Abstract
Infection of cells by foot-and-mouth disease virus (FMDV) results in the rapid inhibition of host cell protein synthesis. This process is accompanied by the early cleavage of the translation initiation factor eIF4G, a component of the cap-binding complex eIF4F. This cleavage is mediated by the leader (L) protease. Subsequently, as the virus proteins accumulate, secondary cleavages of eIF4G occur. Furthermore, eIF4A (46 kDa), a second component of eIF4F, is also cleaved in these later stages of the infection cycle. The 33-kDa cleavage product of eIF4A has lost a fragment from its N terminus. Transient-expression assays demonstrated that eIF4A was not cleaved in the presence of FMDV L or with the poliovirus 2A protease (which also mediates eIF4G cleavage) but was cleaved when the FMDV 3C protease was expressed. The FMDV 3C protease was also shown in such assays to induce cleavage of eIF4G, resulting in the production of cleavage products different from those generated by the L protease. Consistent with these results, within cells infected with a mutant FMDV lacking the L protease or within cells containing an FMDV replicon lacking L-P1 coding sequences it was again shown that eIF4A and eIF4G were cleaved.
Foot-and-mouth disease virus (FMDV), an aphthovirus, is a member of the family Picornaviridae, which also includes the enteroviruses, and rhinoviruses, and cardioviruses. In common with other picornaviruses, the FMDV RNA genome encodes a single large polyprotein which is proteolytically cleaved by internal proteases to produce the mature viral proteins (reviewed in references 2 and 57). FMDV RNA encodes two distinct trans-acting proteases, the leader (L) protease and the 3C protease. The L protein cleaves itself from the P1-2A structural protein precursor, while the 3C protease is responsible for most of the polyprotein cleavages. Poliovirus (PV) (like other enteroviruses) and the rhinoviruses do not produce a leader protein, but these viruses also encode a second protease, 2A, in addition to the 3C protease. The cardioviruses, e.g., encephalomyocarditis virus (EMCV), do produce a leader protein, but this has no proteolytic activity and only the 3C protein has trans-acting protease activity. The FMDV L protease is a member of the papain-like family of cysteine proteases (56), whereas the enterovirus 2A and all 3C proteases are members of the trypsin-like family of proteases (reviewed in reference 58).
The infection of cells by FMDV results in the inhibition of host cell protein synthesis. This process is accompanied by the cleavage of the translation initiation factor eIF4G (formerly termed p220) (42), a scaffold component of the cap-binding complex eIF4F, which also contains eIF4E and eIF4A. Two distinct binding sites on eIF4G for eIF4A, an RNA helicase, have been identified (27), as have distinct sites for interaction with eIF4E, the cap-binding protein (43), and eIF3 (40). Recently, binding sites for the poly(A)-binding protein (29) and the eIF4E kinase termed Mnk-1 (55) on eIF4G have also been identified. Two forms of eIF4A, termed eIF4AI and eIF4AII, which are 91% identical to each other have been found within mammalian cells (46). Furthermore, a second form of mammalian eIF4G, termed eIF4GII, has also been identified (24); this protein is a functional homologue of eIF4GI, although it is only about 50% identical in amino acid sequence.
The cleavage of eIF4G is achieved by the L protein of FMDV (15, 44). The L protein is produced in two forms, termed Lab and Lb, as a result of initiation of protein synthesis at two different AUG codons, 84 nucleotides apart (1, 60). Expression of either species of L results in the cleavage of eIF4G (44). The site in eIF4G cleaved by FMDV L in vitro has been identified as the C-terminal side of residue 479 (36) (residue 635 with the revised numbering system [29]). In PV- or rhinovirus-infected cells eIF4G is also cleaved (19, 20). The 2A proteases from these viruses are responsible for the cleavage of eIF4G (37). The site in eIF4G cleaved by the 2A proteases in vitro is adjacent to residue 486 (39) (residue 642 with the revised numbering system [29]).
Translation of picornavirus RNA is dependent on the presence of an internal ribosome entry site (IRES) element within the 5′ noncoding region (reviewed in references 6 and 31). These elements are about 450 bases in length and direct internal initiation of protein synthesis, which is maintained when cap-dependent protein synthesis is inhibited following the cleavage of eIF4G. The IRES-directed translation requires the same translation initiation factors as cap-dependent protein synthesis, except for the eIF4E and eIF4G components of eIF4F (52). It has been shown that a region of eIF4G (residues 457 to 1396 [now residues 613 to 1560]), like the very similar C-terminal cleavage products generated by the L and 2A proteases, can replace the full-length eIF4G for EMCV IRES-directed translation initiation (10, 47, 53). This region contains the two eIF4A binding sites but lacks the eIF4E binding site. Both cap-dependent protein synthesis and IRES-directed (cap-independent) protein synthesis are inhibited by dominant negative mutants of eIF4A (49), even though the ATP requirement, reflecting RNA unwinding by eIF4A, for IRES-directed translation is low (30).
There is generally a good correlation between loss of cap-dependent protein synthesis and the cleavage of eIF4G within PV-infected cells; however, under certain conditions this correlation can break down (9, 51). This suggests that other mechanisms are involved in the inhibition of host cell protein synthesis. Indeed, in EMCV-infected cells no cleavage of eIF4G occurs (45), but the inhibition of cap-dependent protein synthesis is coincident with the dephosphorylation (and hence activation) of the translational repressor 4E-BP1 (23, 50). The identification of eIF4GII and the observation that its cleavage can have kinetics different from those of eIF4GI within PV-infected or rhinovirus-infected cells (25, 61) further demonstrate the complexity of the shutoff process.
In this study it is demonstrated that the FMDV 3C protease, in addition to the FMDV Lb protease, induces the cleavage of the translation initiation factor eIF4G and furthermore that eIF4A is also cleaved in FMDV-infected cells through the action of the 3C protease alone.
MATERIALS AND METHODS
FMDV infection.
Monolayer cultures of baby hamster kidney (BHK) or Madin-Darby bovine kidney (MDBK) cells were infected with FMDV (O1Kaufbeuren B64 or O1BFS, respectively) in phosphate-buffered saline. After 30 min of virus absorption, virus growth medium (1% serum) was added to the cells, and incubation was continued until the indicated times (time zero was the time when virus was added to the cells). Where appropriate, metabolic labelling with [35S]EXPRESS (NEN) was performed in Met- and Cys-free and serum-free Dulbecco modified Eagle medium for 15 min. Cell extracts were prepared with buffer C (0.125 M NaCl, 20 mM Tris-HCl [pH 8.0], 0.5% NP-40). Aliquots were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (38) (6 or 10% polyacrylamide as indicated), and 35S-labelled proteins were visualized by autoradiography. Alternatively, samples analyzed by SDS-PAGE were transferred onto Immobilon-P (Millipore) and probed with rabbit antibodies specific for eIF4G (1:3,000; kindly provided by N. Sonenberg, Montreal, Canada), eIF4A (1:1,000) or eIF4B (1:1,000; both kindly provided by N. Méthot, Montreal, Canada), or actin (1:1,000; Sigma) or with mouse ascitic fluid containing an anti-eIF2α monoclonal antibody (MAb) (1:1,000; kindly provided by C. G. Proud, Dundee, United Kingdom) or anti-FMDV 3C MAb 1G1 (1:1,000; kindly provided by E. Brocci, Brescia, Italy). Rabbit antisera specific for the N- or C-terminal region of eIF4A (kindly provided by S. J. Morley, Sussex, United Kingdom) were also used at a 1:1,000 dilution. Detection on X-ray film was achieved by using peroxidase-linked donkey anti-rabbit immunoglobulin (Ig) or sheep anti-mouse Ig (1:3,000; Amersham) as appropriate and chemiluminescent reagents (Pierce).
FMDV A12-LLV2 (54) (kindly provided by M. J. Grubman, Plum Island, N.Y.) was used to infect monolayers of BHK cells, and samples were harvested at 8 or 16 h postinfection and analyzed as described above. At the latter time, significant cytopathic effect (CPE) was evident.
Transient-expression assays.
BHK cells were infected with the recombinant vaccinia virus vTF7-3 (22), which expresses the T7 RNA polymerase, and then transfected with plasmid DNA (2 μg) containing a T7 promoter by using Lipofectin (Life Technologies) and Optimem as described previously (5). After 20 h, cell extracts were prepared. SDS-PAGE and immunoblot analysis were performed as described for the FMDV-infected cell extracts.
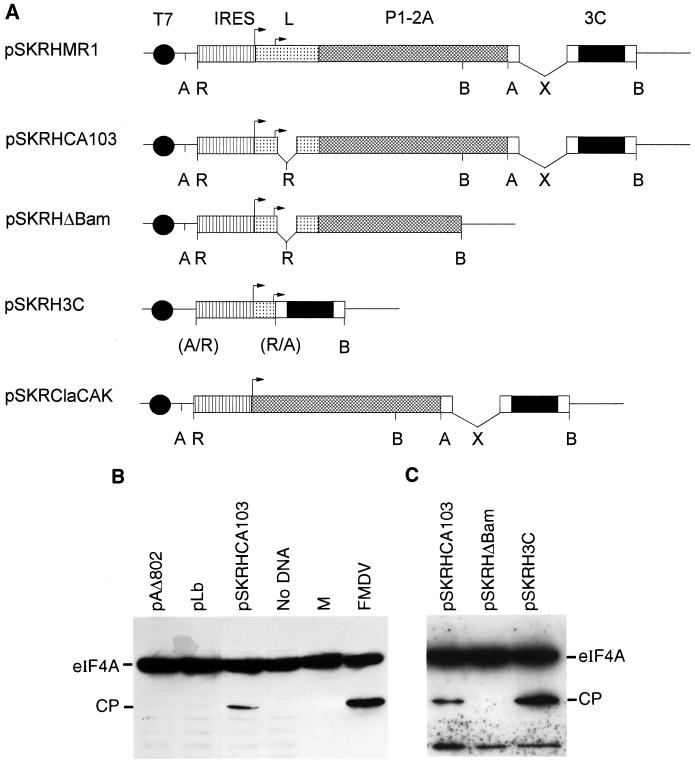
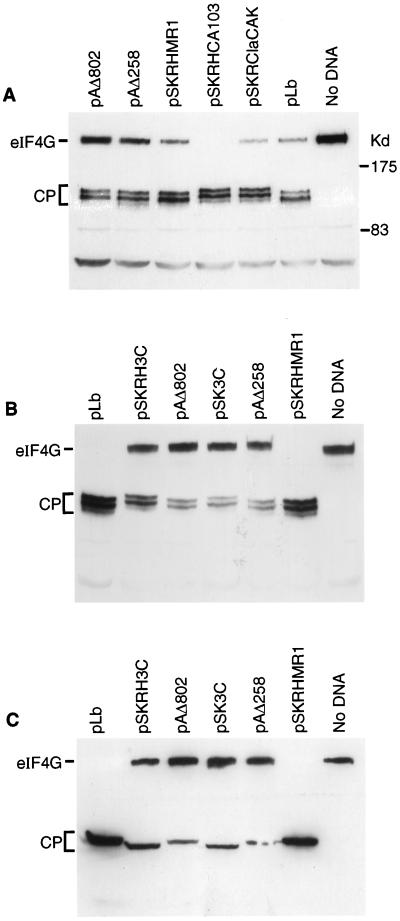
The plasmids used have been described previously (3, 34, 44) or are shown in Fig 3A. The derivatives of pSKRHCA103 were prepared by standard methods (59). The parental plasmid was digested with BamHI or ApaI, and the large fragment was self-ligated to produce pSKRHΔBam (see Fig. 3A) and pSKΔApa (not shown), respectively. The latter plasmid was linearized with ApaI, blunt ended with T4 DNA polymerase, treated with phosphatase, and ligated to the small EcoRI fragment (blunt ended) from pSKRHCA103 to produce pSKRH3C (see Fig. 3A). The plasmid pSKRClaCAK (which has a precise deletion of the L-coding sequence) was produced by a three-way ligation of EcoRI- and SmaI-digested pSK+ (Stratagene) with the ClaI (blunt ended)-EcoRI fragment from pSKRCla (16) containing the FMDV IRES with the large EcoRV-DraI fragment from pCAK (4).
FIG. 3.
Identification of eIF4A-directed protease. (A) Structures of plasmids expressing FMDV cDNA cassettes encoding the indicated proteases. All plasmids contain the T7 promoter and the FMDV IRES element. Restriction enzyme sites: A, ApaI; R, EcoRI; B, BamHI; X, XhoI. Parentheses indicate that the site is lost. Note that plasmids pSKRHMR1 and pSKRHCA103 were reported previously (3) and are included for completeness. (B) BHK cells were infected with vTF7-3 (22) and transfected with the indicated plasmids. After 20 h, cell extracts were prepared and analyzed by SDS-PAGE (10% polyacrylamide) and immunoblotting with anti-eIF4A. Samples from mock (lane M)- or FMDV-infected BHK cells were analyzed on the same gel. (C) vTF7-3-infected HTK-143 cells were transfected with the indicated plasmids (see panel A) and analyzed as for panel B.
FMDV replicon.
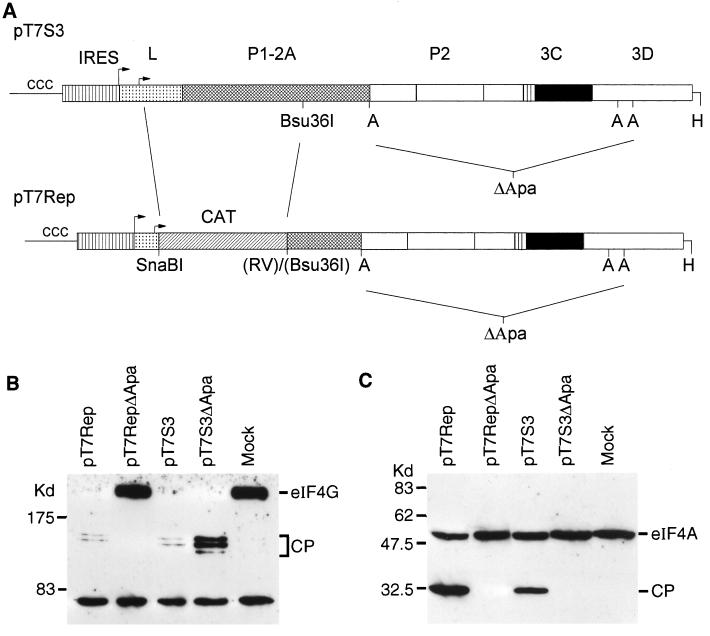
A plasmid (pT7Rep) encoding an FMDV replicon based on the type O infectious-copy cDNA pT7S3 (18) was constructed by deleting sequences between a SnaBI site engineered just 3′ of the Lb initiation codon and a blunt-ended Bsu361 site (within P1). The FMDV sequences were replaced with a PCR-generated fragment including the chloramphenicol acetyltransferase-coding sequence flanked by cleaved SnaBI and EcoRV restriction sites to restore the open reading frame. Replication-defective versions of pT7Rep and pT7S3 were produced by digestion with ApaI and recircularization to produce pT7RepΔApa and pT7S3ΔApa, respectively, which lack a major portion of the nonstructural protein-coding sequence (see Fig. 6A). A full description of these plasmids will be published elsewhere (G. M. McInerney, G. J. Belsham, and A. M. Q. King, unpublished results). RNA transcripts were prepared from the HpaI-linearized forms of these plasmids with T7 RNA polymerase by using an Ambion Megascript kit and introduced into BHK cells by electroporation with a Bio-Rad Gene Pulser. Significant CPE could be observed in cells containing replication-competent RNAs by 6 to 8 h postelectroporation. For these analyses, cell extracts were prepared at 6 h postelectroporation and analyzed for eIF4G and eIF4A as described above.
FIG. 6.
L-deficient FMDV replicon induces specific eIF4A and eIF4G cleavage. (A) The structure of the FMDV replicon (pT7Rep) is indicated together with the parental infectious cDNA pT7S3 (18) and the sequences removed (by digestion of the cDNA with ApaI and religation) to generate the nonreplicating derivatives of the replicon (pT7RepΔApa) and its parent (pT7S3ΔApa). Restriction sites: A, ApaI; H, HpaI; RV, EcoRV. (B and C) RNA transcripts prepared from HpaI-linearized plasmids by using T7 RNA polymerase were electroporated into BHK cells. After 6 h cell extracts were prepared and analyzed for eIF4G (B) and eIF4A (C) as for Fig. 3 and 4.
RESULTS
eIF4A and eIF4G are cleaved in FMDV-infected cells.
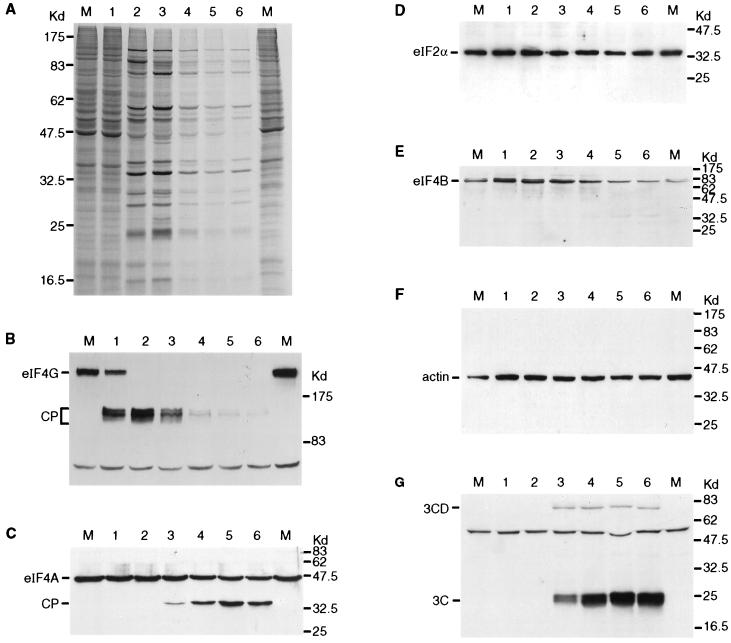
FMDV infection of BHK cells results in the rapid loss of host cell protein synthesis and a switch to the production of virus-encoded proteins (Fig. 1A). It is apparent that the highest rate of viral protein synthesis occurred at about 3 h postinfection in these cells. At later stages of infection there was some decrease in the rate of virus-encoded protein production, although the rate remained significant up to 6 h postinfection, when the experiment terminated. The cells showed significant CPE by about 4 h postinfection. The effect of the FMDV infection on a selection of translation initiation factors was assessed by immunoblotting cell extracts with a panel of specific antibodies. Figure 1B shows the analysis of eIF4G cleavage during this time course. Even at 1 h postinfection a major portion of this protein is cleaved, and by 2 h postinfection cleavage is complete, consistent with previous data (42). However, it is also apparent that subsequently the N-terminal cleavage products of eIF4G detected by this antibody (from rabbit 1620 [14a]) also decayed; thus, from 4 to 6 h postinfection most of these initial cleavage products have been lost. The same extracts were also analyzed for the presence of eIF4A (Fig. 1C). Unexpectedly, it was apparent that this initiation factor was also cleaved during the FMDV infection. The full-length eIF4A migrates at about 46 kDa, and the cleavage product migrates at about 33 kDa. The kinetics of eIF4A cleavage were clearly very different from those observed for eIF4G. The eIF4A cleavage product was observed from about 3 h postinfection and reached a plateau at about 4 to 5 h postinfection, with similar intensities of signal for the cleaved and intact forms of eIF4A. To check that this cleavage was not a reflection of general proteolytic degradation of cellular proteins, eIF2α, eIF4B, and actin were also examined. No significant loss of eIF2α (Fig. 1D) or actin (Fig. 1F) occurred throughout the time course, but a small change in the mobility of eIF4B was observed (Fig. 1E), perhaps reflecting modification of its phosphorylation state, in addition to a moderate decrease in its overall quantity. For reasons that will become apparent below, we also monitored the generation of the FMDV 3C protease (Fig. 1G); it is apparent that significant accumulation of this protein occurred from about 3 h postinfection. Since all picornavirus proteins are produced from a single polyprotein, it is expected that the production of all of the mature virus-encoded proteins will follow similar kinetics, although some particular processing intermediates may have distinct patterns of production.
FIG. 1.
Analysis of translation initiation factors within FMDV-infected cells. BHK cells were infected with FMDV. (A) At the indicated times (hours postinfection), cells were transferred to [35S]EXPRESS-containing medium and incubation was continued for a further 15 min. Cell extracts were prepared and analyzed by SDS-PAGE (10% polyacrylamide). (B to G) Alternatively, cells infected in parallel with those analyzed in panel A were harvested at the same times without metabolic labelling. Cell extracts were analyzed by SDS-PAGE (6% [B] or 10% [C to G] polyacrylamide) and immunoblotting with antibodies specific for the N-terminal region of eIF4G (B), eIF4A (C), eIF2α (D), eIF4B (E), actin (F), or FMDV 3C (G). Mock-infected BHK cell extracts (lanes M) were analyzed in parallel in each case.
To confirm the observation of eIF4A cleavage in FMDV-infected cells, a similar experiment was performed with bovine cells and the O1BFS strain of FMDV. The infection progressed more slowly in these cells. However, the key findings of eIF4G cleavage occurring early in infection followed by a subsequent decay of the initial cleavage products and the incomplete eIF4A cleavage occurring late in infection were again observed (data not shown). BHK cells infected with serotype A and SAT2 FMDVs have also been shown to have the same pattern of eIF4G and eIF4A cleavage (data not shown). Thus, these observations are consistent for different sources of virus and host cell.
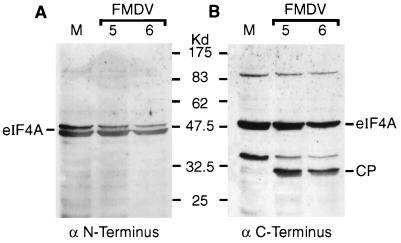
The change in mobility of eIF4A during FMDV infection was assumed to reflect loss of one terminus of the protein. In order to determine which terminus of eIF4A was being removed, samples from mock-infected and FMDV-infected cells were analyzed by immunoblotting with antisera specific for synthetic peptides derived from the N- and C-terminal regions of eIF4A. The N-terminal region-specific antibody recognized only full-length eIF4A (Fig. 2A). However, the C-terminal region-specific antibody recognized both full-length eIF4A and the cleavage product (Fig. 2B), indicating that the cleavage product had lost the original N terminus of eIF4A. Interestingly, the N-terminal region-specific antibody recognized a doublet of eIF4A species, perhaps eIF4AI and eIF4AII (Fig. 2A). However, the C-terminal region-specific antibody and that raised against recombinant eIF4AI specifically recognized only a single species of eIF4A, corresponding to eIF4AI, in uninfected cells (Fig. 1C and 2B). Since these two antisera both recognize the cleavage product, it seems that the fragment observed is derived from eIF4AI. However, this does not exclude eIF4AII also being cleaved, since its cleavage product may not be recognized by any of these antisera.
FIG. 2.
Characterization of eIF4A cleavage product in FMDV-infected cells. BHK cell extracts from mock-infected cells (lane M) or FMDV-infected cells (5 or 6 h postinfection as indicated) were analyzed by SDS-PAGE and immunoblotting with rabbit antisera specific for the N-terminal region peptide SQDSRSDNGPDGMEPEK (A) or the C-terminal region peptide DLPANRENYIHRTGRGGRFGRK (B) of the human eIF4AI sequence (kindly provided by S. J. Morley, University of Sussex). Detection on X-ray film was achieved by using peroxidase-labelled anti-rabbit IgG antibody and chemiluminescence reagents.
FMDV 3C induces cleavage of eIF4A in cells.
Two distinct trans-acting proteases (i.e., L and 3C) are produced by FMDV within cells; thus, it was necessary to use a different assay system to identify which virus-encoded enzyme is responsible for inducing the cleavage of eIF4A. Plasmids carrying cDNA cassettes of either the FMDV polyprotein (Fig. 3A), the FMDV Lb protease (from pLb [44]) or the PV 2A protease (from pAΔ802 [34]) under control of a T7 promoter were transfected into BHK cells infected with the recombinant vaccinia virus vTF7-3 (22), which expresses the T7 RNA polymerase. Cell extracts were prepared, and eIF4A was analyzed by immunoblotting as described above. Neither FMDV Lb nor the PV 2A protease had any effect on eIF4A (Fig. 3B). However, cleavage of eIF4A to generate a cleavage product with migration identical to that detected in FMDV-infected cells was observed in extracts expressing pSKRHCA103 (containing a truncated, inactive version of the Lb protease but with the intact FMDV 3C protease) (Fig. 3B). To confirm the role of 3C, two derivatives of pSKRHCA103 were prepared, namely, pSKRHΔBam (which lacks the 3C protease) and pSKRH3C (which essentially encodes only 3C) (Fig. 3A). These plasmids were also assayed, and as expected, eIF4A cleavage was detected with pSKRHCA103 and pSKRH3C but not with pSKRHΔBam (Fig. 3C).
FMDV 3C induces cleavage of eIF4G.
In similar studies, transfected-cell extracts were also analyzed for the cleavage of eIF4G by the viral proteases. As expected, eIF4G was cleaved by PV 2A (from plasmids pAΔ258 and pAΔ802 [34]) and FMDV L (from pLb [44] and pSKRHMR1 [3] [Fig. 3A]) to produce very similar cleavage products (Fig. 4A). However, unexpectedly, it was also found that plasmids which express the FMDV 3C protease, without the FMDV L protease or with a severely truncated inactive form of the L protease (pSKRClaCAK [Fig. 3A] and pSKRHCA103 [3]) also induced cleavage of eIF4G (Fig. 4A). This 3C-mediated cleavage of eIF4G yielded products, detected by this N-terminal region-specific antibody, which migrated slightly more slowly than those generated by PV 2A or FMDV L protease (Fig. 4A), indicating that a different cleavage site in eIF4G was recognized. When the plasmid included both FMDV L and 3C sequences (i.e., pSKRHMR1), the pattern of eIF4G cleavage products observed was the same as that observed with FMDV Lb alone (Fig. 4A). To analyze this process further, extracts from cells transfected with plasmids encoding FMDV Lb (pLb and pSKRHMR1), FMDV 3C (pSKRH3C and pSK3C), or PV 2A (pAΔ258 and pAΔ802) were analyzed with the anti-eIF4G antibody used above (Fig. 4B) and also with an antibody specific for the C-terminal fragment of eIF4G (Fig. 4C). The results demonstrated that the C terminus of eIF4G generated by the expression of FMDV 3C is smaller than the product induced by FMDV Lb or PV 2A (Fig. 4C; c.f. the larger fragment of eIF4G recognized by using the N-terminal region-specific anti-eIF4G antiserum in Fig. 4B) in cells expressing FMDV 3C. This indicates that the cleavage site generated by FMDV 3C expression lies C terminal of residue 642 (in the revised numbering system).
FIG. 4.
FMDV 3C induces cleavage of eIF4G (A). BHK cells were infected with vTF7-3 (as for Fig. 3) and transfected with the indicated plasmids (Fig. 3A). After 20 h, cell extracts were prepared and analyzed by SDS-PAGE (6% polyacrylamide) and immunoblotting with an N-terminal region-specific anti-eIF4G antiserum. (B and C) In a similar experiment vTF7-3-infected BHK cells were transfected with the indicated plasmids, and cell extracts were analyzed by SDS-PAGE (6% polyacrylamide) and immunoblotting with antisera specific for the N-terminal region of eIF4G (B) or the C-terminal fragment of eIF4G (C).
L protease-independent modification of cellular translation factors within FMDV-infected cells.
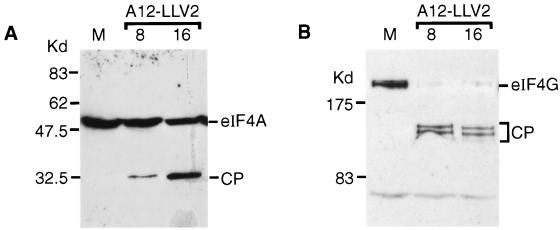
It was important to verify that the results obtained within the transient assay reflected the processes occurring within FMDV-infected cells. Hence, we made use of two different systems to analyze the effect of replicating mutant FMDV genomes which lack the L protease-coding sequence on eIF4G and eIF4A. A mutant of the A12 strain of FMDV, termed A12-LLV2, in which the Lb-coding sequence has been precisely deleted has been described previously (54). This virus is attenuated but does induce a delayed inhibition of host cell protein synthesis, and loss of eIF4G was observed. However, it was possible that this effect was simply a consequence of general protein degradation late in infection. Based on the observations described above, it could be predicted that both eIF4G and eIF4A should be cleaved by FMDV 3C within A12-LLV2-infected cells. To examine this, cells were infected with the mutant virus and cell extracts were prepared at 8 and 16 h postinfection. At the latter time significant CPE was apparent. By using immunoblot analysis, it was apparent that specific degradation of both eIF4G and eIF4A occurred in the A12-LLV2-infected cells, generating the characteristic pattern of cleavage products (Fig. 5).
FIG. 5.
eIF4A and eIF4G cleavage in A12-LLV2 FMDV-infected cells. Mock (lanes M)- or A12-LLV2 FMDV-infected BHK cells were harvested at 8 or 16 h postinfection as indicated. Cell extracts were prepared and analyzed by SDS-PAGE and immunoblotting with anti-eIF4A (A) or anti-eIF4G (B) as for Fig. 3 and 4.
In an alternative approach, a replicon based on the FMDV genome in which the L-coding sequence and part of the P1-coding sequence have been replaced by the chloramphenicol acetyltransferase-coding sequence has been produced. The structure of the replicon is indicated in Fig. 6A. This is the first FMDV-derived replicon reported and will be described in more detail elsewhere (McInerney et al., unpublished results). RNA transcripts from the full-length plasmid (pT7Rep) and from a control plasmid in which much of the replicative protein-coding sequences have been removed (pT7RepΔApa [Fig. 6A]) were produced. In parallel, transcripts from a full-length infectious copy of type O FMDV (pT7S3 [18]) and its replication-defective derivative pT7S3ΔApa (Fig. 6A) were also prepared. All four transcripts were introduced separately into BHK cells by electroporation, and cell extracts were prepared after 6 h. Immunoblot analysis of eIF4G showed that it was cleaved in cells containing the functional (but L-deficient) replicon pT7Rep but not in cells containing the replication- and 3C-deficient version pT7RepΔApa (Fig. 6B). In contrast, in cells containing transcripts which encoded the L protease, the eIF4G was cleaved with or without RNA replication (Fig. 6B). The cleavage of eIF4A was observed in cell extracts containing both of the replication-competent RNAs (Fig. 6C) but not in control cells or when 3C was removed and replication was blocked.
DISCUSSION
It has been known for some time that the picornavirus proteases cleave certain cellular proteins in addition to cleaving the viral polyprotein. The cleavage of eIF4G by PV 2A and FMDV L proteases has been well characterized and is clearly important in the biology of these picornaviruses, since it contributes to the inhibition of host cell protein synthesis.
Analysis of PV-infected cells has also revealed the PV 3C-dependent cleavage of certain transcription factors (13, 14, 62–64) which may be responsible for the virus-induced loss of host cell transcription. Furthermore, the microtubule-associated protein 4 has been shown to be cleaved by PV 3C, which may influence cell morphology (32). The double-stranded RNA-activated protein kinase, PKR, is also degraded during PV infection (7, 8); presumably this assists the virus in avoiding the host defense system, which could inhibit all protein synthesis through the phosphorylation of eIF2α. Recently evidence has been presented that the poly(A)-binding protein is also cleaved late in PV-infected cells (33, 35), and both the 2A and 3C proteases have been implicated in this cleavage event.
Within FMDV-infected cells, cleavage of histone H3 has been observed (21), and it has been suggested this may have a role in the inhibition of host cell transcription. Within cell extracts, it has been shown that recombinant FMDV Lb protease can cleave a variety of substrates (65).
This is the first report that the FMDV 3C protease induces the cleavage of cellular translation initiation factors. The evidence from the transient-expression assays showed that FMDV 3C is able to induce the cleavage of both eIF4A and eIF4G. The delayed appearance of eIF4A cleavage products (c.f. the rapid generation of the eIF4G cleavage products) within FMDV-infected cells is entirely consistent with the kinetics of FMDV 3C accumulation (Fig. 1). This contrasts with the L-mediated cleavage of eIF4G, which occurs before detectable accumulation of virus-encoded proteins. However, the secondary decay of eIF4G cleavage products does occur after 3C protein has significantly accumulated. The secondary decay of eIF4G products in FMDV-infected cells contrasts with the stability of the primary cleavage products in PV-infected and rhinovirus-infected cells (19, 61) and with the complete stability of eIF4G in EMCV-infected cells (45). The data presented here shows that in the presence of both L and 3C proteases, the pattern of eIF4G products observed is very similar to that observed to result from the activity of L alone (Fig. 4). This probably reflects the fact that the L-mediated cleavage of eIF4G occurs at low levels of L protease expression, and by the time sufficient 3C accumulates to achieve significant cleavage, all of the eIF4G has already been cleaved by the L protease. However, it seems possible that the decay of the initial N-terminal eIF4G cleavage products in FMDV-infected cells represents cleavage of these products by 3C alone or possibly by 3C and L together, since secondary cleavages of eIF4G by L have been observed in vitro (40). It should be noted that the mapping of the cleavage site generated by FMDV 3C indicates that it is on the C-terminal side of the site cleaved as a result of FMDV L protease expression. Hence, the loss of the initial cleavage products must represent cleavages at different sites. It is possible that the initial cleavage of eIF4G reveals new cleavage sites within the N-terminal domain which are susceptible to attack by either the L- or 3C-dependent process.
There is still some controversy concerning whether eIF4G cleavage induced by PV 2A expression is a direct event or not. In vitro analysis indicates that direct cleavage is possible (11, 26, 39); however, other studies have indicated the separation of eIF4G cleavage activity from PV 2A (12, 41). It has been proposed that a cellular protease is activated by the viral proteases, since only very low levels of PV 2A and FMDV L are required within infected cells to bring about complete eIF4G cleavage (12) and (Fig. 1 and 6). In vitro, rather high levels of protease are required; however, these levels can be reduced by the inclusion of eIF4E (26). Thus, it may be that the presence of other proteins within the infected cell markedly increases the susceptibility of eIF4G to cleavage by low-level viral protease. It remains to be determined whether the cleavage of eIF4G or eIF4A by FMDV 3C is achieved by a direct or indirect mechanism.
Loss of the N-terminal cleavage products of eIF4G may not have any significant effect on the translation of the viral RNA, since only part of the C-terminal region of eIF4G (residues 480 to 1396 [now residues 636 to 1560]) is required (by analogy to other picornaviruses [10, 47, 53]) for IRES activity. It is apparent that the viral RNA continues to be translated when almost complete loss of the N-terminal cleavage products of eIF4G has occurred (Fig. 1). Presently, the form of eIF4G which remains in these cells is not known. It has been shown that a central region (amino acids 457 to 932 [now residues 613 to 1086]) of eIF4G, which includes a single eIF4A binding site is sufficient to replace eIF4G for the formation of a preinitiation complex on the EMCV IRES (53). Further analysis of the residual form of the eIF4G late in FMDV-infected cells may permit characterization of the minimal element required to support FMDV IRES activity.
The cleavage of eIF4A is a process which may be expected to be unfavorable for the virus, since eIF4A is required for the virus RNA to be translated (49). However, since the cleavage is only partial, this may not be very serious for the cellular translation machinery, since eIF4A is the most abundant of the translation initiation factors. It is possible that partial loss of this protein may reflect the loss of only one of the two eIF4A species, eIF4AI and eIF4AII, present within mammalian cells. As explained above, it seems probable that in our analysis we are monitoring eIF4AI; indeed, we have recently demonstrated cleavage within cells of a tagged cDNA clone of human eIF4AI by FMDV 3C protease (N. Ross-Smith and G. J. Belsham, unpublished results). Furthermore, it may be only the eIF4A associated with the cleaved eIF4G (and/or other proteins, e.g., p97 [28], which also bind eIF4A), or, conversely, the eIF4A that is outside such complexes, that is susceptible to cleavage. If any of these scenarios are correct, then clearly the complete loss of one population of eIF4A may have a significant effect on the cell if the different pools of eIF4A have distinct roles. The data obtained by using the antipeptide antisera specific for the two termini of eIF4A clearly demonstrated that the cleavage product has lost the N terminus of the native molecule (this result is also confirmed by the generation of a 33-kDa cleavage product carrying a C-terminal tag from the eIF4AI clone in the presence of FMDV 3C as discussed above [Ross-Smith and Belsham, unpublished results]). Near the N terminus of eIF4A is one of the conserved ATPase motifs characteristic of the DEAD box family of proteins (48). The change in mobility (from 46 to 33 kDa), if accurately representing the size change, indicates a loss of about 100 amino acids. The loss of this much sequence can be expected to inactivate the protein; however, it should also be borne in mind that although the protein sequence is cut in the presence of FMDV 3C, the protein segments may not dissociate within the cell and the fragments may continue to function together. Alternatively, the cleavage product may act as a dominant negative inhibitor (49) and thus exert a negative effect on viral protein synthesis. It is interesting that no cleavage of eIF4A has been detected in PV-infected or EMCV-infected cells (17; A. Gradi, Y. V. Svitkin, and N. Sonenberg, personal communication). One possible role for a reduction in the capacity of the cell to produce viral protein late in the infection cycle would be to enhance the packaging of RNA transcripts into capsids, rather than the RNA just being used to produce more protein.
It was noted that intact eIF4G was lost late in infection with the leaderless form of FMDV, A12-LLV2 (54), but it was not established whether this was merely a consequence of cellular breakdown. The results obtained here indicate that the FMDV 3C within this mutant virus can induce specific cleavage of eIF4G and also eIF4A. It seems likely that late in infection the accumulation of 3C within the cell exceeds the requirement for polyprotein processing, and hence cleavage of alternative substrates is facilitated.
As noted above, no loss of the initial eIF4G cleavage products occurs in PV-infected or rhinovirus-infected cells (19, 61), and no breakdown of intact eIF4G occurs in EMCV-infected cells (45). Thus, not all picornavirus 3C proteases have the ability to cleave eIF4G or, as described above, eIF4A. Hence, from these results it can be concluded that only the FMDV 3C protease is capable of inducing the cleavage of eIF4A and eIF4G, while the FMDV Lb and PV 2A proteases each mediate cleavage of eIF4G but not eIF4A and other picornavirus 3C proteases cleave neither of these factors within cells.
ACKNOWLEDGMENTS
We thank E. Brocci (Brescia, Italy) for anti-FMDV 3C MAb 1G1, N. Méthot (McGill, Montreal, Canada) for anti-eIF4A and anti-eIF4B antisera, S. J. Morley (Sussex, United Kingdom) for anti-eIF4A peptide antisera, C. G. Proud (Dundee, United Kingdom) for eIF2α antibodies, and N. Sonenberg (McGill) for anti-eIF4G antisera. We also thank T. Jackson (Pirbright, United Kingdom) and M. Grubman (Plum Island, N.Y.) for FMDV stocks and A. M. Q. King for interest and reading of the manuscript. R. A. Seamons provided skilled technical assistance in the early part of this work.
G.M.M. gratefully acknowledges a studentship from the Institute for Animal Health.
REFERENCES
- 1.Belsham G J. Dual initiation sites of protein synthesis on foot-and-mouth disease virus RNA are selected following internal entry and scanning of ribosomes in vivo. EMBO J. 1992;11:1105–1110. doi: 10.1002/j.1460-2075.1992.tb05150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belsham G J. Distinctive features of foot-and-mouth disease virus, a member of the picornavirus family; aspects of virus protein synthesis, protein processing and structure. Prog Biophys Mol Biol. 1993;60:241–260. doi: 10.1016/0079-6107(93)90016-D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belsham G J, Brangwyn J K. A region of the 5′ noncoding region of foot-and-mouth disease virus RNA directs efficient internal initiation of protein synthesis within cells: involvement with the role of L protease in translational control. J Virol. 1990;64:5389–5395. doi: 10.1128/jvi.64.11.5389-5395.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belsham G J, Abrams C A, King A M Q, Roosien J, Vlak J M. Myristoylation of foot-and-mouth disease virus capsid protein precursors is independent of other viral proteins and occurs in both mammalian and insect cells. J Gen Virol. 1991;72:747–751. doi: 10.1099/0022-1317-72-3-747. [DOI] [PubMed] [Google Scholar]
- 5.Belsham G J. Analysis of picornavirus internal ribosome entry site function in vivo. In: Richter J, editor. mRNA formation and function. New York, N.Y: Academic Press; 1997. pp. 323–340. [Google Scholar]
- 6.Belsham G J, Sonenberg N. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol Rev. 1996;60:499–511. doi: 10.1128/mr.60.3.499-511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Black T L, Safer B, Hovanessian A, Katze M G. The cellular 68,000-Mr protein kinase is highly autophosphorylated and activated yet significantly degraded during poliovirus infection: implications for translational regulation. J Virol. 1989;63:2244–2251. doi: 10.1128/jvi.63.5.2244-2251.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Black T L, Barber G N, Katze M G. Degradation of interferon-induced 68,000-Mr protein-kinase by poliovirus requires RNA. J Virol. 1993;67:791–800. doi: 10.1128/jvi.67.2.791-800.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonneau A-M, Sonenberg N. Proteolysis of the p220 component of the cap-binding protein complex is not sufficient for complete inhibition of host cell protein synthesis after poliovirus infection. J Virol. 1987;61:986–991. doi: 10.1128/jvi.61.4.986-991.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borman A M, Kirchweger R, Ziegler E, Rhoads R E, Skern T, Kean K M. eIF4G and its proteolytic cleavage products: effect on initiation of protein synthesis from capped, uncapped and IRES-containing mRNAs. RNA. 1997;3:186–196. [PMC free article] [PubMed] [Google Scholar]
- 11.Bovee M L, Lamphear B, Rhoads R E, Lloyd R E. Direct cleavage of eIF4G by poliovirus 2A protease is inefficient in vitro. Virology. 1998;245:241–249. doi: 10.1006/viro.1998.9172. [DOI] [PubMed] [Google Scholar]
- 12.Bovee M L, Marissen W E, Zamora M, Lloyd R E. The predominant eIF4G-specific cleavage activity in poliovirus-infected HeLa cells is distinct from 2A protease. Virology. 1998;245:229–240. doi: 10.1006/viro.1998.9171. [DOI] [PubMed] [Google Scholar]
- 13.Clark M E, Hammerle T, Wimmer E, Dasgupta A. Poliovirus 3C proteinase converts an active form of transcription factor IIIC to an inactive form: a mechanism for inhibition of host cell polymerase III transcription by poliovirus. EMBO J. 1991;10:2941–2947. doi: 10.1002/j.1460-2075.1991.tb07844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark M E, Lieberman P M, Berk A J, Dasgupta A. Direct cleavage of human TATA-binding protein by poliovirus protease 3C in vivo and in vitro. Mol Cell Biol. 1993;13:1232–1237. doi: 10.1128/mcb.13.2.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Craig A W B, Haghighat A, Yu A T K, Sonenberg N. Interaction of polyadenylate-binding protein with the eIF4G homolgue PAIP enhances translation. Nature. 1998;392:520–523. doi: 10.1038/33198. [DOI] [PubMed] [Google Scholar]
- 15.Devaney M A, Vakharia V N, Lloyd R E, Ehrenfeld E, Grubman M J. Leader protein of foot-and-mouth disease virus is required for cleavage of the p220 component of the cap binding protein complex. J Virol. 1988;62:4407–4409. doi: 10.1128/jvi.62.11.4407-4409.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drew J, Belsham G J. trans complementation of defective foot-and-mouth disease virus internal ribosome entry site elements. J Virol. 1994;68:697–703. doi: 10.1128/jvi.68.2.697-703.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duncan R, Etchison D, Hershey J W B. Protein synthesis eukaryotic initiation factors 4A and 4B are not altered by poliovirus infection of HeLa cells. J Biol Chem. 1983;258:7236–7239. [PubMed] [Google Scholar]
- 18.Ellard F M, Drew J, Blakemore W, Stuart D I, King A M Q. Evidence for the role of histidine 142 of protein 1C in the acid-induced disassembly of foot-and-mouth disease virus capsids. J Gen Virol. 1999;80:1911–1918. doi: 10.1099/0022-1317-80-8-1911. [DOI] [PubMed] [Google Scholar]
- 19.Etchison D, Milburn S, Edery I, Sonenberg N, Hershey J W B. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000 dalton polypeptide associated with eukaryotic initiation factor 3 and a cap-binding complex. J Biol Chem. 1982;257:14806–14810. [PubMed] [Google Scholar]
- 20.Etchison D, Fout S. Human rhinovirus 14 infection of HeLa cells results in the proteolytic cleavage of the p220 cap-binding complex subunit and inactivates globin mRNA translation in vitro. J Virol. 1985;54:634–638. doi: 10.1128/jvi.54.2.634-638.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falk M M, Grigera P R, Bergmann I E, Zibert A, Multhaup G, Beck E. FMDV protease 3C induces specific proteolytic cleavage of host cell histone H3. J Virol. 1990;64:748–756. doi: 10.1128/jvi.64.2.748-756.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gingras A-C, Svitkin Y, Belsham G J, Pause A, Sonenberg N. Activation of the translational repressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus. Proc Natl Acad Sci USA. 1996;93:5578–5583. doi: 10.1073/pnas.93.11.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gradi A, Imataka H, Svitkin Y V, Rom E, Raught B, Morino S, Sonenberg N. A novel functional human eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:334–342. doi: 10.1128/mcb.18.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gradi A, Svitkin Y V, Imataka H, Sonenberg N. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc Natl Acad Sci USA. 1998;95:11089–11094. doi: 10.1073/pnas.95.19.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haghighat A, Svitkin Y, Novoa I, Küechler E, Skern T, Sonenberg N. The eIF4G-eIF4E complex is the target for direct cleavage by the rhinovirus 2A protease. J Virol. 1996;70:8444–8450. doi: 10.1128/jvi.70.12.8444-8450.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imataka H, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol Cell Biol. 1997;17:6940–6947. doi: 10.1128/mcb.17.12.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imataka H, Olsen H S, Sonenberg N. A new translational regulator with homology to eukaryotic translation initiation factor 4G. EMBO J. 1997;16:817–825. doi: 10.1093/emboj/16.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackson R J. The ATP requirement for initiation of eukaryotic translation varies according to the mRNA species. Eur J Biochem. 1991;200:285–294. doi: 10.1111/j.1432-1033.1991.tb16184.x. [DOI] [PubMed] [Google Scholar]
- 31.Jackson R J, Hunt S L, Reynolds J E, Kaminski A. Cap-dependent and cap-independent translation: operational distinctions and mechanistic interpretations. Curr Top Microbiol Immunol. 1995;203:1–30. doi: 10.1007/978-3-642-79663-0_1. [DOI] [PubMed] [Google Scholar]
- 32.Joachims M, Harris K S, Etchison D. Poliovirus protease 3C mediates cleavage of microtubule-associated protein 4. Virology. 1995;211:451–461. doi: 10.1006/viro.1995.1427. [DOI] [PubMed] [Google Scholar]
- 33.Joachims M, Van Breugel P C, Lloyd R E. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J Virol. 1999;73:718–727. doi: 10.1128/jvi.73.1.718-727.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaminski A, Howell M T, Jackson R J. Initiation of encephalomyocarditis virus RNA translation: the authentic initiation site is not selected by a scanning mechanism. EMBO J. 1990;9:3753–3759. doi: 10.1002/j.1460-2075.1990.tb07588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kerekatte V, Keiper B D, Badorff C, Cai A, Knowlton K U, Rhoads R E. Cleavage of poly(A)-binding protein by coxsackievirus 2A protease in vitro and in vivo: another mechanism for host protein synthesis shutoff? J Virol. 1999;73:709–717. doi: 10.1128/jvi.73.1.709-717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirchweger R, Ziegler E, Lamphear B J, Waters D, Liebig H-D, Sommergruber W, Sobrino F, Hohenadl C, Blaas D, Rhoads R E, Skern T. Foot-and-mouth disease virus leader proteinase: purification of the Lb form and determination of its cleavage site on eIF-4γ. J Virol. 1994;68:5677–5684. doi: 10.1128/jvi.68.9.5677-5684.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kräusslich H-G, Nicklin M J, Toyoda H, Etchison D, Wimmer E. Poliovirus protease 2A induces cleavage of eucaryotic initiation factor 4F polypeptide p220. J Virol. 1987;61:2711–2718. doi: 10.1128/jvi.61.9.2711-2718.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 39.Lamphear B J, Yan R Q, Yang F, Waters D, Liebig H D, Klump H, Küechler E, Skern T, Rhoads R E. Mapping of the cleavage site in protein synthesis initiation factor eIF-4γ of the 2A proteases from human coxsackievirus and rhinovirus. J Biol Chem. 1993;268:19200–19203. [PubMed] [Google Scholar]
- 40.Lamphear B J, Kirchweger R, Skern T, Rhoads R E. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 41.Lloyd R E, Toyoda H, Etchison D, Wimmer E, Ehrenfeld E. Cleavage of the cap binding complex polypeptide p220 is not effected by the second poliovirus protease 2A. Virology. 1986;150:299–303. doi: 10.1016/0042-6822(86)90291-6. [DOI] [PubMed] [Google Scholar]
- 42.Lloyd R E, Grubman M, Ehrenfeld E. Relationship of p220 cleavage during picornavirus infection to 2A protease sequences. J Virol. 1988;62:4216–4223. doi: 10.1128/jvi.62.11.4216-4223.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4γ and the translational repressors 4E-binding proteins. Mol Cell Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Medina M, Domingo E, Brangwyn J K, Belsham G J. The two species of the foot-and-mouth disease virus leader protein, expressed individually, exhibit the same activities. Virology. 1993;194:355–359. doi: 10.1006/viro.1993.1267. [DOI] [PubMed] [Google Scholar]
- 45.Mosenkis J, Daniels-McQueen S, Janovec S, Duncan R, Hershey J W B, Grifo J A, Merrick W C, Thach R E. Shutoff of host translation by encephalomyocarditis virus infection does not involve cleavage of the eukaryotic initiation factor 4F polypeptide that accompanies poliovirus infection. J Virol. 1985;54:643–645. doi: 10.1128/jvi.54.2.643-645.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nielsen P J, Trachsel H. The mouse protein synthesis initiation factor 4A gene family includes two related functional genes which are differentially expressed. EMBO J. 1988;7:2097–2105. doi: 10.1002/j.1460-2075.1988.tb03049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohlmann T, Rau M, Pain V M, Morley S J. The C-terminal domain of eukaryotic protein synthesis initiation (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J. 1996;15:1371–1382. [PMC free article] [PubMed] [Google Scholar]
- 48.Pause A, Sonenberg N. Helicases and RNA unwinding in translation. Curr Opin Struct Biol. 1993;3:953–959. [Google Scholar]
- 49.Pause A, Methot N, Svitkin Y, Merrick W C, Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 1994;13:1205–1215. doi: 10.1002/j.1460-2075.1994.tb06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pause A, Belsham G J, Gingras A-C, Donze O, Lin T-A, Lawrence J C, Jr, Sonenberg N. Insulin dependent stimulation of protein synthesis by phosphorylation of a novel regulator of 5′-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 51.Perez L, Carrasco L. Lack of direct correlation between p220 cleavage and the shut off of host translation after poliovirus infection. Virology. 1991;189:178–186. doi: 10.1016/0042-6822(92)90693-j. [DOI] [PubMed] [Google Scholar]
- 52.Pestova T V, Hellen C U T, Shatsky I. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pestova T V, Shatsky I N, Hellen C U T. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Piccone M E, Rieder E, Mason P W, Grubman M J. The foot-and-mouth disease virus leader proteinase is not required for viral replication. J Virol. 1995;69:5376–5382. doi: 10.1128/jvi.69.9.5376-5382.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pyronnet S, Imataka H, Gingras A-C, Fukunaga R, Hunter T, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) recruits Mnk 1 to phosphorylate eIF4E. EMBO J. 1999;18:270–279. doi: 10.1093/emboj/18.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roberts P J, Belsham G J. Identification of critical amino acids within the foot-and-mouth disease virus Leader protein, a cysteine protease. Virology. 1995;213:140–146. doi: 10.1006/viro.1995.1554. [DOI] [PubMed] [Google Scholar]
- 57.Rueckert R R. Picornaviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 609–654. [Google Scholar]
- 58.Ryan M D, Flint M. Virus-encoded proteinases of the picornavirus super-group. J Gen Virol. 1997;78:699–723. doi: 10.1099/0022-1317-78-4-699. [DOI] [PubMed] [Google Scholar]
- 59.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 60.Sangar D V, Newton S E, Rowlands D J, Clarke B E. All FMDV serotypes initiate protein synthesis at two separate AUGs. Nucleic Acids Res. 1987;15:3305–3315. doi: 10.1093/nar/15.8.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Svitkin Y V, Gradi A, Imataka H, Morino S, Sonenberg N. Eukaryotic initiation factor 4GII (eIF4GII), but not eIF4GI, cleavage correlates with inhibition of host cell protein synthesis after human rhinovirus infection. J Virol. 1999;73:3467–3472. doi: 10.1128/jvi.73.4.3467-3472.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yalamanchili P, Harris K, Wimmer E, Dasgupta A. Inhibition of basal transcription by poliovirus: a virus-encoded protease (3Cpro) inhibits formation of TBP-TATA box complex in vitro. J Virol. 1996;70:2922–2929. doi: 10.1128/jvi.70.5.2922-2929.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yalamanchili P, Weidman K, Dasgupta A. Cleavage of transcriptional activator Oct-1 by poliovirus encoded protease 3C. Virology. 1997;239:176–185. doi: 10.1006/viro.1997.8862. [DOI] [PubMed] [Google Scholar]
- 64.Yalamanchili P, Datta U, Dasgupta A. Inhibition of host cell transcription by poliovirus: cleavage of transcription factor CREB by poliovirus-encoded protease 3C pro. J Virol. 1997;71:1220–1226. doi: 10.1128/jvi.71.2.1220-1226.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ziegler E, Borman A M, Kirchweger R, Skern T, Kean K M. Foot-and-mouth disease virus Lb proteinase can stimulate rhinovirus and enterovirus IRES-driven translation and cleave several proteins of cellular and viral origin. J Virol. 1995;69:3465–3474. doi: 10.1128/jvi.69.6.3465-3474.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]