Abstract
Objective:
This study aims to elucidate anti-liver cancer components and potential mechanisms of Curcumae Rhizoma and Hedyotis diffusa Willd (CR-HDW).
Methods:
Effective components and targets of CR-HDW were identified from the Traditional Chinese Medicine Systems Pharmacology (TCMSP) database. Liver cancer-related genes were collected from GeneCards, Gene-Disease Association (DisGeNET), and National Center for Biotechnology Information (NCBI). Protein-protein interaction networks, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment were conducted to analyze the identified genes. Molecular docking was used to simulate binding of the active components and their target proteins. Cell activity assay, western blot, and senescence-associated β-galactosidase (SA-β-gal) experiments were conducted to validate core targets identified from molecular docking.
Results:
Ten active compounds of CR-HDW were identified including quercetin, 3-epioleanic acid and hederagenin. The primary core proteins comprised Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), Protein Kinase B(AKT1), etc. The pathways for Phosphoinositide 3-kinase (PI3K)/ AKT, cellular senescence, Fork head boxO (FOXO) were revealed as important for anti-cancer activity of CR-HDW. Molecular docking demonstrated strong binding between liver cancer target proteins and major active components of CR-HDW. In-vitro experiments confirmed that hederagenin and 3-epioleolic acid inhibited HuH-7 cell growth, reduced expression of PI3K, AKT, and mechanistic target of rapamycin (mTOR) proteins. Hederagenin also induced HuH-7 senescence.
Conclusions:
In summary, The authors’ results suggest that the CR-HDW component (Hederagenin, 3-epoxy-olanolic acid) can inhibit the proliferation of HuH-7 cells by decreasing PI3K, AKT, and mTOR. Hederagenin also induced HuH-7 senescence.
Keywords: curcumae Rhizoma, Hedyotis diffusa willd, liver cancer, hederagenin, 3-epioleolic acid
Introduction
Highlights
Network pharmacology analysis elucidated ten active constituents including hederagenin and 3-epioleolic acid.
One hundred fifty-one potential targets were predicted for the 10 components in the treatment of liver cancer.
Pathways important for cancer proliferation, viral infection, cellular senescence were revealed as critical for anti-tumour activity of the active components.
Hederagenin and 3-epioleolic acid inhibited HuH-7 proliferation, protein expression of PI3K, AKT, and mTOR.
According to data from the WHO, primary liver cancer was the third leading cause of cancer-related deaths worldwide in 2022, with ~865 000 new cases and 760 000 fatalities1. The majority of these cases occurred in East Asia (Mongolia and China), Southeast Asia (Thailand, Cambodia, and Vietnam), and regions and countries in North and West Africa (Egypt and Niger)1. Current treatment modalities encompass surgery, radiotherapy, interventional therapy, targeted drug therapy, immunotherapy, ablation therapy, among others. However, owing to factors such as cancer heterogeneity, drug resistance, and side effects, patient survival rates remain notably low2,3. Hence, there is a pressing need for novel drugs and treatment regimens. In contrast to conventional chemotherapy drugs, traditional Chinese medicine (TCM) has shown promising effects in inhibiting tumour development, mitigating the side effects of radiotherapy and chemotherapy, and enhancing the overall life quality of cancer patients4.
According to TCM theory, the aetiology of liver cancer is attributed to factors such as blood yu (stasis), du (toxicity), and xu (deficiency). Treatment strategies involve approaches such as huo-xue-hua-yu (removing blood stasis), or qing-re-jie-du (clearing heat and detoxifying)5,6. According to TCM principles, huo-xue-hua-yu could enhance blood circulation, thereby inhibiting cancer growth. This aligns with modern medical theories, where tumour development is closely associated with abnormalities in angiogenesis and the coagulation system7,8. A retrospective cohort study on the mortality rate of patients with inoperable giant hepatocellular carcinoma (≥10 cm) treated with TCM formula showed significantly higher 3-year overall survival rates, with a median survival time extended by ~3 months. Core herbals identified from the formula were Hedyotis diffusa Willd (HDW, Chinese BAIHUASHESHECAO) and Curcuma Rhizoma (CR, Chinese herbal medicine EZHU)9. Based on TCM theory, CR functions as xing-qi-san-yu-zhi-tong (promoting qi circulation, resolving blood stasis and relieving pain) and is used primarily for treatment of diseases such as zheng-jia-ji-ju (Substances such as qi, blood, toxins, etc., accumulate to form solid masses). Modern pharmacological studies have shown that the main active compounds of CR possess anti-thrombotic, anti-tumour, anti-bacterial, anti-inflammatory, and antiviral effects10. The extracts of CR inhibited the growth of human liver cancer cell lines (HepG2)11. HDW is known for its efficacy in qing-re-jie-du (clearing heat and detoxification), hua-yu-zhi-tong (resolving blood stasis and relieving pain) and has been used primarily for the treatment of abscesses and swellings5,12. Recent pharmacological research revealed that the primary bioactive compounds of HDW consist of anthraquinones and flavonoids. A lot of publications also found out that HDW exerted anti-angiogenic effects by regulating multiple pathways such as AKT1, IL-6, IL-1β, HIF-1α, and TNF-α13. However, the pharmacological basis and molecular mechanisms of CR and HDW in the treatment of liver cancer have not been fully elucidated.
Network pharmacology utilizes large-scale data and computer technology to investigate the intricate network of interactions between drug molecules and disease entities, including drug targets, pathways, protein-protein interactions, etc.14. The complex composition and diverse targets of CR-HDW present substantial challenges in elucidating the mechanism of action of their active constituents. Network pharmacology provides a novel approach for expediting drug development and comprehending the intricate interconnections between biological systems and diseases15. This study endeavoured to employ a synergistic methodology, integrating network pharmacology, molecular docking, and in-vitro experiments, to elucidate the active compounds, their protein targets, and the mechanism of action of the active compounds in the treatment of liver cancer.
Materials and methods
Cell lines and reagents
The human liver cancer cell line HuH-7 (CL-0120), Dulbecco’s modified eagle medium (DMEM, PM150210), foetal bovine serum (FBS, 164210-50) were purchased from ProCell (China). Hederagenin (HY-N0256) and 3-epioleanolic acid (HY-N4290) were purchased from MedChemExpress (USA). Enhanced Cell Counting Kit 8 (WST-8/CCK-8, E-CK-A362) was purchased from ElabScience (China). Mouse anti-beta-actin monoclonal antibody (3700T), rabbit antibodies for phosphoinositide 3-kinase (PI3K, 4249T ), protein kinase B (AKT, 4691T), heat shock protein 90 alpha family class A member 1 (HSP90AA1, 4877T) and mammalian target of mammalian target of rapamycin (mTOR, 983T) were purchased from Cell Signaling Technology (USA). Senescence-associated β-galactosidase (SA-β-gal) staining kit (C0602), radioimmunoprecipitation assay lysis buffer (RIPA, P0013B), and the bicinchoninic acid (BCA) protein quantification kit (P0012), and electrochemiluminescence (ECL) kit (P0018S) were purchased from Beyotime (China). Varioskan Lux microplate reader was from Thermoscientific and was used to quantify cell viability using CCK-8. ChemiDoc XRS+ was from Bio-Rad (USA) and was used to capture western blotting images.
Stock solutions of hederagenin and 3-epioleanolic acid were prepared by dissolving the chemicals in DMSO to make 10 mM stock solution and stored at −20oC before use. To male working solution, the stock solution was initially diluted 100-fold into DMEM containing 10% FBS to make 100 uM solution and then diluted with DMEM tontaining 10% FBS and 1% DMSO to make different concentrations of compounds for use.
Acquisition of effective components and targets of CR-HDW
Compound data of CR-HDW were retrieved from the TCMSP database (https://old.tcmsp-e.com/index.php) using the keyword “Curcumae Rhizoma and Hedyotis diffusa Willd”. Compounds were selected based on an oral bioavailability (OB) of greater than or equal to 30% and a drug-likeness (DL) of greater than or equal to 0.1816. The SMILES IDs of these compounds were queried in the PubChem database (https://pubchem.ncbi.nlm.nih.gov/)17 to retrieve their chemical information. Subsequently, the SwissTarget Prediction database (http://www.swisstargetprediction.ch/)18 was utilized to predict potential targets with a “probability” value exceeding a specified threshold. Following target prediction, duplicate entries were eliminated, and the UniProt database (https://www.uniprot.org/)19 was used for target protein validation.
Acquisition of disease targets of liver cancer
Using “liver cancer” as the keyword, we conducted gene retrieval in the GeneCards database (https://www.genecards.org/)20, NCBI database (https://www.ncbi.nlm.nih.gov/)21 and DisGeNET database (https://www.disgenet.org/)22 to identify human genes associated with the disease. After combining the retrieved data, duplications were removed, and the resultant set of disease-related genes was used for subsequent analysis.
Construction of protein-protein interaction (PPI) network
The selected drug and disease targets were input into Venny 2.1 software to find common targets. Subsequently, these common targets were imported into the String database (https://stringdb.org/cgi/input.pl)23, specifying the biological species as “Homo sapiens” to construct a PPI network. The PPI network diagram was generated using Cytoscape 3.8.0 (https://cytoscape.org/)24. Topological analysis was performed using the Network Analyzer tool, and key targets were discerned based on degree scores that exceeded the average. The selected key targets were visualized using R 4.0.3.
Gene Ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis
The shared targets of the drugs and diseases were analyzed using GO and KEGG and displayed using the Metascape database (http://metascape.org/gp/index.htmL)25. Retaining significantly enriched terms and pathways with a corrected p value of less than 0.05, the results were further visualized using R 4.1.2. ClusterProfiler, enrichplot, and ggplot2 packages were used to generate bar charts and bubbles for a comprehensive visualization.
Construction of the component-disease-target-pathway network
To unravel the intricate interplay among selected active components, diseases, and their respective targets, a drug-compound-disease-target network was constructed and was visualized using Cytoscape 3.8.0, providing a comprehensive representation of the relationships.
Molecular docking
In the PPI analysis, the top five components exhibiting the highest degree values were identified as the primary active compounds along with their associated core targets, signifying potential receptors. The three-dimensional structures of the main active compounds in CR-HDW were sourced from the PubChem database, whereas the crystal structures of the core target proteins were retrieved from the RCSB PDB database (https://www.rcsb.org/)26. Standard processing of the proteins and ligands was conducted using AutoDock Tools 1.5.7 software, and the files were saved in PDBQT format. Dehydration, hydrogenation, and other requisite operations were performed using PyMOL 2.3.0, with the Autogrid program function utilized to pinpoint active docking sites. Subsequent molecular docking simulations were performed to determine the corresponding binding energies.
Cell culture
Standard cell culture procedures were followed through the experiment. Briefly, the HuH-7 cells were cultured in DMEM supplemented with 10% FBS and 1% antibiotics (penicillin and streptomycin). The culture flasks were maintained in a humidified incubator at 37°C and 5% CO2. Upon cells reaching 70–80% confluence, cells were subcultured for downstream experiment. For subculture, the cells medium was removed, and the cells were washed with PBS and then digested with 0.25% trypsin-EDTA to detach the cells. The digestion was stopped with the addition of a complete culture medium, and the cells were counted.
Cell viability assay
HuH-7 cells in the logarithmic growth phase were seeded in a 96-well plate at a density of 5000 cells per well and cultured overnight before the addition of different concentrations of selected components (Hederagenin, 3-epioleanic acid, and Hederagenin+3-epioleanic acid, respectively). The cells were then cultured for 24, 48, and 72 h. At indicated time, one plate was removed from the incubator, and the cell viability was assayed using CCK-8 approach according to the instructions of the kit. Briefly, the culture medium was aspirated, and 100 ul of CCK-8 working solution was added to each well, and the plate was incubated at 37 °C for 1 h in the dark. The absorbance was measured at 450 nm using a microplate reader Varioskan Lux (ThermoScientific).
SA-β-gal staining
HuH-7 cells in the logarithmic growth phase were seeded into 6-well plates at a density of 10 000 cells per well and cultured overnight. On the next day, the culture medium was removed and replaced with 2 ml of fresh medium containing either hederagenin (50 μM), 3-epioleanic acid (8 μM), or combination of hederagenin (25μM) and 3-epioleanic acid (4 μM). The DMSO (1%) was served as control. The cells were then cultured for another 48 hours. After removing the culture medium, the cells were washed with PBS once and fixed for 15 min with SA-β-gal fixation solution. The cells were then washed three times with PBS and 1 ml of X-Gal solution was added to each well. The plates were wrapped with parafilm and incubated at 37°C overnight in a regular incubator. On the next day, the solution was discarded, and the cells were washed three times with PBS and the cells were observed under microscope and blue cells were counted. Five randomly selected fields per well were observed and counted to calculate the blue cell rate.
Western blotting
For detection of proteins with Western blotting, in general the cells were washed with cold PBS twice, lysed with RIPA for 30 min on ice, and then centrifuged for 15 min at 15 000g and the supernatant was transferred to Eppendorf tube. The protein concentration was determined by BCA method according to the kit instruction. About 50 ug of protein per sample was separated on 10% SDS-PAGE, transferred to PVDF membrane, and blocked with 5% non-fat milk in Tris-buffered saline with Tween 20 (TBST) for one hour. The membrane was then incubated with primary antibody at 4°C overnight, washed three times with TBST, and incubated with HRP-conjugated secondary antibody for one hour at room temperature. After the final wash with TBST for five times, the membrane was covered with ECL solution, and the images were captured with ChemiDOC XRS (Bio-Rad), and the intensity of the band was quantified and normalized to that of beta-actin.
Statistical analysis
The SPSS version 27.0 was used for statistical analysis of the data, and the result was expressed as mean ± standard deviation (mean ± SD). In cases where the data exhibited a normal distribution, intergroup comparisons were conducted utilizing one-way analysis of variance (ANOVA). Subsequent pairwise comparisons were performed using the least significant difference (LSD) test to identify significant differences between groups. Alternatively, if the assumption of normality was violated, non-parametric tests such as the Mann–Whitney U test were utilized for intergroup comparisons. Upon detecting significant differences between groups, multiple comparisons were executed using the Dunn–Sidak correction method. A significance threshold of P less than 0.05 was adopted to denote statistical significance. The graphical representation of statistical findings was accomplished using GraphPad Prism 8 software.
Results
Identification of active compounds of CR-HDW and compound-disease targets
Ten active compounds of CR and HDW were identified (Table 1) using the TCMSP and Swiss Target Prediction databases, corresponding to 337 drug targets. Three compounds were specific for CR (hederagenin, wenjine, bisdemethoxycurcumin, Table 1), three were specific for HDW (2,3-dimethoxy-6-methyanthraquinone, poriferasterol, 2-methoxy-3-methyl-9,10-anthraquinone, Table 1), and four were found in both herbs (stigmasterol, beta-sitosterol, quercetin, 3-epioleanic acid, Table 1). A set of 2605 disease-associated target proteins was acquired using the GeneCards, NCBI, and DisGeNET databases. Using Venny 2.1 software, we identified 151 common targets between CR-HDW active compounds and liver cancer, as illustrated in Fig. 1. These 151 common genes were then used for downstream analysis.
Table 1.
Information related to CR-HDW active ingredients in TCMSP databases
| Number | Molecule ID | Molecule name | OB (%) | DL |
|---|---|---|---|---|
| 1 | MOL000296 | Hederagenin | 36.91 | 0.75 |
| 2 | MOL000906 | Wenjine | 47.93 | 0.27 |
| 3 | MOL000940 | Bisdemethoxycurcumin | 77.38 | 0.26 |
| 4 | MOL001646 | 2,3-dimethoxy-6-methyanthraquinone | 34.86 | 0.26 |
| 5 | MOL001659 | Poriferasterol | 43.83 | 0.76 |
| 6 | MOL001670 | 2-methoxy-3-methyl-9,10-anthraquinone | 37.83 | 0.21 |
| 7 | MOL000449 | Stigmasterol | 43.83 | 0.76 |
| 8 | MOL000358 | Beta-sitosterol | 36.91 | 0.75 |
| 9 | MOL000098 | Quercetin | 46.43 | 0.28 |
| 10 | MOL001663 | 3-epioleanic acid | 32.03 | 0.76 |
CR-HDW, Curcumae Rhizoma and Hedyotis diffusa Willd; DL, drug similarity; OB, oral bioavailability; No. 1–3: chemical composition of CR, No. 4–6: chemical composition of HDW; TCMSP, Traditional Chinese Medicine Systems Pharmacology.
Figure 1.

The Venn diagram of common targets between 10 components of Curcumae Rhizoma and Hedyotis diffusa Willd and liver cancer.
Construction of the PPI network
Using String software, we visually depicted the PPI network (Fig. 2) of the 151 common genes. The result revealed these 151 functional proteins engaged in 2081 interactions, with an average degree value of 27.6. The degree was calculated using the Network Analyzer, and the top 20 targets were graphically delineated using R 4.0.3 (Fig. 3). Their names and functions of the top 20 genes were shown in Table 2. The top five core targets, ranked by degree, were glyceraldehyde-3-phosphate dehydrogenase (GAPDH), AKT serine/threonine kinase 1 (AKT1), epidermal growth factor receptor (EGFR), signal transducer and activator of transcription 3 (STAT3), and heat shock protein HSP90AA1. Other genes of the top 20 were associated with cell cycle, cell proliferation, cancer growth and metastasis, cancer microenvironment (Table 2).
Figure 2.

Protein-protein interaction network. The String was used for construction protein-protein interaction (PPI) network with the identified 151 common genes. The genes with the most PPI were designed as hub genes.
Figure 3.

The top 20 of the core targets.
Table 2.
Characteristics of the top 20 hub gene
| Gene | Name | Degree | Core functions |
|---|---|---|---|
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 111 | Participate in cell energy metabolism and survival27 |
| AKT1 | RAC-alphaserine/threonine protein kinase | 107 | Regulate cell survival, proliferation and metabolism28,29 |
| EGFR | Epidermal growth factor receptor | 84 | Promotes the growth and metastasis of liver cancer cells30 |
| STAT3 | Signal transducer and activator of transcription 3 | 82 | Regulate cell survival, proliferation and immune evasion31,32 |
| HSP90AA1 | Heat shock protein 90 alpha family class A member 1 | 81 | Involved in protein folding and stabilization33 |
| IL-6 | Interleukin-6 | 80 | Inflammatory microenvironment promoting liver cancer34 |
| SRC | Proto-oncogene tyrosine-protein kinase Src | 80 | Promote the invasion, metastasis and anti-apoptosis ability of liver cancer cells35 |
| ESR1 | Oestrogen receptor alpha | 75 | Affects cell differentiation and growth36 |
| mTOR | Mechanistic target of rapamycin kinase | 74 | Regulate cell growth and metabolism34 |
| CCND1 | Cyclin D1 | 73 | Regulates the cell cycle, especially the G1/S transition37,38 |
| MAPK3 | Mitogen-activated protein kinase 3 | 72 | Mediates cell survival, proliferation and differentiation39 |
| MDM2 | E3 ubiquitin-protein ligase Mdm2 | 62 | Responsible for the degradation of p53, affecting cell cycle and apoptosis40,41 |
| ATM | Serine-protein kinase ATM | 58 | In the DNA damage response, maintaining genome stability42 |
| EP300 | Histone acetyltransferase p300 | 57 | Regulate gene expression and affect the proliferation and survival of cells43 |
| MMP9 | Matrix metalloproteinase-9 | 57 | Involved in cell migration and tumour metastasis44,45 |
| PPARG | Peroxisome proliferator-activated receptor gamma | 56 | Regulates adipocyte differentiation and energy metabolism46,47 |
| PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase | 55 | Catalytic subunit of PI3K signalling pathway, involved in cell proliferation and survival48,49 |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 | 55 | Participate in cell proliferation and survival50 |
| AR | Androgen receptor | 49 | Regulate signalling51 |
| CDK4 | Cyclin-dependent kinase 4 | 47 | Promote cell G1/S phase transition52 |
GO and KEGG analyses
By performing GO enrichment analysis on the shared targets of compounds and diseases, we selected terms with adjusted p values less than 0.05. The top ten GO terms were represented visually in a bar chart (Fig. 4). In the Biological Process (GO: BP) category, 1992 terms were identified, with noteworthy terms encompassing cellular response to chemical stimulus, oxidative stress, and G1/S phase transition of the cell cycle. Molecular Function (GO: MF) analysis yielded 140 terms, including several kinase activities, transcription factor, lipid binding, NADP+ metabolism. In the Cell Component (GO: CC) analysis, 69 terms were identified that were primarily enriched in protein kinase complexes, membranes, and chromosomes.
Figure 4.

Bar charts displaying Gene Ontology enrichment of the key genes.
KEGG pathway enrichment analysis resulted in 155 identified signalling pathways. The top 20 pathways were selected and a bubble chart was generated (Fig. 5). The visualization highlighted pathways related to cancers, viral infection, reactive oxygen species (ROS), PI3K-AKT signalling, cellular senescence, Forkhead box O (FoxO) signalling, and hypoxia-inducible factor (HIF-1) signalling, lipid and atherosclerosis, indicating that these pathways might be the critical molecular mechanisms of the CR-HDW in the treatment of liver cancer.
Figure 5.

Kyoto Encyclopedia of Genes and Genomes bubble charts.
Construction of the herb-component-disease-target network
The 10 active components and their shared targets were integrated into Cytoscape 3.8.0 to formulate a visual network diagram illustrating the component-target-disease relationships (Fig. 6). This visualization offers a more intuitive representation of the multi-component, multi-target attributes of TCM, emphasizing the complexity of interaction between the active components of CR-HDW and the cellular targets throughout the therapeutic process.
Figure 6.
Curcumae Rhizoma and Hedyotis diffusa Willd composition-disease-pathway-target network. The green rectangle represents disease, the yellow hexagons represents herbs. The purple rhombus represents 10 active compounds. The red ovals represent 151 common targets between drugs and disease.
Molecular docking
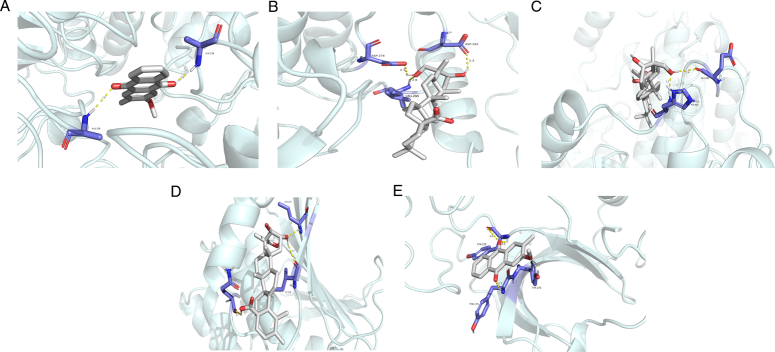
The molecular docking results revealed binding energies, where a smaller value indicated a stronger affinity between a chemical and its target. Binding energies below -4.25 kcal/mol suggest potential ligand-receptor binding, while values below -5.00 kcal/mol indicate a relatively strong binding affinity. The docking analysis produced results for five compound-target pairs (Table 3). The most favourable ligand-protein interactions were visualized using PyMOL software, showing specific binding interactions (Fig. 7). Hederagenin and 2-methoxy-3-methyl-9,10-anthraquinone exhibited robust binding affinities for all five targets (binding energy < −5.00 kcal/mol). Quercetin and bisdemethoxycurcumin demonstrated moderate binding energies for all targets (< −4.25 kcal/mol). Notably, hederagenin and 3-epioleanic acid exhibited strong binding affinities for AKT1 and HSP90AA1.
Table 3.
Binding energies of the top 5 CR-HDW compounds to Munich-related core targets
| Core gene docking score(kcal/mol) | |||||
|---|---|---|---|---|---|
| Molecule name | GAPDH | AKT1 | EGFR | STAT3 | HSP90AA1 |
| Core components | |||||
| Quercetin | −4.04 | −5.09 | −4.71 | −3.39 | −4.70 |
| 2-methoxy-3-methyl-9,10-anthraquinone | −6.39 | −7.68 | −5.94 | −5.68 | −5.48 |
| Bisdemethoxycurcumin | −3.96 | −4.94 | −4.98 | −2.92 | −4.38 |
| 3-epioleanic acid | −4.68 | −6.73 | −4.32 | −3.77 | −7.46 |
| Hederagenin | −6.22 | 7.90 | −7.44 | −6.79 | −6.40 |
AKT1, Akt murine thymoma viral oncogene homologue 1; CR-HDW, Curcumae Rhizoma and Hedyotis diffusa Willd; EGFR, epidermal growth factor receptor; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HSP90AA1, heat shot protein 90 AA1; STAT3, signal transducer and activator of transcription 3.
Figure 7.
The molecular docking of the main components and core targets. (A) 2-methoxy-3-methyl-9,10-anthraquinone-GAPDH. (B) Hederagenin-AKT1 (C) Hederagenin-EGFR. (D) 3-epioleanic acid -HSP90AA1. (E) 2-methoxy-3-methyl-9,10-anthraquinone-AKT1.
Hederagenin and 3-epioleanic acid inhibited HuH-7 cell proliferation
Bioinformatic analysis revealed five active compounds from CR-HDW as the most important compounds. To test whether these compounds truly represent the active compounds of CR-HDW, hederagenin and 3-epioleanolic acid were selected for in-vitro test based on their biological activities (Fig. 5). To achieve this purpose, HuH-7 cells were seeded in 96-well plate and treated with hederagenin and 3-epioleanolic acid at different concentrations for different times. Cell viability was estimated using CCK-8 assay. As shown in Figs. 8A, 3-epioleanolic acid displayed half maximal inhibitory concentration (IC50) values of 18.69, 8.86, and 7.35 μM for HuH-7 cell at 24, 48, and 72 h, respectively. Hederagenin displayed IC50 values of 86.67, 53.60, and 41.93 μM for HuH-7 cell at 24, 48, and 72 h, respectively (Fig. 8). The inhibitory effect of the two compounds on HuH-7 cells showed time and concentration-dependent manner.
Figure 8.

Effect of hederagenin and 3-epioleanolic acid on HuH-7 cell growth. HuH-7 cells were treated with 3-epioleanolic acid (A) or hederagenin (B) with different concentrations and for different times. The cell viability was examined by using CCK-8 and expressed as percentage to the control. Data represents average of three repeats.
Hederagenin-induced Huh-7 senescence
Cellular senescence is important in the treatment of cancer cells, and KEGG pathway enrichment (Fig. 5) revealed cellular senescence as one of the important pathways for the treatment of liver cancer by CR-HDW. To test whether hederagenin and 3-epioleanic acid could induce HuH-7 senescence, the HuH-7 cells were seeded in six-well plate and treated with hederagenin, or 3-epioleanic acid, or their combination for 48 h and stained with X-Gal for senescence marker SA-β-gal. As shown in Figs. 9A and B, while hederagenin treatment displayed over 40% of blue cells, 3-epioleanic acid showed blue-staining cells with no significant increase than that of the control. This suggests that hederagenin possesses the capacity to induce cellular senescence in HuH-7 cells, while 3-epioleanolic acid may lack this ability.
Figure 9.

Cellular senescence induction by hederagenin and 3-epioleanolic acid. HuH-7 cells were treated either with hederagenin (Hed, 50 μM), 3-epioleanolic acid (3-e, 8 μM), combination of Hed (25 μM) and 3-e (4 μM), or DMSO for 48 h, and the senescent cells were stained blue with X-Gal. (A) Representative of the microscope images showing blue staining of senescent cells. (B) Senescent cell rate. The data represent average of five fields per well, three wells per treatment.
Expression of HSP90 and PI3K pathway proteins upon treatment with hederagenin and 3-epioleanic acid
To address whether the predicted PI3K pathway was affected by selected active ingredients, the HuH-7 cells were treated with the compounds alone or in combination for 48 hours and the total cellular proteins were used for the detection of proteins using Western blot. As show Fig. 10, compared with control, both hederagenin and 3-epioleanic acid alone or in combination caused an obvious reduction in PI3K protein with hederagenin showed slight effectiveness than 3-epioleanic acid. When AKT1 was detected, only hederagenin treatment reduced AKT1 level. Importantly, both hederagenin and 3-epioleanic acid greatly reduced mTOR levels (Fig. 10). There was no notable difference in HSP90AA1 protein expression between the control and experimental groups (P> 0.05). Thus, the bioactive constituents hederagenin and 3-epioleanic acid of CR-HDW reduced the expression of target proteins in the PI3K and AKT pathways, with hederagenin exerting a more pronounced effect on PI3K, AKT1, and mTOR, while 3-epioleanolic acid had no effect on AKT1.
Figure 10.

Effect of hederagenin and 3-epioleanolic acid on protein expression in HuH-7 cells. The HuH-7 cells were treated with hederagenin (Hed), 3-epioleanolic acid (3-e), or combination of Hed and 3-e for 48 h, the total proteins were used for western blotting to detect indicated protein and the relative expression level was normalized to beta-actin. (A) Representative western blotting images. (B) Relative protein expression as normalized to the DMSO control. Data were average of three repeats. *P< 0.05; Compared with 3-e group, △ p<0.05 Compared with Hed group, ☆ P< 0.05.
Discussion
The search for better treatment of liver cancer is still a challenge for scientists. The TCM herb pair CR-HDW has been widely used in the clinic to treat different cancers including liver cancer. However, the underlying anti-cancer active compounds and their mechanism of action are still unearthed. In this study, ten compounds were identified as anti-cancer chemicals based on the network pharmacology approach and several pathways important for cancer proliferation and for the treatment by active compounds were enriched using GO and KEGG analysis. Two of the compounds hederagenin (from Curcumae Rhizoma) and 3-epioleanolic acid (from both herbs) were validated for their anti-cancer activity using HuH-7 cells, supporting the hypothesis that active compounds from Curcumae Rhizoma and Hedyotis diffusa Willd had anti-cancer activity according to TCM theory.
In line with our result, previous reports showed that hederagenin had a diverse range of pharmacological activities, encompassing anti-tumour, anti-inflammatory, and antiviral properties53. Its promising potential in anti-tumour therapy has been highlighted, particularly in inducing apoptosis of HepG2 cells via the mitochondrial pathway54. Because of this, hederagenin has been used to serve as a triterpenoid template for the development of novel anti-tumour compounds53,55. Bisdemethoxycurcumin exhibits anti-inflammatory and antioxidant effects through transforming growth factor beta (TGF-β)/Smad, c-Jun N-terminal kinase 1/2 (JNK1/2)- reactive oxygen species (ROS), and nuclear factor kappa-B (NF-κB)56. Limited reports are available on 2,3-Dimethoxy-6-methyanthraquinone and 3-epioleanolic acid.
Shared 151 genes between the ten active compounds and the liver cancer formed PPI networks with key hub genes function in cell cycle-related proteins (CDK4and CCND1), molecular chaperone proteins (HSP90AA1), protein kinases (AKT1, EGFR), and oxidative stress-related proteins (SRC, MMP9). GAPDH, an enzyme in the glycolytic pathway, is a hallmark of cancer metabolism, and GAPDH inhibitors inhibit the growth and metastasis of liver cancer57,58. HSP90AA1, a molecular chaperone protein, aids in the proper folding and stabilization of other proteins33. In hepatocellular carcinoma, HSP90AA1 regulated the abundance of PKM2, promoted cell glycolysis, and inhibited apoptosis59. AKT1 plays a key role in various biological processes such as proliferation, metabolism, and apoptosis60,61. EGFR is implicated in the proliferation, angiogenesis, invasion, metastasis, and apoptosis of tumour cells when overexpressed62,63. The signal transducer and activator of transcription 3 (STAT3) is a central regulator of anti-tumour immune response. Excessive activation of STAT3 reduces the expression of immune-stimulating factors and has profound immunomodulatory effects64.
A pivotal characteristic of tumour cells is the dysregulation of cell cycle control, leading to uncontrolled cell division, abnormal replication of genetic material, aberrant expression of cell cycle proteins, failure of cell cycle checkpoints, inactivation of tumour suppressor genes, and activation of oncogenes65–67. Our and KEGG analysis GO enrichment analysis revealed protein kinases, cell cycle regulation, oxidative stress as important cellular process and pathways such as PI3K-Akt, FOXO, HIF-1, and cellular senescence are critical for anti-tumour mechanisms of the compounds from CR-HDW. Particularly, senescent cells cease to proliferate, a characteristic that has been explored for cancer treatment. Tumour development68,69. Research is actively exploring ways to leverage the mechanisms of cellular senescence in liver cancer treatment70.
Typically, external signals such as growth factors activate PI3K on the cell surface. PI3K converts phosphatidylinositol 4,5-bisphosphate into phosphatidylinositol 3,4,5-trisphosphate, which activates AKT. Activated AKT then inhibits the tuberous sclerosis complex 1/2 (TSC1/2) complex, activating mTOR complex 1 (mTORC1), which regulates protein synthesis and cell growth71. Inhibiting the PI3K-Akt signalling pathway has potential therapeutic efficacy in suppressing liver cancer growth72–74.
Our in-vitro study demonstrated that hederagenin and 3-epioleanolic acid inhibited HuH-7 cell growth, and this inhibitory effect may associated with reduced protein levels of PI3K, AKT, and mTOR. Hederagenin also induced the expression of cellular senescence marker SA-β-gal (Fig. 9), suggesting that cellular senescence may play a role in the treatment of liver cancer by CR-HDW.
Although we showed some evidence to support the notion that compounds from CR-HDW have anti-cancer activity, our research had limitations. First, some active compounds may not collected in the databases; second, molecular docking prediction is still not satisfactory to identify the direct target of a compound, it only predicts potential interactions based on binding affinity. Extensive experiment approaches are required to validate unequivocally the direct target of a compound. So, we are not confident about the predicted targets of the selected compounds. Further experimental verification is required to confirm our predictions and findings.
Conclusions
In summary, our results demonstrate that CR-HDW components hederagenin and 3-epioleanolic acid inhibited proliferation and induced senescence in HuH-7 cells, concomitantly reducing the protein expression of PI3K and AKT, an important molecule for cancer cell growth. Future investigations with more compounds and animal studies will add to our understanding of the TCM theory and advance clinic application to benefit our patients.
Ethical approval
This study exclusively conducted cell experiments, thus obviating the need for ethical review involving animals or patients.
Consent
Not applicable—this study exclusively conducted cell experiments, thus obviating the need for ethical review involving patients.
Sources of funding
This study is funded by the Hunan Provincial Science and Technology Department project (No.2020SK3034) and National Natural Science Foundation of China (No.81803993).
Author contribution
Conceptualization: S.T., T.T. Data curation: G.Z., D.O., X.L. Funding acquisition: J.C. Investigation: S.T., T.T. Software: S.T., T.T. Supervision: J.C. Visualization: S.T., T.T. Writing—original draft: S.T., T.T. Writing—review and polishing: J.C., X.L.
Conflicts of interest disclosure
The author(s) of this work have nothing to disclose.
Research registration unique identifying number (UIN)
Not applicable—not required for this study.
Guarantor
Jianzhong Cao is Guarantor.
Data availability statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Provenance and peer review
Not commissioned, externally peer-reviewed.
Footnotes
S.T. and T.T. contributed equally to this article and should be considered as the co-first author.
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Published online 23 April 2024
Contributor Information
Songyan Tie, Email: t20213804@163.com.
Tianhao Tong, Email: t38572922@stu.hnucm.edu.cn.
Gangxiang Zhan, Email: zgx@stu.hnucm.edu.cn.
Xin Li, Email: lixin20082005@163.com.
Dan Ouyang, Email: 1527459187@qq.com.
Jianzhong Cao, Email: 20183217@stu.hnucm.edu.cn.
References
- 1. International Agency for Research on Cancer. Global cancer burden growing, amidst mounting need for services. Saudi Med J. 2024;45:326–327. [Google Scholar]
- 2. Brown ZJ, Tsilimigras DI, Ruff SM, et al. Management of hepatocellular carcinoma: a review. JAMA Surg 2023;158:410–420. [DOI] [PubMed] [Google Scholar]
- 3. Vogel A, Cervantes A, Chau I, et al. Corrigendum to “Hepatocellular carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up” [Annals of Oncology 2018;29 suppl. 4:v238-iv255]. Ann Oncol 2022;33:666. [DOI] [PubMed] [Google Scholar]
- 4. Wang S, Long S, Deng Z, et al. Positive role of chinese herbal medicine in cancer immune regulation. Am J Chin Med 2020;48:1577–1592. [DOI] [PubMed] [Google Scholar]
- 5. Wang X, Wang N, Cheung F, et al. Chinese medicines for prevention and treatment of human hepatocellular carcinoma: current progress on pharmacological actions and mechanisms. J Integr Med 2015;13:142–164. [DOI] [PubMed] [Google Scholar]
- 6. Liao X, Bu Y, Jia Q. Traditional Chinese medicine as supportive care for the management of liver cancer: past, present, and future. Genes Dis 2020;7:370–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lu X, Li B. Exploration of the effect and mechanism of activating blood circulation and stasis-removing therapy on tumor metastasis. Chin J Integr Med 2009;15:395–400. [DOI] [PubMed] [Google Scholar]
- 8. Xu GL, Geng D, Xie M, et al. Chemical composition, antioxidative and anticancer activities of the essential oil: Curcumae Rhizoma-Sparganii Rhizoma, a traditional herb pair. Molecules 2015;20:15781–15796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen SL, Ho CY, Lin WC, et al. The characteristics and mortality of chinese herbal medicine users among newly diagnosed inoperable huge hepatocellular carcinoma (≥10 cm) patients: a retrospective cohort study with exploration of core herbs. Int J Environ Res Public Health 2022;19:12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li Z, Hao E, Cao R, et al. Analysis on internal mechanism of zedoary turmeric in treatment of liver cancer based on pharmacodynamic substances and pharmacodynamic groups. Chin Herb Med 2022;14:479–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Abdel-Lateef E, Mahmoud F, Hammam O, et al. Bioactive chemical constituents of Curcuma longa L. rhizomes extract inhibit the growth of human hepatoma cell line (HepG2). Acta Pharm 2016;66:387–398. [DOI] [PubMed] [Google Scholar]
- 12. Kocarnik JM, Compton K, Dean FE, et al. Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life Years for 29 Cancer Groups From 2010 to 2019: A Systematic Analysis for the Global Burden of Disease Study 2019. JAMA Oncol 2022;8:420–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu H, Zhang L, Wang C, et al. Network pharmacology analysis and experimental verification on antiangiogenesis mechanism of hedyotis diffusa willd in liver cancer. Evid Based Complement Alternat Med 2023;2023:1416841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhao L, Zhang H, Li N, et al. Network pharmacology, a promising approach to reveal the pharmacology mechanism of Chinese medicine formula. J Ethnopharmacol 2023;309:116306. [DOI] [PubMed] [Google Scholar]
- 15. Li H, Li T, Quang D, et al. Network propagation predicts drug synergy in cancers. Cancer Res 2018;78:5446–5457. [DOI] [PubMed] [Google Scholar]
- 16. Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform 2014;6:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim S, Chen J, Cheng T, et al. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res 2021;49(D1):D1388–D1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res 2019;47(W1):W357–W364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. UniProt Consortium. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 2019;47(D1):D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Safran M, Dalah I, Alexander J, et al. GeneCards Version 3: the human gene integrator. Database (Oxford) 2010;2010:baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sayers EW, Beck J, Bolton EE, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 2021;49(D1):D10–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Piñero J, Bravo À, Queralt-Rosinach N, et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res 2017;45(D1):D833–D839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47(D1):D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Doncheva NT, Morris JH, Gorodkin J, et al. Cytoscape StringApp: network analysis and visualization of proteomics data. J Proteome Res 2019;18:623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Burley SK, Bhikadiya C, Bi C, et al. RCSB Protein Data Bank (RCSB.org): delivery of experimentally-determined PDB structures alongside one million computed structure models of proteins from artificial intelligence/machine learning. Nucleic Acids Res 2023;51(D1):D488–D508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Talwar D, Miller CG, Grossmann J, et al. The GAPDH redox switch safeguards reductive capacity and enables survival of stressed tumour cells. Nat Metab 2023;5:660–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Basu A, Lambring CB. Akt isoforms: a family affair in breast cancer. Cancers (Basel) 2021;13:3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang X, Feng Y, Liu Y, et al. Fuzheng Jiedu Xiaoji formulation inhibits hepatocellular carcinoma progression in patients by targeting the AKT/CyclinD1/p21/p27 pathway. Phytomedicine 2021;87:153575. [DOI] [PubMed] [Google Scholar]
- 30. Zhang H, Deng T, Liu R, et al. Exosome-delivered EGFR regulates liver microenvironment to promote gastric cancer liver metastasis. Nat Commun 2017;8:15016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hashimoto S, Hashimoto A, Muromoto R, et al. Central roles of STAT3-mediated signals in onset and development of cancers: tumorigenesis and immunosurveillance. Cells 2022;11:2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xie L, Zeng Y, Dai Z, et al. Chemical and genetic inhibition of STAT3 sensitizes hepatocellular carcinoma cells to sorafenib induced cell death. Int J Biol Sci 2018;14:577–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shi W, Feng L, Dong S, et al. FBXL6 governs c-MYC to promote hepatocellular carcinoma through ubiquitination and stabilization of HSP90AA1. Cell Commun Signal 2020;18:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rose-John S, Jenkins BJ, Garbers C, et al. Targeting IL-6 trans-signalling: past, present and future prospects. Nat Rev Immunol 2023;23:666–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Weng YS, Chiang IT, Tsai JJ, et al. Lenvatinib synergistically promotes radiation therapy in hepatocellular carcinoma by inhibiting Src/STAT3/NF-κB-mediated epithelial-mesenchymal transition and metastasis. Int J Radiat Oncol Biol Phys 2023;115:719–732. [DOI] [PubMed] [Google Scholar]
- 36. Tolaney SM, Toi M, Neven P, et al. Clinical significance of PIK3CA and ESR1 mutations in circulating tumor DNA: analysis from the MONARCH 2 study of abemaciclib plus fulvestrant. Clin Cancer Res 2022;28:1500–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu J, Lin J, Wang X, et al. CCND1 amplification profiling identifies a subtype of melanoma associated with poor survival and an immunosuppressive tumor microenvironment. Front Immunol 2022;13:725679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu SY, Lan SH, Liu HS. Degradative autophagy selectively regulates CCND1 (cyclin D1) and MIR224, two oncogenic factors involved in hepatocellular carcinoma tumorigenesis. Autophagy 2019;15:729–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sun H, Qian X, Yang W, et al. Novel prognostic signature based on HRAS, MAPK3 and TFRC identified to be associated with ferroptosis and the immune microenvironment in hepatocellular carcinoma. Am J Transl Res 2022;14:6924–6940. [PMC free article] [PubMed] [Google Scholar]
- 40. Wang W, Qin JJ, Rajaei M, et al. Targeting MDM2 for novel molecular therapy: Beyond oncology. Med Res Rev 2020;40:856–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhu H, Gao H, Ji Y, et al. Targeting p53-MDM2 interaction by small-molecule inhibitors: learning from MDM2 inhibitors in clinical trials. J Hematol Oncol 2022;15:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hopkins JL, Lan L, Zou L. DNA repair defects in cancer and therapeutic opportunities. Genes Dev 2022;36:278–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rubio K, Molina-Herrera A, Pérez-González A, et al. EP300 as a molecular integrator of fibrotic transcriptional programs. Int J Mol Sci 2023;24:12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang HL, Thiyagarajan V, Shen PC, et al. Anti-EMT properties of CoQ0 attributed to PI3K/AKT/NFKB/MMP-9 signaling pathway through ROS-mediated apoptosis. J Exp Clin Cancer Res 2019;38:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Huang H. Matrix metalloproteinase-9 (MMP-9) as a cancer biomarker and MMP-9 biosensors: recent advances. Sensors (Basel) 2018;18:3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chu S, Yang Y, Nazar M, et al. miR-497 regulates LATS1 through the PPARG pathway to participate in fatty acid synthesis in bovine mammary epithelial cells. Genes (Basel) 2023;14:1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. To JC, Chiu AP, Tschida BR, et al. ZBTB20 regulates WNT/CTNNB1 signalling pathway by suppressing PPARG during hepatocellular carcinoma tumourigenesis. JHEP Rep 2021;3:100223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tiwari A, Iida M, Kosnopfel C, et al. Blocking Y-box binding protein-1 through simultaneous targeting of PI3K and MAPK in triple negative breast cancers. Cancers (Basel) 2020;12:2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhao T, Guo BJ, Xiao CL, et al. Aerobic exercise suppresses hepatocellular carcinoma by downregulating dynamin-related protein 1 through PI3K/AKT pathway. J Integr Med 2021;19:418–427. [DOI] [PubMed] [Google Scholar]
- 50. Ting HK, Chen CL, Meng E, et al. Inflammatory regulation by TNF-α-activated adipose-derived stem cells in the human bladder cancer microenvironment. Int J Mol Sci 2021;22:3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu G, Ouyang X, Sun Y, et al. The miR-92a-2-5p in exosomes from macrophages increases liver cancer cells invasion via altering the AR/PHLPP/p-AKT/β-catenin signaling. Cell Death Differ 2020;27:3258–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fassl A, Geng Y, Sicinski P. CDK4 and CDK6 kinases: from basic science to cancer therapy. Science 2022;375:eabc1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xie W, Fang X, Li H, et al. Advances in the anti-tumor potential of hederagenin and its analogs. Eur J Pharmacol 2023;959:176073. [DOI] [PubMed] [Google Scholar]
- 54. Liu Z, Tan X, Peng L, et al. Hederagenin induces apoptosis of human hepatoma HepG2 cells via the mitochondrial pathway. Comb Chem High Throughput Screen 2023;27:1495–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rodríguez-Hernández D, Demuner AJ, Barbosa LC, et al. Hederagenin as a triterpene template for the development of new antitumor compounds. Eur J Med Chem 2015;105:57–62. [DOI] [PubMed] [Google Scholar]
- 56. Gao TH, Liao W, Lin LT, et al. Curcumae rhizoma and its major constituents against hepatobiliary disease: Pharmacotherapeutic properties and potential clinical applications. Phytomedicine 2022;102:154090. [DOI] [PubMed] [Google Scholar]
- 57. Lin XT, Zhang J, Liu ZY, et al. Elevated FBXW10 drives hepatocellular carcinoma tumorigenesis via AR-VRK2 phosphorylation-dependent GAPDH ubiquitination in male transgenic mice. Cell Rep 2023;42:112812. [DOI] [PubMed] [Google Scholar]
- 58. Ganapathy-Kanniappan S, Kunjithapatham R, Geschwind JF. Glyceraldehyde-3-phosphate dehydrogenase: a promising target for molecular therapy in hepatocellular carcinoma. Oncotarget 2012;3:940–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Xu Q, Tu J, Dou C, et al. HSP90 promotes cell glycolysis, proliferation and inhibits apoptosis by regulating PKM2 abundance via Thr-328 phosphorylation in hepatocellular carcinoma. Mol Cancer 2017;16:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang R, Akhtar N, Wani AK, et al. Discovering deleterious single nucleotide polymorphisms of human AKT1 oncogene: an in silico study. Life (Basel) 2023;13:1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Xu Z, Xu M, Liu P, et al. The mTORC2-Akt1 cascade is crucial for c-myc to promote hepatocarcinogenesis in mice and humans. Hepatology 2019;70:1600–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Khemlina G, Ikeda S, Kurzrock R. The biology of Hepatocellular carcinoma: implications for genomic and immune therapies. Mol Cancer 2017;16:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sabbah DA, Hajjo R, Sweidan K. Review on epidermal growth factor receptor (EGFR) structure, signaling pathways, interactions, and recent updates of EGFR inhibitors. Curr Top Med Chem 2020;20:815–834. [DOI] [PubMed] [Google Scholar]
- 64. Zou S, Tong Q, Liu B, et al. Targeting STAT3 in cancer immunotherapy. Mol Cancer 2020;19:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Liu J, Peng Y, Wei W. Cell cycle on the crossroad of tumorigenesis and cancer therapy. Trends Cell Biol 2022;32:30–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. He S, Chen M, Lin X, et al. Triptolide inhibits PDGF-induced proliferation of ASMCs through G0/G1 cell cycle arrest and suppression of the AKT/NF-κB/cyclinD1 signaling pathway. Eur J Pharmacol 2020;867:172811. [DOI] [PubMed] [Google Scholar]
- 67. Knudsen ES, Kumarasamy V, Nambiar R, et al. CDK/cyclin dependencies define extreme cancer cell-cycle heterogeneity and collateral vulnerabilities. Cell Rep 2022;38:110448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhang M, Serna-Salas S, Damba T, et al. Hepatic stellate cell senescence in liver fibrosis: characteristics, mechanisms and perspectives. Mech Ageing Dev 2021;199:111572. [DOI] [PubMed] [Google Scholar]
- 69. Chen HA, Ho YJ, Mezzadra R, et al. Senescence rewires microenvironment sensing to facilitate antitumor immunity. Cancer Discov 2023;13:432–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang C, Vegna S, Jin H, et al. Inducing and exploiting vulnerabilities for the treatment of liver cancer. Nature 2019;574:268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sun EJ, Wankell M, Palamuthusingam P, et al. Targeting the PI3K/Akt/mTOR pathway in hepatocellular carcinoma. Biomedicines 2021;9:1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Paskeh M, Ghadyani F, Hashemi M, et al. Biological impact and therapeutic perspective of targeting PI3K/Akt signaling in hepatocellular carcinoma: Promises and Challenges. Pharmacol Res 2023;187:106553. [DOI] [PubMed] [Google Scholar]
- 73. Wang W, Dong X, Liu Y, et al. Itraconazole exerts anti-liver cancer potential through the Wnt, PI3K/AKT/mTOR, and ROS pathways. Biomed Pharmacother 2020;131:110661. [DOI] [PubMed] [Google Scholar]
- 74. Yang J, Pi C, Wang G. Inhibition of PI3K/Akt/mTOR pathway by apigenin induces apoptosis and autophagy in hepatocellular carcinoma cells. Biomed Pharmacother 2018;103:699–707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.