Summary
3T3-L1 is a model cell line which can be differentiated from preadipocytes into mature adipocytes. Here, we present a protocol for changing gene expression in 3T3-L1 (pre)adipocytes using small interfering RNA (siRNA)-mediated knockdown. We describe steps to perform the knockdown of a certain gene prior to differentiation (day 4) to analyze the impact on adipogenesis. We then detail procedures for knockdown on day 8 of differentiation to study the role of a certain gene in mature adipocyte function.
For complete details on the use and execution of this protocol, please refer to Kaczmarek et al.1
Subject areas: Cell culture, Metabolism, Molecular Biology, Gene Expression
Graphical abstract

Highlights
-
•
Differentiation of 3T3-L1 preadipocytes to mature adipocytes
-
•
Optimization and validation of transfection methods and reagents for 3T3-L1
-
•
siRNA-mediated knockdown strategies in 3T3-L1 preadipocytes and mature adipocytes
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
3T3-L1 is a model cell line which can be differentiated from preadipocytes into mature adipocytes. Here, we present a protocol for changing gene expression in 3T3-L1 (pre)adipocytes using small interfering RNA (siRNA)-mediated knockdown. We describe steps to perform the knockdown of a certain gene prior to differentiation (day -4) to analyze the impact on adipogenesis. We then detail procedures for knockdown on day 8 of differentiation to study the role of a certain gene in mature adipocyte function.
Before you begin
The fibroblast-like cell line 3T3-L1 is widely used for studying adipogenesis and (pre)adipocyte function in vitro. Adipogenesis from fibroblast like 3T3-L1 preadipocytes to mature adipocytes can be induced by adding 3-isobutyl-1-methylxanthine (IBMX), dexamethasone, and insulin.2,3 IBMX inhibits phosphodiesterases leading to increased intracellular cAMP levels and, thereby, PKA activity.4 The synthetic glucocorticoid dexamethasone activates transcription factors of the C/EBP family, which are crucial for the onset of adipogenesis.5 Insulin induces glucose uptake and fatty acid synthesis as well as transcription factors such as SREBP-1c and PPARγ.6 To further induce differentiation, rosiglitazone, an agonist of PPARγ, the master regulator of adipogenesis, can be added. Even though rosiglitazone is not mandatory for adipogenesis, several publications as well as our own experience demonstrate that differentiation is more pronounced and reliable when adding rosiglitazone.7 To investigate the role of a certain gene onto (pre)adipocyte function or adipogenesis, we use small interfering RNA (siRNA)-mediated knockdown (KD) of this gene. As KD efficiency declines during differentiation due to siRNA degradation, we describe a protocol for siRNA-mediated KD in 3T3-L1 preadipocytes as well as in adipocytes.
Note: If not stated otherwise, reagents can be substituted with alternatives from different vendors. Working under sterile conditions using safety cabinets is crucial during every step of this protocol. PBS and all media should be warmed up to 37°C before use.
Cell culture of 3T3-L1 cells
Timing: ∼1 h
The protocol below describes steps for splitting and seeding 3T3-L1 cells. The volumes used are adjusted to cells seeded in a 175 cm2 flask. General information about cell culture handling is described by J.A. Ryan.8
Note: Cells need to be splitted on a regular basis (every other/third day) since confluency higher than 80% (7,000 cells/cm2) will reduce the adipogenic capacity.
-
1.
Discard the old media and wash cells with 5 mL PBS.
Note: Using PBS without Ca2+ and Mg2+ is crucial. The wash step not only removes non-viable cells, but also any traces of FBS, calcium, and magnesium that might inhibit the action of EDTA in the dissociation reagent.
-
2.Trypsinate the cells.
-
a.Add 2 mL of 0.25% Trypsin/EDTA and incubate for 1–2 min (37°C, 5% CO2).
-
b.Remove the cells from the flask bottom by tapping it with the flat hand.
-
c.Resuspend cells in 8 mL of culture media (CM, composition is shown in materials and equipment section).
-
d.Centrifuge cell suspension for 5 min at 200 × g and 21°C with an acceleration and a deceleration rate of 9.
-
a.
-
3.Count cells.
-
a.Discard the supernatant and resuspend cells in 2 mL of CM.
-
b.Count the cells using a Neubauer cell counting chamber.
-
a.
Note: Counter staining with Trypan Blue is not necessary. Washing of the cells (before you begin, cell culture of 3T3-L1 cells, Step 1) is sufficient to remove non-viable cells.
-
4.Dilute cells in 20 mL CM/ flask.
-
a.Cultivation for 48 h: 2.50 × 105 cells/flask.
-
b.Cultivation for 72 h: 1.25 × 105 cells/flask.
-
a.
Note: By moving the flask, the cells will be distributed evenly.
-
5.
Incubate cells for 2–3 days (37°C, 5% CO2).
Note: As contamination with mycoplasma not only reduces proliferation but also differentiation of 3T3-L1 cells, we recommend regular testing for mycoplasma every other week using the MycoAlert PLUS detection Kit. If contamination of 3T3-L1 was detected cells, should be treated with BM-Cyclin for two weeks before further usage.
Differentiation of 3T3-L1
Timing: ∼14 days
The cell line 3T3-L1 is commonly used as a model to study adipogenesis from preadipocytes to mature adipocytes.9,10 This process is induced by adding different substances leading to morphological changes including the characteristic accumulation of lipid droplets.1,11,12,13,14 For differentiation of 3T3-L1 cells, we recommend to use 3T3-L1 cells in a passage number below 25.
-
6.Day 0 of cell culture/ day --4 of differentiation.Optional: Day --4 of differentiation is flexible from day --3 to day --5.
-
a.Wash, trypsinate, and count the cells as described in the before you begin section (cell culture of 3T3-L1 cells).
-
b.Calculate the number of cells necessary for the desired well format according to the following table.
Well format Media/Well Cell count/well Cell density 6-well plate 2.00 mL 1 × 105 cells 104 cells/cm2 12-well plate 1.00 mL 5 × 104 cells 1.3 × 104 cells/cm2 24-well plate 0.50 mL 3 × 104 cells 1.6 × 104 cells/cm2 48-well plate 0.25 mL 2 × 104 cells 2.3 × 104 cells/cm2 96-well plate 0.10 mL 1 × 104 cells 3.0 × 104 cells/cm -
c.Prepare a master mix for all wells containing cells and media.
-
d.Seed the cells into a well format of interest.
-
a.
-
7.Day 2 of cell culture/ day −2 of differentiation.
-
a.Verify 100% confluency.
-
b.Incubate for further 2 days (37°C, 5% CO2).
-
a.
Optional: Renew CM if necessary.
Note: Renewing CM is only necessary if the pH is below 7 indicated by an orange color. Contact inhibition for 2 additional days is necessary for 3T3-L1 cells for a complete proliferation stop and successful differentiation.
-
8.Day 4 of cell culture/ day 0 of differentiation (Figure 1).
-
a.Change media to differentiation media 1 (DM1, composition is shown in materials and equipment section).
-
b.Incubate for 3 days (37°C, 5% CO2).
-
a.
-
9.Day 7 of cell culture/ day 3 of differentiation (Figure 1).
-
a.Change media to differentiation media 2 (DM2, composition is shown in materials and equipment section).
-
b.Incubate for 3 days (37°C, 5% CO2).
-
a.
-
10.Day 10 of cell culture/ day 6 of differentiation (Figure 1).
-
a.Change media to CM (composition is shown in materials and equipment section).
-
b.Incubate for 2 days (37°C, 5% CO2).
-
a.
Note: Media will become viscous. Be careful when removing media.
-
11.Day 12 of cell culture/ day 8 of differentiation (Figure 1).
-
a.Renew CM (composition is shown in materials and equipment section).
-
b.Incubate for 2 days (37°C, 5% CO2).
-
a.
-
12.
Day 14 of cell culture/ day 10 of differentiation (Figure 1) 3T3-L1 cells are fully differentiated into mature adipocytes. A detailed description about morphological differences and changes in expression of marker genes can be found in Figure 1.
Figure 1.
Differentiation of 3T3-L1 preadipocytes to mature adipocytes
While differentiating 3T3-L1 preadipocytes into mature adipocytes, microscopic pictures were taken before media change. On day 4 of cell culture/ day 0 of differentiation, we can observe 3T3-L1 preadipocytes with a fibroblast-like phenotype. 3 days later (day 7 of cell culture/ day 3 of differentiation), the filaments are getting shorter and the maturing adipocytes rounder. On day 10 of cell culture/ day 6 of differentiation, we see the formation of the first lipid droplets in the round shaped premature adipocytes. The cells on day 12 of cell culture/ day 8 of differentiation show enlarging lipid droplets. This process is supported with an additional 2 days of differentiation (until day 14 of cell culture/ day 10 of differentiation). During the process of differentiation, the expression of several marker genes, like Pparg, Lep and AdipoQ, is changing. Pparg, the master regulator of adipogenesis,15,16 and the adipokines Lep and AdipoQ17 are continuously higher expressed during differentiation. In mature adipocytes their expression peaks. Scale bar: 50 μm.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| 1x D-PBS | Thermo Fisher Scientific | 14190144 |
| AnnexinV | Biomol | ABD-20092 |
| ATPLite | PerkinElmer | 6016941 |
| BM-Cyclin | Sigma-Aldrich | 10799050001 |
| CaCl2 | Sigma-Aldrich | C3306 |
| Caspase-3/7 Assay | Promega | G7790 |
| Dexamethasone | Sigma-Aldrich | D4902 |
| Dulbecco’s modified Eagle’s medium (DMEM) | Thermo Fisher Scientific | 41966029 |
| Fetal bovine serum (FBS) | Thermo Fisher Scientific | 10270106 |
| HEPES | Thermo Fisher Scientific | 15630056 |
| Hoechst | Sigma-Aldrich | B2261 |
| IBMX | Sigma-Aldrich | I5879 |
| Insulin from bovine pancreas | Sigma-Aldrich | I6634 |
| Lipofectamine 2000 | Invitrogen | 11668 |
| Lipofectamine RNAiMAX | Invitrogen | 13778 |
| MTT | Sigma-Aldrich | M-2128 |
| MycoAlert PLUS detection kit | Lonza | LT07-701 |
| NaCl | Carl Roth | 9265.2 |
| Neon Transfect Kit | Thermo Fisher Scientific | MPK10096 |
| OptiMEM | Thermo Fisher Scientific | 31985070 |
| Penicillin-Streptomycin (10,000 U/mL) | Thermo Fisher Scientific | 15140122 |
| Propidium iodide | Sigma-Aldrich | P-4170 |
| Rosiglitazone | Sigma-Aldrich | R2408 |
| ROTI fect | Carl Roth | P001.4 |
| Trypsin-EDTA (0.25%), phenol red | Thermo Fisher Scientific | 25200056 |
| Critical commercial assays | ||
| Luna Universal qPCR master mix | New England Biolabs | M3003 |
| ReliaPrep RNA Miniprep systems | Promega | Z6012 |
| SuperScript II reverse transcriptase | Invitrogen | 18064071 |
| Experimental models: Cell lines | ||
| 3T3-L1 cells | ATCC | CL-173; RRID:CVCL_0123 |
| Oligonucleotides | ||
| Non coding (siNC), rCrGrUrUrArArUrCrGrCrGrUrArUrArArUrArCrGrCrGrUAT |
OriGene Technologies GmbH | SR30004 |
| siFzd5, rGrCrArCrUrArArGrArCrGrGrArCrArArGrCrUrArGrArGAA |
OriGene Technologies GmbH | SR417789, siRNA C |
| Fluorescent-labeled transfection control siRNA duplex | OriGene Technologies GmbH | SR30002 |
| Oligo dT, TTTTTTTTTTTTTTTTTTTTTTTVN |
Microsynth Seqlab | Custom synthesis |
| Random hexamer, NNNNNN |
Microsynth Seqlab | Custom synthesis |
| Software and algorithms | ||
| BioRender | BioRender | https://biorender.com/ |
| GraphPad Prism | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Other | ||
| 12-well plate | Greiner Bio-One | 665180 |
| 175 cm2 cell culture flask | Greiner Bio-One | 658175 |
| 24-well plate | TPP | 92024 |
| 48-well plate | Greiner Bio-One | 677165 |
| 50 mL tubes | Sarstedt GmbH | 62547254 |
| 6-well plate | Greiner Bio-One | 657160 |
| 96-well plate | Greiner Bio-One | 665180 |
| 96-well qPCR plate | Bio-Rad | HSP9601 |
| Centrifuge | Thermo Fisher Scientific | Megafuge 16R |
| CO2 incubator for cell culture | Thermo Fisher Scientific | Heraeus Kendro HeraCell |
| Electroporation system | Thermo Fisher Scientific | Neon Transfection System |
| Filtropur S 0.2 | Sarstedt GmbH | 831826001 |
| Improved Neubauer counting chamber | Th. Geyer GmbH | 6261165 |
| Inverted microscope | Nikon | TS100 |
| Microseals | Bio-Rad | MSB1001 |
| Safety cabinets | Thermo Fisher Scientific | HERAsafe KS 12 |
Materials and equipment
Culture media (CM)
| Reagent | Final concentration | Volume |
|---|---|---|
| DMEM | 89% (v/v) | 44.5 mL |
| FBS | 10% (v/v) | 5.0 mL |
| Penicillin-Streptomycin (Pen/Strep) | 1% (v/v) | 0.5 mL |
| Total | N/A | 50.0 mL |
Note: Media can be stored up to 3 months at 4°C. Pen/Strep is not essential for cultivation of 3T3-L1 cells but can be added without significantly affecting cell growth. Furthermore, we did not observe differences in 3T3-L1 differentiation with and without Pen/Strep. We recommend adding Pen/Strep when the cells are handled by several individuals. FBS was not heat-inactivated prior to use.
Differentiation media 1 (DM1)
| Reagent | Stock concentration | Final concentration | Volume |
|---|---|---|---|
| CM | - | - | 50 mL |
| Insulin | 2 mg/mL | 1 μg/mL | 25.0 μL |
| Dexamethasone | 10 mM | 0.25 μM | 1.25 μL |
| IBMX | 500 mM | 500 μM | 50.0 μL |
| Rosiglitazone | 20 mM | 2 μM | 5.0 μL |
| Total | N/A | N/A | 50.0 mL |
Note: DM1 must be sterile filtered (0.2 μm filter) before usage and can be stored up to 2 weeks at 4°C. Prewarming of DMEM to 37 °C before mixing the media is crucial, as IBMX precipitates when added to cold media. Rosiglitazone is not mandatory for differentiation of 3T3-L1 cells into adipocytes; however, it was shown that esp. cells of higher passage have a significantly better lipid accumulation when rosiglitazone was added.
Differentiation media 2 (DM2)
| Reagent | Stock concentration | Final concentration | Volume |
|---|---|---|---|
| CM | - | - | 50 mL |
| Insulin | 2 mg/mL | 1 μg/mL | 25.0 μL |
| Total | N/A | N/A | 50.0 mL |
Note: DM2 must be sterile filtered (0.2 μm filter) before usage and can be stored up to 2 weeks at 4°C.
Step-by-step method details
While establishing a transfection protocol for 3T3-L1 to generate siRNA-mediated KD, we analyzed different protocols for electroporation and lipofection with various reagents. Electroporation with the Neon Transfection System (Invitrogen) results in good transfection efficiencies of fluorescent-labeled transfection control siRNA in 3T3-L1 at day -4 of differentiation. However, for electroporation of partly or fully differentiated 3T3-L1 adipocytes detachment of the cells is mandatory. Unfortunately, cell adherence afterwards is insufficient. Testing the transfection efficiency of various lipofection reagents (ROTI®Fect (Carl Roth), Lipofectamin 2000 (Invitrogen), Lipofectamine MessengerMAX (Invitrogen), and Lipofectamin RNAiMAX (Invitrogen)) with fluorescent-labeled transfection control siRNA in 3T3-L1 (pre)adipocytes, we found Lipofectamin RNAiMAX to be the most suitable product. Transfection efficiency for 3T3-L1 with Lipofectamine RNAiMAX and the fluorescent-labeled transfection control siRNA is shown in Figure 2.
Figure 2.
Determination of transfection efficiency
3T3-L1 cells were transfected with fluorescent-labeled transfection control siRNA using Lipofectamine RNAiMAX on day 0 of cell culture/ day -4 of differentiation or on day 12 of cell culture/ day 8 of differentiation. Transfection efficiency was determined on day 0 or day 10 of differentiation and normalized against total cell count (n = 4). Given is the mean ± SEM of the indicated number of independent experiments.
To analyze the function of a certain gene in mature 3T3-L1 adipocytes, KD in preadipocytes (day 0 of cell culture/ day -4 of differentiation) is not suitable due to possible interference with adipogenesis and siRNA degradation over time. To determine a suitable time point for KD to analyse a certain gene in adipocyte function, two different KD strategies are available: 1) after reaching maturity on day 14 of cell culture/ day 10 of differentiation or 2) two days prior to maturity on day 12 of cell culture/ day 8 of differentiation before conducting further experiments two days later. To reduce stress to the cells KD one day prior to assay is not recommended. Different groups claim 3T3-L1 to be mature at different days after differentiation initiation.1,15,16,18 According to our protocol the process of adipogenesis is fully activated after day 10 of cell culture/ day 6 of differentiation and, thereafter, 3T3-L1 cells are steadily accumulating lipids and increase the expression of marker genes. Further, Malekpour-Dehkordi et al. showed that prolonged incubation of 3T3-L1 adipocytes for additional 30 days leads to an even higher lipid accumulation into the cells.19 Thus, maturity of 3T3-L1 is an artificially set time point after adding differentiation media. Since we conducted KD-independent experiments like stimulations and other treatments on day 14 of cell culture/ day 10 of differentiation in our lab, we decided on KD at day 12 of cell culture/ day 8 of differentiation.1 The transfection efficiency of this KD strategy with fluorescent-labeled transfection control siRNA is shown in Figure 2.
Note: If not stated otherwise, reagents can be substituted with alternatives from different vendors. Working under sterile conditions using safety cabinets is crucial during every step of this protocol. PBS and all media should be warmed up to 37°C before use.
Transfection, KD; day -4 of differentiation
Timing: 45 min + 20–24 h incubation
3T3-L1 is a model cell line with adipogenic potential.9,10 To identify the role of a certain gene onto adipogenesis or onto preadipocyte function siRNA-mediated KD of this gene on day -4 of differentiation can be performed.1,11,13
Note: This step is carried out on day 0 of cell culture/ day -4 of differentiation. After this protocol, one can continue with the differentiation (day 2 of cell culture/ day -2 of differentiation) as described in the before you begin section (differentiation of 3T3-L1).
-
1.Preparation of the transfection mix for KD on day 0 of cell culture/ day -4 of differentiation. The volumes of all reagents are described per well.Note: Always prepare a transfection master mix for better reproducibility. We highly recommend on using Lipofectamine RNAiMAX instead of other lipofection reagents.
Plate format 96-Well 48-Well 24-Well 12-Well 6-Well OptiMEM 17.60 μL 58.7 μL 120.0 μL 200.0 μL 500 μL siRNA (5 μM Stock) 0.22 μL 0.7 μL 1.4 μL 2.4 μL 6 μL Lipofectamine RNAiMAX 0.18 μL 0.6 μL 1.2 μL 2.0 μL 5.0 μL -
a.Mix the indicated amount of OptiMEM and siRNA.
-
b.Add Lipofectamine RNAiMAX slowly to avoid bubbles and gently invert the mixture.
-
c.Add transfection mix into each well.
-
d.Incubate for 20 min (37°C, 5% CO2).Optional: Use a fluorescently labeled siRNA in the experiment to ensure successful transfection. Fluorescence can be detected the day after lipofection.
-
a.
-
2.Seeding of cells and lipofection.
-
a.Add the appropriate cell number diluted in the indicated amount of OptiMEM to the wells containing the transfection mix. The volumes of OptiMEM and cell counts are given per well.
Plate format 96-Well 48-Well 24-Well 12-Well 6-Well Cell count/well 9,000 cells 30,000 cells 60,000 cells 100,000 cells 250,000 cells OptiMEM 90 μL 300 μL 600 μL 1000 μL 2500 μL Cell density 27,000 cells/cm2 35,000 cells/cm2 32,000 cells/cm2 26,000 cells/cm2 26,000 cells/cm2 -
b.Incubate for 20–24 h (37°C, 5% CO2).
-
a.
-
3.
Addition of CM as indicated in the table below. The volumes of CM are given per well.
| Plate format | 96-Well | 48-Well | 24-Well | 12-Well | 6-Well |
|---|---|---|---|---|---|
| CM | 200 μL | 600 μL | 1200 μL | 2000 μL | 5000 μL |
-
4.
Either continuing with the differentiation, day 2 of cell culture/ day -2 of the differentiation (before you begin, differentiation of 3T3-L1) or analysis of desired preadipocyte function 48 h later.
Transfection, KD; day 8 of differentiation
Timing: 45 min + 6 h incubation
As KD in preadipocytes (day 0 of cell culture/ day -4 of differentiation) might interfere with adipogenesis or siRNA is degraded during the process of differentiation, this KD strategy is not suitable for analyzing the function of a certain gene in adipocytes. Instead, KD of this gene can be conducted at day 12 of cell culture/ day 8 of differentiation.
Note: This step is carried out on day 12 of cell culture/ day 8 of differentiation described in the before you begin section (differentiation of 3T3-L1).
-
1.
Differentiate the cells according to the protocol ‘differentiation of 3T3-L1’ in the before you begin section until day 12 of cell culture/ day 8 of differentiation.
-
2.Prepare the transfection mix according to the table below. The volumes of all reagents are given per well.Note: Always prepare a transfection master mix for better reproducibility. We highly recommend using RNAiMAX instead of other lipofection reagents.
Plate format 96-Well 48-Well 24-Well 12-Well 6-Well OptiMEM 35 μL 80 μL 160.0 μL 400.0 μL 1000 μL siRNA (5 μM Stock) 0.22 μL 0.5 μL 1.0 μL 2.5 μL 6.25 μL Lipofectamine RNAiMAX 0.18 μL 0.4 μL 0.8 μL 2.0 μL 5.0 μL -
a.Mix OptiMEM and siRNA.
-
b.Add Lipofectamine RNAiMAX slowly to prevent bubbles and invert gently.
-
c.Incubate for 20 min (37°C, 5% CO2).Optional: Use a fluorescently labeled siRNA in the experiment to ensure successful transfection. Fluorescence can be detected the day after lipofection.
-
a.
-
3.Lipofection.
-
a.Remove the old media from the cells.Note: Cells should be 100% confluent. About 80% of the cells should be almost fully differentiated adipocytes.
-
b.Add transfection mix to the cells.
-
c.Incubate for 6 h (37°C, 5% CO2).
-
a.
-
4.Addition of CM.
-
a.Carefully add CM to the cells according to the table below. The volumes of CM are given per well:Note: 3T3-L1 adipocytes are easily detached; therefore, careful addition of CM is necessary.
Plate format 96-Well 48- Well 24-Well 12-Well 6-Well CM 150 μL 300 μL 600 μL 1500 μL 3750 μL -
b.Incubate for 2 days (37°C, 5% CO2).
-
a.
-
5.
Day 14 of the cell culture/ day 10 of the differentiation: Differentiation of the cells is completed and further experiments can take place.
Expected outcomes
3T3-L1 cells can be differentiated from preadipocytes to adipocytes (Figure 1). During adipogenesis 3T3-L1 change their morphology as well as the expression levels of several marker genes (e.g., Pparg, Lep, AdipoQ).
To evaluate the role of a certain gene in (pre)adipocyte function and adipogenesis, KD studies of this gene can be performed. Importantly, transfection efficiency of the method of choice was determined before conducting KD studies (Figure 2). Following, it is mandatory to evaluate the effect of the lipofection on cell viability. Thus, 3T3-L1 WT were compared to siNC transfected cells for apoptosis, necrosis, cell count, ATP amount and metabolic activity (Figure 3). To validate whether transfection and, thereby, the KD of a certain gene was sufficient, gene expression of this gene needs to be determined using qPCR. Alternatively, protein level of this gene can be determined. However, designing and validating a qPCR primer is cheaper and less time consuming than antibody validation. Exemplarily, we reduced gene expression of Fzd5 (siFzd5) on day 0 of cell culture/ day -4 of differentiation and day 12 of cell culture/ day 8 of differentiation before comparing it to control transfected cells (siNC) (Figure 4).
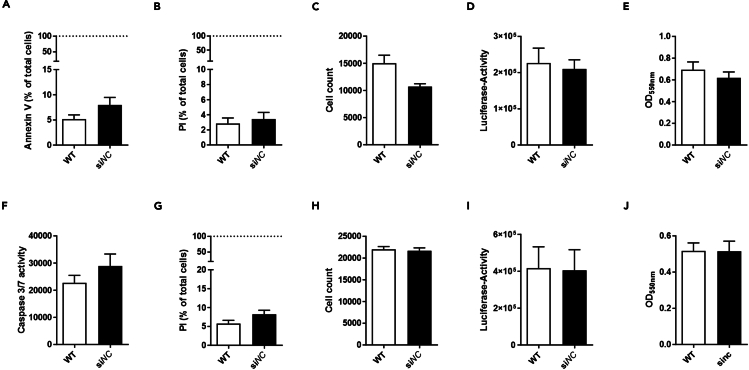
Figure 3.
Assessing the viability of 3T3-L1 (pre)adipocytes after lipofection with siNC in comparison to wild-type cells
3T3-L1 (pre)adipocytes were transfected on day 0 of cell culture/ day -4 of differentiation (A-E, preadipocytes) or on day 12 of cell culture/ day 8 of differentiation (F-J, mature adipocytes) before viability was determined on day 4 of cell culture/ day 0 of differentiation or day 14 of cell culture/ day 10 of differentiation. While analyzing viability in preadipocytes, staining for A) apoptosis (n = 4), B) necrosis (n = 4) and C) cell count (n = 4), revealed no differences between WT and siNC. Further, D) ATP amount (n = 4) and E) metabolic activity (n = 4), showed no changes comparing WT and siNC. In mature adipocytes F) apoptosis, G) necrosis, H) cell count, I) ATP amount as well as J) metabolic activity were not influenced by lipofection either. Apoptosis, necrosis, and cell count were stained using AnnexinV, propidium iodide (PI) and Hoechst33342 (in 10 mM HEPES, 140 mM NaCl, 2.5 mM CaCl2). In mature adipocytes background fluorescence interfered with AnnexinV staining. Instead, caspase-3/7 activity was analyzed. ATP amount was determined by using ATPLite and metabolic activity by using MTT. Given is the mean ± SEM of the indicated number of independent experiments.1
Figure 4.
Verification of gene silencing efficiency after siRNA-mediated KD in 3T3-L1 (pre)adipocytes
(A) siRNA-mediated KD of Fzd5 (siFzd5) was performed in 3T3-L1 preadipocytes on day 0 of cell culture/ day -4 of differentiation. KD efficiency was monitored during differentiation of the cells from preadipocytes (day 4 of cell culture/ day 0 of differentiation) to mature adipocytes (day 14 of cell culture/ day 10 of differentiation) every other day (n ≥ 4). KD efficiency declines during differentiation due to siRNA degradation and cannot be detected from day 10 of cell culture/ day 8 of differentiation on. The time frame of effective KD before siRNA degradation is siRNA-specific. ΔCt values were determined using Actb20 as housekeeping gene (Ct = 15.08 ± 0.20). B) KD on day 12 of cell culture/ day 8 of differentiation reduces Fzd5 gene expression by 70% compared to control (siNC) (n = 7). ΔCt values were determined using Actb as housekeeping gene (Ct = 14.71 ± 0.22). Given is the mean ± SEM of the indicated number of independent experiments. KD efficiency is given relative to control-transfected cells (siNC). Statistical significance was tested using multiple t-tests (A) and paired student’s t-test (B). ∗p < 0.05, ∗∗∗p < 0.001.1
Limitations
This protocol shows how to perform siRNA-mediated KD at different stages (day -4 or day 8 of differentiation) during the differentiation of 3T3-L1 preadipocytes to mature adipocytes. The KD on day 0 of cell culture/ day -4 of differentiation can be used to analyze the impact of a certain gene onto preadipocyte function and adipogenesis. Moreover, KD on day 12 of cell culture/ day 8 of differentiation can be used to study the role of a certain gene in adipocyte function.
The main limitation of this assay is that siRNA-mediated KD is only reducing not depleting gene expression leading to residual gene expression in the cells. Moreover, the KD on day 0 of cell culture/ day -4 of differentiation is performed at the beginning of the whole differentiation process with a duration of 14 days. Due to siRNA degradation over time, the KD is (most often) not stable during the whole differentiation process and the effect of a certain gene in later adipogenesis cannot be analyzed properly. As 3T3-L1 is a mouse-derived cell line comparability to human cells is limited. Further, 3T3-L1 is an immortalized cell line, which might not depict the in vivo or ex vivo effects. A more physiological alternative is the stromal vascular fraction (SVF) of adipose tissue (AT). However, SVF is a co-culture of various cells (e.g., preadipocytes, immune cells), which are extracted from AT after sacrificing mice. This makes the SVF not only less accessible compared to 3T3-L1 cells, but also raises ethical questions. Thus, we recommend validating data generated in 3T3-L1 in the SVF.
Troubleshooting
Problem 1
Transfection efficiency of 3T3-L1 cells assessed using fluorescent-labeled siRNA is not sufficient (step-by-step method details or expected outcomes, Figure 2).
Potential solution
-
•
When using lipofection avoid media containing FBS. Use OptiMEM as suggested in this protocol or DMEM without FBS.
OR
-
•
Potential mycoplasma contamination can interfere with transfection. Test for mycoplasma as described in Step 5 of cell culture of 3T3-L1 cells section.
OR
-
•
Another lipofection reagent than Lipofectamine RNAiMax was used. We highly recommend using RNAiMAX as a transfection reagent for siRNA-mediate KD in 3T3-L1, because those cells are difficult to transfect and we verified, that this reagent leads to the sufficient transfection efficiency after lipofection on day -4 and day 8 of differentiation. However, we could not test every available lipofection reagent, therefore, if another reagent is chosen, transfection efficiency needs to be verified using a fluorescent-labeled siRNA.
Problem 2
When analyzing KD efficiency, no downregulation of the gene of interest is observed (expected outcomes, Figure 4).
Potential solution
-
•
Assess transfection efficiency using a fluorescent-labeled control siRNA. If you cannot detect fluorescent-labeled control siRNA in your cells we refer to problem 1.
OR
-
•
Try different siRNAs from several distributors specific for your gene of interest.
Problem 3
3T3-L1 differentiation prior to KD on day 8 is insufficient (step-by-step method details, transfection, KD; day 8 of differentiation).
Potential solution
-
•
Prepare differentiation media freshly and repeat differentiation protocol.
OR
-
•
If 3T3-L1 cells have a high passage (>25), thaw fresh cells and repeat differentiation.
OR
-
•
If 3T3-L1 reached confluency over 80% (8,500 cells/cm2) during expansion, the adipogenic potential might be lost. Thaw fresh cells and repeat differentiation.
OR
-
•
3T3-L1 cells are contaminated with mycoplasma. Test for mycoplasma like described in Step 5 of cell culture of 3T3-L1 cells section.
Problem 4
IBMX precipitates after adding to media (Materials and equipment, Differentiation media 1 (DM1)).
Potential solution
-
•
Repeat the preparation of DM1 and preheat media to 37°C before adding IBMX.
Problem 5
Transfection with control siRNA affects cell viability, adherence and/or adipogenesis (expected outcomes, Figure 3).
Potential solution
-
•
Potential mycoplasma contamination can interfere with transfection. Test for mycoplasma as described in Step 5 of cell culture of 3T3-L1 cells section.
OR
-
•
If 3T3-L1 cells have a high passage (>25), thaw fresh cells and repeat differentiation.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Doreen Thor (Doreen.Thor@medizin.uni-leipzig.de).
Technical contact
Technical questions on executing this protocol should be directed to and will be answered by the technical contact, Isabell Kaczmarek (Isabell.Kaczmarek@medizin.uni-leipzig.de).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
The graphical abstract was created using BioRender.com. Our research related to this topic is funded by the European Social Funds, European Union (D.T.).
Author contributions
Conceptualization, M.S., D.T., and I.K.; methodology, D.T. and I.K.; investigation, D.T. and I.K.; writing – original draft, M.S. and I.K.; writing – review and editing, M.S., D.T., and I.K.; funding acquisition, D.T.; resources, D.T.; supervision, D.T. and I.K.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Doreen Thor, Email: doreen.thor@medizin.uni-leipzig.de.
Isabell Kaczmarek, Email: isabell.kaczmarek@medizin.uni-leipzig.de.
References
- 1.Kaczmarek I., Wower I., Ettig K., Kuhn C.K., Kraft R., Landgraf K., Körner A., Schöneberg T., Horn S., Thor D. Identifying G protein-coupled receptors involved in adipose tissue function using the innovative RNA-seq database FATTLAS. iScience. 2023;26 doi: 10.1016/j.isci.2023.107841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green H., Kehinde O. An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell. 1975;5:19–27. doi: 10.1016/0092-8674(75)90087-2. [DOI] [PubMed] [Google Scholar]
- 3.Russell T.R., Ho R. Conversion of 3T3 fibroblasts into adipose cells: triggering of differentiation by prostaglandin F2alpha and 1-methyl-3-isobutyl xanthine. Proc. Natl. Acad. Sci. USA. 1976;73:4516–4520. doi: 10.1073/pnas.73.12.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang D.-C., Tsay H.-J., Lin S.-Y., Chiou S.-H., Li M.-J., Chang T.-J., Hung S.-C. cAMP/PKA regulates osteogenesis, adipogenesis and ratio of RANKL/OPG mRNA expression in mesenchymal stem cells by suppressing leptin. PLoS One. 2008;3 doi: 10.1371/journal.pone.0001540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zubiría M.G., Giordano A.P., Gambaro S.E., Alzamendi A., Frontini-López Y.R., Moreno G., Spinedi E., Giovambattista A. Dexamethasone primes adipocyte precursor cells for differentiation by enhancing adipogenic competency. Life Sci. 2020;261 doi: 10.1016/j.lfs.2020.118363. [DOI] [PubMed] [Google Scholar]
- 6.Cignarelli A., Genchi V.A., Perrini S., Natalicchio A., Laviola L., Giorgino F. Insulin and Insulin Receptors in Adipose Tissue Development. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20030759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zebisch K., Voigt V., Wabitsch M., Brandsch M. Protocol for effective differentiation of 3T3-L1 cells to adipocytes. Anal. Biochem. 2012;425:88–90. doi: 10.1016/j.ab.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 8.Ryan J.A. 1994. Understanding and Managing Cell Culture Contamination (Corning Incorporated) [Google Scholar]
- 9.Green H., Meuth M. An established pre-adipose cell line and its differentiation in culture. Cell. 1974;3:127–133. doi: 10.1016/0092-8674(74)90116-0. [DOI] [PubMed] [Google Scholar]
- 10.Green H., Kehinde O. Sublines of mouse 3T3 cells that accumulate lipid. Cell. 1974;1:113–116. doi: 10.1016/0092-8674(74)90126-3. [DOI] [Google Scholar]
- 11.Buerger F., Müller S., Ney N., Weiner J., Heiker J.T., Kallendrusch S., Kovacs P., Schleinitz D., Thiery J., Stadler S.C., Burkhardt R. Depletion of Jmjd1c impairs adipogenesis in murine 3T3-L1 cells. Biochim. Biophys. Acta, Mol. Basis Dis. 2017;1863:1709–1717. doi: 10.1016/j.bbadis.2017.05.011. [DOI] [PubMed] [Google Scholar]
- 12.Suchý T., Kaczmarek I., Maricic T., Zieschang C., Schöneberg T., Thor D., Liebscher I. Evaluating the feasibility of Cas9 overexpression in 3T3-L1 cells for generation of genetic knock-out adipocyte cell lines. Adipocyte. 2021;10:631–645. doi: 10.1080/21623945.2021.1990480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suchý T., Zieschang C., Popkova Y., Kaczmarek I., Weiner J., Liebing A.-D., Çakir M.V., Landgraf K., Gericke M., Pospisilik J.A., et al. The repertoire of Adhesion G protein-coupled receptors in adipocytes and their functional relevance. Int. J. Obes. 2020;44:2124–2136. doi: 10.1038/s41366-020-0570-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scott M.A., Nguyen V.T., Levi B., James A.W. Current methods of adipogenic differentiation of mesenchymal stem cells. Stem Cells Dev. 2011;20:1793–1804. doi: 10.1089/scd.2011.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arimura N., Horiba T., Imagawa M., Shimizu M., Sato R. The peroxisome proliferator-activated receptor gamma regulates expression of the perilipin gene in adipocytes. J. Biol. Chem. 2004;279:10070–10076. doi: 10.1074/jbc.M308522200. [DOI] [PubMed] [Google Scholar]
- 16.Chawla A., Schwarz E.J., Dimaculangan D.D., Lazar M.A. Peroxisome proliferator-activated receptor (PPAR) gamma: adipose-predominant expression and induction early in adipocyte differentiation. Endocrinology. 1994;135:798–800. doi: 10.1210/endo.135.2.8033830. [DOI] [PubMed] [Google Scholar]
- 17.Palhinha L., Liechocki S., Hottz E.D., Pereira J.A.d.S., de Almeida C.J., Moraes-Vieira P.M.M., Bozza P.T., Maya-Monteiro C.M. Leptin Induces Proadipogenic and Proinflammatory Signaling in Adipocytes. Front. Endocrinol. 2019;10:841. doi: 10.3389/fendo.2019.00841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zieger K., Weiner J., Krause K., Schwarz M., Kohn M., Stumvoll M., Blüher M., Heiker J.T. Vaspin suppresses cytokine-induced inflammation in 3T3-L1 adipocytes via inhibition of NFκB pathway. Mol. Cell. Endocrinol. 2018;460:181–188. doi: 10.1016/j.mce.2017.07.022. [DOI] [PubMed] [Google Scholar]
- 19.Malekpour-Dehkordi Z., Mohiti-Ardakani J., Nourbakhsh M., Teimourian S., Naghiaee Y., Hemati M., Jafary F. Gene expression profile evaluation of integrins in 3T3-L1 cells differentiated to adipocyte, insulin resistant and hypertrophied cells. Gene. 2019;710:406–414. doi: 10.1016/j.gene.2019.06.019. [DOI] [PubMed] [Google Scholar]
- 20.Zhang J., Tang H., Zhang Y., Deng R., Shao L., Liu Y., Li F., Wang X., Zhou L. Identification of suitable reference genes for quantitative RT-PCR during 3T3-L1 adipocyte differentiation. Int. J. Mol. Med. 2014;33:1209–1218. doi: 10.3892/ijmm.2014.1695. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.