Abstract
Influenza virus ribonucleoproteins (RNPs) were reconstituted in vivo from cloned cDNAs expressing the three polymerase subunits, the nucleoprotein (NP), and short template RNAs. The structure of purified RNPs was studied by electron microscopy and image processing. Circular and elliptic structures were obtained in which the NP and the polymerase complex could be defined. Comparison of the structure of RNPs of various lengths indicated that each NP monomer interacts with approximately 24 nucleotides. The analysis of the amplification of RNPs with different lengths showed that those with the highest replication efficiency contained an even number of NP monomers, suggesting that the NP is incorporated as dimers into newly synthesized RNPs.
The genome of influenza A virus consists of eight ribonucleoprotein complexes (vRNPs) containing a single-stranded RNA segment of negative polarity associated to nucleoprotein (NP) molecules and bound to the polymerase. This enzyme is a heterotrimer formed by the PB1, PB2, and PA proteins (11, 12, 25, 29), all of them being required for efficient RNA transcription and replication (52) (B. Perales, unpublished data). Both transcription and replication take place in the nucleus of the infected cells (24, 27). The replication of viral RNA (vRNA) involves the generation of a full-length RNA copy of positive polarity that is encapsidated with NP molecules and complexed with the polymerase (cRNP). These cRNPs serve as intermediates for the synthesis of vRNA progeny molecules (22). For RNA transcription, capped primers generated from cellular hnRNAs by a cap-stealing mechanism (35) are elongated by copying the vRNA template. The termination and polyadenylation signal consists of an oligo(U) sequence located close to the 5′ terminus of the vRNA templates (59, 64) next to the panhandle structure (38). These processes require the interaction of the polymerase with the conserved 5′-terminal sequences of the template (58, 61, 62).
The polymerase domains involved in intersubunit interactions have been identified (21, 51, 53, 73, 78), as well as the sequences in PB1 that bind the vRNA template (19, 36) and the cRNA template (20). The PB1 protein contains amino acid motifs present in other RNA-dependent RNA polymerases (56), whose mutation abolishes the transcriptional activity (5). The PB2 subunit is involved in the initiation of viral transcription (3, 51). It is a cap-binding protein (6, 70, 74) and contains the cap-dependent endonuclease activity (37). The biochemical role of the PA subunit is still uncertain. The phenotypes of temperature-sensitive mutants (reviewed in reference 39) suggest its involvement in vRNA synthesis. The PA subunit is a phosphoprotein (68) whose expression by transfection leads to the degradation of coexpressed proteins (67, 69).
The structure of the RNPs present in influenza virions has been studied by electron microscopy (9, 23, 28, 57). They consist of a ribbon-like cord, held together at its ends and folded back to form a coiled structure with a terminal loop. The available evidence indicates that each unit in the ribbon is a molecule of NP, and the polymerase is present at one end of the supercoil (46) and helps in keeping the ends linked together (32). The main component of the RNP is the NP, a basic protein capable of binding RNA without sequence specificity (1, 4) in such a way that the sugar-phosphate backbone is protected from modification (4). Complexes of viral RNA and NP molecules reconstituted in vitro show structural and biochemical properties similar to those of natural RNPs (31, 76). Purified NP, which is essentially RNA-free, is also able to self-assemble to generate oligomers and coiled structures analogous to RNPs (66).
Up to now, the flexibility and heterogeneity of the RNPs have prevented electron microscopy from getting medium- to high-resolution information by averaging techniques, and thus only their general morphological features are available. In this report we present an optimized procedure to reconstitute in vivo viral RNPs from cloned genes that allowed the purification of essentially single-size classes of mini-RNPs. The analysis of such specimens by electron microscopy, combined with classification and averaging techniques, revealed the presence of the NP monomers and the polymerase complex. Furthermore, combination of these structural data with the replication properties of reconstituted mini-RNPs with different sizes suggests that the NP molecules are incorporated as dimers into progeny RNPs.
MATERIALS AND METHODS
Biological materials.
The COS-1 cell line (18) was provided by Y. Gluzman and was cultivated as described previously (47). The vaccinia recombinant virus vTF7-3 (16) was a gift of B. Moss. The origin of plasmids pGPB1, pGPB2, pGPA, and pGNPpolyA has been described previously (44, 52). Plasmid 2.0 (2) was kindly provided by A. Ball. Plasmid pNS3, containing the NS sequence of influenza A/Victoria/3/75 under the T7 promoter, was provided by A. Portela. An anti-PB1 protein serum was prepared by immunization of rabbits with purified PB1 protein obtained by isolation from sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels. The antiserum specific for NP protein (1) was a gift of A. Portela. The origin of anti-PB2 and anti-PA monoclonal antibodies has been described (3).
Mutant construction.
The plasmids used for in vivo transcription of influenza virus model vRNAs were constructed as follows. First, the intermediate cloning vector pUC19RT was generated by inserting the SmaI-XbaI fragment of plasmid 2.0, which contains the cDNA copy of the hepatitis δ virus ribozyme and the T7 RNA polymerase terminator (2), into pUC19. To construct plasmids pT7NSCAT-RT and pT7NS-RT, PCR fragments were amplified by using as templates plasmids pIVACAT1/S (54) and pNS3, respectively. The primers used were 5′-AGCAAAAGCAGG-3′, which is complementary to the 3′ end of the NS RNA segment, and 5′-GCCTGGTACC-TAATACGACTCACTATA-AGTAGAAACAAGG-3′, which contains an Asp718 restriction site, the T7 RNA polymerase promoter (underlined) and the 5′-terminal sequence of the NS segment. Finally, the PCR fragments were digested with Asp718 restriction nuclease and ligated with the SmaI/Asp718-digested pUC19RT. Plasmid pT7NSΔCAT-RT was constructed from pT7NSCAT-RT by deletion of the BsmI restriction fragment internal to the cat gene. To generate a library of deletions of plasmid pT7NS-RT, the plasmid was digested with nucleases MunI and XcmI, treated with Bal31 nuclease for various times, and then self-ligated. The NS sequence from individual plasmid clones was determined, and the plasmids were used for reconstitution of RNPs as described below.
Transfection.
Cultures of COS-1 cells were infected with vTF7-3 virus at a multiplicity of infection of 5 PFU per cell. After virus adsorption for 1 h at 37°C, the cultures were washed with Dulbecco modified Eagle medium (DMEM) and transfected with a mixture of plasmids containing (for 100-mm dishes) pGPB1 (3 μg), pGPB2 (3 μg), pGPA (0.6 μg), pGNPpolyA (12 μg), and either pT7NSΔCAT-RT or pT7ΔNSRT clones (12 μg). The DNA mixtures were diluted to 0.5 ml in DMEM and, in a separate tube, cationic liposomes (2 μl/μg of DNA) were diluted to 0.5 ml in DMEM. The contents of both tubes were mixed, kept at room temperature for 15 min, and added to the culture plates containing 4 ml of DMEM. When RNA was transfected instead of a ribozyme construct, the procedure was as described earlier (52). After 24 h of incubation at 37°C, the medium was replaced by 10 ml of DMEM containing 2% fetal bovine serum and incubated for further 24 h. Cationic liposomes were prepared as described previously (65).
RNP purification.
Cultures infected and transfected as indicated above were collected 48 h after transfection and lysed for 2 h at 0°C in buffer A (10 mM Tris-HCl–1 mM EDTA–7.5 mM ammonium sulfate–0.025% NP-40–1 mM dithiothreitol, pH 7.9). After centrifugation at 10,000 × g for 30 s at 4°C, the extract was centrifuged on a 20 to 35% glycerol gradient in TN buffer (150 mM NaCl–50 mM Tris-HCl, pH 7.8) for 17 h at 35,000 rpm and 4°C in an SW41 rotor. Fractions containing active RNPs were pooled and centrifuged in a step glycerol gradient in TN buffer (48) for 8 h at 55,000 rpm and 4°C in an SW55 rotor. Active fractions were pooled and used for further analyses.
RNA analyses.
The synthesis of vNSZ and cNSZ probes was carried out as described earlier (52). They were used for dot hybridization as described previously (43). In vitro RNA synthesis reactions were performed as indicated previously (51). To identify the active RNP fractions in the glycerol gradients, aliquots of each one were used for in vitro RNA synthesis, the products were precipitated with 10% trichloroacetic acid (TCA) and filtered in a dot blot apparatus, and the membrane was autoradiographed. To assay the relative amplification of deletion library clones, extracts of infected and transfected cells were used for in vitro RNA synthesis, and the purified RNA products were separated by denaturing polyacrylamide-urea gel electrophoresis (51). The RNA bands were quantitated in a phosphorimager, and the data were corrected by use of the relative RNA lengths of the clones. When indicated, the synthesized RNA was separated by chromatography in oligo(dT) cellulose as described earlier (51).
Protein analyses.
Western blotting was carried out as described elsewhere (43). In brief, cell extracts were separated by SDS-PAGE and transferred to Immobilon filters, and the membranes were saturated with 3% bovine serum albumin for 1 h at room temperature. The filters were incubated with either anti-PB1 serum (1:500 dilution), anti-NP serum (1:300 dilution), or anti-PA monoclonal antibodies or anti-PB2 monoclonal antibodies (1:40 dilution of culture supernatants) for 1 h at room temperature. The filters were washed two times for 30 min with phosphate-buffered saline containing 0.25% Tween 20 and were incubated with a 1:10,000 dilution of goat anti-rabbit immunoglobulin G (IgG) or goat anti-mouse IgG conjugated to horseradish peroxidase. Finally, the filters were washed two times for 30 min as above and developed by enhanced chemiluminescence.
Electron microscopy and image processing.
Samples were adhered to carbon-coated collodion grids previously glow discharged in low air pressure. In some experiments, the RNPs were previously hybridized with a positive-polarity oligonucleotide specific for the vRNA template (hybridization for 10 min with a 100-fold excess of oligonucleotide in 150 mM NaCl–50 mM Tris-HCl [pH 7.5]) in order to increase the adhesion to the grids. Staining was performed with 2% uranyl acetate. For visualization of RNP-antibody complexes, RNPs adsorbed onto grids were washed twice with 150 mM NaCl–50 mM Tris-HCl (pH 7.5) and incubated for 1 h at 37°C with a 1:1,000 dilution of monoclonal antibody PB2-25 (3). After a washing with the same buffer, the sample was stained as indicated above. Transmission electron micrographs were recorded by using a low-dosage protocol at an approximate magnification of ×60,000 in a JEOL 1200 EXII microscope. Micrographs were digitized at 4 Å/pixel by using an Eikonix 1412 digital camera. Images corresponding to RNPs (130 by 130 pixels) were extracted, centered, and aligned by using a free-pattern algorithm (42, 49). Due to the heterogeneity of the images, the aligned projections of the RNPs were subjected to self-organizing Kohonen maps (33, 41). After this classification, homogeneous populations were obtained and averaged. The resolution of the average images was estimated by the spectral signal-to-noise ratio method (75). Final average images were filtered to the resolution obtained in each case, as indicated in the figure legends.
RESULTS
Purification of biologically active viral RNPs reconstituted from cloned genes.
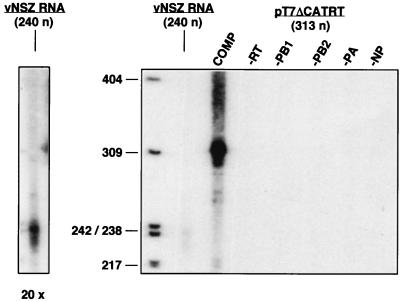
Several experimental approaches have been used to reconstitute in vivo active influenza virus RNPs from cloned genes. The expression of the polymerase subunits and the NP has been obtained by infection with recombinant vaccinia viruses (26), by infection with simian virus 40 recombinant viruses (10), by use of polymerase II-driven constructs (30, 55), and by transfection into vaccinia T7 virus-infected cells (44; reviewed in reference 45). Usually, an in vitro transcript modeled on a vRNA segment has been provided by direct RNA transfection (10, 26), by a construct driven by polymerase I (15), and terminated by a ribozyme sequence (55). These approaches have allowed the determination of the viral components essential for activity and the characterization of mutants affected in the polymerase subunits and the NP (reviewed in reference 60). However, none of these experimental systems has permitted researchers to purify and analyze biochemically the reconstituted RNPs. To improve the previous methods, we transfected a plasmid construct capable of generating the vRNA model transcript intracellularly (pT7NSΔCAT-RT), instead of a synthetic viral model RNA. Plasmid pT7NSΔCAT-RT contains a 313-nucleotide (nt) model viral cDNA driven by the T7 promoter and followed by a hepatitis δ virus ribozyme and a T7 terminator. This change, as well as the optimization of the experimental conditions indicated in Materials and Methods, led to a significant increase in the yield of active RNPs. A comparison of the results obtained with previous conditions, in which a 240-nt vRNA was directly transfected, and present conditions is shown in Fig. 1 and indicates that the increase in active RNPs obtained was at least 20-fold. It is clear that no transcription activity in vitro was obtained when the vRNA template or the ribozyme construct were omitted (Fig. 1, −RT). As expected, the reconstitution of active RNPs was strictly dependent on the expression of the NP and each polymerase subunit (Fig. 1).
FIG. 1.
Reconstitution of influenza virus mini-RNPs. Cultures of COS-1 cells were infected with vaccinia T7 virus and transfected with pGPB1, pGPB2, pGPA, and pGNPpolyA plasmids. The viral template was provided by simultaneous transfection of pT7NSΔCAT-RT plasmid or by delayed transfection of vNSZ RNA, as indicated in Materials and Methods. After incubation for 48 h, the viral RNPs were extracted and used for in vitro RNA synthesis by using ApG as primer. The RNA product was isolated and analyzed by electrophoresis on sequencing gels. The product obtained after transfection of pT7NSΔCAT-RT (313 nt) is shown (COMP), as well as the product generated by transfection of vNSZ RNA (240 nt) and those obtained when each of the elements of the systems was omitted. The panel to the left shows a 20× overexposure of the lane containing the vNSZ RNA product. The lengths of molecular weight markers are indicated to the left in nucleotides.
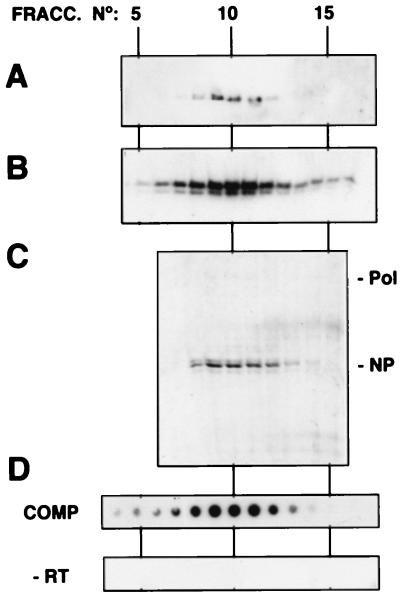
In view of the efficient RNP reconstitution obtained, we attempted their biochemical purification according to established procedures, i.e., successive centrifugation on velocity and density glycerol gradients (see Materials and Methods). The analyses of the last step in the purification are presented in Fig. 2. The Western blot signals obtained with anti-PB1 serum (Fig. 2A), anti-PB2 and anti-PA (not shown), and anti-NP (Fig. 2B) were all localized to fractions 9 to 13 in the density gradient. These fractions were shown to contain essentially NP and polymerase subunits when analyzed by Coomassie blue staining (Fig. 2C). Moreover, the in vitro transcription activity of these fractions strictly correlated with the presence of NP and polymerase (Fig. 2D, COMP). No in vitro transcription activity was detectable in the corresponding fractions of the gradient obtained from a parallel purification of RNPs reconstituted in the absence of the ribozyme construct (Fig. 2D, −RT). Correspondingly, neither NP nor polymerase protein was detectable by Western blotting in these fractions (data not shown). In summary, purified preparations of reconstituted RNPs were obtained by use of the methodology described.
FIG. 2.
Purification of influenza virus mini-RNPs reconstituted in vivo. The viral RNPs reconstituted in vivo as indicated in Fig. 1 were purified by two successive glycerol gradients as indicated in Materials and Methods. The analyses corresponding to the fractionation of the second gradient are presented. Aliquots of each fraction were processed for Western blotting by using anti-PB1 (A) or anti-NP antibodies (B) or were analyzed by SDS-PAGE and Coomassie blue staining (C). The activity of each fraction was determined by in vitro transcription and TCA precipitation, filtration on a dot blot apparatus, and autoradiography (D, COMP). As a control, the activity of a parallel gradient with a sample in which no template was transfected (−RT) was determined. Pol and NP indicate the positions of the polymerase proteins and the NP in the gel, respectively.
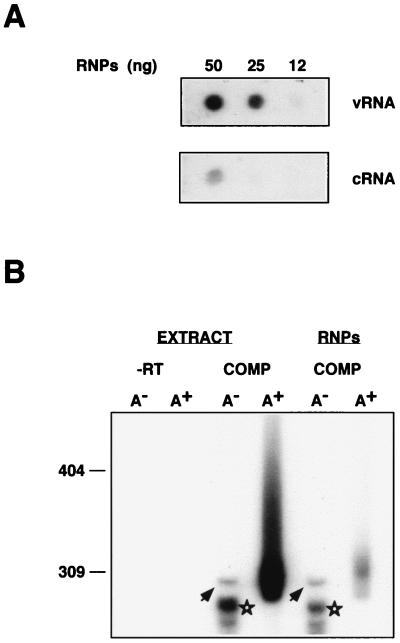
To characterize the purified RNPs, their RNA was extracted and analyzed by dot blot hybridization with positive- or negative-polarity riboprobes. As indicated in Fig. 3A, the reconstituted RNPs contained mostly vRNA; i.e., they were vRNPs, with a minor proportion of cRNA. To ascertain whether the purified RNPs represent active complexes, their in vitro products were characterized by oligo(dT) chromatography and gel electrophoresis in parallel to those produced by the cell extracts. The results are presented in Fig. 3B. Like the cell extract, the purified vRNPs synthesized mRNA (Fig. 3B, RNPs, A+), as well as a nonpolyadenylated full-length product (Fig. 3B, arrows) and nonpolyadenylated mRNA (Fig. 3B, stars) (50, 51).
FIG. 3.
Characterization of influenza virus RNPs reconstituted in vivo. Influenza virus RNPs were reconstituted and purified as indicated in Fig. 1 and 2. (A) The RNA contained in the purified RNPs was isolated and analyzed by dot blot hybridization by using positive (vRNA)- and negative (cRNA)-polarity riboprobes. (B) Either cellular extracts from infected and transfected cells (EXTRACT) or purified RNPs (RNPs) were used for in vitro transcription by using ApG as primer. The in vitro product was purified, fractionated on oligo(dT) cellulose into poly(A)− (A−) and poly(A)+ (A+) RNA, and analyzed on a sequencing gel. The products obtained when the complete reconstitution system was used are shown (COMP), as well as a control in which plasmid pT7NSΔCAT-RT was omitted (−RT). The arrowheads indicate the band corresponding to a full-length product. The stars show mRNA products deficient in polyadenylation. Numbers to the left refer to the mobility of molecular weight markers (in nucleotides).
Morphology of reconstituted RNPs.
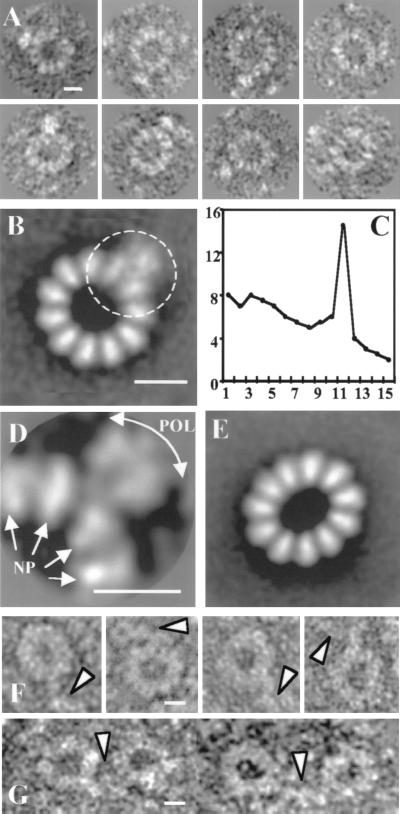
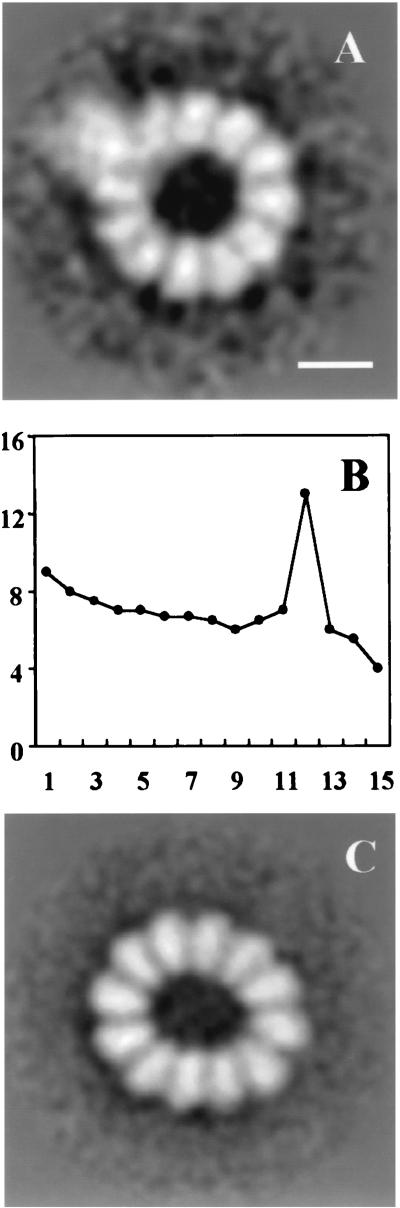
The availability of purified, transcriptionally active vRNPs reconstituted with a short model RNA (313 nt) allowed their structural characterization by electron microscopy. Preparations of vRNPs were visualized after negative staining, and the images of individual particles were classified and averaged as described in Materials and Methods. Two size classes were distinguished: a predominant one containing 11 repetitive units and a minor one containing 10 units and representing about 30% of the population. A second classification was performed with the 11-mer size class, and two conformations were identified: circular and elliptic. A gallery of individual images of the circular 11-mer size class is shown in Fig. 4A. The average image of the population with a circular conformation shows a circular shape, with a single peak of rotational symmetry at value 11 (32% of the population; 296 images) (Fig. 4B and C) and contain 11 morphological units that could correspond to NP molecules, with average sizes of 56 and 76 Å (external base height). Moreover, a conspicuous mass was localized to the external side of the circle, a finding consistent with the presence of the polymerase complex (see below). The average image of the population with ellipsoidal conformations (68% of the population; 641 images; Fig. 4E) confirmed the presence of 11 NP units but failed to show the polymerase. This result could be due to a stronger weight of the ellipticity over the polymerase boundary in the aligning procedure, leading to the positive enhancement of the NP units in the ring and discarding the polymerase image. Only if the polymerase had been placed in a fixed position with respect to the ellipse axis would a positive averaging have taken place. Since this was not the case, the polymerase image was averaged out.
FIG. 4.
Structure of influenza virus NSΔCAT RNP. Purified NSΔCAT RNPs were analyzed by electron microscopy after negative staining. The photographic plates were digitized, and 1,282 images from individual particles were stored and classified. Each homogeneous class was processed as described in Materials and Methods. (A) Gallery of circular RNPs. (B) Average image of the population of circular RNPs (average of 156 images; resolution, 30 Å). (C) Percentage of the total rotational power of the images presented in panel B plotted for the first 15 harmonics. (D) Average image with an alignment procedure that was centered on the polymerase complex (average of 55 images; resolution, 35 Å). (E) Average image of the population of ellipsoid RNPs (average of 871 images). (F) Gallery of images from RNP–anti-PB2 complexes. (G) Gallery of images from RNP–anti-PB2 complex dimers. Bar, 15 nm.
The identification of the outer morphological units as the polymerase complex was carried out by electron microscopy of vRNPs complexed with anti-PB2 monoclonal antibodies. As shown in Fig. 4F, images consistent with antibody molecules labeling the presumptive polymerase complex could be detected in individual vRNPs (arrowheads). Furthermore, polymerase complexes of two vRNPs were sometimes found linked by interaction to an individual antibody molecule (Fig. 4G, arrowheads). The variability of the images obtained for the RNPs suggested some degree of flexibility in the relative positions of the NP and the polymerase. To generate an improved image of the latter, the area of each image containing the polymerase (Fig. 4B, circle) was independently processed by alignment centered on the polymerase complex. In this way, an enhanced image was obtained, and three domains could be distinguished within the polymerase complex (Fig. 4D).
The replication efficiency of reconstituted RNPs is length dependent.
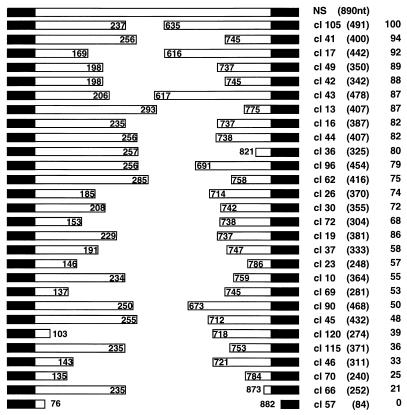
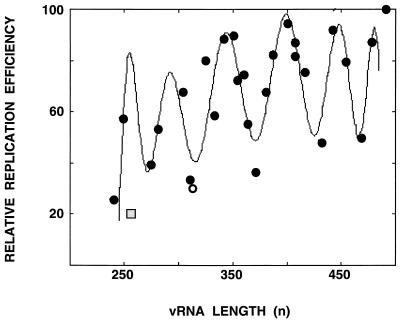
To test the possibility that the vRNA length would impose restrictions on the amplification of the vRNP, in line with well-known cases of paramyxoviruses (7; reviewed in reference 34), we took advantage of the strict correlation between vRNP amplification in vivo and the in vitro activity of the corresponding cell extracts (data not shown). The experimental approach taken involved the generation of a library of deleted constructs from plasmid pT7NS-RT by restriction at internal sites of the NS gene and Bal31 treatment. The clones obtained were sequenced and used to reconstitute vRNPs, and the activity in vitro of the corresponding cell extracts was tested as indicated in Materials and Methods. A diagram of the structure of the clones and their relative transcriptional activities is presented in Fig. 5. No specific sequence of the NS gene could be correlated to a high biological activity of the reconstituted vRNPs. Clones that had lost portions of the conserved segment termini were not active (Fig. 5, clone 57). Likewise, deletion of sequences close to the termini affected the activity of the reconstituted vRNP (Fig. 5, clone 66), in agreement with previous results (77). When the relative activity of the various deleted clones was represented as a function of their length, the graph presented in Fig. 6 was obtained. A clear dependence of the length is apparent, and the pitch of the sinusoidal graph had an average value of 48 nt. Since the length of viral RNA complexed to each NP molecule was estimated to be on the order of 20 nt (8), these results suggest that the entry of the NP in the replicating RNP occurs as a dimer. Clone 66, containing a deletion of sequences proximal to the segment terminus, had an activity lower than expected for its length (Fig. 6, shaded square), confirming that it lacks sequences required for optimal RNP amplification.
FIG. 5.
Relative amplification efficiency of influenza virus vRNPs reconstituted from templates of various lengths. Diagram of the structure of the NS segment (NS) and the collection of deletion derivatives, indicating the sequences deleted, the transcript length (in parentheses), and the relative activity in the in vitro transcription assay (taking clone 105 as a reference). The black regions at the ends represent the sequences conserved among all influenza virus RNA segments.
FIG. 6.
Length dependence of the efficiency of amplification of influenza virus RNPs. Graphic representation of the data presented in Fig. 5. The curve was adjusted to a polynomic function by using the program MATLAB 2.0.
Viral RNPs with highest efficiency of replication contain an even number of NP molecules.
It is worth mentioning that clone pT7NSΔCAT-RT, whose structure has been analyzed above, corresponds to a minimum in the sinusoidal graph and predominantly contains 11 NP molecules. These facts suggest that vRNPs with optimal capacity to amplify in vivo would have a length sufficient to hold an even number of NP molecules. To test this prediction, the vRNPs reconstituted with clone 49 (350 nt) were analyzed by electron microscopy and image processing as indicated above for NSΔCAT RNPs. The classification of the images obtained revealed that 80% of the particles contained 12 NP molecules and minor populations of 11-mers and 10-mers (10% each). A fraction of the 12-mers (28% of the population) were circular structures with a single peak of rotational symmetry at value 12 (Fig. 7A). In addition to the NP monomers, the presence of the polymerase complex was apparent. As with the vRNPs derived from pT7NSΔCAT-RT plasmid, clone 49 vRNPs also contained ellipsoid particles with 12 NP monomers (77% of the population) (Fig. 7C), in which the polymerase image was averaged out due to the reasons given above.
FIG. 7.
Structure of clone 49 RNP. Purified clone 49 RNPs were analyzed by electron microscopy after negative staining. The photographic plates were digitized, and 386 images from individual particles were stored and classified. Each homogeneous class was processed as described in Materials and Methods. (A) Average image of the population of circular RNPs (average of 44 images; resolution, 35 Å). (B) Percentage of the total rotational power of the images presented in panel A plotted for the first 15 harmonics. (C) Average image of the population of ellipsoid RNPs (average of 336 images). Bar, 15 nm.
DISCUSSION
Improved reconstitution efficiency allows biochemical and structural analyses of recombinant influenza virus RNPs.
The reconstitution of influenza virus RNPs requires the expression of the three subunits of the polymerase and the NP, in addition to the viral RNA template (26). Several experimental approaches have led to the generation of active RNPs (reviewed in reference 45), with transfection into vaccinia virus T7-infected cells being the most efficient and effective of these approaches (44). In this study we have improved this reconstitution system and the extraction of the RNPs. Thus, instead of transfecting a model vRNA into cells already expressing the viral core proteins (44, 51), we cotransfected the viral cDNAs together with a ribozyme construct capable of intracellularly generating the viral model template. As indicated in Fig. 1, an approximately 20-fold-higher yield of active, soluble RNPs was obtained. Such a result opened the way to the biochemical purification of the reconstituted RNPs (Fig. 2). It is interesting that the generation of recombinant RNPs was strictly dependent on the activity of the polymerase, since transfection of a deletion mutant of PB2 protein, which is inactive in viral transcription and replication but still capable of entering into the polymerase complex (51), did not allow the generation of biochemically detectable RNPs (data not shown). This result implies that the simple reconstitution of the RNPs from the T7-directed transcript is not sufficient to account for the accumulation of active RNPs. Rather, amplification mediated by the viral replication machinery is required. This conclusion, together with the fact that most of the RNPs generated in vivo are vRNPs, i.e., contain vRNA (Fig. 3), indicates that a situation similar to the virus infection is reproduced in the transfected cells: a small amount of the T7-directed transcript (paternal vRNA) forms active vRNPs that lead to the generation of a small amount of cRNPs (replication intermediate). These cRNPs are used as templates for the efficient accumulation of progeny vRNPs. Thus, the reconstituted RNPs only differ from authentic ones in their length.
The average image of a reconstituted RNP reveals its structural elements, the polymerase and the nucleoprotein.
The structure of cellular ribonucleoprotein particles has been analyzed by electron microscopy and image processing with great success. By using these techniques, the ribosome has been studied as a single-particle, noncrystalline sample, and progressively improved data has led to three-dimensional reconstructions with a resolution of 15 to 25 Å (13, 40). Likewise, the structures of spliceosomal A complex and the large nuclear RNP particle have been determined, although to a lower resolution (17, 71). However, very few studies of this type have been carried out with RNPs from negative-strand RNA viruses (14), and none have been carried out with influenza virus RNPs. The main reason for this lack of data might be the flexible nature of viral RNPs, which preclude the application of image-processing tools. In addition, the influenza virus RNPs are heterogeneous in size, a fact that may contribute to the complexity of the problem. The reconstitution of mini-RNPs from cloned DNA and their purification, albeit to a low concentration (Fig. 1 and 2), has allowed the use of essentially single size class specimens with sufficient rigidity to apply image-processing techniques. The analysis of such images and their classification showed the presence of a proportion of circular structures made up of 11 identical elements that should correspond to NP monomers and an external morphological unit representing the polymerase complex (Fig. 4). A 31% fraction of the population had only 10 NP monomers, in addition to the polymerase (data not shown). Interestingly, the polymerase complex could be directly detected by electron microscopy for the first time, albeit so far at low resolution, and the image obtained is consistent with the presence of the three subunits (Fig. 4). Particle-to-particle differences in staining and/or the flexibility of the polymerase bound to the panhandle in relation to the ring of NP monomers might be the reason for the low resolution obtained and the small apparent mass of the polymerase complex. Progress in this regard is envisaged in the use of even smaller reconstituted RNPs.
Two additional features of the collection of images obtained provide information in regard to the generation of the normal, coiled structures seen in virion RNPs (28). First, a large fraction of the structures were not strict circles but ellipsoids (Fig. 4 and 7). These elliptic structures might represent initial stages of the helicity. Second, in this context it is worth mentioning that the proportion of the elliptic structures was larger for the RNP reconstituted from clone 49 (350 nt) than from pT7NSΔCAT-RT plasmid (313 nt) and, furthermore, only helical structures could be detected for clone 41-derived RNPs (400 nt) (data not shown).
The replication efficiency of reconstituted RNPs fluctuates with their length, with a pitch of 48 nt.
The stoichiometry of NP versus RNA could be calculated from the structure of the RNP derived from pT7NSΔCAT-RT plasmid. Thus, if we assume that the polymerase covers about 12 to 15 nt from each RNA end as it interacts with the panhandle structure (32, 72), 26 to 28 nt of RNA should be interacting with each NP monomer, in good agreement with previous estimates obtained by chemical analyses (8). However, a large fraction of the RNPs generated contained 10 NP monomers instead of 11, a result that suggested that this RNP might not have the optimal length for amplification. Keeping in mind the strict requirement for precise length that some paramyxoviruses show (the rule of six) (7, 34), we tested whether recombinant templates with different lengths might show a similar length dependence, a presumable “rule of 26 to 28.” A strict restriction of amplification was not expected, since influenza virus polymerase interacts directly with the ends of the RNA template (19, 36, 72) and should be expected to provide a source of flexibility to the structure. Indeed, a fluctuation of the amplification efficiency of the RNPs was observed as a function of their template's length but, surprisingly, a pitch of 48 nt was detected (Fig. 6). The low relative amplification capacity of the RNP derived from pT7NSΔCAT-RT plasmid and the presence of a large proportion of 10-mers suggested that an even number of NP monomers should be present in an RNP to allow for its optimal amplification. Such an assumption was confirmed by the presence of 12 NP monomers in clone 49 RNPs, which are highly efficient in amplification (Fig. 6 and 7). The lengths of the complete viral RNA segments are not always multiples of 48, probably because the longer RNAs permit enough flexibility to the structure. However, the increments in length among the various RNA segments are frequently approximate multiples of 48. Similarly, the lengths of defective-interfering RNAs isolated from infected cells are frequently, but not always, approximate multiples of 48 (28). For defective-interfering RNAs, however, other restrictions might also operate, such as the relative positions of appropriate sequences for the jumping of the polymerase within the RNP (28).
The molecular basis of the fluctuation of the amplification efficiency with a pitch of 48 is unknown at present. It is tempting to speculate that the NP molecules are incorporated into replicating progeny RNAs as dimers. If this were the case, the terminal dimer entering at the 3′ end of the RNA molecule would be restricted when the length of the RNA deviated from a multiple of 48, forcing the entry of a monomeric NP molecule instead of a dimer or skipping the incorporation of the last dimer. In our experiments, the RNP derived from pT7NSΔCAT-RT plasmid incorporates 11 NP monomers but frequently is terminated at the stage of 10 monomers (i.e., 5 dimers). In line with this speculation, dimeric forms of NP have been detected in influenza virus-infected cells (63). However, no evidence for dimers of NP was found by electron microscopy of purified protein (66).
In summary, application of image-processing techniques to electron microscopy images of in vivo-reconstituted influenza virus RNPs has provided the first images of the viral polymerase complex and helped to obtain a more precise picture of the NP monomer. Moreover, the amplification characteristics of mini-RNPs of various lengths suggested that the NP molecule is incorporated in progeny RNPs in the form of dimers and indicated that each NP monomer covers around 24 nt of the template RNA.
ACKNOWLEDGMENTS
We are indebted to J. A. Melero, A. Nieto, and A. Portela for their critical comments on the manuscript. We thank A. Ball, B. Moss, P. Palese, and A. Portela for providing biological materials. The technical assistance of Y. Fernández and J. Fernández is gratefully acknowledged. We thank Carlos Oscar Sánchez for help with the MATLAB program.
J. Ortega was a fellow of Instituto de Estudios Turolenses. This work was supported by Programa Sectorial de Promoción General del Conocimiento (grants PB97-1160 and PB96-0818).
REFERENCES
- 1.Albo C, Valencia A, Portela A. Identification of an RNA binding region within the N-terminal third of the influenza A virus NP polypeptide. J Virol. 1995;69:3799–3806. doi: 10.1128/jvi.69.6.3799-3806.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball L A. Cellular expression of a functional nodavirus RNA replicon from vaccinia virus vectors. J Virol. 1992;66:2335–2345. doi: 10.1128/jvi.66.4.2335-2345.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bárcena J, Ochoa M, de la Luna S, Melero J A, Nieto A, Ortín J, Portela A. Monoclonal antibodies against influenza virus PB2 and NP polypeptides interfere with the initiation step of viral mRNA synthesis in vitro. J Virol. 1994;68:6900–6909. doi: 10.1128/jvi.68.11.6900-6909.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baudin F, Bach C, Cusack S, Ruigrok R W. Structure of influenza virus RNP. I. Influenza virus nucleoprotein melts secondary structure in panhandle RNA and exposes the bases to the solvent. EMBO J. 1994;13:3158–3165. doi: 10.1002/j.1460-2075.1994.tb06614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biswas S K, Nayak D P. Mutational analysis of the conserved motifs of influenza A virus polymerase basic protein 1. J Virol. 1994;68:1819–1826. doi: 10.1128/jvi.68.3.1819-1826.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaas D, Patzelt E, Keuchler E. Identification of the cap binding protein of influenza virus. Nucleic Acids Res. 1982;10:4803–4812. doi: 10.1093/nar/10.15.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calain P, Roux L. The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J Virol. 1993;67:4822–4830. doi: 10.1128/jvi.67.8.4822-4830.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Compans R W, Chopin P W. Reproduction of myxoviruses. In: Fraenkel-Conrat H, Wagner R R, editors. Comprehensive virology. New York, N.Y: Plenum Press; 1975. pp. 179–252. [Google Scholar]
- 9.Compans R W, Content J, Duesberg P H. Structure of the ribonucleoprotein of influenza virus. J Virol. 1972;4:795–800. doi: 10.1128/jvi.10.4.795-800.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de la Luna S, Martín J, Portela A, Ortín J. Influenza virus naked RNA can be expressed upon transfection into cells co-expressing the three subunits of the polymerase and the nucleoprotein from SV40 recombinant viruses. J Gen Virol. 1993;74:535–539. doi: 10.1099/0022-1317-74-3-535. [DOI] [PubMed] [Google Scholar]
- 11.Detjen B M, St. Angelo C, Katze M G, Krug R M. The three influenza virus polymerase (P) proteins not associated with viral nucleocapsids in the infected cell are in the form of a complex. J Virol. 1987;61:16–22. doi: 10.1128/jvi.61.1.16-22.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Digard P, Blok V C, Inglis S C. Complex formation between influenza virus polymerase proteins expressed in Xenopus oocytes. Virology. 1989;171:162–169. doi: 10.1016/0042-6822(89)90523-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dube P, Wieske M, Stark H, Schatz M, Stahl J, Zemlin F, Lutsch G, Van Heel M. The 80S rat liver ribosome at 25 Å resolution by electron cryomicroscopy and angular reconstitution. Structure. 1998;6:389–399. doi: 10.1016/s0969-2126(98)00040-9. [DOI] [PubMed] [Google Scholar]
- 14.Egelman E H, Wu S S, Amrein M, Portner A, Murti G. The Sendai virus nucleocapsid exists in at least four different helical states. J Virol. 1989;63:2233–2243. doi: 10.1128/jvi.63.5.2233-2243.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flick R, Neumann G, Hoffmann E, Neumeier E, Hobom G. Promoter elements in the influenza vRNA terminal structure. RNA. 1996;2:1046–1057. [PMC free article] [PubMed] [Google Scholar]
- 16.Fuerst T R, Earl P L, Moss B. Use of a hybrid vaccinia virus-T7 RNA polymerase system for expression of target genes. Mol Cell Biol. 1987;7:2538–2544. doi: 10.1128/mcb.7.7.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furman E, Glitz D G. Purification of the spliceosome A-complex and its visualization by electron microscopy. J Biol Chem. 1995;270:15515–15522. doi: 10.1074/jbc.270.26.15515. [DOI] [PubMed] [Google Scholar]
- 18.Gluzman Y. SV40 transformed simian cells support the replication of early SV40 mutants. Cell. 1981;23:175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- 19.González S, Ortín J. Characterization of the influenza virus PB1 protein binding to vRNA: two separate regions of the protein contribute to the interaction domain. J Virol. 1999;73:631–637. doi: 10.1128/jvi.73.1.631-637.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.González S, Ortín J. Distinct regions of influenza virus PB1 polymerase subunit recognize vRNA and cRNA templates. EMBO J. 1999;18:3767–3775. doi: 10.1093/emboj/18.13.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.González S, Zürcher T, Ortín J. Identification of two separate domains in the influenza virus PB1 protein responsible for interaction with the PB2 and PA subunits: a model for the viral RNA polymerase structure. Nucleic Acids Res. 1996;24:4456–4463. doi: 10.1093/nar/24.22.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hay A J. Characterization of influenza virus RNA complete transcripts. Virology. 1982;116:517–522. doi: 10.1016/0042-6822(82)90144-1. [DOI] [PubMed] [Google Scholar]
- 23.Heggeness M H, Smith P R, Ulmanen I, Krug R M, Chopin P W. Studies on the helical nucleocapsid of influenza virus. Virology. 1982;118:466–470. doi: 10.1016/0042-6822(82)90367-1. [DOI] [PubMed] [Google Scholar]
- 24.Herz C, Stavnezer E, Krug R M, Gurney T. Influenza virus, an RNA virus, synthesizes its messenger RNA in the nucleus of infected cells. Cell. 1981;26:391–400. doi: 10.1016/0092-8674(81)90208-7. [DOI] [PubMed] [Google Scholar]
- 25.Honda A, Mukaigawa J, Yokoiyama A, Kato A, Ueda S, Nagata K, Krystal M, Nayak D P, Ishihama A. Purification and molecular structure of RNA polymerase from influenza virus A/PR8. J Biochem Tokyo. 1990;107:624–628. doi: 10.1093/oxfordjournals.jbchem.a123097. [DOI] [PubMed] [Google Scholar]
- 26.Huang T S, Palese P, Krystal M. Determination of influenza virus proteins required for genome replication. J Virol. 1990;64:5669–5673. doi: 10.1128/jvi.64.11.5669-5673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson D A, Caton A J, McCready S J, Cook P R. Influenza virus RNA is synthesized at fixed sites in the nucleus. Nature. 1982;296:366–368. doi: 10.1038/296366a0. [DOI] [PubMed] [Google Scholar]
- 28.Jennings P A, Finch J T, Winter G, Robertson J S. Does the higher order structure of the influenza virus ribonucleoprotein guide sequence rearrangements in influenza viral RNA? Cell. 1983;34:619–627. doi: 10.1016/0092-8674(83)90394-x. [DOI] [PubMed] [Google Scholar]
- 29.Kato A, Mizumoto K, Ishihama A. Purification and enzymatic properties of an RNA polymerase-RNA complex from influenza virus. Virus Res. 1985;3:115–127. doi: 10.1016/0168-1702(85)90002-4. [DOI] [PubMed] [Google Scholar]
- 30.Kimura N, Mishida M, Nagata K, Ishihama A, Oda K, Nakada S. Transcription of a recombinant influenza virus RNA in cells that can express the influenza virus RNA polymerase and nucleoprotein genes. J Gen Virol. 1992;73:1321–1328. doi: 10.1099/0022-1317-73-6-1321. [DOI] [PubMed] [Google Scholar]
- 31.Kingsbury D W, Jones I M, Murti K G. Assembly of influenza ribonucleoprotein in vitro using recombinant nucleoprotein. Virology. 1987;156:396–403. doi: 10.1016/0042-6822(87)90419-3. [DOI] [PubMed] [Google Scholar]
- 32.Klumpp K, Ruigrok R W, Baudin F. Roles of the influenza virus polymerase and nucleoprotein in forming a functional RNP structure. EMBO J. 1997;16:1248–1257. doi: 10.1093/emboj/16.6.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kohonen T. The self organizing map. Proc IEEE. 1980;78:1464–1480. [Google Scholar]
- 34.Kolakofsky D, Pelet T, Garcin D, Hausmann S, Curran J, Roux L. Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited. J Virol. 1998;72:891–899. doi: 10.1128/jvi.72.2.891-899.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krug R M, Broni B A, Bouloy M. Are the 5′-ends of influenza viral mRNAs synthesized in vivo donated by host mRNAs? Cell. 1979;18:329–334. doi: 10.1016/0092-8674(79)90052-7. [DOI] [PubMed] [Google Scholar]
- 36.Li M L, Ramírez B C, Krug R M. RNA-dependent activation of primer RNA production by influenza virus polymerase: different regions of the same protein subunit constitute the two required RNA-binding sites. EMBO J. 1998;17:5844–5852. doi: 10.1093/emboj/17.19.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Licheng S, Summers D F, Peng Q, Galarza J M. Influenza A virus polymerase subunit PB2 is the endonuclease which cleaves host cell mRNA and functions only as the trimeric enzyme. Virology. 1995;208:38–47. doi: 10.1006/viro.1995.1127. [DOI] [PubMed] [Google Scholar]
- 38.Luo G X, Luytjes W, Enami M, Palese P. The polyadenylation signal of influenza virus RNA involves a stretch of uridines followed by the RNA duplex of the panhandle structure. J Virol. 1991;65:2861–2867. doi: 10.1128/jvi.65.6.2861-2867.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahy B W J. Mutants of influenza virus. In: Palese P, Kingsbury D W, editors. Genetics of influenza viruses. Vienna, Austria: Springer-Verlag; 1983. pp. 192–253. [Google Scholar]
- 40.Malhotra A, Penczek P, Agrawal R K, Gabashvili I S, Grassucci R A, Junemann R, Burkhardt N, Nierhaus K H, Frank J. Escherichia coli 70 S ribosome at 15 Å resolution by cryo-electron microscopy: localization of fMet-tRNAfMet and fitting of L1 protein. J Mol Biol. 1998;280:103–116. doi: 10.1006/jmbi.1998.1859. [DOI] [PubMed] [Google Scholar]
- 41.Marabini R, Carazo J M. Pattern recognition and classification of images of biological macromolecules using artificial neural networks. Biophys J. 1994;66:1804–1814. doi: 10.1016/S0006-3495(94)80974-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marco S, Chagoyen M, de la Fraga L G, Carazo J M, Carrascosa J L. A variant of the “random approximation” of the reference-free alignment algorithm. Ultramicroscopy. 1996;66:5–10. [Google Scholar]
- 43.Marión R M, Zürcher T, de la Luna S, Ortín J. Influenza virus NS1 protein interacts with viral transcription-replication complexes in vivo. J Gen Virol. 1997;78:2447–2451. doi: 10.1099/0022-1317-78-10-2447. [DOI] [PubMed] [Google Scholar]
- 44.Mena I, de la Luna S, Albo C, Martín J, Nieto A, Ortín J, Portela A. Synthesis of biologically active influenza virus core proteins using a vaccinia-T7 RNA polymerase expression system. J Gen Virol. 1994;75:2109–2114. doi: 10.1099/0022-1317-75-8-2109. [DOI] [PubMed] [Google Scholar]
- 45.Mena I, de la Luna S, Martín J, Albó C, Perales B, Nieto A, Portela A, Ortín J. Systems to express recombinant RNA molecules by the influenza A virus polymerase in vivo. In: Adolph K W, editor. Methods in molecular genetics. Molecular virology techniques, part B. Orlando, Fla: Academic Press; 1995. pp. 329–342. [Google Scholar]
- 46.Murti K G, Webster R G, Jones I M. Localization of RNA polymerases of influenza viral ribonucleoproteins by immunogold labeling. Virology. 1988;164:562–566. doi: 10.1016/0042-6822(88)90574-0. [DOI] [PubMed] [Google Scholar]
- 47.Ortín J, Nájera R, López C, Dávila M, Domingo E. Genetic variability of Hong Kong (H3N2) influenza viruses: spontaneous mutations and their location in the viral genome. Gene. 1980;11:319–331. doi: 10.1016/0378-1119(80)90072-4. [DOI] [PubMed] [Google Scholar]
- 48.Parvin J D, Palese P, Honda A, Ishihama A, Krystal M. Promoter analysis of influenza virus RNA polymerase. J Virol. 1989;63:5142–5152. doi: 10.1128/jvi.63.12.5142-5152.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Penczek P, Radermacher M, Frank J. Three-dimensional reconstruction of single particles embedded in ice. Ultramicroscopy. 1992;40:33–53. [PubMed] [Google Scholar]
- 50.Perales B. Reconstitución in vivo e in vitro de los procesos de transcripción y replicación del virus de la gripe a partir de genes clonados: análisis mutacional de la subunidad PB2 de la polimerasa viral. Madrid, Spain: Universidad Autónoma de Madrid; 1997. [Google Scholar]
- 51.Perales B, de la Luna S, Palacios I, Ortín J. Mutational analysis identifies functional domains in the influenza A PB2 polymerase subunit. J Virol. 1996;70:1678–1686. doi: 10.1128/jvi.70.3.1678-1686.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perales B, Ortín J. The influenza A virus PB2 polymerase subunit is required for the replication of viral RNA. J Virol. 1997;71:1381–1385. doi: 10.1128/jvi.71.2.1381-1385.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pérez D R, Donis R O. A 48-amino-acid region of influenza A virus PB1 protein is sufficient for complex formation with PA. J Virol. 1995;69:6932–6939. doi: 10.1128/jvi.69.11.6932-6939.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Piccone M E, Fernandez S A, Palese P. Mutational analysis of the influenza virus vRNA promoter. Virus Res. 1993;28:99–112. doi: 10.1016/0168-1702(93)90129-b. [DOI] [PubMed] [Google Scholar]
- 55.Pleschka S, Jaskunas R, Engelhardt O G, Zürcher T, Palese P, Garcia Sastre A. A plasmid-based reverse genetics system for influenza A virus. J Virol. 1996;70:4188–4192. doi: 10.1128/jvi.70.6.4188-4192.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1990;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pons M W, Schulze I T, Hirst G K. Isolation and characterization of the ribonucleoprotein of influenza virus. Virology. 1969;39:250–259. doi: 10.1016/0042-6822(69)90045-2. [DOI] [PubMed] [Google Scholar]
- 58.Poon L L, Pritlove D C, Sharps J, Brownlee G G. The RNA polymerase of influenza virus, bound to the 5′ end of virion RNA, acts in cis to polyadenylate mRNA. J Virol. 1998;72:8214–8219. doi: 10.1128/jvi.72.10.8214-8219.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Poon L L M, Pritlove D C, Fodor E, Brownlee G G. Direct evidence that the poly(A) tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template. J Virol. 1999;73:3473–3476. doi: 10.1128/jvi.73.4.3473-3476.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Portela, A., T. Zürcher, A. Nieto, and J. Ortín. Replication of orthomyxoviruses. Adv. Virus Res., in press. [DOI] [PubMed]
- 61.Pritlove D C, Poon L L, Fodor E, Sharps J, Brownlee G G. Polyadenylation of influenza virus mRNA transcribed in vitro from model virion RNA templates: requirement for 5′ conserved sequences. J Virol. 1998;72:1280–1286. doi: 10.1128/jvi.72.2.1280-1286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pritlove D C, Poon L L M, Devenish L J, Leahy M B, Brownlee G G. A hairpin loop at the 5′ end of influenza A virus virion RNA is required for synthesis of poly(A)+ mRNA in vitro. J Virol. 1999;73:2109–2114. doi: 10.1128/jvi.73.3.2109-2114.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Prokudina K E, Semenova N P. Intracellular oligomerization of influenza virus nucleoprotein. Virology. 1996;223:51–56. doi: 10.1006/viro.1996.0454. [DOI] [PubMed] [Google Scholar]
- 64.Robertson J S, Schubert M, Lazzarini R A. Polyadenylation sites for influenza mRNA. J Virol. 1981;38:157–163. doi: 10.1128/jvi.38.1.157-163.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rose J K, Buonocore L, Whitt M A. A new cationic liposome reagent mediating nearly quantitative transfection of animal cells. BioTechniques. 1991;10:520–525. [PubMed] [Google Scholar]
- 66.Ruigrok R W, Baudin F. Structure of influenza virus ribonucleoprotein particles. II. Purified RNA-free influenza virus ribonucleoprotein forms structures that are indistinguishable from the intact influenza virus ribonucleoprotein particles. J Gen Virol. 1995;76:1009–1014. doi: 10.1099/0022-1317-76-4-1009. [DOI] [PubMed] [Google Scholar]
- 67.Sanz-Ezquerro J J, de la Luna S, Ortín J, Nieto A. Individual expression of influenza virus PA protein induces degradation of coexpressed proteins. J Virol. 1995;69:2420–2426. doi: 10.1128/jvi.69.4.2420-2426.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sanz-Ezquerro J J, Fernández Santarén J, Sierra T, Aragón T, Ortega J, Ortín J, Smith G L, Nieto A. The PA influenza polymerase subunit is a phosphorylated protein. J Gen Virol. 1998;79:471–478. doi: 10.1099/0022-1317-79-3-471. [DOI] [PubMed] [Google Scholar]
- 69.Sanz-Ezquerro J J, Zürcher T, de la Luna S, Ortin J, Nieto A. The amino-terminal one-third of the influenza virus PA protein is responsible for the induction of proteolysis. J Virol. 1996;70:1905–1911. doi: 10.1128/jvi.70.3.1905-1911.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shi L, Galarza J M, Summers D F. Recombinant-baculovirus-expressed PB2 subunit of the influenza A virus RNA polymerase binds cap groups as an isolated subunit. Virus Res. 1996;42:1–9. doi: 10.1016/0168-1702(96)01289-0. [DOI] [PubMed] [Google Scholar]
- 71.Sperling R, Koster A J, Melamed B C, Rubinstein A, Angenitzki M, Berkovitch Y Z, Sperling J. Three-dimensional image reconstruction of large nuclear RNP (lnRNP) particles by automated electron tomography. J Mol Biol. 1997;267:570–583. doi: 10.1006/jmbi.1997.0898. [DOI] [PubMed] [Google Scholar]
- 72.Tiley L S, Hagen M, Mathews J T, Krystal M. Sequence-specific binding of the influenza virus RNA polymerase to sequences located at the 5′-end of the viral RNAs. J Virol. 1994;68:5108–5116. doi: 10.1128/jvi.68.8.5108-5116.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Toyoda T, Adyshev D M, Kobayashi M, Iwata A, Ishihama A. Molecular assembly of the influenza virus RNA polymerase: determination of the subunit-subunit contact sites. J Gen Virol. 1996;77:2149–2157. doi: 10.1099/0022-1317-77-9-2149. [DOI] [PubMed] [Google Scholar]
- 74.Ulmanen I, Broni B A, Krug R M. The role of two of the influenza virus core P proteins in recognizing cap 1 structures (m7GpppNm) on RNAs and in initiating viral RNA transcription. Proc Natl Acad Sci USA. 1981;78:7355–7359. doi: 10.1073/pnas.78.12.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Unser M, Trus B L, Steven A C. A new resolution criterion based on spectral signal-to-noise method ratios. Ultramicroscopy. 1987;23:39–52. doi: 10.1016/0304-3991(87)90225-7. [DOI] [PubMed] [Google Scholar]
- 76.Yamanaka K, Ishihama A, Nagata K. Reconstitution of influenza virus RNA-nucleoprotein complexes structurally resembling native viral ribonucleoprotein cores. J Biol Chem. 1990;265:11151–11155. [PubMed] [Google Scholar]
- 77.Zheng H, Palese P, Garcia Sastre A. Nonconserved nucleotides at the 3′ and 5′ ends of an influenza A virus RNA play an important role in viral RNA replication. Virology. 1996;217:242–251. doi: 10.1006/viro.1996.0111. [DOI] [PubMed] [Google Scholar]
- 78.Zürcher T, de la Luna S, Sanz-Ezquerro J J, Nieto A, Ortín J. Mutational analysis of the influenza virus A/Victoria/3/75 PA protein: studies of interaction with PB1 protein and identification of a dominant negative mutant. J Gen Virol. 1996;77:1745–1749. doi: 10.1099/0022-1317-77-8-1745. [DOI] [PubMed] [Google Scholar]