Abstract
Background
TNFRSF4 plays a significant role in cancer progression, especially in hepatocellular carcinoma (HCC). This study aims to investigate the prognostic value of TNFRSF4 expression in patients with HCC and to develop a predictive pathomics model for its expression.
Methods
A cohort of patients with HCC retrieved from the TCGA database was analyzed using RNA-seq analysis to determine TNFRSF4 expression and its impact on overall survival (OS). Additionally, hematoxylin-eosin staining analysis was performed to construct a pathomics model for predicting TNFRSF4 expression. Then, pathway enrichment analysis was conducted, immune checkpoint markers were investigated, and immune cell infiltration was examined to explore the underlying biological mechanism of the pathomics score.
Results
TNFRSF4 expression was significantly higher in tumor tissues than in normal tissues. TNFRSF4 expression also exhibited significant correlations with various clinical variables, including pathologic stage III/IV and R1/R2/RX residual tumor. Furthermore, elevated TNFRSF4 expression was associated with unfavorable OS. Interestingly, in the subgroup analysis, elevated TNFRSF4 expression was identified as a significant risk factor for OS in male patients. The newly developed pathomics model successfully predicted TNFRSF4 expression with good performance and revealed a significant association between high pathomics scores and worse OS. In male patients, high pathomics scores were also associated with a higher risk of mortality. Moreover, pathomics scores were also involved in specific hallmarks, immune-related characteristics, and apoptosis-related genes in HCC, such as epithelial-mesenchymal transition, Tregs, and BAX expression.
Conclusions
Our findings suggest that TNFRSF4 expression and the newly devised pathomics scores hold potential as prognostic markers for OS in patients with HCC. Additionally, gender influenced the association between these markers and patient outcomes.
Keywords: Hepatocellular carcinoma, TNFRSF4, Prognostic factor, Pathomics, Minimum redundancy maximum relevance, Recursive feature elimination, Gradient boosting model
1. Introduction
Hepatocellular carcinoma (HCC) is a primary liver malignancy and ranks as the third leading cause of cancer-related deaths worldwide [1]. Despite the presence of effective treatment methods such as surgical resection and liver transplantation for patients with HCC, postoperative recurrence remains common [2]. Furthermore, current traditional prognostic indicators—i.e., clinical, pathological characteristics, diagnostic markers like alpha-fetoprotein (AFP), and imaging techniques (e.g., CT, MRI, ultrasound)—are not sufficient to meet the clinical demands of precision medicine [3]. Therefore, it is imperative to identify improved prognostic markers to guide clinical decision-making and identify high-risk patient subgroups [4].
TNF receptor superfamily member 4 (TNFRSF4), also known as OX40, is a co-stimulatory receptor expressed on activated T cells [5]. A previous study has suggested that this protein could inhibit cellular apoptosis via the PI3K/AKT signaling pathway [6]. Another study involving 1699 patients with HCC from six international multicenter cohorts found that the expression of the immune checkpoint TNFRSF4 was associated with infiltration of immune cells such as intra-tumoral B cells, CD4+/CD8+ T cells, and neutrophils, displaying an "immune hot" tumor phenotype that is more responsive to immune checkpoint inhibitors [7]. Therefore, investigating the role of TNFRSF4 expression as a prognostic marker in HCC could provide valuable insights into the disease's underlying biology and potentially guide therapeutic strategies.
Currently, TNFRSF4 expression can be detected through various approaches. However, these methods have limitations in terms of accuracy, cost, and practicality. For instance, peripheral blood cytokine detection provides real-time information but is expensive and fails to reflect the actual tumor condition. Techniques such as qPCR, RNA-sequencing (RNA-seq), Western blotting analysis, and flow cytometry for mRNA and protein levels require fresh tissue specimens, which are challenging to collect and can be influenced by operator and antibody effects. Furthermore, detection based on paraffin-embedded tissue specimens, including immunohistochemistry, fluorescence, and high-throughput sequencing, also has limitations related to operators, antibodies, and cost. Hematoxylin-eosin (H&E) staining is routinely used for clinical diagnosis and provides easily accessible imaging data [8]. Moreover, advancements in artificial intelligence (AI) have revolutionized the field of pathology [9], exemplified by the emerging field of pathomics. This AI-driven approach converts pathological images into high-throughput data [10], offering the potential for predicting treatment outcomes, disease prognosis, and molecular expression in various cancers [[11], [12], [13]]. Specifically, in the context of HCC, pathomics analysis has yielded promising results by uncovering prognostic signatures and gene mutations [14,15]. Furthermore, pathomics models have demonstrated the capability to predict RNA-seq expression from whole slide images [16]. These findings suggest that AI analysis of H&E staining slides could facilitate the detection of TNFRSF4 expression.
In light of the above, this study proposed an innovative approach to predict the expression of TNFRSF4 in HCC tissues using pathomics technology. Simultaneously, bioinformatics analysis was conducted to explore the molecular mechanisms underlying the newly devised pathomics score.
2. Materials and methods
2.1. Data collection and processing
RNA-seq data from the TCGA-LIHC [liver hepatocellular carcinoma] project were downloaded and organized from the TCGA database (https://portal.gdc.cancer.gov) [17]. The data were obtained using the STAR [Spliced Transcripts Alignment to a Reference] pipeline, which is a widely used tool for aligning RNA-seq reads to a reference genome [18]. RNA-seq data were retrieved in the TPM [Transcripts Per Million] format, which provides normalized expression values. Besides, clinical information was collected, including ablation embolization, AFP, age, gender, hepatic inflammation, histologic grade, pathologic stage, pharmaceutical therapy, residual tumor, and vascular invasion.
After obtaining the RNA-seq and clinical data, a rigorous sample selection and filtering process was performed. We specifically used 01A tumor tissue samples for our study. In cases where there were duplicates, we utilized the average value to represent each patient's transcriptional level, ensuring that each patient was represented by a single data point in our analysis. In total, we obtained data from 377 liver cancer patients in the TCGA database. We then selected patients who met the following criteria: (1) newly diagnosed and untreated, (2) overall survival time greater than one month, and (3) available RNA-seq data. Exclusion criteria included samples with incomplete survival status or survival time data, survival times of less than a month, and lacking essential clinical data. After applying these filters, we included 295 patients in our study. Additionally, we retrieved high-quality pathological images for 339 cases from the TCGA-LIHC pathology database. The intersection of patients with available clinical data, high-quality pathological images, and RNA-seq data comprised a subset of 267 individuals.
2.2. TNFRSF4 expression and prognostic analysis
Differences in TNFRSF4 expression between tumor and normal tissues were determined using the Wilcoxon rank-sum test and visualized via the “ggplot2” package of R v4.2.1. The R package "survminer" was employed to determine the optimal cutoff value for TNFRSF4 expression. The cutoff value of 1587 was selected to classify patients into TNFRSF4 high- and low-expression groups. Then, the correlation between TNFRSF4 expression and clinical variables was assessed. Furthermore, "survminer" was utilized to summarize and visualize the results of survival analysis performed using the R package "survival". Log-rank test was conducted to determine the significance of differences in overall survival (OS) between groups.
To assess the impact of these factors on OS, a univariate Cox regression analysis was conducted using the R packages “survival” and “forestplot”. Hazard ratios (HRs) with corresponding 95 % confidence intervals (CIs) were calculated to estimate the strength and direction of the association. The significance level was set at α = 0.05. To account for potential confounding variables, a multivariate analysis was performed.
To investigate the prognostic effect of TNFRSF4 expression in different subgroups of patients with HCC, subgroup analyses were performed using univariate Cox regression analysis with the “survival” R package. The TNFRSF4 high- and low-expression groups were compared within each subgroup. Furthermore, the potential interaction between TNFRSF4 expression and other covariates was examined using the "cmprsk" package in R. Moreover, Likelihood ratio tests were performed to assess the significance of the interactions.
2.3. Image acquisition, segmentation, and feature extraction
Histopathological tissue sections embedded in formalin and paraffin, in SVS format, were obtained by downloading pathological images from the TCGA database. The images were captured at either 20 × or 40 × magnifications [19,20].
The tissue regions of the pathological slides were obtained using the OTSU algorithm sourced from the OpenCV library (https://opencv.org/). The OTSU algorithm, also referred to as the maximum interclass variance method, effectively performs image thresholding by dividing the image into two distinct components: the undesired background and the essential tissue region for subsequent analysis [21]. For images at 40 × magnification, segmentation involved partitioning the images into multiple sub-images of 1024 × 1024 pixels in size, whereas images at 20 × magnification were divided into sub-images of 512 × 512 pixels in size, which were subsequently upsampled to 1024 × 1024 pixels. Following segmentation, the pathologists reviewed the segmented sub-images to exclude those with deficient image quality, such as those with contamination, blurriness, or blank areas exceeding 50 %. A total of 20 random sub-images were selected from each pathological image for further analysis.
The PyRadiomics open-source package (https://pyradiomics.readthedocs.io/en/latest/) was employed to standardize the sub-images and extract a comprehensive set of 93 original features, encompassing both first- and second-order features. In addition, high-order features such as Wavelet (LL, LH, HL, HH), LoG (kernel size: 1, 2, 3, 4, 5), Square, SquareRoot, Logarithm, Exponential, Gradient, and LBP2D were extracted. This resulted in a total of 1488 features. To ensure robustness, features of 10 sub-images were individually extracted for each patient, and the corresponding average values were subsequently calculated. These average values served as the histopathological features for each sample, thereby facilitating subsequent data analysis [12,22,23].
2.4. Dataset partitioning
The dataset was randomly divided into training and validation sets in a 7:3 ratio using the “caret” package in R. The histopathological features extracted by the “pyradiomics” package, comprising a total of 1488 features, were normalized by z-score within the training set. Subsequently, the validation set was standardized using the mean and standard deviation values obtained from the training set. To analyze intergroup differences in clinical variables between the training and validation sets, the “CBCgrps” package in R was utilized.
2.5. Feature selection
To improve the analysis outcomes and reduce dataset dimensionality, a feature selection process was conducted. First, features with zero variance were removed as they lacked discriminatory information. Next, to reduce redundancy and multicollinearity, one feature from each highly correlated pair (correlation coefficient >0.9) was removed. To determine the optimal feature subset, the minimum Redundancy Maximum Relevance (mRMR) and Recursive Feature Elimination (RFE) algorithms were employed, using the “mRMRe” package in R [24]. These algorithms select features with high relevance to the target variable while minimizing redundancy.
2.6. Model construction and evaluation
The “caret” package in R was utilized to construct a gradient boosting model (GBM) for predicting TNFRSF4 expression based on the selected pathomics features. GBM is a popular ensemble machine learning algorithm known for its predictive performance. In this study, it operates by training a sequence of weak classifiers, each informed by the negative gradient of the current model's loss function, and sequentially integrates these trained classifiers in an additive manner to construct the predictive model. We chose GBM for its robustness and effectiveness in handling various types of data, including non-linear and complex interactions between features. Additionally, GBM has been shown to perform well with imbalanced datasets, which is particularly relevant to biomedical research where certain classes of data may be underrepresented. The advantages of using GBM in our pathomics model include its ability to model complex interactions between features, handle missing data, and provide importance scores for each feature, thereby providing useful insights into the underlying biological mechanisms. Moreover, GBM tends not to require extensive data preprocessing, such as normalization or scaling, and is less sensitive to outliers in the data than other algorithms. In our application, the GBM model was tuned using a grid search approach to optimize hyperparameters, including the number of trees, tree depth, learning rate, and subsample size. The final model was then cross-validated to ensure generalizability and prevent overfitting. The performance of the developed GBM model was assessed using various evaluation metrics. The classification ability of the model was evaluated by plotting a receiver operating characteristic (ROC) curve using the “pROC” package in R. The ROC curve helps in assessing the trade-off between sensitivity and specificity at various thresholds. In addition to the ROC curve, the precision-recall (PR) curve was employed to evaluate the performance of the predictive model. The PR curve demonstrates the relationship between precision (positive predictive value) and recall (sensitivity) across different classification thresholds. The PR curve can provide a more comprehensive evaluation in scenarios where class imbalance exists. To evaluate the calibration of the predictive model, a calibration curve was plotted using the “rms” package in R. Additionally, the Hosmer-Lemeshow goodness-of-fit test was conducted using the “ResourceSelection” package to assess the calibration of the predictive model. To demonstrate the potential clinical utility of the predictive model, a decision curve analysis (DCA) was performed using the “rmda” package. The DCA curve illustrates the net benefit of the predictive model in a clinical context.
2.7. Pathomics score analysis
The model provided a probability value, herein referred to as the “pathomics score”. To investigate the differences in pathomics scores between TNFRSF4 high- and low-expression groups, statistical analysis using the “ggpubr” package in R was performed.
To further analyze the relationship between the pathomics score and various clinical variables, a cutoff value for the pathomics score using the “survminer” package in R was determined. The cutoff value was set at 0.373, which divided the patients into two groups: the high-pathomics score group (n = 158) and the low-pathomics score group (n = 109).
To assess the correlation between the pathomics score and different clinical variables, the “survminer” package in R was used. Kaplan-Meier survival curves were plotted to demonstrate the relationship between different groups for each variable (such as pathomics score) and patient survival. Log-rank test was conducted to examine the statistical significance of survival differences between the groups.
The impact of the pathomics score and clinical variables on OS was assessed via univariate and multivariate Cox regression analyses using the R packages “survival” and “forestplot”. To investigate the prognostic effect of the pathomics score in different subgroups of patients with HCC, subgroup analyses were performed using univariate Cox regression analysis with the “survival” package in R. The high- and low-pathomics score groups were compared within each subgroup. Furthermore, the potential interaction between the pathomics score and other covariates was examined using the "cmprsk" package in R. Likelihood ratio tests were performed to assess the significance of the interactions.
2.8. Gene set enrichment analysis (GSEA)
After dividing the samples into high- and low-pathomics score groups, a differential gene analysis was conducted to explore the differences between these groups. GSEA was employed to identify significantly enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) and Hallmark gene sets within either the high- or low-pathomics score group [25]. The KEGG gene set contains a collection of known biological pathways, while the Hallmark gene set represents a curated set of gene sets that reflect specific biological states or signaling pathways. To visualize the enriched pathways from the GSEA analysis, the top 20 pathways ranked by their enrichment scores were selected for visualization.
2.9. Correlation analysis of immune checkpoint expression, immune cell infiltration, and apoptosis-related gene expression
The Spearman correlation analysis was performed to assess the relationship between the pathomics score and the expression of immune checkpoint markers.
To evaluate immune cell infiltration in each sample, the RNA-seq expression matrix was uploaded to the CIBERSORTx website (https://cibersortx.stanford.edu/). This database utilizes computational algorithms to estimate the proportions of immune cell types within a given sample [26]. Correlation analysis between the pathomics score and immune cell infiltration was conducted using the “corrplot” package in R.
To explore the differential expression of apoptosis-related genes between the high- and low-pathomics score groups, a statistical analysis using the Wilcoxon rank-sum test was performed.
2.10. Statistical analysis
All statistical analyses were performed using R v4.1.0 and its relevant packages. Descriptive statistics were used to summarize the patient characteristics and clinical variables. Student's t-test or Mann-Whitney U test was applied for continuous variables, and chi-square or Fisher's exact test was used for categorical variables, as appropriate. Statistical significance was defined as P < 0.05.
3. Results
3.1. Study cohort and data integrity
The analysis flowchart in Fig. 1. To ensure the integrity and relevance of the data for our study, rigorous inclusion and exclusion criteria were applied. This yielded a final set of 295 samples for RNA-seq analysis and 267 samples for pathology image analysis. Subsequent analysis showed a significantly higher TNFRSF4 expression in tumor tissues than in normal tissues (Fig. 2A). Based on a cutoff value of 1.587 for TNFRSF4 expression, the patients were divided into two groups: TNFRSF4 high-expression group (n = 128) and TNFRSF4 low-expression group (n = 167). The distribution of vascular invasion showed a significant difference between the TNFRSF4 high-expression group and the TNFRSF4 low-expression group (P = 0.006, Table 1).
Fig. 1.
Analysis flowchart.
Fig. 2.
Expression and prognosis value of TNFRSF4 and other clinical variables. A, TNFRSF4 expression differences between hepatocellular carcinoma (HCC) tissues and normal tissues. B–H, Kaplan-Meier analysis demonstrating a significant association between worsened overall survival (OS) and high expression of TNFRSF4 (B), unknown ablation embolization (C), unknown AFP (D), unknown hepatic inflammation (E), pathologic stage III/IV (F), R1/R2/RX residual tumor (G), and unknown vascular invasion (H). ***, P < 0.001.
Table 1.
The association of TNFRSF4 and clinical variables.
| Variables | Total (n = 295) | Low (n = 167) | High (n = 128) | p |
|---|---|---|---|---|
| Gender, n (%) | 0.055 | |||
| Female | 92 (31) | 44 (26) | 48 (38) | |
| Male | 203 (69) | 123 (74) | 80 (62) | |
| Age, n (%) | 1 | |||
| ∼59 | 142 (48) | 80 (48) | 62 (48) | |
| 60∼ | 153 (52) | 87 (52) | 66 (52) | |
| AFP, n (%) | 0.277 | |||
| ∼399 | 159 (54) | 89 (53) | 70 (55) | |
| 400∼ | 74 (25) | 47 (28) | 27 (21) | |
| Unknown | 62 (21) | 31 (19) | 31 (24) | |
| Pathologic_stage, n (%) | 0.796 | |||
| I/II | 220 (75) | 126 (75) | 94 (73) | |
| III/IV | 75 (25) | 41 (25) | 34 (27) | |
| Histologic_grade, n (%) | 0.482 | |||
| G1/G2 | 183 (62) | 107 (64) | 76 (59) | |
| G3/G4 | 112 (38) | 60 (36) | 52 (41) | |
| Ablation_embolization, n (%) | 0.251 | |||
| NO | 224 (76) | 127 (76) | 97 (76) | |
| Unknown | 50 (17) | 25 (15) | 25 (20) | |
| YES | 21 (7) | 15 (9) | 6 (5) | |
| Vascular_invasion, n (%) | 0.006 | |||
| None | 167 (57) | 108 (65) | 59 (46) | |
| Micro/Macro | 88 (30) | 40 (24) | 48 (38) | |
| Unknown | 40 (14) | 19 (11) | 21 (16) | |
| Hepatic_inflammation, n (%) | 0.18 | |||
| None | 101 (34) | 62 (37) | 39 (30) | |
| Mild/Severe | 100 (34) | 59 (35) | 41 (32) | |
| Unknown | 94 (32) | 46 (28) | 48 (38) | |
| Pharmaceutical_therapy, n (%) | 0.851 | |||
| NO | 265 (90) | 151 (90) | 114 (89) | |
| YES | 30 (10) | 16 (10) | 14 (11) | |
| Residual_tumor, n (%) | 0.64 | |||
| R0 | 273 (93) | 153 (92) | 120 (94) | |
| R1/R2/RX | 22 (7) | 14 (8) | 8 (6) |
3.2. Prognostic significance of TNFRSF4 expression
A Kaplan-Meier curve was used to illustrate the relationship between several factors (including TNFRSF4 expression and clinical variables) and OS in the studied cohort. Elevated TNFRSF4 expression was found to be a prognostic marker for unfavorable OS (Fig. 2B). The results showed a median OS time of 84.7 months for the TNFRSF4 low-expression group, as well as 46.2 months for the TNFRSF4 high-expression group. Moreover, unknown ablation embolization (Fig. 2C), unknown AFP (Fig. 2D), unknown hepatic inflammation (Fig. 2E), pathological Stage III/IV (Fig. 2F), R1/R2/RX residual tumor (Fig. 2G), and unknown vascular invasion (Fig. 2H) were significantly correlated with a deterioration in OS, suggesting their potential as predictive biomarkers in patients with HCC. However, No correlation between age, gender, histologic grade, pharmaceutical therapy, and OS was observed (Fig. S1).
3.3. Risk factors for overall survival
Univariate Cox regression analysis revealed multiple risk factors for OS. High TNFRSF4 expression was shown to be a risk factor for OS (HR = 1.939, 95 % CI 1.307−2.877, P = 0.001), even after adjustment for multiple factors (HR = 1.803, 95 % CI 1.173−2.773, P = 0.007). Additionally, pathological Stage III/IV was identified as a risk factor for OS in the univariate analysis (HR = 2.679, 95 % CI 1.802−3.982, P < 0.001), which remained after adjustment for multiple factors (HR = 2.614, 95 % CI 1.697−4.025, P < 0.001). Further, the presence of residual tumor R1/R2/RX emerged as a risk factor for OS (HR = 2.631, 95 % CI 1.398−4.953, P = 0.003), which also remained after adjustment for multiple factors (HR = 2.271, 95 % CI 1.086−4.749, P = 0.029) (Fig. 3).
Fig. 3.
Comprehensive univariate and multivariate Cox regression analyses of the correlation between TNFRSF4 expression and clinical variables with OS. Each risk factor's hazard ratio (HR), 95 % confidence interval (95 % CI), and the corresponding statistical significance (P-value) are represented.
3.4. Gender-specific analysis
In the subgroup analysis, elevated TNFRSF4 expression was identified as a significant risk factor for OS in males (HR = 2.542, 95 % CI 1.538−4.2, P < 0.001) but not in females (HR = 1.142, 95 % CI 0.624−2.092, P = 0.67). Furthermore, a significant interaction was observed between gender and the association of TNFRSF4 expression with patient OS (P = 0.047) (Fig. 4).
Fig. 4.
Subgroup analysis of TNFRSF4 expression and overall survival (OS) stratified by ten clinical variables.
3.5. Data preprocessing for HCC pathological images
HCC histopathological images were obtained from the TCGA database (Fig. 5A) and subjected to segmentation, preprocessing, and feature extraction (Fig. 5B). The dataset was then split into a training set (188 cases) and a validation set (79 cases) in a 7:3 ratio.
Fig. 5.
Representative examples of histopathological hematoxylin-eosin (H&E)-stained images and patch segmentation. A, Histopathological H&E slides showing tissue morphology. B, Visualization of the image patch segmentation, demonstrating the subdivision of the image into smaller patches.
To assess the comparability between the training and validation sets, an intergroup differences analysis was conducted. The results demonstrated an insignificant difference for each clinical variable (P > 0.05), indicating similar baseline characteristics of patients in both sets. This similarity assures comparability between the two sets (Table 2).
Table 2.
The clinical variables in training set and test set.
| Variables | Total (n = 267) | Train (n = 188) | Validation (n = 79) | p |
|---|---|---|---|---|
| TNFRSF4, n (%) | 1 | |||
| Low | 153 (57) | 108 (57) | 45 (57) | |
| High | 114 (43) | 80 (43) | 34 (43) | |
| Gender, n (%) | 0.669 | |||
| Female | 81 (30) | 59 (31) | 22 (28) | |
| Male | 186 (70) | 129 (69) | 57 (72) | |
| Age, n (%) | 0.153 | |||
| ∼59 | 129 (48) | 85 (45) | 44 (56) | |
| 60∼ | 138 (52) | 103 (55) | 35 (44) | |
| AFP, n (%) | 0.731 | |||
| ∼399 | 138 (52) | 99 (53) | 39 (49) | |
| 400∼ | 69 (26) | 46 (24) | 23 (29) | |
| Unknown | 60 (22) | 43 (23) | 17 (22) | |
| Pathologic_stage, n (%) | 0.623 | |||
| I/II | 203 (76) | 145 (77) | 58 (73) | |
| III/IV | 64 (24) | 43 (23) | 21 (27) | |
| Histologic_grade, n (%) | 0.053 | |||
| G1/G2 | 164 (61) | 123 (65) | 41 (52) | |
| G3/G4 | 103 (39) | 65 (35) | 38 (48) | |
| Ablation_embolization, n (%) | 0.413 | |||
| NO | 203 (76) | 146 (78) | 57 (72) | |
| Unknown | 45 (17) | 28 (15) | 17 (22) | |
| YES | 19 (7) | 14 (7) | 5 (6) | |
| Vascular_invasion, n (%) | 0.182 | |||
| Micro/Macro | 76 (28) | 56 (30) | 20 (25) | |
| None | 153 (57) | 110 (59) | 43 (54) | |
| Unknown | 38 (14) | 22 (12) | 16 (20) | |
| Hepatic_inflammation, n (%) | 0.665 | |||
| Mild/Severe | 86 (32) | 61 (32) | 25 (32) | |
| None | 93 (35) | 68 (36) | 25 (32) | |
| Unknown | 88 (33) | 59 (31) | 29 (37) | |
| Pharmaceutical_therapy, n (%) | 0.828 | |||
| NO | 240 (90) | 168 (89) | 72 (91) | |
| YES | 27 (10) | 20 (11) | 7 (9) | |
| Residual_tumor, n (%) | 0.722 | |||
| R0 | 246 (92) | 172 (91) | 74 (94) | |
| R1/R2/RX | 21 (8) | 16 (9) | 5 (6) | |
| OS, n (%) | 0.847 | |||
| Alive | 173 (65) | 123 (65) | 50 (63) | |
| Dead | 94 (35) | 65 (35) | 29 (37) | |
| OS.time, Median (Q1,Q3) | 20.7 (12.3, 42.77) | 20.95 (12.72, 46.79) | 20.5 (12.07, 36.17) | 0.415 |
Furthermore, the mRMR and RFE methods were utilized to screen relevant features. These methods independently identified the top 30 features based on their respective criteria (Fig. 6A). To determine the most robust and informative set of features, the intersection of the top 30 features for both the mRMR and RFE methods was selected. As a result, four features were obtained, which demonstrated consistent importance across both methods (Fig. 6B).
Fig. 6.
Analysis of feature selection and model performance. A, Top 30 features identified by mRMR and RFE methods independently. B, Final set of four features consistently identified as important by both methods. C, Precision-recall (PR) curves showing the model's predictive performance in the training set, with an area under the curve (AUC) value of 0.773. D, Receiver operating characteristic (ROC) curves depicting the model's performance in the training set, yielding an AUC value of 0.787. E, Calibration curve and Hosmer-Lemeshow goodness-of-fit test indicating the consistency between predicted probabilities of high TNFRSF4 expression and actual values. F, Decision curve analysis (DCA) displaying the clinical utility of the model. G, Distribution of pathomics scores in the TNFRSF4 high- and low-expression groups in the training set. ****, P < 0.0001.
3.6. Pathomics model and its performance
To predict TNFRSF4 expression, a pathomics model that demonstrated good predictive performance was constructed. As shown in the PR curves, the model achieved an area under the curve (AUC) value of 0.773 in the training set (Fig. 6C) and 0.638 in the validation set (Fig. 7A). Similarly, based on the ROC curves, the model achieved an AUC value of 0.787 in the training set (Fig. 6D) and 0.723 in the validation set (Fig. 7B). Additionally, the calibration curve and Hosmer-Lemeshow goodness-of-fit test indicated consistency between the predicted probabilities of high gene expression by the predictive model and the actual values (P > 0.05) (Fig. 6, Fig. 7C). This indicates that the model accurately predicts high expression of TNFRSF4. Furthermore, the DCA curve demonstrated a high degree of clinical utility of the model, as depicted in Fig. 6, Fig. 7D.
Fig. 7.
Validation of model performance and clinical correlations. A, Precision-recall (PR) curves illustrating the model's performance in the validation set, with an area under the curve (AUC) value of 0.638. B, Receiver operating characteristic (ROC) curves representing the model's performance in the validation set, resulting in an AUC value of 0.723. C, Calibration curve and Hosmer-Lemeshow goodness-of-fit test showing the consistency between predicted probabilities of high gene expression and actual values in the validation set. D, Decision curve analysis (DCA) demonstrating the clinical utility of the model in terms of decision-making. E, Distribution of pathomics scores in the TNFRSF4 high- and low-expression groups in the validation set. F, Kaplan-Meier curve analysis demonstrating a significant association between high pathomics scores and worse overall survival (OS). ***, P < 0.001.
3.7. Pathomics score characteristics
The pathomics score distribution significantly differed between TNFRSF4 high- and low-expression groups, both in the training and validation sets. In the TNFRSF4 high-expression group, the pathomics scores were higher (Fig. 6, Fig. 7E). Moreover, no significant differences in the distribution of clinical variables were observed between the high- and low-pathomics score groups, as shown in Table 3. This suggests that the difference in pathomics scores is primarily attributed to the expression of TNFRSF4.
Table 3.
The association of pathomics score and clinical variables.
| Variables | Total (n = 267) | Low (n = 109) | High (n = 158) | p |
|---|---|---|---|---|
| Gender, n (%) | 0.141 | |||
| Female | 81 (30) | 39 (36) | 42 (27) | |
| Male | 186 (70) | 70 (64) | 116 (73) | |
| Age, n (%) | 0.59 | |||
| ∼59 | 129 (48) | 50 (46) | 79 (50) | |
| 60∼ | 138 (52) | 59 (54) | 79 (50) | |
| AFP, n (%) | 0.399 | |||
| ∼399 | 138 (52) | 60 (55) | 78 (49) | |
| 400∼ | 69 (26) | 29 (27) | 40 (25) | |
| Unknown | 60 (22) | 20 (18) | 40 (25) | |
| Pathologic_stage, n (%) | 0.635 | |||
| I/II | 203 (76) | 85 (78) | 118 (75) | |
| III/IV | 64 (24) | 24 (22) | 40 (25) | |
| Histologic_grade, n (%) | 0.099 | |||
| G1/G2 | 164 (61) | 60 (55) | 104 (66) | |
| G3/G4 | 103 (39) | 49 (45) | 54 (34) | |
| Ablation_embolization, n (%) | 0.593 | |||
| NO | 203 (76) | 86 (79) | 117 (74) | |
| Unknown | 45 (17) | 17 (16) | 28 (18) | |
| YES | 19 (7) | 6 (6) | 13 (8) | |
| Vascular_invasion, n (%) | 0.056 | |||
| None | 153 (57) | 69 (63) | 84 (53) | |
| Micro/Macro | 76 (28) | 31 (28) | 45 (28) | |
| Unknown | 38 (14) | 9 (8) | 29 (18) | |
| Hepatic_inflammation, n (%) | 0.304 | |||
| None | 93 (35) | 43 (39) | 50 (32) | |
| Mild/Severe | 86 (32) | 30 (28) | 56 (35) | |
| Unknown | 88 (33) | 36 (33) | 52 (33) | |
| Pharmaceutical_therapy, n (%) | 0.146 | |||
| NO | 240 (90) | 102 (94) | 138 (87) | |
| YES | 27 (10) | 7 (6) | 20 (13) | |
| Residual_tumor, n (%) | 0.668 | |||
| R0 | 246 (92) | 99 (91) | 147 (93) | |
| R1/R2/RX | 21 (8) | 10 (9) | 11 (7) |
Furthermore, the median OS in the high-pathomics score group was 52 months compared to 82.87 months in the low-pathomics score group. The Kaplan-Meier curve analysis demonstrated a significant association between high pathomics scores and worse OS (Fig. 7F). This indicates that patients with high pathomics scores have a poorer prognosis than those with low pathomics scores.
The results of the univariate Cox regression analysis revealed a high pathomics score as a risk factor for OS (HR = 1.579, 95 % CI 1.029−2.422, P = 0.036). After adjustment for multiple factors, the high pathomics score remained a significant risk factor for OS (HR = 1.94, 95 % CI 1.206−3.123, P = 0.006). Additionally, independent risk factors for OS included pathologic stage III/IV (HR = 2.546, 95 % CI 1.634−3.967, P < 0.001), histologic grade G3/G4 (HR = 1.618, 95 % CI 1.033−2.532, P = 0.035), and residual tumor R1/R2/RX (HR = 3.293, 95 % CI 1.557−6.962, P = 0.002) (Fig. 8).
Fig. 8.
Comprehensive univariate and multivariate Cox regression analyses of the correlation between pathomics scores and clinical variables with overall survival (OS). Each risk factor's hazard ratio (HR), 95 % confidence interval (CI), and the corresponding statistical significance (P-value) are represented.
In the subgroup analysis, an elevated pathomics score was identified as a significant risk factor for OS in male patients (HR = 2.437, 95 % CI 1.319−4.501, P = 0.004). Furthermore, the interaction test indicated a significant interaction between gender and the association of the pathomics score with patient OS (P = 0.012) (Fig. 9).
Fig. 9.
Subgroup analysis of pathomics scores and overall survival (OS) stratified by ten clinical variables.
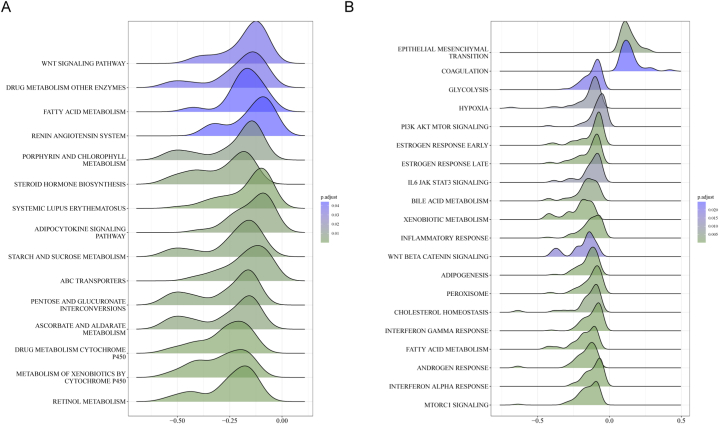
In the KEGG gene set, the differentially expressed genes in the low-pathomics score group were significantly enriched in signaling pathways such as the WNT pathway (Fig. 10A). Conversely, in the Hallmark gene set, the differentially expressed genes in the high-pathomics score group were significantly enriched in signaling pathways associated with epithelial-mesenchymal transition (EMT). By contrast, the differentially expressed genes in the low-pathomics score group were significantly enriched in the PI3K-AKT-mTOR signaling pathway (Fig. 10B).
Fig. 10.
Enrichment analysis of differentially expressed genes between low- and high-pathomics score groups. A, Enrichment analysis of the differentially expressed genes in the low-pathomics score group using the KEGG gene set. B, Enrichment analysis of the differentially expressed genes in the high-pathomics score group using the Hallmark gene set.
A significant positive correlation was observed between the pathomics score and the expression of immune checkpoint markers, including TNFRSF4 (Fig. 11A). Additionally, we investigated the immune cell infiltration in HCC and observed a significant positive correlation between the pathomics score and the extent of T regulatory cell (Tregs) infiltration (Fig. 11B). Lastly, in the high-pathomics score group, a significant upregulation in the expression of BAX and BIRC3 was observed (Fig. 11C). These findings suggest a potential association between the pathomics score and immune-related characteristics in HCC.
Fig. 11.
Correlation analysis of pathomics score with immune microenvironment and apoptosis. A, Positive correlation between the pathomics score and the expression of immune checkpoint markers. B, Positive correlation between the pathomics score and immune cell infiltration. C, Apoptosis-related gene expression between low- and high-pathomics score groups. *, P < 0.05; **, P < 0.01.
4. Discussion
HCC is the most common type of liver cancer that exhibits aggressive growth and poor prognosis [27]. The present study investigated the implications of TNFRSF4 expression in patients with HCC and developed a pathomics model to predict its expression. Our results demonstrated that TNFRSF4 expression and pathomics scores were risk factors for OS in patients with HCC. Moreover, pathomics scores were related to the immune microenvironment and apoptosis in HCC. The findings might provide insights into potential prognostic markers and therapeutic targets for HCC.
Our results demonstrated that elevated TNFRSF4 expression was associated with a significant reduction in OS in the HCC population. This finding supports previous research that has indicated TNFRSF4 expression as a prognostic marker in various cancers, including HCC [28,29]. The association between TNFRSF4 expression and OS remained significant even after adjusting for multiple clinical factors, indicating that TNFRSF4 expression can independently predict patient outcomes. These results are consistent with previous studies that have reported TNFRSF4 expression as an independent risk factor for poor survival in patients with cancer [30,31]. Moreover, our subgroup analysis revealed a significant interaction between gender and the association of TNFRSF4 expression with OS in patients with HCC. Specifically, elevated TNFRSF4 expression was found to be a significant risk factor for OS in male patients but not in female patients. This gender-specific difference in the prognostic value of TNFRSF4 expression may be attributed to hormonal and molecular differences between men and women [32].
The mechanism via which TNFRSF4 may contribute to HCC progression and worse outcomes in patients could be multifactorial. First, the activation of TNFRSF4 can lead to the proliferation of effector T cells, which, in a cancerous environment, could contribute to an immunosuppressive tumor microenvironment through the expansion of Tregs [33]. Our study observed increased TNFRSF4 expression correlating with the presence of Tregs, suggesting that TNFRSF4 may facilitate an immune escape mechanism for HCC. Second, pathway enrichment analysis revealed associations between TNFRSF4 expression and genes involved in EMT. EMT is a process via which epithelial cells lose their cell polarity and adhesion, gaining migratory and invasive properties and contributing to cancer metastasis [34]. The link between TNFRSF4 expression and EMT markers might shed light on the aggressive behavior of HCC in patients with high TNFRSF4 levels. Furthermore, our study found a significant correlation between TNFRSF4 expression and apoptosis-related genes, such as BAX. The balance between cell proliferation and apoptosis is critical to cancer development. Thus, the observed correlation suggests that TNFRSF4 may be involved in the dysregulation of apoptosis in HCC.
Pathomics is a thriving research field. In the context of HCC, significant research achievements have been made utilizing AI algorithms. Cheng et al. [35] developed four different deep-learning models to assist pathologists in classifying HCC nodular lesions. Saillard et al. [36] employed whole slide images and established two deep-learning algorithms to predict the prognosis of patients with HCC after surgical resection. Chen et al. [37] utilized a multiple-instance learning model to evaluate the microvascular invasion status of patients with HCC on H&E-stained histopathology slide images. Through establishing a pathomics model for predicting TNFRSF4 expression, this study investigated the impact of pathomics scores on OS in patients with HCC. Our results showed that high pathomics scores were associated with a worse prognosis, consistent with previous studies suggesting the prognostic value of pathomics scores in different cancers [38,39]. Furthermore, the association between pathomics scores and OS remained significant after adjusting for multiple clinical factors, highlighting the independent prognostic value of pathomics scores in HCC. Similarly, the subgroup analysis revealed a significant interaction between gender and the association of pathomics scores with OS.
Our study also explored the underlying molecular pathways associated with different pathomics scores in HCC. The enrichment analysis revealed that differentially expressed genes in the high-pathomics score group were significantly associated with EMT, a critical process involved in cancer metastasis and drug resistance [40]. This finding suggests that high pathomics scores may reflect a more aggressive phenotype characterized by increased EMT. Moreover, differentially expressed genes in the low-pathomics score group were enriched in the PI3K-AKT-mTOR signaling pathway, which has been implicated in HCC progression and represents a potential therapeutic target [41,42].
Furthermore, our study identified a significant positive correlation between pathomics scores and immune checkpoint marker expression, particularly TNFRSF4. This finding suggests that high pathomics scores may be associated with immune evasion and immunosuppression in HCC. Notably, an increase in Tregs infiltration was observed in patients with high pathomics scores. Tregs play a crucial role in immune tolerance and tumor immune escape, which can contribute to tumor progression and poor prognosis [43,44]. The immune-related characteristics associated with pathomics scores support the growing interest in immunotherapy as a potential treatment strategy for HCC [45]. Moreover, the elevated apoptosis-related genes in the high-pathomics score group indicate its potential involvement in cellular apoptosis.
While our study provides important insights into the implications of TNFRSF4 expression and pathomics scores in HCC, the following limitations should be acknowledged. First, our study relied on publicly available datasets, which may introduce selection bias and limit the generalizability of our findings. Second, the retrospective nature of the study limits our ability to establish causality between TNFRSF4 expression, pathomics scores, and patient outcomes. Prospective studies with larger cohorts are needed to confirm our findings and evaluate the clinical utility of TNFRSF4 expression and pathomics scores as prognostic markers in HCC.
5. Conclusions
In conclusion, our study demonstrated that elevated TNFRSF4 expression and high pathomics scores are associated with unfavorable outcomes in patients with HCC. TNFRSF4 expression and pathomics scores may serve as independent prognostic markers and guide personalized treatment strategies in HCC. Further research is warranted to validate our findings and explore the mechanisms underlying the association between TNFRSF4 expression, pathomics scores, and patient outcomes in HCC.
Funding
This work was supported by the Talent Support Project of Shaanxi Provincial People's Hospital (#2021JY-02), the National Nature Science Foundation of China (#82103371), and the Program of Shaanxi Administration of Traditional Chinese Medicine (SZY-KJCYC-2023-051).
Ethics statement
Informed consent and approval by an ethics committee were not needed for this study because this study utilizes data from the publicly available TCGA database, which houses anonymized patient data. Since all patient data were previously collected with informed consent and ethical approval, no additional ethics approval was necessary for this secondary analysis.
Data availability statement
Data will be made available upon reasonable request.
CRediT authorship contribution statement
Zhaoyong Yan: Writing – review & editing, Writing – original draft, Visualization, Formal analysis, Data curation, Conceptualization. Xiang Li: Writing – review & editing, Writing – original draft, Visualization, Formal analysis, Data curation, Conceptualization. Zeyu Li: Methodology, Formal analysis, Data curation. Sinan Liu: Methodology, Formal analysis, Data curation. Hulin Chang: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Resources, Project administration, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e31882.
Abbreviations
- HCC
Hepatocellular carcinoma
- OS
Overall survival
- AFP
Alpha-fetoprotein
- TNFRSF4
TNF receptor superfamily member 4
- RNA-seq
RNA-sequencing
- H&E
Hematoxylin-eosin
- AI
Artificial intelligence
- HRs
Hazard ratios
- CIs
Confidence intervals
- GBM
Gradient boosting model
- ROC:
Receiver operating characteristic
- PR
Precision-recall
- DCA
Decision curve analysis
- GSEA
Gene Set Enrichment Analysis
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- AUC
Area under the curve
- EMT
Epithelial-mesenchymal transition
- Tregs
T regulatory cell
Appendix A. Supplementary data
The following is the Supplementary data to this article:
No significant associations between worsened overall survival (OS) and age (A), gender (B), histologic grade (C), or pharmaceutical therapy (D).
References
- 1.Vogel A., et al. Hepatocellular carcinoma. Lancet. 2022;400(10360):1345–1362. doi: 10.1016/S0140-6736(22)01200-4. [DOI] [PubMed] [Google Scholar]
- 2.Yang J.D., et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019;16(10):589–604. doi: 10.1038/s41575-019-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ganesan P., Kulik L.M. Hepatocellular carcinoma: new developments. Clin. Liver Dis. 2023;27(1):85–102. doi: 10.1016/j.cld.2022.08.004. [DOI] [PubMed] [Google Scholar]
- 4.Pinero F., Dirchwolf M., Pessoa M.G. Biomarkers in hepatocellular carcinoma: diagnosis, prognosis and treatment response assessment. Cells. 2020;9(6) doi: 10.3390/cells9061370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Webb G.J., Hirschfield G.M., Lane P.J., Ox40 OX40L and autoimmunity: a comprehensive review. Clin. Rev. Allergy Immunol. 2016;50(3):312–332. doi: 10.1007/s12016-015-8498-3. [DOI] [PubMed] [Google Scholar]
- 6.Wu L.Y., et al. Recombinant OX40 attenuates neuronal apoptosis through OX40-OX40L/PI3K/AKT signaling pathway following subarachnoid hemorrhage in rats. Exp. Neurol. 2020;326 doi: 10.1016/j.expneurol.2020.113179. [DOI] [PubMed] [Google Scholar]
- 7.Liu Z., et al. Tumor suppressor gene mutations correlate with prognosis and immunotherapy benefit in hepatocellular carcinoma. Int Immunopharmacol. 2021;101(Pt B) doi: 10.1016/j.intimp.2021.108340. [DOI] [PubMed] [Google Scholar]
- 8.de Haan K., et al. Deep learning-based transformation of H&E stained tissues into special stains. Nat. Commun. 2021;12(1):4884. doi: 10.1038/s41467-021-25221-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niazi M.K.K., Parwani A.V., Gurcan M.N. Digital pathology and artificial intelligence. Lancet Oncol. 2019;20(5):e253–e261. doi: 10.1016/S1470-2045(19)30154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang Y., et al. Emerging role of deep learning-based artificial intelligence in tumor pathology. Cancer Commun. 2020;40(4):154–166. doi: 10.1002/cac2.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu K., Hu J. Classification of acute myeloid leukemia M1 and M2 subtypes using machine learning. Comput. Biol. Med. 2022;147 doi: 10.1016/j.compbiomed.2022.105741. [DOI] [PubMed] [Google Scholar]
- 12.Nishio M., et al. Homology-based image processing for automatic classification of histopathological images of lung tissue. Cancers. 2021;13(6) doi: 10.3390/cancers13061192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banna G.L., et al. The promise of digital biopsy for the prediction of tumor molecular features and clinical outcomes associated with immunotherapy. Front. Med. 2019;6:172. doi: 10.3389/fmed.2019.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nam D., et al. Artificial intelligence in liver diseases: improving diagnostics, prognostics and response prediction. JHEP Rep. 2022;4(4) doi: 10.1016/j.jhepr.2022.100443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen M., et al. Classification and mutation prediction based on histopathology H&E images in liver cancer using deep learning. npj Precis. Oncol. 2020;4:14. doi: 10.1038/s41698-020-0120-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmauch B., et al. A deep learning model to predict RNA-Seq expression of tumours from whole slide images. Nat. Commun. 2020;11(1):3877. doi: 10.1038/s41467-020-17678-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z., Jensen M.A., Zenklusen J.C. A practical guide to the cancer genome atlas (TCGA) Methods Mol. Biol. 2016;1418:111–141. doi: 10.1007/978-1-4939-3578-9_6. [DOI] [PubMed] [Google Scholar]
- 18.Dobin A., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L., et al. Histopathological image and gene expression pattern analysis for predicting molecular features and prognosis of head and neck squamous cell carcinoma. Cancer Med. 2021;10(13):4615–4628. doi: 10.1002/cam4.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng H., et al. Integration of histopathological images and multi-dimensional omics analyses predicts molecular features and prognosis in high-grade serous ovarian cancer. Gynecol. Oncol. 2021;163(1):171–180. doi: 10.1016/j.ygyno.2021.07.015. [DOI] [PubMed] [Google Scholar]
- 21.Wang X., et al. Weakly supervised deep learning for whole slide lung cancer image analysis. IEEE Trans. Cybern. 2020;50(9):3950–3962. doi: 10.1109/TCYB.2019.2935141. [DOI] [PubMed] [Google Scholar]
- 22.Saednia K., et al. Quantitative digital histopathology and machine learning to predict pathological complete response to chemotherapy in breast cancer patients using pre-treatment tumor biopsies. Sci. Rep. 2022;12(1):9690. doi: 10.1038/s41598-022-13917-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H., et al. Integrative analysis of histopathological images and genomic data in colon adenocarcinoma. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.636451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Jay N., et al. mRMRe: an R package for parallelized mRMR ensemble feature selection. Bioinformatics. 2013;29(18):2365–2368. doi: 10.1093/bioinformatics/btt383. [DOI] [PubMed] [Google Scholar]
- 25.Subramanian A., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steen C.B., et al. Profiling cell type abundance and expression in bulk tissues with CIBERSORTx. Methods Mol. Biol. 2020;2117:135–157. doi: 10.1007/978-1-0716-0301-7_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chakraborty E., Sarkar D. Emerging therapies for hepatocellular carcinoma (HCC) Cancers. 2022;14(11) doi: 10.3390/cancers14112798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma H., et al. Identification and validation of TNFRSF4 as a high-profile biomarker for prognosis and immunomodulation in endometrial carcinoma. BMC Cancer. 2022;22(1):543. doi: 10.1186/s12885-022-09654-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang D., et al. Elevated expression of TNFRSF4 impacts immune cell infiltration and gene mutation in hepatocellular carcinoma. Cancer Biomark. 2023;36(2):147–159. doi: 10.3233/CBM-210538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y., et al. Novel prognostic model based on immune signature for head and neck squamous cell carcinoma. BioMed Res. Int. 2020;2020 doi: 10.1155/2020/4725314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gamaleldin M.A., Imbaby S.A.E. The role of tumor necrosis factor receptor superfamily member 4 (TNFRSF4) gene expression in diagnosis and prognosis of acute myeloid leukemia. Mol. Biol. Rep. 2021;48(10):6831–6843. doi: 10.1007/s11033-021-06682-6. [DOI] [PubMed] [Google Scholar]
- 32.Pi S., et al. Body composition and risk of liver cancer: a population-based prospective cohort study on gender difference. Front. Nutr. 2023;10 doi: 10.3389/fnut.2023.1102722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weinberg A.D., Vella A.T., Croft M. OX-40: life beyond the effector T cell stage. Semin. Immunol. 1998;10(6):471–480. doi: 10.1006/smim.1998.0146. [DOI] [PubMed] [Google Scholar]
- 34.Manfioletti G., Fedele M. Epithelial-mesenchymal transition (EMT) Int. J. Mol. Sci. 2023;24(14) doi: 10.3390/ijms241411386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng N., et al. Deep learning-based classification of hepatocellular nodular lesions on whole-slide histopathologic images. Gastroenterology. 2022;162(7):1948–1961 e7. doi: 10.1053/j.gastro.2022.02.025. [DOI] [PubMed] [Google Scholar]
- 36.Saillard C., et al. Predicting survival after hepatocellular carcinoma resection using deep learning on histological slides. Hepatology. 2020;72(6):2000–2013. doi: 10.1002/hep.31207. [DOI] [PubMed] [Google Scholar]
- 37.Chen Q., et al. Deep learning for evaluation of microvascular invasion in hepatocellular carcinoma from tumor areas of histology images. Hepatol Int. 2022;16(3):590–602. doi: 10.1007/s12072-022-10323-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo C., et al. Predicting the recurrence and overall survival of patients with glioma based on histopathological images using deep learning. Front. Neurol. 2023;14 doi: 10.3389/fneur.2023.1100933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qu W.F., et al. Development of a deep pathomics score for predicting hepatocellular carcinoma recurrence after liver transplantation. Hepatol Int. 2023;17(4):927–941. doi: 10.1007/s12072-023-10511-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pastushenko I., Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29(3):212–226. doi: 10.1016/j.tcb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 41.Wang W., et al. Itraconazole exerts anti-liver cancer potential through the Wnt, PI3K/AKT/mTOR, and ROS pathways. Biomed. Pharmacother. 2020;131 doi: 10.1016/j.biopha.2020.110661. [DOI] [PubMed] [Google Scholar]
- 42.Wu Y., et al. PI3K/AKT/mTOR pathway-related long non-coding RNAs: roles and mechanisms in hepatocellular carcinoma. Pharmacol. Res. 2020;160 doi: 10.1016/j.phrs.2020.105195. [DOI] [PubMed] [Google Scholar]
- 43.Shao Q., et al. Tissue Tregs and maintenance of tissue homeostasis. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.717903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li C., et al. Regulatory T cells in tumor microenvironment: new mechanisms, potential therapeutic strategies and future prospects. Mol. Cancer. 2020;19(1):116. doi: 10.1186/s12943-020-01234-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Foerster F., et al. Emerging immunotherapy for HCC: a guide for hepatologists. Hepatology. 2022;75(6):1604–1626. doi: 10.1002/hep.32447. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
No significant associations between worsened overall survival (OS) and age (A), gender (B), histologic grade (C), or pharmaceutical therapy (D).
Data Availability Statement
Data will be made available upon reasonable request.