Abstract
Objective:
The purpose of this study is to comparatively analyze the anticancer properties of Tetrahydrocannabinol (THC), Cannabidiol (CBD), and Tetrahydrocannabivarin (THCV) using In silico tools.
Methods:
Using SwissADME and pkCSM, the physicochemical and pharmacokinetics properties of the cannabinoids were evaluated. Protox-II was utilized for the assessment of their cytotoxicity. The chemical-biological interactions of the cannabinoids were also predicted using the Way2Drug Predictive Server which comprises Acute Rat Toxicity, Adver-Pred, CLC-Pred, and Pass Target Prediction.
Results:
Both physicochemical and drug-likeness analysis using SwissADME favored THCV due to high water solubility and lower MLOGP value. On the other hand, ADMET assessment demonstrated that THC and CBD have good skin permeability while both THC and THCV exhibited better BBB permeability and have low inhibitory activity on the CYP1A2 enzyme. Furthermore, toxicity predictions by Protox-II revealed that CBD has the lowest probability of hepatotoxicity, carcinogenicity, and immunotoxicity. Contrarily, it has the highest probability of being inactive in mutagenicity and cytotoxicity. Additionally, CLC results revealed that CBD has the highest probability against lung carcinoma. The rat toxicity prediction showed that among the cannabinoids, THCV had the lowest LD50 concentration in rat oral and IV.
Conclusion:
Overall, in silico predictions of the three cannabinoid compounds revealed that they are good candidates for oral drug formulation. Among the three cannabinoids, THCV is an excellent anticancer aspirant for future chemotherapy with the most favorable results in drug-likeness and ADMET analysis, pharmacological properties evaluation, and cytotoxicity assessment results. Further study on bioevaluation of compounds is needed to elucidate their potential pharmacological activities.
Key Words: Virtual screening, ADME, anticancer drugs, cannabinoids, cytotoxicity
Introduction
Even with technological advancements in healthcare that have dramatically improved the survival rates of patients across the globe, cancer has remained one of the top leading causes of death worldwide [1]. Cancer epidemiology has been associated with carcinogenic effects of various infectious agents for the past thirty years, according to a study conducted by Parkin (2006) [2]. In their research, current estimates imply that specific infectious agents cause 20% of all cancers in which H. pylori plays a significant role in stomach cancer development, accounting for 63% of all stomach cancers and 5.5% of all cancers. The second is HPV, which accounts for 100% of all cervix cancer and 4.5% of the global cancer burden. Aside from the inaccessibility of treatment plans and drug discovery in many countries due to wealth gaps among nations, one of the biggest challenges that researchers face today is the rising emergence of multidrug relapse and resistance. Natural bioactive compounds from plants have been gaining popularity and attention for their various anticancer activities as a source of treatment alternative; in support of this, some studies revealed that these plant-based bioactive compounds could optimize chemotherapy’s effectiveness and, in some cases, combat some of the side effects of the drugs used as chemotherapeutic agents [3].
Cannabis sativa L. is a known ancient herbal medicine that is a rich source of phytocannabinoids. The endocannabinoid system consists of receptors, ligands, and enzymes responsible for various physiological and pathological processes that interact with phytocannabinoids and synthetic cannabinoids, which affect the progression of diseases such as cancer. In recent years, there have been extensive studies on cannabinoids as a potential anticancer agent [4]. Phytocannabinoids have anticancer properties, and studies have shown that combining cannabinoids with leukemia cells in vitro increases their cytotoxic effects [5]. Furthermore, the most effective components were used in anti-leukemia drugs, such as cytarabine and vincristine, which produced an effect superior to that achieved if the components were utilized individually [5].
Virtual screening (VS) of prospective anticancer agents is a well-established computational approach adopted by cancer research groups and pharmaceutical industries worldwide to identify novel hit compounds and improve the success of drug discovery and development processes. To address the cancer drug discovery crisis, this study explored predictive in silico tools, such as publicly available databases and accessible web resources, to perform comparative analyses on the anticancer properties of the cannabinoids tetrahydrocannabinol (THC), cannabidiol (CBD), and tetrahydrocannabivarin (THCV), which have been reported to have inhibitory activities against cancer cell lines in vitro [6, 7]. Furthermore, considering the strong influence of pharmacokinetic studies on drug development, the calculated in silico ADME (absorption, distribution, metabolism, and excretion) and cytotoxicity profiles of the selected compounds were summarized (Figure 1), which may be helpful in drug design and drug safety assessment. The findings of this investigation encourage future research into the potential of the bioactive constituents of Cannabis sativa L. to uncover promising and novel anticancer therapies.
Figure 1.
Schematic Representation of Anticancer Properties Assessment of Phytocannabinoids (THC, CBD, & THCV) Compounds Using in silico Approach
Materials and Methods
Data Collection of Molecular Compounds
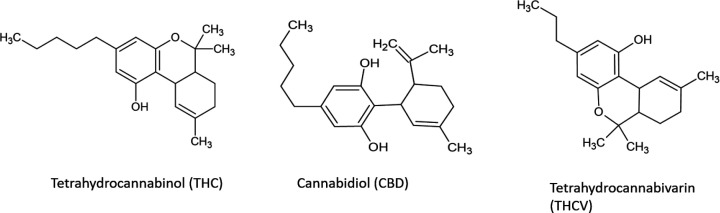
The PubChem (https://pubchem.ncbi.nlm.nih.gov/) database was used to obtain the simplified molecular-input line-entry system (SMILES) sequence of Tetrahydrocannabinol (THC), Cannabidiol (CBD), Tetrahydrocannabivarin (THCV) compounds (Figure 2). The obtained SMILES sequences will then be subjected to different in silico screening for their potential anticancer properties.
Figure 2.
Molecular Structures of THC, CBD, and THCV
Drug-likeness and pharmacokinetic prediction (ADMET) analysis
For a compound to be developed and used as a drug, a crucial step in the Computer-Aided Drug Design (CADD) pipeline is pre-clinical optimization, which includes physicochemical properties, absorption, distribution, metabolism, and excretion (ADME) prediction, and in silico toxicity evaluation. A number of these well-validated tools have been made accessible on web servers such as SwissADME (https://swissadme.ch) and pkCSM-pharmacokinetics (http://biosig.unimelb.edu.au/pkcsm/) that share the objective of predicting ADMET parameters from molecular structure but differ in their computational approaches. SwissADME® is a free online software utilized for the computation of parameters such as physicochemical properties, pharmacokinetics, drug-likeness, and medicinal chemistry of THC, CBD, and THCV compounds. pkCSM (Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures), on the other hand, is another computational method used for the prediction and optimization of pharmacokinetics and toxicity properties of the compounds. As stated above, the canonical SMILES (Simplified Molecular Input Line Entry System) of each compound obtained from the PubChem database were imported to Swiss ADME and pkCSM to predict and assess physicochemical and pharmacokinetics (ADMET) properties. These parameters determined whether the biologically active compounds have the qualities to be orally bioavailable. Additionally, the ADMET/pharmacokinetic assessment was carried out based on the Brain or Intestinal Estimated permeation method (BOILED-Egg) model, which predicts projections for passive human gastrointestinal absorption (HIA) and blood-brain barrier (BBB) permeability that will calculate the lipophilicity and polarity of small molecules.
Pharmacological Properties and Cytotoxicity Assessment of Cannabinoids
The cytotoxicity of active cannabinoids THC, CBD, and THCV was significant in assessing the compounds’ anticancer effect in silico and their biological and pharmacological properties. Initially, in ProTox-II Toxicity Webserver (https://tox-new.charite.de/protox_II/), canonical SMILES sequences of each compound from the PubChem database were input on the server—automatically generating a variety of predicted toxicity endpoints, including acute toxicity, hepatotoxicity, cytotoxicity, carcinogenicity, mutagenicity, immunotoxicity, adverse outcomes (Tox21) pathways, and toxicity targets. In the case of the study, all parameters were considered in obtaining a wide range of cytotoxic properties; that is, to be further analyzed whether the compound is toxic or relatively safe with regards to their molecular mechanism for drug discovery.
In assessing the chemical-biological interactions of THC, CBD, and THCV, Way2Drug Predictive Server (http://www.way2drug.com/passonline/) services were utilized, such as Acute Rat Toxicity, Adver-Pred, Cell Line Cytotoxicity Prediction (CLC-Pred), and Pass Target Prediction. Acute Rat Toxicity Prediction (http://www.way2drug.com/gusar/acutoxpredict.html) involves the General Unrestricted Structure-Activity Relationship (GUSAR) software wherein training sets were generated using data from the SYMYX MDL Toxicity Database containing data on 100,000 chemical structures, as well as data on acute rat toxicity expressed by LD50 values (log10 (mmol/kg)). In AdverPred-Web Service (http://www.way2drug.com/adverpred/), adverse drug effects (e.g., arrhythmia, cardiac failure, hepatotoxicity, myocardial infarction, nephrotoxicity) were predicted, generating the structure-activity relationships of compounds with its potency for drug discovery. In addition, Cell Line Cytotoxicity Prediction (CLC-Pred) Web Service (http://www.way2drug.com/cell-line/) was used to assess further and predict the cytotoxicity of the cannabinoids against tumor and non-tumor cell lines. Finally, the Pass Target Prediction (http://www.way2drug.com/passtargets/) server was used to predict biological activity and interaction with molecular targets. Prior to toxicity and cell line prediction, the structures of the investigated compounds were uploaded using canonical SMILES (simplified molecular-input line-entry system) format. Whereas, for adverse effects prediction of the compounds, the ‘sdf’ format was uploaded to the ADVERPred Predictive Server cannot read and process the acquired canonical SMILES sequences.
Results
Computer-aided drug design (CADD) methodologies are becoming increasingly important, particularly in the field of drug discovery, where they are critical in identifying promising drug candidates. These computational methods are useful for limiting the use of animal models in pharmacological research, and for assisting the rational design of novel and safe drug candidates at a lower cost and in less time.
Drug-likeness and pharmacokinetic prediction (ADMET) analysis
The structural features of the bioactive compounds THC, CBD, and THCV found in Cannabis sativa L. were entered into the ADME/Tox web server tools SwissADME and pkCSM using their canonical SMILES retrieved from the PubChem database. Results obtained from the output panels of SwissADME were used to evaluate their general characteristics, physicochemical properties (Table 1), lipophilicity and water solubility characteristics (Tables 2 and 3), drug-likeness rule and bioavailability score (Table 4), medicinal chemistry friendliness (Table 5), bioavailability radar for drug-likeness (Figure 3), and BOILED-Egg model for GIT absorption and BBB permeation assessment (Figure 4). The vital pharmacokinetic properties provided by pkCSM web tool were selected to represent the ADME/Tox profiles of THC, CBD, and THCV compounds (Table 6).
Table 1.
Physicochemical Properties of THC, CBD, and THCV
| Molecule | Fraction Csp3a,b |
No. H-bond acceptors a,b |
No. H-bond donors a,b |
MR a,b | TPSA (0Å2) a,b |
Molecular Weight (MW) |
|---|---|---|---|---|---|---|
| THC | 0.62 | 2 | 1 | 97.91 | 29.46 | 314.46 |
| CBD | 0.52 | 2 | 2 | 99.85 | 40.46 | 314.46 |
| THCV | 0.58 | 2 | 1 | 88.3 | 29.46 | 286.41 |
Fraction Csp3: ratio of sp3 hybridized carbons over the total carbon count of the molecule, H-bond: hydrogen bond, MR: molecular refractivity, TPSA: topological polar surface area bOptimal range: HBA ≤ 10, HBD ≤ 5, MR ≤ 130 TPSA ≤150 Å2
Table 2.
Lipophilicity Characteristics of THC, CBD, and THCV
| Molecule | iLOGP | XLOGP3 | WLOGP | MLOGP | SILICOS-IT Log P | Consensus Log Pa |
|---|---|---|---|---|---|---|
| THC | 3.9 | 6.97 | 5.74 | 4.39 | 5.41 | 5.28 |
| CBD | 3.9 | 6.52 | 5.85 | 4.31 | 5.42 | 5.2 |
| THCV | 3.47 | 5.89 | 4.96 | 3.94 | 4.62 | 4.58 |
aOptimal range: log P ≤ 5
Table 3.
Water Solubility Characteristics of THC, CBD, and THCV
| Molecule | ESOL | Ali | SILICOS- IT | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Log S (ESOL)b | Solubility | Classa | Log S (Ali)c | Solubility | Class a | Log S (SILICOS- IT)d |
Solubility | Class a | ||||
| mg/ml | mol/mL | mg/mL | mol/mL | mg/mL | mol/mL | |||||||
| THC | -6.11 | 2.44E-04 | 7.77E-07 | PS | -7.4 | 1.24E-05 | 3.96E-08 | PS | -5.93 | 3.96E-04 | 1.17E-06 | MS |
| CBD | -5.69 | 6.36E-04 | 2.02E-07 | MS | -7.17 | 2.14E-05 | 6.81E-08 | PS | -5.41 | 1.21E-04 | 3.86E-06 | MS |
| THCV | -5.41 | 1.12E-03 | 3.93E-06 | MS | -6.28 | 1.50E-04 | 5.23E-07 | PS | -5.13 | 2.10E-03 | 7.35E-06 | MS |
aSolubility class, PS- Poorly soluble, S- Soluble, MS- Moderately soluble, VS- Very soluble; bESOL model Log S scale, insoluble ≤ -10, poorly soluble ≤ -6, moderately soluble ≤ -4, soluble ≤ -2, very soluble ≤ 0; cAli model Log S scale, insoluble ≤ -10, poorly soluble ≤ -6, moderately soluble ≤ -4, soluble ≤ -2, very soluble ≤ 0; dSILICOS-IT model Log S scale, insoluble ≤ -10, poorly soluble ≤ -6, moderately soluble ≤ -4, soluble ≤ -2, very soluble ≤ 0
Table 4.
Drug-Likeness Rule and Bioavailability Scores of THC, CBD, and THCV
| Molecule | Lipinskia | Ghoseb | Veberc | Egand | Mueggee | Bioavailability Scoref |
|---|---|---|---|---|---|---|
| THC | Yes; 1 violation: MLOG>4.15 | No; 1 violation: WLOGP>5.6 | Yes | Yes | No; 1 violation: XLOGP3>5 | 0.55 |
| CBD | Yes; 1 violation: MLOG>4.15 | No; 1 violation: WLOGP>5.6 | Yes | Yes | No; 1 violation: XLOGP3>5 | 0.55 |
| THCV | Yes; 0 violation | Yes | Yes | Yes | No; 1 violation: XLOGP3>5 | 0.55 |
aLipinski filter (Pfizer) qualifying range, MW ≤ 500, MLOGP ≤ 4.15, N or O ≤ 10, NH or OH ≤ 5; bGhose filter qualifying range, MW 160 to 480 Da, WlogP –0.4 to 5.6, MR 40 to 130, total no. of atom 20-70; cVerber filter qualifing range, No. of rotatable bonds ≤ 10, TPSA ≤ 140Å2, HBD and HBA ≤ 12; dEgan filter qualifying range, WLOGP ≤ 5.88, TPSA ≤ 131.6; eMuegge filter qualifying range, MW 200 to 600 Da, XLOGP –2 to 5, TPSA ≤ 150, no. of rings ≤ 7, no. of carbon atoms > 4, number of heteroatoms >1, no. of rotatable bonds ≤ 15, HBA ≤ 10, HBD ≤ 5; fBioavailability score, at least 0.10
Table 5.
Medicinal Chemistry Properties of THC, CBD, and THCV
| Molecule | PAINS | Brenka | Leadlikenessb | Synthetic Accessibilityc |
|---|---|---|---|---|
| THC | 0 | 1 alert: isolated alkene | No; 1 violation: XLOGP3>3.5 | 4.27 |
| CBD | 0 | 1 alert: isolated alkene | No; 1 violation: XLOGP3>3.5 | 4.05 |
| THCV | 0 | 1 alert: isolated alkene | No; 1 violation: XLOGP3>3.5 | 4.05 |
aBrenk model restrictions, ClogP/ClogD 0 to 4, HBD and HBA < 4 to 7, no. of heavy atoms 10 to 27, no. of rotatable bonds < 8, ring system < 5, no ring systems with fused rings > 2; bLeadlikeness considerations, MW 100 to 350 Da, ClogP 1 to 3.0; cSA scores, range from 1 (very easy to synthesize) to 10 (difficult to synthesize)
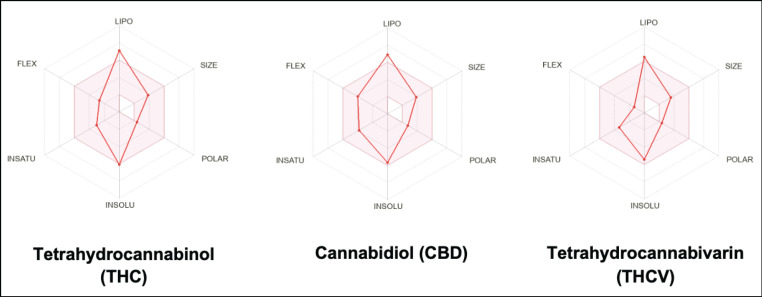
Figure 3.
Schematic Diagram of Bioavailability Radar for Drug likeness of THC, CBD, and THCV (lipophilicity: XLOGP3 between +5.89 and +6.97, size: MW between 286.41 and 314.46 g/mol, polarity: TPSA between 29.46 and 40.46 Å2, solubility: log S not higher than 6, saturation: fraction of carbons in the sp3 hybridization not less than 0.25, and flexibility: no more than 9 rotatable bonds)
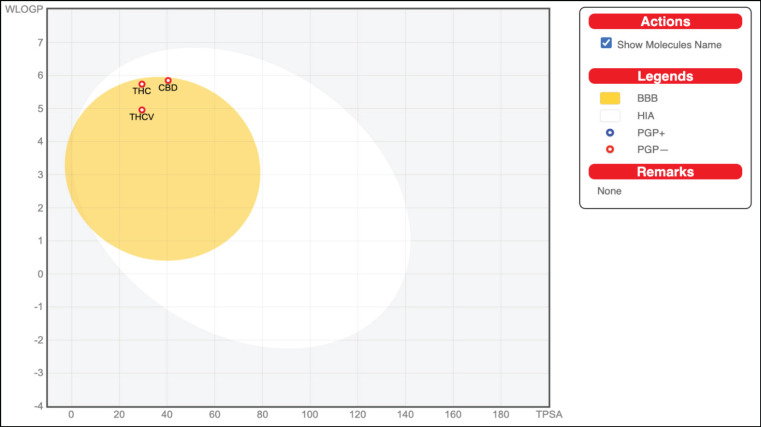
Figure 4.
Schematic Representation of Perceptive Evaluation of Passive Gastrointestinal Absorption (HIA) and Blood-Brain Barrier (BBB) Penetration of THC, CBD, and THCV in the WLOGP-versus-TPSA using BOILED-Egg. High probability of GIT passive absorption is represented by the white region, whereas the yellow region (yolk) represents the high probability of BBB penetration. In addition, the blue dot indicator of the molecule shows that the molecule is actively effluxed by P-glycoprotein, represented as (PGP+), whereas the red color indicator shows the non-substrate of P-gp, represented as (PGP−)
Table 6.
Pharmacokinetic Profile and Toxicity Prediction of THC, CBD, and THCV
| Parameter | THC | CBD | THCV |
|---|---|---|---|
| Absorption (A) | |||
| Water solubility (log mol/L) | -6.275 | -4.901 | -5.506 |
| Caco-2 permeability (log Papp, cm/s) | 1.519 | 1.79 | 1.52 |
| Intestinal absorption (human) % | 93.091 | 90.657 | 93.772 |
| Skin permeability (log Kp) | -2.538 | -2.795 | -2.441 |
| BioS (from SwissADME) | 0.55 | 0.55 | 0.55 |
| Distribution (D) | |||
| VDss (human) (log L/kg) | 0.977 | 0.939 | 0.888 |
| BBB permeability (log BB) | 0.448 | -0.113 | 0.32 |
| BBB perm. (SwissADME) | Yes | Yes | Yes |
| Metabolism (M) | |||
| CYP1A2 inhibitor | No | No | No |
| CYP2C19 inhibitor | Yes | Yes | Yes |
| CYP2C9 inhibitor | Yes | Yes | Yes |
| CYP2D6 inhibitor | Yes | Yes | Yes |
| CYP3A4 inhibitor | No | Yes | No |
| Excretion (E) | |||
| Total clearance | 0.883 | 1.092 | 0.827 |
| Renal OCT2 substrate | No | No | No |
| Toxicity (T) | |||
| Ames test | No | No | No |
| Hepatotoxicity | No | No | No |
| Oral rat acute toxicity (LD50, in mg/kg) | 482 | 500 | 482 |
The chemical structures of THC, CBD, and THCV compounds investigated in this study are shown in Figure 2. THC and THCV are structurally similar, with the exception that THCV has a shorter side chain and a propyl (-C3) side chain (-C5) instead of a pentyl side chain. When compared to CBD, THC contains a cyclic ring, whereas CBD has hydroxyl group. The bioavailability radar provides an overview of a compound’s drug-likeness which is predicted based on 6 physicochemical parameters: lipophilicity, size, polarity, solubility, flexibility, and saturation. Except for their affinity in a lipid environment, as demonstrated by the out-of-range red point of their distorted hexagon for lipophilicity, all compounds had physicochemical properties within the favorable range (pink zone).
The BOILED-Egg depicted in Figure 4 illustrates the human intestinal absorption (HIA) and the brain access or penetration of the molecules. All of the compounds (THC, CBD, and THCV) were found within the yellow region of the egg (yolk). This suggests that these compounds have a high probability of being absorbed by the gastrointestinal tract and permeating into the brain. Moreover, PGP-, represented by red dots, denotes substances that are believed to be non-efflux from the CNS by P-gp, implying that all investigated compounds are non-substrates of P-gp.
Table 1 summarizes the general characteristics of THC, CBD, and THCV. THC and CBD share the same molecular weight of 314.46 Da while THCV is 286.41 Da. The obtained molecular weight for each compound was considered acceptable as specified by L-Ro5 to be deemed an orally accessible bioactive drug.
The TPSA for THC and THCV was 40.46 Å2, while for CBD was 29.46 Å2. TPSA values lower than 20-150 Å2 indicate either excellent intestinal permeability or oral absorption, and the compounds were not strongly acidic.
THC, CBD, and THCV had Consensus LogP values of 5.28, 5.20, and 4.58, respectively. The optimal range for lipophilicity should be equal to or less than 5. THC was determined to be the compound to be the most lipophilic. THC and CBD were found to have lipophilicity values that exceeded the optimal range, which could lead to a high rate of insolubility and toxicity.
The Estimated SOLubility (ESOL) was computed directly from the structure of THC, CBD, and THCV, and the obtained Log S values were -6.11, -5.69, and -5.41, respectively. For the SILICOS-IT, the Log S values for the compounds above were -5.93, -5.41, and -5.13, respectively. Both ESOL and SILICOS-IT all within the recommended range of solubility. The calculated Log S values for Ali, were -7.40, -7.17, and -6.28, did not fall within the recommended range, suggesting that these compounds are not highly soluble. Lipinski, Ghose, Veber, Egan, Muegge Filters, and the Bioactivity Score were used to assess each compound’s drug-likeness. In Lipinski and Ghose Filters, THC and CBD were found to violate the optimal values for MLOG and WLOGP, respectively. All compounds passed the Veber and Egan criteria but failed the Muegge criteria. The bioactivity score for each compound is 0.55. All of the compounds passed the Pan-assay Interference compounds (PAINS) with 0 violations, Brenk filters with 1 alert of isolated alkene, and 1 violation for leadlikeness. All compounds passed the L-Ro5 for the number of HBDs (≤5), HBAs (≤10), and rotatable bonds (≤10) but violated the criteria for leadlikeness (cLogP).
The ADMET properties of THC, CBD, and THCV are shown in Table 6. The absorption level of the substances was predicted using Caco-2 permeability, intestinal absorption (human), skin permeability, and P-glycoprotein substrate or inhibitor. THC, CBD, THCV were predicted to have high Caco-2-permeability with CBD having the highest. In terms of human intestine absorption, none of the three compounds performed poorly, with absorption rates ranging from 90 to 93%. THC, CBD, and THCV resulted in log Kp values of -2.538 log Kp, -2.795 log Kp, and -2.441 log Kp, respectively, suggesting a good skin permeability. The distribution of substances was characterized using the distribution volume (VDss), fraction unbound (human), CNS permeability, and blood–brain barrier membrane permeability (logBB). The distribution volumes of THC, CBD, and THCV were 0.977, 0.939, and 0.888, respectively, indicating a good distribution. On the other hand, the permeability of Blood-Brain Barrier (BBB) membrane for THC (0.448) and THCV (0.320), implying that they can able to cross the BBB easily, with the exception of cannabidiol (-0.113). CYP-mediated metabolic properties of THC, CBD, and THCV showed that all compounds are inhibitors for CYP219, CYP2C9, and CYP2D6 and non-inhibitors for CYP1A2. Moreover, all compounds except CBD are non-inhibitors for CPY3A4. Excretion parameters predicted from total clearance and renal OCT2 substrate. The total clearance of THC generated a value of 0.883, 1.092 for CBD, and 0.827 for THCV. None of the compounds were renal OCT2 substrates. The results also suggest that THC, CBD, and THCV were non-toxic according to the AMES test, and non-hepatotoxic, based on the hepatoxicity test. The oral rate of acute toxicity for THC, CBD, and THCV resulted in 482, 500, and 482, respectively.
Pharmacological Properties and Cytotoxicity Assessment of Cannabinoids
The canonical SMILES of the compounds THC, CBD, and THCV were submitted to ProTox-II and different Way2Drug predictive tools namely Acute Rat Toxicity Prediction, ADVER-Pred, CLC-Pred, and Pass Target Prediction was utilized to predict and assess their pharmacological properties and cytotoxicity against human cell lines. Protox-II webserver provided a prediction of the compounds’ toxic doses (LD50 in mg/kg) and toxicity classes (Table 7). Way2Drug predictive tools in silico prediction of rat LD50 values (Table 8), adverse drug effects (Table 9), human cell line cytotoxicity (Table 10), and biological activities and molecular targets (Table 11).
Table 7.
Protox-II Toxicity Estimations on Cannabinoids: THC, CBD, and THCV
| THC | CBD | THCV | |||||
|---|---|---|---|---|---|---|---|
| Predicted LD50 | 482 mg/kg | 500 mg/kg | 482 mg/kg | ||||
| Predicted Toxicity Class | 4 | 4 | 4 | ||||
| Toxicity Model Report | |||||||
| Classification | Target | Prediction | Prob. | Prediction | Prob. | Prediction | Prob. |
| Toxicity end points | Hepatotoxicity | Inactive | 0.93 | Inactive | 0.79 | Active | 0.85 |
| Toxicity end points | Carcinogenicity | Inactive | 0.86 | Inactive | 0.66 | Active | 0.84 |
| Toxicity end points | Immunotoxicity | Active | 0.99 | Active | 0.93 | Active | 0.95 |
| Toxicity end points | Mutagenicity | Inactive | 0.77 | Inactive | 0.85 | Inactive | 0.74 |
| Toxicity end points | Cytotoxicity | Inactive | 0.84 | Inactive | 0.87 | Inactive | 0.83 |
Table 8.
Rat Acute Toxicity Prediction of THC, CBD, and THCV by GUSAR
| THC | CBD | THCV | |
|---|---|---|---|
| Rat IP LD50 Log10 (mmol/kg) | 0,038 in AD | 0,038 in AD | 0,154 in AD |
| Rat IV LD50 Log10 (mmol/kg) | -0,912 in AD | -0,912 in AD | -0,841 in AD |
| Rat Oral LD50 Log10 (mmol/kg) | 0,405 in AD | 0,405 in AD | 0,365 in AD |
| Rat SC LD50 Log10 (mmol/kg) | -0,256 in AD | -0,256 in AD | -0,681 out of AD |
| Rat IP LD50 (mg/kg) | 343,300 in AD | 343,300 in AD | 408,100 in AD |
| Rat IV LD50 (mg/kg) | 38,530 in AD | 38,350 in AD | 41,350 in AD |
| Rat Oral LD50 (mg/kg) | 799,200 in AD | 799,200 in AD | 663,300 in AD |
| Rat SC LD50 (mg/kg) | 174,500 in AD | 174, 500 in AD | 59,660 out of AD |
| Rat IP LD50 Classification | Class 4 in AD | Class 4 in AD | Class 4 in AD |
| Rat IV LD50 Classification | Class 3 in AD | Class 3 in AD | Class 4 in AD |
| Rat Oral LD50 Classification | Class 4 in AD | Class 4 in AD | Class 4 in AD |
| Rat SC LD50 Classification | Class 4 in AD | Class 4 in AD | Class 3 out of AD |
Table 9.
ADVERPred Prediction Results of THC, CBD, and THCV
| THC | ||
|---|---|---|
| Paa | Pib | Side effects |
| 0.092 | 0.663 | Nephrotoxicity |
| 0.063 | 0.660 | Cardiac failure |
| 0.063 | 0.669 | Myocardial infarction |
| 0.053 | 0.773 | Arrhythmia |
| CBD | ||
| Paa | Pib | Side effects |
| 0.128 | 0.522 | Arrhythmia |
| 0.110 | 0.583 | Nephrotoxicity |
| 0.109 | 0.468 | Cardiac failure |
| 0.073 | 0.617 | Myocardial infarction |
| CBD | ||
| Paa | Pib | Side effects |
| 0.128 | 0.522 | Arrhythmia |
| 0.110 | 0.583 | Nephrotoxicity |
| 0.109 | 0.468 | Cardiac failure |
| 0.073 | 0.617 | Myocardial infarction |
aPa, probable activity; bPi, probable inactivity
Table 10.
In silico Prediction of Cytotoxic Effect of THC, CBD, and THCV Compounds on Cancer Cell Lines
| THC | |||||
|---|---|---|---|---|---|
| Cancer cell line | |||||
| Paa | Pib | Cell-line | Cell-line full name | Tissue | Tumor type |
| 0.341 | 0.048 | U-251 | Glioma | Brain | Glioma |
| 0.321 | 0.034 | MKN-7 | Gastric carcinoma | Stomach | Carcinoma |
| 0.319 | 0.064 | HOP-18 | Non-small cell lung carcinoma | Lung | Carcinoma |
| 0.344 | 0.096 | A2058 | Melanoma | Skin | Melanoma |
| 0.389 | 0.155 | DMS-114 | Lung carcinoma | Lung | Carcinoma |
| Non-tumor cell line | |||||
| 0.328 | 0.048 | WI-38 VA13 | Embryonic lung fibroblast | Lung | |
| 0.246 | 0.008 | PrEC | Prostate epithelial cell | Prostate | |
| 0.217 | 0.11 | MRC5 | Embryonic lung fibroblast | Lung | |
| 0.1 | 0.035 | HaCaT | Keratinocyte | Skin | |
| 0.143 | 0.099 | IMR-90 | Embryonic lung fibroblast | Lung | |
| CBD | |||||
| Cancer-cell line | |||||
| Paa | Pib | Cell-line | Cell-line full name | Tissue | Tumor type |
| 0.542 | 0.004 | MDA-MB-453 | Breast adenocarcinoma | Breast | Adenocarcinoma |
| 0.372 | 0.021 | 8505C | Thyroid gland undifferentiated (anaplastic) carcinoma | Thyroid | Carcinoma |
| 0.385 | 0.056 | NALM-6 | Adult B acute lymphoblastic leukemia | Haematopoietic and lymphoid tissue | Leukemia |
| 0.418 | 0.107 | DMS-114 | Lung carcinoma | Lung | Carcinoma |
| 0.308 | 0.042 | MKN-7 | Gastric carcinoma | Stomach | Carcinoma |
| Non-tumor cell line | |||||
| 0.252 | 0.072 | WI-38-VA13 | Embryonic lung fibroblast | Lung | |
| 0.144 | 0.076 | PrEC | Prostate epithelial cell | Prostate | |
| 0.099 | 0.036 | HaCaT | Keratinocyte | Skin | |
| THCV | |||||
| Cancer cell line | |||||
| Paa | Pib | Cell-line | Cell-line full name | Tissue | Tumor type |
| 0.369 | 0.04 | HOP-18 | Non-small cell lung carcinoma | Lung | Carcinoma |
| 0.317 | 0.058 | U-251 | Glioma | Brain | Glioma |
| 0.318 | 0.059 | M19-MEL | Melanoma | Skin | Melanoma |
| 0.276 | 0.07 | MKN-7 | Gastric carcinoma | Stomach | Carcinoma |
| 0.234 | 0.034 | BGC-823 | Stomach adenocarcinoma | Stomach | Carcinoma |
| Non-tumor cell line | |||||
| 0.364 | 0.041 | WI-38 VA13 | Embryonic lung fibroblast | Lung | |
| 0.258 | 0.005 | PrEC | Prostate epithelial cell | Prostate | |
| 0.262 | 0.076 | MRC5 | Embryonic lung fibroblast | Lung | |
| 0.095 | 0.038 | HaCaT | Keratinocyte | Skin | |
aPa, probable activity; bPi, probable inactivity
Table 11.
Direct and Mediated Protein Targets of THC, CBD, and THCV
| THC | ||
|---|---|---|
| Direct Interaction | ||
| Target Name | Confidence | ChEMBL ID |
| Cannabinoid CB2 receptor | 0.4686 | (CHEMBL253) |
| Cannabinoid CB1 receptor | 0.4431 | (CHEMBL218) |
| Cytochrome P450 2C9 | 0.3556 | (CHEMBL 3397) |
| Topoisomerase II-α, | 0.3056 | (CHEMBL1806) |
| Estrogen receptor-β | 0.1988 | (CHEMBL242) |
| Mediated Interaction | ||
| Glycine receptor subunit α-1 | 0.5569 | (CHEMBL5845) |
| Cannabinoid CB2 receptor | 0.5351 | (CHEMBL253) |
| Cannabinoid CB1 receptor | 0.3306 | (CHEMBL218) |
| Serotonin 1e (5-HT1e) receptor | 0.2906 | (CHEMBL2182) |
| Nuclear receptor subfamily o group B member 1 | 0.2284 | (CHEMBL1795094) |
| CBD | ||
| Direct Interaction | ||
| Target Name | Confidence | ChEMBL ID |
| Phosphatidylinoitol-4-phosphate-5-kinasetype 1γ | 0.7681 | (CHEMBL1908383) |
| Cannabinoid CB1 receptor | 0.6356 | (CHEMBL218) |
| Serine/Threonine-protein kinase 32A | 0.6151 | (CHEMBL6150) |
| Rhodopsin kinase | 0.5937 | (CHEMBL5607) |
| Cannabinoid CB2 receptor | 0.5239 | (CHEMBL253) |
| Mediated Interaction | ||
| Cannabinoid CB2 receptor | 0.7529 | (CHEMBL253) |
| Glycine receptor subunit α-1 | 0.6822 | (CHEMBL218) |
| Tyrosine-protein kinase FYN | 0.4922 | (CHEMBL3397) |
| Cannabinoid CB1 receptor | 0.4637 | (CHEMBL218) |
| N-arachidonyl glycine receptor | 0.4088 | (CHEMBL2384898) |
| THCV | ||
| Direction Interaction | ||
| Target Name | Confidence | ChEMBL ID |
| Phosphatidylinoitol-4-phosphate-5kinasetype 1γ | 0.8084 | (CHEMBL1908383) |
| Rhodopsin kinase | 0.6532 | (CHEMBL5607) |
| Serine/Threonine-protein kinase 32A | 0.6438 | (CHEMBL6150) |
| Cannabinoid CB1 receptor | 0.6241 | (CHEMBL218) |
| Tyrosine-protein kinase Srms | 0.5934 | (CHEMBL5703) |
| Mediated Interaction | ||
| Glycine receptor subunit α-1 | 0.6769 | (CHEMBL218) |
| Cannabinoid CB2 receptor | 0.6167 | (CHEMBL253) |
| Tyrosine-protein kinase FYN | 0.4396 | (CHEMBL3397) |
| Cannabinoid CB1 receptor | 0.4253 | (CHEMBL218) |
| N-arachidonyl glycine receptor | 0.3361 | (CHEMBL218) |
Computational Toxicity Estimations of Compounds using Protox-II
The toxicity of THC, CBD, and THCV are illustrated in Table 7. The median lethal dose (LD50) is the dose at which 50% of test subjects die after being exposed to a substance. All cannabis drugs have an LD50 value of (300 < LD50 ≤ 2000), which ranges from toxic dosages of 482 mg/kg to 500 mg/kg. As a result, these cannabinoid pharmaceuticals fall within Class 4 toxicity, which is detrimental when taken orally. With a probability ranging from 0.93-0.99, all compounds are considered active in the toxicity endpoint under the immunotoxicity parameter. However, assessing the other parameters connected to cancer, such as hepatotoxicity, carcinogenicity, mutagenicity, and cytotoxicity of the three cannabinoids compounds, generated inactive results or no effects if used as drugs.
Biological Activity of THC, CBD, and THCV
Cannabis sativa L. is considered one of the most controversial herbal medicines globally. They are mainly composed of biologically active secondary metabolites known as phenolic compounds, specifically cannabinoids, such as THC, CBD, and THCV (Helcman and Šmejkal, 2021) [8]. Their complex biological activities, as shown in the adverse drug effects and cell line cytotoxicity, ADMET, PAINS, and its low toxicity as demonstrated in GUSAR analysis, reveal that they have the potential to be used for therapies in treating various diseases. However, considering that some cannabinoids, including THC and THCV, is responsible for their psychoactive effects, it has been a concern in the research community on whether or not their applicability outweighs their cons for the welfare of its users.
Acute Rat Toxicity
The median lethal dose for acute rat toxicity (LD50) is required to categorize chemicals based on the potential hazard posed to human health after acute exposure (Gadaleta et al., 2019) [9]. This is crucial in determining the adverse effects of drugs upon administration and is also essential in giving out information such as safe acute dosage for humans, target potential organs for toxicity, the difference of toxicity among species, and the time course of drug-induced clinical observations. The in silico prediction of LD50 values of acute rat toxicity from THC, CBD, and THCV showed four results based on the different types of administration by GUSAR software. From Table 8, it can be seen that the highest LD50 levels are seen among all cannabinoids in Rat Oral, and the lowest are seen in Rat IV, indicating which routes are most potent for these compounds. For the classification of LD50 of the rat the following range is used as a reference for understanding its toxicity: Class 1: Extreme toxicity (LD50 of ≤5 for oral, ≤50 for dermal, ≤100 for gases, and ≤0.05 for vapors); Class 2: High toxicity (LD50 of >5- ≤50 for oral,>50- ≤200 for dermal, >100- ≤500 for gases, and >0.05 - ≤2.0 for vapors); Class 3: Moderate toxicity (LD50 of >50- ≤300 for oral,>200- ≤1000 for dermal, >500- ≤2500 for gases, and >2.0- ≤10.0 for vapors); and Class 4: Low toxicity (LD50 of >300- ≤2000 for oral,>1000- ≤2000 for dermal, >2500- ≤5000 for gases, and >10.0- ≤20.0 for vapors) (ChemSafetyPro, 2018).
Adverse Drug Effects
The results on adverse drug effects of THC, CBD, and THCV showed Pa and Pi values corresponding to the probable activity or inactivity of the following toxic effects: nephrotoxicity, cardiac failure, myocardial infarction, and arrhythmia. As shown in Table 9, the estimated Pi values (probable inactivity) are higher than Pa values (probable activity). It can be inferred that THC, CBD, and THCV are unlikely to cause adverse side effects on the cardiovascular and hepatobiliary systems. THC and THCV had the highest activity probability to cause nephrotoxicity (Pa= 0.092; Pi= 0.663 and Pa=0.080; Pi=0.708, respectively), yet it has the least probability for arrhythmia (Pa=0.053; Pi 0.883 and Pa=0.048; Pi=0.792, respectively). In comparison, CBD had the highest probable activity for arrhythmia (Pa=0.128; Pi=0.522), although it is least probable in myocardial infarction (Pa=0.073; Pi=0.617).
Cell Line Cytotoxicity
Human cell line cytotoxicity of THC, CBD, and THCV was predicted using the Pa>Pi threshold in CLC-Pred. Pa and Pi values correspond to the probability of the compounds being active and inactive in corresponding cancer and non-tumor cell lines. Results also indicated the target tissues and the tumor type, which could develop within the cell caused by the cannabinoids.
The probability values were observed to be Pa>Pi, indicating high probable activity of the compounds against human cell lines. As depicted in Table 10, THC had the highest probability towards the cancer cell line, Lung Carcinoma (DMS-114), non-tumor cell line, and embryonic lung fibroblast (WI-38 VA13), with the Pa and Pi values of 0.389 and 0.328, respectively. On the other hand, CBD expresses its active cytotoxicity to the cancer cell line, Breast adenocarcinoma (MDA-MB-453), with a Pa value of 0.542. At the same time, CBD acts upon the non-tumor cell line, Embryonic lung fibroblast (WI-38-VA13), with a Pi value of 0.252. Subsequently, THCV had the highest cytotoxicity to the cancer cell line, non-small cell lung carcinoma (HOP-18), non-tumor cell line, and Embryonic lung fibroblast (WI-38 VA13). THCV’s Pa and Pi values against these cell lines were 0.369 and 0.364, respectively. Results show that the high Pa and Pi values of THC, CBD, and THCV specifically target lung tissues.
Direct and Mediated Protein Target Interactions
The Direct and Mediated Protein Targets of THC, CBD, and THCV predicted by Pass Target Prediction are presented in Table 11. This analyzes the biological potential and activity of the compounds. Based on the results obtained, THC has shown to have a direct interaction with the Cannabinoid CB2 receptor at a p-value of 0.4686 and a mediated interaction with Glycine receptor subunit alpha-1 at 0.5569. On the other hand, CBD has a direct interaction with Phosphatidylinositol-4-phosphate-5 type-1 gamma at a p-value of 0.7681 and a mediated interaction with Cannabinoid CB2 receptor at 0.7529. Lastly, the THCV showed a direct interaction with Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma at a p-value of 0.8084 and a mediated interaction with Glycine receptor subunit alpha-1 at 0.6769.
Discussion
Cannabis sativa L. is an indigenous plant originating from Central Asia. It has been used for medicinal uses for centuries and is a significant source of phytocannabinoids. Today, its compounds are being used as antiemetic drugs for cancer patients. In the United States, patients with advanced stages of cancer are given cannabis medicine as part of palliative care to manage symptoms such as chronic pain and nausea and to stimulate appetite. A cross-sectional study by the National Cancer Institute in 2017 reported that 24% of adult cancer patients in Washington are active users of cannabis to relieve cancer treatment-related symptoms, whereas 74% expressed interest in gaining more information about cannabis usage from cancer providers [10]. THC, CBD, and THCV are some of the most studied and abundant bioactive compounds found in Cannabis sativa L. These compounds have been demonstrated by in vitro studies to attenuate cell proliferation, invasion, and angiogenesis of human cancer cell lines such as human glioma [11], breast, and prostate cancers [12].
Moreover, they have 23 shown antitumor activity in numerous rodent models, which further establishes their potential as anticancer agents.
In silico approaches such as CADD and virtual screening are utilized to undertake ADME/Tox studies since it allows for efficient screening and synthesizing of a large number of drug-like compounds in a shorter period. Vimblastine, vincristine, taxol, podophyllotoxin, captothecin and cytarabine are some examples of successful anticancer agents developed through CADD techniques [13]. In the present study, the ADMET properties, toxicity profiles, and pharmacological activities of THC, CBD, and THCV were predicted using several free web servers to provide an initial evaluation of prospective drug candidates in the hit-to-lead stage in drug discovery and later, lead-optimization.
There is scant research and data on the physicochemical and pharmacokinetic properties of cannabinoids in humans; therefore, there is not enough understanding of factors that influence their drug action and effectiveness. SwissADME is a free, simple, robust, and accurate web tool to determine the ADME parameters of small compounds. The physicochemical properties, lipophilicity, water solubility, pharmacokinetics, drug likeness, and medicinal chemistry properties of THC, CBD, and THCV were predicted to assess their potential as anticancer agents and integrate these findings in modern drug development. SwissADME provides a one-panel-per-molecule output that comprises all the physical and chemical information of the molecules entered. Although there are variations in the computed findings, the projected physicochemical characteristics of THC, CBD, and THCV follow a regular pattern, with little difference in each parameter.
The freely available pkCSM-pharmacokinetics web tool relies on graph-based signatures and experimental data to predict and optimize small-molecule ADME/Tox properties [14]. The investigated compounds (THC, CBD, and THCV) must be non-carcinogenic and non-hepatotoxic. Accordingly, the pkCSM web server along with SwissADME were utilized to estimate the pharmacokinetics and toxicity of the aforementioned compounds. The pkCSM server makes it possible to predict ADME/Tox properties quickly and accurately.
SwissADME was used to predict the physicochemical properties of THC, CBD, and THCV. The transportation of an unmetabolized drug from the site of delivery to the body’s circulatory system is known as drug absorption. Some patients opt for their chemotherapeutics to be delivered orally, instead of intravenously due to low cost and lack of infusion-related inconveniences [15]. However, in terms of pharmacokinetic considerations, this route of administration does not allow for fast and efficient absorption since they are affected by the first-pass metabolism that occurs in the intestine and liver [16]. In addition, cancer drugs generally have low and highly variable oral bioavailability, rendering the drug ineffective. The bioavailability radar of the screened cannabinoids is depicted in Figure 3, which provides a graphical depiction of the drug-likeness of the submitted molecules. The physicochemical properties that characterize the oral druggability of the selected cannabinoids include lipophilicity, size, polarity, solubility, and saturation. The pink region in the diagram represents the ideal range for these parameters. The aforementioned properties were analyzed based on the guidelines of Lipinski’s rule-of-five (L-Ro5) [17] for drug-likeness. L-Ro5 is a set of criteria for determining whether a compound has the qualities to be regarded as a good oral drug.
The general characteristics of the compounds shown in Table 2 revealed that all compounds had MW ranging from 286.41 to 314.46 Da, which is within the acceptable MW (≤ 500 Da) set by L-Ro5 to be considered an orally available bioactive drug. Lipophilicity describes the permeability of compounds through biological systems. The lipophilicity property is measured by the partition coefficient P (log Po/w) of the compounds between n-octanol and water systems [18]. In SwissADME, lipophilicity is estimated as consensus Log P (Table 4), which is the average value of all Log P evaluated with various lipophilicity criteria. Among the compounds screened, only THCV met the lipophilicity cutoff (log P ≤ 5) set by L-Ro5 with a value of 4.58. In contrast, THC and CBD with 5.28 and 5.20 lipophilicity values, respectively, were out-of-range. High lipophilicity indicates a high insolubility rate and poor oral absorption. In addition, the water solubility of the compounds is ranked from highly water-soluble (THCV) to least water-soluble or insoluble (THC and CBD), respectively, in fats, oil, lipids, and nonpolar solvents [19].
The topological predicted polar surface area (TPSA) was 29.26 Å2 for THC and THCV and 40.46 Å2 for CBD. TPSA values should be between 20 to 130 Å2 to have a high probability of oral bioavailability [20]. In addition, Lipinski (1997) [17] suggested that less than 140-150 Å2 TPSA commonly exhibits an adequate intestinal permeability, which means that the obtained TPSA for the said compounds was not highly acidic. The penetration of these compounds into the cell membranes is not restricted. All compounds were found to satisfy L-Ro5 for the number of HBDs (≤5), HBAs (≤10), and rotatable bonds (≤10) (Table 3), but all compounds violated the cLogP [17] (Table 4). Nevertheless, the results obtained from L-Ro5 may indicate the potential for increased oral absorption [21]. For saturation, the ratio of sp3 hybridized carbons to total molecule count of fraction Csp3 should be at least 0.25 [14], in which all compounds met with values ranging from 0.52 to 0.62 (Table 3). Another parameter assessed to determine the oral bioavailability is molar refractivity (MR) as described by the Ghose filter [22] and molar solubility (log S). According to L-Ro5, MR should be between 40-130 for drug-likeness [17], while log S should not exceed 6 [23]. The cannabinoids had MR values ranging from 88.30 to 99.85 (Table 3). On the other hand, based on the calculations by the ESOL, Ali, and SILICOS-IT model, as shown in Table 5, the cannabinoids had log S values are within the accepted range; however, the aqueous solubility equation adapted from Ali et al. (2012) [24] shows that all compounds are poorly soluble.
The absorption properties of each compound based on computed lipophilicity and polarity of small molecules were depicted by the Brain or IntestinaL EstimateD permeation method or BOILED-Egg model shown in Figure 4. The BOILED-Egg model is an accurate predictive method to evaluate the passive gastrointestinal absorption (HIA) and blood-brain barrier permeation (BBB) of drug-like molecules [25]. Molecules found in the white region have a high probability for HIA, while the yellow region or yolk has a high probability for brain penetration [25]. Results showed that the positioning of the compounds was seen to be close and similar to one another. THC, CBD, and THCV were found on the yolk or yellow region of the BOILED-egg model, implying that they exhibit good BBB permeation and high GIT absorption. In addition, the blue dots represent P-gp substrates (PGP+) and the red dots for P-gp non-substrate (PGP-) [25]. All compounds were represented by a red dot, indicating that they are non-P-glycoprotein (Pgp) substrates; therefore, they will not be subjected to being actively effluxed out of the cells.
Drug-likeness and medicinal chemistry properties of THC, CBD, and THCV are shown in Tables 6 and 7. SwissADME gives access to five different rule-based filters: Lipinski (Pfizer) filter, Ghose (Amgen) filter, Veber (GSK) filter, Egan (Pharmacia) filter, and Muegge (Bayer) filter [14]. These filters come with a diverse range of properties inside of which the molecule is defined as drug-like. Most of the compounds followed the filtered rule invoked in SwissADME, and the violation exhibited by the molecules are minimal. All compounds did not have any PAINS alert and passed L-Ro5 for drug-likeness; however, THC and CBD had one violation due to their MLOGP values exceeding the lipophilicity threshold of >4.15. To broaden lead optimization prospects, the Brenk filter considered molecules that are smaller and less hydrophobic, as well as those that are not characterized by L-Ro5 [26]. Compounds with potentially mutagenic, reactive, and unfavorable groups are excluded [26]. All compounds had one alert in Brenk filter due to the presence of isolated alkene in their compounds. Moreover, all compounds failed the leadlikeness criteria due to a violation in their out-of-range XLOGP3 values (should be in the range –0.7 to +6.0). Finally, the synthetic accessibility (SA) score estimates the easiness of the compounds to be synthesized. SA scores range from 1 (very easy to synthesize) to 10 (very difficult to synthesize) (Daina et al., 2017) [14]. The cannabinoids had SA scores ranging from 4.05 to 4.27, suggesting that they could be relatively easy to synthesize.
The absorption, distribution, metabolism, and excretion (ADME) profiles of THC, CBD, and THCV according to pkCSM web server are presented in Table 8. Absorption is the movement of a drug into the systemic circulation from an extravascular administration site. Compounds with values less than (more negative than) −6 in the pkCSM Log mol/L scale are considered poorly soluble [27]. Water solubility (log mol/L) values for the compounds THC, CBD, and THCV were predicted to range from −4.901 to −6.275, with CBD and THCV having high solubility and THC having low solubility, indicating poor absorption and elimination by the urinary tract, as shown in Table 8. The data shows high negative values, which indicate the presence of moderately to poorly soluble compounds. The pkCSM solubility predictions are consistent with results obtained from SwissADME.
The Caco-2 cell line, which is made up of human epithelial colorectal adenocarcinoma cells, is widely used as an in vitro model of the human intestinal mucosa for predicting drug absorption by measuring the log of the evident permeability coefficient (log Papp; log cm/s). A compound with a log Papp value >0.90 cm/s is considered to have high Caco-2 permeability [28]. Table 8 shows that all phytocannabinoids have a high Caco-2 permeability. All compounds have nearly the same Caco-2 permeability value, which is predicted to be 1.5 cm/s, except for cannabidiol (1.7 cm/s).
Furthermore, all compounds have a good profile of absorption rate in the intestine. Interestingly, the human intestine absorption (HIA) values are high, indicating that phytocannabinoids derivatives have a greater than 90% chance of being absorbed by the human intestine, with tetrahydrocannabivarin having the highest probability (93.772) and cannabidiol having the lowest (90.657). According to Cerqueira et al. (2015) [29], the recommended value of the skin permeability (log Kp) for a drug molecule, an important consideration for improving drug efficacy and is particularly pertinent in developing transdermal drug delivery is set at more than -2.5 cm/h. All compounds have log Kp values ranging from -2.4 to -2.7 cm/h. THC and CBD compounds are predicted to have good skin permeability with 2.538 cm/h and 2.795 cm/h, respectively. THCV, on the other hand, has a lower skin permeability of 2.441 cm/h. The bioavailability score of 0.55 confirms that all investigated compounds have good absorption since they may have more than 10% of bioavailability in the rat [30].
The volume of distribution at steady state (VDss) and the blood−brain barrier (BBB) are two important parameters to consider when evaluating a drug’s ability to be distributed in the body. The higher the VD is, the larger the amount of a drug is distributed to tissue rather than plasma. Pires et al. (2015) [28] reported that a compound has good distribution if its VDss value is greater than 0.45. All the compounds have VDss values that are twice as high as the recommended value. In terms of the BBB, which defines a drug’s ability to cross into the brain while improving efficacy (fewer side effects), a compound is capable of passing through the blood-brain barrier when log BB is greater than 0.3. The pkCSM web server reveals that THC and THCV can easily cross the BBB except for CBD. Since the log BB value of CBD is less than 0.3, hence, it can only penetrate the blood-brain barrier moderately. BBB permeability results for THC and THCV are higher than 0.3, suggesting that this compound has a high probability of being absorbed by the gastrointestinal tract and permeating into the brain. Testing with pKCSM gives results that are slightly different from SwissADME, especially on the distribution parameters.
Metabolism was also predicted using the pkCSM web server in accordance with the inhibition of the main cytochromes (CYP) of the P450 superfamily, namely CYP1A2, CYP2C19, CYP2C9, CYP2D6, and CYP3A4. A common mechanism for metabolism-based drug–drug interactions is CYP enzyme inhibition, which involves competition with another drug for the same enzyme binding site. All clinically used drugs, including several anticancer agents, are impaired by enzyme inhibition, resulting in higher plasma levels of drugs that influence the therapeutic outcome. The effect is reduced if the drug is a prodrug. Therefore, inhibition of CYPs can result in drug toxicity or ineffectiveness [31]. According to Thorn et al. (2012) [32], CYP1A2 is involved in the metabolism of xenobiotics in the body. In contrast, CYP2C19 is responsible for the metabolism of several drugs as well as the detoxification of carcinogens and bioactivation of some environmental procarcinogens [33]. CYP2C9, on the other hand, is the primary enzyme that metabolizes drugs with a narrow therapeutic index [34]. CYP2D6 is highly polymorphic, and its metabolism is variable; people with reduced or no activity of this enzyme would be at risk of reduced efficacy of drugs or present adverse effects [35]. Lastly, CYP3A4 is responsible for the metabolism of drugs, carcinogens, steroids, and eicosanoids [36]. Based on the results obtained, THC, CBD, and THCV were all predicted to be CYP2C19, CYP2C9, and CYP2D6 inhibitors. Additionally, the results of metabolism prediction for THC and THCV are promising, as all compounds were found to have a low likelihood of inhibiting the CYP1A2 enzyme. The findings also showed that both THC and THCV compounds are non-inhibitors of CYP3A4 enzyme, except for CBD, which was found to be an inhibitor of the said enzyme.
The excretion profiles of the three compounds were predicted using total clearance and OCT2 (organic cation transporter2) substrate renal. Total clearance (log ml/min/kg) is a combination of hepatic and renal clearance. OCT2, on the other hand, is a renal uptake transporter that plays a vital role in drug uptake, disposition, and clearance of drugs in the kidneys. Hence, the total clearance is proportional to renal OCT2. OCT2 evaluation of a candidate compound’s transfer provides information on both its clearance and potential contraindications [37]. The pKCSM prediction results (Table 8) show that the total clearance of cannabidiol is the highest, followed by tetrahydrocannabinol, while tetrahydrocannabivarin has the lowest. This means that tetrahydrocannabivarin has the highest bioavailability. The findings also revealed that none of the compounds are OCT2 substrates. From the results above, it can be inferred that these phytocannabinoids are excreted through the kidney via a mechanism other than OCT2.
Toxicity refers to how much a substance can harm an organism or its organs, such as cells and tissues (Duran-Iturbide et al., 2020) [38]. Vital parameters such as the Ames test, hepatotoxicity, and oral rat acute toxicity LD50 values were used to predict the toxicity levels of THC, CBD, and THCV. The toxicity results show that none of the compounds are mutagenic and are unlikely to be hepatotoxic. On the other hand, all compounds obtained LD50 values that are in the range of 482-500 mg/kg/day, which fall into toxicity class 4 of the Globally Harmonized System (300 < Category 4 ≤ 2000 mg/kg/day), indicating that they are slightly toxic if swallowed and thus can be considered safe [9].
The pre-assessment of the toxic properties of a compound is essential for drug discovery, especially on the increasing number of compounds and their mixtures. There is also increased exposure of humans to chemicals, particularly to the abused usage of cannabinoids. THC, CBD, and THCV are the major psychoactive components of marijuana plants. Hence, Protox-II was utilized to predict the toxicity of these cannabinoid compounds. Protox-II results exhibited that it has active effects on immunotoxicity; three compounds are positive to have an activity on immune cells. Several pieces of literature supported the immunotoxicity effects where the amount of various immune system cells was reduced and the immune response of mice to influenza virus infection after the administration of the cannabinoids. In addition, pregnant mice were also subjected to a cannabinoid compound and noticed the immunosuppressive effect on the fetus. These immunotoxicity effects are due to the ligation of G protein-coupled receptors, CB 1 and 2. Varying expression of the receptors modifies the response to immune stimulation, although CB2 has a more significant expression than CB1.
Therefore, in silico results from Protox-II were confirmed by several in vitro experiments performed. The Protox-II results also demonstrated that CBD has immunotoxicity effects, supported by the study by Zwicker et al. (2015) [39]. In their study, the immunotoxic potential of CBD was evaluated using in vitro approach. Based on the proteomic, functional, and metabolomic data acquired, it was discovered that CBD has immunotoxic effects on T cells. According to Lee et al. (2008) [40], immune cell apoptosis is caused by the formation of reactive oxygen species that is induced by the application of CBD.
On the other hand, these cannabinoid compounds have shown inactive results on hepatotoxicity, carcinogenicity, mutagenicity, and cytotoxicity. In a study by Stohs & Ray (2020) [41], where epilepsy patients received a higher oral dose of the drug with cannabinoid compounds for 48 weeks, four patients withdrew from the study stating the reasons of diarrhea, fatigue, somnolence, and convulsions. Nevertheless, patients did not mention any elevated serum aminotransferases or hepatoxicity. Therefore, based on in vitro studies performed, there is a low incidence of hepatotoxicity caused by cannabinoids in low and therapeutic doses. Furthermore, cannabinoid compounds are also found to be inactive with carcinogenicity, as supported by the study of Melamede (2005) [42], discovering that cannabinoids initiate tumor regression and inhibit angiogenic factors in mice. In addition, in vitro studies in treating glioma have resulted to depreciating levels of VEGF with the administration of cannabinoids. This is due to the special immune regulatory activity of cannabinoids that down-regulates immunologically produced free radical by promoting Th2 immune cytokine profile.
Among the compounds, CBD has the highest probability of being inactive in mutagenicity, with a value of 0.85 compared to 0.77 for THC and 0.74 for THCV. The lack of mutagenic effects of CBD is also evident in the study of Jones et al. (2012) [43], wherein CBD was administered in rats. There has been no observation of significant toxicity, genotoxicity, or mutagenicity in vivo. As for THC, it has no mutagenic properties in vitro, according to Zimmerman et al. (1978) [44]. After exposure to THC to the cultured fibroblasts derived from healthy individuals and Xeroderma pigmentosum patients, the amount of chromosomal breakage and chromatid exchanges did not appear to have increased. Similarly, THCV was found to be non-mutagenic in the Ames test prediction [45]. Nonetheless, in terms of cytotoxicity, the results show that the three compounds are all inactive. Cannabinoids have selective cytotoxic properties against tumor cells while also preventing the apoptosis of healthy, normal cells [46]. Several studies in different cancer cell lines have demonstrated that these cannabinoids activate autophagy and ultimately causes cell death [47, 48]. Therefore, based on the predicted results generated from Protox-II that are supported by in vitro and in vivo studies, these cannabinoid compounds are not toxic and safe for cancer treatment uses.
Both cannabinoid and non-cannabinoid molecules contribute significantly to the cannabis extract’s pharmacological profile. Research among phytoplants contributed to developing the defined chemotypes of high-yield cannabis cultivars. Cannabis sativa L. is herbal medicine from which cannabinoid compounds like THC, CBD, and THCV can be derived. This herbaceous plant has at least 554 compounds, including about 150 terpenophenolic active molecules and phytocannabinoids that are only exclusive to this plant. Aside from this, it also contains various phytomolecular families such as oxylipins, flavonoids, and terpenoids. This has been a subject of interest mainly due to its pharmaceutical applications since it has metabolites that exhibit potent bioactivities on human health. The cannabis extracts contain large amounts of pharmacological properties, and two major cannabinoid molecules are responsible for this, namely cannabidiol (CBD) and tetrahydrocannabinol (THC) [49]. CBD is the primary molecule found in hemp, while THC is the main component of drug-type cannabis. THC is responsible for psychotropic activity, while CBD lacks this and is reported to significantly reduce adverse effects like anxiety and psychosis [50].
On the other hand, THCV is a minor cannabinoid that is known to stimulate alertness and euphoria among its users. Although cannabinoids are mostly known for their psychoactive components, non-psychoactive compounds such as CBD give new light to future research. They have been found moderately effective in clinical trials of multiple brain injury, neuropathic pain, multiple sclerosis, and arthritis [50].
LD50 is the drug’s concentration at which half of the cells undergo death. This implies that the higher amount of LD50 means lesser drug action making it necessary for there to be a higher drug concentration to cause death in cells or cause harmful effects. On the other hand, lower LD50 values mean that the drug action is more potent at lower concentrations.
The pharmacokinetics of CBD is complex, and the bioavailability of oral CBD is low across species due to extensive Phase I metabolism. Comprehensive studies in animals, including rodents, indicate that much of the administered CBD is excreted intact or as its glucuronide [51]. In acute rat toxicity, CBD falls mainly in Class 4 (Rat IP LD50, Rat IV LD50, and Rat Oral LD50). With this, intake by oral and intraperitoneal CBD is classified as non-toxic. However, in Rat SC LD50, it is in Class 3, which is categorized as slightly toxic upon intravenous intake.
For the rat toxicity prediction of CBD and THC, The LD50 value at rat oral is 799,200 mg/kg, making its concentration comparatively low than THCV. Moreover, THCV has the lowest LD50 concentration in rat oral which is 663,300 mg/kg. In LD50 concentrations, CBD and THC yield the highest value at rat IV-0,912 mg/kg) while THCV is within the rat IV (-0,841 mg/kg). The classification of the routes for the cannabinoids shows that THC has moderate toxicity in Rat IV and low toxicity in Rat IP, Oral, and SC. CBD has the same toxicity classification as THC. On the other hand, THCV is moderately toxic when administered subcutaneously compared to when it is administered intravenously, orally, or intraperitoneally as these routes have a classification of low toxicity. Based on these data, it can be inferred that cannabinoids, when administered intravenously, cause more toxicity compared to other routes at a given concentration as it is considered as least harmful when administered orally [52].
ADVER-Pred is a free online service for in silico prediction of adverse pharmacological effects of drug candidates, including arrhythmia, cardiac failure, hepatotoxicity, myocardial infarction, and nephrotoxicity on the cardiovascular and hepatobiliary systems. The estimated Pa and Pi values predict their probable activity or inactivity to show toxic effects. Compounds with Pa > Pi or high Pa and Pa-Pi values are considered active and have a greater probability of activity in in vivo studies or clinical trials [53]. Results shown in Table 11 revealed that all compounds had higher Pi values and lower Pa values, indicating that THC, CBD, and THCV are unlikely to lead to such adverse effects; thus, these compounds are designated to the subset of “inactive” in the PASS (Prediction of Activity Spectra for Substances) training set consisting of drug-like biologically active compounds. “Inactive” compounds in the PASS training set refer to those that lack definitive information about their biological activity found in an authoritative data source [54].
Pi values influence how these “inactive” correlate to their minimal adversity property. CBD had the highest probable activity (Pa= 0.128) among the compounds, which can lead to arrhythmia. In comparison, THCV had the highest probability of being inactive (Pi = 0.792), also corresponding to arrhythmia. THCV had the lowest activity probability of arrhythmia (Pa = 0.048) whereas CBD had the lowest inactivity probability of cardiac failure (Pi = 0.468).
Despite their high in silico predicted probability of inactivity values, there have been reports showing that there is still a possibility for THC, CBD, and THCV to be active upon intake and exert negative effects on human health. Findings from Holland (2020) [55] reported that THC and CBD cause no minimal side effects such as increased heart rate, coordination problems, fatigue, and diarrhea. However, studies have shown that high dosages of cannabinoids, particularly THCV, have a profound effect on specific brain nerve cells, which could lead to long-term complications [56]. Hence, it can be deduced that excessive intake of THC and CBD could potentially lead to adversity affecting the brain. Despite these reported side effects, cannabinoids are still medically used to treat related disorders due to their vast therapeutic properties.
CLC-pred web service was accessed to predict the cytotoxicity of drug-like compounds against tumor and normal human cell lines [57]. Pa>Pi threshold was used to determine active compounds. Table 12 reveals that CBD has the highest probability for their top cell lines with a Pa value of 0.542 and Pi value of 0.004 against breast adenocarcinoma (MDA-MB-453). This is comparatively high compared to the probability of THC against Glioma (U-251) with only 0.341 as its Pa value (Pi value = 0.048) and the probability of THCV against non-small cell lung carcinoma (HOP-18) with a Pa value of 0.369 (Pi value = 0.040). Following this, CBD has a probability of only 0.372 (Pi value = 0.021) and 0.385 (Pi value = 0.056) against thyroid gland undifferentiated (anaplastic) carcinoma (8505C) and adult B acute lymphoblastic leukemia (NALM-6), respectively. THC also has a probability of 0.321 (Pi value = 0.034) and 0.319 (Pi value = 0.064) against gastric carcinoma (MKN-7) and non-small cell lung carcinoma (HOP-18), making these its top 2 and 3 cell lines. On the other hand, THCV was found to have a probability of 0.317 (Pi value = 0.058) against Glioma (U-251) and a probability of 0.318 (Pi value = 0.059) against Melanoma (M19-MEL). Comparing the top three results of these cannabinoids, CBD having the highest probability against adenocarcinoma infers that it shows promising results and potential as an alternative component for cancer treatment. In addition to this, in terms of addressing Glioma, THC is slightly more effective against this disease than THCV. There is a slight difference between the probability of the two, with THC having a higher probability against the said disease.
The Pi values signify each cannabinoid’s probability of inactivity. Looking at the data gathered, CBD, which has the highest Pa value compared to THC and THCV, also has the lowest Pi value among the top three cancer cell lines. This further proves that, to an extent, CBD has active components that can be effective against breast adenocarcinoma. It is also the only cannabinoid that has exemplified a Pa value of greater than 0.5, which means that CBD is the only cannabinoid with a considerably high probability of action against the disease compared to the rest of the data in the table.
Although some inferences can be made from this study’s cell line cytotoxicity prediction, further investigation and research are recommended to determine their bioactive components, applicability, and efficiency against diseases like cancer, which could pave the way for future clinical studies and the development of treatment plans.
Pass Target Prediction predicts the biological potential of THC, CBD, and THCV by analyzing their biological activity, pharmacological effects, mechanism of action, toxic and adverse effects, interactions with metabolic enzymes and transporters, and influence on gene expression (Filimonov et al., 2014) [58]. The structure of THC, CBD, and THCV is represented in PASS predicting proteins with possible direct and mediated target interactions. The result in PASS prediction comes in the form of confidence p-values for each class [59].
The predicted results of THC produced a list of proteins that have direct interactions with the compound in descending order Cannabinoid CB2 receptor, Cannabinoid CB1 receptor, Cytochrome P450 2C9, DNA Topoisomerase II-a, and Estrogen receptor-b. Meanwhile, the list of proteins that have mediated interactions with THC is Glycine receptor subunit a-1, Cannabinoid CB2 receptor, Cannabinoid CB1 receptor, Serotonin 1e (5HT1e) receptor, and Nuclear receptor subfamily o group B member 1. Whereas the predicted results of CBD produced a list of proteins that have direct interactions with the compound in descending order are as follows: Phosphatidylinoitol-4-phosphate-5-kinasetype 1g, Cannabinoid CB1 receptor, Serine/threonine-protein kinase 32A, Rhodopsin kinase, and Cannabinoid CB2 receptor. Meanwhile, the list of proteins that have mediated interactions with CBD is Cannabinoid CB2 receptor, Glycine receptor subunit a-1, Tyrosine-protein kinase FYN, Cannabinoid CB1 receptor, and N-arachidonyl glycine receptor. Lastly, the predicted results of THCV produced a list of proteins that has direct interactions with the compound also in descending order are Phosphatidylinoitol-4-phosphate-5-kinasetype 1g, Rhodopsin kinase, Serine/threonine-protein kinase 32A, Cannabinoid CB1 receptor, and tyrosine-protein kinase Srms. Correspondingly, the list of proteins that have mediated interactions with THCV is Glycine receptor subunit a-1, Cannabinoid CB2 receptor, Tyrosine-protein kinase FYN, Cannabinoid CB1 receptor, and N-arachidonyl glycine receptor.
Among the direct protein targets involved in THC, the Cannabinoid CB2 receptor had the highest confidence of 0.4686. According to Vučković et al. (2018) [60], THC acts as a partial agonist toward the Cannabinoid CB2 receptor, which, regardless of the quantity of cannabinoid used, cannot fully activate the said receptor—hence affecting their binding interactions. Although, as presented in hyperalgesia models, THC binds to the Cannabinoid CB2 receptor, which indicates the relation of analgesic qualities with the attenuation of nerve growth factor and consequent inhibition of adenylyl cyclase [61]. On the other hand, CBD has been demonstrated to be a promising therapeutic and pharmacological medication target, with both direct and indirect interactions. Phosphatidylinoitol-4-phosphate-5-kinasetype 1g had the highest confidence interaction of 0.7681 out of the five proteins that can interact with CBD directly. D’Souza and Epand (2013) [62] found that phosphatidyl is substantially enriched with acyl chains, supporting this finding. Furthermore, the Cannabinoid CB2 receptor had the highest binding affinity with CBD, with 0.7529. The functional relevance of the Cannabinoid CB2 receptor in neuropathic pain and other neuroinflammatory illnesses underscores the therapeutic promise of CB2 receptor-targeting medicines [63]. Lastly, the compound THCV directly interacted with Phosphatidylinoitol-4-phosphate-5-kinasetype 1g with a confidence p-value of 0.8084. It is known that THCV is a homolog of THC where it only differs by a propyl side chain. It appears that it is a potential treatment for neurological and psychiatric conditions. Furthermore, it has a mediated interaction with Glycine receptor subunit α-1 at 0.6769. The interaction of THCV and glycine receptors may exhibit similar or more potent neuroprotective properties like THC and CBD. Moreover, THCV is a homolog of THC where it only differs at a propyl side chain. Hence, THCV acts as a CB1 receptor agonist that showed promise as an antiepileptic agent and protected neurons [64].
In conclusion, the physicochemical properties obtained from in silico ADME indicate that CBD had significant cell line toxicity and direct and mediated protein target interaction results, but had a poor oral absorption due to its insolubility, inactivity in mutagenicity, an inhibitor of only the CYP3A4 enzyme; THC exhibited excellent HIA and BBB, low possibility of blocking the CYPA2 enzyme, show a violation in drug-likeness analysis, insolubility, resulting in poor oral absorption, and higher toxicity than THCV. Among the three cannabinoids, THCV was a potential candidate for chemotherapeutic drugs obtaining the most favorable results in drug-likeness analysis, physicochemical properties, acute rat toxicity, and ADMET results.
To further improve this study and evaluate the effects of THC, CBD, and THCV, Biopharmaceutics Drug Disposition Classification System (BDDCS) must be performed to distinguish the transporters that participate in the drug absorption and elimination pathway. Furthermore, molecular docking analysis is also recommended to determine the binding affinity of the ligand and the structure of the protein-ligand interaction, which is vital for lead optimization. Due to the broad list of cannabinoid compounds with anticancer potential, there is a struggle to choose the cannabinoid with optimal anticancer development. Thus, a systematic investigation and more comprehensive ADMET profiles and different anticancer mechanisms must be conducted to generate broader antitumor spectra on cannabinoids to address this problem.
Author Contribution Statement
The authors confirm their contribution to the paper as follows: study conceptualization and design, AM Labrador and MKP Devanadera; organization and collection of data, AA Gallardo, LAJ Gomez, MR Gutierrez, MIH Liquido, and EDAB Maglaqui; analysis and interpretation of results, AA Gallardo, PAO Delos Reyes, and AMG Manalo; draft manuscript preparation, SAA Dones, MMU Dumbrique, LAJ Gomez, HE Lim, and MML Macawile; and proofreading and editing of the manuscript, MR Gutierrez, and AM Labrador. All authors reviewed, agreed, and approved the final version of the manuscript.
Acknowledgements
General
The authors would also like to express their gratitude to the academic staff of the internship committee of the Mammalian Tissue Culture Laboratory of the University of Santo Tomas Research Center for Natural and Applied Sciences through the Department of Biochemistry, Faculty of Pharmacy, the University of Santo Tomas for providing the training for the use of in silico tools used in this study.
Data Availability
Available through the corresponding author.
Study Registration
It is not registered as a clinical study or meta-analysis study.
Conflict of Interest
All authors declared no conflict of interest.
References
- 1.Nagai H, Kim YH. Cancer prevention from the perspective of global cancer burden patterns. J Thorac Dis. 2017;9(3):448–51. doi: 10.21037/jtd.2017.02.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parkin DM. The global health burden of infection‐associated cancers in the year 2002. Int J Cance. 2006;118(12):3030–44. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 3.Subramaniam S, Selvaduray KR, Radhakrishnan AK. Bioactive compounds: Natural defense against cancer? Biomolecules. 2019;9(12) doi: 10.3390/biom9120758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dariš B, Tancer Verboten M, Knez Ž, Ferk P. Cannabinoids in cancer treatment: Therapeutic potential and legislation. Bosn J Basic Med Sci. 2019;19(1):14–23. doi: 10.17305/bjbms.2018.3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scott KA, Dalgleish AG, Liu WM. Anticancer effects of phytocannabinoids used with chemotherapy in leukaemia cells can be improved by altering the sequence of their administration. Int J Oncol. 2017;51(1):369–77. doi: 10.3892/ijo.2017.4022. [DOI] [PubMed] [Google Scholar]
- 6.Suriyachan K TS. In vitro anti-proliferative activity of cannabis extract on human cancer cell lines. Journal of The Department of Medical Services. 2021;46(3):23–8. [Google Scholar]
- 7.Hosami F, Ghadimkhah MH, Salimi V, Ghorbanhosseini SS, Tavakoli-Yaraki M. The strengths and limits of cannabinoids and their receptors in cancer: Insights into the role of tumorigenesis-underlying mechanisms and therapeutic aspects. Biomed Pharmacother. 2021;144:112279. doi: 10.1016/j.biopha.2021.112279. [DOI] [PubMed] [Google Scholar]
- 8.Helcman M, Šmejkal K. Biological activity of cannabis compounds: A modern approach to the therapy of multiple diseases. Phytochemistry Reviews. 2021:1–42. [Google Scholar]
- 9.Gadaleta D, Vuković K, Toma C, Lavado GJ, Karmaus AL, Mansouri K, et al. Sar and qsar modeling of a large collection of ld50 rat acute oral toxicity data. J Cheminform. 2019;11(1):1–16. doi: 10.1186/s13321-019-0383-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pergam SA, Woodfield MC, Lee CM, Cheng GS, Baker KK, Marquis SR, Fann JR. Cannabis use among patients at a comprehensive cancer center in a state with legalized medicinal and recreational use. Cancer. 2017;123(22):4488–97. doi: 10.1002/cncr.30879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pagano C, Navarra G, Coppola L, Bifulco M, Laezza C. Molecular mechanism of cannabinoids in cancer progression. Int J Mol Sci. 2021;22:7. doi: 10.3390/ijms22073680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Petrocellis L, Ligresti A, Schiano Moriello A, Iappelli M, Verde R, Stott CG, et al. Non-thc cannabinoids inhibit prostate carcinoma growth in vitro and in vivo: Pro-apoptotic effects and underlying mechanisms. Br J Pharmacol. 2013;168(1):79–102. doi: 10.1111/j.1476-5381.2012.02027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Araújo RSA, da Silva-Junior EF, de Aquino TM, Scotti MT, Ishiki HM, Scotti L, Mendonça-Junior FJB. Computer-aided drug design applied to secondary metabolites as anticancer agents. Curr Top Med Chem. 2020;20(19):1677–703. doi: 10.2174/1568026620666200607191838. [DOI] [PubMed] [Google Scholar]
- 14.Daina A, Michielin O, Zoete V. Swissadme: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stuurman FE, Nuijen B, Beijnen JH, Schellens JH. Oral anticancer drugs: Mechanisms of low bioavailability and strategies for improvement. Clin Pharmacokinet. 2013;52(6):399–414. doi: 10.1007/s40262-013-0040-2. [DOI] [PubMed] [Google Scholar]
- 16.Parodi A, Buzaeva P, Nigovora D, Baldin A, Kostyushev D, Chulanov V, et al. Nanomedicine for increasing the oral bioavailability of cancer treatments. J Nanobiotechnology. 2021;19(1):1–19. doi: 10.1186/s12951-021-01100-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46(1-3):3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 18.Daina A, Michielin O, Zoete V. Ilogp: A simple, robust, and efficient description of n-octanol/water partition coefficient for drug design using the gb/sa approach. J Chem Inf Model. 2014;54(12):3284–301. doi: 10.1021/ci500467k. [DOI] [PubMed] [Google Scholar]
- 19.Zafar F, Gupta A, Thangavel K, Khatana K, Sani AA, Ghosal A, et al. Physicochemical and pharmacokinetic analysis of anacardic acid derivatives. ACS Omega. 2020;5(11):6021–30. doi: 10.1021/acsomega.9b04398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dahlin JL, Inglese J, Walters MA. Mitigating risk in academic preclinical drug discovery. Nat Rev Drug Discov. 2015;14(4):279–94. doi: 10.1038/nrd4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doak BC, Over B, Giordanetto F, Kihlberg J. Oral druggable space beyond the rule of 5: Insights from drugs and clinical candidates. Chem Biol. 2014;21(9):1115–42. doi: 10.1016/j.chembiol.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 22.Ghose AK, Viswanadhan VN, Wendoloski JJ. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery 1 A qualitative and quantitative characterization of known drug databases. J Comb Chem. 1999;1(1):55–68. doi: 10.1021/cc9800071. [DOI] [PubMed] [Google Scholar]
- 23.Delaney JS. Esol: Estimating aqueous solubility directly from molecular structure. J Chem Inf Comput Sci. 2004;44(3):1000–5. doi: 10.1021/ci034243x. [DOI] [PubMed] [Google Scholar]
- 24.Ali J, Camilleri P, Brown MB, Hutt AJ, Kirton SB. Revisiting the general solubility equation: In silico prediction of aqueous solubility incorporating the effect of topographical polar surface area. Journal of chemical information and modeling. 2012;52(2):420–8. doi: 10.1021/ci200387c. [DOI] [PubMed] [Google Scholar]
- 25.Daina A, Zoete V. A boiled-egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem. 2016;11(11):1117–21. doi: 10.1002/cmdc.201600182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brenk R, Schipani A, James D, Krasowski A, Gilbert IH, Frearson J, Wyatt PG. Lessons learnt from assembling screening libraries for drug discovery for neglected diseases. ChemMedChem. 2008;3(3):435–44. doi: 10.1002/cmdc.200700139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Domínguez-Villa FX, Durán-Iturbide NA, Ávila-Zárraga JG. Synthesis, molecular docking, and in silico adme/tox profiling studies of new 1-aryl-5-(3-azidopropyl)indol-4-ones: Potential inhibitors of sars cov-2 main protease. Bioorg Chem. 2021;106:104497. doi: 10.1016/j.bioorg.2020.104497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pires DE, Blundell TL, Ascher DB. Pkcsm: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J Med Chem. 2015;58(9):4066–72. doi: 10.1021/acs.jmedchem.5b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cerqueira NM, Gesto D, Oliveira EF, Santos-Martins D, Brás NF, Sousa SF, et al. Receptor-based virtual screening protocol for drug discovery. Arch Biochem Biophys. 2015;582:56–67. doi: 10.1016/j.abb.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 30.Martin YC. A bioavailability score. Journal of medicinal chemistry. 2005;48(9):3164–70. doi: 10.1021/jm0492002. [DOI] [PubMed] [Google Scholar]
- 31.Manikandan P, Nagini S. Cytochrome p450 structure, function and clinical significance: A review. Curr Drug Targets. 2018;19(1):38–54. doi: 10.2174/1389450118666170125144557. [DOI] [PubMed] [Google Scholar]
- 32.Thorn CF, Aklillu E, Klein TE, Altman RB. Pharmgkb summary: Very important pharmacogene information for cyp1a2. Pharmacogenet Genomics. 2012;22(1):73–7. doi: 10.1097/FPC.0b013e32834c6efd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, Yang H, Wu Z, Wang T, Li W, Tang Y, Liu G. In silico prediction of blood–brain barrier permeability of compounds by machine learning and resampling methods. ChemMedChem. 2018;13(20):2189–201. doi: 10.1002/cmdc.201800533. [DOI] [PubMed] [Google Scholar]
- 34.Daly AK, Rettie AE, Fowler DM, Miners JO. Pharmacogenomics of cyp2c9: Functional and clinical considerations. J Pers Med. 2017;8:1. doi: 10.3390/jpm8010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Del Tredici AL, Malhotra A, Dedek M, Espin F, Roach D, Zhu GD, et al. Frequency of cyp2d6 alleles including structural variants in the united states. Front Pharmacol. 2018;9:305. doi: 10.3389/fphar.2018.00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dresser GK, Spence JD, Bailey DG. Pharmacokinetic-pharmacodynamic consequences and clinical relevance of cytochrome p450 3a4 inhibition. Clin Pharmacokinet. 2000;38(1):41–57. doi: 10.2165/00003088-200038010-00003. [DOI] [PubMed] [Google Scholar]
- 37.Pratama MRF, Poerwono H, Siswodiharjo S. Admet properties of novel 5-o-benzoylpinostrobin derivatives. J Basic Clin Physiol Pharmacol. 2019;30:6. doi: 10.1515/jbcpp-2019-0251. [DOI] [PubMed] [Google Scholar]
- 38.Durán-Iturbide NA, Díaz-Eufracio BI, Medina-Franco JL. In silico adme/tox profiling of natural products: A focus on biofacquim. ACS Omega. 2020;5(26):16076–84. doi: 10.1021/acsomega.0c01581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zwicker P SN, Niehs S, Methling K, Wurster M, Bernhardt J, et al. An in vitro approach for evaluating the immunotoxic potential of cannabidiol. Toxicology Letters. 2015;238(2):S221. [Google Scholar]
- 40.Lee CY, Wey SP, Liao MH, Hsu WL, Wu HY, Jan TR. A comparative study on cannabidiol-induced apoptosis in murine thymocytes and el-4 thymoma cells. Int Immunopharmacol. 2008;8(5):732–40. doi: 10.1016/j.intimp.2008.01.018. [DOI] [PubMed] [Google Scholar]
- 41.Stohs S, Ray SD. Is cannabidiol hepatotoxic or hepatoprotective: A review. Toxicology Research and Application. 2020;4:2397847320922944. [Google Scholar]
- 42.Melamede R. Cannabis and tobacco smoke are not equally carcinogenic. Harm Reduct J. 2005;2:21. doi: 10.1186/1477-7517-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones NA, Glyn SE, Akiyama S, Hill TD, Hill AJ, Weston SE, et al. Cannabidiol exerts anti-convulsant effects in animal models of temporal lobe and partial seizures. Seizure. 2012;21(5):344–52. doi: 10.1016/j.seizure.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 44.Zimmerman AM, Stich H, San R. Nonmutagenic action of cannabinoids in vitro. Pharmacology. 1978;16(6):333–43. doi: 10.1159/000136789. [DOI] [PubMed] [Google Scholar]
- 45.Altyar AE, Youssef FS, Kurdi MM, Bifari RJ, Ashour ML. The role of cannabis sativa l As a source of cannabinoids against coronavirus 2 (sars-cov-2): An in silico study to evaluate their activities and admet properties. Molecules. 2022;27(9) doi: 10.3390/molecules27092797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bogdanović V, Mrdjanović J, Borišev I. A review of the therapeutic antitumor potential of cannabinoids. J Altern Complement Med. 2017;23(11):831–6. doi: 10.1089/acm.2017.0016. [DOI] [PubMed] [Google Scholar]
- 47.Salazar M, Carracedo A, Salanueva ÍJ, Hernández-Tiedra S, Lorente M, Egia A, et al. Cannabinoid action induces autophagy-mediated cell death through stimulation of er stress in human glioma cells. J Clin Invest. 2009;119(5):1359–72. doi: 10.1172/JCI37948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Armstrong JL, Hill DS, McKee CS, Hernandez-Tiedra S, Lorente M, Lopez-Valero I, et al. Exploiting cannabinoid-induced cytotoxic autophagy to drive melanoma cell death. J Invest Dermatol. 2015;135(6):1629–37. doi: 10.1038/jid.2015.45. [DOI] [PubMed] [Google Scholar]
- 49.Walsh KB, McKinney AE, Holmes AE. Minor cannabinoids: Biosynthesis, molecular pharmacology and potential therapeutic uses. Front Pharmacol. 2021;12:777804. doi: 10.3389/fphar.2021.777804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klein TW, Newton CA. Therapeutic potential of cannabinoid-based drugs. Adv Exp Med Biol. 2007;601:395–413. doi: 10.1007/978-0-387-72005-0_43. [DOI] [PubMed] [Google Scholar]
- 51.Harvey DJ. Metabolism and pharmacokinetics of the cannabinoids. In: Physiology of substande abuse, Watson RR., editors. Biochemistry and Physiology of substande abuse. Boca Raton FL: CRC Press; 1991. pp. 279–365. [Google Scholar]
- 52.Lucas CJ, Galettis P, Schneider J. The pharmacokinetics and the pharmacodynamics of cannabinoids. Br J Clin Pharmacol. 2018;84(11):2477–82. doi: 10.1111/bcp.13710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ivanov SM, Lagunin AA, Rudik AV, Filimonov DA, Poroikov VV. Adverpred-web service for prediction of adverse effects of drugs. J Chem Inf Model. 2018;58(1):8–11. doi: 10.1021/acs.jcim.7b00568. [DOI] [PubMed] [Google Scholar]
- 54.Pogodin PV, Lagunin AA, Rudik AV, Filimonov DA, Druzhilovskiy DS, Nicklaus MC, Poroikov VV. How to achieve better results using pass-based virtual screening: Case study for kinase inhibitors. Front Chem. 2018;6:133. doi: 10.3389/fchem.2018.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holland K. What’s the difference between cbd and thc? Healthline. 2019 [Google Scholar]
- 56.Kasten CR, Zhang Y, Boehm SL. Acute cannabinoids produce robust anxiety-like and locomotor effects in mice, but long-term consequences are age- and sex-dependent. Front Behav Neurosci. 2019;13:32. doi: 10.3389/fnbeh.2019.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lagunin AA, Dubovskaja VI, Rudik AV, Pogodin PV, Druzhilovskiy DS, Gloriozova TA, et al. Clc-pred: A freely available web-service for in silico prediction of human cell line cytotoxicity for drug-like compounds. PLoS One. 2018;13(1):e0191838. doi: 10.1371/journal.pone.0191838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Filimonov D, Lagunin A, Gloriozova T, Rudik A, Druzhilovskii D, Pogodin P, Poroikov V. Prediction of the biological activity spectra of organic compounds using the pass online web resource. Chem Heterocycl Compd. 2014;50:444–57. [Google Scholar]
- 59.Lampa S, Alvarsson J, Arvidsson Mc Shane S, Berg A, Ahlberg E, Spjuth O. Predicting off-target binding profiles with confidence using conformal prediction. Front Pharmacol. 2018;9:1256. doi: 10.3389/fphar.2018.01256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vučković S, Srebro D, Vujović KS, Vučetić Č, Prostran M. Cannabinoids and pain: New insights from old molecules. Front Pharmacol. 2018;9:1259. doi: 10.3389/fphar.2018.01259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Capodice JL, Kaplan SA. The endocannabinoid system, cannabis, and cannabidiol: Implications in urology and men’s health. Curr Urol. 2021;15(2):95–100. doi: 10.1097/CU9.0000000000000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.D’Souza K, Epand RM. Enrichment of phosphatidylinositols with specific acyl chains. Biochim Biophys Acta. 2014;1838(6):1501–8. doi: 10.1016/j.bbamem.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 63.Bie B, Wu J, Foss JF, Naguib M. An overview of the cannabinoid type 2 receptor system and its therapeutic potential. Curr Opin Anaesthesiol. 2018;31(4):407–14. doi: 10.1097/ACO.0000000000000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.da Silva AL, Silva TL, Borges LL. Investigation of molecular mechanisms of cannabinoids with neuromodulator activity using in silico tools. Revista Brasileira Militar De Ciências. 2021;7:19. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Available through the corresponding author.