Abstract
Germinal centers (GCs) or analogous secondary lymphoid microstructures (SLMs) are thought to have evolved in endothermic species. However, living representatives of their ectothermic ancestors can mount potent secondary antibody responses upon infection or immunization, despite the apparent lack of SLMs in these cold-blooded vertebrates. How and where adaptive immune responses are induced in ectothermic species in the absence of GCs or analogous SLMs remain poorly understood. Here, we infected a teleost fish (trout) with the parasite Ichthyophthirius multifiliis (Ich) and identified the formation of large aggregates of highly proliferating IgM+ B cells and CD4+ T cells, contiguous to splenic melanomacrophage centers (MMCs). Most of these MMC-associated lymphoid aggregates (M-LAs) contained numerous antigen (Ag)–specific B cells. Analysis of the IgM heavy chain CDR3 repertoire of microdissected splenic M-LAs and non–M-LA areas revealed that the most frequent B cell clones induced after Ich infection were highly shared only within the M-LAs of infected animals. These M-LAs represented highly polyclonal SLMs in which Ag-specific B cell clonal expansion occurred. M-LA–associated B cells expressed high levels of activation-induced cytidine deaminase and underwent significant apoptosis, and somatic hypermutation of Igμ genes occurred prevalently in these cells. Our findings demonstrate that ectotherms evolved organized SLMs with GC-like roles. Moreover, our results also point to primordially conserved mechanisms by which M-LAs and mammalian polyclonal GCs develop and function.
INTRODUCTION
Immunoglobulin (Ig)–based adaptive immunity in jawed vertebrates emerged more than 500 million years ago (1, 2). Elasmobranchs are the oldest vertebrate group with living representatives that bear bona fide Igs, a spleen, B cells, and T cells (3). The immune system has since evolved into the highly compartmentalized and exquisitely fine-tuned adaptive response seen in modern day mammals (2, 4). This includes the appearance of class-switch recombination, which first arose in the ancestors of amphibians (5), and organized secondary lymphoid microstructures (SLMs) such as germinal centers (GCs), which first arose in the ancestors of birds (4, 6). It is thought that organized SLMs evolved to maximize encounters between antigen (Ag), Ag-presenting cells, and B/T lymphocytes to efficiently sustain the B cell clonal expansion and affinity maturation processes required to fine-tune adaptive antibody (Ab) responses. Despite their lack of GCs and SLMs, cold-blooded species, including teleost fish, can mount potent Ag-specific secondary Ab responses upon infection or immunization (1). How and where adaptive immune responses are induced in these species in the absence of GCs or analogous SLMs are not well understood.
All ectothermic jawed vertebrates express several classes of Igs, although the specific gene isotypes differ between groups (1, 3). Ig rearrangement machinery is largely conserved across these species, and their primary Ig repertoires are highly diversified, similar to those of birds and mammals (3). Titers of systemic Ag-specific Ig of jawed ectotherms, which are mainly IgM in fish or IgY in amphibians and reptiles, significantly increase in infection or immunization (1). Although Ig-specific titers can increase significantly upon reinfection or boosting, Ab affinity maturation remains relatively poor (3). This has been connected to the lack of GCs in ectotherms, leading to the theory that selection of high-affinity B cells by somatic hypermutation (SHM) may be compromised in these animals (3). However, all jawed vertebrates contain genes homologous to activation-induced cytidine deaminase (AID), the enzyme that mediates both class-switch recombination and SHM (3, 7). AID expression has also been reported in the spleens of fish and amphibians, although its localization and expression patterns at the cellular level remain poorly understood (7, 8). The presence of AID in these species supports the ability of their Ig genes to undergo SHM (9, 10).
Although the induction of SLMs has never been described in fish, Ig+ clusters of cells have been observed in proximity to melanomacrophage centers (MMCs), which are aggregates of dark, pigmented phagocytes mainly found in the head kidneys (HKs) and spleens of cold-blooded vertebrates (11–14). MMCs increase in size after infection and trap Ag (15), suggesting that these teleost MMCs may represent functional analogs of GCs or lymphoid follicles (15, 16). However, no visible tissue organization (e.g., B cell–T cell zones) has yet been identified in ectotherms.
In this study, we investigated whether ectothermic vertebrates have evolved lymphoid microstructures analogous to GCs to support Ab responses. We identified that induction of systemic Ab responses in teleost fish occurs in primordially organized and highly polyclonal SLMs, where structurally and functionally analogous processes to those of GCs occur.
RESULTS
Infection induces IgM+ B cell proliferation and aggregation adjacent to MMCs
To identify sites of immune responses within the infected teleost spleen, we searched for 5-ethynyl-2′-deoxyuridine+ (EdU+) areas that could indicate lymphocyte proliferation and expansion. Trout (Oncorhynchus mykiss) were infected with the parasite Ichthyophthirius multifiliis (Ich), which induces strong systemic IgM responses and IgT-driven mucosal responses (17–20). At 2 weeks after secondary infection, we identified aggregates of dark-colored melanomacrophages that represent MMCs (fig. S1) (15), which were significantly larger in infected animals than noninfected controls (Fig. 1, A to C). Cell proliferation was significantly higher in infected spleens compared with controls (Fig. 1, A, B, and D) and was mainly concentrated in areas surrounding MMCs (Fig. 1E). Furthermore, the number of EdU+ cells inside of MMCs was significantly lower in control spleens than in infected fish (Fig. 1, B and E). We observed significant numbers of IgM+ B cell (IgMhi) aggregates in proximity to MMCs in infected spleens (Fig. 1, B and F, and fig. S1B). Conversely, B cell aggregates associated with MMCs in control animals were much smaller or absent and contained very few IgMhi B cells (Fig. 1, A and F, and fig. S1A). In both infected and control fish, IgM+ B cells outside MMC areas were for the most part IgMlow (Fig. 1, A, B, and F, and fig. S1).
Fig. 1. Intense IgM+ B cell proliferation occurs near splenic MMCs upon infection.
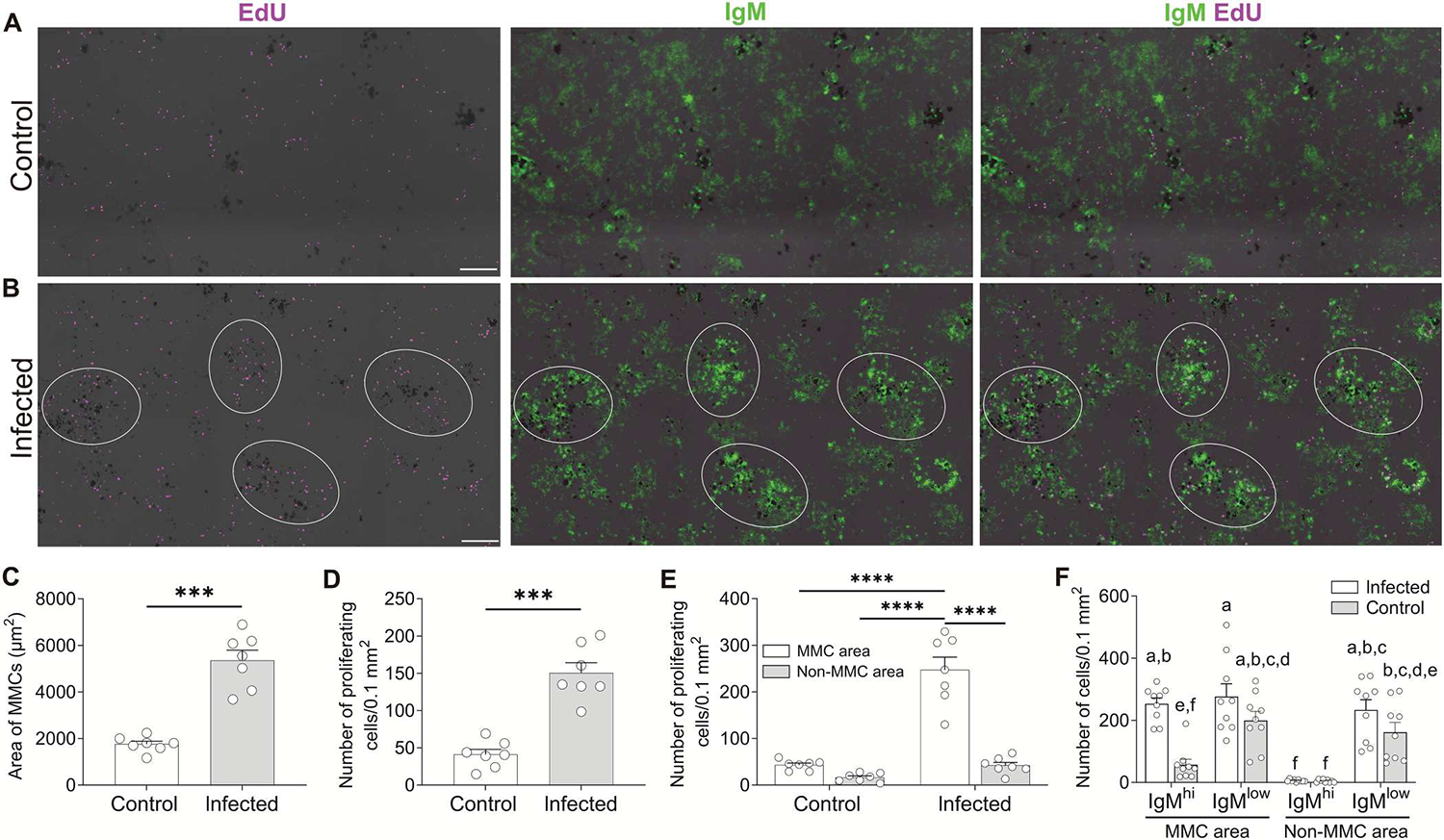
(A and B) IF analysis of EdU incorporation by cells of spleens from control (A) or infected fish (B). Spleen cryosections were stained for EdU (magenta) and IgM (green). Groups of dark cells in all images represent MMCs. White circles in (B) outline MMC-adjacent areas with high cell proliferation. Scale bars, 100 μm. (C and D) Area of MMCs, defined as the total surface of melanomacrophages (C) and number of proliferating cells (D) calculated from spleen cryosections of control or infected fish (n = 7). (E) Number of proliferating cells in MMC and non-MMC areas of spleen cryosections from control or infected fish (n = 7). (F) Number of IgMhi and IgMlow B cells in MMC and non-MMC areas of spleen cryosections from control or infected fish (n = 9). Data in (C) to (F) are representative of at least three independent experiments (means and SEM). Empty circles represent values from individual fish. Statistical analyses were performed by unpaired Student’s t test (C and D) or two-way ANOVA, followed by Tukey’s post hoc test (E and F). ***P < 0.001 and ****P < 0.0001. Plot bars of (F) not sharing common letters are significantly different (P < 0.05).
Red and white pulp areas were intermingled throughout the spleen and not separated into distinct regions (fig. S1), as previously reported (21–23). MMCs in infected fish were more frequently associated with white pulp regions and were populated with more lymphocytic cells when compared with control fish (fig. S1). Together, these results indicate that the large proliferative responses observed in areas adjacent to MMCs in infected animals are sites of immune activation in splenic tissue.
Large aggregates of highly proliferating IgM+ B cells and CD4+ T cells are induced adjacent to MMCs upon infection
To further evaluate whether MMC-adjacent IgM+ B cell aggregates represent inductive sites of immune responses, we next assessed whether CD4+ T cells were also present among these IgM+ B cell aggregates because organized SLMs in mammals are typically characterized by the presence of closely associated B and T cell zones (24, 25). To identify CD4+ T cells, we generated an antitrout CD4 polyclonal Ab (pAb) (fig. S2). We found that upon infection, a majority of MMC areas containing IgM+ B cell aggregates were in proximity to CD4+ T cell zones (Fig. 2, A and B). In many instances, B cell– and T cell–like zones were defined [Fig. 2, A(i) and B(i)], whereas in others, boundaries between B cell and T cell aggregates were more diffuse [fig. S3, A(i) and B(i)]. We have named these organized lymphoid tissue areas, which include both the MMCs and their associated B/T cell aggregates, “MMC-associated lymphoid aggregates” (M-LAs). Within M-LAs from infected fish, significantly more IgM+ B cells were in contact with MMCs compared with CD4+ T cells [Fig. 2, A(i), B(i), and D]. M-LAs from infected fish contained more proliferating cells [Fig. 2, A(ii) and B(ii)] than those of control animals [Fig. 2, C(ii) and E]. Accordingly, a large percentage of IgM+ B cells [Fig. 2, A(iii and iv), B(iii and iv), and E] and CD4+ T cells [Fig. 2, A(iii and v), B(iii and v), and E] within these M-LAs were proliferating, suggesting the expansion of Ag-specific B and T cells in these areas. Although control fish also contained M-LAs, they were much smaller in size [Figs. 2C (i) and 3A] and contained far fewer proliferating T and B cells [Fig. 2, C(iii to v) and E]. Together, these data suggest that induction of systemic IgM responses occurs in M-LAs.
Fig. 2. Aggregates of highly proliferating splenic IgM+ B cells and CD4+ T cells are induced near MMCs upon infection.
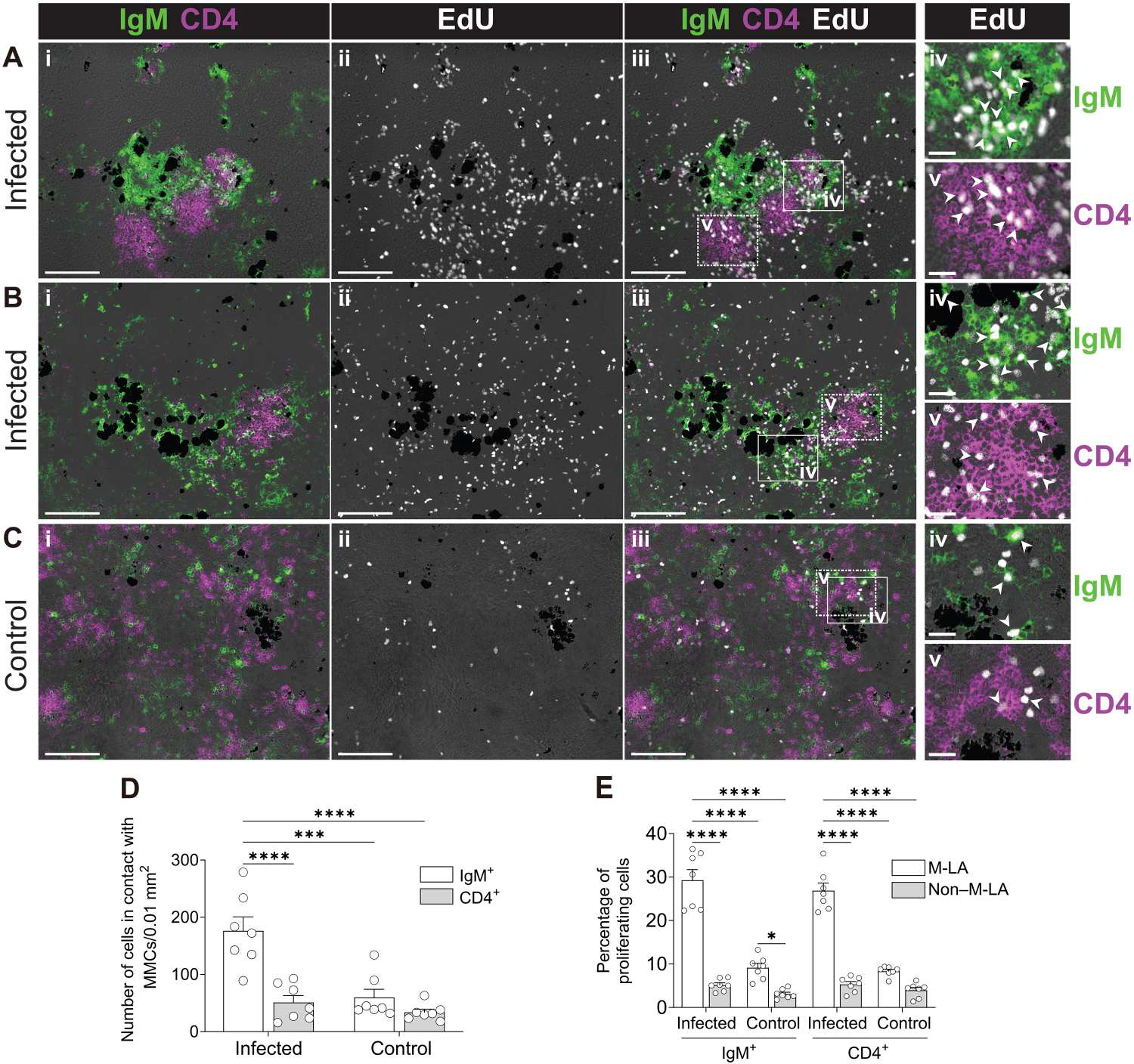
(A to C) IF analysis of EdU incorporation by IgM+ B cells and CD4+ T cells from M-LAs of the spleen from infected (A and B) and control fish (C). Spleen cryosections were stained for IgM (green), CD4 (magenta), and EdU (white). (A to C) Image in (i) shows a representative M-LA stained for IgM (green) and CD4 (magenta); (ii and iii) show the same tissue area of (i) stained for EdU (white) (ii), IgM (green), CD4 (magenta), and EdU (white) (iii); (iv and v) show enlarged images of the areas outlined in (iii) displaying proliferating (EdU+) IgM+ B cells (iv) and proliferating (EdU+) CD4+ T cells (v). White arrowheads point to examples of proliferating IgM+ B cells and CD4+ T cells. Scale bars, 100 μm (i to iii) and 20 μm (iv and v). Groups of dark cells in all images represent MMCs. Data are representative of at least three independent experiments. (D) Number of IgM+ B cells and CD4+ T cells in contact with MMCs of spleen cryosections from control or infected fish. (E) Percentage of proliferating IgM+ B cells and CD4+ T cells in M-LA and non–M-LA areas of spleen cryosections from control or infected fish. Data are representative of at least three independent experiments (means and SEM). Empty circles represent values from individual fish (n = 7). Statistical analyses were performed by two-way ANOVA, followed by Tukey’s post hoc test. *P < 0.05, ***P < 0.001, and ****P < 0.0001.
M-LAs from immunized fish contain significant numbers of Ag-specific IgM+ B cells
The strong proliferation of IgM+ B cells observed in M-LAs from infected fish suggested the presence of Ag-specific B cells in these tissue microstructures. To evaluate this hypothesis, we modified a reported method developed in rats (26) to detect Ag on B cells from tissue sections and adapted it to our spleen cryosections (fig. S4). To detect sufficient Ag-specific B cells, we used the T cell–dependent Ag [dinitropheny–keyhole limpet hemocyanin (DNP-KLH)], which is known to induce strong IgM responses in rainbow trout (27). At 2 weeks after boost, high DNP-specific IgM endpoint titers (102,400 to 409,600) were detected in sera. Detection of DNP–phycoerythrin (PE) Ag was negligible from all M-LAs or non–M-LA areas of control fish (Fig. 3, A and D). DNPKLH–induced M-LAs (Fig. 3B) contained large aggregates of IgM+ B cells in proximity to both MMCs and CD4+ T cell zones (Fig. 3, B and C). In immunized spleens, DNP-PE Ag colocalized with M-LA–associated IgM+ B cells (M-LA B cells) for the most part, which represented 98.3 ± 1.2% of all the Ag-positive cells (Fig. 3, B and C). Moreover, a large proportion of the M-LA B cells were Ag positive, although Ag signal was residual on CD4+ T cells and negligible in non–M-LA areas of the same fish (Fig. 3, B to D). In immunized fish, a large percentage of M-LAs (59.8 ± 8.5%) contained significant proportions of Ag-specific B cells (Fig. 3, B and E), although tissue areas from MMCs not associated with B and T cell aggregates (Fig. 3B, yellow circles) were devoid of Ag binding in these fish. Flow cytometric analysis confirmed the increases in splenic Ag-specific IgM+ B cells in the spleens of immunized fish, although negligible Ag binding was detected on CD4+ T cells (fig. S5, A and C) or in IgM+ and CD4+ T cells from control fish (fig. S5, B and C). Overall, these results support the notion that Ag-specific clonal B cell expansion occurs within M-LAs.
Fig. 3. Ag-specific B cells are mostly localized within M-LAs of immunized fish.
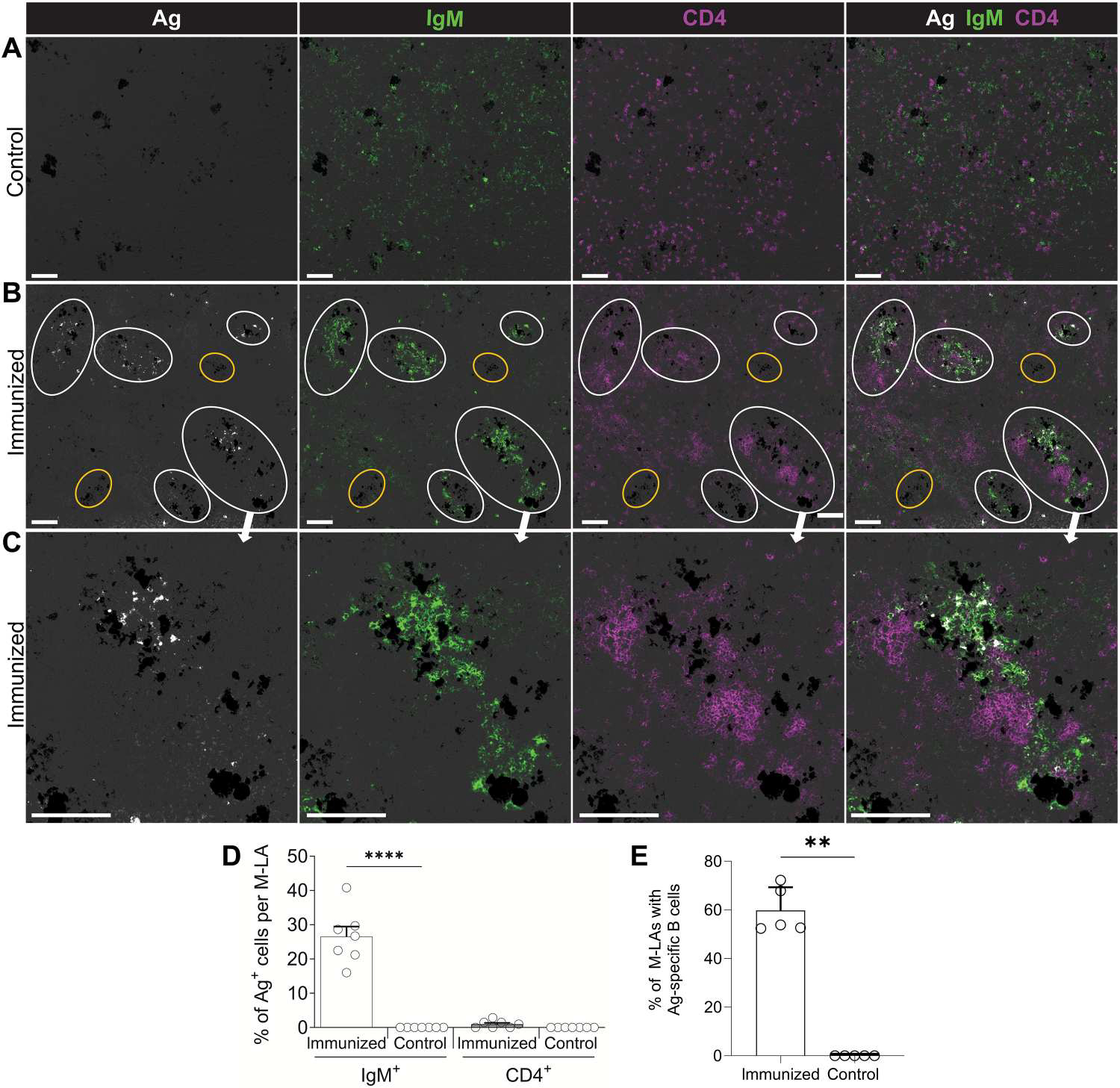
(A to C) IF analysis of Ag-specific cells in the spleens of control fish (A) and immunized fish (B and C). Spleen cryosections were probed with DNP-PE (white), followed by detection of IgM (green) and CD4 (magenta). White circles outline M-LAs containing Ag-specific B cells, whereas yellow circles outline MMCs lacking B and T cell zones and Ag-specific cells. Scale bars, 100 μm. Groups of dark cells in all images represent MMCs. (D) Percentage of IgM+ B cells and CD4+ T cells positive for DNP-PE in M-LAs and non–M-LA areas from immunized and control fish. Empty circles represent values from individual fish (n = 7). (E) Percentage of M-LA areas containing a high proportion (>10%) of Ag-specific B cells from immunized and control fish. Empty circles represent values from individual fish (n = 5). Data are representative of at least three independent experiments (means and SEM). Statistical analyses were performed by one-way ANOVA, followed by Tukey’s post hoc test (D) and Mann-Whitney test (E). **P < 0.01 and ****P < 0.0001.
CDR3 repertoire analysis demonstrates that M-LAs from infected fish share a large proportion of their most frequent CDR3s
We next determined whether the proliferating Ag-specific IgM+ B cells in M-LAs from infected fish were undergoing clonal expansion and selection. To this end, we analyzed the IgM heavy chain (IgHμ) repertoire of Ich-infected or control fish by sequencing the expressed IgHμ complementarity-determining region 3 (CDR3) region of laser capture microdissected M-LAs and non–M-LA areas (fig. S6).
We first compared the degree of overlap or sharing of the CDR3 sequences obtained from M-LA and non–M-LA areas of the spleens of immunized animals. To this end, we determined to what extent the most frequent CDR3s (top 1000 and top 50 CDR3s) obtained from each M-LA, and non–M-LA areas, were present in other M-LA or non–M-LA areas of the same spleen (Fig. 4A). A large proportion (~248 to 436) of the top 1000 CDR3 sequences from each M-LA were also detected in other M-LAs in a representative fish [Fig. 4A(i)] [e.g., 330 of the top 1000 CDR3s found in M-LA1 were also identified in M-LA2 (Fig. 4A(i)]. Conversely, a significantly lower proportion (~91 to 247) of these top 1000 CDR3s found in M-LAs were present in non–M-LA areas, and an even lower fraction (~28 to 169) of the top 1000 CDR3s from each of the non–M-LA areas was detected in the other non–M-LA regions [Fig. 4A(i)]. This pattern of CDR3 sharing was also observed when analyzing the top 50 CDR3s of M-LA and non–M-LA areas of the same fish spleen [Fig. 4A(i)]. As an example of this CDR3 overlap, we found that 32 of the top 50 CDR3s found in M-LA1 were also identified in M-LA2 [Fig. 4A(ii)]. To evaluate whether the high sharing of the most frequent CDR3s among the M-LAs of infected fish was statistically significant, we computed the shared proportions of the top 50 CDR3s among all M-LAs and non–M-LA areas from several infected and control fish (Fig. 4B). The degree of sharing of the top 50 CDR3s among the different M-LA and non–M-LA areas reached its highest level (~46%) only when comparing pairwise of all M-LAs from infected fish [i.e., top 50 CDR3s from M-LAs found in all CDR3s from M-LAs (indicated by the magenta arrow in y axis of Fig. 4B)], in line with our observation from a representative fish (Fig. 4A). This finding indicates that the highest degree of cooccurrence (or sharing) of the most frequent CDR3s induced upon infection was observed mainly in the M-LAs of each of these animals. An appreciable degree of sharing of the most frequent CDR3s was also observed among M-LAs from control fish (Fig. 4B), although it was more infrequent than that of M-LAs from infected fish. This CDR3 overlap in M-LAs from control fish may reflect a residual response due to prior exposure of these fish to other immunogens.
Fig. 4. IgHμ repertoire analysis shows that M-LAs from infected fish share a significant proportion of their most frequent CDR3s.
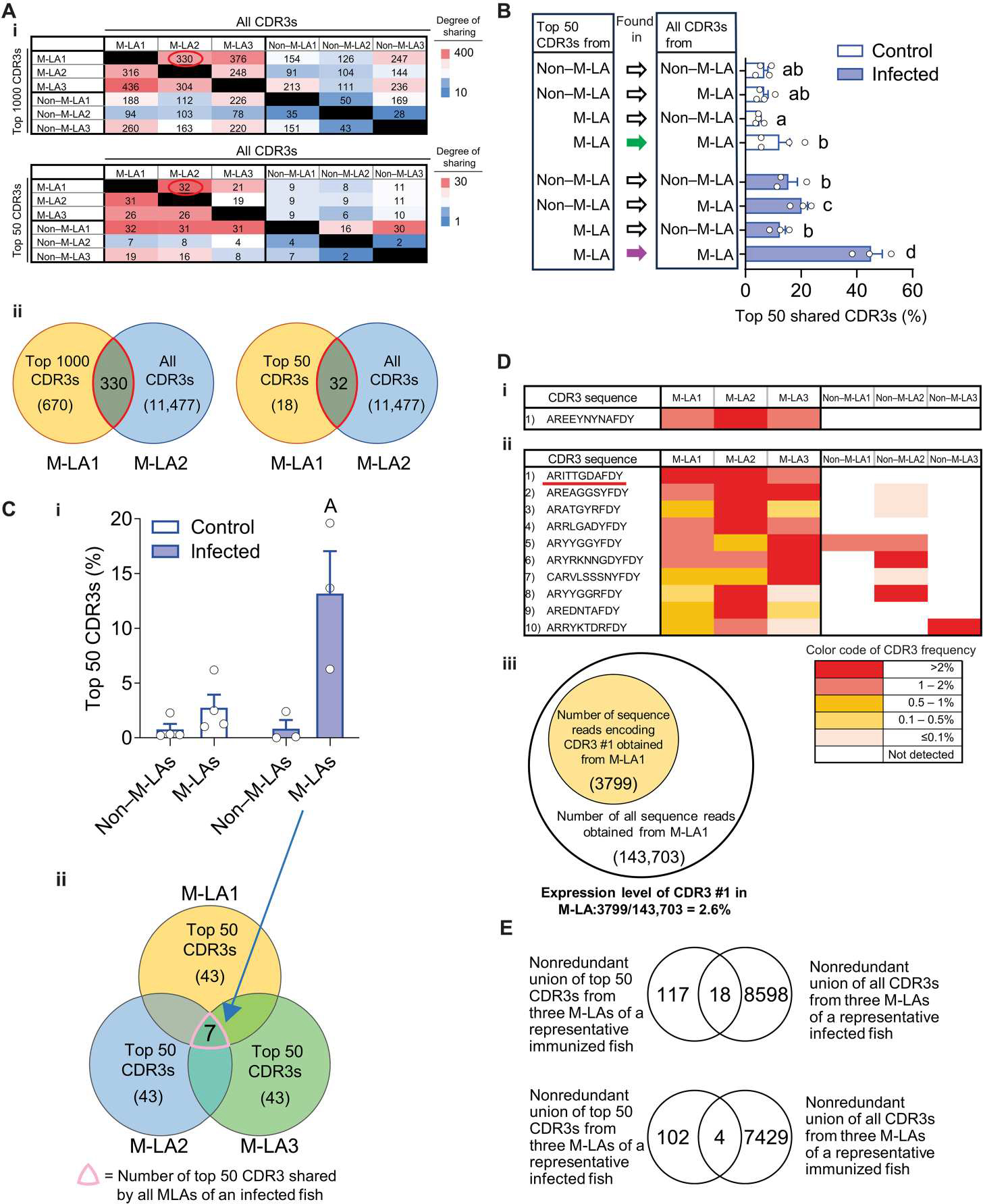
M-LAs and non–M-LA areas from the spleens of infected and control fish were microdissected, and their IgHμ clonal composition was determined by CDR3 repertoire analysis. (A) Pairwise comparison of the most frequent CDR3s from three M-LAs and three non–M-LA areas of a representative infected fish. (i) Matrix displaying the numbers of the most frequent CDR3s [top1000 (top) or top 50 (bottom)] from M-LA or non–M-LA areas (rows) present in (i.e., shared by) each M-LA or non–M-LA area (columns). Heatmap shows graded colors from intense red (high CDR3 sharing) to intense blue (low CDR3 sharing). Graphical illustrations (ii) of representative matrix values [red-circled numbers in (i)]. Left illustration shows that 330 CDR3s are shared between the top 1000 CDR3s from M-LA1 and all identified CDR3s (11477) from M-LA2, whereas the right illustration shows that 18 CDR3s are shared between the top 50 CDR3s from M-LA1 and all identified CDR3s (11477) from M-LA2. (B) Bar plots represent statistics of the same analysis as in (A) for several fish and show the mean percentage of the top 50 CDR3s from M-LAs or non–M-LA areas found in (i.e., shared by) M-LAs or non–M-LA areas from the same spleen. Plot bars show mean values, and error bars show SEM. Control fish (white bars), n = 4; infected fish (blue bars), n = 3. Empty circles represent values from individual fish. Statistical analyses were performed by two-way ANOVA, followed by Tukey’s post hoc test. Plot bars not sharing common letters are significantly different (P < 0.05). Green and magenta arrows point to the top 50 CDR3s from M-LAs found in all CDR3s from M-LAs of control and infected fish, respectively. (C) Proportion of the top 50 CDR3s shared by all M-LAs or all non–M-LA areas within the same spleen. Values in bar plot (i) represent the mean % of the top 50 CDR3s shared by all M-LAs or non–M-LA areas of the same spleen from infected (n = 3) and control fish (n = 4), and error bars show SEM. Empty circles represent values from individual fish. Values were generated from the same datasets as for (B). Statistical analyses were performed by two-way ANOVA, followed by Tukey’s post hoc test; plot bar with the letter A above is significantly different with the other groups (P < 0.05). (ii) depicts a graphical illustration representing the number of top 50 CDR3s (7) shared by three M-LAs of a representative fish, thus indicating that those three M-LAs share 14% of their top 50 CDR3s. (D) Color-coded representation of the frequency (i.e., expression level) of the top 50 CDR3s (left column) found in all M-LAs from one representative control (i) or infected (ii) fish. Graphical illustration (iii) representing the number of Illumina sequence reads (3799) for CDR3 #1 [underlined in (D, ii)] detected in M-LA1 [i.e, that number of sequence reads represent 2.6% of all the CDR3 reads (143,703) sequenced from M-LA1 of a representative infected fish]. (E) Top panel depicts a Venn diagram showing the intersect between the top 50 CDR3s detected in three M-LAs of a representative immunized DNP-KLH fish and all CDR3s detected in three M-LAs from an infected fish, whereas the bottom panel depicts a Venn diagram showing the intersection between the top 50 CDR3s detected in three M-LAs from a representative infected fish and all CDR3s detected in three M-LAs from a fish immunized with DNP-KLH.
We next evaluated whether the overlap of the top frequent CDR3s induced in the response was widespread across M-LAs throughout the same spleen. We selected the top 50 CDR3s of each of the three M-LAs analyzed from the spleen of each infected or control fish and calculated which proportion of these CDR3s was concurrently detected in all three M-LAs (Fig. 4C). The same calculation was also done for the top 50 CDR3 sets of each of the three non–M-LA areas analyzed from the same fish. The mean proportion of the top 50 CDR3s common to all three analyzed M-LAs was increased in the infected group and was more than 10-fold higher than the proportion of top 50 CDR3s common to all non–M-LA areas [Fig. 4C(i)]. As an example of this CDR3 overlap among M-LAs of infected fish, we found that a total of seven top 50 CDR3s (i.e., ~14% of these CDR3s) were shared by all three M-LAs of the same spleen from a representative infected fish [Fig. 4C(ii)]. In conclusion, this significant sharing of the most frequent CDR3 sets among all M-LAs of the same spleen in each of the infected fish suggests an expansion of these Ag-specific B cell clones.
We next assessed to what degree the shared top 50 CDR3s found in all M-LAs were expressed across individual M-LAs and non–M-LA areas from the spleen of a representative infected and control fish. We found that only one top 50 CDR3 was shared by all three analyzed M-LAs from the spleen of a representative control animal [Fig. 4D(i)]. In contrast, we found 10 top 50 CDR3s present in all three analyzed M-LAs from the infected spleen [Fig. 4D(ii)]. Furthermore, 7 of 10 of these CDR3s were expressed at a frequency higher than 0.5% in all three M-LAs. None of these CDR3s were concurrently detected in the non–M-LA areas of the same spleen. For example, the number of Illumina sequence reads detected for CDR3#1 in M-LA1 (3799 reads) represents 2.6% of all the CDR3 reads (143,703 reads) sequenced from M-LA1 of that representative fish [Fig. 4D(iii)], whereas CDR3#1 reads were not detected in any of the non–M-LA areas of the same fish [Fig. 4D(ii)]. Overall, these data suggest that highly expressed CDR3s correspond to Ag-specific B cell clonal expansions.
To further support the idea that the most abundant CDR3s induced in the M-LAs of infected fish are specific for the Ag (i.e., Ich parasite), we evaluated whether a different Ag (DNP-KLH) could induce a completely different set of CDR3 sequences. The overlap in CDR3s obtained by the two different Ag exposures was marginal (Fig. 4E). No public responses (public CDR3 expansions) to Ich were detected in the M-LA or non–M-LA areas of infected animals.
Overall, our results indicate that a large fraction of the most frequent CDR3s from a given M-LA of an infected fish are shared and highly expressed in all M-LAs from the same fish. This IgHμ repertoire convergence only in M-LAs of infected fish points to the induction of Ag-specific B cell expansions upon infection.
M-LA B cells express high levels of AID and undergo significant apoptosis
Given that M-LAs in infected fish contain clonally expanded Ag-specific B cells, we next investigated AID expression, which, in mammals, is expressed at its highest levels in GC B cells (25, 28). AID expression was detectable and mainly localized in M-LAs (outlined in white) of infected fish (Fig. 5B), where ~43 ± 4% of M-LAs contained significant numbers of AID+ cells (Fig. 5C). In contrast, AID expression was minimal in the M-LAs or non–M-LA areas of control fish (Fig. 5, A and C). A large percentage of M-LA B cells were AID+ (Fig. 5E), whereas a low proportion of non–M-LA B cells in the same fish expressed AID (Fig. 5, B and D).
Fig. 5. IgM+ B cells from M-LAs of infected fish express high levels of AID.
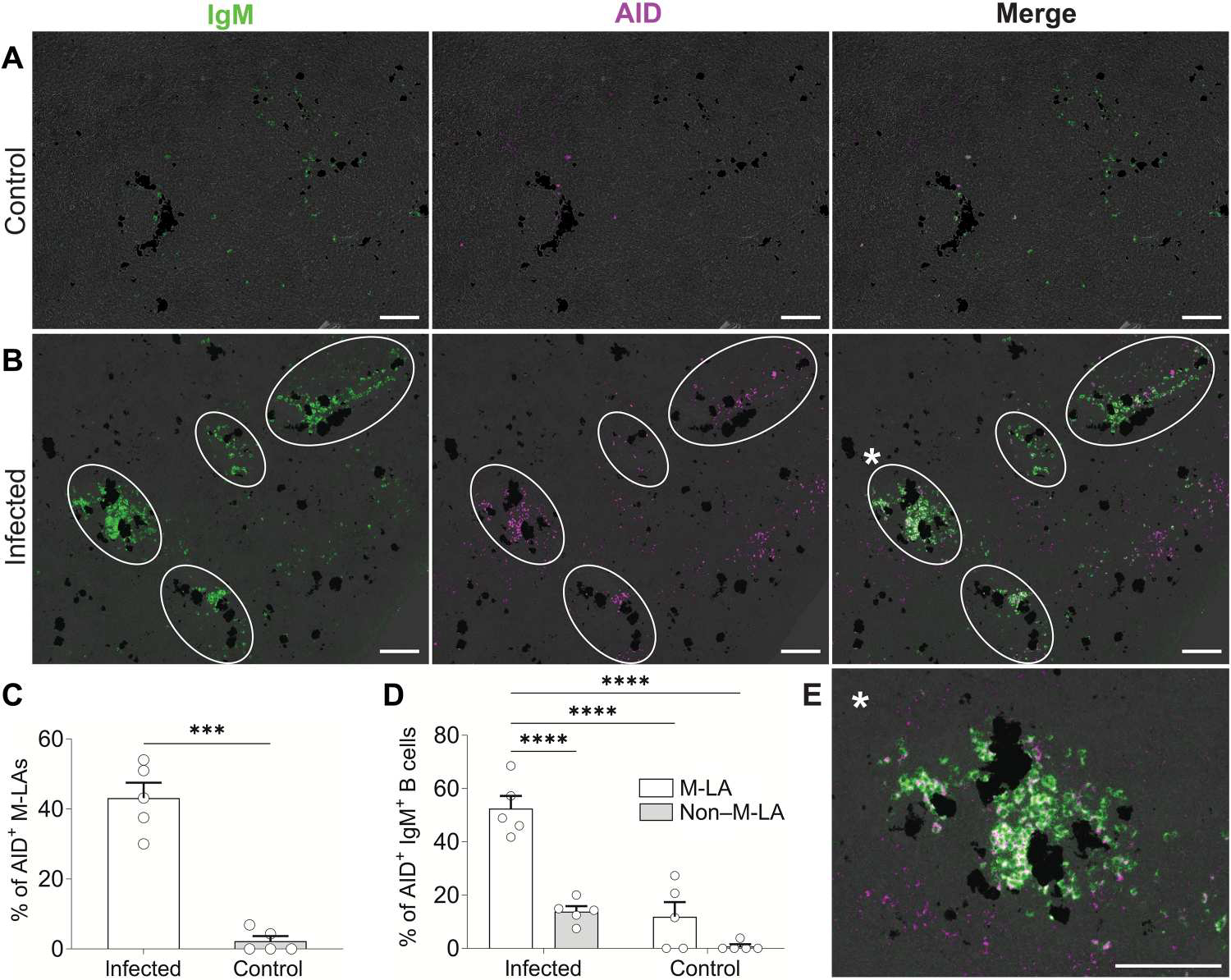
(A and B) Representative double in situ hybridization images of trout AID and IgM from the spleens of control (A) or infected fish (B). Spleen cryosections were hybridized with IgM (green) and AID (magenta) antisense probes. White circles represent M-LAs containing IgM+ and AID+ cells. AID+IgM+ cells are seen as white cells in merged images. (C) Percentage of M-LAs from infected and control fish containing significant numbers of AID+ cells. M-LAs were considered AID+ when containing more than 10 AID+ cells per M-LA. (D) Percentage of AID+IgM+ B cells from M-LA and non–M-LA areas of infected and control fish. (E) Enlarged M-LA from (B) [enlarged M-LA from merged image of (B) is marked with a white asterisk]. Scale bars, 100 μm. Groups of dark cells in all images represent MMCs. Data are representative of at least three independent experiments (means and SEM). Empty circles represent values from individual fish (n = 5). Statistical analyses were performed by unpaired Student’s t test (C) or two-way ANOVA, followed by Tukey’s post hoc test (D). ***P < 0.001 and ****P < 0.0001.
In mammals, AID is responsible for a large percentage of apoptosis in GC B cells, particularly in the dark zone (25, 28), and is considered a fundamental mechanism to counteract the large degree of B cell proliferation within GCs (25, 28). Given the high rate of M-LA B cell proliferation in infected fish (Fig. 2E) combined with their expression of AID (Fig. 5, B and D), we next determined whether a proportion of M-LA B cells were apoptotic. Activated caspase 3 (casp3) staining was used to detect apoptosis and was mainly detected within M-LA B cells of infected animals (Fig. 6, A to D). About 75% of M-LAs from infected fish contained casp3+ B cells, compared with only ~34% of M-LAs from control fish (Fig. 6E). Casp3 in B cells displayed a typical punctuated staining (Fig. 6, A and B), as previously reported (29). The highest proportion of casp3+ B cells was found within M-LAs of infected fish, which contained more than double of that found in control animals (Fig. 6D), whereas the proportion of casp3+ B cells in non–M-LA areas was small in both control and infected fish (Fig. 6D). In addition, the proportion of casp3+ CD4+ T cells was minimal both in M-LA and in non–M-LA areas of both infected and control fish (Fig. 6, A to D). These findings suggest that AID expression in the spleens of infected fish is concentrated on M-LA B cells and that apoptosis of these cells is a mechanism to offset their high rates of proliferation in these species.
Fig. 6. M-LA B cells undergo significant apoptosis upon infection.
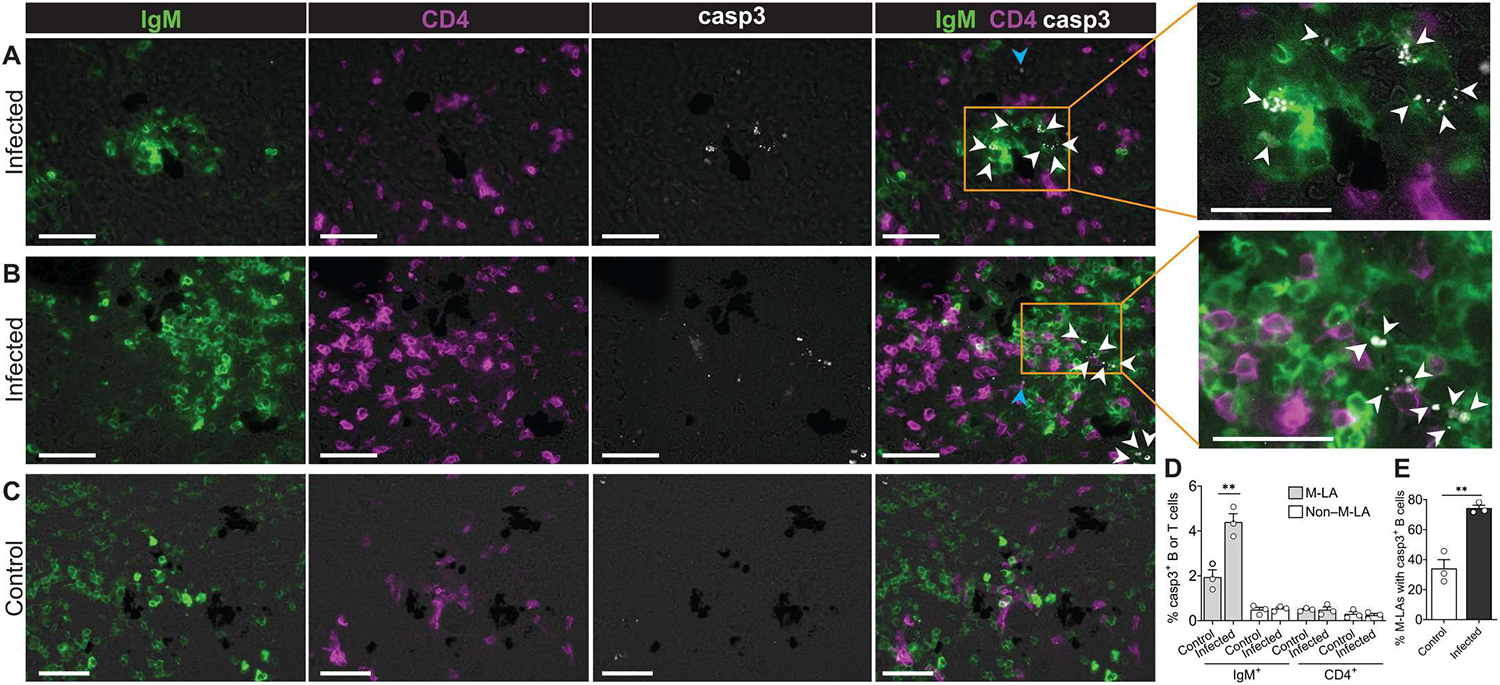
IF analysis of activated casp3 staining in M-LA and non–M-LA areas of the spleen from infected and control fish. (A to C) Representative M-LAs from infected (A and B) and control (C) fish stained for IgM (green), CD4 (magenta), and casp3 (white). The right panels of (A) and (B) represent enlarged images from areas outlined in orange. Scale bars, 50 μm. White triangles point to casp3+IgM+ cells, whereas blue triangles point to casp3+CD4−IgM− cells. (D) Percentage of casp3+IgM+ and casp3+CD4+ cells in M-LA and non–M-LA areas from control and infected fish. (E) Percentage of M-LAs with casp3+ B cells in control and infected fish. Data are representative of at least three independent experiments. The bars of the graphs represent the mean (n = 3), and the error bars show the SEM (D and E); empty circles represent values from individual fish. Statistical analyses were performed by unpaired Student’s t test. **P < 0.01.
SHM occurs prevalently in M-LA–associated B cells
SHM of Ig genes in warm-blooded vertebrates is known to occur mainly in GC B cells (25). We next explored whether AID expression in M-LA B cells was associated with the occurrence of SHM of IgHμ V genes in these cells (Fig. 7). To look for evidence of SHM, we assessed IgHμ V sequence variation among sequences expressed by B cells belonging to the same clonal sets (Fig. 7A). Variation within sequences was measured by evaluating the sequence changes within the CDR1–frame region 2 (FR2)–CDR2-FR3 region of their V domain. Thereafter, the variation rate in a clonal set was computed after comparison with the most frequent sequence of that specific clonal set (Fig. 7A). Because CDR3 repertoire analysis revealed that the most frequent clonal sets were shared among M-LAs of the spleen of an immunized fish (Fig. 4), we reasoned that evidence of SHM could most likely be detected when comparing sequence variation within clonal sets shared by splenic M-LAs from these fish. To this end, sequence variation was analyzed in clonal sets shared among M-LA and non–M-LA areas of the same spleen from immunized and control fish, respectively. Sequence variation was highest in the shared clonal sets of M-LAs from both control and immunized fish (Fig. 7B). In comparison with M-LAs, sequence variation in clonal sets from non–M-LA areas was significantly lower than that of M-LAs, and we did not find significant differences when comparing sequence variation of shared versus nonshared clonal sets in these areas (Fig. 7B). Although the rate of variation was roughly similar within clonal sets shared across M-LAs from control and infected fish, the proportion of shared B cell clones was much higher in M-LAs from infected fish, as seen in Fig. 4. Accordingly, hypermutated B cell clones were found more abundantly in M-LAs from infected fish than M-LAs from control animals. As an example of sequence variation in a clonal set shared by three different M-LAs of an infected fish, we depict a dominant clonal set tree (Fig. 7C) and show multiple sequence alignments of a subcluster of the tree displaying accumulation of mutations (Fig. 7D). The sequence alignments show a significant amount of base nonsilent mutations in several clonal set variants (d-h sequences in top panel of Fig. 7D), which lead to changes in amino acid composition of these variants (bottom panel of Fig. 7D). Overall, these data indicate that upon infection, SHM of Igμ genes occurs for the most part in M-LA B cells and is prevalent in Ig clonal sets specifically shared by the different splenic M-LAs of the same individual.
Fig. 7. IgHμ V gene sequences are somatically hypermutated prevalently in clonal sets shared among M-LAs from the same spleen.
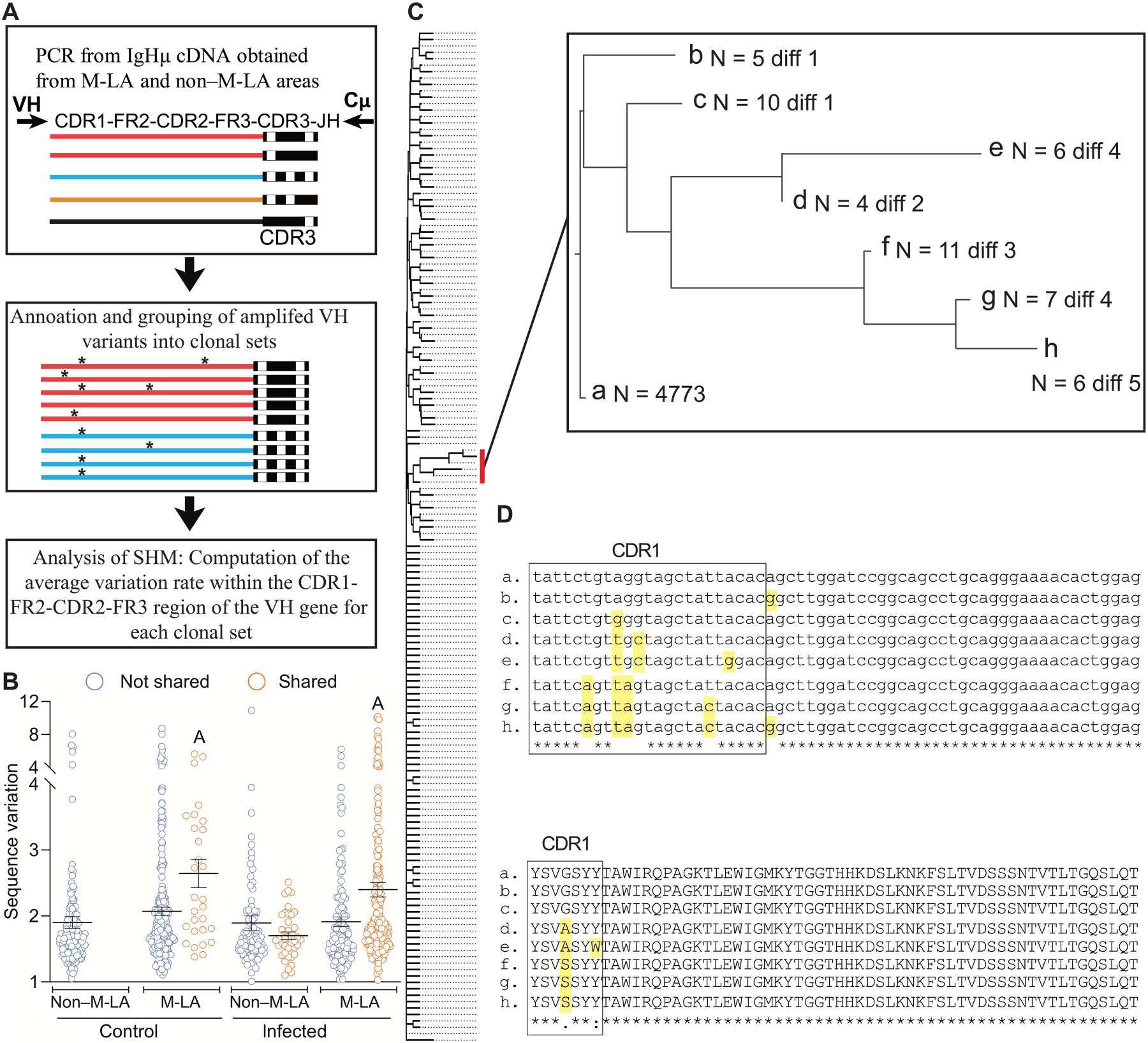
(A) Pipeline of analysis of sequence variation within clonal sets from M-LA and non–M-LA areas of control and infected fish. Clonal sets are defined as sets of sequences with the same CDR3 and the same VH gene annotation. Different color lines in top panel denote different VH genes. Black rectangles at the end of each line represent the V(D)J junction, whereas the different number and/or position of the white squares within them denote sequence differences in the V(D)J junction. In the middle panel, two different clonal sets (one in red and the other in blue lines, respectively) are represented. Asterisks denote sequence differences of individual VH variants within each clonal set. (B) Sequence variation was calculated for shared and nonshared clonal sets from M-LAs and non–M-LA areas of the same spleen from control (n = 4) and infected fish (n = 3). Variation is represented as the proportion (per 1000) of nucleotides divergent from the most frequent sequence of the clonal set. Horizontal lines represent means (±SEM), and empty circles represent the sequence variation value found in a clonal set. Scatter groups with the letter A above are significantly different from the other groups (two-way ANOVA, Tukey test, P < 0.05). (C) Example of a tree of sequences of a clonal set from an M-LA of an infected fish (this clonal set is defined by the expression of the IGHV6-4 gene and its CDR3 sequence). This clonal set comprised 9174 sequences (representing 1102 unique sequence variants). The neighbor joining tree was computed by Clustal Omega and comprises all sequences found more than five times (i.e., 309 sequence variants). A magnification of a tree branch showing accumulation of mutations is shown on the right. “N” is the number of sequences found in the dataset for each variant, whereas “diff” refers to the number of nucleotide differences to the reference sequence [sequence “a” of (D)]. (D) Multiple alignment of nucleotide and amino acid sequences of tree branch shown in (C). Mutations are highlighted in yellow. The CDR1 is boxed.
DISCUSSION
In this study, we addressed how and where adaptive immune responses are induced in species where GCs or analogous SLMs have not been described. It is well accepted that the spleen of cold-blooded jawed vertebrates represents a secondary lymphoid organ in which B and T cell responses have been shown to occur, and, thus, it is believed that secondary immune responses are induced in this organ (3). However, whether B cell–T cell interactions leading to such responses occur stochastically in random areas of the spleen or in a specific splenic microenvironment is a question that has remained elusive. Supporting the later scenario, our results indicate that induction of systemic Ab responses in teleost fish occurs in primordially organized and highly polyclonal SLMs that are structurally and functionally analogous to GCs. More specifically, our unbiased strategy to identify inductive sites of the immune response in the fish spleen led to the discovery of SLMs, which we termed M-LAs. These M-LAs are inducible tissue microstructures composed of MMCs and their associated B and T cell aggregates. Upon infection or immunization, M-LA–associated B and T cells proliferate, whereas M-LA B cells express high levels of AID and undergo significant apoptosis. Critically, Ag-specific B cell clonal expansion and SHM processes mostly occur in the induced M-LAs and not in the surrounding non–M-LA areas.
Although there are a number of structural and functional similarities between M-LAs and GCs, there also exist significant differences, most critically the following: (i) T cell zones are inducible only in M-LAs; (ii) M-LAs lack the light and dark zones found in GCs; (iii) GCs are localized within highly organized lymphoid areas (i.e., lymph nodes and tertiary lymphoid organs), but M-LAs are not; (iv) GCs contain follicular dendritic cells (FDCs) that retain Ag, whereas in M-LAs, this role might be played by MMCs. We cannot rule out that some of the above stated differences between M-LAs and GCs may reflect the current limitations imposed by our animal model, including the absence of Ab reagents specific for fish immune molecules that can distinguish other key leukocyte markers (i.e., macrophages) and subsets of B cells in various states of differentiation (i.e., plasmablast, plasma cells, and memory B cells).
From an evolutionary perspective, we propose that the most primordial SLMs primarily evolved to promote processes of Ag-specific B cell clonal expansion, which, in turn, led to the generation of highly amplified Ag-specific Ab responses. In early jawed vertebrates, these SLMs arose as open, polyclonal, and rudimentarily organized SLMs (analogous to M-LAs). Because of their primitive organization [and consistent with the low-affinity Ab responses reported for cold-blooded species (17)], these primeval SLMs or M-LA–like structures would contain mostly low-affinity B cell clones because of their inability to select high-affinity ones. Over evolutionary time and with the acquisition of novel immunological innovations (i.e., FDCs, dark/light GC zones, more efficient AID, and lymph nodes), SLMs of early endotherms evolved to fine-tune Ab responses through their new capacity to select high-affinity B cell clones, which led to vast improvements in the affinity of the resulting Ab responses. Further evolution of these more highly organized SLMs in mammals probably led to the emergence of GCs with two main types of clonal composition, the newly identified polyclonal GCs (30) and the well-known homogeneous GCs composed of only a few selected and expanded high-affinity B cell clones. It is possible that polyclonal GCs and M-LAs have derived from primordial M-LA–like structures that emerged in the common ancestors of teleosts and mammals. Alternatively, polyclonal GCs and M-LAs arose independently through convergent evolutionary processes.
The induced B and T cell aggregates in these M-LAs showed different degrees of organization. It is possible that the less-organized B cell–T cell zones in some of these M-LAs represent intermediate stages of M-LA maturation. Although control fish also contained M-LAs, they were much smaller in size, and clear B cell and T cell zones were for the most part absent. In control fish, these small M-LAs could be aging M-LAs induced by previous antigenic exposure or M-LAs in the process of shutting down, similar to what has been described for mammalian GCs (31). Alternatively, they may represent active M-LAs induced by low concentrations of environmental Ag, auto-Ag, or Ag derived from microbiota translocating from mucosal areas. In that regard, it has been shown that Gram-negative microbiota from the guts of humans and mice continuously translocate at low levels into the bloodstream, thereby inducing significant specific systemic Ab responses (32). We have previously reported that a large proportion of the trout microbiota are coated by IgM (33). Thus, microbiota-specific B cells might be induced in M-LAs possibly by translocated microbiota. Further work is needed to evaluate these possibilities.
Although the induction of M-LAs or similar structures has never been described in fish, it has been proposed that fish MMCs could represent functional analogs of GCs (15, 16), in part because of their reported role in trapping Ag. Accordingly, melanomacrophages from these centers might play a role analogous to that of GC-associated FDCs, and, thus, MMCs could function in the selection of cognate B cells upon antigenic challenge. In support of this hypothesis, we saw large numbers of M-LA B cells, but not CD4+ T cells, in contact to MMCs. Selection of Ag-specific B cells could lead to clonal expansion of these selected B cell clones, as shown in this study. Upon infection, expansion of B cells in some teleosts has also been observed in mucosal lymphoid tissues lacking MMCs (19, 34, 35). Thus, the possibility exists that these B cell–containing areas without MMCs may also represent inductive sites of the immune response. For example, the presence of a novel organized nasopharynx-associated lymphoid tissue (O-NALT) in teleosts was recently identified (35). Upon viral infection, IgM+ B cell numbers increased in the O-NALT, and this was accompanied by up-regulation of aicda and cxcr4 transcripts in the same tissue and in the tip of the NALT, which is considered a nonorganized lymphoid tissue. The authors concluded that O-NALT expresses molecular markers that resemble mammalian GC reactions, and further work is warranted to evaluate whether O-NALT and other teleost lymphoid regions devoid of MMCs may functionally behave as GC analogs. In contrast to teleosts, some cold-blooded species contain B cell follicles, and those regions could represent tissue areas where adaptive immune responses are induced. For instance, sharks have previously been shown to contain B cell follicles (3), and a recent study reported that nurse shark B cell follicles constitute organized B cell selection sites where immune responses appear to be induced (36). B cell follicles have also been described in the adult Xenopus spleen, and although they are not present in all amphibian species (3), they appear to contain most of the cellular and molecular machinery required for the induction of adaptive immune responses (37).
The CDR3 repertoire data reported here support the notion that fish M-LAs are highly polyclonal structures where processes of Ag-specific B cell clonal expansion occur and indicate that sharing of the most frequent CDR3s among M-LAs of Ag-stimulated animals is an important feature of fish M-LAs. These findings support the notion of a high degree of reseeding and/or exchanges of common Ag-specific B cell clones among M-LAs induced during the same response. These properties of fish M-LAs show interesting commonalities with mammalian GCs operating in polyclonal settings. These GCs are seeded by several hundred naïve B cells, in particular when complex Ags are used (30). Furthermore, early secondary GCs after boost are reseeded and diversified by a substantial number of naïve B cells in addition to memory B cells (38, 39). In such scenarios, the resulting GC reaction ends up with clonally diverse (polyclonal) GCs that include not only a few highly expanded high-affinity B cell clones but also a large number of medium- and low-affinity B cell clones (38) and even B cell clones with no specificity for the Ag used for immunization (40). Moreover, and similar to what we show here for M-LAs, it has been reported that polyclonal GCs induced in the same response also share a significant number of their most frequent B cell clones (30), although only pairs of GCs were compared, and the degree of clonotype sharing between those GCs was significantly lower than what we typically see when comparing the herein analyzed fish M-LAs. Whereas M-LAs resemble those newly described polyclonal GCs, M-LAs would not contain medium- or high-affinity B cell clones based on previous studies, showing that affinity maturation of the Ab response in teleosts is poor (17, 41). After infection, we did not detect an M-LA in which the most frequent clones were not shared with other M-LAs. The latter strongly suggests that polyclonal M-LAs are the norm in fish, whereas mammals contain those recently found polyclonal GCs in addition to the well-known clonally homogenous GCs that contain only a few selected and highly expanded high-affinity B cell clones because of intraclonal competition and as a result of clonal burst processes (25).
We found that AID expression was mainly localized in M-LA B cells, a situation resembling that of mammalian AID that is known to be expressed at its highest levels in GC B cells (25). It is known that within GCs, AID is responsible for 75% of apoptosis in dark zone B cells (28) and that apoptosis is considered an important mechanism required to counteract the large degree of B cell proliferation (25). Here, we show that significant B cell apoptosis was only detected within M-LAs of infected animals, thus suggesting that AID may also be a significant inducer of B cell apoptosis within M-LAs and that apoptosis in these microstructures is an important mechanism required to offset the high proliferation levels of M-LA B cells. We found that the percentage of B cells entering apoptosis in M-LAs was similar to that reported for GCs (2.6 to 5.7%).
Although affinity maturation of Ig responses is generally poor in fish and other poikilotherms, evidence of SHM of Ag receptor genes has been reported in several species (9, 42–46). The high levels of AID expression detected in M-LA B cells of infected fish suggested that SHM could be taking place within these cells. In support, our data show that M-LAs display significantly higher sequence variation in the CDR1-FR3 region of the V domain compared with non–M-LA areas, thus indicating the occurrence of SHM within these lymphoid microstructures. Although CDR3 sharing analysis between M-LAs in the same spleen indicated that large B cell clones after immunization likely move between these areas, our SHM studies also showed that shared lineages are the ones displaying the highest sequence variation. This prevalent concentration of SHM on shared lineages of M-LAs has not been described in any other vertebrate lymphoid microstructure, including GCs. A recent study found strong evidence of SHM taking place in MMCs from the HK in zebrafish (47). However, it remains to be determined whether SHM occurred in resident HK B cells responding to Ag or whether SHM occurred in plasmablast/plasma-like cells that migrated into the HK from inductive lymphoid tissue localized in the spleen or other lymphoid organs.
In conclusion and in contrast to the current dogma stating the absence of GCs or analogous organized SLMS in cold-blooded vertebrates (3), here, we show the existence of inducible teleost M-LAs with structural and functional features analogous to those of endothermic GCs. Further research is warranted to evaluate the evolutionary origins of M-LAs and GCs and how these structures shape the ultimate diversity of Ab responses in polyclonal settings. Last, it is also worth considering that because the overall structure of M-LAs is less organized than that of GCs, M-LAs might be more evolutionarily related to immature tertiary lymphoid organs (TLOs), which contain loosely organized B and T cell zones and lack GCs (48). We propose that comparative studies of fish M-LAs (or equivalent cold-blooded SLM structures) may point to primordially conserved principles and/or convergent processes by which M-LAs, TLOs, and the newly found polyclonal GCs are formed and regulated.
MATERIALS AND METHODS
Study design
The objective of this study was to determine how and where adaptive immune responses are induced in vertebrate species that lack GCs or analogous lymphoid structures. Immune responses were induced by parasite infection or through immunization with DNP-KLH. We used immunofluorescence (IF) microscopy to search the spleen for areas of Ag-specific B cell and T cell proliferation that led to the identification of M-LAs. To determine the clonality of B cells, we analyzed the IgHμ CDR3 repertoire of microdissected splenic M-LAs and non–M-LA areas from infected and control fish. Through in situ hybridization studies, we also determined whether M-LA B cells expressed AID, whereas through IF microscopy, we analyzed the percentages of these cells undergoing apoptosis. To evaluate whether SHM occurs in M-LA B cells, we calculated the sequence variation of clonal set variants found within microdissected splenic M-LAs and non–M-LA areas. Animal numbers per group are indicated in the respective figure legends. Sample sizes were chosen according to previous comparable studies conducted in our laboratory. Investigators were not blinded to groups. No data were excluded.
Fish husbandry
Rainbow trout (O. mykiss) were obtained from Troutlodge (Summer, WA) and were maintained as previously described by us (18). Fish (13 to 17 g) were acclimatized for 2 weeks at 15°C in an aerated recirculating aquarium with internal biofilters and fed daily with dry pellets at 1% biomass/day. All animal procedures were approved by the Institutional Animal Care and Use Committees of the University of Pennsylvania.
Ich parasite isolation and infection
Isolation of Ich parasite and Ich infections was performed as previously reported by us (18, 19). Briefly, for primary Ich infections, fish (n = 7) were exposed to Ich theronts (~1000 theronts per fish), and 8 to 10 days after exposure, infection was visually confirmed with the typical white spots (Ich trophonts) covering the skin and gills of the fish. Once the Ich trophonts detached from fish and no visible white spots were detected (~14 days after exposure), fish were transferred and quarantined in an Ich-free flow through tank for 2 weeks. Reinfection (secondary Ich exposure) of quarantined fish was conducted following the aforementioned protocol 4 weeks after the first exposure. Mock-infected fish (control fish) were kept in an Ich-free water tank following the same timelines used for Ich primary and secondary exposures. Both reinfected and control fish were euthanized 14 days after reinfection or mock infection, respectively, with an overdose (300 mg/liter) of tricaine methanesulfonate (MS-222, Syndel) as previously reported by us (18, 19). The spleens and sera from euthanized animals were collected as described below for further experiments. All analyses in this manuscript were performed on samples from reinfected fish and their respective mock-infected control animals.
Fish immunizations
Fish were immunized by intraperitoneal injection with 100 μg of DNP-KLH [Millipore; 1 mg/ml in phosphate-buffered saline (PBS), pH 7.2] emulsified in Freund’s complete adjuvant (Sigma-Aldrich). Four weeks after primary immunization, fish were boosted with the same amount of DNP-KLH emulsified in Freund’s incomplete adjuvant (Sigma-Aldrich). Two weeks after the booster immunization, fish were euthanized as described above. Control fish received 100 μl of PBS mixed with the aforementioned adjuvants and were sampled following the same protocol. Sera and spleen tissue samples were collected as previously described by us (18, 19) or further experiments. All analyses below were performed on samples from boosted fish and their respective control animals.
Specificity analysis of guinea pig antitrout CD4 pAb by flow cytometry
Splenic leukocyte suspensions were obtained as previously described (19, 49). To evaluate the specificity of the newly produced guinea pig (gp) antitrout CD4 pAb, splenic leukocyte suspensions (1 × 106) were double-stained with gp antitrout CD4 pAb (1 μg/ml) and rat antitrout CD4 monoclonal Ab (mAb) (clone 4.2.12, rat IgG2b; 5 μg/ml) (49). Primary Abs were then detected with Alexa Fluor 647–conjugated goat anti-gp IgG (Jackson ImmunoResearch) and PE-conjugated goat anti-rat IgG2b (Invitrogen) at 1 μg/ml each. To elucidate whether the gp antitrout CD4 pAb showed any potential cross-reactivity IgM+ B cells, splenic leukocytes (1 × 106) were stained with both antitrout CD4 pAb (1 μg/ml) and biotinylated antitrout IgM mAb (1 μg/ml). Primary Abs were thereafter detected with Alexa Fluor 647–conjugated goat anti-gp IgG (Jackson ImmunoResearch) and Brilliant Violet 421 Streptavidin (BioLegend) at 1 μg/ml each. As isotype-matched controls for primary Abs, we used gp IgG purified from preimmune sera (1 μg/ml) and rat IgG2b (clone RTK4530, BioLegend; 5 μg/ml). Flow cytometry was performed using a FACSCanto cell analyzer (BD Biosciences), and data were analyzed using FlowJo software (FlowJo LLC).
Detection of Ag-specific B cells by IF microscopy and flow cytometry
The strategy of the methodology to detect Ag-specific B cells on spleen cryosections is shown in fig. S4. Ag-specific B cells were detected by IF microscopy on spleen cryoblocks of DNP-KLH–boosted fish and the respective control fish. Spleen cryoblocks were sectioned at a thickness of 10 μm and then fixed for 10 min in 2% paraformaldehyde. The sections were thereafter permeabilized in 0.5% Tween 20 (Promega) in PBS (PBS-Tween; pH 7.2) for 15 min at room temperature (RT) and then blocked with 2% bovine serum albumin (BSA; Gold Biotechnology) in PBS (PBS-BSA; pH 7.2) for 30 min. To detect Ag-specific B cells, sections were then probed with DNP-PE (Millipore; 50 μg/ml) for 2 hours at RT. After incubation with DNP-PE, sections were washed three times with PBS and stained with antitrout IgM mAb and antitrout CD4 pAb, followed by Alexa Fluor 488–conjugated goat anti-mouse IgG1 (Invitrogen) and Alexa Fluor 647–conjugated goat anti-gp IgG (Invitrogen), respectively, as described above. Cell nuclei were stained with 4′,6-diamidino-2-phenylindole (1 μg/ml) before mounting with Fluoroshield. Images were acquired and analyzed using a Leica DM6000 fluorescence microscope or Leica SP8 confocal microscope and LAS X software. The percentage of M-LAs areas containing a high proportion of Ag-specific B cells from immunized and control fish was found by quantifying the percentage of M-LAs containing >10% of Ag-specific B cells in the spleens of infected and control fish.
To detect Ag-specific B cells by flow cytometry, spleen leukocytes from control and boosted fish were obtained as described above and stained with DNP-PE (50 μg/ml) for 30 min at 4°C in Dulbecco’s modified Eagle’s medium (DMEM, Thermo Fisher Scientific). Thereafter, cells were washed three times with DMEM and costained with mouse antitrout IgM mAb (1 μg/ml) and gp antitrout CD4 pAb (1 μg/ml) as described above. Positively stained IgM+ and CD4+ cells were detected with Alexa Fluor 488–conjugated goat anti-mouse IgG1 (Invitrogen) and Alexa Fluor 647–conjugated goat anti-gp IgG (Invitrogen), respectively, at 2.5 μg/ml each. Flow cytometry was performed using a FACSCanto cell analyzer (BD Biosciences), and data were analyzed using FlowJo software (FlowJo LLC).
LCM of M-LA and non–M-LA areas and RNA extraction
The strategy of the methodology to identify and microdissect M-LA and non–M-LA areas of spleen cryoblocks is depicted in fig. S6. Cryoblocks of the spleen from Ich-reinfected fish, DNP-KLH–boosted fish, and the respective control fish were serially sectioned (17 to 20 sections) at a thickness of 10 μm and mounted onto PEN membrane slides (Leica). M-LA and non–M-LA areas in these sections were identified as described in fig. S6 (A and B). Sections containing the chosen M-LA and non–M-LA areas (corresponding to sections 2 to 6 from fig. S6B) were then fixed in ethanol (Sigma-Aldrich) and briefly washed in nuclease-free water (Sigma-Aldrich). The fixed sections were thereafter stained with HistoGene staining solution (Applied Biosystems) and dehydrated in a series of ethanol (70, 90, and 100%). At this point, sections were ready for microdissection with LCM system (LMD7000, Leica). Using the Leica software from LCM apparatus, we quantified the exact area that was harvested for M-LAs, and these data were then used to harvest the same amount of area from non–M-LA regions of the same spleen section. The microdissected tissues were collected in a microtube containing buffer RLT from the RNeasy Micro Kit (QIAGEN), and total RNA from M-LA and non–M-LA microdissected sections was extracted using the RNeasy Micro Kit according to the manufacturer’s instructions. To minimize RNA degradation, all procedures were performed quickly and under ribonuclease-free conditions.
Production of gp antitrout CD4 pAbs
To detect trout CD4+ T cells on tissue cryosections by IF microscopy and flow cytometry, we produced a pAb against trout CD4-2b. To this end, recombinant trout CD4-2b (CD4) was produced as a CD4/Fc fusion protein (Fc from mouse IgG2b) and purified as previously described by us (20). The recombinant protein was used to raise pAbs in gps by Cocalico Biologicals Inc. according to their standard protocols. gp CD4-specific serum IgG titers from immunized animals were determined by enzyme-linked immunosorbent assay as described in (20). Purification of IgG from gp serum was carried out as previously described by us (20) with some protocol modifications. Briefly, total gp IgG was purified from antisera or preimmune sera using a HiTrap protein A column (GE Healthcare) according to the manufacturer’s instructions. The purified total IgG from antisera was then passed through an in-house mouse IgG2b-affinity column to remove pAbs recognizing Fc from mouse IgG2b. The resulting flow-through fraction was then applied to an in-house CD4/Fc fusion protein-affinity column to purify CD4-specific gp IgG. Specificity of affinity-purified gp antitrout CD4 IgG against trout CD4 was determined by enzyme-linked immunosorbent assay, flow cytometry, and IF microscopy following the same methodologies described by us in (20, 49).
In situ hybridization
An 820–base pair (bp) fragment of trout AID cDNA (GenBank accession number: XM_036961711.1) and a 1742-bp fragment of the constant region of trout IgHμ cDNA (GenBank accession number: S63348.1) were subcloned into a pGEM-T easy vector (Promega) according to the manufacturer’s instructions. The AID probes (table S2) were synthesized using digoxigenin (DIG) RNA Labeling Kit (Roche), and the IgHμ probes (table S2) were synthesized using biotin RNA labeling Mix (Roche) and stained following the manufacturer’s instructions. Spleen cryoblocks were sectioned at a thickness of 10 μm, then fixed for 2 hours at 4°C in 4% paraformaldehyde and washed three times with PBS. The sections were permeabilized by incubating in 0.5% PBS-Tween for 15 min at RT and then acetylated. After acetylation, the sections were washed with PBS, and then prehybridization was carried out in hybridization solution [4× saline sodium citrate buffer (SSC; Corning Life Sciences), 40% formamide (WAKO), 10% dextran sulfate sodium (WAKO), 2× Denhardt’s solution (WAKO), yeast tRNA (0.5 mg/ml; Thermo Fisher Scientific, and 1× ribonuclease inhibitor (Sigma-Aldrich)] for 10 min at RT. Hybridization was performed with DIG-labeled and biotinylated RNA probes (final concentration, 1.1 μg/ml) in hybridization solution for 18 hours at 50°C in a humidified chamber. After hybridization, the slides were washed as follows: 2× SSC for 5 min at RT, 2× SSC containing 10% formamide for 20 min at 50°C, twice with 1× SSC for 30 min at 50°C, and PBS for 10 min at RT. Before immunostaining, the slides were blocked with 2% PBS-BSA. Anti-digoxigenin-AP (alkaline phosphatase), Fab fragments (Roche) at a dilution of 1:250, and streptavidin Alexa Fluor 488 (BioLegend) with blocking solution were applied and incubated overnight at 4°C. Subsequently, the slides were washed three times for 5 min with 0.05% PBS-Tween at RT and lastly for 5 min in PBS at RT. Hybridized signals were visualized using ImmPACT Vector Red AP Substrate kit (Vector Laboratories). Cell nuclei were stained with 4′,6-diamidino-2-phenylindole (1 μg/ml) before mounting with Fluoroshield, and images were acquired and analyzed using a Leica DM6000 fluorescence microscope or Leica SP8 confocal microscope and LAS X software.
cDNA synthesis, reverse transcription polymerase chain reaction, and Illumina sequencing of IgHμ CDR3s repertoire
To analyze the expressed IgHμ CDR3 repertoire from LCM of M-LAs and non–M-LA areas from the spleens of Ich-reinfected fish, DNP-KLH–boosted fish, and the respective control fish, we specifically amplified IgHμ cDNA transcripts from the total RNA preparations obtained from each microdissected area as described above. To this end, first-strand cDNA was synthesized using 10 to 20 pg of total RNA with SuperScript IV reverse transcriptase (Thermo Fisher Scientific) using the IGMR1 primer (table S1). For IgHμ CDR3 analysis, the first-strand cDNA was amplified with IGV5 (table S1) and IGC3 primers (table S1) using Platinum Hot Start PCR Master Mix (Thermo Fisher Scientific) for 30 cycles. These PCR products therefore contained the end of the FR3, the CDR3, and the IGHJ segment of IgHμ. Illumina adaptors were then introduced in a second PCR (10 cycles) using the Nextera XT Index Kit v2 (Illumina). These PCR products were measured with Qubit (Thermo Fisher Scientific), combined, and purified twice with the QIAGEN PCR clean up kit. These libraries were then sequenced with MiSeq Reagent Kits v3 (Illumina) (150 cycles) according to the manufacturer’s instructions. CDR3-IMGT sequences [positions 105 to 117 in IMGT numbering (i.e., position following the Cys104 to position preceding J-Trp118 in IgHμ sequences)] from each sample were extracted and organized in a database.
Analysis of SHM
To look for evidence of SHM in IgHμ V sequences, we amplified the IgVHμ region containing CDR1-FR2-CDR2-FR3-CDR3-IGHJ, using as a template the IgHμ cDNAs from the same M-LA and non–M-LA areas obtained in the previous section (CDR3 repertoire analysis). To this end, we used a forward VH-specific primer cocktail (table S1) covering all IGHV subgroups, although Cμ1_R (primer in the first constant domain of IgHμ) was used as a reverse primer (table S1). After amplification, MiSeq Illumina libraries were constructed from these amplified PCR products and sequenced as described in the above section using MiSeq Reagent Kits v3 (Illumina). The obtained sequences were analyzed by IMGT/HighV-QUEST based on our rainbow trout annotation of the IgH locus (50, 51). IGHV gene annotation proposed by IMGT HighV-QUEST was validated when the sequences were >97.5% identical to the closest germline sequence in the aforementioned IMGT database. The nonvalidated sequences (i.e., gene/allele variation) were discarded to reduce the probability of confusion between sequence variation due to hypermutation detected in the validated sequences and that derived from gene/allele variation arisen during evolution. Among sequences kept for further analysis (i.e., validated sequences), those with identical CDR3-IMGT (i.e., sequences with the same nucleotide sequence encoding residues 105 to 117, between conserved Cys104 and Trp118 of the V domain) and with the same IGHV gene annotation were grouped, thus becoming a “clonal set” [as described in (52)]. Within each clonal set, reads that deviated by more than five nucleotides from the most frequent sequence of the clonal set were filtered out, again to reduce the risk of including divergent germline variants produced during evolution. This highly conservative approach for producing sequence lineages excluded both SHM located in the CDR3 and potential heavily mutated sequences, but it offers a maximum guarantee of avoiding variations due to illicit grouping of related distinct genes. Sequence variation (i.e., SHM) was assessed within each clonal set containing more than 30 sequences and was computed as the number of nucleotides deviating from the most frequent sequence of the clonal set divided by the total number of nucleotides. Sequence variation was computed both in shared and nonshared clonal sets of M-LAs or non–M-LA areas within the spleen of the same fish. Sharing of clonal sets in different M-LAs or non–M-LA areas was defined as the presence in these areas of at least one sequence of the clonal set (i.e., if three M-LAs were obtained from the same spleen, then a clonal set was considered shared when at least one of its sequences was identified in each of the three M-LAs). In contrast to these shared clonal sets, we considered nonshared clonal sets in which their sequence variants could only be found in one of either the M-LA or non–M-LA areas of the same spleen.
Statistical analysis
The sample size and number of independent experiments are indicated in the figure captions. Histological evaluations and cell counting were performed by two independent researchers. No data were excluded. Unpaired Student’s t test and one-way and two-way analyses of variance (ANOVAs) followed by Tukey’s multiple comparison test were performed in Prism (GraphPad) for analysis of differences between groups. P values of 0.05 or less were considered statistically significant.
Supplementary Material
Acknowledgments:
J.O.S. dedicates this manuscript to the loving memory of his father, who recently passed away. His incessant encouragement, love, and quest for continuous improvement have deeply influenced J.O.S.’s life, including this work. We thank S. Prouty and the Penn Skin Biology and Diseases Resource–based Center, funded by NIH/NIAMS grant P30-AR069589 and the University of Pennsylvania Perelman School of Medicine, for the help provided with the LCM; G. Ruthel of PennVet Imaging Core and Nihon University general research institute for help with microscope analysis; the personnel of the University Laboratory Animal Resources (ULAR) of the University of Pennsylvania for invaluable help in the daily husbandry of our rainbow trout; A. Carty for the excellent veterinary care of the fish; and D. Stefanovski (School of Veterinary Medicine, University of Pennsylvania) and A. Serradell (University of Las Palmas, Spain) for advice in statistical analysis of this study. Y.S. is a Japan Society for the Promotion of Science (JSPS) Overseas Research Fellow.
Funding:
This work was supported by National Institutes of Health 2R01GM085207-09 (to J.O.S.), U.S. Department of Agriculture grant USDA-NIFA 2020-06446 (to J.O.S.), U.S. Department of Agriculture grant USDA-NIFA 2021-06961 (to J.O.S.), Japan Society for the Promotion of Science Overseas Research Fellowship (to Y.S.), Japan Society for the Promotion of Science KAKENHI 20 K22594 (to Y.S.), Japan Society for the Promotion of Science KAKENHI 21H02288 (to Y.S. and F.T.), Japan Society for the Promotion of Science KAKENHI 20KK0144 (to Y.S. and F.T.), and National Natural Science Foundation of China 31802337 (to Y.D.).
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability:
All data are available in the main text or the Supplementary Materials. Sequencing data used in this work are deposited at the BioProject National Center for Biotechnology Information database with the SRA accession number PRJNA854339 (www.ncbi.nlm.nih.gov/bioproject/854339). The newly generated gp antitrout CD4 pAbs can be provided by J.O.S. pending a completed material transfer agreement. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
REFERENCES AND NOTES
- 1.Parra D, Takizawa F, Sunyer JO, Evolution of B cell immunity. Annu. Rev. Anim. Biosci. 1, 65–97 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hirano M, Das S, Guo P, Cooper MD, The evolution of adaptive immunity in vertebrates. Adv. Immunol. 109, 125–157 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Flajnik MF, A cold-blooded view of adaptive immunity. Nat. Rev. Immunol. 18, 438–453 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hofmann J, Greter M, Du Pasquier L, Becher B, B-cells need a proper house, whereas T-cells are happy in a cave: The dependence of lymphocytes on secondary lymphoid tissues during evolution. Trends Immunol. 31, 144–153 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Zarrin AA, Alt FW, Chaudhuri J, Stokes N, Kaushal D, Du Pasquier L, Tian M, An evolutionarily conserved target motif for immunoglobulin class-switch recombination. Nat. Immunol. 5, 1275–1281 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Yasuda M, Taura Y, Yokomizo Y, Ekino S, A comparative study of germinal center: Fowls and mammals. Comp. Immunol. Microbiol. Infect. Dis. 21, 179–189 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Barreto VM, Magor BG, Activation-induced cytidine deaminase structure and functions: A species comparative view. Dev. Comp. Immunol. 35, 991–1007 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Marr S, Morales H, Bottaro A, Cooper M, Flajnik M, Robert J, Localization and differential expression of activation-induced cytidine deaminase in the amphibian Xenopus upon antigen stimulation and during early development. J. Immunol. 179, 6783–6789 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Yang F, Waldbieser GC, Lobb CJ, The nucleotide targets of somatic mutation and the role of selection in immunoglobulin heavy chains of a teleost fish. J. Immunol. 176, 1655–1667 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Marianes AE, Zimmerman AM, Targets of somatic hypermutation within immunoglobulin light chain genes in zebrafish. Immunology 132, 240–255 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Press CM, Dannevig BH, Landsverk T, Immune and enzyme histochemical phenotypes of lymphoid and nonlymphoid cells within the spleen and head kidney of Atlantic salmon (Salmo salar L.). Fish Shellfish Immunol. 4, 79–93 (1994). [Google Scholar]
- 12.Bermudez R, Vigliano F, Marcaccini A, Sitja-Bobadilla A, Quiroga MI, Nieto JM, Response of Ig-positive cells to Enteromyxum scophthalmi (Myxozoa) experimental infection in turbot, Scophthalmus maximus (L.): A histopathological and immunohistochemical study. Fish Shellfish Immunol. 21, 501–512 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Steinel NC, Bolnick DI, Melanomacrophage centers as a histological indicator of immune function in fish and other poikilotherms. Front. Immunol. 8, 827 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gyimesi ZS, Howerth EW, Severe melanomacrophage hyperplasia in a crocodile lizard, Shinisaurus crocodilurus: A review of melanomacrophages in ectotherms. J. Herpetol. Med. Surg. 14, 19–23 (2004). [Google Scholar]
- 15.Agius C, Roberts RJ, Melano-macrophage centres and their role in fish pathology. J. Fish Dis. 26, 499–509 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Ferguson HW, The relationship between ellipsoids and melano-macrophage centres in the spleen of turbot (Scophthalmus maximus). J. Comp. Pathol. 86, 377–380 (1976). [DOI] [PubMed] [Google Scholar]
- 17.Sunyer JO, Boudinot P, B-cell responses and antibody repertoires in teleost fish: From Ag receptor diversity to immune memory and vaccine development, in Principles of Fish Immunology: From Cells and Molecules to Host Protection, Buchmann K, Secombes CJ, Eds. (Springer, 2022), pp. 253–278. [Google Scholar]
- 18.Xu Z, Takizawa F, Casadei E, Shibasaki Y, Ding Y, Sauters TJC, Yu Y, Salinas I, Sunyer JO, Specialization of mucosal immunoglobulins in pathogen control and microbiota homeostasis occurred early in vertebrate evolution. Sci. Immunol. 5, eaay3254 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Z, Takizawa F, Parra D, Gomez D, von Gersdorff Jorgensen L, LaPatra SE, Sunyer JO, Mucosal immunoglobulins at respiratory surfaces mark an ancient association that predates the emergence of tetrapods. Nat. Commun. 7, 10728 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang YA, Salinas I, Li J, Parra D, Bjork S, Xu Z, LaPatra SE, Bartholomew J, Sunyer JO, IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat. Immunol. 11, 827–835 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sales CF, Silva RF, Amaral MG, Domingos FF, Ribeiro RI, Thomé RG, Santos HB, Comparative histology in the liver and spleen of three species of freshwater teleost. Neotrop. Ichthyol. 15, 10.1590/1982-0224-20160041, (2017). [DOI] [Google Scholar]
- 22.Steiniger BS, Human spleen microanatomy: Why mice do not suffice. Immunology 145, 334–346 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fänge R, Nilsson S, The fish spleen: Structure and function. Experientia 41, 152–158 (1985). [DOI] [PubMed] [Google Scholar]
- 24.Garside P, Ingulli E, Merica RR, Johnson JG, Noelle RJ, Jenkins MK, Visualization of specific B and T lymphocyte interactions in the lymph node. Science 281, 96–99 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Victora GD, Nussenzweig MC, Germinal centers. Annu. Rev. Immunol. 40, 413–442 (2022). [DOI] [PubMed] [Google Scholar]
- 26.Mizutani Y, Tsuge S, Shiogama K, Shimomura R, Kamoshida S, Inada K, Tsutsumi Y, Enzyme-labeled antigen method: Histochemical detection of antigen-specific antibody-producing cells in tissue sections of rats immunized with horseradish peroxidase, ovalbumin, or keyhole limpet hemocyanin. J. Histochem. Cytochem. 57, 101–111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cossarini-Dunier M, Desvaux FX, Dorson M, Variability in humoral responses to DNP-KLH of rainbow trout (Salmo gairdneri). Comparison of antibody kinetics and immunoglobulins spectrotypes between normal trouts and trouts obtained by gynogenesis or self-fertilization. Dev. Comp. Immunol. 10, 207–217 (1986). [DOI] [PubMed] [Google Scholar]
- 28.Mayer CT, Gazumyan A, Kara EE, Gitlin AD, Golijanin J, Viant C, Pai J, Oliveira TY, Wang Q, Escolano A, Medina-Ramirez M, Sanders RW, Nussenzweig MC, The micro-anatomic segregation of selection by apoptosis in the germinal center. Science 358, eaao2602 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sepahi A, Kraus A, Casadei E, Johnston CA, Galindo-Villegas J, Kelly C, Garcia-Moreno D, Munoz P, Mulero V, Huertas M, Salinas I, Olfactory sensory neurons mediate ultrarapid antiviral immune responses in a TrkA-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 116, 12428–12436 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tas JMJ, Mesin L, Pasqual G, Targ S, Jacobsen JT, Mano YM, Chen CS, Weill J-C, Reynaud C-A, Browne EP, Meyer-Hermann M, Victora GD, Visualizing antibody affinity maturation in germinal centers. Science 351, 1048–1054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arulraj T, Binder SC, Robert PA, Meyer-Hermann M, Germinal centre shutdown. Front. Immunol. 12, 705240 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng MY, Cisalpino D, Varadarajan S, Hellman J, Warren HS, Cascalho M, Inohara N, Nunez G, Gut microbiota-induced immunoglobulin G controls systemic infection by symbiotic bacteria and pathogens. Immunity 44, 647–658 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salinas I, Fernandez-Montero A, Ding Y, Sunyer JO, Mucosal immunoglobulins of teleost fish: A decade of advances. Dev. Comp. Immunol. 121, 104079 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Z, Parra D, Gómez D, Salinas I, Zhang Y-A, von Gersdorff Jørgensen L, Heinecke RD, Buchmann K, La Patra S, Sunyer JO, Teleost skin, an ancient mucosal surface that elicits gut-like immune responses. Proc. Natl. Acad. Sci. U.S.A. 110, 13097–13102 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia B, Dong F, Casadei E, Rességuier J, Ma J, Cain KD, Castrillo PA, Xu Z, Salinas I, A novel organized nasopharynx-associated lymphoid tissue in teleosts that expresses molecular markers characteristic of mammalian germinal centers. J. Immunol. 209, 2215–2226 (2022). [DOI] [PubMed] [Google Scholar]
- 36.Matz H, Taylor RS, Redmond AK, Hill TM, Daniels RR, Beltran M, Henderson NC, Macqueen DJ, Dooley H, Organized B cell sites in cartilaginous fishes reveal the evolutionary foundation of germinal centers. Cell Rep. 42, 112664 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neely HR, Guo J, Flowers EM, Criscitiello MF, Flajnik MF, “Double-duty” conventional dendritic cells in the amphibian Xenopus as the prototype for antigen presentation to B cells. Eur. J. Immunol. 48, 430–440 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mesin L, Schiepers A, Ersching J, Barbulescu A, Cavazzoni CB, Angelini A, Okada T, Kurosaki T, Victora GD, Restricted clonality and limited germinal center reentry characterize memory B cell reactivation by boosting. Cell 180, 92–106.e11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turner JS, Zhou JQ, Han J, Schmitz AJ, Rizk AA, Alsoussi WB, Lei T, Amor M, McIntire KM, Meade P, Strohmeier S, Brent RI, Richey ST, Haile A, Yang YR,Klebert MK, Suessen T, Teefey S, Presti RM, Krammer F, Kleinstein SH, Ward AB, Ellebedy AH, Human germinal centres engage memory and naive B cells after influenza vaccination. Nature 586, 127–132 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gregoire C, Spinelli L, Villazala-Merino S, Gil L, Holgado MP, Moussa M, Dong C, Zarubica A, Fallet M, Navarro JM, Malissen B, Milpied P, Gaya M, Viral infection engenders bona fide and bystander subsets of lung-resident memory B cells through a permissive mechanism. Immunity 55, 1216–1233.e9 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ye J, Kaattari IM, Kaattari SL, The differential dynamics of antibody subpopulation expression during affinity maturation in a teleost. Fish Shellfish Immunol. 30, 372–377 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Wilson M, Hsu E, Marcuz A, Courtet M, Du Pasquier L, Steinberg C, What limits affinity maturation of antibodies in Xenopus--the rate of somatic mutation or the ability to select mutants? EMBO J. 11, 4337–4347 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hinds-Frey KR, Nishikata H, Litman RT, Litman GW, Somatic variation precedes extensive diversification of germline sequences and combinatorial joining in the evolution of immunoglobulin heavy chain diversity. J. Exp. Med. 178, 815–824 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diaz M, Greenberg AS, Flajnik MF, Somatic hypermutation of the new antigen receptor gene (NAR) in the nurse shark does not generate the repertoire: Possible role in antigen-driven reactions in the absence of germinal centers. Proc. Natl. Acad. Sci. U.S.A. 95, 14343–14348 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee SS, Tranchina D, Ohta Y, Flajnik MF, Hsu E, Hypermutation in shark immunoglobulin light chain genes results in contiguous substitutions. Immunity 16, 571–582 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Jiang N, Weinstein JA, Penland L, White III RA, Fisher DS, Quake SR, Determinism and stochasticity during maturation of the zebrafish antibody repertoire. Proc. Natl. Acad. Sci. U. S.A. 108, 5348–5353 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waly D, Muthupandian A, Fan C-W, Anzinger H, Magor BG, Immunoglobulin VDJ repertoires reveal hallmarks of germinal centers in unique cell clusters isolated from zebrafish (Danio rerio) lymphoid tissues. Front. Immunol. 13, 7032 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neyt K, Perros F, GeurtsvanKessel CH, Hammad H, Lambrecht BN, Tertiary lymphoid organs in infection and autoimmunity. Trends Immunol. 33, 297–305 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takizawa F, Magadan S, Parra D, Xu Z, Korytar T, Boudinot P, Sunyer JO, Novel teleost CD4-bearing cell populations provide insights into the evolutionary origins and primordial roles of CD4+ lymphocytes and CD4+ macrophages. J. Immunol. 196, 4522–4535 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Magadan S, Krasnov A, Hadi-Saljoqi S, Afanasyev S, Mondot S, Lallias D, Castro R, Salinas I, Sunyer JO, Hansen J, Koop BF, Lefranc MP, Boudinot P, Standardized IMGT® nomenclature of salmonidae IGH genes, the paradigm of Atlantic salmon and rainbow trout: From genomics to repertoires. Front. Immunol. 10, 2541 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Magadan S, Mondot S, Palti Y, Gao G, Lefranc MP, Boudinot P, Genomic analysis of a second rainbow trout line (Arlee) leads to an extended description of the IGH VDJ gene repertoire. Dev. Comp. Immunol. 118, 103998 (2021). [DOI] [PubMed] [Google Scholar]
- 52.Ott JA, Castro CD, Deiss TC, Ohta Y, Flajnik MF, Criscitiello MF, Somatic hypermutation of T cell receptor α chain contributes to selection in nurse shark thymus. eLife 7, e28477 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeLuca D, Wilson M, Warr GW, Lymphocyte heterogeneity in the trout, Salmo gairdneri, defined with monoclonal antibodies to IgM. Eur. J. Immunol. 13, 546–551 (1983). [DOI] [PubMed] [Google Scholar]
- 54.Lambert V, Lecomte J, Hansen S, Blacher S, Gonzalez M-LA, Struman I, Sounni NE, Rozet E, de Tullio P, Foidart JM, Rakic J-M, Noel A, Laser-induced choroidal neovascularization model to study age-related macular degeneration in mice. Nat. Protoc. 8, 2197–2211 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Ke F, Benet ZL, Maz MP, Liu J, Dent AL, Kahlenberg JM, Grigorova IL, Germinal center B cells that acquire nuclear proteins are specifically suppressed by follicular regulatory T cells. eLife 12, e83908 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen E, Rabe H, Luo L, Wang L, Wang Q, Yin J, Yang X, Liu W, Sido JM, Nakagawa H, Ao L, Kim H-J, Cantor H, Leavenworth JW, Control of germinal center localization and lineage stability of follicular regulatory T cells by the Blimp1 transcription factor. Cell Rep. 29, 1848–1861.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the Supplementary Materials. Sequencing data used in this work are deposited at the BioProject National Center for Biotechnology Information database with the SRA accession number PRJNA854339 (www.ncbi.nlm.nih.gov/bioproject/854339). The newly generated gp antitrout CD4 pAbs can be provided by J.O.S. pending a completed material transfer agreement. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.


