Abstract
The chaperone protein EROS (“Essential for Reactive Oxygen Species”) was recently discovered in phagocytes. EROS was shown to regulate the abundance of the ROS-producing enzyme NADPH oxidase isoform 2 (NOX2) and to control ROS-mediated cell killing. Reactive oxygen species are important not only in immune surveillance, but also modulate physiological signaling responses in multiple tissues. The roles of EROS have not been previously explored in the context of oxidant-modulated cell signaling. Here we show that EROS plays a key role in ROS-dependent signal transduction in vascular endothelial cells. We used siRNA-mediated knockdown and developed CRISPR/Cas9 knockout of EROS in human umbilical vein endothelial cells (HUVEC), both of which cause a significant decrease in the abundance of NOX2 protein, associated with a marked decrease in RAC1, a small G protein that activates NOX2. Loss of EROS also attenuates receptor-mediated hydrogen peroxide (H2O2) and Ca2+ signaling, disrupts cytoskeleton organization, decreases cell migration, and promotes cellular senescence. EROS knockdown blocks agonist-modulated eNOS phosphorylation and nitric oxide (NO●) generation. These effects of EROS knockdown are strikingly similar to the alterations in endothelial cell responses that we previously observed following RAC1 knockdown. Proteomic analyses following EROS or RAC1 knockdown in endothelial cells showed that reduced abundance of these two distinct proteins led to largely overlapping effects on endothelial biological processes, including oxidoreductase, protein phosphorylation, and endothelial nitric oxide synthase (eNOS) pathways. These studies demonstrate that EROS plays a central role in oxidant-modulated endothelial cell signaling by modulating NOX2 and RAC1.
Keywords: Endothelial cells, Redox signaling, EROS, NOX2, RAC1, Reductive stress
Graphical abstract
Highlights
-
•
The chaperone protein EROS (“Essential for Reactive Oxygen Species”) is abundant in endothelial cells and regulates oxidant-modulated signal transduction.
-
•
EROS is required for the expression of the NADPH oxidase isoform2 (NOX2) and the small GTPase RAC1.
-
•
EROS deletion impedes cell proliferation, migration and promotes cellular senescence in human umbilical vein endothelial cells.
-
•
EROS is essential for critical oxidant signaling, posttranslational modifications and nitric oxide bioavailability.
-
•
EROS deletion disrupts the pro-oxidative NOX2/RAC1 axis, leading to cytosolic reductive stress and resulting in endothelial cell dysfunction.
1. Introduction
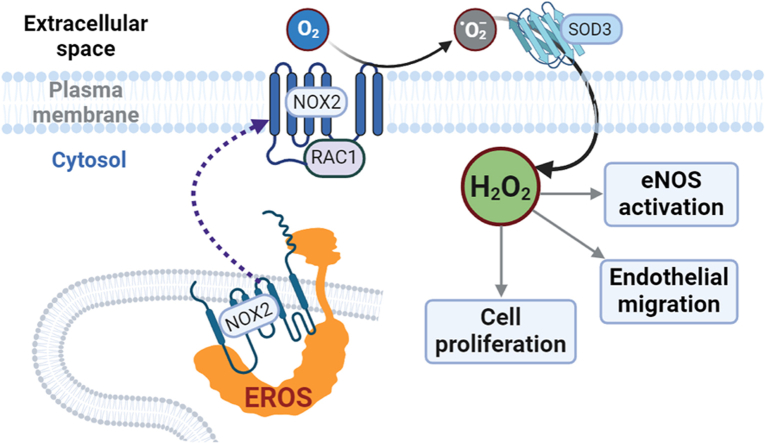
Reactive oxygen species (ROS) in mammalian cells have long been viewed as cytotoxic molecules, either as part of the chemical armamentarium used by immune cells to kill microbes, or as the dreaded mediators of pathological oxidative distress seen in inflammatory disease states such as atherosclerosis or neurodegeneration. But more recently, the stable ROS hydrogen peroxide (H2O2) has been found to facilitate critical physiological roles, regulating a broad range of cellular pathways that modulate cell signaling, differentiation, migration, and growth [1]. The balance of H2O2 physiological oxidative “eustress” and pathological oxidative distress is tightly regulated by a network of oxidases and reductases that reversibly modify protein cysteine residues in cells [1]. The pathways governing the balance between pathological oxidative distress and physiological oxidative eustress are incompletely understood. The recently identified protein EROS (Essential for Reactive Oxygen Species) plays a key role in phagocyte-mediated cytotoxicity, but the role of EROS in oxidative eustress pathways has not been previously defined.
EROS is a chaperone protein that is required for the maturation and stabilization of the ROS generating enzyme NADPH oxidase isoform 2 (NOX2) [2,3]. NOX2 was first characterized in activated phagocytes, where the enzyme generates high amounts of ROS to combat bacterial infections. Phagocyte NOX2 protein levels are markedly lower in EROS-knockout mice, and these knockout mice die from bacterial infections because their phagocytes are unable to generate ROS to kill invading bacteria. Importantly, patients with EROS mutations are susceptible to bacterial infections, again associated with NOX2 deficiency in phagocytes caused by the loss of EROS function [4,5]. The role of EROS in non-hematopoietic cells remains to be defined.
In endothelial cells, NOX2 plays a central role in oxidant-modulated cellular pathways [6]. Endothelial NOX2 generates low levels of ROS that regulate physiological responses including signal transduction, cell adhesion, angiogenesis, migration, and vascular permeability [7]. Endothelial H2O2 signaling requires the small GTPase RAC1 in response to agonist binding to cell surface receptors, RAC1 forms a complex with NOX2 that generates ROS [8] and activates the key signaling protein endothelial nitric oxide synthase (eNOS) and other critical phosphoproteins in these cells [[9], [10], [11], [12], [13], [14]]. Nitric oxide (NO●) formed by eNOS causes vascular smooth muscle cell relaxation and modulates vital endothelial functions [15]. eNOS enzyme function is highly dependent on the cellular redox state, and redox imbalances attenuate both eNOS activity and NO● bioavailability [12,16]. The development of sensitive fluorescent biosensors for H2O2 and NO● has allowed the real-time imaging of the intracellular dynamics of H2O2 and NO● following receptor activation [17,18]. Using the novel H2O2 biosensor Hyper7, we recently discovered that NOX2 and RAC1 play a critical role in receptor-mediated H2O2 generation in human umbilical vein endothelial cells (HUVEC) [6]. The present studies have explored the roles of EROS in redox related pathways in endothelial cells. We show here that EROS is required for NOX2-and RAC1-dependent H2O2 generation and present data suggesting that EROS knockdown results in a cytosolic redox imbalance towards a reductive state that affects vital endothelial cell functions.
2. Results
2.1. EROS modulates critical agonist-dependent signaling pathways
To investigate the function of EROS in endothelial cells, we developed EROS “knockout” and “knockdown” approaches in HUVEC. Using either lentiviral CRISPR/Cas9 knockout or siRNA knockdown methods enabled us to study the effects of complete EROS deletion and shorter-term downregulation of EROS, respectively. For the knockout approach we designed 2 different single-guide RNAs for EROS (EROS SG1 and EROS SG2) which were inserted into a lentiviral CRISPR/Cas9 construct carrying a mCherry tag to identify infected cells (Fig. S1A and Table S1). After infecting HUVEC with these EROS-specific lentiviruses, we found that all cells expressed mCherry (Fig. S1B). As a control for lentiviral EROS SG infections, we used a control Cas9 lentivirus (CAS9, Control) and quantitated EROS protein abundance in cells infected with this construct. Robust protein abundance of EROS was verified in control cells, but no detectable EROS signal was observed in both knockout lines (Figs. S1C and S4E). We also performed siRNA-mediated EROS knockdown by transfecting HUVEC with duplex siRNA constructs; 72 h later, we quantitated EROS mRNA levels using qRT-PCR and measured protein abundance in immunoblots. We found that siRNA-mediated EROS knockdown led to a marked decrease in EROS abundance compared to HUVEC transfected with control duplex siRNA (Figs. S1D–F).
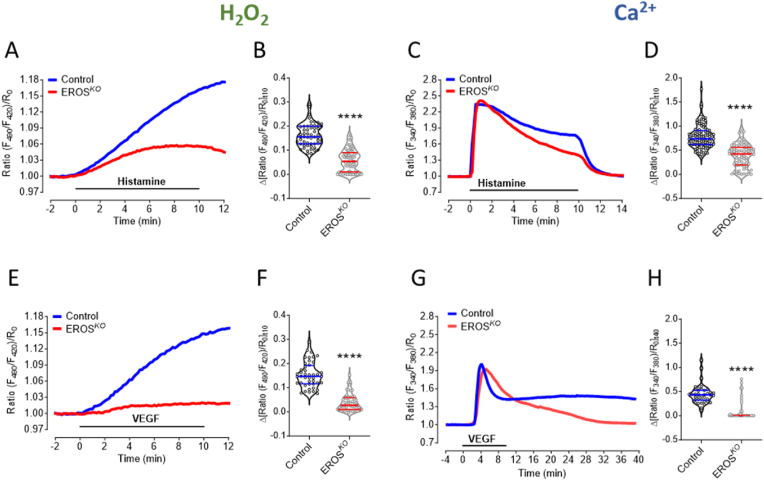
We used the H2O2 biosensor HyPer7 to explore receptor-modulated H2O2 signaling in HUVEC [6], and quantitated intracellular H2O2 increases in response to the G-protein coupled receptor (GPCR) agonist histamine and to the receptor tyrosine kinase (RTK) agonist VEGF. We discovered that knockout of EROS in HUVEC markedly attenuated receptor-modulated H2O2 responses (Fig. 1A, B, E, F and Figs. S2B and C); basal H2O2 levels were not affected by EROS knockout (Fig. S2A). The effects of EROS knockout are similar to the inhibitory effects of siRNA-mediated NOX2 or RAC1 knockdown on receptor-modulated H2O2 that we reported previously [6]. Histamine and VEGF also elicit Ca2+ responses in HUVEC [19], and we next studied the effects of EROS knockout on receptor-stimulated intracellular Ca2+ in real time using the calcium dye Fura2. EROS knockout had no substantive effect on basal intracellular Ca2+ levels (Fig. S2D), and the initial responses to histamine or VEGF stimulation were not different between EROS knockout and control cells (Fig. 1C–G). However, the return to baseline of the receptor-mediated increases in intracellular Ca2+ was much more rapid following EROS knockout in response to both agonists (Fig. 1C, D, G, H and Figs. S2E and F). These results indicate that EROS plays a key role in both histamine- and VEGF-stimulated H2O2 and Ca2+ signaling in HUVEC.
Fig. 1.
EROS knockout modulates agonist-induced H2O2and Ca2+signaling
A. Real time imaging of H2O2 level in response to histamine was measured in CRISPR/Cas9 transduced EROS knockout (EROS SG, red curve) vs. lentiviral CAS9 transduced HUVEC (Control, blue curve). B. Detected H2O2 levels are significantly lower in EROS SG cells (n = 74) compared to Control (n = 55) 10 min after histamine stimulation. C. Ca2+ measurements under same experimental condition as shown in A. D. Histamine treated EROS SG cells (n = 78) show lower Ca2+ levels than Control (n = 93) 10 min after histamine stimulation. E. Average curves of HyPer7 signals in response to VEGF in Control (blue curve) and EROS SG cells (red curve). F. Values of EROS SG (n = 53) and Control (n = 44) 10 min after VEGF treatment. G. Average curves of Ca2+ imaging experiments in Control (blue curve) and EROS SG (red curve) cells stimulated for 10 min with VEGF. H. Statistical analysis of Control (n = 38) and EROS SG (n = 52) indicates decreased Ca2+ levels in EROS SG cells 30 min after VEGF treatment. Violin plots are marked with median and first and third quartiles for the graphed data, ****P < 0.0001 using unpaired t-test. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
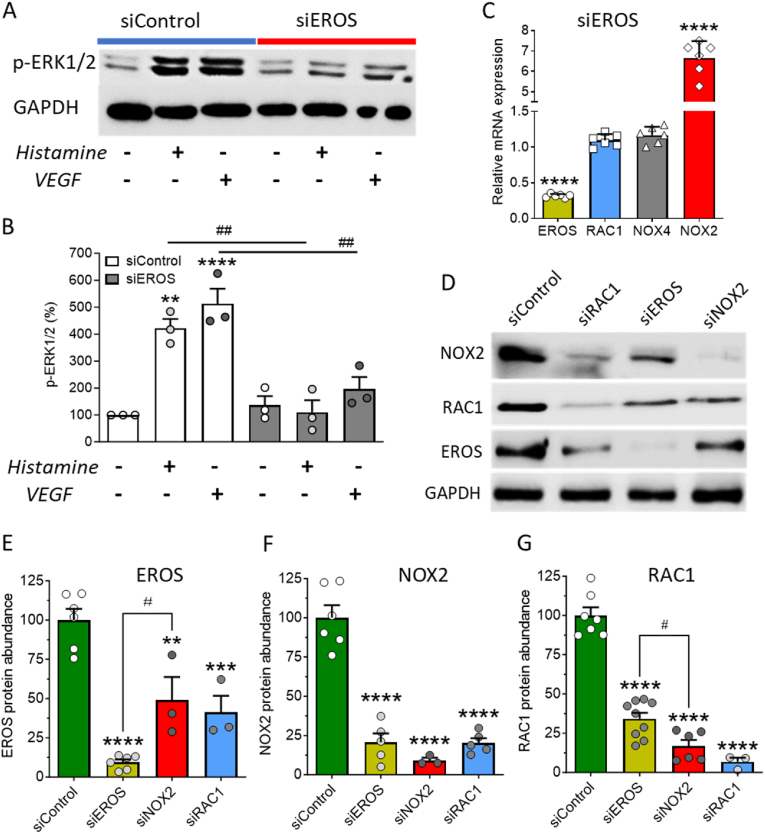
Many receptor-modulated protein kinases are dependent on oxidant signaling [20]. To elucidate the role of EROS in the regulation of receptor-modulated phosphorylation pathways in HUVEC, we next studied the effects of EROS knockdown on ERK1/2 phosphorylation in response to histamine or VEGF [6,19,21]. We found that agonist-stimulated ERK1/2 phosphorylation was blocked by EROS knockdown (Fig. 2A and B and Figs. S3A and B), as well as by siRNA-mediated knockdown of RAC1 (Figs. S3C and D). Taken together, these observations underscore the critical role of EROS for endothelial cell signaling and suggest that EROS may regulate similar signaling pathways as RAC1.
Fig. 2.
Effect of EROS knockdown on ERK1/2 phosphorylation and abundance of EROS, NOX2 and RAC1
A. Representative immunoblot shows ERK1/2 phosphorylation in response to histamine and VEGF stimulation in siControl transfected HUVEC, which is strongly attenuated in EROS knockdown cells. B. Statistical analysis of ERK1/2 phosphorylation (n = 3 for all conditions). C. Quantitative RT-PCR of HUVEC mRNA downregulating EROS reveals low EROS (yellow bar, n = 6), unchanged RAC1 (blue bar, n = 6) as well as NOX4 (gray bar, n = 6) and high NOX2 transcripts (red bar, n = 6). D. Western blots of protein lysates from HUVEC transfected with either siControl, siRAC1, siEROS or siNOX2 probed with antibodies for NOX2, RAC1, EROS and GAPDH. E-G. EROS, NOX2 and RAC1 are significantly downregulated within all single-guided siRNA samples, siEROS (yellow bars), siNOX2 (red bars) and siRAC1 (blue bars) compared to siControl (green bars). All values are presented as means ± SEM, **P < 0.01, ***P < 0.001 and ****P < 0.0001 compared to untreated siControl HUVEC, #P < 0.05 of EROS siRNA vs. NOX2 siRNA and ##P < 0.01 compared to same treatments, either histamine or VEGF, following siRNA knockdown of RAC1 vs. Control siRNA using 1-way ANOVA. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.2. EROS regulates expression of NOX2 and RAC1
We previously found that HUVEC only contain the NOX isoforms NOX2 and NOX4 [6,22]. To test the effects of EROS on the abundance of NOX2 and RAC1, we first performed qRT-PCR experiments after siRNA-mediated EROS knockdown. EROS knockdown cells had no effect on RAC1 transcript abundance, but we found that the NOX2 transcript level increased by > 6-fold (Fig. 2C). Previous studies in phagocytes have shown that EROS deletion leads to NOX2 protein mis-folding and subsequent degradation [3,4]. We probed immunoblots following siRNA-mediated EROS knockdown and found, in contrast to the marked increase in NOX2 mRNA, that NOX2 protein levels were markedly decreased (Fig. 2D–F and Fig. S4A). We then discovered that RAC1 abundance was also significantly lower following EROS knockdown (Fig. 2D–G and Figs. S4A and C). Conversely, NOX2 and RAC1 knockdown led to a marked decrease in EROS protein abundance (Fig. 2D and E). In similar fashion, we also found that EROS knockout mediated by two different single-guide RNAs, EROS SG1 and SG2, led to a marked reduction in NOX2 and RAC1 protein abundance (Figs. S4D–G). Moreover, RAC1 knockdown also resulted in NOX2 downregulation (Fig, 2D, F and Fig. S4B) and NOX2 knockdown led to a decrease in RAC1 abundance (Fig, 2D, G and Fig. S4C).
Since the NADPH oxidase isoform NOX4 is also expressed in HUVEC, we tested the effect of EROS or NOX4 knockdown on EROS, NOX2, NOX4 and RAC1 protein levels (Fig. S5A). We observed that the siRNA-mediated NOX4 knockdown had no impact on the protein abundance of NOX2, RAC1 or EROS. Similarly, EROS knockdown did not affect NOX4 protein abundance but again showed marked downregulation of NOX2 and RAC1 (Fig. S5B). In summary, these data indicates that EROS, NOX2 and RAC1 are strongly interrelated and depend on each other's expression independently of NOX4 protein abundance in HUVEC.
2.3. Knockdown of EROS affects actin polymerization, cell migration, proliferation and leads to cell senescence
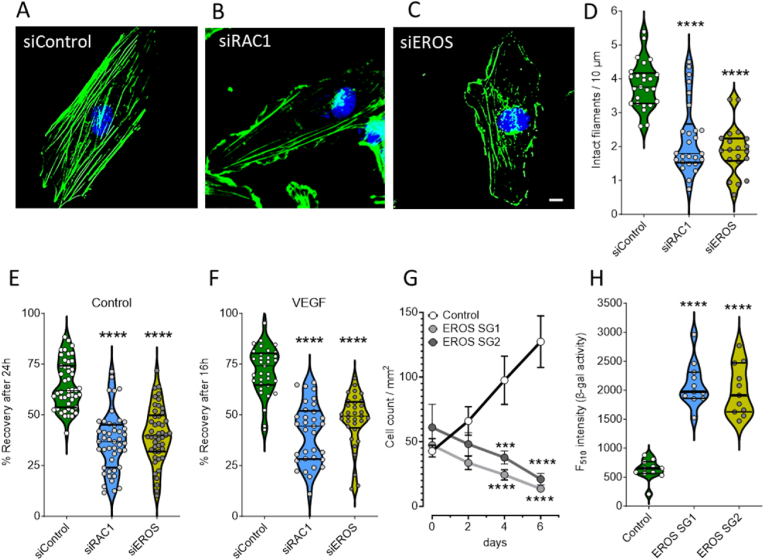
We next conducted experiments to examine the effects of EROS knockdown on endothelial cell function. We first explored the effects of siRNA-mediated EROS- or RAC1-knockdown on the integrity of the F-actin cytoskeletal organization by immunostaining HUVEC with fluorescent-labelled actin binding phalloidin (Fig. 3A–C). To quantitate the occurrence of actin filaments, we performed LineScan analysis (Figs. S6A and B) and found that the presence of intact actin fibers in EROS and RAC1 downregulated cells were markedly reduced (Fig. 3D). We next addressed the effect of a disrupted cytoskeleton on cell motility by performing migration assays in HUVEC following siRNA-mediated EROS- or RAC1-knockdown. Since migration of endothelial cells has been reported to accelerate in response to VEGF [23,24], we tested the effects of VEGF on cell motility following EROS or RAC1 downregulation. We found that VEGF increased the rate of endothelial cell migration, while knockdown of EROS and RAC1 resulted in attenuated cell motility independent of the presence of VEGF (Fig. 3E and F). To explore the impact of EROS deficiency on cell growth, we monitored the proliferation of the EROS knockout cells for 6 days and found that cell proliferation was significantly attenuated compared to control cells (Fig. 3G). We next performed a cell senescence assay in HUVEC following EROS knockout (Fig. S6C) and documented strikingly higher activities of the senescence marker β-galactosidase compared to control cells (Fig. 3H). Taken together, these results demonstrate that loss of EROS in HUVEC has functional consequences similar to those previously observed in RAC1-depleted endothelial cells [23], in both cases leading to disrupted cytoskeletal organization, inhibition of cell migration and proliferation, and increased cellular senescence.
Fig. 3.
Loss of EROS disrupts endothelial cell function
A-C. Representative images of siRNA transfected HUVEC with either siControl, siRAC1 or siEROS show staining of F-actin binding fluorescent dye conjugated phalloidin (green) and nuclear counterstain with DAPI (blue). Scale bar is 10 μm. D. Quantitative analysis of intact actin filaments as performed shown in Figs. S5A and B indicates disorganized cytoskeleton in HUVEC downregulating EROS (yellow plot, n = 20) as well as RAC1 (blue plot, n = 26) versus siControl (green plot, n = 25). E-F. Statistical evaluation of scratch area recovery in Control (E) or under VEGF-treated conditions (F) indicates lower migratory activities in HUVEC following knockdown of RAC1 (blue plots) as well as EROS (yellow plots) versus siControl (green plots). G. CAS9 (Control) and EROS SG1 or SG2 transduced HUVEC were seeded to ∼20 % confluence in endothelial cell growth medium and monitored at day 0, 2, 4 and 6. Graph shows progression of cell proliferation in Control cells (black line, n = 8), while both EROS SG populations (dark and light gray lines, n = 8 each) over time even decreased in cell number counted per mm2. H. Statistical analysis of fluorescence-based β-galactosidase activity indicates similar degree of senescence progression in EROS SG1 (blue plot, n = 11) as well as EROS SG2 (yellow plot, n = 11) compared to CAS9 infected cells (Control, green plot, n = 11). For all panels when indicated, ***P < 0.001 and ****P < 0.0001 (1way ANOVA for equal variance). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.4. EROS and RAC1 downregulation reveal similar quantitative protein regulatory effects
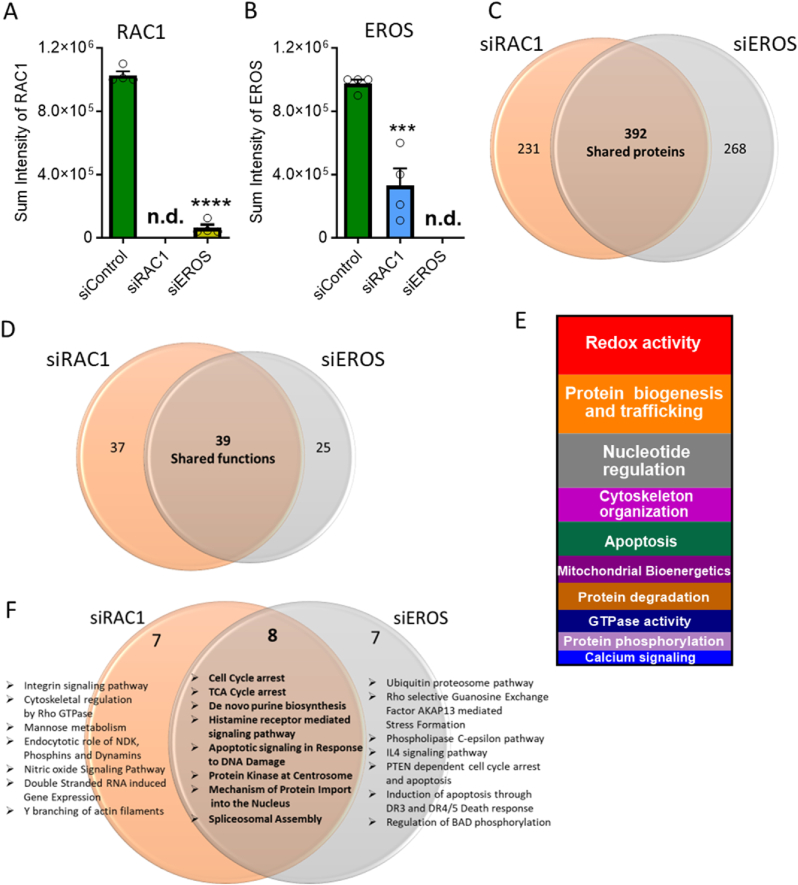
Given the functional similarities of EROS and RAC1 downregulation on HUVEC cellular responses, we performed comparative proteomics following siRNA-mediated knockdowns of EROS and RAC1. The specific knockdowns of EROS and RAC1 protein levels were verified by mass spectrometry (MS) (Fig. 4A and B), consistent with our findings in immunoblot analyses (Fig. 2D, E, G). MS identified significant changes in the HUVEC proteome driven by siRNA-mediated knockdown of EROS or siRAC1. The alterations in cellular protein levels were highly similar following siRNA-mediated knockdown of RAC1 and EROS (Fig. 4C and Figs. S7A, S7B, S8; Data S1). Analysis of these proteomic perturbations also revealed large overlaps in the biological processes that are affected by RAC1 or EROS knockdown (Fig. 4D and Data S2), as revealed by Gene Ontology analyses of biological functions (Fig. S7C; Data S3) or cellular pathways (Fig. 4F and Data S4). These proteomic analyses also identified negative regulation of oxidoreductase and kinase activities, protein changes associated with cytoskeletal organization and Ca2+ signaling as well as induction of apoptotic pathways. Other pathways and biological processes that were similarly affected following both EROS and RAC1 knockdown included proteins involved in cell cycle arrest, ER stress, and mitochondrial disorders, as well as negative regulation of endothelial nitric oxide synthase (eNOS) activity.
Fig. 4.
EROS and RAC1 knockdown show comparable endothelial proteomic regulation
A. Detected sum intensities of RAC1 peptides by MS in HUVEC transfected with either siControl, siEROS or siRAC1. B. Detected sum intensities of EROS peptides by mass spectrometry in HUVEC transfected with either Control, EROS or RAC1 siRNA transfection. C. Venn diagram shows numbers of altered protein expression in siRAC1 and siEROS samples. Expression levels equal or lower than 0.5-fold were considered as downregulated proteins and levels equal or higher than 2-fold as upregulated protein changes compared with siControl. Changes in the proteome of individual knockdown largely overlaps (expressed as shared proteins). All altered proteins following siRNA-medicated knockdown of RAC1 or EROS were regulated in the same direction (detailed regulation of individual protein expression levels is provided in Data S1). D. Gene ontology annotated biological processes following siRNA-mediated knockdown of EROS or RAC1 indicates similar overlap in functional changes (for detailed lists of shared and unique functions see Data S2). E. Vertical slice chart presents Gene ontology analysis of shared processes within EROS or RAC1 siRNA-mediated downregulation distributed according to their different functional categories (Data S3). F. Gene ontology pathway analysis using R studio highlights most affected shared and unique pathways (Data S4 lists unique and shared top hits of pathways, protein interactions, phosphatases and kinome enrichments). ****P < 0.0001 (1way ANOVA for equal variance).
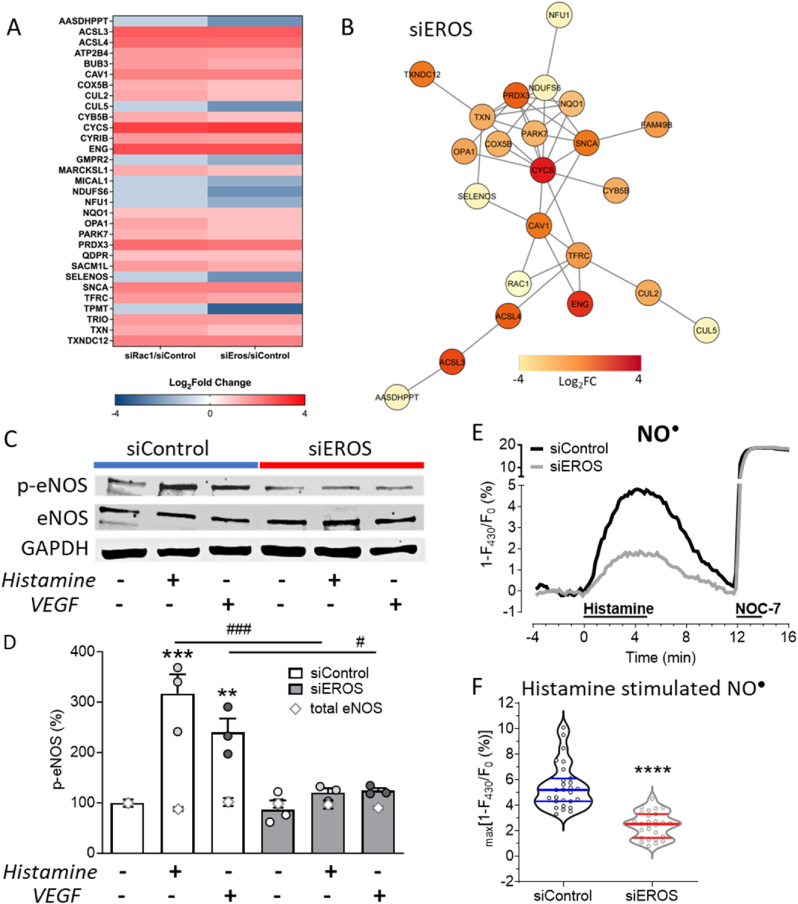
To assess the consequences of these diverse EROS and RAC1 dependent functional changes on the protein level, we analyzed the biological processes associated with these protein alterations (Fig. 4E and Data S5) and extracted well-characterized key proteins (Fig. 5A and Table S2, see for full names and related references therein). We identified proteins such as MICAL1, MARCKSL1, TRIO or CYRIB, which are key regulators of actin polymerization. Changes in the expression levels of these proteins may disrupt cytoskeletal organization and migration. Other protein regulations were associated with inhibition of cell growth (CUL5) and eNOS activity (CAV1, ENG, CYB5B), senescence (NDUFS6), cell cycle arrest (BUB3), ER stress (SACM1L, TXNDC12), apoptosis (CYCS) or defects in lipid (ACSL3, ACSL4), purine (TPMT, AASDHPPT, GMPR2, TRIO) and mitochondrial (COX5B, NFU1, OPA1, SNCA, PRDX3) metabolism. Upregulation of the reductive stress responsive scaffold E3 ligase Cullin2 (CUL2) points to a loss of physiologically relevant oxidative reactions and modifications [25]. Thus, the regulation of oxidoreductase active proteins such as TXN, PRDX3, NQO1, OPA1, COX5B, ENG, QDPR, PARK7 or SELENOS suggest similar changes in protein abundance that are due to reductive stress responses. Network analysis revealed that most of these proteins (Table S2) are connected upon EROS-mediated RAC1 downregulation (Fig. 5B). Taken together, these data indicate that siRNA-mediated EROS and RAC1 knockdown have strikingly similar effects on HUVEC proteins and pathways.
Fig. 5.
EROS knockdown induces reductive stress associated protein regulation resulting in diminished nitric oxide formation
A. Heat map of critical siEROS and siRAC1 regulated shared proteins identified in functional analysis. B. Network analysis shows most significant protein interactions. The network has been refined with a highest confidence cut off of 0.900. Nodes are colored based on calculated fold change. C. Representative immunoblots show abundances of eNOS phosphorylation (p-eNOS, Ser1177), total eNOS (eNOS) and GAPDH following transfection with Control or EROS siRNA in HUVEC after stimulation with histamine or VEGF vs. untreated. D. Analysis of p-eNOS blots reveals p-eNOS Ser1177 phosphorylation in siControl HUVEC stimulated with histamine or VEGF for 10 min, which is blocked in EROS downregulated HUVEC. Total eNOS (gray open rhombic characters) is unchanged among samples. All values are presented as mean ± SEM, **P < 0.01 and ***P < 0.001 compared to untreated Control siRNA and #P < 0.05 or ###P < 0.001 compared to same treatments, either histamine or VEGF, of siEROS vs. siControl cells using 1way ANOVA. E. Average curves of NO● imaging experiments in Control (black curve) and EROS (gray curve) siRNA-transfected cells stimulated with histamine and NOC-7 as indicated. F. Statistical analysis of Control (n = 27) and EROS siRNA-transfected (n = 27) cells indicates decreased NO● levels following siRNA-mediated EROS knockdown, ****P < 0.0001 using unpaired t-test.
2.5. EROS is essential for endothelial nitric oxide formation
Our proteomic analyses implicated inhibition of eNOS (endothelial nitric oxide synthase) pathways following either RAC1 or EROS knockdown. We have previously established the critical role of RAC1 in eNOS activation, but the effects of EROS on eNOS pathways have been entirely unknown. We found that siRNA-mediated knockdown of EROS did not affect total eNOS protein levels (eNOS). However, siRNA-mediated knockdown of EROS attenuated both histamine- and VEGF-stimulated eNOS phosphorylation (Fig. 5C–D). In order to directly measure the inhibitory effect of EROS knockdown on eNOS activity for NO● biosynthesis, we performed real-time imaging experiments using the NO● biosensor C-geNOp (Fig. 5E). We found that histamine-stimulated NO● formation was markedly attenuated following siRNA-mediated EROS knockdown (Fig. 5F). These results confirm that EROS knockdown blocks receptor-mediated eNOS activation and NO● formation in HUVEC.
3. Discussion
These studies have examined the role of EROS in endothelial cells, and have identified new roles for this recently-identified protein that was first characterized in phagocytes as a critical chaperone for the ROS-generating enzyme NOX2 [2]. Here we have shifted the focus to vascular endothelial cells (HUVEC), where the roles of EROS have not been previously characterized. We used the H2O2 biosensor HyPer7 to quantitate receptor modulated H2O2 signals in response to histamine or VEGF. As we previously established for NOX2 and RAC1 knockdown cells [6], here we found that CRISPR/Cas9-mediated knockout of EROS markedly inhibited receptor-modulated H2O2 responses. We also showed that EROS in HUVEC has a similar regulatory role in stabilizing NOX2 as in phagocytes. However, basal H2O2 levels were unchanged following EROS knockdown. Although HyPer7 is currently considered the most sensitive H2O2 probe, sensing might be below its detection limit due to the low cytosolic H2O2 concentration in resting cells [26]. Alternatively, the unchanged basal HyPer7 ratios may reflect the many other sources that contribute to the basal level of H2O2 in cells. This data at least indicate that apparently higher basal ratios as seen in oxidative stressed cell models [27] can be excluded for EROS knockout cells (Fig. S2A). In addition, we discovered that EROS knockout mitigates sustained cytosolic Ca2+ signaling in response to histamine or VEGF and MS identified alterations of numerous Ca2+ regulating proteins (Data S3; Calcium signaling). In EROS deficient bone marrow-derived macrophages, the Ca2+ flux-mediating purinergic receptors, P2X7 and P2X1 showed a decrease in abundance-in addition to a marked reduction in NOX2 [3]. However, we did not find any evidence for the expression of P2X7 or P2X1 receptors in HUVEC, and we focused our studies on the Ca2+-mobilizing agonists VEGF and histamine. Many Ca2+-dependent receptors and channels are redox sensitive, and in turn, ROS-generated enzymes can be modulated by Ca2+. The precise molecular pathways whereby EROS affects Ca2+ signaling in endothelial cells remains to be determined.
Oxidant signaling by H2O2 promotes key posttranslational phosphorylations, including effects on the MAPK pathway, whereas reducing agents inhibit MAP kinases [6,21,23,28]. Mass spectroscopic (MS) analysis following siRNA-mediated EROS knockdown revealed negative regulation of protein phosphorylation, kinase activity (Data S2), and enrichment of phosphatases (Data S4; Phosphatases). Accordingly, when we probed for archetypal phosphoproteins such as ERK1/2 Thr202/Tyr204 and eNOS Ser1177 in response to histamine and VEGF, agonist-modulated phosphorylation of these residues was blocked in EROS deficient HUVEC (Fig. 2, Fig. 5C). These results might be due to the disturbed H2O2 signaling (Fig. 1) which may explain the antiproliferative effects in EROS deficient endothelial cells (Fig. 3G).
Since EROS was originally found to control NOX2 abundance in phagocytes, we verified the same effect on the NOX2 protein level in EROS knockdown cells. In contrast to the decrease in NOX2 protein, we found that the abundance of NOX2 transcripts were markedly increased. Discordances between protein and mRNA expression have been reported as proteome signatures of redox stress [29]. Indeed, EROS may have broader effects on gene transcription. It is also possible that the striking increase in NOX2 mRNA reflects a compensatory increase in NOX2 transcription following the marked decrease in NOX2 protein in EROS knockdown cells. Because our previous study revealed the crucial role of RAC1 in signal transduction involving NOX2 activation [6] and a recent study implicated RAC1 in feedforward activation of NOX2 leading to ROS production [30], we additionally probed for RAC1 abundance. While RAC1 transcript levels were unchanged following siRNA-mediated EROS knockdown, RAC1 protein was markedly downregulated (Fig. 3C–G). Notably, EROS knockout in phagocytes does not lead to changes in abundance of this small GTPase, perhaps reflecting the fact that phagocytes predominantly express the RAC2 isoform [2,3]. The effects of EROS knockdown on RAC1 protein in endothelial cells may reflect the fact that NOX2 is the only NADPH oxidase isoform that binds RAC1 in these cells. By contrast, phagocytes express both NOX1 and NOX2, both of which bind RAC1. Importantly, we found that individually knocking down either EROS, NOX2 or RAC1 in endothelial cells led to a decrease in the protein abundance of all the others. However, EROS knockdown had no effect on the abundance of NOX4, which does not interact with RAC1 [31], and downregulation of NOX4 did not affect protein abundances in either EROS, NOX2 or RAC1 (Fig. S5). It is also possible that EROS is modulating other proteins and pathways controlling endothelial function. Taken together, these data demonstrate that EROS and RAC1 are closely interrelated and are indispensable for the expression of NOX2 (Fig. 2D–G). In contrast, the distinct NADPH oxidase isoform NOX4 is not involved in this interplay.
Our proteomic approach has expanded the view of protein expression changes following EROS or RAC1 knockdown. Among ∼2500 identified proteins, more than a quarter showed significant changes in their abundance, of which more than 10 % were associated with the cytoskeleton (Data S3; Cytoskeleton organization). F-actin polymerization is highly sensitive and dependent on intracellular H2O2, which regulates the reversible formation of disulfide bonds within actin molecules that are required for cross-links between actin filaments [32]. RAC1 activity may control actin polymerization, thereby regulating angiogenesis and chemotaxis [23,33]. The pro-oxidative and -angiogenic effects of RAC1 on cytoskeleton organization of various cell types including platelets, fibroblasts and endothelial cells have been extensively studied, demonstrating the regulatory effects of actin-dependent lamellipodia and filopodia formation that promote endothelial migration and microtubule formation [8,23,34]. Staining of F-actin in EROS-deficient endothelial cells reveals disrupted cytoskeletal fibers, associated with an attenuation of cell migration (Fig. 3A and B). This is consistent with recent findings indicating that RAC1 can serve to coordinate cell motility and cell cycle progression [35]. Proteomic analyses following siRNA-mediated EROS or RAC1 knockdown identifies changes associated with cell cycle arrest and induction of apoptotic pathways. These proteomic findings match well with functional data showing that EROS knockout in HUVEC has antiproliferative effects and is associated with markers of cell senescence. Taken together, these data suggest that EROS and RAC1 might be essential for the maintenance of the cellular redox balance required for cell growth, migration, and cell cycle progression.
Mitochondria represent another major source of ROS, while maintaining an antioxidative environment within the organelle [36]. NOX-mediated H2O2 generation may be a principal source for redox signaling which in turn may also affect redox dependent processes of other cell organelles such as endoplasmic reticulum (ER) protein folding and mitochondrial metabolism [37,38]. Following EROS knockdown, mass spectrometry identified changes in biological processes such as regulation of response to ER stress, protein catabolic processes or mitochondrial electron transport chain (Data S2) as well as enrichments in TCA cycle and apoptotic pathways (Fig. 4F and Data S4). Thiol oxidation of cysteine residues is a prerequisite for intact protein folding within the ER predominantly required for maturation of proteins of the secretory pathway [39,40]. Moreover, recruitment of the reductive stress response scaffold CUL2, which gets upregulated in EROS downregulated cells, is associated with a decrease in mitochondrial respiration [25,41]. Although several oxidoreductase regulations observed in EROS knockdown endothelial cells point to ER stress and mitochondrial disorder (Table S2, see related references therein), further studies are required to unveil the mechanisms whereby EROS-mediated redox homeostasis may affect ER and mitochondrial metabolism.
Proteomic analysis also unveiled a negative regulation of nitric oxide synthase activity after siRNA-mediated EROS knockdown (Data S2). We have previously reported that RAC1 is required for agonist-promoted activation of eNOS [[9], [10], [11], [12], [13], [14]]. In similar fashion, siRNA-mediated EROS knockdown in HUVEC blocks agonist-dependent phosphorylation of eNOS and attenuates agonist-stimulated NO● formation (Fig. 5C–F). These data indicate that NOX2 is required to maintain the cellular redox balance needed for proper eNOS function [42]. Thus, eNOS activation and NO● formation are highly dependent on H2O2 signaling, which is blocked following EROS knockdown in endothelial cells.
These studies have established that EROS plays a critical role in endothelial cell redox balance, and our results show that EROS modulates the key pro-oxidative RAC1/NOX2 axis in endothelial cells. Oxidative stress plays a central role in the progression of many diseases, yet reductive stress is being increasingly implicated in disease pathogenesis. This study shows the dramatic consequences of redox balance caused by deletion of the chaperone EROS and may pave the way for further investigations into the various aspects and mechanisms underlying the role of reductive stress in vascular disease states.
4. Materials and methods
4.1. Materials
HEK293T cells (293T), Dulbecco's Modified Eagle Medium with high glucose (DMEM), Fetal Bovine Serum (FBS), Penicillin-Streptomycin-Glutamine 100 × (PenStrep), Trypsin-EDTA 0.05 % (Trypsin), Phosphate buffered Saline (PBS), Pierce™ Phosphatase Inhibitor Mini Tablets (phosphatase inhibitor), Restore™ PLUS Western Blot Stripping Buffer (Stripping buffer), Opti-MEM™ I Reduced Serum Medium (OptiMEM), LentiArray™ CAS9 Lentivirus (CAS9 LV), CellEvent™ Senescence Green Detection Kit (Senescence kit), AlexaFluor™ 488 or 568 Phalloidin (Phalloidin Green or Red), Fluoromount™-G Mounting Medium with DAPI (Fluoromount-DAPI), Hoechst 33342 (Hoechst), Lipofectamine™ 3000 Transfection Reagent (LF3000), High-Capacity cDNA Reverse Transcription Kit (RT Kit), Pierce™ BCA Protein Assay Kit (BCA assay) and synthesis of Custom Oligos (Primers) were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Steriflip® Vacuum-driven Filtration System (0.45 μm PES filter), Histamine dihydrochloride (histamine), l-ascorbic acid (vitamin C), Bovine Serum Albumin (BSA), Ammonium Persulfate, Triton™X-100 (X100), Ammonium iron(II)sulfate hexahydrate (Mohr's salt), HEPES, Sodium chloride (NaCl), Calcium chloride dihydrate (CaCl2) Potassium chloride (KCl), Magnesium chloride hexahydrate (MgCl2), Sodium hydroxide (NaOH), d-glucose and CAPSO were from Sigma Aldrich (St. Louis, MO, USA). All other reagents were obtained from other suppliers as indicated.
4.2. Cell culture
These studies were performed using the pooled human umbilical vein endothelial cell (HUVEC) strain CC-2519 lot #18TL14996 (Lonza, Walkersville, MD, USA) at passages 3-7. For accelerated growth HUVEC were cultured in Endothelial Cell Growth Medium-V2 (ECGM-V2, #213-500, Cell Applications Inc., San Diego, CA, USA) containing serum and growth factors. Prior to all experiments HUVEC were cultured in ECGM without serum and VEGF (#211-500, Cell Applications Inc.) for at least 48 h 293T cells were cultured in DMEM supplemented with 10 % FBS and 1 % PenStrep. All cells were maintained under standard conditions in a humidified incubator (37 °C, 5 % CO2, 95 % air).
4.3. Generation of EROS knockout cells
Manufacturing of Lentiviral (LV) CRISPR/Cas9 mediated single guide RNA (SG) constructs against EROS was done as previously described [43]. In brief, EROS SG1 or 2 primers (10 μM) were annealed in a reaction with a 10 × T4 ligation buffer and T4 PNK (New England Biolabs, NEB, Ipswich, MA, USA) for 30 min at 37 °C. Double stranded SG sequences were then ligated in a BsmBI (NEB) digested LentiCRISPRv2-mCherry linearized vector (AddGene #99154) and transformed in stable competent E. coli (NEB). Positive clones were sequenced for correct SG insertion and the resulting plasmids (EROS SG1 and SG2 vectors) were then co-transfected with the packaging plasmids pVSVg (AddGene #8454) and psPAX2 (AddGene #12260) into 2 × 106 293T in a ratio of 3:6:4 using PolyJet™ transfection reagent (SignaGen, Frederick, MD, USA) in antibiotic-free DMEM. Media were exchanged 18 and 32 h post transfection by collecting and combining supernatants. Lentiviral supernatants were filtered through 0.45 μm PES filters and lentiviral particles were concentrated with Lenti concentrator (OriGene, Rockville, MD, USA). Lentiviral titers were determined with Lenti-X™ GoStix™ Plus (TaKaRa, San Jose, CA, USA) and concentrations were calculated for both EROS SG LV at ∼1 × 106 IFU/ml. HUVEC were infected at a multiplicity of infection (MOI) of 2 with lentiviral particles encoding for EROS SG1, EROS SG2 or a CAS9 LV representing a CAS9 expressing Control. Lentiviral transduction of EROS guide RNAs in HUVEC was further evaluated by fluorescence detection of mCherry positive cells (Fig. S1B).
4.4. Transduction of siRNA and genetically encoded biosensors
All siRNAs were generated (Horizon Discovery, Waterbeach, UK) according to unique mRNA target sequences (Table S1). For individual knockdown of target genes respective siRNA was mixed with PepMute transfection reagent in PepMute siRNA buffer (SignaGen) at a concentration of 40 nM. siRNA transduction was performed in HUVEC suspended in Opti-MEM supplemented with 2 % heat-inactivated FBS (HyClone, Cytiva, Marlborough, MA, USA) as described for the PepMute siRNA reverse transfection method (SignaGen). siRNA transfection mix was exchanged with ECGM 6 h after transfection. All siRNA-mediated knockdown experiments were done 3 days after transfection. All siRNA constructs used here have been validated for “knockdown” efficiency and for the absence of off-target effects (6). Moreover, results using the new duplex siRNAs for EROS were reproduced in CRISPR-Cas9-mediated knockdown experiments.
For H2O2 measurements endothelial cells were infected 24 h after siRNA transfection with an adenovirus AV5 (Viraquest, North Liberty, IA, USA) construct for the H2O2 probe HyPer7-NES at a multiplicity of infection of 10. Plasmid encoding for cytosolic C-geNOp-NES (NGFI, Graz, Austria) was transiently transfected on the day after siRNA transfection using Opti-MEM containing 2 % FBS and LF3000 transfection reagent according to the manufacturers’ protocol. Medium was exchanged with ECGM 6 h after transfection and cells were maintained in the incubator for another 2 days before imaging experiments.
4.5. Quantitative real time PCR
For quantitative real time PCR (qRT-PCR), total RNA was isolated from HUVEC using the RNeasy Plus Universal Mini Kit (Qiagen Inc., Germantown, MD, USA), and reverse transcription was performed in a EdvoCycler™ Jr. Personal PCR Machine (Edvotek Inc., Washington, DC, USA) using the RT kit. The resulting cDNA was then applied in Real Time PCR experiments using iTaq™ Universal SYBR® Green One-Step Kit (Bio-Rad, Hercules, CA, USA) and respective primers for target genes (Table S1) were added to the reaction. Real time PCR measurements were done on a StepOnePlus™ Real-Time PCR System (Thermo Fisher Scientific) and the relative expression (Ct values) was normalized with RNA polymerase II subunit A (Rpol2) as a housekeeping gene.
4.6. Immunochemical procedures
All Western blot experiments were performed 3 days after siRNA transfection. For agonist treatment of cells for immunoblot analyses, media was exchanged to either ECGM containing 15 μM histamine, 10 ng/ml vascular endothelial growth factor (VEGF, R&D Systems, Minneapolis, MN, USA) or ECGM alone (untreated). After cell treatments, cell lysates were collected using RIPA buffer (BostonBioProducts Inc., Ashland, MA, USA) supplemented with phosphatase and protease inhibitors (Roche Diagnostics Corp., Indianapolis, IN, USA) and quantified with BCA assay. Proteins were separated on 10 % SDS-PAGE and blotted on nitrocellulose membranes (Bio-Rad). Immunoblots for detection of p44/42 Mitogen-activated protein kinases specific phosphorylation sites (p-ERK1/2, Thr202/Tyr204) and phosphorylated endothelial nitric oxide synthase (p-eNOS, Ser1177) were incubated in 5 % BSA using respective phospho-specific primary antibodies (CST, Cell Signaling Technologies, Danvers, MA, USA) in a dilution of 1:1000. For quantification of protein expression levels following primary antibodies were diluted 1:1000 in 5 % non-fat dry milk (milk, Bio-Rad): C17orf62 (EROS, Atlas Antibodies AB, Lund, Sweden), gp91phox (NOX2, ABclonal, Woburn, MA, USA), NOX4 (NOX4, mAB 47-6) [44,45], Cdc42 (RAC1, CST), total eNOS (eNOS, CST) and GAPDH (CST). CAS9 was labelled with secondary anti-mouse antibody (CST) and all other proteins with secondary anti-rabbit antibody (CST) diluted 1:2000 in 5 % milk or 5 % BSA.
For staining of actin filament structures siRNA transfected HUVEC were seeded and cultivated on glass coverslips (Ø = 12 mm). The medium was discarded, and the cells were washed twice with room temperature PBS. Cells were fixed with 4 % formaldehyde in PBS (Wako, Osaka, Japan) for 30 min at 4 °C, permeabilized with 0.1 % X100 in PBS and subsequently blocked in PBS containing 2 % BSA for at least 45 min. To visualize the localization of actin, the cells were incubated with Phalloidin Green or Red diluted 1:100 in PBS with 1 % BSA for 60 min at room temperature. Samples were then mounted with Fluoromount-DAPI on microscopic slides, dried in the dark overnight and sealed with nail polish. For quantification of intact actin filaments images taken from phalloidin stained cytoskeletal structures were binarized and analyzed using the integrated LineScan tool of MetaMorph version 7.10.5.476 (Molecular Devices LLC., San Jose, CA, USA). Occurrence of intact actin filaments were calculated and expressed in number of filaments per 10 μm as illustrated (Figs. S5A and B).
4.7. Fluorescent dye and iron(II) loading
Prior to Ca2+ measurements, endothelial cells were loaded with ECGM containing 3.3 μM Fura2 for 40 min at 37 °C. Fura2 medium was then replaced with fresh ECGM, and cells were placed for an hour in the incubator. Loading of HUVEC with the nuclear labelling dye Hoechst was performed in the same way.
For β-galactosidase activity assays (to measure cell senescence), we used a green fluorescence-based senescence kit according to the manufacturer's protocol. CAS9 (Control) or EROS SG transduced HUVEC were plated on 8 well ibiTreat μ-Slides (ibidi, Fitchburg, WI, USA). Cells were fixed with 4 % paraformaldehyde and incubated with senescence green buffer in the dark for 2 h at 37 °C. Quantitative measurements of green fluorescence intensities are directly proportional to β-galactosidase activities within single cells enabling an evaluation of cell senescence.
For analyses of NO● using the Fe(II)-sensitive NO● biosensor geNOp, HUVEC expressing the cyan fluorescent NO● sensitive biosensor C-geNOp-NES in the cytosol were loaded with Fe(II) as follows: Mohr's salt and vitamin C were dissolved in Ca2+ buffer at a molarity of 10 mM each. After sterile filtration, the resulting Fe(II) solution was mixed with prewarmed ECGM in a ratio of 1:3 and the Fe(II) containing medium mix was added to C-geNOp transfected HUVEC and placed for 10 min in the incubator. Fresh medium was then replaced, and cells rested for 2 h in the incubator prior to NO● imaging experiments.
4.8. Live cell imaging
Live-cell fluorescent recordings were performed on an inverted wide-field fluorescent microscope (IX80, Olympus, Waltham, MA, USA) equipped with a motorized sample stage (Prior, Rockland, MA, USA), a Lumen 200 Fluorescence Illumination System (Prior), and a charge-coupled device camera (Hamamatsu, Bridgewater, NJ, USA). Real-time H2O2 measurements were performed with a 20 × oil immersion objective (PlanSApo, Olympus). For all real-time measurements HUVEC were grown on 30 mm glass coverslips (Bioptechs, Butler, PA, USA) that were mounted in a perfusion chamber (NGFI, Graz, Austria). Using a gravity-based perfusion system cells were perfused with a physiological HEPES-buffered solution composed of (in mM) 140 NaCl, 5 KCl, 2 CaCl2, 1 MgCl2, 10 d-glucose and 1 HEPES, adjusted to pH 7.45 with NaOH enabling monitoring of biosensor intensity changes over time in response to either 15 μM histamine or 10 ng/ml VEGF. Ratiometric HyPer7 was alternately excited at 420 and 490 nm (Semrock, Rochester, NY, USA) using an optical filter wheel (Sutter Instruments, Novato, CA, USA) and emission was recorded at 542 nm (Semrock) using Metafluor Software (Molecular Devices, San Jose, CA, USA).
All data collected from real-time imaging were background subtracted. Normalized ratios of HyPer7 were calculated from resulting emission values according to the formula R = [(F490/F420)/R0] were R0 represents basal HyPer7 ratio. Ca2+ imaging was done after Fura2 loading and filter settings for imaging were at excitations of 340 and 380 nm and emission of 510 nm (Chroma technologies, Rockingham, VT, USA). Fura2 ratios were normalized using the formula R = [(F340/F380)/R0]. Fe(II) loaded cytosolic C-geNOp-NES expressing HUVEC were excited at 430 nm (Semrock) and emissions were collected at 472 nm (Semrock). Changes of intensiometric cyan fluorescence were calculated in % using the formula ΔF = 1−(F/F0) × 100. The NO● donor NOC-7 (Santa Cruz Biotechnology, Dallas, TX, USA) was used as a positive control for C-geNOp NO● bioimaging. Stains for phalloidin were visualized with a 60 × oil immersion objective (UPlanSApo, Olympus) using dichroic filter sets of GFP for Phalloidin Green (U-M41017, Chroma) or of OFP for Phalloidin Red (SP Gold-B OMF, Semrock). Hoechst and DAPI were excited with a filter at 360 and captured using a filter at 460 nm (Semrock). Excitation and emission filters for imaging mCherry were at 586 and 610 nm (Chroma technologies), respectively.
4.9. Proliferation and migration assay
Measurements of assays for monitoring of cell proliferation and migration were based on differential interference contrast (DIC) images, which were taken from pre-marked regions enabling recordings of identical image areas using a 4 × air objective (UPlanPLN, Olympus). For the proliferation assay CAS9 and EROS SG transduced HUVEC were seeded on ibiTreat 8-well slides (ibidi) at low (∼20 %) confluence and DIC images were recorded after 6 h (day 0), 2 days, 4 days, and 6 days. Cell numbers from each sample were counted in the same areas of 1 mm2 enabling a determination of cell growth rate over time.
For monitoring migration, siRNA treated HUVEC were seeded at high (∼80 %) confluence and monolayer were scratched with a P200 pipet. Medium was subsequently exchanged with or without 10 ng/ml VEGF and images of marked regions were immediately taken at time 0 and 24 or 16 h after scratch, respectively. The migration rate was then calculated as described before [16].
4.10. Proteomic LC-MS procedures
siRNA transfected HUVEC were suspended and lysed with RIPA buffer. Total protein was TCA precipitated and resolubilized in RapiGestSF (Waters, Milford, MA, USA). Samples were then reduced and alkylated as described before [46]. 10 μl (20 ng/μl) of sequencing-grade trypsin (Promega, Madison, WI, USA) was spiked into 300 μl PBS and the samples were left in a 37 °C chamber overnight. Samples were acidified with 20 μl 20 % formic acid solution and then desalted by STAGE tip [47].
The samples were reconstituted before analysis in 10 μl of HPLC solvent A. A nano-scale reverse-phase HPLC capillary column was generated by packing 2.6 μm C18 spherical silica beads into a fused silica capillary (100 μm inner diameter x ∼30 cm length). After equilibration of the column, each sample was loaded with a Famos auto sampler (LC Packings, San Francisco, CA, USA). Peptides were eluted in solvent B (97.5 % acetonitrile, 0.1 % formic acid). After elution, the peptides were detected via an electrospray ionization attached to a LTQ Orbitrap Velos Elite ion-trap mass spectrometer (Thermo Fisher Scientific) to get detected.
Proteins were annotated from raw data files using Proteome Discoverer (v 2.2) and searched against the SwissProt Homo sapiens database using Sequest (Thermo Fisher Scientific) with the subsequent search parameters: fixed modification, Carbamidomethyl (C); variable modifications, deamidation (NQ), oxidation (M); peptide mass tolerance 5 ppm; fragment mass tolerance 0.8 Da; enzyme, trypsin; max 2 missed cleavages. Proteins were selected based on a threshold range of 0 (min) to 3 (max) unique peptides during comparison among samples. All databases include a revised version of all the sequences and the data was filtered for a false peptide discovery rate between one and two percent.
4.11. Proteomic data analysis
Annotated proteins were further analyzed by R package gprofiler2 for over-representation analysis of biological functions and pathways. Heat map was constructed using ComplexHeatmap package (version 2.10.0) [48,49]. Network visualizations and analysis were performed by CytoscapeR via R Cy3 [50]. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE66 partner repository with the dataset identifier PXD052412. Shared protein alterations within siEROS and siRAC1 transfected HUVEC or those occurring in common processes were further sorted into 13 functional categories by evaluating the UniProt information on protein functions and GO annotations. Categories were defined by the main individual protein functions as following: Redox activity, Protein biogenesis and trafficking, Nucleotide regulation, Cytoskeleton organization, Apoptosis, Mitochondrial Bioenergetics, Protein degradation, GTPase activity, Protein phosphorylation, Calcium signaling, Lipid metabolism, Immune response, and Unknown or other functions.
4.12. Statistical analysis
Data were analyzed using GraphPad Prism software version 10.1 (GraphPad Software, San Diego, CA, USA) and presented as mean ± standard error of mean (SEM) of independent experiments (n) throughout the whole manuscript. For comparisons between two groups, two-tailed Student t-test was used, and for comparison across multiple groups, one- or 2-way analysis of variance (ANOVA) with Tukey's Multiple Comparison was utilized as indicated. A p value between 0.01 and 0.05 was considered significant and indicated with “*”, p between 0.001 and 0.01 shown as “**”; p < 0.001 with “***”; and p < 0.0001 with “****”. Data shown are either average or representative curves of at least three independent experiments, including analyses from imaging, immunoblot, migration, proliferation, senescence assay, qRT-PCR and proteomic experiments.
CRediT authorship contribution statement
Markus Waldeck-Weiermair: Writing – original draft, Visualization, Validation, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Apabrita A. Das: Visualization, Validation, Methodology, Investigation, Formal analysis, Data curation. Taylor A. Covington: Methodology, Investigation. Shambhu Yadav: Investigation. Jonas Kaynert: Investigation. Ruby Guo: Investigation. Priyanga Balendran: Visualization, Investigation. Venkata Revanth Thulabandu: Investigation. Arvind K. Pandey: Investigation. Fotios Spyropoulos: Investigation. David C. Thomas: Writing – review & editing, Visualization, Conceptualization. Thomas Michel: Writing – review & editing, Resources, Project administration, Methodology, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare no competing financial interests.
Acknowledgments
We thank Drs. Jennifer L. Meitzler and James L. Doroshow for providing the NOX4 specific antibody. This work was supported by National Institutes of Health Grants R21 AG063073, R33 HL157918, and R01 HL152173 (to T.M.); by K08 HL168240 and 5T32HL007609-34 (to F.S.); and by the Austrian Science Foundation (FWF) Grant J4466 (to M.W.-W.). Graphical abstract was created in biorender.com.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2024.103214.
Contributor Information
Markus Waldeck-Weiermair, Email: mwaldeck-weiermair@bwh.harvard.edu.
David C. Thomas, Email: tdct2@cam.ac.uk.
Thomas Michel, Email: thomas_michel@hms.harvard.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Sies H. Hydrogen peroxide as a central redox signaling molecule in physiological oxidative stress: oxidative eustress. Redox Biol. 2017;11:613–619. doi: 10.1016/j.redox.2016.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomas D.C., Clare S., Sowerby J.M., Pardo M., Juss J.K., Goulding D.A., van der Weyden L., Storisteanu D., Prakash A., Espéli M., Flint S., Lee J.C., Hoenderdos K., Kane L., Harcourt K., Mukhopadhyay S., Umrania Y., Antrobus R., Nathan J.A., Adams D.J., Bateman A., Choudhary J.S., Lyons P.A., Condliffe A.M., Chilvers E.R., Dougan G., Smith K.G.C. Eros is a novel transmembrane protein that controls the phagocyte respiratory burst and is essential for innate immunity. J. Exp. Med. 2017;214:1111–1128. doi: 10.1084/jem.20161382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Randzavola L.O., Mortimer P.M., Garside E., Dufficy E.R., Schejtman A., Roumelioti G., Yu L., Pardo M., Spirohn K., Tolley C., Brandt C., Harcourt K., Nichols E., Nahorski M., Woods G., Williamson J.C., Suresh S., Sowerby J.M., Matsumoto M., Santos C.X.C., Kiar C.S., Mukhopadhyay S., Rae W.M., Dougan G.J., Grainger J., Lehner P.J., Calderwood M.A., Choudhary J., Clare S., Speak A., Santilli G., Bateman A., Smith K.G.C., Magnani F., Thomas D.C. EROS is a selective chaperone regulating the phagocyte NADPH oxidase and purinergic signalling. Elife. 2022;11:1–31. doi: 10.7554/eLife.76387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas D.C., Charbonnier L.M., Schejtman A., Aldhekri H., Coomber E.L., Dufficy E.R., Beenken A.E., Lee J.C., Clare S., Speak A.O., Thrasher A.J., Santilli G., Al-Mousa H., Alkuraya F.S., Chatila T.A., Smith K.G.C. EROS/CYBC1 mutations: decreased NADPH oxidase function and chronic granulomatous disease. J. Allergy Clin. Immunol. 2019;143:782–785.e1. doi: 10.1016/j.jaci.2018.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnadottir G.A., Norddahl G.L., Gudmundsdottir S., Agustsdottir A.B., Sigurdsson S., Jensson B.O., Bjarnadottir K., Theodors F., Benonisdottir S., Ivarsdottir E.V., Oddsson A., Kristjansson R.P., Sulem G., Alexandersson K.F., Juliusdottir T., Gudmundsson K.R., Saemundsdottir J., Jonasdottir A., Jonasdottir A., Sigurdsson A., Manzanillo P., Gudjonsson S.A., Thorisson G.A., Magnusson O.T., Masson G., Orvar K.B., Holm H., Bjornsson S., Arngrimsson R., Gudbjartsson D.F., Thorsteinsdottir U., Jonsdottir I., Haraldsson A., Sulem P., Stefansson K. A homozygous loss-of-function mutation leading to CYBC1 deficiency causes chronic granulomatous disease. Nat. Commun. 2018;9:1–9. doi: 10.1038/s41467-018-06964-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waldeck-Weiermair M., Yadav S., Kaynert J., Thulabandu V.R., Pandey A.K., Spyropoulos F., Covington T., Das A.A., Krüger C., Michel T. Differential endothelial hydrogen peroxide signaling via Nox isoforms: critical roles for Rac1 and modulation by statins. Redox Biol. 2022;58 doi: 10.1016/j.redox.2022.102539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Negri S., Faris P., Moccia F. Reactive oxygen species and endothelial ca2+ signaling: brothers in arms or partners in crime? Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22189821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan W., Palmby T.R., Gavard J., Amornphimoltham P., Zheng Y., Gui J.S. An essential role for Rac1 in endothelial cell function and vascular development. Faseb. J. 2008;22:1829–1838. doi: 10.1096/fj.07-096438. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez E., Nagiel A., Lin A.J., Golan D.E., Michel T. Small interfering RNA-mediated down-regulation of caveolin-1 differentially modulates signaling pathways in endothelial cells. J. Biol. Chem. 2004;279:40659–40669. doi: 10.1074/jbc.M407051200. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez E., Kou R., Michel T. Rac1 modulates sphingosine 1-phosphate-mediated activation of phosphoinositide 3-kinase/Akt signaling pathways in vascular endothelial cells. J. Biol. Chem. 2006;281:3210–3216. doi: 10.1074/jbc.M510434200. [DOI] [PubMed] [Google Scholar]
- 11.Kou R., Michel T. Epinephrine regulation of the endothelial nitric-oxide synthase: roles of RAC1 and β3-adrenergic receptors in endothelial no signaling. J. Biol. Chem. 2007;282:32719–32729. doi: 10.1074/jbc.M706815200. [DOI] [PubMed] [Google Scholar]
- 12.Levine Y.C., Li G.K., Michel T. Agonist-modulated regulation of AMP-activated protein kinase (AMPK) in endothelial cells: evidence for an AMPK → Rac1 → Akt → endothelial nitric-oxide synthase pathway. J. Biol. Chem. 2007;282:20351–20364. doi: 10.1074/jbc.M702182200. [DOI] [PubMed] [Google Scholar]
- 13.Hess C.N., Kou R., Johnson R.P., Li G.K., Michel T. ADP signaling in vascular endothelial cells: ADP-dependent activation of the endothelial isoform of nitric-oxide synthase requires the expression but not the kinase activity of AMP-activated protein kinase. J. Biol. Chem. 2009;284:32209–32224. doi: 10.1074/jbc.M109.032656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalwa H., Sartoretto J.L., Sartoretto S.M., Michel T. Angiotensin-II and Marcks: a hydrogen peroxide- and Rac1-dependent signaling pathway in vascular endothelium. J. Biol. Chem. 2012;287 doi: 10.1074/jbc.M112.381517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heiss C., Rodriguez-Mateos A., Kelm M. Central role of eNOS in the maintenance of endothelial homeostasis. Antioxidants Redox Signal. 2015;22:1230–1242. doi: 10.1089/ars.2014.6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pandey A.K., Waldeck-Weiermair M., Wells Q.S., Xiao W., Yadav S., Eroglu E., Michel T., Loscalzo J. Expression of CD70 modulates nitric oxide and redox status in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2022;42:1169–1185. doi: 10.1161/ATVBAHA.122.317866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eroglu E., Gottschalk B., Charoensin S., Blass S., Bischof H., Rost R., Madreiter-Sokolowski C.T.C.T., Pelzmann B., Bernhart E., Sattler W., Hallstrom S., Malinski T., Waldeck-Weiermair M., Graier W.F.W.F., Malli R. Development of novel FP-based probes for live-cell imaging of nitric oxide dynamics. Nat. Commun. 2016;7:1–11. doi: 10.1038/ncomms10623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pak V.V., Ezeriņa D., Lyublinskaya O.G., Pedre B., Tyurin-Kuzmin P.A., Mishina N.M., Thauvin M., Young D., Wahni K., Martínez Gache S.A., Demidovich A.D., Ermakova Y.G., Maslova Y.D., Shokhina A.G., Eroglu E., Bilan D.S., Bogeski I., Michel T., Vriz S., Messens J., Belousov V.V. Ultrasensitive genetically encoded indicator for hydrogen peroxide identifies roles for the oxidant in cell migration and mitochondrial function. Cell Metabol. 2020;31:642–653.e6. doi: 10.1016/j.cmet.2020.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faviaa A., Desiderib M., Gambaraa G., D'Alessioa A., Ruas M., Esposito B., Del Bufalo D., Parrington J., Ziparoa E., Palombia F., Galioned A., Filippini A. VEGF-induced neoangiogenesis is mediated by NAADP and two-pore channel-2 -dependent ca2+signaling. Proc. Natl. Acad. Sci. U.S.A. 2014;111:4706–4715. doi: 10.1073/pnas.1406029111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griendling K.K., Sorescu D., Lassègue B., Ushio-Fukai M. Modulation of protein kinase activity and gene expression by reactive oxygen species and their role in vascular physiology and pathophysiology. Arterioscler. Thromb. Vasc. Biol. 2000;20:2175–2183. doi: 10.1161/01.ATV.20.10.2175. [DOI] [PubMed] [Google Scholar]
- 21.Song M., Finley S.D. ERK and Akt exhibit distinct signaling responses following stimulation by pro-angiogenic factors. Cell Commun. Signal. 2020;18:1–19. doi: 10.1186/s12964-020-00595-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Buul J.D., Fernandez-Borja M., Anthony E.C., Hordijk P.L. Expression and localization of NOX2 and NOX4 in primary human endothelial cells. Antioxidants Redox Signal. 2005;7 doi: 10.1089/ars.2005.7.308. [DOI] [PubMed] [Google Scholar]
- 23.Nohata N., Uchida Y., Stratman A.N., Adams R.H., Zheng Y., Weinstein B.M., Mukouyama Y.S., Gutkind J.S. Temporal-specific roles of Rac1 during vascular development and retinal angiogenesis. Dev. Biol. 2016;411:183–194. doi: 10.1016/j.ydbio.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Cerezo A.B., Hornedo-Ortega R., Álvarez-Fernández M.A., Troncoso A.M., García-Parrilla M.C. Inhibition of VEGF-induced VEGFR-2 activation and HUVEC migration by melatonin and other bioactive indolic compounds. Nutrients. 2017;9 doi: 10.3390/nu9030249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manford A.G., Rodríguez-Pérez F., Shih K.Y., Shi Z., Berdan C.A., Choe M., Titov D.V., Nomura D.K., Rape M. A cellular mechanism to detect and alleviate reductive stress. Cell. 2020;183:46–61.e21. doi: 10.1016/j.cell.2020.08.034. [DOI] [PubMed] [Google Scholar]
- 26.Kritsiligkou P., Shen T.K., Dick T.P. A comparison of Prx- and OxyR-based H2O2 probes expressed in S. cerevisiae. J. Biol. Chem. 2021;297 doi: 10.1016/j.jbc.2021.100866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoehne M.N., Jacobs L.J.H.C., Lapacz K.J., Calabrese G., Murschall L.M., Marker T., Kaul H., Trifunovic A., Morgan B., Fricker M., V Belousov V., Riemer J. Spatial and temporal control of mitochondrial H 2 O 2 release in intact human cells. EMBO J. 2022;41:1–16. doi: 10.15252/embj.2021109169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saeedi Saravi S.S., Eroglu E., Waldeck-Weiermair M., Sorrentino A., Steinhorn B., Belousov V., Michel T. Differential endothelial signaling responses elicited by chemogenetic H2O2 synthesis. Redox Biol. 2020;36 doi: 10.1016/j.redox.2020.101605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sunny S., Jyothidasan A., David C.L., Parsawar K., Veerappan A., Jones D.P., Pogwizd S., Rajasekaran N.S. Tandem mass tagging based identification of proteome signatures for reductive stress cardiomyopathy. Frontiers in Cardiovascular Medicine. 2022;9 doi: 10.3389/fcvm.2022.848045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoang H.M., Johnson H.E., Heo J. Rac-dependent feedforward autoactivation of NOX2 leads to oxidative burst. J. Biol. Chem. 2021;297 doi: 10.1016/j.jbc.2021.100982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geiszt M. NADPH oxidases: new kids on the block. Cardiovasc. Res. 2006;71:289–299. doi: 10.1016/j.cardiores.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 32.Rouyère C., Serrano T., Frémont S., Echard A. Oxidation and reduction of actin: origin, impact in vitro and functional consequences in vivo. Eur. J. Cell Biol. 2022;101 doi: 10.1016/j.ejcb.2022.151249. [DOI] [PubMed] [Google Scholar]
- 33.Marston D.J., Anderson K.L., Swift M.F., Rougie M., Page C., Hahn K.M., Volkmann N., Hanein D. High Rac1 activity is functionally translated into cytosolic structures with unique nanoscale cytoskeletal architecture. Proc. Natl. Acad. Sci. U.S.A. 2019;116:1267–1272. doi: 10.1073/pnas.1808830116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mehidi A., Rossier O., Schaks M., Chazeau A., Binamé F., Remorino A., Coppey M., Karatas Z., Sibarita J.B., Rottner K., Moreau V., Giannone G. Transient activations of Rac1 at the lamellipodium tip trigger membrane protrusion. Curr. Biol. 2019;29:2852–2866.e5. doi: 10.1016/j.cub.2019.07.035. [DOI] [PubMed] [Google Scholar]
- 35.Hirata H., Dobrokhotov O., Sokabe M. Coordination between cell motility and cell cycle progression in keratinocyte sheets via cell-cell adhesion and Rac1. iScience. 2020;23 doi: 10.1016/j.isci.2020.101729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duan J., Zhang T., Gaffrey M.J., Weitz K.K., Moore R.J., Li X., Xian M., Thrall B.D., Qian W.J. Stochiometric quantification of the thiol redox proteome of macrophages reveals subcellular compartmentalization and susceptibility to oxidative perturbations. Redox Biol. 2020;36 doi: 10.1016/j.redox.2020.101649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pecchillo Cimmino T., Ammendola R., Cattaneo F., Esposito G. NOX dependent ROS generation and cell metabolism. Int. J. Mol. Sci. 2023;24:1–16. doi: 10.3390/ijms24032086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poet G.J., Oka O.B., van Lith M., Cao Z., Robinson P.J., Pringle M.A., Arnér E.S., Bulleid N.J. Cytosolic thioredoxin reductase 1 is required for correct disulfide formation in the ER. EMBO J. 2017;36 doi: 10.15252/embj.201695336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kosuri P., Alegre-Cebollada J., Feng J., Kaplan A., Inglés-Prieto A., Badilla C.L., Stockwell B.R., Sanchez-Ruiz J.M., Holmgren A., Fernández J.M. Protein folding drives disulfide formation. Cell. 2012;151:794–806. doi: 10.1016/j.cell.2012.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhattarai K.R., Riaz T.A., Kim H.R., Chae H.J. The aftermath of the interplay between the endoplasmic reticulum stress response and redox signaling. Exp. Mol. Med. 2021;53:151–167. doi: 10.1038/s12276-021-00560-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao W., Loscalzo J. Metabolic responses to reductive stress. Antioxidants Redox Signal. 2020;32:1330–1347. doi: 10.1089/ars.2019.7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sies H., Mailloux R., Jakob U. Fundamentals of redox regulation in biology. Nat. Rev. Mol. Cell. Biol. Apr 30 2024 doi: 10.1038/s41580-024-00730-2. Epub ahead of print. PMID: 38689066. [DOI] [PubMed] [Google Scholar]
- 43.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meitzler J.L., Makhlouf H.R., Antony S., Wu Y., Butcher D., Jiang G., Juhasz A., Lu J., Dahan I., Jansen-Dürr P., Pircher H., Shah A.M., Roy K., Doroshow J.H. Decoding NADPH oxidase 4 expression in human tumors. Redox Biol. 2017;13:182–195. doi: 10.1016/j.redox.2017.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diebold B.A., Garrett Wilder S., De Deken X., Meitzler J.L., Doroshow J.H., Mccoy J.W., Zhu Y., Lambeth J.D. Guidelines for the detection of NADPH oxidases by immunoblot and RT-qPCR HHS public access. Methods Mol. Biol. 2019;1982:191–229. doi: 10.1007/978-1-4939-9424-3_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yadav S., Waldeck-Weiermair M., Spyropoulos F., Bronson R., Pandey A.K., Das A.A., Sisti A.C., Covington T.A., Thulabandu V., Caplan S., Chutkow W., Steinhorn B., Michel T. Sensory ataxia and cardiac hypertrophy caused by neurovascular oxidative stress in chemogenetic transgenic mouse lines. Nat. Commun. 2023;14 doi: 10.1038/s41467-023-38961-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rappsilber J., Ishihama Y., Mann M. Stop and Go Extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 48.Huber W., Carey V.J., Long L., Falcon S., Gentleman R. Graphs in molecular biology. BMC Bioinf. 2007;8 doi: 10.1186/1471-2105-8-S6-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32 doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 50.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software Environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13 doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.