Abstract
Sleeping disease (SD) is currently a matter of concern for salmonid fish farmers in most parts of the world. A viral etiology of SD has recently been suspected, since virus-like particles have been observed in infected rainbow trout cells. In salmonid-derived cell lines, the maximal rate of virus production was observed at 10°C, while little virus was produced at 14°C. Through biochemical, physicochemical, and morphological studies, SD virus (SDV) was shown to be an enveloped virus of roughly 60 nm in diameter. The genome consists of 12 kb of RNA, with the appearance of a 26S subgenomic RNA during the time course of SDV replication. The screening of a random-primed cDNA library constructed from the genomic RNA of semipurified virions facilitated the identification of a specific SDV cDNA clone having an open reading frame related to the alphavirus E2 glycoproteins. To extend the comparison between SDV structural proteins and the alphavirus protein counterparts, the nucleotide sequence of the total 4.1-kb subgenomic RNA has been determined. The 26S RNA encodes a 1,324-amino-acid polyprotein exhibiting typical alphavirus structural protein organization. SDV structural proteins showed several remarkable features compared to other alphaviruses: (i) unusually large individual proteins, (ii) very low homology (ranging from 30 to 34%) (iii) an unglycosylated E3 protein, and (iv) and E1 fusion domain sharing mutations implicated in the pH threshold. Although phylogenetically related to the Semliki Forest virus group of alphaviruses, SDV should be considered an atypical member, able to naturally replicate in lower vertebrates.
Sleeping disease (SD) syndrome of farmed freshwater rainbow trout has been observed in France for many years (4). The most characteristic sign of the disease is the unusual behavior of the fish, which stay on their side at the bottom of the tank. Histological observations of diseased fish showed a chronological appearance of lesions in the pancreas, in the heart, and in the muscle at the last stage of the disease (5, 6). Transmission of SD may occur through contact with contaminated tissue from fish that have SD (5). A viral etiology of SD was suspected, since virus-like particles were observed in purified homogenates from kidneys of diseased fish (3). However, all attempts to isolate a viral agent on commonly available fish cell lines by inoculating organ homogenates from diseased fish remained unsuccessful until recently (7). Isolation of SD virus (SDV) in cell culture was successfully achieved by direct inoculation of salmonid cell lines (CHSE-214 and RTG-2) with plasma from infected fish.
The characterization of SDV was successfully achieved by optimizing viral production in tissue culture and by studying several physicochemical features of this virus. Data include the type of nucleic acid, size, and organization of the SDV genome. The viral genome has been shown to be an RNA molecule of roughly 12 kb. A cDNA library has been constructed, and the nucleotide sequencing of recombinant cDNA clones definitively classified SDV as a member of the important genus Alphavirus of the family Togaviridae. The nucleotide sequence data presented in this report emphasize the unusual characteristics of SDV compared to other alphaviruses. The generation of a specific SDV anti-E2 rabbit antiserum from recombinant protein provides a powerful tool for further epidemiologic studies aiming to evaluate the impact of this virus in the field.
MATERIALS AND METHODS
Virus and cells.
The rainbow trout-derived cell line RTG-2 (26) was maintained in Eagle's minimum essential medium (MEM) (Sigma) buffered at pH 7.4 with Tris-HCl and supplemented with 10% fetal bovine serum (FBS) (Boehringer Mannheim), 50 IU of penicillin ml−1, and 50 μg of streptomycin ml−1. Cells were propagated in 75- and 150-cm2 plastic cell culture flasks at 20°C. The SDV isolate used in this study was originally obtained from kidney tissues of infected rainbow trout and adapted in CHSE-214 cells (7). Other viruses used included viral hemorrhagic septicemia virus (VHSV) (French strain 07-71) and infectious pancreatic necrosis virus (IPNV) (French strain Sp), which were isolated in our institute, and salmonid herpesvirus 1 (SaHV1), which was provided through the courtesy of R. P. Hedrick, University of California, Davis.
Virus titration.
Freshly trypsinized RTG-2 cells were grown overnight at 20°C in six-well plates. Inoculum for which the titer was to be determined was diluted from 10−2 to 10−7 in Tris-buffered MEM (pH 7.4) and used to infect cells. Following adsorption, 2 ml of a 0.3% melted agarose solution in Tris-buffered MEM containing 2% FBS was added to each well. Plates were incubated at 10°C for 9 days. Agarose was then removed, and either cells were stained with neutral red and visible plaques were directly counted or cells were overinfected with the lytic VHSV and left at 14°C for 2 days. Cells were then fixed with methanol and stained with crystal violet solution. VHSV-resistant cell foci were then directly counted.
Electron microscopic examination.
SDV-infected cells were fixed in 4% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.8) and then postfixed in 1% cacodylate-buffered osmium tetroxide. After dehydration, fixed material was embedded in resin and thin sections were mounted onto copper grids. Sample grids were stained in 2% uranyl acetate. Grids were examined with a Philips TEM 300 at 60 kV.
Physicochemical studies.
Sensitivity of SDV to lipid solvent was checked by adding 0.5 ml of chloroform to 1 ml of SDV supernatant. The mixture was shaken for 30 min at 4°C and then centrifuged at 400 × g for 1 min to remove the chloroform. Titers of infectious viral particles were then determined on RTG-2 cells. IPNV (nonenveloped) and VHSV (enveloped) were also included as nonsensitive and sensitive virus controls, respectively. The influence of pH on SDV stability was evaluated by adjusting an SDV suspension to pH 3.0 or 11 and incubating for 4 h at 4°C before determining titers of infectious viral particles on RTG-2 cells. IPNV and VHSV were used as resistant and sensitive control viruses, respectively. The stability of SDV to temperature was also investigated by incubating SDV suspension aliquots for 60 min at temperatures of 10 to 50°C and then transferring them at 4°C prior to virus titration. Titers of infectious viral particles were then determined on RTG-2 cells.
Nature of the SDV genome.
Determination of the presence of RNA or DNA within the SDV genome was examined in growing SDV in the presence of 5-bromo-2′-deoxyuridine (BrdU) (Sigma) with and without thymidine (THY) (Sigma). Groups of four wells in each of three 24-well plates containing RTG-2 cells were inoculated with 100 μl of 100-fold dilutions of SDV and allowed to absorb for 1 h at 10°C. To each individual well was added culture medium with or without 1 mM BrdU or BrdU plus THY (1 mM). An RNA virus (IPNV) and a DNA virus (SaHV-1) were included as controls. The plates were incubated for 14 days at 10°C (SDV and SaHV-1) or for 3 days at 14°C (IPNV), and examined for cytopathic effect (CPE). Titers of supernatants of each well were then determined.
Metabolic [5,6-3H]uridine labeling of SDV-infected cells.
Freshly trypsinized RTG-2 cells were grown overnight in 75-cm2 plastic cell culture flasks. SDV was allowed to absorb for 1 h at 10°C, and then culture medium containing 2% FBS was added and the cells were incubated at 10°C for 3 days. The medium was removed, replaced with fresh medium containing 2% FBS and actinomycin D (0.5 mg/ml), and incubated for 2 h at 10°C before [5,6-3H]uridine (final concentration, 100 μCi/ml) was added to infected and uninfected cells. At 7 days after the radiolabeling, supernatants and cells were collected and processed for RNA extraction.
RNA preparation and electrophoresis.
Supernatants from infected cells were clarified for 15 min at 3,000 × g and then precipitated overnight at 4°C with polyethylene glycol 6000 (Sigma; 10% final concentration) (16). Pellets were collected by centrifugation at 10,000 × g for 30 min. The pellets were resuspended in TNE (50 mM Tris [pH 7.4] 100 mM NaCl, 1 mM EDTA), and viral RNA was extracted following SDS-proteinase K treatment for 15 min at 37°C. RNA was recovered following phenol-chloroform extraction and ethanol precipitation. Total RNAs were recovered from infected and mock-infected cells by using the Stratagene RNA isolation kit and the manufacturer's procedure. Labeled RNA pellets were resuspended in denaturing buffer (1× MOPS [morpholinepropanesulfonic acid], 6.4% formaldehyde, 50% formamide), left for 15 min at 65°C, and cooled on ice. RNAs were separated on a Tris-borate-EDTA–0.8% agarose gel. An RNA ladder (RNA low-range marker; Gibco-BRL) was used as a size marker. The gel was stained with Radiant Red RNA stain (Bio-Rad) and fixed with methanol during 30 min, processed for fluorography with 1% 2,5-diphenyloxazol in methanol, and then dried. The gel was subjected to autoradiography.
cDNA synthesis and cloning.
SDV virions were pelleted by ultracentrifugation for 90 min at 35,000 rpm, and viral genomic RNA was extracted by the QIAAMP viral RNA kit procedure (Qiagen, Courtaboeuf, France). The eluted RNA was denatured with hydroxymethyl mercury before cDNA synthesis. Random cDNA synthesis was done by the procedure described previously (20). Blunt-ended cDNA was ligated into EcoRV-digested pBluescript plasmid and used to transform the XL1 Blue competent Escherichia coli strain. Recombinant clones were randomly selected, and DNA was sequenced. The sequencing reactions were carried out on an ABI 373A DNA automatic sequencer with the DyeDeoxy Terminator-Prism (Applied Biosystems Division of Perkin-Elmer). A search for homologies against data banks was made by using the BLAST series program (1). From one cDNA clone (clone 19 [cl19]) exhibiting homologies with the E2 gene of alphaviruses, a specific oligonucleotide, SDV3 (5′GGATCCATTCAGATGTGGCGTTGCTATGG3′) was derived and used in a 5′ RACE (rapid amplification of cDNA ends) system (Life Technologies, Gibco-BRL). Larger PCR products were gel purified and subcloned in the pGEM-T vector (Promega). At least three independent clones were entirely sequenced by using universal primers (T7 and SP6) and by primer walking.
The cDNA corresponding to the 3′ part of the SDV 26S subgenomic RNA was obtained with a dT15 linker (GAGA-XhoI) primer (Stratagene) to initiate the cDNA reaction, and PCR was conducted with a specific E2-derived primer and a GAGA-XhoI oligonucleotide. Conditions for PCR were one step at 94°C for 3 min; then 35 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 2 min; and a final extension step at 72°C for 7 min. PCR products were gel purified and subcloned in the pGEM-T vector (Promega). At least three independent clones were entirely sequenced.
SDV E2 antiserum.
The insert of SDV cl19 was excised from pBluescript and subcloned into the pET14b procaryotic expression vector (Novagen), and the construct was transferred in E. coli BL21(DE3) strain. By using the pET14b expression vector, six histidine residues are fused at the N-terminal ends of the recombinant proteins, enabling their purification by metal chelation affinity chromatography. Expression of recombinant proteins was induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) into log-phase cultures. An aliquot was analyzed on a sodium dodecyl sulfate (SDS)–12% polyacrylamide gel and visualized by Coomassie blue staining. Protein of the expected molecular weight was purified by using His-bind resin columns (Novagen). Briefly, the cell pellet from 50 ml of induced culture was resuspended in 20 ml of buffer A (5 mM imidazole, 500 mM NaCl, 20 mM Tris-HCl [pH 7.9]) containing 200 U of benzonase (Merck) and sonicated. Following centrifugation, the resulting pellet was resuspended in 5 ml of buffer B containing 6 M urea. Solubilized proteins were loaded on His-bind resin columns (Novagen) preequilibrated in buffer B plus 6 M urea. Following washes and elution in buffer C (400 mM imidazole, 500 mM NaCl, 20 mM Tris-HCl [pH 7.9]), purified recombinant protein was recovered, dialyzed against phosphate-buffered saline (PBS), and used to immunize a rabbit three times every 3 weeks (roughly 200 μg/injection). Following the final boost, the rabbit was bled.
Reactivity of the SDV rabbit anti-E2 serum.
Proteins of concentrated SDV virions and lysate of E. coli expressing ΔE2 were separated on an SDS–10% polyacrylamide gel and electrotransferred onto a ProBlott membrane (Applied Biosystems) in CAPS (3-[cyclohexylamino]-1-propanesulfonic acid) buffer containing 10% methanol. The membrane was blocked overnight in 3% bovine serum albumin in PBS and then incubated with anti-E2 serum diluted to 1:500 in PBS–3% bovine serum albumin containing 0.05% Tween 20. The membrane was washed and incubated with an alkaline phosphatase-conjugated anti-rabbit immunoglobulin (BYOSIS). Detection of bound antibodies was accomplished by using the Gibco-BRL nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate detection kit.
Nucleotide sequence accession numbers.
The nucleotide sequences of the SDV cDNA cl19 and of 26S subgenomic RNA have been deposited in the EMBL database under accession no. AJ007631.1 and AJ238578, respectively.
RESULTS
SDV optimally replicates at very low temperature.
The improvement of the viral yield in cell culture was a prerequisite to any further investigations. Among the numerous fish cell lines available in our lab, only two salmonid-derived cells (RTG-2 and CHSE-214) exhibited a visible CPE in some areas of the cell monolayer following SDV infection (data not shown). The CPE consisted of small groups of refringent cells with either a twisted or stretched shape, or with a rounder appearance for some other cells. The more reliable method to determine the titer of the viral production was a reverse plaque titration assay in which SDV-infected cells were overinfected with the lytic VHSV. Cell clusters, corresponding to SDV-infected cells protected from VHSV infection by the heterologous interference phenomenon (10), could be accurately counted.
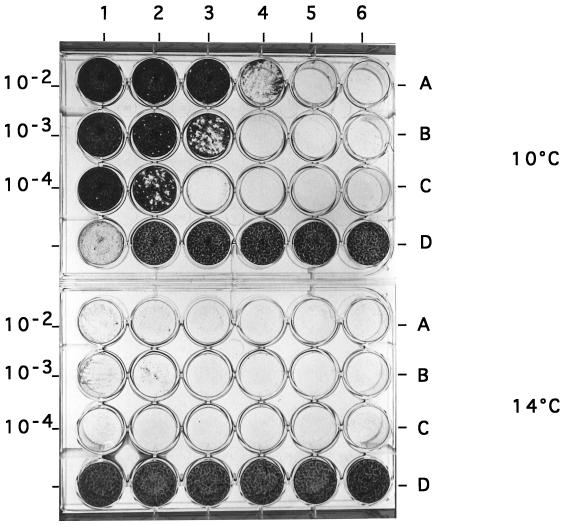
The maximum rate of virus replication was observed at 10°C; the titer routinely ranged from 106 to 107 PFU/ml, whereas at 14°C, the titer was dramastically reduced, since only few plaques were observed (Fig. 1). The strict temperature dependence was not due to heat lability of the virus, since incubation of SDV before infection at temperatures of as high as 37°C had no effect on the viral production yield (Table 1). The other parameters tested relative to the SDV growth were the pH and viral yield. Infectivity was totally lost following exposure at pH 3.0 and was only reduced at pH 11.0 (Table 1). The viral yield was optimal at 10 days postinfection (p.i.).
FIG. 1.
Comparison of SDV growth in RTG2 cells at 10 and 14°C. (Rows A to C) RTG-2 cells were infected with serial dilutions of SDV (1 to 10−5 [columns 1 to 6, respectively]) starting from the inoculum concentration indicated on the left. Plates were incubated at 10°C (upper) or 14°C (lower). At 7 days p.i. SDV-infected cells were overinfected with VHSV, and 3 days later they were overlaid with agarose. SDV-infected cell foci were visualized following crystal violet staining. (Rows D) Uninfected cells (except column 1 at 10°C, where cells were infected only with VHSV).
TABLE 1.
Stability of SDV exposed to chloroform or various temperatures and pHs
| Virus | Titera (log10 PFU/ml) under the indicated conditions
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Chloroform
|
pH
|
Temp (°C)
|
||||||||
| − | + | 3 | 7 | 11 | 10 | 15 | 25 | 37 | 45 | |
| SDV | 5.7 | 2.4 | 1 | 5.7 | 4.3 | 5.3 | 5.3 | 5.3 | 5.0 | 0 |
| VHSV | 7.7 | <1.0 | 3 | 7.7 | 4 | NDb | ND | ND | ND | ND |
| IPNV | 8.2 | 7.9 | 8.2 | 8.2 | ND | ND | ND | ND | ND | ND |
Each experiment was done in triplicate.
ND, not done.
SDV is an enveloped virus.
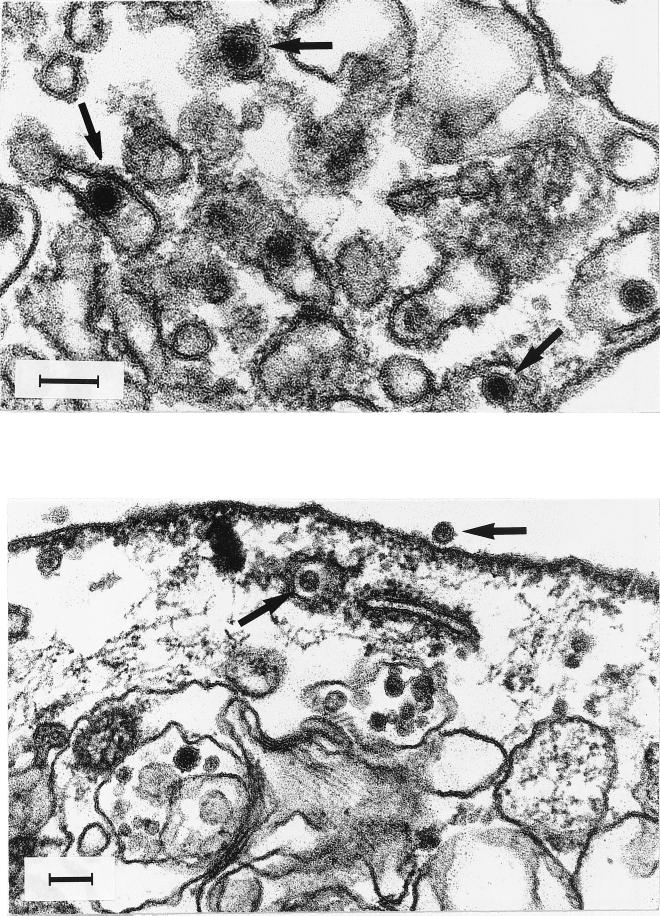
Numerous virus-like, spherical particles, homogenous in size (60 ± 5 nm) were observed in the cell cytoplasm of damaged infected cells sampled at 10 days p.i. (Fig. 2). The particles show an inner core of indefinite structure apparently surrounded by a bilayered lipidic membrane with short spikes visible on the viral membrane surface. The majority of the viral particles accumulated in cytoplasmic vesicles. An inoculum of SDV was subjected to chloroform treatment before infection of cells to confirm that SDV was an enveloped virus. Table 1 summarizes the results of that experiment, in which chloroform treatment was done in parallel with VHSV, a salmonid rhabdovirus, and IPNV, a salmonid birnavirus, as controls sensitive and resistant to chloroform treatment, respectively. A significant decrease in infectivity following exposure of SDV to chloroform was observed, demonstrating the presence of a lipid-containing envelope in the viral particle.
FIG. 2.
SDV ultrastructural morphology. RTG-2 infected cells were fixed in 1.25% glutaraldehyde, postfixed in 1% OsO4, and embedded in Epon by conventional techniques. Thin sections were stained with uranyl acetate and examined with an electron microscope. Arrows indicates viral particles. Bars, 100 nm.
SDV has an RNA genome.
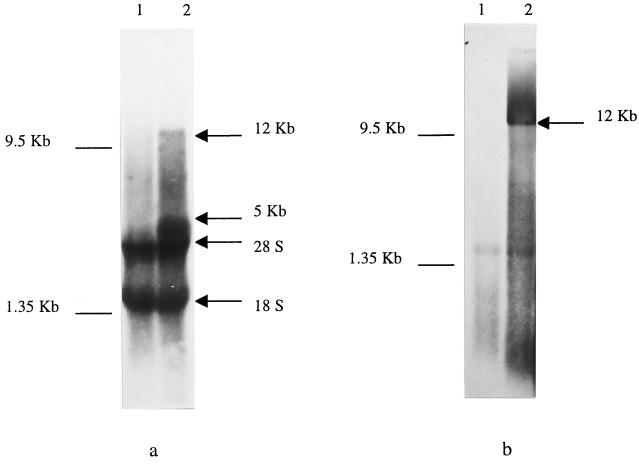
Further characterization of SDV involved the determination whether DNA or RNA is in the SDV genome. Initially, an inhibitor of DNA virus replication was used to evaluate the effect on SDV replication. SDV replication was not affected when BrdU was added to the culture medium, whereas the replication of the DNA virus control (SaHV-1) was inhibited (Table 2). The data strongly suggested that the SDV genome consisted of RNA. The size and type (segmented or not) of that RNA genome was determined by using labeled RNAs from SDV-infected cells and from concentrated supernatant which were separated on a gel and visualized following fluorography. The comparison of the RNAs extracted from the mock-infected (Fig. 3a, lane 1) and infected (Fig. 3b, lane 2) cells indicated the presence of two additional bands at approximately 5 and 12 kb in the total RNA extracted from infected cells. Analysis of RNA extracted from infected cell supernatant also revealed the presence of a 12-kb viral band (Fig. 3b, lane 2). The detection of a 12-kb viral genomic RNA and a putative 5-kb subgenomic viral RNA strongly supports the possibility that SDV belongs to the Togaviridae (21).
TABLE 2.
SDV replication in the presence of BrdU
| Virus | Titera (log10 PFU/ml) with:
|
||
|---|---|---|---|
| No addition | 1 mM BrdU | 1 mM BrdU + THY | |
| SDV | 5.4 | 5.7 | 5.8 |
| SaHV-1 | 2 | 0 | 2 |
| IPVN | 5.7 | 5.5 | 6.5 |
Each experiment was done in triplicate.
FIG. 3.
Characterization of the SDV genome. (a) Total RNA from metabolically [5,6-3H]uridine-labeled mock-infected (lane 1) or SDV-infected (lane 2) RTG-2 cells separated on a nondenaturing agarose gel. (b) RNA extracted from supernatant of mock-infected (lane 1) or SDV-infected (lane 2) cells.
Molecular cloning of the SDV genome.
Definitive characterization and grouping of SDV as a member of the Togaviridae were accomplished through the construction of a random-primed cDNA library from viral genomic RNA. Recombinant cDNA clones were randomly selected, and cDNA inserts were used as probes against the SDV RNA genome. A positive signal was observed for one SDV cDNA clone (cl19) (Fig. 4, lane 2). The cDNA of cl19 was sequenced and compared to known sequences in data banks by using BLAST programs (1). Significant homologies were detected at both the nucleotide and amino acid levels with part of the gene and the encoded glycoprotein E2 of all the alphavirus isolates sequenced so far (data not shown).
FIG. 4.
Slot blot hybridization. RNA extracted from mock-infected (lane 1) or SDV-infected (lane 2) cell supernatants was spotted onto a nitrocellulose membrane and subjected to hybridization with 32P-radiolabeled cl19 cDNA insert.
The complete 26S subgenomic RNA was cloned in a multistep procedure (Fig. 5), starting from total RNA extracted from SDV-infected cells. A GAGA-XhoI-anchored dT15 primer (Stratagene) was used to initiate the reverse transcription from the poly(A) tail. Specific SDV cDNA was amplified by PCR with a specific SDV primer derived from cl19 and a GAGA-XhoI primer. A 2-kbp PCR product covering part of E2, the 6,000-molecular-weight protein (6K), E1, and the 3′ untranslated region was obtained. The 5′ untranslated region together with the capsid (C) and the E3 regions were recovered by 5′ RACE with the 5′-end sequence of cl19 as a cDNA primer. Additionally, by using specific primers, the complete coding region of the 26S RNA was recovered by reverse transcription-PCR as two overlapping cDNA clones, clP-C/E2 and clP-E2/E1 (Fig. 5). These clones were sequenced to ascertain that no artifactual sequences were added during the 5′ RACE process.
FIG. 5.
Cloning and sequencing strategy for SDV. The initial SDV cl19 cDNA, cDNA clones clT-E2 and clT-ntr [generated by 5′ RACE with cl19-derived and oligo(dT)-anchored primers, respectively], and cDNA clones cl-C/E2 and cl-E2/E1 (generated through specific reverse transcription-PCR) are shown. Three clones from each series were sequenced by using universal primers and by primer walking.
Compilation of sequencing data allowed the determination of the total sequence of the SDV subgenomic RNA encoding the structural proteins.
Relationships between SDV and other alphaviruses.
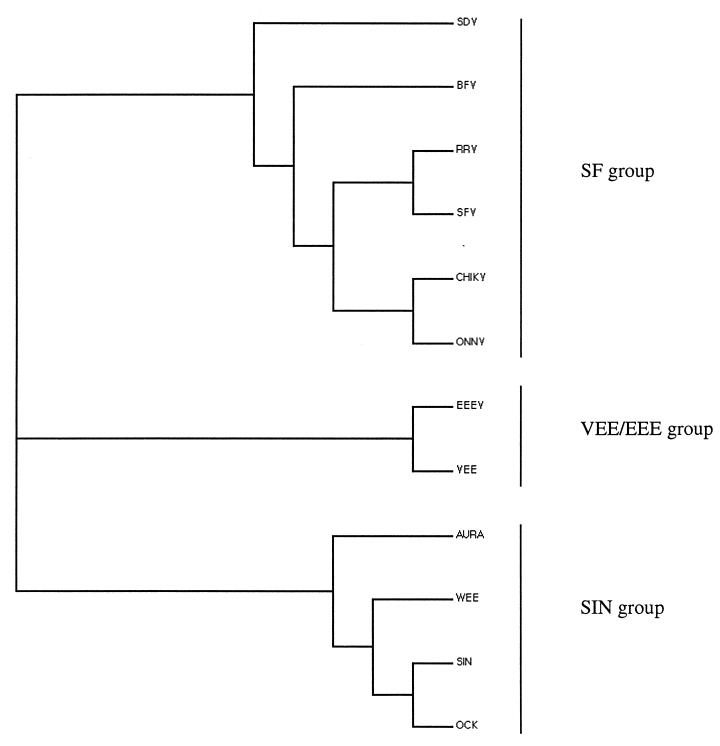
The translated sequence of the 3′-terminal 4,179 nucleotides of the SDV genome is shown in Fig. 6. This sequence starts in the region encoding the carboxy terminus of the nonstructural protein nsp4 and continues through a junction untranslated region to the start codon of the subgenomic mRNA which encodes a 1,324-amino-acid p130 polyprotein. The overall organization of the SDV polyprotein is similar to that for other alphaviruses, as observed by the presence of proteins similar to capsid, E3, E2, 6K, and E1, from the amino terminus to the carboxy terminus of the polyprotein. The overall identity of the SDV polyprotein with alphaviruses was 30 to 32%, and the similarity was 47 to 50%, depending on the alphavirus used for comparison. A phylogenic tree was constructed (Fig. 7), which emphasizes that SDV is more related to the Semliki Forest virus group of alphaviruses than to the others. Individual SDV structural proteins have the lowest percentages of homology among the alphaviruses and exhibit some remarkable features. A unique feature is the putative size of each individual processed protein, which is larger than for the other alphaviruses (Table 3).
FIG. 6.
Translated nucleotide sequence of SDV 26S RNA and the extreme C terminus of nsp4. The nucleotide sequence is numbered from the 3′ end of nsp4 gene. Amino acids are numbered from the first Met capsid residue. Putative polyprotein cleavage site are indicated by bent arrows. Potential glycosylation sites are indicated by arrows. Amino acid residues with dots correspond to the transmembrane region. Underlined residues correspond to the E1 fusion domain. Asterisks indicate catalytic amino acid triad residues.
FIG. 7.
Phylogenic tree for the structural polyproteins of various alphaviruses. The phylogenic tree was generated by using the ClustalW program (11) and drawn with the TreeView software. Alphaviruses and accession numbers: SDV, AJ238578; Barmah forest virus (BFV), AAB40702; Ross river virus (RRV), K00046; Semliki forest virus (SFV), S42462; Chikungunya virus (CHIKV), AAA53256; O'Nyong-Nyong virus (ONNV), M20303; EEEV, X63135; VEE virus, L04598; Aura virus (AURA), S78478; WEE virus, J03854; Sindbis virus (SIN), M13818; Ockelbo virus (OCK), P27285.
TABLE 3.
Comparison of SDV structural proteins and 26S untranslated regions with those of other alphaviruses
| Virusa | No. of nucleotides in junction regionb | No. of amino acids in the following structural protein:
|
No. of nucleotides in 3′ untranslated regionc | ||||
|---|---|---|---|---|---|---|---|
| Capsid | E3 | E2 | 6K | E1 | |||
| SDV | 39 | 285 | 71 | 438 | 68 | 461 | 90 |
| BFV | 32 | 253 | 69 | 421 | 58 | 439 | 445 |
| RRV | 47 | 270 | 64 | 426 | 56 | 438 | 524 |
| EEE | 66 | 260 | 60 | 425 | 57 | 441 | 361 |
| CHIK | NDd | 261 | 64 | 427 | 57 | 435 | 138 |
| ONN | 48 | 260 | 64 | 376 | 57 | 439 | 425 |
| AURA | 53 | 267 | 61 | 425 | 54 | 438 | 465 |
| SFV | 41 | 267 | 66 | 426 | 56 | 438 | 264 |
| SIN | 48 | 264 | 64 | 426 | 55 | 439 | 322 |
| VEE | 38 | 275 | 59 | 423 | 56 | 442 | 121 |
| WEE | 42 | 259 | 60 | 423 | 55 | 439 | 303 |
Abbreviations are as described in the legend to Fig. 7.
Sequence between the 26S RNA 5′ and 3′ ends.
The 3′ untranslated region is the sequence preceding the poly(A) tail, including the E1 stop codon.
ND, not determined.
Capsid protein.
The SDV nucleocapsid is 285 amino acid residues long (calculated molecular weight of 31,600), slightly larger than the other alphavirus capsids. The characteristic catalytic triad amino acid residues forming the serine protease active site i.e., H141, D163, and S215 for Sindbis virus (23), are found at H162, D184, and S236, respectively, for SDV. The putative autocleavage site is located at W285 at a consensus site, (E/P) W ↓(S/T). The N-terminal domain from R68 to K113 is very rich in K, R, and P residues, similar to the case for other alphavirus capsid proteins.
The domain described by Wengler et al. (24) that acts as a ribosome binding region cannot be found in the SDV core protein. Whether this element is different for binding to fish cell ribosomes is unknown.
E3 signal peptide and E2 glycoprotein.
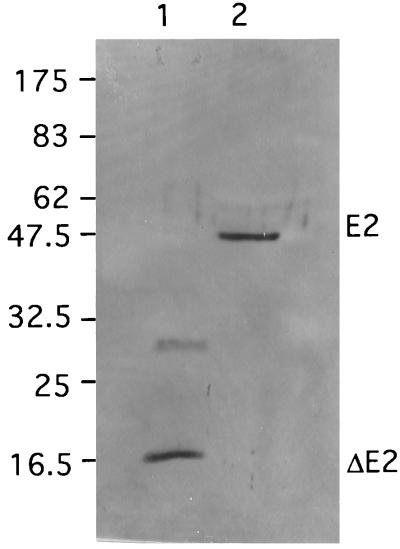
The signal peptide E3 and the E2 glycoprotein are respectively, 71 and 438 amino acids long and have respective calculated molecular weights of 7,900 and 47,000. The site of processing between E3 and E2 by a furin-like host cell protease is thought to be located in the consensus region RKKR ↓X (17). Surprisingly, no N-glycosylation site can be found in the SDV E3 protein. The lack of an N-glycosylation site does not appear to affect the maturation and cleavage of E2, since an SDV E2 having the expected molecular weight was detected on virions by Western blotting with anti-E2 SDV antiserum (Fig. 8, lane 2). A variant of the Western equine encephalitis (WEE) virus in which E3 is not glycosylated has been shown to grow poorly in BHK cells (22); whether the lack of SDV E3 glycosylation may explain the relatively poor growth of SDV in lower vertebrate cells remains to be determined.
FIG. 8.
Immunodetection of SDV E2 glycoprotein. The partial SDV E2 protein produced in E. coli and concentrated SDV virions were separated on an SDS–12% polyacrylamide gel, transferred onto a polyvinylidene difluoride membrane, and incubated with a rabbit E2 antiserum. Lane 1, recombinant E. coli ΔE2 protein; lane 2, SDV virions. Positions of molecular weight markers (in thousands) are indicated on the left.
The E2 protein has a single N-linked glycosylation site at N319 from the amino terminus of the E2 protein. A highly hydrophobic region, presumably representing the transmembrane domain, was detected at the carboxy end from amino acid residue 380 to 404, followed by 34 residues of cytoplasmic tail domain encompassing, near the end, 2 cysteine residues. These two Cys residues are conserved in all alphaviruses and have been predicted to be the acylation sites by the addition of palmytic acid (12). The motif TPY is strictly conserved in the E2 endo domains of all alphaviruses; this TPY motif has been shown to be a site for phosphorylation and to play a role in capsid binding to the E2 endo domain (15). The cytoplasmic tail of the SDV E2 glycoprotein does not have such a TPY motif but rather has one tyrosine and three threonine residues, located at different places, which may serve as phosphorylation sites.
SDV 6K equivalent and E1 glycoprotein.
The short protein is 68 amino acids long and has a calculated molecular weight of 7,500, an unusually large size for a “6K” protein. The putative signalase cleavage site (A↓ YE) at the carboxy end of 6K is very conserved among all alphaviruses and was also found for SDV. The mature E1 product is 462 amino acid long and possesses a unique N-linked glycosylation site at N35. N35 is a unique position compared to the case for other alphaviruses, where the N-glycosylation site is usually located at least 130 amino acid residues from the amino terminus. The transmembrane domain is presumably located at amino acid residues 430 to 450, followed by a short cytoplasmic tail very rich in charged amino acids (K and R). A unique feature for SDV E1 is the putative fusion domain (Fig. 9), which is highly conserved among alphaviruses and also for SDV, with the exception of two particular glycine residues which have been described by Levi-Mintz and Kielan (14). When these Gly residues are replaced, the threshold of the pH for fusion is shifted to a more acidic range. Both glycine residues are replaced in SDV, by N94 and A102 respectively. It is expected that the fusion process for SDV takes place more efficiently at a pH ranging from 5.0 to 5.4.
FIG. 9.
Multiple-sequence alignment of the E1 fusion domain. The SDV E1 putative fusion domain was aligned with some alphavirus E1 glycoproteins by using Multalin software (9). Abbreviations and accession numbers are as described in the legend to Fig. 7. Asterisks and colons represent identical and similar amino acids, respectively. Residues in boldface are the invariant amino acids in all viruses except SDV (see text).
Untranslated regions of the 26S RNA.
The nucleotide sequence corresponding to the junction region between the nsp4 end and the beginning of the structural proteins has been compared to the equivalent regions of other alphaviruses. That region is expected to be more or less conserved, since it contains important regulatory elements for transcription of the subgenomic mRNA. Figure 10a shows that with the exception of three nucleotide changes invariant in all alphaviruses, there is an overall conservation of the consensus junction region in SDV.
FIG. 10.
Multiple-sequence alignment of the junction region and the 3′ untranslated region. (a) Nucleotide sequences of the junction regions of SDV and several alphaviruses were aligned. Nucleotides conserved with those of SDV are indicated with dots. The three invariant nucleotides which are modified in the SDV sequence are indicated with asterisks. (b) Alignment of consensus 19-nucleotide 3′-end sequences of SDV and several alphaviruses (19). Dots indicate conserved nucleotides. Dashes have been introduced to optimize the alignment. Abbreviations are as described in the legend to Fig. 7. BEB, Bebaru; UNA, Una; BCR, Buggy Creek; MID, Middelburg.
Similarly, the extreme 3′ end of the SDV subgenomic RNA was compared to the ends of several alphaviruses, since it is hypothesized that this region serves as a promoter for initiation of minus-strand RNA synthesis on the plus-strand template. As shown in the multiple-sequence alignment in Fig. 10b, a significant conservation of this region exists in SDV, although there are three additional nucleotides before the poly(A) tail. The untranslated region is rather short (90 nucleotides), one of the shorter described. No repeated sequence elements can be detected in this region, but these elements do not seem to be essential for alphavirus growth in vitro (13).
DISCUSSION
In the present study, we have for the first time characterized and assigned to the alphavirus subfamily the viral agent (SDV) responsible for SD in rainbow trout. The finding that SDV replicated in cell culture at low temperature substantiated the temperature dependence of SDV growth in fish. Natural outbreaks of SD and experimental disease transmission to rainbow trout appeared almost exclusively at 10°C. The strict temperature growth restraint was not related to stability of the SDV particles, since following incubation at 37°C, SDV was still infectious. It can be hypothesized that temperatures higher than 10°C may affect the correct folding of viral proteins and most probably the glycoproteins during the time course of viral replication and particle assembly through the replication cycle in cells. Similar observations were made for VHSV. VHSV is capable of replicating at temperatures higher than 20°C; however, the VHSV glycoprotein is no longer expressed at the cell surface but remains fully expressed in the cytoplasm (M. Béarzotti and M. Brémont, unpublished data).
The CPE induced in SDV-infected tissue culture cells, even 10 days p.i., remained incomplete and was confined to localized areas of the cell monolayer when a low multiplicity of infection (0.1) was used. The lack of obvious CPE at a low multiplicity of infection may explain why during the past 10 years no virus was recovered from SD outbreaks, even though several attempts to isolate a virus were made.
SD has been estimated to affect roughly 30% of the fish farms in Brittany; however, that estimate is unreliable, since it has been mainly supported by the observations of the particular behavior of fish in tanks and by some histological studies of moribund fish, for which it is sometimes difficult to discriminate between the similar pancreatic lesions induced by SDV and IPNV, a salmonid birnavirus. The generation of specific SDV polyclonal antibodies, such as anti-E2, which reacts against SDV-infected cells as soon as 6 days p.i. (data not shown), will permit future clarification of the situation in the field and facilitate a survey of the epidemiologic status of SDV in fish farms.
Several lines of evidence suggest that SDV belongs to the Togaviridae, including (i) electron microscopic examinations of infected cells, which revealed numerous enveloped viral particles of roughly 60 nm in diameter, and (ii) studies on the viral genome, which demonstrated the presence of a 12-kb genomic RNA and a 5-kb subgenomic RNA in infected cells.
The cloning and nucleotide sequencing of the 4.1 kb at the 3′ end of the SDV genome confirmed that SDV belongs to the Alphavirus genus. The organization of the polyprotein is similar to that for other alphaviruses, and putative cleavage sites for the processing of the structural proteins were conserved in SDV. The main differences were in the sizes of the individual proteins, which were larger than those of other alphaviruses, and in the low percentage of amino acid identity between SDV and other alphavirus proteins, with the exception of some functional residues such as the serine-like protease catalytic triad (H, D, and S) in the capsid protein. Untranslated regions at the 5′ and 3′ ends of the subgenomic RNA are, in contrast, rather conserved, which emphasizes the role of these regions in the replication process of the alphaviruses. Although alphaviruses are usually transmitted by arthropods (8), it is unlikely that a mosquito serves as a reservoir for SDV, since direct transmission from fish to fish has been experimentally demonstrated by cohabition experiments (2).
Very recently, an alphavirus, salmon pancreas disease virus (SPDV), infecting Salmon salar has been described (18, 25). SDV and SPDV seem to be closely related, and it will be of interest to carefully compare SDV and SPDV sequences to evaluate if these novel atypical alphaviruses may represent a new group in the Alphavirus genus, specific for lower vertebrates, which could represent the alphaviruses ancestors, since, interestingly, Eastern equine encephalitis (EEE) virus, WEE virus, and Venezuelan equine encephalitis (VEE) virus are able to replicate in salmonid cell lines (27).
ACKNOWLEDGMENTS
We are grateful to F. Baudin-Laurencin (AFSSA Brest, Plouzané, France), C. Chastel (Université de Bretagne, Brest, France), and P. de Kinkelin (INRA, Jouy-en-Josas, France) for helpful suggestions during the study. Scott Kramer (INRA, Jouy-en-Josas, France) is gratefully acknowledged for critical reading of the manuscript. We are also grateful to P. Vende for the nucleotide sequencing data.
S.V. is a Ph.D. student financially supported by AFSSA and INRA.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boucher P. Ph.D. thesis. Rennes, France: Université de Rennes I; 1995. [Google Scholar]
- 3.Boucher P, Castric J, Baudin-Laurencin F. Observation of virus-like particles in rainbow-trout Oncorhynchus mykiss infected with sleeping disease virulent material. Bull Eur Assoc Fish Pathol. 1994;14:215–216. [Google Scholar]
- 4.Boucher P, Baudin-Laurencin F. Sleeping disease of salmonids. Bull Eur Assoc Fish Pathol. 1994;14:179–180. [Google Scholar]
- 5.Boucher P, Le Ven A, Baudin-Laurencin F. Transmission experimentale de la maladie du sommeil: histologie et caracterisation de l'agent infectieux. Nouvelles Sci Technol. 1995;13:115–117. [Google Scholar]
- 6.Boucher P, Baudin-Laurencin F. Sleeping disease and pancreas disease comparative histopathology and acquired cross-protection. J Fish Dis. 1996;19:303–310. [Google Scholar]
- 7.Castric J, Baudin-Laurencin F, Brémont M, Jeffroy J, Le Ven A, Béarzotti M. Isolation of the virus responsible for sleeping-disease in experimentally infected rainbow-trout Oncorhynchus Mykiss. Bull Eur Assoc Fish Pathol. 1997;17:27–30. [Google Scholar]
- 8.Chamberlain R W. Epidemiology of arthropod-borne togaviruses: the role of arthropods as hosts and vectors and of vertebrate hosts in natural transmission cycles. In: Schlesinger R W, editor. The togaviruses: biology, structure, replication. Orlando, Fla: Academic Press; 1980. pp. 175–227. [Google Scholar]
- 9.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukusho A, Ogawa N, Yammamoto H, Sawada M, Sazawa H. Reverse plaque formation by hog cholera virus of the GPE-strain inducing heterologous interference. Infect Immun. 1976;14:332–336. doi: 10.1128/iai.14.2.332-336.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higgins D, Thompson J, Gibson T, Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ivanova L, Schlesinger M J. Site-directed mutations in the Sindbis virus E2 glycoprotein identify palmitoylation sites and affect virus budding. J Virol. 1993;67:2546–2551. doi: 10.1128/jvi.67.5.2546-2551.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuhn R J, Hong Z, Strauss J H. Mutagenesis of the 3′ nontranslated region of Sindbis virus RNA. J Virol. 1990;64:1465–1476. doi: 10.1128/jvi.64.4.1465-1476.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levi-Mintz P, Kielan M. Mutagenesis of the putative fusion domain of the Semliki Forest virus spike protein. J Virol. 1991;65:4292–4300. doi: 10.1128/jvi.65.8.4292-4300.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu L N, Lee H, Hernandez R, Brown D T. Mutations in the endo domain of Sindbis virus glycoprotein E2 block phosphorylation, reorientation of the endo domain, and nucleocapsid binding. Virology. 1996;222:236–246. doi: 10.1006/viro.1996.0414. [DOI] [PubMed] [Google Scholar]
- 16.Magar R, Lecomte J. Comparison of methods for concentration and purification of bovine viral diarrhea virus. J Virol Methods. 1987;16:271–279. doi: 10.1016/0166-0934(87)90012-7. [DOI] [PubMed] [Google Scholar]
- 17.Mayne J T, Rice C M, Strauss E G, Hunkapiller M W, Strauss J H. Biochemical studies of the maturation of the small Sindbis virus glycoprotein E3. Virology. 1984;134:338–357. doi: 10.1016/0042-6822(84)90302-7. [DOI] [PubMed] [Google Scholar]
- 18.Nelson R T, McLoughlin M F, Rowley H M, Platten M A, McCormick J I. Isolation of a toga-like virus from farmed Atlantic salmon Salmo salar with pancreas disease. Dis Aquat Org. 1995;22:25–32. [Google Scholar]
- 19.Pfeffer M, Kinney R M, Kaaden O R. The alphavirus 3′-nontranslated region: size heterogeneity and arrangement of repeated sequence elements. Virology. 1998;240:100–108. doi: 10.1006/viro.1997.8907. [DOI] [PubMed] [Google Scholar]
- 20.Rutledge R G, Seligy V I, Cote M J, Dimock K, Lewin L L, Tenniswood M P. Rapid synthesis and cloning of complementary DNA from any RNA molecule into plasmid and phage M13 vectors. Gene. 1988;68:151–158. doi: 10.1016/0378-1119(88)90607-5. [DOI] [PubMed] [Google Scholar]
- 21.Schlesinger S, Schlesinger M J. Togaviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. Philadelphia, Pa: Lipincott-Raven Publishers; 1996. pp. 825–841. [Google Scholar]
- 22.Simizu B, Hashimoto K, Ishida I. A variant of western equine encephalitis virus with nonglycosylated E3 protein. Virology. 1983;125:99–106. doi: 10.1016/0042-6822(83)90066-1. [DOI] [PubMed] [Google Scholar]
- 23.Strauss J H, Strauss E G. Alphavirus proteinases. Semin Virol. 1990;1:347–356. [Google Scholar]
- 24.Wengler G, Wurkner D, Wengler G. Identification of a sequence element in the alphavirus core protein which mediates interaction of cores with ribosomes and the disassembly of cores. Virology. 1992;191:880–888. doi: 10.1016/0042-6822(92)90263-o. [DOI] [PubMed] [Google Scholar]
- 25.Weston J H, Welsh M D, McLoughlin M F, Todd D. Salmon pancreas disease virus, an alphavirus infecting farmed atlantic salmon, salmo salar L. Virology. 1999;256:188–195. doi: 10.1006/viro.1999.9654. [DOI] [PubMed] [Google Scholar]
- 26.Wolf K, Quimby M C. Established eurythermic line of fish cells in vitro. Science. 1962;135:1065–1066. doi: 10.1126/science.135.3508.1065. [DOI] [PubMed] [Google Scholar]
- 27.Wolf K, Mann J A. Poikilotherm vertebrate cell lines and viruses: a current listing for fishes. In Vitro. 1980;16:168–179. doi: 10.1007/BF02831507. [DOI] [PubMed] [Google Scholar]