and spleen explants (~50 mg each) from control and survivor fish were cultured in medium (400 μl) for 7 d. Immunoblot analysis of IgT-, IgM- and IgD-specific binding to F. columnare in the culture medium of skin (A), head kidney (B), and spleen (C) (dilution 1:2) from control and survivor fish. (D–F) IgT-, IgM- and IgD-specific binding to F. columnare in dilutions of culture medium from skin (D), head kidney (E), and spleen (F) from control and survivor fish (n = 12 per group) measured by densitometric analysis of immunoblots and presented as relative values to those of control fish. Data are representative of at least three independent experiments (mean ± SEM). *p < 0.05, **p < 0.01, ***p < 0.001 by unpaired Student t test.
pIgR in trout skin after bacterial infection
Previous studies have indicated that rainbow trout pIgR may take part in the skin mucosal immune responses to parasitic infection by mediating the transepithelial transport of secretory Ig (9). Thus, we hypothesized that pIgR might also mediate the transport of IgT to the skin mucus during immune responses to F. columnare. In this study, using immunofluorescence analysis, we observed that some of the skin epithelial cells from control fish stained by the anti-trout pIgR Ab (Fig. 7A; isotype control Ab staining is shown in Supplemental Fig. 3B). Importantly, the number of pIgR+ cells significantly increased in the epidermis of infected (~1.6-fold) and survivor fish (~2.4-fold) (Fig. 7A, 7B) when compared with those of control fish. Using qRT-PCR, the expression of pIgR was found to be significantly upregulated in the skin of infected (~3-fold) and survivor fish (~6.6-fold), respectively, when compared with that of control fish (Fig. 7C). Moreover, the concentration of pIgR in skin mucus after bacterial infection were also detected and calculated by Western blot analysis. Our results showed that the relative fold changes of pIgR in infected and survivor fish were significantly increased when compared with that of control fish (~1.7-fold for infected group and ~3.4-fold for survivor group; Fig. 7C).
FIGURE 7.
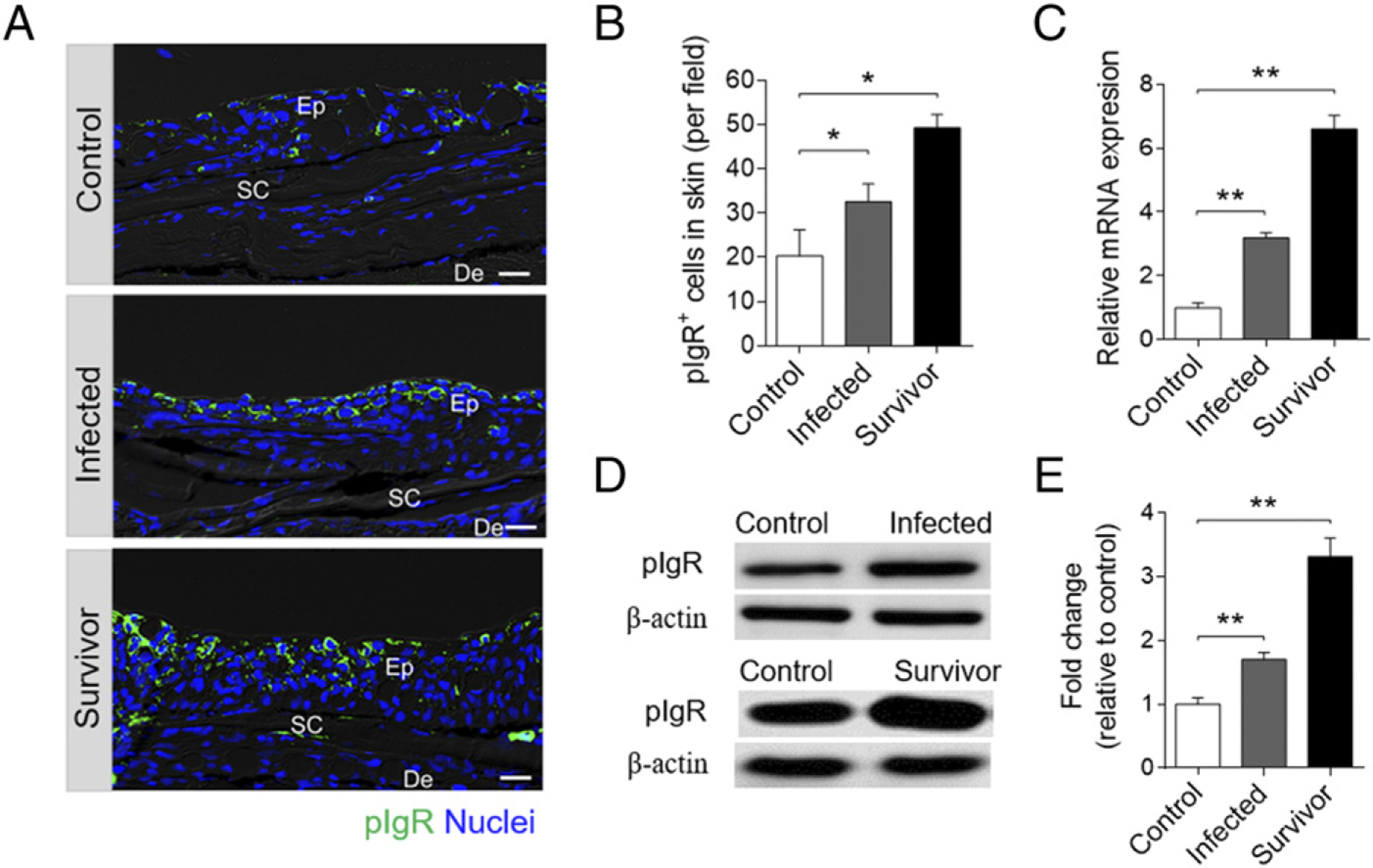
Accumulation of pIgR and pIgR+ cells in the skin of rainbow trout infected with F. columnare. (A) Differential interference contrast images of immunofluorescence staining on trout skin paraffinic sections from control fish, infected, and survival fish stained for pIgR (green) and nuclei with DAPI (blue) (n = 12). Scale bar, 20 μm. (B) The number of pIgR+ cells in skin paraffin sections of control, infected, and survivor fish are counted (n = 12; original magnification ×20). (C) Relative mRNA expression of pIgR in the skin of control fish, infected, and survivor fish were detected by qRT-PCR (n = 6). (D) Immunoblot analysis of pIgR concentration in skin mucus from control, infected, and survivor fish following F. columnare infection. (E) Relative concentration of pIgR from infected and survivor trout skin mucus to that of control fish (n = 12). Data are representative of at least three independent experiments (mean ± SEM). *p < 0.05, **p < 0.01 by unpaired Student t test. De, dermis; Ep, epidermis; Sc, scale.
Discussion
Fish skin is the outermost organ of the organism’s body, which is continuously exposed to a wide array of potentially harmful insults in the aquatic environment. This constant exposure to microbes has led to the evolution of a mucosal immune network in the teleost SALT that ensures that adequate immune responses are mounted upon antigenic challenge while maintaining overall tissue and microbiota homeostasis (16, 48–50). However, until now, little was known with regards to the evolution of skin mucosal B cell and Ab responses to bacterial pathogens in early vertebrates. Most of the literature in that area for teleost fish predates the discovery of IgT as the key mucosal Ig in these species (10). In that regard, some studies in several teleosts have reported low levels of pathogen-specific IgM titers upon bacterial infection (51, 52). Moreover, whether the skin mucosa behaves as an inductive or effector site of mucosal Ig responses has never been investigated. In this study, we report for the first time, to our knowledge, that in a teleost, infection with F. columnare, a widespread fish bacterial pathogen, can elicit local skin proliferative IgT+ B cell responses as well as significant bacteria-specific mucosal IgT titers.
Columnaris disease, caused by the Gram-negative bacterium F. columnare, severely impacts the global production of many fish species (34, 53). As a mucosal bacterial pathogen, F. columnare infection results in skin epithelial erosion, gill necrosis, and ulcers, with a high degree of mortality (34, 53). To evaluate mucosal immune responses to F. columnare, we exposed fish to this pathogen following a well-established waterborne challenge model with a novel fluorescent (GFP) strain of F. columnare (39). Importantly, GFP–F. columnare exhibited similar growth and virulence properties as the wild-type bacteria, and thus, this newly constructed GFP–F. columnare represents a new reagent that is expected to significantly advance the study of host–F. columnare interactions. At 4 dpi, clinical signs were detected in the bacteria-infected rainbow trout, and qRT-PCR and immunofluorescence further proved the successful invasion of GFP–F. columnare into rainbow trout. It is worth pointing out that the use of our unique GFP–F. columnare strain was extremely instrumental in assessing the capacity of this pathogen to intrude into the skin epidermis of fish under the experimental conditions tested. Importantly, upon infection, we observed significantly decreased numbers of mucus cells and thickness of the epidermis in the trout skin. Thus, our results suggest that the F. columnare successful invasion relies on destroying the structure of the epidermal layer of fish skin, which is in line with what has been shown in previous studies (36, 54, 55). Considering that the taxonomy of F. columnare is complex and likely composed of multiple species with different host association (56, 57), it is possible that the pathology induced by F. columnare G4 in rainbow trout is different of that induced by other F. columnare strains that are isolated from colder temperatures. Thus, further experiments are warranted to evaluate potential differences in the pathologies induced by different strains of F. columnare derived from hot and cold temperature waters. In agreement with the noteworthy histopathological changes in trout skin, we found that 15 immune-related genes were significantly upregulated in trout skin as early as 1 dpi when compared with those in the head kidney and spleen, which indicates that the immune responses in trout skin are activated significantly earlier than in systemic lymphoid tissues, thus supporting further the notion of the skin acting as the first line of defense against bacterial infections in fish. Importantly, both innate and adaptive immune molecules were induced in the skin upon F. columnare infection, including antibacterial peptides, cytokines, chemokines, complement factors, and Igs. Thus, our results provide a general picture of the predominant immune responses that take place in the trout skin after F. columnare infection. Of these responses, it is worth pointing out that we found increased transcript levels of cathelicidin 1 and proinflammatory cytokines, such as IL-8 and TNF-α, in trout skin immediately after F. columnare infection. Moreover, the anti-inflammatory cytokine IL-10 was also induced after bacterial infection. Combined with previous studies (58–61), our results suggest that F. columnare infection induces a significant inflammatory reaction as well as strong immune responses in the fish skin epidermis early upon infection. In contrast, these types of responses occur in the dermis of mammals (16, 62), probably reflecting the different physiological needs of fish and mammalian skin tissues with regards to the water or terrestrial environments where these species live.
The use of GFP–F. columnare enabled us to find that the skin epithelium of trout previously exposed and thus immune to F. columnare, cannot get intruded upon re-exposure to this pathogen. These findings suggested that local humoral immunity might be elicited in trout SALT, thus contributing to protection upon reinfection. In this study, we first showed significant increases in the concentration of IgT but not IgM in the skin mucus of infected and survivor fish exposed to F. columnare. These data correlated with the large accumulation of IgT+ but not IgM+ B cells in the skin epidermis of the same animals, despite of the fact that a high IgM transcript level was detected in skin at 14 dpi. The different performance between IgM mRNA expression level and protein concentration or B cells numbers may be caused by many factors. For instance, there were many complicated and varied posttranscriptional mechanisms involved in turning mRNA into protein that were not yet sufficiently well defined to be able to compute protein concentrations from mRNA. In this regard, not all of the IgM protein translated by mRNA in cells was secreted and transported by pIgR into skin mucus upon bacterial infection. Additionally, proteins might differ substantially in their in vivo half-lives (63). Interestingly, the changes of Igs protein level and B cells paralleled those obtained in a previous study, in which fish surviving from a parasite infection exhibited large accumulations of IgT and IgT+ B cells in the skin epidermis (9). It is worth mentioning that upon bacterial infection, several mammalian MALTs have been shown to have similar significant accumulations of IgA+ B cells (64–68). In humans, several studies have described a correlation between resistance to Vibrio cholerae infection and high specific sIgA titers (69, 70). Importantly, to our knowledge, our findings show for the first time the detection of bacteria-specific IgT titers in fish, mainly in the skin mucus and to a much lesser degree in the serum of survivor fish. Although the kinetics of IgM production in the skin following bacterial infection had been investigated in some species (71–74), until now, the role of sIgT in skin bacterial infections had not been addressed, probably because of the lack of specific Ab reagents recognizing IgT (75, 76). Moreover, we hardly found any bacteria-specific IgM titers in skin mucus of infected and survivor fish. They were instead detected almost exclusively in the serum in very high levels. Therefore, we can preliminarily conclude that skin IgT and IgT+ B cell responses against F. columnare are specifically confined in its mucosa, whereas IgM responses are overwhelmingly found in the serum and thus of systemic nature. Interestingly, previous studies in trout with a similar bacterial pathogen (F. psychrophilum) have shown that bath exposure of trout to this pathogen could not elicit bacteria-specific IgM responses in the trout skin mucus (24), as we have also found for F. columnare. Thus, the authors speculated that perhaps IgT is the Ig involved in skin responses against F. psychrophilum, a hypothesis that seems to be supported by our data, although that hypothesis remains to be investigated.
The accumulation of IgT+ B cells and high bacteria-specific IgT titers observed in the skin mucosa from infected and survivor fish led us to hypothesize that the local proliferation and induction of IgT+ B cells and IgT Ab responses, respectively, upon bacterial infection. Confirming this hypothesis, we found strong local proliferative IgT+ B cell responses in the teleost skin but not in the head kidney of the same fish, suggesting that the accumulation of IgT+ B cells in their skin is due to local proliferation rather than migration of these cells from the systemic immune organs. We cannot rule out, however, the possibility that these IgT+ B cells are induced in other mucosal surfaces and, upon migration to the SALT, start proliferating. Future work will have to address the aforementioned possibility. Critically, the production of significant titers of bacteria-specific IgT in skin explant cultures confirmed that these IgT Abs are produced locally and suggest the presence of specific plasma cells in the local skin mucosa. Overall, considering that F. columnare first invades and stays in the skin epidermis, it is unlikely that the inductive site is a systemic lymphoid tissue, thus our results imply that trout SALT is an inductive site of the observed IgT mucosal immune responses. We have previously shown similar results in the mucosa of the gill and nose when exposed to pathogenic challenge (7, 8). It is also worth noting that similar results have been shown in mucosal inductive sites of some mammalian MALTs, including the gut-associated lymphoid tissue, nasopharynx-associated lymphoid tissue, and bronchus-associated lymphoid tissue, in which a local mucosal response governed by a mucosal Ig (sIgA) was strongly induced upon bacterial infection, and in contrast, the systemic response is dominated by IgG/IgM (77–79).
A key feature of pIgR is the mediation of the transepithelial transport of secretory Igs into the mucosal surfaces in both mammals and teleost (2, 80). Previous studies have shown that the putative trout secretory component of pIgR is associated with mucosal IgT and IgM in the gut (10), skin (9), gills (7), and nasal mucus (8), contributing to the Ig response to parasite infection. In this study, we found that trout pIgR was mainly expressed in the epithelial layer of the skin from control, infected, and survivor fish. Critically, in this study, we see a significant increase in the numbers of pIgR+ cells and transcript levels of pIgR in the skin from bacteria-infected trout when compared with that of control fish. Interestingly, similar results have been reported in mammals; for example, the expression of pIgR was found upregulated in germ-free mice implanted with Bacteroides thetaiotaomicron (81). In addition, pIgR expression can be upregulated in HT-29 cells by reovirus (dsRNA) (82) and bacteria (Enterobacteriaceae) with LPS on the surface (83). Thus, our results indicate such increases in the levels of pIgR upon pathogenic challenge is an evolutionarily conserved feature from fish to mammals that probably has the role of increasing the transport rate of mucosal Igs to mucosal surfaces upon pathogenic insult.
In conclusion, to our knowledge, our study provides the first evidence of local B cell proliferation and IgT secretion in teleost skin, which suggests that teleost SALT is an inductive site of mucosal B cell and Ig responses. Based on our studies, we propose a model in which, upon bacterial infection, epidermis damage occur, leading to an upregulation of immune-related genes in the skin as well as to processes of IgT+ B cell activation and proliferation within the SALT, resulting in the production of bacteria-specific IgT (Fig. 8). Thus, from an evolutionary viewpoint, our results reinforce the formerly stated view (84) that, regardless of their phylogenetic origin and tissue localization, specialized mucosal Igs (i.e., IgA and IgT) of all vertebrate MALT-containing surfaces function under the guidance of primordially conserved principles. Moreover, our findings in this phylogenetically primitive vertebrate strongly suggest a universal requirement for vertebrate MALT-containing mucosal surfaces to use a dedicated Ig for the maintenance of homeostasis. Additionally, because many fish pathogens enter the host through the skin mucosa, the knowledge derived from our findings also has special relevance from a more practical perspective, as it may lead to the development of fish vaccines that can effectively induce bacteria-specific IgT immune responses in skin.
FIGURE 8.
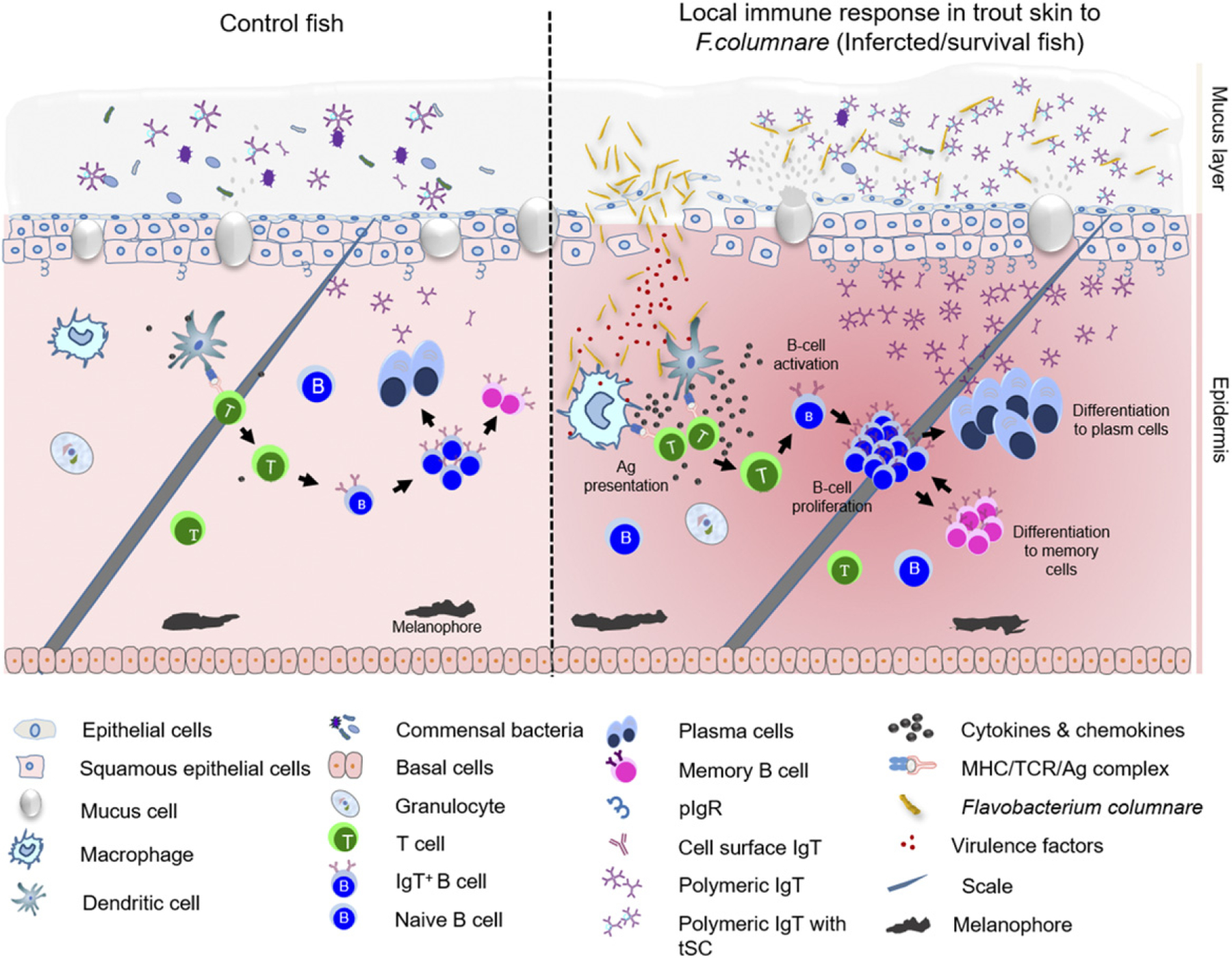
Proposed model of local IgT and IgT+ B cell induction in the skin after F. columnare infection. The presumed structure of skin displaying mucus layer, epithelial layer, and dermis. The proposed model contains two partitions: control (left) and infected/survivor (right). Induction of local IgT responses in the trout skin based on our findings. On the left is the scheme of a typical skin structure in control (naive) teleost fish. The number of IgT+ B cells in control fish skin is low. IgT are produced by IgT-secreting B cells and transported from the epithelium into the skin mucus layer via pIgR. The sIgT coats the majority of microbiota in the skin surface. In the skin epidermis, abundant mucus cells are present, and their production contains antibacterial molecules to protect host against pathogens. Upon F. columnare invasion, their Ag can be taken up by APCs and presented to naive CD4+ T cells. IgT+ B cells are then activated by Ag-specific CD4+ T cells and start to proliferate locally and differentiate into plasma cells in skin epithelial layer. These differentiated IgT-secreting cells can produce large amounts of pathogen-specific IgT, which can be transported by pIgR into skin mucus layer, where those pathogen-specific IgT can specifically bind to the bacteria F. columnare. Alternatively, some IgT+ plasma cells may differentiate further into memory IgT+ B cells. When F. columnare invade the host again, the memory IgT+ B cells would directly proliferate and differentiate into plasma cells and then rapidly produce specific IgT to binding F. columnare.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Key Research and Development Program of China (2018YFD0900503 and 2018YFD0900400) to Z.X. and the National Natural Science Foundation of China (31873045 and 32073001) to Z.X. The anti–rainbow trout IgD mAb was provided by the U.S. Veterinary Immune Reagent Network, which was funded by the U.S. Department of Agriculture, National Institute of Food and Agriculture (Award 2010-65121-20649), National Science Foundation Grant NSF-IOS-1457282 to J.O.S., and the U.S. Department of Agriculture Grants USDA-NIFA-2016-09400 and USDA-NRI-2013-01107 to J.O.S. This work was also supported by National Institutes of Health Grant 2R01GM085207-09 to J.O.S. and Japan Society for the Promotion of Science KAKENHI Grants JP19K21158 and JP20K06230 to F.T.
Abbreviations used in this article
- AB
Alcian blue
- DEG
differentially expressed gene
- dpi
day postinfection
- EdU
5-ethynyl-20-deoxyuridine
- FDR
false discovery rate
- GFP–F. columnare
GFP-expressing F. columnare
- MALT
mucosal-associated lymphoid tissue
- pAb
polyclonal Ab
- pIgR
poly-Ig receptor
- qRT-PCR
quantitative real-time PCR
- RNA-seq
RNA-sequencing
- SALT
skin-associated lymphoid tissue
- sIgA
secreted IgA
- sIgT
secreted IgT
Footnotes
The RNA sequences presented in this article have been submitted to the National Center for Biotechnology Information Sequence Read Archive (https://www.ncbi.nlm.nih.gov) under accession number PRJNA674968.
The online version of this article contains supplemental material.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Fillatreau S, Six A, Magadan S, Castro R, Sunyer JO, and Boudinot P. 2013. The astonishing diversity of Ig classes and B cell repertoires in teleost fish. Front. Immunol. 4: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salinas I, Zhang YA, and Sunyer JO. 2011. Mucosal immunoglobulins and B cells of teleost fish. Dev. Comp. Immunol. 35: 1346–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zapata A, Diez B, Cejalvo T, Gutiérrez-de Frías C, and Cortés A. 2006. Ontogeny of the immune system of fish. Fish Shellfish Immunol. 20: 126–136. [DOI] [PubMed] [Google Scholar]
- 4.Bromage ES, Kaattari IM, Zwollo P, and Kaattari SL. 2004. Plasmablast and plasma cell production and distribution in trout immune tissues. J. Immunol. 173: 7317–7323. [DOI] [PubMed] [Google Scholar]
- 5.Solem ST, and Stenvik J. 2006. Antibody repertoire development in teleosts--a review with emphasis on salmonids and Gadus morhua L. Dev. Comp. Immunol. 30: 57–76. [DOI] [PubMed] [Google Scholar]
- 6.Perdiguero P, Martín-Martín A, Benedicenti O, Díaz-Rosales P, Morel E, Muñoz-Atienza E, García-Flores M, Simón R, Soleto I, Cerutti A, and Tafalla C. 2019. Teleost IgD+IgM− B cells mount clonally expanded and mildly mutated intestinal IgD responses in the absence of lymphoid follicles. Cell Rep. 29: 4223–4235.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Z, Takizawa F, Parra D, Gómez D, von Gersdorff Jørgensen L, LaPatra SE, and Sunyer JO. 2016. Mucosal immunoglobulins at respiratory surfaces mark an ancient association that predates the emergence of tetrapods. Nat. Commun. 7: 10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu YY, Kong W, Yin YX, Dong F, Huang ZY, Yin GM, Dong S, Salinas I, Zhang YA, and Xu Z. 2018. Mucosal immunoglobulins protect the olfactory organ of teleost fish against parasitic infection. PLoS Pathog. 14: e1007251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu Z, Parra D, Gómez D, Salinas I, Zhang YA, von Gersdorff Jørgensen L, Heinecke RD, Buchmann K, LaPatra S, and Sunyer JO. 2013. Teleost skin, an ancient mucosal surface that elicits gut-like immune responses. Proc. Natl. Acad. Sci. USA 110: 13097–13102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang YA, Salinas I, Li J, Parra D, Bjork S, Xu Z, LaPatra SE, Bartholomew J, and Sunyer JO. 2010. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat. Immunol. 11: 827–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu YY, Kong WG, Xu HY, Huang ZY, Zhang XT, Ding LG, Dong S, Yin GM, Dong F, Yu W, et al. 2019. Convergent evolution of mucosal immune responses at the buccal cavity of teleost fish. iScience 19: 821–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong WG, Yu YY, Dong S, Huang ZY, Ding LG, Cao JF, Dong F, Zhang XT, Liu X, Xu HY, et al. 2019. Pharyngeal immunity in early vertebrates provides functional and evolutionary insight into mucosal homeostasis. J. Immunol. 203: 3054–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Z, Takizawa F, Casadei E, Shibasaki Y, Ding Y, Sauters TJC, Yu Y, Salinas I, and Sunyer JO. 2020. Specialization of mucosal immunoglobulins in pathogen control and microbiota homeostasis occurred early in vertebrate evolution. Sci. Immunol. 5: eaay3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Taniguchi T, Takita T, and Ali AB. 2003. A study on the epidermal structure of Periophthalmodon and Periophthalmus mudskippers with reference to their terrestrial adaptation. Ichthyol. Res. 50: 310–317. [Google Scholar]
- 15.Streilein JW 1985. Circuits and signals of the skin-associated lymphoid tissues (SALT). J. Invest. Dermatol. 85(Suppl): 10s–13s. [DOI] [PubMed] [Google Scholar]
- 16.Kabashima K, Honda T, Ginhoux F, and Egawa G. 2019. The immunological anatomy of the skin. Nat. Rev. Immunol. 19: 19–30. [DOI] [PubMed] [Google Scholar]
- 17.Tong PL, Roediger B, Kolesnikoff N, Biro M, Tay SS, Jain R, Shaw LE, Grimbaldeston MA, and Weninger W. 2015. The skin immune atlas: three-dimensional analysis of cutaneous leukocyte subsets by multiphoton microscopy. J. Invest. Dermatol. 135: 84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ángeles Esteban M, and Cerezuela R. 2015. Fish mucosal immunity: skin. In Mucosal Health in Aquaculture. Beck BH, and Peatman E, eds. Academic Press, San Diego, CA, p. 67–92. [Google Scholar]
- 19.Ángeles Esteban M 2012. An overview of the immunological defenses in fish skin. ISRN Immunol. 2012: 1–29. [Google Scholar]
- 20.Brandtzaeg P 2009. Mucosal immunity: induction, dissemination, and effector functions. Scand. J. Immunol. 70: 505–515. [DOI] [PubMed] [Google Scholar]
- 21.Wright PF 2011. Inductive/effector mechanisms for humoral immunity at mucosal sites. Am. J. Reprod. Immunol. 65: 248–252. [DOI] [PubMed] [Google Scholar]
- 22.Saitoh-Inagawa W, Hiroi T, Yanagita M, Iijima H, Uchio E, Ohno S, Aoki K, and Kiyono H. 2000. Unique characteristics of lacrimal glands as a part of mucosal immune network: high frequency of IgA-committed B-1 cells and NK1.1+ alphabeta T cells. Invest. Ophthalmol. Vis. Sci. 41: 138–144. [PubMed] [Google Scholar]
- 23.Brandtzaeg P 2013. Secretory immunity with special reference to the oral cavity. J. Oral Microbiol. 5: 20401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makesh M, Sudheesh PS, and Cain KD. 2015. Systemic and mucosal immune response of rainbow trout to immunization with an attenuated Flavobacterium psychrophilum vaccine strain by different routes. Fish Shellfish Immunol. 44: 156–163. [DOI] [PubMed] [Google Scholar]
- 25.Bernardet J-F, Segers P, Vancanneyt M, Berthe F, Kersters K, and Vandamme P. 1996. Cutting a Gordian knot: emended classification and description of the genus Flavobacterium, emended description of the family Flavobacteriaceae, and proposal of Flavobacterium hydatis nom. nov. (basonym, Cytophaga aquatilis Strohl and Tait 1978). Int. J. Syst. Bacteriol. 46: 128–148. [Google Scholar]
- 26.Plumb J 1999. Health Maintenance and Culture of Microbial Diseases of Cultured Fishes. Iowa State University Press, Ames, IA. [Google Scholar]
- 27.Bernardet J-F, and Bowman J. 2006. The prokaryotes: a handbook on the biology of bacteria. Proteobacteria: delta and Epsilon subclasses. Deeply Rooted Bacteria 7: 481–532. [Google Scholar]
- 28.Decostere A, Haesebrouck F, and Devriese LA. 1998. Characterization of four Flavobacterium columnare (Flexibacter columnaris) strains isolated from tropical fish. Vet. Microbiol. 62: 35–45. [DOI] [PubMed] [Google Scholar]
- 29.Figueiredo HC, Klesius PH, Arias CR, Evans J, Shoemaker CA, Pereira DJ Jr., and Peixoto MT. 2005. Isolation and characterization of strains of Flavobacterium columnare from Brazil. J. Fish Dis. 28: 199–204. [DOI] [PubMed] [Google Scholar]
- 30.Morley NJ, and Lewis JW. 2010. Consequences of an outbreak of columnaris disease (Flavobacterium columnare) to the helminth fauna of perch (Perca fluviatilis) in the Queen Mary reservoir, south-east England. J. Helminthol. 84: 186–192. [DOI] [PubMed] [Google Scholar]
- 31.Řehulka J, and Minařík B. 2007. Blood parameters in brook trout Salvelinus fontinalis (Mitchill, 1815), affected by columnaris disease. Aquacult. Res. 38: 1182–1197. [Google Scholar]
- 32.Soto E, Mauel MJ, Karsi A, and Lawrence ML. 2008. Genetic and virulence characterization of Flavobacterium columnare from channel catfish (Ictalurus punctatus). J. Appl. Microbiol. 104: 1302–1310. [DOI] [PubMed] [Google Scholar]
- 33.Suomalainen LR, Bandilla M, and Valtonen ET. 2009. Immunostimulants in prevention of columnaris disease of rainbow trout, Oncorhynchus mykiss (Walbaum). J. Fish Dis. 32: 723–726. [DOI] [PubMed] [Google Scholar]
- 34.Declercq AM, Haesebrouck F, Van den Broeck W, Bossier P, and Decostere A. 2013. Columnaris disease in fish: a review with emphasis on bacterium-host interactions. Vet. Res. (Faisalabad) 44: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li N, Zhu Y, LaFrentz BR, Evenhuis JP, Hunnicutt DW, Conrad RA, Barbier P, Gullstrand CW, Roets JE, Powers JL, et al. 2017. The type IX secretion system is required for virulence of the fish pathogen Flavobacterium columnare. Appl. Environ. Microbiol. 83: e01769–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie HX, Nie P, Chang MX, Liu Y, and Yao WJ. 2005. Gene cloning and functional analysis of glycosaminoglycan-degrading enzyme chondroitin AC lyase from Flavobacterium columnare G4. Arch. Microbiol. 184: 49–55. [DOI] [PubMed] [Google Scholar]
- 37.Li N, Qin T, Zhang XL, Huang B, Liu ZX, Xie HX, Zhang J, McBride MJ, and Nie P. 2015. Gene deletion strategy to examine the involvement of the two chondroitin lyases in Flavobacterium columnare virulence. Appl. Environ. Microbiol. 81: 7394–7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tongsri P, Meng K, Liu X, Wu Z, Yin G, Wang Q, Liu M, and Xu Z. 2020. The predominant role of mucosal immunoglobulin IgT in the gills of rainbow trout (Oncorhynchus mykiss) after infection with Flavobacterium columnare. Fish Shellfish Immunol. 99: 654–662. [DOI] [PubMed] [Google Scholar]
- 39.Xu HY, Dong F, Zhai X, Meng KF, Han GK, Cheng GF, Wu ZB, Li N, and Xu Z. 2020. Mediation of mucosal immunoglobulins in buccal cavity of teleost in antibacterial immunity. Front. Immunol. 11: 562795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bader JA, Shoemaker CA, and Klesius PH. 2003. Rapid detection of columnaris disease in channel catfish (Ictalurus punctatus) with a new species-specific 16-S rRNA gene-based PCR primer for Flavobacterium columnare. J. Microbiol. Methods 52: 209–220. [DOI] [PubMed] [Google Scholar]
- 41.Zhang X, Ding L, Yu Y, Kong W, Yin Y, Huang Z, Zhang X, and Xu Z. 2018. The change of teleost skin commensal microbiota is associated with skin mucosal transcriptomic responses during parasitic infection by Ichthyophthirius multifillis. Front. Immunol. 9: 2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liao Y, Smyth GK, and Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. [DOI] [PubMed] [Google Scholar]
- 44.McCarthy DJ, Chen Y, and Smyth GK. 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40: 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang DW, Sherman BT, and Lempicki RA. 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeLuca D, Wilson M, and Warr GW. 1983. Lymphocyte heterogeneity in the trout, Salmo gairdneri, defined with monoclonal antibodies to IgM. Eur. J. Immunol. 13: 546–551. [DOI] [PubMed] [Google Scholar]
- 47.Ramirez-Gomez F, Greene W, Rego K, Hansen JD, Costa G, Kataria P, and Bromage ES. 2012. Discovery and characterization of secretory IgD in rainbow trout: secretory IgD is produced through a novel splicing mechanism. J. Immunol. 188: 1341–1349. [DOI] [PubMed] [Google Scholar]
- 48.Baldissera MD, Souza CF, Dias JB, Da Silva TO, Tavares GC, Valladão GMR, da Silva AS, Verdi CM, Santos RCV, Vencato M, et al. 2020. Branchial bioenergetics dysfunction as a relevant pathophysiological mechanism in freshwater silver catfish (Rhamdia quelen) experimentally infected with Flavobacterium columnare. Microb. Pathog. 138: 103817. [DOI] [PubMed] [Google Scholar]
- 49.Byrd AL, Belkaid Y, and Segre JA. 2018. The human skin microbiome. Nat. Rev. Microbiol. 16: 143–155. [DOI] [PubMed] [Google Scholar]
- 50.Belkaid Y, and Tamoutounour S. 2016. The influence of skin microorganisms on cutaneous immunity. Nat. Rev. Immunol. 16: 353–366. [DOI] [PubMed] [Google Scholar]
- 51.Gomez D, Sunyer JO, and Salinas I. 2013. The mucosal immune system of fish: the evolution of tolerating commensals while fighting pathogens. Fish Shellfish Immunol. 35: 1729–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yin X, Mu L, Fu S, Wu L, Han K, Wu H, Bian X, Wei X, Guo Z, Wang A, and Ye J. 2019. Expression and characterization of Nile tilapia (Oreochromis niloticus) secretory and membrane-bound IgM in response to bacterial infection. Aquaculture 508: 214–222. [Google Scholar]
- 53.Castro R, Abós B, González L, Granja AG, and Tafalla C. 2017. Expansion and differentiation of IgM+ B cells in the rainbow trout peritoneal cavity in response to different antigens. Dev. Comp. Immunol. 70: 119–127. [DOI] [PubMed] [Google Scholar]
- 54.Austin DA, Jordan EM, and Austin B. 2003. Recovery of an unusual Gram-negative bacterium from ulcerated rainbow trout, Oncorhynchus mykiss (Walbaum), in Scotland. J. Fish Dis. 26: 247–249. [DOI] [PubMed] [Google Scholar]
- 55.Declercq AM, Chiers K, Van den Broeck W, Dewulf J, Eeckhaut V, Cornelissen M, Bossier P, Haesebrouck F, and Decostere A. 2015. Interactions of highly and low virulent Flavobacterium columnare isolates with gill tissue in carp and rainbow trout. Vet. Res. (Faisalabad) 46: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kayansamruaj P, Dong HT, Hirono I, Kondo H, Senapin S, and Rodkhum C. 2017. Comparative genome analysis of fish pathogen Flavobacterium columnare reveals extensive sequence diversity within the species. Infect. Genet. Evol. 54: 7–17. [DOI] [PubMed] [Google Scholar]
- 57.LaFrentz BR, García JC, Waldbieser GC, Evenhuis JP, Loch TP, Liles MR, Wong FS, and Chang SF. 2018. Identification of four distinct phylogenetic groups in Flavobacterium columnare with fish host associations. Front. Microbiol. 9: 452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Katzenback BA 2015. Antimicrobial peptides as mediators of innate immunity in teleosts. Biology (Basel) 4: 607–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang TT, Song XH, Bao GM, Zhao LX, Yu X, and Zhao J. 2013. Molecular characterization, expression analysis, and biological effects of interleukin-8 in grass carp Ctenopharyngodon idellus. Fish Shellfish Immunol. 35: 1421–1432. [DOI] [PubMed] [Google Scholar]
- 60.Peng Y, Cai X, Zhang G, Wang J, Li Y, Wang Z, Wang B, Xiong X, Wu Z, and Jian J. 2017. Molecular characterization and expression of interleukin-10 and interleukin-22 in golden pompano (Trachinotus ovatus) in response to Streptococcus agalactiae stimulus. Fish Shellfish Immunol. 65: 244–255. [DOI] [PubMed] [Google Scholar]
- 61.Hong S, Li R, Xu Q, Secombes CJ, and Wang T. 2013. Two types of TNF-α exist in teleost fish: phylogeny, expression, and bioactivity analysis of type-II TNF-α3 in rainbow trout Oncorhynchus mykiss. J. Immunol. 191: 5959–5972. [DOI] [PubMed] [Google Scholar]
- 62.Bose M, and Farnia P. 1995. Proinflammatory cytokines can significantly induce human mononuclear phagocytes to produce nitric oxide by a cell maturation-dependent process. Immunol. Lett. 48: 59–64. [DOI] [PubMed] [Google Scholar]
- 63.Greenbaum D, Colangelo C, Williams K, and Gerstein M. 2003. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 4: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tamura S, Iwasaki T, Thompson AH, Asanuma H, Chen Z, Suzuki Y, Aizawa C, and Kurata T. 1998. Antibody-forming cells in the nasal-associated lymphoid tissue during primary influenza virus infection. J. Gen. Virol. 79: 291–299. [DOI] [PubMed] [Google Scholar]
- 65.Fagarasan S 2006. Intestinal IgA synthesis: a primitive form of adaptive immunity that regulates microbial communities in the gut. Curr. Top. Microbiol. Immunol. 308: 137–153. [DOI] [PubMed] [Google Scholar]
- 66.Cerutti A, and Rescigno M. 2008. The biology of intestinal immunoglobulin A responses. Immunity 28: 740–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Onodera T, Takahashi Y, Yokoi Y, Ato M, Kodama Y, Hachimura S, Kurosaki T, and Kobayashi K. 2012. Memory B cells in the lung participate in protective humoral immune responses to pulmonary influenza virus reinfection. Proc. Natl. Acad. Sci. USA 109: 2485–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Milligan GN, and Bernstein DI. 1995. Generation of humoral immune responses against herpes simplex virus type 2 in the murine female genital tract. Virology 206: 234–241. [DOI] [PubMed] [Google Scholar]
- 69.Uddin T, Harris JB, Bhuiyan TR, Shirin T, Uddin MI, Khan AI, Chowdhury F, LaRocque RC, Alam NH, Ryan ET, et al. 2011. Mucosal immunologic responses in cholera patients in Bangladesh. Clin. Vaccine Immunol. 18: 506–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Johnson RA, Uddin T, Aktar A, Mohasin M, Alam MM, Chowdhury F, Harris JB, LaRocque RC, Bufano MK, Yu Y, et al. 2012. Comparison of immune responses to the O-specific polysaccharide and lipopolysaccharide of Vibrio cholerae O1 in Bangladeshi adult patients with cholera. Clin. Vaccine Immunol. 19: 1712–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zilberg D, and Klesius PH. 1997. Quantification of immunoglobulin in the serum and mucus of channel catfish at different ages and following infection with Edwardsiella ictaluri. Vet. Immunol. Immunopathol. 58: 171–180. [DOI] [PubMed] [Google Scholar]
- 72.Coscia MR, Simoniello P, Giacomelli S, Oreste U, and Motta CM. 2014. Investigation of immunoglobulins in skin of the Antarctic teleost Trematomus bernacchii. Fish Shellfish Immunol. 39: 206–214. [DOI] [PubMed] [Google Scholar]
- 73.Guardiola FA, Cuesta A, Arizcun M, Meseguer J, and Esteban MA. 2014. Comparative skin mucus and serum humoral defence mechanisms in the teleost gilthead seabream (Sparus aurata). Fish Shellfish Immunol. 36: 545–551. [DOI] [PubMed] [Google Scholar]
- 74.Valdenegro-Vega VA, Crosbie P, Vincent B, Cain KD, and Nowak BF. 2013. Effect of immunization route on mucosal and systemic immune response in Atlantic salmon (Salmo salar). Vet. Immunol. Immunopathol. 151: 113–123. [DOI] [PubMed] [Google Scholar]
- 75.Piazzon MC, Galindo-Villegas J, Pereiro P, Estensoro I, Calduch-Giner JA, Gómez-Casado E, Novoa B, Mulero V, Sitja A`-Bobadilla, and J. Pérez-Sánchez. 2016. Differential modulation of IgT and IgM upon parasitic, bacterial, viral, and dietary challenges in a perciform fish. Front. Immunol. 7: 637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu J, Yu Y, Huang Z, Dong S, Luo Y, Yu W, Yin Y, Li H, Liu Y, Zhou X, and Xu Z. 2019. Immunoglobulin (Ig) heavy chain gene locus and immune responses upon parasitic, bacterial and fungal infection in loach, Misgurnus anguillicaudatus. Fish Shellfish Immunol. 86: 1139–1150. [DOI] [PubMed] [Google Scholar]
- 77.Woof JM, and Kerr MA. 2006. The function of immunoglobulin A in immunity. J. Pathol. 208: 270–282. [DOI] [PubMed] [Google Scholar]
- 78.Lamm ME 1997. Interaction of antigens and antibodies at mucosal surfaces. Annu. Rev. Microbiol. 51: 311–340. [DOI] [PubMed] [Google Scholar]
- 79.Randall TD 2010. Bronchus-associated lymphoid tissue (BALT) structure and function. Adv. Immunol. 107: 187–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mostov KE 1994. Transepithelial transport of immunoglobulins. Annu. Rev. Immunol. 12: 63–84. [DOI] [PubMed] [Google Scholar]
- 81.Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, and Gordon JI. 2001. Molecular analysis of commensal host-microbial relationships in the intestine. Science 291: 881–884. [DOI] [PubMed] [Google Scholar]
- 82.Pal K, Kaetzel CS, Brundage K, Cunningham CA, and Cuff CF. 2005. Regulation of polymeric immunoglobulin receptor expression by reovirus. J. Gen. Virol. 86: 2347–2357. [DOI] [PubMed] [Google Scholar]
- 83.Bruno ME, Rogier EW, Frantz AL, Stefka AT, Thompson SN, and Kaetzel CS. 2010. Regulation of the polymeric immunoglobulin receptor in intestinal epithelial cells by Enterobacteriaceae: implications for mucosal homeostasis. Immunol. Invest. 39: 356–382. [DOI] [PubMed] [Google Scholar]
- 84.Sunyer JO 2013. Fishing for mammalian paradigms in the teleost immune system. Nat. Immunol. 14: 320–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


