Abstract
Strain ZW T0_25T was isolated from an onion sample (Allium cepa var. Hytech F1) within a storage trial and proofed to be a novel, aerobic, Gram-stain negative, rod-shaped bacterial strain. Analyses of the 16S rRNA gene sequence and of the whole draft genome sequences, i.e., digital DNA–DNA hybridization (dDDH), Average Nucleotide Identity (ANI) and Average Amino Acid Identity (AAI) showed that this strain represents a new species of the genus Bosea. The genome size of strain ZW T0_25T is 6.19 Mbp, and the GC content is 66.9%. As whole cell sugars, rhamnose, ribose and glucose were identified. Ubiquinone Q-10 is the major respiratory quinone with 97.8%. Polar lipids in strain ZW T0_25T are composed of one phosphatidylethanolamine, one phosphatidylglycerol, one aminophospholipid, two aminolipids, one glycolipid and two phospholipids whereas the fatty acid profile predominantly consists of C18:1 w7c (63.3%), C16:1 w7c (19.5%) and C16:0 (7.1%). Phenotypic traits were tested in the wet lab as well as predicted in silico from genome data. Therefore, according to this polyphasic approach, the new name Bosea rubneri sp. nov. with the type strain ZW T0_25T (= DSM 116094 T = LMG 33093 T) is proposed.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00284-024-03717-6.
Introduction
The genus Bosea was proposed in 1996 for a new bacterial strain that was isolated from agricultural soil around Calcutta, India [1] and named in honour of J.C. Bose, the founder of the Bose Institute, where the type strain of the type species was isolated [1]. The genus Bosea belongs to the alphaproteobacteria family Boseaceae [2] and at time of writing, the genus comprises 12 species, (I) B. caraganae [3], (II) B. eneae [4], (III) B. lathyri [5], (IV) B. lupini [5], (V) B. massiliensis [4], (VI) B. minatitlanensis [6], (VII) B. psychrotolerans [7], (VIII) B. robiniae [5], (IX) B. spartocytisi [8], (X) B. thiooxidans [1], (XI) B. vaviloviae [9] and (XII) B. vestrisii [4]. The type strains of half of the 12 species of Bosea were isolated from root nodules of different plants belonging to the family Fabaceae [3, 5, 8, 9]. Four type strains were isolated from fresh water, i.e., three of them from hospital water supply [4] and one from Lake Michigan [7]. In addition, one type strain was isolated from an anaerobic sludge blanket reactor fed with wastewater [6] whereas the type strain of the type species [1] was isolated from soil as mentioned above. Strain ZW T0_25T was isolated from an onion sample in November 2020 alongside a collection of 316 bacterial strains within a study with the objective to investigate the bacterial microbiota of different onion varieties. Among this bacterial collection, a number of 13 strains including ZW T0_25T could not be identified by 16S rRNA gene sequencing or MALDI-TOF analysis. However, whole-genome shotgun sequencing of these 13 bacterial strains revealed that these strains including strain ZW T0_25T represent potential new species of different genera. One of these strains, ZW T2_19T has been previously described as Rathayibacter rubneri sp. nov. [10].
Materials and Methods
Isolation of Bacterial Strain ZW T0_25T, Culture Conditions and Morphology
Strain ZW T0_25T was isolated from an onion (Allium cepa var. Hytech F1) which was harvested, air-dried and selected for storage at 2 °C. The isolation was performed as published previously [10]. Briefly, about a quarter of an onion bulb was minced, serially diluted in quarter-strength Ringer’s solution, plated on Standard Nutrient I agar (Merck KGaA, Darmstadt, Germany) and incubated at 30 °C. Strains were streaked out until purity and cryopreserved with 15% glycerol. Mid-exponential to stationary phase cells were visualized under a phase-contrast microscope (Leica), and motility was tested with hanging-drop method. In addition, cell morphology was investigated by scanning electron microscopy (SEM) as described previously [11]. For SEM, bacterial cells were grown on Standard Nutrient I agar for 48 h. Furthermore, the cell diameters and cell lengths of 50 cells of strain ZW T0_25T were measured using ObjectJ in ImageJ 1.53q [12].
Biochemical Characteristics
Growth was tested at 7, 24, 37, and 42 °C in Standard Nutrient I broth as well as at 24 °C, 100 rpm in Standard Nutrient I broth supplemented with 1, 2, 3, 4, 5, 6.5, 8, and 10% NaCl and 0.01, 0.02, or 0.03% potassium tellurite. In addition, growth was tested at pH 5, pH 6, pH 7, pH 8 and pH 9 according to [13] at 24 °C, 100 rpm and in MRS broth supplemented with 1% ox bile. Haemolysis was investigated on blood agar (bioMérieux, Nürtingen, Germany). The presence of catalase was tested according to standard microbiological methods as described in [4, 14], while presence of cytochrome oxidase was tested using Bactident Oxidase test strips (Merck KGaA) according to the instructions of the manufacturer. Biochemical characteristics of strain ZW T0_25T were determined using API 20NE, API 50CH, API Coryne, API ZYM (bioMérieux) at 24 °C according to the manufacturer’s instructions. Chemotaxonomic analyses of strain ZW T0_25T were carried out by the Identification Service of the DSMZ (German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany) including analyses of respiratory quinones [15, 16], fatty acids [15, 17–21], whole cell sugars [22, 23] and polar lipids [24, 25]. For this purpose, the biomass of 10 L liquid culture (Standard Nutrient I broth, 24 °C, 110 rpm, 24 h) of strain ZW T0_25T was collected by centrifugation (9,622 × g; 10 min; 4 °C) and the total moist biomass of strain ZW T0_25T (6.06 g) was sent on dry ice to the DSMZ.
16S rRNA Gene Sequencing, Genome Sequencing and Analyses
For 16S rRNA gene sequencing, isolation of bacterial DNA of strain ZW T0_25T, 16S rRNA gene amplification and analysis were carried out as described previously [26]. Briefly, the bacterial DNA was isolated using the Blood and Tissue Kit (Qiagen), and the almost complete 16S rRNA gene was amplified using the primers 16Sseq fw (5’-ATA GTT TGA TCM TGG CTC AG-3’) and 16Sseq rev (5’-GGN TAC CTT GTT ACG ACT TC-3’). The accession number for the 16S rRNA gene sequence of strain ZW T0_25T at GenBank/EMBL/DDBJ is OR512845. Phylogenetic trees (maximum likelihood, maximum parsimony and neighbour joining) of strain ZW T0_25T and type strains of the genus Bosea were built using BioNumerics (version 8.1, Applied Maths). The 16S rRNA gene sequences of the type strains were obtained from NCBI (https://www.ncbi.nlm.nih.gov). In addition, pairwise nucleotide similarities of the 16S rRNA gene sequences were calculated using EZBioCloud [27]. The whole draft genome sequence of strain ZW T0_25T was sequenced on an Illumina MiSeq (Illumina Nextera XT DNA Library Prep Kit, 250 bp paired-end reads) by our group. The reads were assembled de novo using SPAdes genome assembler v. 3.13.1. [28]. This whole-genome shotgun project has been deposited at DDBJ/ENA/GenBank under the accession JAWDID000000000. The version described in this paper is version JAWDID010000000. In addition, digital DNA–DNA hybridization (dDDH) was calculated by using Type (Strain) Genome Server (TYGS) [29, 30], average nucleotide identity (ANI) was calculated as OrthoANIu values by using EzBioCloud [27, 31], and AAI (Average Amino Acid Identity) was determined by using AAI calculator [32]. A Genome BLAST Distance Phylogeny (GBDP) tree (whole-genome sequence based) of strain ZW T0_25T and type strains of Bosea was calculated by using TYGS [29]. The software package Traitar [33] and the bioinformatic tool Protologger [34] running on the galaxy platform [35] were used for phenotype and protologue prediction from the genome data, respectively. ResFinder (version 4.1) [36–38] was used for prediction of antimicrobial resistances.
Results and Discussion
Morphology
After 48 h growth at 30 °C on Standard Nutrient I agar, strain ZW T0_25T occurred as smooth, round and cream-coloured colonies with a diameter of about 4 mm. Strain ZW T0_25T occurred as motile, small rods under the phase-contrast microscope. The rod-shaped morphology was confirmed by SEM (Fig. 1). The mean diameter and mean cell length of strain ZW T0_25T were 0.40 ± 0.05 µm and 1.16 ± 0.23 µm, respectively. This corresponds well to the cell width of 0.4 to 0.85 µm and cell length of 1.1 to 3 µm as reported for other type strains of the genus Bosea [1, 3, 5–7, 9].
Fig. 1.
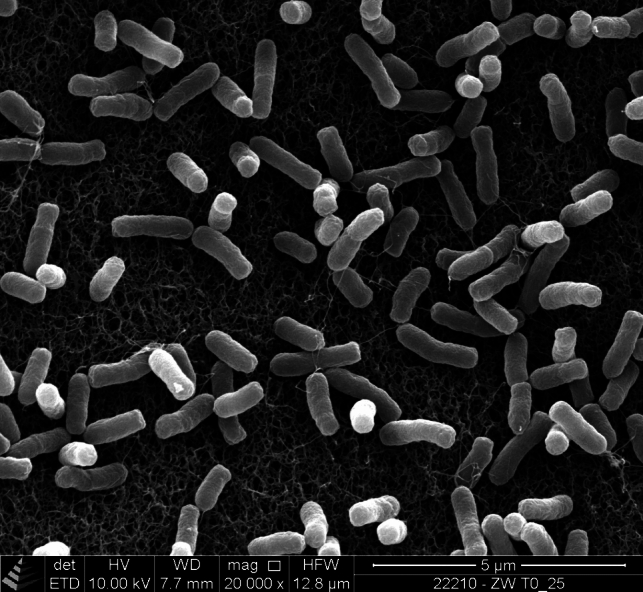
Scanning electron micrograph of cells of strain ZW T0_25T. Bacterial cells were grown on Standard Nutrient I agar for 48 h. Bars, 5 μm
Phylogenetic and Genome Analyses
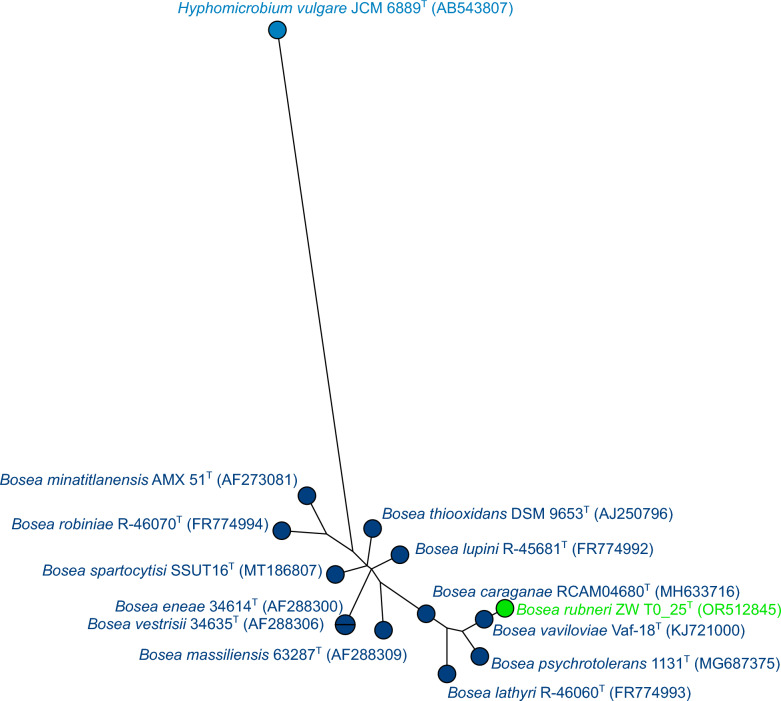
The 16S rRNA gene sequence (1059 bp, Genbank accession number OR512845) of strain ZW T0_25T was used for BLASTn search and was classified as belonging to the genus Bosea. Figure 2 shows a maximum-likelihood tree based on the 16S rRNA gene sequences of all type strains of Bosea and shows clearly that strain ZW T0_25T belongs to the genus Bosea but clusters apart from the other type strains. The cluster analysis was repeated with two additional clustering methods, namely neighbour joining and maximum parsimony (Figs. S1 and S2), and confirmed that strain ZW T0_25T belongs to the genus Bosea with a similarity value of 97.99% to the nearest neighbours according to 16S phylogeny, i.e., the type strains of B. eneae and B. vestrisii (Fig. S1). Pairwise nucleotide similarity comparisons of the 16S rRNA gene sequences using EZBioCloud [27] of strain ZW T0_25T to the type strains of all Bosea species showed that approximately half of the values are below the threshold value for species delimitation of 98.7% [39] (Table S1). Despite the other half had values above the threshold, this also happens in the pairwise comparisons of the type strains of other Bosea species (Table S1). Another example where this commonly happens is the genus Bradyrhizobium [40]. In any case, the threshold value of 16S rRNA gene similarities is only one part of the picture for a new species description and should be accompanied by other criteria like DDH, ANI as well as discriminant phenotypic properties [39].
Fig. 2.
Analysis of 16S rRNA gene sequences of strain ZW T0_25T and all twelve type strains of Bosea. Hyphomicrobium vulgare JCM 6889T was used as an outgroup. The tree was built using maximum likelihood with Jukes–Cantor as the evolutionary model (BioNumerics, version 8.1; Applied Maths). Square-root scaling was used to display branch length
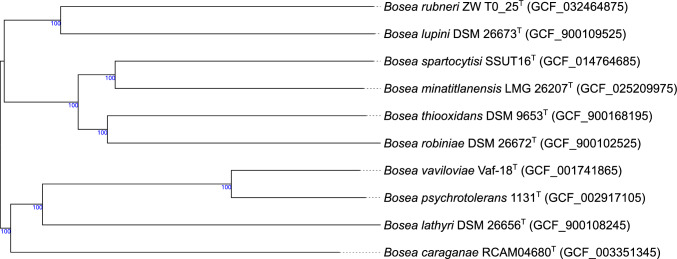
Characteristics of the genome sequences (e.g., genome size, GC% content and number of proteins) of strain ZW T0_25T and nine type strains of Bosea for which the whole genomes were available, were obtained by the TYGS [29] and are summarized in Table 1. The genome characteristics of strain ZW T0_25T were well comparable to genome characteristics of the type strains of Bosea. The genome size of ZW T0_25T has a length of 6.19 Mbp, and the genome sizes of Bosea type strains range from 4.71 to 6.72 Mbp. The genome of ZW T0_25T shows 66.9 GC%, and Bosea type strains show 64.8 to 68.6 GC%. A number of 5910 proteins is predicted for ZW T0_25T and proteins of Bosea type strains range from 4507 to 6078 proteins. These characteristics of the genome of ZW T0_25T were confirmed as analysed by Protologger [34]. In addition, the results of dDDH, ANI and AAI values are included in Table 1. The dDDH values of strain ZW T0_25T compared to the type strains of the genus Bosea were clearly below the threshold of 70% for species delimitation [41, 42] with the highest similarity of strain ZW T0_25T to the type strain B. lupini with a dDDH value of 27.1%. All OrthoANIu values of strain ZW T0_25T compared to the type strains of Bosea were below the 95–96% cut-off value for species delimitation [43] and similar to the results of dDDH: The highest similarity compared to strain ZW T0_25T was observed for the type strain B. lupini with a value of 83.66%. All AAI values of strain ZW T0_25T compared to the type strains of Bosea were below the 95–96% cut-off value for species delimitation as proposed by [44]. Again, the highest similarity compared to strain ZW T0_25T was observed for the type strain B. lupini with a value of 81.42%. Comparable to the results of dDDH, ANI and AAI, the GBDP tree of strain ZW T0_25T and type strains of Bosea confirmed that the nearest neighbour of strain ZW T0_25T was the type strain of B. lupini (Fig. 3).
Table 1.
Whole draft genome characteristics, values of digital DNA–DNA hybridization (dDDH), average nucleotide identity (ANI) and average amino acid identity (AAI) of ZW T0_25T and type strains of the genus Bosea
| Characteristic | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Genome length (Mbp)# | 6.19 | 6.17 | 5.92 | 6.13 | 4.71 | 6.39 | 5.38 | 5.92 | 5.37 | 6.72 |
| GC%# | 66.9 | 66.9 | 64.8 | 66.7 | 68.6 | 65.8 | 66.3 | 66.4 | 67.4 | 65.4 |
| Number of proteins# | 5910 | 5639 | 5477 | 5921 | 4507 | 5899 | 5006 | 5600 | 5054 | 6078 |
| dDDH [%]# versus ZW T0_25T | – | 24.4 | 23.4 | 27.1 | 23.7 | 23.6 | 23.2 | 23.3 | 23.8 | 23.8 |
| OrthoANIu [%]‡ versus ZW T0_25T | – | 81.02 | 79.69 | 83.66 | 80.50 | 80.14 | 79.94 | 79.55 | 80.41 | 80.01 |
| AAI [%]§ vs ZW T0_25T | – | 74.56 | 75.18 | 81.42 | 73.78 | 74.38 | 74.12 | 72.70 | 75.03 | 74.54 |
Strain: 1, B. rubneri sp. nov. ZW T0_25T (GCF_032464875); 2, B. caraganae RCAM04680T (GCF_003351345); 3, B. lathyri DSM 26656 T (GCF_900108245); 4, B. lupini LMG 26383 T (GCF_900109525); 5, B. minatitlanensis LMG 26207 T (GCF_025209975); 6, B. psychrotolerans 1131T (GCF_002917105); 7, B. robiniae DSM 26672 T (GCF_900102525); 8, B. spartocytisi SSUT16T (GCF_014764685); 9, B. thiooxidans DSM 9653 T (GCF_900168195); 10, B. vaviloviae Vaf-18 T (GCF_001741865)
For the species B. eneae, B. massiliensis and B. vestrisii no genome sequences were available at Genbank
#Values were obtained from TYGS [29, 30]. For dDDH formula d4 is given
‡Values were obtained from EzBioCloud [27, 31]
§Values were obtained from [32]
Fig. 3.
Genome BLAST Distance Phylogeny (GBDP) tree (whole-genome sequence based) of strain ZW T0_25T and all nine type strains of Bosea. The tree was built using the type strain genome server (TYGS) [29]. Tree inferred with FastME 2.1.6.1 [45] from GBDP distances calculated from genome sequences. The branch lengths are scaled in terms of GBDP distance formula d5. The numbers above branches are GBDP pseudo-bootstrap support values > 60% from 100 replications, with an average branch support of 91.3%. The tree was rooted at the midpoint [46]
In Silico and In Vivo Phenotypical and Biochemical Analyses
The in silico prediction for the phenotype of strain ZW T0_25T as predicted by Traitar [33] was well comparable to the predictions for the other Bosea type strains (Table S2). Phenotypic characteristics that are positive for strain ZW T0_25T according to both predictors in Traitar are aerobe, bacillus-similar morphology, Gram-negative and motile as well as catalase and oxidase positive which fits very well to the results of the wet lab analysis. Furthermore, strain ZW T0_25T was predicted to be bile-susceptible, to grow in KCN and to be colistin-polymyxin susceptible. In addition, growth on ordinary blood agar was predicted by Traitar, which was confirmed in the laboratory: Strain ZW T0_25T grows well on blood agar and shows γ-haemolysis. Moreover, the optimal growth conditions in Standard Nutrient I liquid medium for strain ZW T0_25T were observed to be at 24 °C. Furthermore, strain ZW T0_25T did not grow in MRS supplemented with 1% ox bile within 7 days which agrees with the Traitar prediction. In addition, in Traitar no growth was predicted in presence of 6.5% NaCl which was confirmed in the wet lab as growth was tested at 1 up to 10% NaCl and strain ZW T0_25T did only grow up to 1% NaCl. Growth of ZW T0_25T was observed at pH 7 and pH 8. No growth was observed in Standard Nutrient I at 42 °C nor in Standard Nutrient I supplemented with 0.01, 0.02 or 0.03% potassium tellurite at 24 °C. However, growth was observed in Standard Nutrient I without supplements at 7 °C after 7 days which is comparable to B. psychrotolerans which grows at temperatures higher than 5 °C [7]. In the API 20NE test, ZW T0_25T was positive for the assimilation of potassium gluconate and weakly positive for the assimilation of adipic acid. The assimilation of potassium gluconate has also been described for the type strains of B. spartocytisi, B. lupini, B. lathyri, B. robiniae, B. eneae, B. massiliensis and B. vestrisii as well as for the type strain of the type species B. thiooxidans [4, 7, 8]. Assimilation of adipic acid is also described for the type strains of B. thiooxidans, B. massiliensis, B. vestrisii, B. eneae, B. lupini and B. lathyri [4, 5]. Strain ZW T0_25T did not ferment any of the substrates (carbohydrates and derivatives) tested with API 50CH. This is in good agreement with the literature as B. minatitlanensis, B. eneae, B. masssiliensis, B. vestrisii and B. thiooxidans are also described to give negative results of all substrates tested with API 50CH [4, 6]. According to API Coryne and API ZYM, the following enzymatic activities were detected: pyrrolidonyl arylamidase, alkaline phosphatase, esterase, esterase lipase, acid phosphatase, leucine arylamidase and naphtol-AS-BI-phosphohydrolase. In addition, acetate production from acetyl-CoA, propionate production from propanoyl-CoA, L-cysteine and acetate production from sulphide and L-serine, as well as L-glutamate production from ammonia were predicted by Protologger. According to ResFinder and Protologger, no antibiotic resistance was detected in the genome of strain ZW T0_25T.
Analyses of Respiratory Quinones, Fatty Acids, Whole Cell Sugars and Polar Lipids
For strain ZW T0_25T, ubiquinone Q-10 was detected as the major quinone via HPLC–DAD and confirmed by mass spectrometry with 97.8%. In addition, minor amounts of Q-9 and Q-11 were detected with 0.7 and 1.6%, respectively. To the best of our knowledge, respiratory quinone data are only available for the type strains of B. thiooxidans [1], B. psychrotolerans, B. vaviloviae and B. lathyri [7]. A predominance of Q-10 with more than 97% has been detected in those four species of Bosea and in addition in ZW T0_25T. Furthermore, minor amounts of Q-11 are present in all Bosea species. In addition, Q-9 has been detected in ZW T0_25T in minor amounts (0.7%) which is described for the first time to be present in a species belonging to the genus Bosea. The polar lipid profile of strain ZW T0_25T was composed of one phosphatidylethanolamine, one phosphatidylglycerol, one aminophospholipid, two aminolipids, one glycolipid and two phospholipids (Fig. S3). To the best of our knowledge, polar lipids were described only for the type species B. thiooxidans [1] and for B. psychrotolerans, B. vaviloviae and B. lathyri [7]. Phosphatidylethanolamine and phosphatidylglycerol have been described for the abovementioned species of Bosea and are also present in ZW T0_25T. In addition, two aminolipids and one unidentified glycolipid have been detected in ZW T0_25T and are also described for B. vaviloviae and B. lathyri. The most noticeable difference in the polar lipid profile of strain ZW T0_25T is the absence of phosphatidylcholine, diphosphatidylglycerol and phosphatidylmonoethylamine compared to the other species of Bosea. The fatty acid profile of strain ZW T0_25T predominantly consisted of C18:1 w7c (63.3%), C16:1 w7c (19.5%) and C16:0 (7.1%) (Table S3). A high occurrence of C18:1 w7c was already stated as a feature in the description of the genus Bosea [1] which has been reported for all other type strains of Bosea species so far [3–9]. It is noticeable that no presence of C19:0 cyclo w8c was detected in strain ZW T0_25T as this feature can be found in all other type strains of Bosea. In addition, the analysis of the whole-cell hydrolysate of strain ZW T0_25T showed the presence of rhamnose, ribose and glucose as major whole cell sugars with traces of mannose.
Conclusion
In this study, a polyphasic approach including genomic as well as phenotypic and biochemical analysis was used to investigate the taxonomical position of strain ZW T0_25T. The genomic analysis comprised i.a. 16S rRNA gene comparisons, dDDH, ANI and AAI and phylogenomic clustering with closely related type strains of the genus Bosea. The results of this study indicate that strain ZW T0_25T represents the type strain of a novel species within the genus Bosea, for which the name Bosea rubneri sp. nov. is proposed.
Description of Bosea rubneri sp. nov.
Bosea rubneri sp. nov. (rub´ne.ri. N.L. gen. n. rubneri, of Rubner, referring to Max Rubner, a German physiologist after whom the Max Rubner-Institute was named, and where the type strain was isolated).
Cells occur as motile, Gram-stain negative and small, single rods. Mean size of cells is 1.16 µm in length and 0.40 µm in width. Colonies (diameter ≈ 4 mm) occur smooth, round and cream coloured after 48 h on Standard Nutrient I agar at 30 °C. Oxidase positive and catalase positive. Growth was observed at temperatures between 7 and 37 °C. The optimal growth temperature is 24 °C. Growth occurs in the presence of up to 1% NaCl and at pH 7 and pH 8 at 24 °C. No gas formation in Standard Nutrient I broth. It shows γ-haemolysis on blood agar. Cells are capable of producing pyrrolidonylarylamidase, alkaline phosphatase, esterase, esterase lipase, leucine arylamidase, acid phosphatase, naphthol-AS-BI-phosphohydrolase and β-glucosidase. Cells were able to assimilate L-arabinose, potassium gluconate and adipic acid. No other positive reactions are observed using API 20NE, API 50CH, API Coryne and API ZYM. The major respiratory quinone is Q-10. Rhamnose, ribose and glucose are detected as major whole cell sugars. The polar lipids are one phosphatidylethanolamine, one phosphatidylglycerol, one aminophospholipid, two aminolipids, one glycolipid and two phospholipids.
The type strain ZW T0_25T was isolated from a bulb of an onion hybrid race (Allium cepa var. Hytech F1) grown in Kleinhohenheim (Germany). Strain ZW T0_25T has been deposited as DSM 116094T and LMG 33098T. The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequences of strain ZW T0_25T is OR512845. The whole-genome shotgun project of strain ZW T0_25T has been deposited at DDBJ/ENA/GenBank under the accession JAWDID000000000. The version described in this paper is version JAWDID010000000. The genome size of the type strain is 6.19 Mbp and the GC content is 66.9%.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Firstly, we thank Lea Böckstiegel and Dr. Christoph Weinert for the wonderful idea to conduct microbiological analyses of the onion samples. In addition, we thank Jenny Burke-Murphy for excellent technical assistance regarding the isolation and characterization of all bacterial isolates including strain ZW T0_25T. Furthermore, we thank Lilia Rudolf and Stephanie Stricker for excellent additional technical assistance. Moreover, we thank Simone Brümmer and Gunilla Breutmann for excellent support with the scanning electron microscope. In addition, we thank Professor Bernhard Schink for valuable advice concerning the new bacterial name. Furthermore, we thank Dr. Maria Romo Perez and Prof. Dr. Christian Zörb (University of Hohenheim, Faculty of Agriculture, Department of Quality of Plant Products, 340e, Stuttgart) for providing the onion bulbs from the organic field trials performed within the scope of the ZwiebOEL project. Finally, we thank the Belgian Coordinated Collection of Microorganisms (BCCM) and the German Collection of Microorganisms and Cell Cultures (DSMZ) for strain deposition.
Author Contributions
DS and MH designed research, performed the polyphasic taxonomy and deposition, and drafted the manuscript. AM contributed with literature research of related type strains. CG performed MALDI analyses. BH performed SEM analysis. SK provided resources. All authors read, revised and approved the final manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was part of the project ‘ZwiebOEL: Exploration of the potential of old onion landraces for organic farming (project number 2819OE019)’ which was funded by the German Federal Ministry of Food and Agriculture (BMEL) within the framework ‘Federal Scheme for Organic Farming and Other Forms of Sustainable Agriculture (BÖL).’
Declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Research Involving Human and/or Animal Participants
This article does not contain any studies with animals or human participants.
Consent for Publication
All authors accepted to publish the submitted manuscript.
Footnotes
The accession numbers for the 16S rRNA and whole-genome sequences of strain ZW T0_25T (= DSM 116094 T = LMG 33093 T) at GenBank/EMBL/DDBJ are OR512845 and JAWDID000000000, respectively.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Das SK, Mishra AK, Tindall BJ, Rainey FA, Stackebrandt E. Oxidation of thiosulfate by a new bacterium, Bosea thiooxidans (strain BI-42) gen. nov., sp. nov.: analysis of phylogeny based on chemotaxonomy and 16S ribosomal DNA sequencing. Int J Syst Bacteriol. 1996;46:981–987. doi: 10.1099/00207713-46-4-981. [DOI] [PubMed] [Google Scholar]
- 2.Hördt A, López MG, Meier-Kolthoff JP, Schleuning M, Weinhold L-M, Tindall BJ, Gronow S, Kyrpides NC, Woyke T, Göker M. Analysis of 1,000+ type-strain genomes substantially improves taxonomic classification of alphaproteobacteria. Front Microbiol. 2020;11:468. doi: 10.3389/fmicb.2020.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sazanova AL, Safronova VI, Kuznetsova IG, Karlov DS, Belimov AA, Andronov EE, Chirak ER, Popova JP, Verkhozina AV, Willems A, Tikhonovich IA. Bosea caraganae sp. nov. a new species of slow-growing bacteria isolated from root nodules of the relict species Caragana jubata (Pall.) Poir. originating from Mongolia. Int J Syst Evol Microbiol. 2019;69:2687–2695. doi: 10.1099/ijsem.0.003509. [DOI] [PubMed] [Google Scholar]
- 4.La Scola B, Mallet MN, Grimont PAD, Raoult D (2003) Bosea eneae sp. nov., Bosea massiliensis sp. nov. and Bosea vestrisii sp. nov., isolated from hospital water supplies, and emendation of the genus Bosea (Das et al. 1996). Int J Syst Evol Microbiol 53:15–20. 10.1099/ijs.0.02127-0 [DOI] [PubMed]
- 5.De Meyer SE, Willems A. Multilocus sequence analysis of Bosea species and description of Bosea lupini sp. nov., Bosea lathyri sp. nov. and Bosea robiniae sp. nov., isolated from legumes. Int J Syst Evol Microbiol. 2012;62:2505–2510. doi: 10.1099/ijs.0.035477-0. [DOI] [PubMed] [Google Scholar]
- 6.Ouattara AS, Assih EA, Thierry S, Cayol JL, Labat M, Monroy O, Macarie H. Bosea minatitlanensis sp. nov., a strictly aerobic bacterium isolated from an anaerobic digester. Int J Syst Evol Microbiol. 2003;53:1247–1251. doi: 10.1099/ijs.0.02540-0. [DOI] [PubMed] [Google Scholar]
- 7.Albert RA, McGuine M, Pavlons SC, Roecker J, Bruess J, Mossman S, Sun S, King M, Hong S, Farrance CE, Danner J, Joung Y, Shapiro N, Whitman WB, Busse HJ. Bosea psychrotolerans sp. nov., a psychrotrophic alphaproteobacterium isolated from Lake Michigan water. Int J Syst Evol Microbiol. 2019;69:1376–1383. doi: 10.1099/ijsem.0.003319. [DOI] [PubMed] [Google Scholar]
- 8.Pulido-Suárez L, Flores-Félix JD, Socas-Pérez N, Igual JM, Velázquez E, Péix Á, León-Barrios M. Endophytic Bosea spartocytisi sp. nov. coexists with rhizobia in root nodules of Spartocytisus supranubius growing in soils of Teide National Park (Canary Islands) Syst Appl Microbiol. 2022;45:126374. doi: 10.1016/j.syapm.2022.126374. [DOI] [PubMed] [Google Scholar]
- 9.Safronova VI, Kuznetsova IG, Sazanova AL, Kimeklis AK, Belimov AA, Andronov EE, Pinaev AG, Chizhevskaya EP, Pukhaev AR, Popov KP, Willems A, Tikhonovich IA. Bosea vaviloviae sp. nov., a new species of slow-growing rhizobia isolated from nodules of the relict species Vavilovia formosa (Stev.) Fed. Antonie Van Leeuwenhoek. 2015;107:911–920. doi: 10.1007/s10482-015-0383-9. [DOI] [PubMed] [Google Scholar]
- 10.Stoll DA, Grimmler C, Hetzer B, Kulling SE, Huch M. Rathayibacter rubneri sp. nov. isolated from Allium cepa var. Rijnsburger, an onion landrace. Int J Syst Evol Microbiol. 2023;73:005811. doi: 10.1099/ijsem.0.005811. [DOI] [PubMed] [Google Scholar]
- 11.Stoll DA, Danylec N, Soukup ST, Hetzer B, Kulling SE, Huch M. Adlercreutzia rubneri sp. nov., a resveratrol-metabolizing bacterium isolated from human faeces and emended description of the genus Adlercreutzia. Int J Syst Evol Microbiol. 2021;71:004987. doi: 10.1099/ijsem.0.004987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Breznak JA, Costilow RN. Physicochemical factors in growth. In: Reddy CA, Beveridge TJ, Breznak JA, Marzluf GA, Schmidt TM, Snyder LR, editors. Methods for general and molecular microbiology. New York: Wiley; 2007. pp. 309–329. [Google Scholar]
- 14.Taylor WI, Achanzar D. Catalase test as an aid to the identification of Enterobacteriaceae. Appl Microbiol. 1972;24:58–61. doi: 10.1128/am.24.1.58-61.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vieira S, Huber KJ, Neumann-Schaal M, Geppert A, Luckner M, Wanner G, Overmann J. Usitatibacter rugosus gen. nov., sp. nov. and Usitatibacter palustris sp. nov., novel members of Usitatibacteraceae fam. Nov. within the order Nitrosomonadales isolated from soil. Int J Syst Evol Microbiol. 2021;71:004631. doi: 10.1099/ijsem.0.004631. [DOI] [PubMed] [Google Scholar]
- 16.Schumann P, Kalensee F, Cao J, Criscuolo A, Clermont D, Köhler JM, Meier-Kolthoff JP, Neumann-Schaal M, Tindall BJ, Pukall R (2021) Reclassification of Haloactinobacterium glacieicola as Occultella glacieicola gen. nov., comb. nov., of Haloactinobacterium album as Ruania alba comb. nov, with an emended description of the genus Ruania, recognition that the genus names Haloactinobacterium and Ruania are heterotypic synonyms and description of Occultella aeris sp. nov., a halotolerant isolate from surface soil sampled at an ancient copper smelter. Int J Syst Evol Microbiol 71:004769. 10.1099/ijsem.0.004769 [DOI] [PubMed]
- 17.Sasser M (1990) MIDI technical note 101: Identification of bacteria by gas chromatography of cellular fatty acids. MIDI, Newark, pp. 1–7 https://www.researchgate.net/publication/303137579_MIDI_technical_note_101_Identification_of_bacteria_by_gas_chromatography_of_cellular_fatty_acids_MIDI_Newark
- 18.Moss CW, Lambert-Fair MA. Location of double bonds in monounsaturated fatty acids of Campylobacter cryaerophila with dimethyl disulfide derivatives and combined gas chromatography-mass spectrometry. J Clin Microbiol. 1989;27:1467–1470. doi: 10.1128/jcm.27.7.1467-1470.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harvey DJ. Picolinyl esters as derivatives for the structural determination of long chain branched and unsaturated fatty acids. Biomed Mass Spectrom. 1982;9:33–38. doi: 10.1002/bms.1200090107. [DOI] [Google Scholar]
- 20.Spitzer V. Structure analysis of fatty acids by gas chromatography–low resolution electron impact mass spectrometry of their 4,4-dimethyloxazoline derivatives–a review. Prog Lipid Res. 1996;35:387–408. doi: 10.1016/s0163-7827(96)00011-2. [DOI] [PubMed] [Google Scholar]
- 21.Yu QT, Liu BN, Zhang JY, Huang ZH. Location of methyl branchings in fatty acids: fatty acids in uropygial secretion of Shanghai Duck by GC-MS of 4,4-dimethyloxazoline derivatives. Lipids. 1988;23:804–810. doi: 10.1007/BF02536225. [DOI] [PubMed] [Google Scholar]
- 22.Schumann P. Peptidoglycan structure. In: Rainey F, Oren A, editors. Methods in microbiology. London: Academic Press; 2011. pp. 101–129. [Google Scholar]
- 23.Staneck JL, Roberts GD. Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol. 1974;28:226–231. doi: 10.1128/am.28.2.226-231.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 25.Tindall BJ, Sikorski J, Smibert RA, Krieg NR. Phenotypic characterization and the principles of comparative systematics. In: Reddy CA, Beveridge TJ, Breznak JA, Marzluf GA, Schmidt TM, Snyder LR, editors. Methods for general and molecular microbiology. New York: Wiley; 2007. pp. 330–393. [Google Scholar]
- 26.Danylec N, Goebl A, Stoll DA, Hetzer B, Kulling SE, Huch M. Rubneribacter badeniensis gen. nov., sp. nov. and Enteroscipio rubneri gen. nov., sp. nov., new members of the Eggerthellaceae isolated from human faeces. Int J Syst Evol Microbiol. 2018;68:1533–1540. doi: 10.1099/ijsem.0.002705. [DOI] [PubMed] [Google Scholar]
- 27.Yoon S-H, Ha S-M, Kwon S, Lim J, Kim Y, Seo H, Chun J. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meier-Kolthoff JP, Göker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun. 2019;10:2182. doi: 10.1038/s41467-019-10210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meier-Kolthoff JP, Carbasse JS, Peinado-Olarte RL, Göker M. TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2021;50:D801–D807. doi: 10.1093/nar/gkab902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoon SH, Ha SM, Lim J, Kwon S, Chun J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek. 2017;110:1281–1286. doi: 10.1007/s10482-017-0844-4. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez-R LM, Konstantinidis KT. The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Preprints. 2016;4:e1900v1901. doi: 10.7287/peerj.preprints.1900v1. [DOI] [Google Scholar]
- 33.Weimann A, Mooren K, Frank J, Pope PB, Bremges A, McHardy AC, Segata N. From genomes to phenotypes: traitar, the microbial trait analyzer. mSystems. 2016;1:e00101–e00116. doi: 10.1128/mSystems.00101-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hitch TCA, Riedel T, Oren A, Overmann J, Lawley TD, Clavel T. Automated analysis of genomic sequences facilitates high-throughput and comprehensive description of bacteria. ISME Commun. 2021;1:16. doi: 10.1038/s43705-021-00017-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The Galaxy Community The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res. 2022;50:W345–W351. doi: 10.1093/nar/gkac247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, Philippon A, Allesoe RL, Rebelo AR, Florensa AF, Fagelhauer L, Chakraborty T, Neumann B, Werner G, Bender JK, Stingl K, Nguyen M, Coppens J, Xavier BB, Malhotra-Kumar S, Westh H, Pinholt M, Anjum MF, Duggett NA, Kempf I, Nykäsenoja S, Olkkola S, Wieczorek K, Amaro A, Clemente L, Mossong J, Losch S, Ragimbeau C, Lund O, Aarestrup FM. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. 2020;75:3491–3500. doi: 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinf. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zankari E, Allesøe R, Joensen KG, Cavaco LM, Lund O, Aarestrup FM. PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J Antimicrob Chemother. 2017;72:2764–2768. doi: 10.1093/jac/dkx217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yarza P, Yilmaz P, Pruesse E, Glockner FO, Ludwig W, Schleifer KH, Whitman WB, Euzeby J, Amann R, Rossello-Mora R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol. 2014;12:635–645. doi: 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]
- 40.Sun L, Zhang Z, Dong X, Tang Z, Ju B, Du Z, Wang E, Xie Z. Bradyrhizobium aeschynomenes sp. nov., a root and stem nodule microsymbiont of Aeschynomene indica. Syst Appl Microbiol. 2022;45:126337. doi: 10.1016/j.syapm.2022.126337. [DOI] [PubMed] [Google Scholar]
- 41.Richter M, Rossello-Mora R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 43.Lee I, Ouk Kim Y, Park S-C, Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66:1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]
- 44.Konstantinidis KT, Tiedje JM. Towards a genome-based taxonomy for prokaryotes. J Bacteriol. 2005;187:6258–6264. doi: 10.1128/jb.187.18.6258-6264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lefort V, Desper R, Gascuel O. FastME 2.0: a comprehensive, accurate, and fast distance-based phylogeny inference program. Mol Biol Evol. 2015;32:2798–2800. doi: 10.1093/molbev/msv150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Farris JS. Estimating phylogenetic trees from distance matrices. Am Nat. 1972;106:645–668. doi: 10.1086/282802. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.