Abstract
The p53 tumor suppressor protein represents a target for viral and cellular oncoproteins, including adenovirus gene products. Recently, it was discovered that several proteins with structural and functional homologies to p53 exist in human cells. Two of them were termed p51 and p73. We have shown previously that the E1B 55-kDa protein (E1B-55 kDa) of adenovirus type 5 (Ad5) binds and inactivates p53 but not p73. Further, p53 is rapidly degraded in the presence of E1B-55 kDa and the E4orf6 protein of this virus. Here, it is demonstrated that p51 does not detectably associate with E1B-55 kDa. While p53 is relocalized to the cytoplasm by E1B-55 kDa, p51's location is unaffected. Finally, p51 retains its full transcriptional activity in the presence of E1B-55 kDa. Apparently, p51 does not represent a target of Ad5 E1B-55 kDa, suggesting that the functions of p51 are distinct from p53-like tumor suppression. E1B-55 kDa from highly oncogenic adenovirus type 12 (Ad12) was previously shown to surpass the oncogenic activity of Ad5 E1B-55 kDa in various assay systems, raising the possibility that Ad12 E1B-55 kDa might target a broader range of p53-like proteins. However, we show here that Ad12 E1B-55 kDa also inhibits p53's transcriptional activity without measurably affecting p73 or p51. Moderate inhibition of p51's transcriptional activity was observed in the presence of the E4orf6 proteins from Ad5 and Ad12. p53 and Ad12-E1B-55 kDa colocalize in the nucleus and also in cytoplasmic clusters when transiently coexpressed. Finally, E1B-55 kDa and E4orf6 of Ad12 mediate rapid degradation of p53 with an efficiency comparable to that of the Ad5 proteins in human and rodent cells. Our results suggest that E1B-55 kDa of either virus type has similar effects on p53 but does not affect p73 and p51.
The p53 gene is subject to the most common genetic alteration in human malignancies, and it plays a central role in tumor suppression, growth regulation, and apoptosis induction (23). The p53 protein's intracellular levels and activities are upregulated by genotoxic stress, and p53 was therefore termed “the guardian of the genome” (22). The p53 protein functions as a transcription factor, binding the DNAs of various cellular promoters and stimulating transcription. While p53's properties were previously believed to be unique, it was recently found that several p53 homologues are encoded by the human genome (4, 6, 20, 32, 42, 43, 50, 59), as reviewed in reference 19. Two of these proteins were termed p73 (20) and p51 (32). Gene products virtually identical to p51 were termed p63 (53) and KET (42). These proteins not only contain strong sequence homologies to p53 but also activate transcription from p53-responsive promoters (18, 20, 32). While several splicing variants of p73 (6, 20) and p51 (32, 50) exist, one of these forms in each case (termed p73β [18, 37] and p51A [32], respectively) was observed to stimulate transcription more actively than the other splicing variants. Despite the functional and structural similarities between p53 and its homologues, it remains an open question whether p53 homologues perform a role that can fully substitute for p53. Genetic studies on tumor material suggest that the gene encoding p73 does not necessarily display the features of a bona fide tumor suppressor gene (27, 31, 47, 48). In similar studies on p51, point mutations within the coding region of p51 were found in human epidermal tumors, albeit with low frequency (32). Mice lacking the p51/p63 gene have been developed. These mice are born alive but have severe developmental defects (29, 54) and therefore may not live until the formation of tumors, even if tumor development was facilitated by the absence of p51. Therefore, the role of p51 in cancer needs to be evaluated by different approaches. Another way to study the role of p53 homologues as tumor suppressors consists of the assessment of their role in virus-induced tumor formation. All known DNA tumor viruses have devised a strategy to inhibit p53. However, most viral p53 antagonists did not turn out to target p73 in addition to p53 (8, 28, 33, 37). We and others therefore suggested that p53 is central to tumor suppression, whereas p73 could act primarily on different aspects of growth regulation, e.g., during embryonic development. In the case of p51, no data on its ability to interact with oncoproteins have been reported so far. Therefore, the question arises whether p51's function is more closely related to the role of p53 or of p73.
The adenovirus type 5 (Ad5) E1B 55-kDa protein (E1B-55 kDa) forms a specific complex with p53 (39). Therefore, Ad5 E1B-55 kDa blocks p53-mediated transcription, an activity that directly correlates with the transforming potential of E1B-55 kDa (55). Ad5 E1B-55 kDa binds to the amino-terminal portion of p53 that is responsible for transcriptional activation (26), and it sequesters p53 to characteristic cytoplasmic clusters (5, 57). While none of these effects were observed when p53 was replaced by p73 (28, 37), the question remained whether Ad5 E1B-55 kDa and E4-34 kDa might affect p51.
Human adenoviruses were initially classified according to their abilities to induce malignant tumors in rodents (44). Ad5 belongs to a group with low oncogenicity, whereas adenovirus type 12 (Ad12) strongly induces tumors in animals. One important difference between the two viruses was found in the E1B-55 kDa proteins: studies using chimeric viruses revealed that the Ad12 E1B-55 kDa promoted tumor formation in athymic mice more strongly than its Ad5 homologue (3). Ad12 E1B-55 kDa also delays senescence (13), and it induces chromosomal fragile sites (1) in a p53-dependent fashion (24). The properties of the E1B-55 kDa proteins from Ad5 and Ad12 differ in several important features. While Ad5 E1B-55 kDa physically interacts with p53, attempts to copurify Ad12 E1B-55 kDa and p53 resulted in weak (15) or absent (58) detectable association. Whereas E1B-55 kDa of Ad5 localizes almost exclusively in cytoplasmic clusters within transformed cells (57), Ad12 E1B-55 kDa is found predominantly in the nuclei of these cells (58). Most importantly, however, the E1B-55 kDa proteins from Ad5 and Ad12 were found to have markedly different transforming properties when coexpressed with other oncogenes. Based on these differences, it has been proposed that Ad12 E1B-55 kDa might modulate p53 activity in a manner that is fundamentally different from the interaction between Ad5 E1B-55 kDa and p53 (51). Given the more recent discovery of p53 homologues, an attractive possibility to explain the stronger oncogenic potential of Ad12 E1B-55 kDa is the hypothesis that Ad12 E1B-55 kDa might interact with a broader spectrum of p53-like proteins than does Ad5 E1B-55 kDa.
Within the Ad5 system, E1B-55 kDa is not the only factor that inactivates p53. Instead, E1B-55 kDa and the E4orf6 protein cooperate to downregulate p53 activity. The two viral proteins are known to form a complex with each other (38), and the E4orf6 protein relocalizes E1B-55 kDa from the cytoplasm to the nucleus when the two proteins are coexpressed in primate cells (14). However, colocalization of E1B-55 kDa with E4orf6 was no longer observed in murine and most other rodent cells, suggesting that primate cells might contain a factor required for the intracellular association of E1B-55 kDa and E4orf6 (14). At least in human cells, the two Ad5 proteins mediate the rapid intracellular degradation of p53 (34, 37, 45). The destabilization of p53 is a feature that was also found for other oncoproteins, such as the E6 proteins of oncogenic human papillomaviruses (41, 52) and the cellular mdm2 protein (16, 21, 36).
In this study, we asked (i) whether adenovirus oncoproteins target p51 in addition to p53 and (ii) whether the features and specificity of p53 inactivation, relocalization, and degradation are maintained between the E1B-55 kDa and E4orf6 proteins of weakly oncogenic Ad5 and highly oncogenic Ad12. Our results show that p51 is not targeted by E1B-55 kDa of either virus. Further, despite the previously reported differences between the E1B-55 kDa proteins of the two viruses, Ad12 E1B-55 kDa and E4orf6 inactivate and destabilize p53 with an efficiency and specificity comparable to those of the Ad5 proteins.
MATERIALS AND METHODS
Cell culture and transfections.
All cells were maintained in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine serum. Transfections were carried out with FuGene 6 (Roche) according to the manufacturer's instructions.
Plasmids.
Expression plasmids for Ad5 E1B-55 kDa and E4orf6 were described previously (10, 37). To generate FLAG-tagged Ad5 E1B-55 kDa, the hemagglutinin (HA) tag within pCGNE1B (37) was replaced by a FLAG tag by using a PCR-based cloning strategy. Similarly, an expression vector for nontagged Ad5 E1B-55 kDa was generated in the same plasmid background. An expression plasmid for Ad12 E1B-55 kDa was obtained from T. van Laar, A. Zantema, and A. van der Eb (51). To express HA-tagged Ad12 E1B-55 kDa, the corresponding coding sequence was amplified by using the primers CGCGGTACCTGATGGAGCGAGAAATCCCACCTG and CGCGGATCCTCAGTTGTCGTCTTCATCACTTG and cloned into the vector pCGN (49, 60) by using KpnI and BamHI. An expression plasmid for Ad12 E4orf6 was obtained by amplifying the coding sequence from Ad12 genomic DNA (a gift from J. Schroer and W. Doerfler) with the primers CGCGGATCCGTCGACACCATGCAGCGCGACAGACGGTATCGC and CGCGGATCCTCAGTGTCCATCAGCCGCCCAAGG (for nontagged E4orf6) and cloned into the vector pCMVneoBam (2) by using BamHI. To append an HA tag to the C terminus of E4orf6, the same procedure was carried out with CGCGGATCCTCAGCTTGCGTAATCCGGTACATCGTAAGGGTAGTGTCCATCAGCCGCCCAAGG as the second primer. Plasmids that allow the intracellular expression as well as in vitro transcription-translation of nontagged or carboxy-terminally tagged p73β were described previously (37). An expression plasmid for p51A was obtained from S. Ikawa (32). To allow better expression, the coding sequence of p51A was amplified by using the primers CGCGGATCCGCCACCATGTCCCAGAGCACACAGACAAATG and CGCTCTAGAGGGTCAGCTTGCGTAATCCGGTACATCGTAAGGGTATGGGTACACTGATCGGTTTGGG (appending on HA tag to the C terminus of p51A) and cloned into the vector pcDNA3 (Invitrogen) by using BamHI and XbaI. The vector also contained the 5′ untranslated region of lamin to increase translational efficiency. All constructs were verified by sequencing.
Coimmunoprecipitation.
p53 and its homologues were generated and labeled with [35S]methionine by transcription and translation in vitro (TNT T7-system; Promega), and the amounts of the proteins were normalized after autoradiographic quantitation. The proteins were assayed for association with Ad5 E1B-55 kDa as described previously (37, 55). Briefly, 293 cells that constitutively express Ad5 E1B-55 kDa (106 per assay) were harvested in 100 μl of lysis buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 5 mM EDTA, 0.05% bovine serum albumin, 0.01% NP-40) and mechanically disrupted with a syringe. The soluble fraction was incubated with the normalized amount of reticulocyte lysate containing in vitro-translated p53 or its homologues at 30°C for 30 min, followed by immunoprecipitation with the monoclonal anti-E1B-55 kDa antibody 2A6 (40) and four washing steps with lysis buffer. p53 and its homologues coprecipitated along with E1B-55 kDa were resolved by sodium dodecyl sulfate-polyacrylamide electrophoresis (SDS-PAGE) and detected by autoradiography with the Bioimager detection system (Fuji).
Immunofluorescence.
Cells were transfected as described above (but all quantities were reduced by a factor of 4), and the cells were seeded on plastic slides (Nunc) suitable for microscopy. Transfected cells were fixed with paraformaldehyde (4% in phosphate-buffered saline [PBS]; 15 min), permeabilized with Triton X-100 (0.2% in PBS; 25 min), and incubated with antibody as described previously (7). The FLAG tag was stained with a polyclonal rabbit antibody (D-8; Santa Cruz). The HA tag was stained with a murine monoclonal antibody (HA.11; Babco). To detect E1B-55 kDa, the murine monoclonal antibody 2A6 (40) was used. The p53 protein was stained with a polyclonal rabbit antibody (FL-393; Santa Cruz). Primary mouse antibodies were visualized by use of secondary antibodies coupled to fluorescein isothiocyanate (Jackson). Primary rabbit antibodies were detected with a Texas Red-labeled secondary antibody (Jackson). Prior to mounting (Fluoprep; bioMérieux), the cell nuclei were briefly stained with 4′,6-diamidino-2-phenylindole (DAPI).
Immunoblots.
Proteins were separated on SDS–10% polyacrylamide gels and transferred to nitrocellulose, followed by incubation with antibodies diluted in PBS containing 5% milk powder and 0.05% Tween 20 and chemiluminescence detection (Pierce) of peroxidase-coupled secondary antibody (Jackson). Antibody Pab1801 to p53 was from Calbiochem. Antibody HA.11 against the HA tag was from Babco. All antibodies were diluted 1:1,000, except for HA.11, which was diluted 1:50,000.
Pulse-chase analysis.
Pulse-chase analysis of p53 stability was carried out essentially as described previously (37). H1299 cells (5 × 105 per lane) were transfected with expression constructs for p53 and adenovirus oncoproteins and starved for 30 min in starvation medium (DMEM lacking methionine and cysteine). The cells were then subjected to incubation with 35S-labeled amino acids (Promix; Amersham) diluted 1:30 in starvation medium. After 10 min, the medium was changed to complete DMEM containing 10% fetal bovine serum. After various chase times, the cells were lysed, and p53 was immunoprecipitated as described previously (35) with the murine monoclonal antibody 421 (Calbiochem) against p53.
Luciferase assays.
H1299 cells (2 × 105 per assay) were transfected. Luciferase activities were determined 18 h later by using a premanufactured assay system (Promega).
RESULTS
The Ad5 E1B-55 kDa protein binds p53 but not p51.
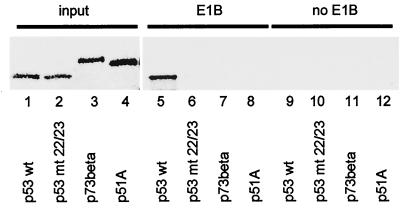
First, we determined whether p53 and its homologues bind to E1B-55 kDa of Ad5 with different efficiencies. Among the alternatively spliced forms of p73 and p51, the species p73β and p51A have the strongest transcriptional activities (references 32 and 37 and our unpublished observations). This prompted us to focus our studies on p73β and p51A. Wild-type p53 (Fig. 1, lane 1), a mutant p53 protein with the mutations L22Q and W23S (known to abolish E1B-binding) (26) (lane 2), the beta form of p73 (lane 3), and p51A (lane 4) were generated by transcription and translation in vitro with rabbit reticulocyte lysate. The proteins were then incubated with cell lysate from 293 cells (containing E1B-55 kDa) (lanes 5 to 8) or H1299 cells (lacking E1B-55 kDa) (lanes 9 to 12). This was followed by immunoprecipitation with a monoclonal antibody against E1B-55 kDa, gel electrophoresis, and autoradiography. Only p53 itself was specifically precipitated by this antibody in the presence (Fig. 1, lane 5) but not in the absence (lane 9) of E1B-55 kDa, and not when amino acids 22 and 23 were mutated (lane 6). Neither p73 (lane 7) nor p51 (lane 8) detectably coprecipitated with E1B-55 kDa. This suggests that E1B-55 kDa fails to associate efficiently with p51.
FIG. 1.
Association of p53 but not p51 with E1B-55 kDa from 293 cells. p53 and its homologues (as indicated) were generated and radioactively labeled by in vitro transcription and translation (lanes 1 to 4). The proteins (10 times the amount shown in lanes 1 to 4) were incubated with a lysate of 293 cells that constitutively express E1B-55 kDa. This was followed by immunoprecipitation with a monoclonal antibody against E1B-55 kDa (clone 2A6). The precipitated material was resolved by SDS-PAGE and visualized by autoradiography (lanes 5 to 8). As a control, an analogous experiment was carried out with a lysate from H1299 cells that do not contain adenovirus proteins (lanes 9 to 12). wt, wild type; mt 22/23, L22Q/W23S mutant.
Ad5 E1B-55 kDa does not affect the nuclear location of p51.
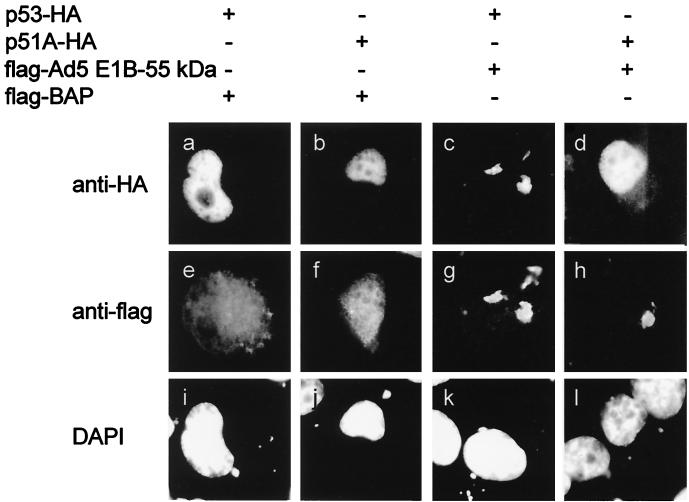
Next, we assessed whether the lack of detectable in vitro association between Ad5 E1B-55 kDa and p51 also results in a failure of E1B-55 kDa to relocalize p51 in vivo. Expression constructs for p53 and p51A that yielded carboxy-terminally HA-tagged versions of each protein were generated, thus allowing direct comparison of the signals obtained. These constructs were cotransfected with expression plasmids for a FLAG-tagged control protein (bacterial alkaline phosphatase; IBI Kodak) or FLAG-tagged E1B-55 kDa into H1299 cells, a human lung adenocarcinoma-derived cell line lacking endogenous p53. At 18 h after transfection, the cells were fixed and stained for the HA tag (Fig. 2a to d) and the flag-tag (Fig. 2e to h). In addition, the nuclei of the cells were visualized by using DAPI (Fig. 2i to l). In the absence of Ad5 E1B-55 kDa, both p53 and p51 were detected almost exclusively in the nuclei of the transfected cells (Fig. 2a and b). When E1B-55 kDa was coexpressed with these proteins (Fig. 2g and h), p53 colocalized with E1B-55 kDa in cytoplasmic clusters (Fig. 2c). In contrast, the nuclear localization of p51 remained unchanged (Fig. 2d). This suggests that within transfected cells, E1B-55 kDa does not associate with p51 to an extent comparable to that with p53.
FIG. 2.
Relocalization of p53 but not p51 by Ad5 E1B-55 kDa. H1299 cells (a human cell line lacking p53) were transiently transfected with plasmids (100 ng) expressing HA-tagged p53 or p51A. In addition, expression plasmids (400 ng) for FLAG-tagged Ad5 E1B-55 kDa or FLAG-tagged bacterial alkaline phosphatase (BAP) were cotransfected as indicated. The cells were fixed and stained simultaneously with murine anti-HA antibody and rabbit anti-FLAG antibody, followed by secondary antibodies labeled with different fluorescent dyes. The same areas were visualized for expression of HA-tagged (a to d) and FLAG-tagged (e to h) proteins. In addition, the cells were stained with DAPI (i to l) to show the location of cell nuclei.
Impact of Ad5 E1B-55 kDa and E4orf6 on the transcriptional activities of p53 and p51.
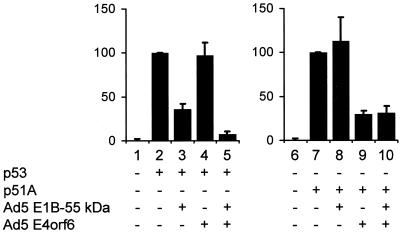
Finally, we asked whether adenovirus oncoproteins can modulate the transcriptional activities of p53 and p51. To determine this, H1299 cells were transfected with expression plasmids for p53 and p51. A reporter construct containing a portion of the murine mdm2 intron that confers transcriptional response to p53 upstream of the luciferase coding region (pBP100luc [12]) was cotransfected. Along with these plasmids, expression constructs for the Ad5-derived oncoproteins E1B-55 kDa and E4orf6 (or the corresponding empty expression vectors) were transfected. Luciferase activity was determined 20 h later. Luciferase expression was markedly enhanced by p53 and also by p51 (Fig. 3, compare bars 1 and 2 as well as bars 6 and 7, respectively). In the case of p53, this activity was decreased by E1B-55 kDa (bar 3), was not decreased by E4orf6 (bar 4), but was more profoundly increased by the combination of E1B-55 kDa and E4orf6 (bar 5), as reported previously (37). In contrast, p51-driven luciferase expression was not diminished in the presence of E1B-55 kDa (Fig. 3, bar 8). However, a moderate reduction in p51's activity was observed in the presence of E4orf6, regardless of E1B-55 kDa expression (bars 9 and 10). These results indicate that E1B-55 kDa does not significantly affect p51, while E4orf6 might have some inhibitory effect on p51's transcriptional activity.
FIG. 3.
Inhibitory effects of Ad5 oncoproteins on p53 and p51. H1299 cells were transfected with expression plasmids for p53 (50 ng) or p51A (50 ng), along with the luciferase reporter plasmid pBP100luc (100 ng) and expression plasmids for Ad5 E1B-55 kDa (500 ng) and Ad5 E4orf6 (900 ng) as indicated. As controls, the corresponding empty vector plasmids were transfected instead. The cells were harvested 18 h after transfection, and the relative amount of expressed luciferase was determined. The luciferase activities measured with p53 alone (bar 2) or p51A alone (bar 7) were set 100%, and the other values were normalized accordingly. Error bars represent the standard errors that were calculated from at least three independent experiments.
Impact of Ad12 E1B-55 kDa and E4orf6 on the transcriptional activities of p53 and its homologues p73 and p51.
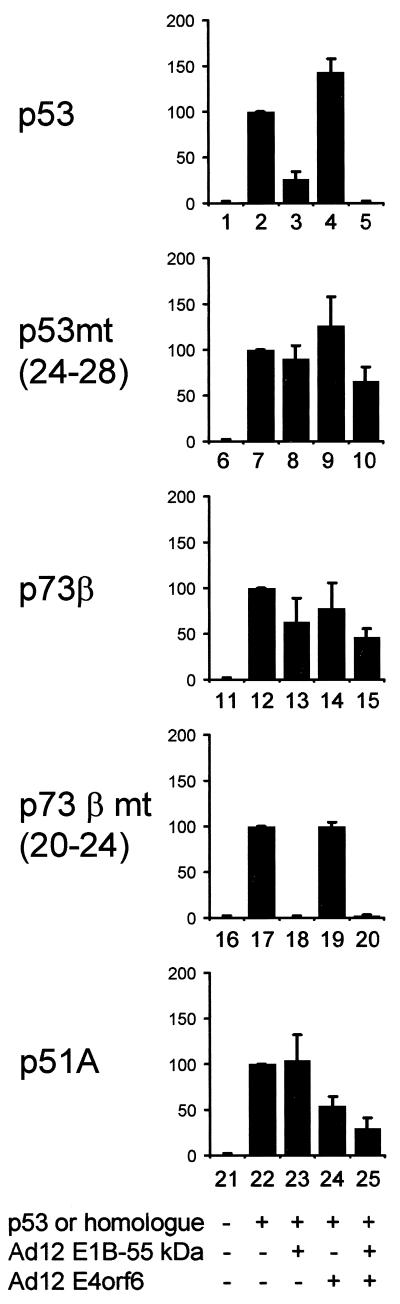
The oncogenic potential of adenovirus in animals strongly depends on the virus type under study. While Ad5 induces tumors with low efficiency or not at all, Ad12 is known to be highly oncogenic (reference 44 and references therein). One possibility to explain this difference is the hypothesis that a different spectrum of cellular growth-regulatory proteins can be targeted by the viral oncoproteins. Therefore, we tested to what extent E1B-55 kDa and E4orf6 from adenovirus type 12 may affect the transcriptional activities of p53 in comparison to its homologues. Expression plasmids for Ad12 E1B-55 kDa and E4orf6 were coexpressed in H1299 cells along with expression plasmids for p53, p73, and p51 and a p53-responsive reporter plasmid. At 18 h after transfection, luciferase activity was determined. p53-driven luciferase expression was inhibited by Ad12 oncoproteins in a pattern very similar to that obtained with Ad5 proteins (Fig. 4, bars 1 to 5). While Ad12 E1B-55 kDa reduced p53-mediated transcription (compare bars 2 and 3), Ad12 E4orf6 did not show such an effect (bar 4). However, in the presence of both E1B-55 kDa and E4orf6, p53 activity was reduced more severely than it was by E1B-55 kDa alone. In the Ad5 system, replacing five amino acids near the amino terminus of p53 (residues 24 to 28) by its homologous sequence from p73 (residues 20 to 24) rendered the protein resistant against inhibition by adenovirus oncoproteins (37). The same turned out to be true when Ad12 proteins were expressed, and no significant inhibition of luciferase expression was observed (Fig. 4, bars 6 to 10). Next, we tested whether the reporter expression driven by p73β might be inhibited in the presence of Ad12 oncoproteins. As shown in Fig. 4 (bars 11 to 15), there was no reduction in activity that was comparable to the effects observed with p53. However, when five amino acids from p53 (residues 24 to 28) were introduced into p73 near the amino terminus of the protein (replacing residues 20 to 24), this rendered the chimeric p73 protein's activity highly sensitive to Ad12 E1B-55 kDa (bar 18). However, this reduction was not further enhanced by E4orf6, even when smaller amounts of E1B-55 kDa expression plasmid were transfected (Fig. 4, bar 20, and data not shown). Finally, the transcriptional activity of p51A in the presence of Ad12 oncoproteins was quantitated. As shown in Fig. 4 (bar 23), no effect of E1B-55 kDa on p51A's activity was observed. However, there was some reduction in p51A's activity when Ad12 E4orf6 was expressed (bars 24 and 25), an effect similar to that seen with Ad5 E4orf6 (Fig. 3, bars 9 and 10). In summary, the p53 homologues under study did not respond to the expression of Ad12 oncoproteins to an extent comparable to that for p53 itself, even though a moderate reduction in activity was observed when p51A was coexpressed with E4orf6 from Ad5 or Ad12.
FIG. 4.
Inhibitory effects of Ad12 oncoproteins on p53, p73, and p51. H1299 cells were transfected with expression plasmids for p53 (50 ng), p73β (20 ng), or p51A (50 ng), along with the luciferase reporter plasmid pBP100luc (100 ng) and expression plasmids for Ad12 E1B-55 kDa (500 ng) and Ad5 E4orf6 (900 ng) as indicated. The cells were subjected to luciferase assays, and the results are presented as described in the legend to Fig. 3. mt, mutant.
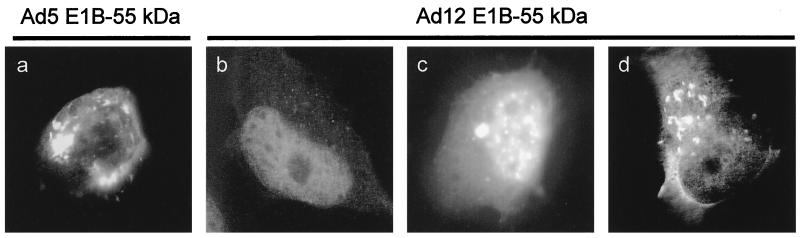
Ad12 E1B-55 kDa localizes to the cytoplasm when expressed at high levels.
To assess the possible intracellular association of Ad12 E1B-55 kDa with p53 in vivo, the intracellular localizations of both proteins were monitored. As a first step to do this, the Ad12 E1B-55 kDa was transiently expressed in H1299 cells with an epitope tag attached to the amino terminus. The transfected cells were immunostained with an antibody to the epitope tag. For comparison, the same procedure was carried out with an expression plasmid for epitope-tagged Ad5 E1B-55 kDa (9). As shown in Fig. 5a, Ad5 E1B-55 kDa is found in cytoplasmic clusters that have been previously described (40). In addition, a small proportion of the protein was frequently detected in the nucleoplasm of the transfected cells (Fig. 5a). When the experiment was performed with Ad12 E1B-55 kDa, the protein was found in the nuclei of cells that stained weakly positive for the epitope tag (Fig. 5b). This was the expected pattern based on previously reported observations (58). However, in cells that strongly expressed the protein, cytoplasmic clusters containing Ad12 E1B-55 kDa were consistently observed (Fig. 5c), and the cytoplasmic staining sometimes even exceeded the nuclear signal (Fig. 6c and e). Similar results were obtained when nontagged Ad12 E1B-55 kDa was expressed and the detection was performed with a monoclonal antibody (8A9B9; a generous gift from T. van Laar) against this protein (data not shown).
FIG. 5.
Intracellular localization of Ad12 E1B-55 kDa. H1299 cells were transfected with expression plasmids for HA-tagged E1B-55 kDa from Ad5 (a) or Ad12 (b to d). The cells were immunostained with a mouse monoclonal antibody against the HA tag (clone HA.11), followed by a fluorescein isothiocyanate-labeled secondary antibody. In the case of Ad12 E1B-55 kDa, cells expressing small (a), moderate (b), or large (c) amounts of the protein are shown. Note that a ca. threefold-shorter exposure time was chosen for panel d.
FIG. 6.
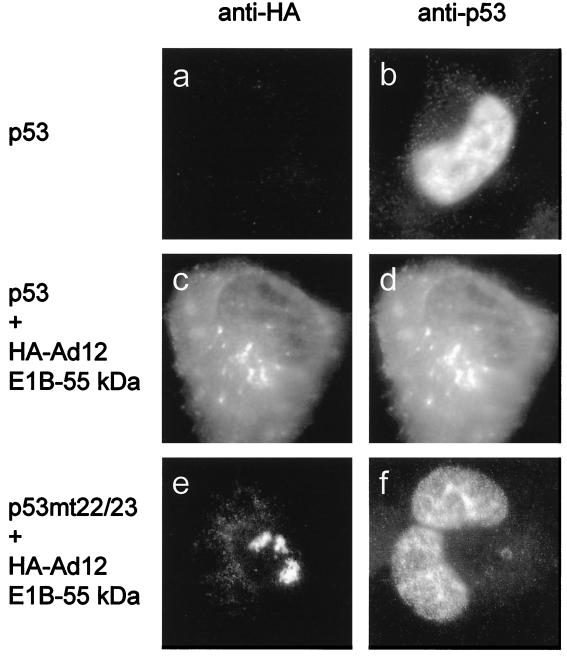
Intracellular colocalization of p53 and Ad12 E1B-55 kDa. H1299 cells were transiently transfected with plasmids (400 ng) expressing HA-tagged Ad12 E1B-55 kDa. In addition, expression plasmids (100 ng) for wild-type or mutant (mt) (L22Q/W23S) p53 were cotransfected as indicated. As a control, the corresponding empty vector was transfected instead of the E1B expression plasmid (a and b). The cells were fixed and stained simultaneously with murine anti-HA antibody and rabbit anti-p53 antibody, followed by secondary antibodies labeled with different fluorescent dyes. Cells with strong expression of Ad12 E1B-55 kDa were monitored. The same areas were visualized for expression of E1B-55 kDa (a, c, and e) and p53 (b, d, and f). Note that in panel e, a cell that expressed Ad12 E1B-55 kDa almost exclusively in the cytoplasm was chosen. This pattern was not always observed (compare Fig. 5) but is shown here to facilitate the observation of any colocalization with p53.
Ad12 E1B-55 kDa relocalizes p53.
Based on the fact that Ad12 E1B-55 kDa can localize in cytoplasmic clusters, we asked whether Ad12 E1B-55 kDa, like its homologue from Ad5, might relocalize p53. To test this, p53 was expressed either along with an empty expression plasmid (Fig. 6a and b) or together with epitope-tagged Ad12 E1B-55 kDa. This was followed by double immunostaining for p53 and the epitope tag. When expressed in the absence of viral proteins, p53 remained predominantly nuclear (Fig. 6b). However, in those cells that contained Ad12 E1B-55 kDa in cytoplasmic clusters, p53 was found to colocalize with it (compare Fig. 6c and d). The same experiment was performed with the L22Q/W23S mutant of p53, which is known not to interact with Ad5 E1B-55 kDa (26) (Fig. 1, compare lanes 2 and 6). In the cells where Ad12 E1B-55 kDa was found in the cytoplasm, this mutant of p53 remained in the nucleus and no colocalization was evident (Fig. 6e and f). These results argue that Ad12 E1B-55 kDa colocalizes with p53 in cytoplasmic clusters and may directly associate with an amino-terminal sequence element of p53 that is also required for the direct interaction of Ad5 E1B-55 kDa with p53.
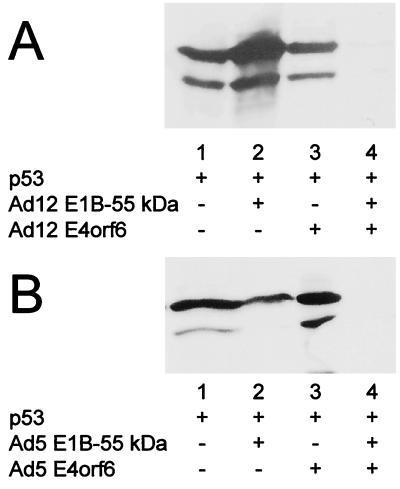
Ad12 E1B-55 kDa and E4orf6 cooperate to reduce p53 levels.
It was previously proposed that Ad12 E1B-55 kDa might not directly contact p53 and interfere with p53's activity by a mechanism that is distinct from the effect of Ad5 E1B-55 kDa (51). On the other hand, the intracellular colocalization of Ad12 E1B-55 kDa (Fig. 6) suggests that the mechanisms of p53 inactivation might be more closely related in the two virus types than previously anticipated. To investigate this in further detail, we determined whether Ad12 E1B-55 kDa and E4orf6 might induce the intracellular destabilization of p53, like the corresponding Ad5 oncoproteins. First, we coexpressed p53 with Ad12 E1B-55 kDa and E4orf6, followed by Western blot detection of p53. While p53 levels were comparable when no adenovirus protein (Fig. 7A, lane 2), Ad12 E1B-55 kDa (lane 3), or Ad12 E4orf6 (lane 4) was coexpressed individually, the amount of p53 was drastically reduced in the presence of both E1B-55 kDa and E4orf6 (lane 5) and was detected only on longer exposures (Fig. 7B, lane 2). This is in perfect analogy to the results previously obtained with the Ad5 oncoproteins (37). Indeed, when p53 was coexpressed along with Ad12 E1B-55 kDa and Ad5 E4orf6, a similar reduction in p53 amounts was found (data not shown), further arguing that the mechanism of cooperation between the two proteins is conserved between the highly oncogenic virus and the nononcogenic virus. When the experiment was carried out with the L22Q/W23S mutant of p53, no significant reduction of the p53 levels was observed (Fig. 7B, lanes 3 to 6). This argues that the intracellular association of p53 with Ad12 E1B-55 kDa is a requirement for the suppression of p53 levels in the presence of Ad12 E1B-55 kDa and Ad12 E4orf6, a situation that again parallels the effects observed with Ad5 oncoproteins (37).
FIG. 7.
Reduction of p53 levels by Ad12 oncoproteins. H1299 cells were transfected with expression plasmids for p53 (wild type or mutant [mt] L22Q/W23S; 300 ng), Ad12 E1B-55 kDa (300 ng), and Ad12 E4orf6 (900 ng) or the corresponding vector constructs, as indicated below the lanes. At 24 h after transfection, the cells were harvested and subjected to SDS-PAGE and Western blotting. p53 was visualized with a monoclonal antibody (clone 1801), followed by a peroxidase-coupled secondary antibody and chemiluminescence detection. Panel B shows a longer exposure than panel A, allowing the detection of residual p53 in the presence of both adenovirus oncoproteins.
E1B-55 kDa and E4orf6 of Ad12 destabilize p53.
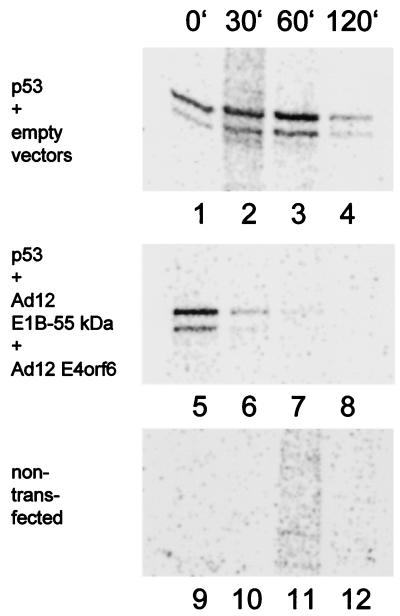
We speculated that the reduction of p53 levels in the presence of Ad12 E1B-55 kDa and Ad12 E4orf6 was due to destabilization of p53. To test this, p53 was coexpressed with the Ad12 oncoproteins (Fig. 8, lanes 5 to 8) or with the corresponding empty vector constructs (lanes 1 to 4), followed by a short (10-min) pulse of radioactive protein labeling. The cells were harvested either immediately (Fig. 8, lanes 1, 5, and 9) or after further incubation in complete medium without radioactively labeled amino acids for various periods of time. After harvesting, p53 was immunoprecipitated and visualized on an SDS-polyacrylamide gel by autoradiography. While p53 remained relatively stable when expressed alone (Fig. 8, lanes 1 to 4), it was rapidly degraded in the presence of Ad12 E1B-55 kDa and E4orf6 (lanes 5 to 8). Thus, Ad12 E1B-55 kDa and E4orf6 reduce p53 levels by triggering p53's intracellular degradation.
FIG. 8.
Destabilization of p53 by Ad12 oncoproteins. Expression plasmids for the p53 (600 ng), Ad12 E1B-55 kDa (300 ng), and Ad12 E4orf6 (600 ng) or empty vector constructs were transfected into H1299 cells as indicated. After 24 h, the cells were labeled with [35S]methionine and [35S]cysteine for 10 min and then incubated in nonradioactive medium (chase). After the time points indicated (minutes), the cells were harvested and subjected to immunoprecipitation with monoclonal antibody Pab421 directed against p53, followed by SDS-PAGE and autoradiography. Note that the signal intensities obtained with p53 alone initially increase, possibly reflecting the incorporation of radioactively labeled amino acids that were internalized into the cells but not yet assembled into protein at the start of the chase.
Reduction of p53 levels by Ad5 and Ad12 E1B-55 kDa and E4orf6 occurs in rodent cells.
The differences in oncogenicity between Ad5 and Ad12 were determined mainly in rodents and rodent cells (44). In most rodent cells, E1B-55 kDa and E4orf6 of Ad5 were reported not to colocalize (14) while both of the homologous Ad12 proteins are found in the nucleus when expressed in small to moderate amounts (30, 51). Therefore, we considered the possibility that oncoproteins from the two viruses might differ in their ability to cooperatively destabilize p53 in rodent cells. To test this, we cotransfected epitope-tagged p53 (to distinguish it from endogenous p53) with E1B-55 kDa and E4orf6 of Ad5 and Ad12 into murine NIH 3T3 cells. Subsequently, p53 levels were determined by Western blot analysis. The pattern of p53 levels did not significantly differ from the situation found in human cells: in the presence of Ad12 or Ad5 E1B-55 kDa and E4orf6 in combination, the amount of p53 was suppressed below detectability (Fig. 9). The same pattern was observed when hamster BHK cells were used (data not shown). We conclude that E1B-55 kDa and E4orf6 from each virus still cooperate to destabilize p53 in rodent cells, even though in these cells, the Ad5 proteins did not detectably colocalize in previous studies (14).
FIG. 9.
Reduction of p53 levels by adenovirus oncoproteins in murine cells. NIH 3T3 cells were transfected with expression plasmids for HA-tagged p53, Ad12 (A) or Ad5 (B) E1B-55 kDa (300 ng), and E4orf6 (900 ng) or the corresponding vector constructs, as indicated below the lanes. At 24 h after transfection, the cells were harvested, and p53 was detected by Western blotting with an antibody to the HA tag.
Destabilization by Ad12 E1B-55 kDa and E4orf6 does not affect p73.
Finally, we asked whether destabilization by Ad12 oncoproteins is limited to p53. H1299 cells were transfected with expression plasmids for Ad12 oncoproteins and p73 or p53. All p53 family members were carboxy-terminally HA tagged to allow direct comparison of protein levels. The intracellular p73 levels were not affected by Ad12 E1B-55 kDa and E4orf6 (Fig. 10, compare lanes 1 and 2). When amino acids 20 to 24 of p73 were replaced by the homologous p53 sequence, this rendered p73-mediated transcription inhibitable by Ad12 E1B-55 kDa (Fig. 4, bars 16 to 20). Nonetheless, this chimera was resistant to destabilization mediated by Ad12 oncoproteins (Fig. 10, lanes 3 and 4). In contrast, p53 levels were strongly suppressed by Ad12 E1B-55 kDa and E4orf6 (compare lanes 5 and 6). Thus, destabilization by Ad12 oncoproteins is specific for p53 and apparently requires different portions of p53 in addition to the amino-terminal putative E1B-binding domain.
FIG. 10.
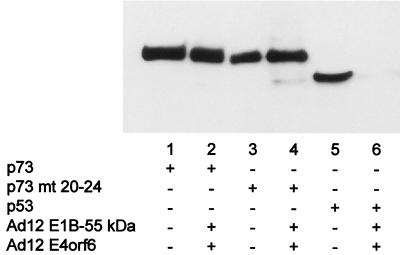
Reduction of p53 levels but not p73 levels by Ad12 oncoproteins. H1299 cells were transfected with expression plasmids for HA-tagged p53, HA-tagged p73β (wild type or with amino acids 20 to 24 replaced by the corresponding p53 sequence [mt 20-24]; 300 ng), nontagged Ad12 E1B-55 kDa (300 ng), and non-tagged Ad12 E4orf6 (900 ng) or the corresponding vector constructs, as indicated below the lanes. At 24 h after transfection, the cells were harvested and subjected to SDS-PAGE and Western blotting. p53 and p73 were visualized with a monoclonal antibody against the HA tag (clone HA.11), followed by a peroxidase-coupled secondary antibody and chemiluminescence detection.
DISCUSSION
Our results strongly suggest that p51, like p73, does not represent a target of the adenovirus E1B-55 kDa oncoprotein. The E1B proteins of differentially oncogenic adenoviruses reveal the same preference for p53 while “neglecting” its homologues. Further, the cooperation between E1B-55 kDa and E4orf6 to destabilize p53 is observed with adenoviruses of different oncogenicities, despite the previously reported differences in the mechanisms of p53 inactivation by Ad5 and Ad12 E1B-55 kDa.
There was a moderate but significant reduction of p51 activity in the presence of E4orf6 from either virus type. Such an inhibition by E4orf6 alone has previously reported for p53 (10) and p73 (17). Another group has described inhibition of p73 but not p53 by E4orf6 (46). In our hands, the transcriptional activities of p53 and p73 were not compromised by E4orf6 alone (this report and reference 37), while p51 is inhibited to some extent by E4orf6. It remains to be determined what factor(s) in the experimental systems used by the different research groups results in the observed discrepancies in transcriptional inhibition and whether this inhibition is of physiological relevance during virus infection and virus-mediated oncogenesis. In these natural settings, E4orf6 is coexpressed with E1B-55 kDa, with the latter reaching its peak levels earlier than the former (44). Further, stably transfected cells expressing E4orf6 are not viable unless they are expressing E1B-55 kDa in addition (our unpublished observations). The cooperation between the two viral oncoproteins reduces p53-mediated transcription to a far greater extent than E4orf6 reduces any p53 family member's activity. This cooperative effect of both proteins, however, was shown to be strictly limited to p53, sparing p73 and p51. Hence, it still appears that adenoviruses evolved to target p53 with strong preference over its homologues p73 and p51. Similar results were obtained with the large T antigen of simian virus 40. This protein also interacts with and inhibits p53 but not p73 or p51 (reference 8 and our unpublished observations).
E1B-55 kDa and E4orf6 of Ad5 have been shown to colocalize in the nuclei of primate cells. While E4orf6 relocalizes E1B-55 kDa from the cytoplasm to the nucleus (14), E1B-55 kDa enables E4orf6 to shuttle between the nucleus and cytoplasm (9). In contrast, the two proteins fail to colocalize detectably in murine cells and most other rodent cells (14). Since colocalization in these cells can be achieved by fusing the transfected rodent cells to primate cells, it was hypothesized that a cellular factor that mediates the association of E1B-55 kDa with E4orf6 might exist in primate but not rodent cells. This factor is apparently not p53, since the viral proteins were found to colocalize in primate cells that contain very low levels of p53 (HeLa cells [14]) or lack p53 entirely (H1299 cells [our unpublished observations]). Nonetheless, we show here that the two viral proteins cooperate in rodent cells to reduce p53 levels. Possibly, only small fractions of the two proteins associate in these cells, and this minor population, albeit insufficient for detection by immunostaining, might be sufficient for p53 degradation. Alternatively or in addition, the two viral proteins could transiently associate and form a trimeric complex with p53 which is disrupted rapidly upon degradation of p53.
The hypothesis that viruses of different oncogenic potential might target different spectra of p53 family members seemed attractive. However, both viruses evolved inhibitory mechanisms that are highly specific for p53. Why did the viruses not adopt similar mechanisms to downregulate the transcriptional activities of p51 and p73? Despite their lack of ability to inhibit p51 and p73, the E1B-55 kDa from either virus, along with E1A, has considerable transforming activity. This phenomenon can be explained in two different, but not mutually exclusive, ways. One possibility is that p73 and p51 lack a function retained in p53 that is crucial for tumor suppression. For instance, it has been reported that p73 shows somewhat different preferences for certain promoters compared to p53 (61). Alternatively, it is possible that while p53 and its homologues could largely substitute for each other, only the levels of p53 are upregulated during virus infection and/or genotoxic stress. In any case, the specificity of viral oncoproteins for p53 strongly suggests a unique role of p53 as a “guardian of the genome” (22).
In the case that the principal functions of p51, p73, and possibly other p53 homologues do not consist of genome stabilization and tumor suppression, what else might be the reason for their evolutionary conservation? Possibly, the proteins may act as regulators of cell differentiation and/or apoptosis during embryonic development. Their activities might be regulated not only by expression levels but also by the proportions of different splicing variants (6, 53), some of which are apparently not transcriptionally active. An important role of p51/p63 was recently shown by gene targeting in mice (29, 54). p51−/− mice show severe defects in limb development, while the p53−/− genotype does not grossly affect embryonic development (11).
The specificity for p53 family members does not detectably differ between the E1B-55 kDa proteins of Ad5 and Ad12. Nonetheless, these proteins were shown to behave differently during tumor formation in animals (3) and during transformation of cells in tissue culture (51). Another explanation for this difference might be that the E1B-55 kDa proteins of both viruses target p53 but that Ad12 E1B does so by a different and possibly more efficient mechanism. This hypothesis was previously proposed (51) and was based on the following observations: (i) the E1B-55 kDa proteins from the two viruses differ in their transforming efficiencies when combined with various other oncoproteins; (ii) Ad5 E1B-55 kDa is largely cytoplasmic, whereas Ad12 E1B-55 kDa resides predominantly in the nucleus; and (iii) the interaction between p53 and Ad5 E1B-55 kDa is readily detectable, while p53 coprecipitates with Ad12 E1B-55 kDa weakly or not at all. Despite these differences, many features of p53 inactivation are conserved between the two viruses' oncoproteins: (i) p53 is stabilized when coexpressed with E1B-55 kDa from either virus for extended periods of time (58); (ii) E1B-55 kDa from either virus colocalizes with p53 in cytoplasmic clusters, at least when expressed to high levels; (iii) mutational analysis shows that E1B-55 kDa from each virus requires a region near the amino terminus of p53 for this colocalization; and (iv) E1B-55 kDa and E4orf6 from each virus cooperate to destabilize p53, and this destabilization again requires the intact amino-terminal domain of p53. Taken together, these findings suggest that Ad12 E1B-55 kDa, like Ad5 E1B-55 kDa, directly associates with the amino terminus of p53. This association, along with the presence of a repression domain within E1B-55 kDa (56), would be sufficient to explain the inhibition of p53's transcriptional activity.
Thus, there are two open questions concerning oncogenic transformation by Ad12 E1B-55 kDa. First, it is unclear why this protein apparently associates with p53 in vivo but does so only poorly, if at all, in vitro. Possibly, the putative complex of Ad12 E1B-55 kDa and p53 contains other cellular components that cannot be solubilized during cell lysis. Thus, the complex might fall apart when trying to coprecipitate the proteins. The second question that remains is why the transforming properties of the Ad5 and Ad12 E1B-55 kDa proteins are different when assayed together with other oncogene products (51). The answer may be that these proteins have additional transforming activities not directly related to transcriptional inactivation of p53. Such activities might involve the alteration of genomic stability, as has been reported for Ad12 E1B-55 kDa (24, 25). In the case of tumorigenesis in animals, another layer of complexity is represented by the immune response to the virus-transformed cells. This could be affected by any sequence element divergent between the two virus types, depending on the animal's major histocompatibility complex haplotypes.
Viruses have adopted and refined strategies to manipulate the cell's growth and survival over an extensive period of evolution. Viral oncoproteins thus continue to represent valuable tools in defining the cell's growth-regulatory mechanisms. The exact definition of their cellular targets constitutes an important step in the evaluation of cellular growth regulators. In the case of p53 and its homologues, future studies are required to elucidate the regulation of other p53 family members by viral oncoproteins. The differential interaction of p53, p73, and p51 with adenovirus proteins strongly suggests a superior role of p53 in tumor suppression.
ACKNOWLEDGMENTS
We thank H.-D. Klenk and R. Arnold for their generous support; T. van Laar and T. Shenk for antibodies; S. Ikawa, T. van Laar, A. Zantema, and A. van der Eb for plasmids; W. Doerfler for Ad12; and C. König and S. Weigel for helpful discussions.
This work was supported by the German Research Foundation and the P. E. Kempkes Foundation. S.W. received a fellowship from the Hoechst scholarship foundation, and M.D. was a recipient of the Stipendium für Infektionsbiologie from the German Cancer Research Center.
REFERENCES
- 1.Bailey A D, Li Z, Pavelitz T, Weiner A M. Adenovirus type 12-induced fragility of the human RNU2 locus requires U2 small nuclear RNA transcriptional regulatory elements. Mol Cell Biol. 1995;15:6246–6255. doi: 10.1128/mcb.15.11.6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker S J, Markowitz S, Fearon E R, Willson J K, Vogelstein B. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science. 1990;249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 3.Bernards R, Schrier P I, Bos J L, Van der Eb A J. Role of adenovirus types 5 and 12 early region 1b tumor antigens in oncogenic transformation. Virology. 1983;127:45–53. doi: 10.1016/0042-6822(83)90369-0. [DOI] [PubMed] [Google Scholar]
- 4.Bian J, Sun Y. p53CP, a putative p53 competing protein that specifically binds to the consensus p53 DNA binding sites: a third member of the p53 family? Proc Natl Acad Sci USA. 1997;94:14753–14758. doi: 10.1073/pnas.94.26.14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blair Zajdel M E, Blair G E. The intracellular distribution of the transformation-associated protein p53 in adenovirus-transformed rodent cells. Oncogene. 1988;2:579–584. [PubMed] [Google Scholar]
- 6.De Laurenzi V, Costanzo A, Barcaroli D, Terrinoni A, Falco M, Annicchiarico-Petruzzelli M, Levrero M, Melino G. Two new p73 splice variants, gamma and delta, with different transcriptional activity. J Exp Med. 1998;188:1763–1768. doi: 10.1084/jem.188.9.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dobbelstein M, Arthur A K, Dehde S, van Zee K, Dickmanns A, Fanning E. Intracistronic complementation reveals a new function of SV40 T antigen that co-operates with Rb and p53 binding to stimulate DNA synthesis in quiescent cells. Oncogene. 1992;7:837–847. [PubMed] [Google Scholar]
- 8.Dobbelstein M, Roth J. The large T antigen of simian virus 40 binds and inactivates p53 but not p73. J Gen Virol. 1998;79:3079–3083. doi: 10.1099/0022-1317-79-12-3079. [DOI] [PubMed] [Google Scholar]
- 9.Dobbelstein M, Roth J, Kimberly W T, Levine A J, Shenk T. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 1997;16:4276–4284. doi: 10.1093/emboj/16.14.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dobner T, Horikoshi N, Rubenwolf S, Shenk T. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science. 1996;272:1470–1473. doi: 10.1126/science.272.5267.1470. [DOI] [PubMed] [Google Scholar]
- 11.Donehower L A, Harvey M, Slagle B L, McArthur M J, Montgomery C A, Jr, Butel J S, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 12.Freedman D A, Epstein C B, Roth J C, Levine A J. A genetic approach to mapping the p53 binding site in the MDM2 protein. Mol Med. 1997;3:248–259. [PMC free article] [PubMed] [Google Scholar]
- 13.Gallimore P H, Lecane P S, Roberts S, Rookes S M, Grand R J, Parkhill J. Adenovirus type 12 early region 1B 54K protein significantly extends the life span of normal mammalian cells in culture. J Virol. 1997;71:6629–6640. doi: 10.1128/jvi.71.9.6629-6640.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodrum F D, Shenk T, Ornelles D A. Adenovirus early region 4 34-kilodalton protein directs the nuclear localization of the early region 1B 55-kilodalton protein in primate cells. J Virol. 1996;70:6323–6335. doi: 10.1128/jvi.70.9.6323-6335.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grand R J A, Grant M L, Gallimore P H. Enhanced expression of p53 in human cells infected with mutant adenoviruses. Virology. 1994;203:229–240. doi: 10.1006/viro.1994.1480. [DOI] [PubMed] [Google Scholar]
- 16.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 17.Higashino F, Pipas J M, Shenk T. Adenovirus e4orf6 oncoprotein modulates the function of the p53-related protein, p73. Proc Natl Acad Sci USA. 1998;95:15683–15687. doi: 10.1073/pnas.95.26.15683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jost C A, Marin M C, Kaelin W G., Jr p73 is a human p53-related protein that can induce apoptosis. Nature. 1997;389:191–194. doi: 10.1038/38298. [DOI] [PubMed] [Google Scholar]
- 19.Kaelin W G., Jr The emerging p53 gene family. J Natl Cancer Inst. 1999;91:594–598. doi: 10.1093/jnci/91.7.594. [DOI] [PubMed] [Google Scholar]
- 20.Kaghad M, Bonnet H, Yang A, Creancier L, Biscan J C, Valent A, Minty A, Chalon P, Lelias J M, Dumont X, Ferrara P, McKeon F, Caput D. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 21.Kubbutat M H, Jones S N, Vousden K H. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 22.Lane D P. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 23.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 24.Li Z, Yu A, Weiner A M. Adenovirus type 12-induced fragility of the human RNU2 locus requires p53 function. J Virol. 1998;72:4183–4191. doi: 10.1128/jvi.72.5.4183-4191.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao D, Yu A, Weiner A M. Coexpression of the adenovirus 12 E1B 55 kDa oncoprotein and cellular tumor suppressor p53 is sufficient to induce metaphase fragility of the human RNU2 locus. Virology. 1999;254:11–23. doi: 10.1006/viro.1998.9512. [DOI] [PubMed] [Google Scholar]
- 26.Lin J, Chen J, Elenbaas B, Levine A J. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 27.Mai M, Yokomizo A, Qian C, Yang P, Tindall D J, Smith D I, Liu W. Activation of p73 silent allele in lung cancer. Cancer Res. 1998;58:2347–2349. [PubMed] [Google Scholar]
- 28.Marin M C, Jost C A, Irwin M S, DeCaprio J A, Caput D, Kaelin W G., Jr Viral oncoproteins discriminate between p53 and the p53 homolog p73. Mol Cell Biol. 1998;18:6316–6324. doi: 10.1128/mcb.18.11.6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mills A A, Zheng B, Wang X J, Vogel H, Roop D R, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 30.Moore M, Horikoshi N, Shenk T. Oncogenic potential of the adenovirus E4orf6 protein. Proc Natl Acad Sci USA. 1996;93:11295–11301. doi: 10.1073/pnas.93.21.11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nomoto S, Haruki N, Kondo M, Konishi H, Takahashi T. Search for mutations and examination of allelic expression imbalance of the p73 gene at 1p36.33 in human lung cancers. Cancer Res. 1998;58:1380–1383. [PubMed] [Google Scholar]
- 32.Osada M, Ohba M, Kawahara C, Ishioka C, Kanamaru R, Katoh I, Ikawa Y, Nimura Y, Nakagawara A, Obinata M, Ikawa S. Cloning and functional analysis of human p51, which structurally and functionally resembles p53. Nat Med. 1998;4:839–843. doi: 10.1038/nm0798-839. [DOI] [PubMed] [Google Scholar]
- 33.Prabhu N S, Somasundaram K, Satyamoorthy K, Herlyn M, El-Deiry W S. p73, unlike p53, suppresses growth and induces apoptosis of human papillomavirus E6-expressing cancer cells. Int J Oncol. 1998;13:5–9. doi: 10.3892/ijo.13.1.5. [DOI] [PubMed] [Google Scholar]
- 34.Querido E, Marcellus R C, Lai A, Charbonneau R, Teodoro J G, Ketner G, Branton P E. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J Virol. 1997;71:3788–3798. doi: 10.1128/jvi.71.5.3788-3798.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roth J, Dittmer D, Rea D, Tartaglia J, Paoletti E, Levine A J. p53 as a target for cancer vaccines: recombinant canarypox virus vectors expressing p53 protect mice against lethal tumor cell challenge. Proc Natl Acad Sci USA. 1996;93:4781–4786. doi: 10.1073/pnas.93.10.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roth J, Dobbelstein M, Freedman D A, Shenk T, Levine A J. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roth J, König C, Wienzek S, Weigel S, Ristea S, Dobbelstein M. Inactivation of p53 but not p73 by adenovirus type 5 E1B 55-kilodalton and E4 34-kilodalton oncoproteins. J Virol. 1998;72:8510–8516. doi: 10.1128/jvi.72.11.8510-8516.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarnow P, Hearing P, Anderson C W, Halbert D N, Shenk T, Levine A J. Adenovirus early region 1B 58,000-dalton tumor antigen is physically associated with an early region 4 25,000-dalton protein in productively infected cells. J Virol. 1984;49:692–700. doi: 10.1128/jvi.49.3.692-700.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarnow P, Ho Y S, Williams J, Levine A J. Adenovirus E1b-58kd tumor antigen and SV40 large tumor antigen are physically associated with the same 54 kd cellular protein in transformed cells. Cell. 1982;28:387–394. doi: 10.1016/0092-8674(82)90356-7. [DOI] [PubMed] [Google Scholar]
- 40.Sarnow P, Sullivan C A, Levine A J. A monoclonal antibody detecting the adenovirus type 5-E1b-58Kd tumor antigen: characterization of the E1b-58Kd tumor antigen in adenovirus-infected and -transformed cells. Virology. 1982;120:510–517. doi: 10.1016/0042-6822(82)90054-x. [DOI] [PubMed] [Google Scholar]
- 41.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 42.Schmale H, Bamberger C. A novel protein with strong homology to the tumor suppressor p53. Oncogene. 1997;15:1363–1367. doi: 10.1038/sj.onc.1201500. [DOI] [PubMed] [Google Scholar]
- 43.Senoo M, Seki N, Ohira M, Sugano S, Watanabe M, Tachibana M, Tanaka T, Shinkai Y, Kato H. A second p53-related protein, p73L, with high homology to p73. Biochem Biophys Res Commun. 1998;248:603–607. doi: 10.1006/bbrc.1998.9013. [DOI] [PubMed] [Google Scholar]
- 44.Shenk T. Adenoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2111–2148. [Google Scholar]
- 45.Steegenga W T, Riteco N, Jochemsen A G, Fallaux F J, Bos J L. The large E1B protein together with the E4orf6 protein target p53 for active degradation in adenovirus infected cells. Oncogene. 1998;16:349–357. doi: 10.1038/sj.onc.1201540. [DOI] [PubMed] [Google Scholar]
- 46.Steegenga W T, Shvarts A, Riteco N, Bos J L, Jochemsen A G. Distinct regulation of p53 and p73 activity by adenovirus E1A, E1B, and e4orf6 proteins. Mol Cell Biol. 1999;19:3885–3894. doi: 10.1128/mcb.19.5.3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sunahara M, Ichimiya S, Nimura Y, Takada N, Sakiyama S, Sato Y, Todo S, Adachi W, Amano J, Nakagawara A. Mutational analysis of the p73 gene localized at chromosome 1p36.3 in colorectal carcinomas. Int J Oncol. 1998;13:319–323. [PubMed] [Google Scholar]
- 48.Takahashi H, Ichimiya S, Nimura Y, Watanabe M, Furusato M, Wakui S, Yatani R, Aizawa S, Nakagawara A. Mutation, allelotyping, and transcription analyses of the p73 gene in prostatic carcinoma. Cancer Res. 1998;58:2076–2077. [PubMed] [Google Scholar]
- 49.Tanaka M, Herr W. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell. 1990;60:375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- 50.Trink B, Okami K, Wu L, Sriuranpong V, Jen J, Sidransky D. A new human p53 homologue. Nat Med. 1998;4:747–748. doi: 10.1038/nm0798-747. [DOI] [PubMed] [Google Scholar]
- 51.van den Heuvel S J, van Laar T, The I, van der Eb A J. Large E1B proteins of adenovirus types 5 and 12 have different effects on p53 and distinct roles in cell transformation. J Virol. 1993;67:5226–5234. doi: 10.1128/jvi.67.9.5226-5234.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Werness B A, Levine A J, Howley P M. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 53.Yang A, Kaghad M, Wang Y, Gillett E, Fleming M D, Dotsch V, Andrews N C, Caput D, McKeon F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 54.Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson R T, Tabin C, Sharpe A, Caput D, Crum C, McKeon F. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 55.Yew P R, Berk A J. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature. 1992;357:82–85. doi: 10.1038/357082a0. [DOI] [PubMed] [Google Scholar]
- 56.Yew P R, Liu X, Berk A J. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 1994;8:190–202. doi: 10.1101/gad.8.2.190. [DOI] [PubMed] [Google Scholar]
- 57.Zantema A, Fransen J A, Davis-Olivier A, Ramaekers F C, Vooijs G P, DeLeys B, Van der Eb A J. Localization of the E1B proteins of adenovirus 5 in transformed cells, as revealed by interaction with monoclonal antibodies. Virology. 1985;142:44–58. doi: 10.1016/0042-6822(85)90421-0. [DOI] [PubMed] [Google Scholar]
- 58.Zantema A, Schrier P I, Davis-Olivier A, van Laar T, Vaessen R T, van der Eb A. Adenovirus serotype determines association and localization of the large E1B tumor antigen with cellular tumor antigen p53 in transformed cells. Mol Cell Biol. 1985;5:3084–3091. doi: 10.1128/mcb.5.11.3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zeng X, Levine A J, Lu H. Non-p53 p53RE binding protein, a human transcription factor functionally analogous to P53. Proc Natl Acad Sci USA. 1998;95:6681–6686. doi: 10.1073/pnas.95.12.6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu H, Shen Y, Shenk T. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J Virol. 1995;69:7960–7970. doi: 10.1128/jvi.69.12.7960-7970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu J, Jiang J, Zhou W, Chen X. The potential tumor suppressor p73 differentially regulates cellular p53 target genes. Cancer Res. 1998;58:5061–5065. [PubMed] [Google Scholar]