Abstract
Error-prone DNA synthesis by retroviral reverse transcriptases (RTs) is a major contributor to variation in retroviral populations. Structural features of retroviral RTs that are important for accuracy of DNA synthesis in vivo are not known. To identify structural elements of murine leukemia virus (MLV) RT important for fidelity in vivo, we developed a D17-based encapsidating cell line (ANGIE P) which is designed to express the amphotropic MLV envelope. ANGIE P also contains an MLV-based retroviral vector (GA-1) which encodes a wild-type bacterial β-galactosidase gene (lacZ) and a neomycin phosphotransferase gene. Transfection of ANGIE P cells with wild-type or mutated MLV gag-pol expression constructs generated GA-1 virus that was able to undergo only one cycle of viral replication upon infection of D17 cells. The infected D17 cell clones were characterized by staining with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), and the frequencies of inactivating mutations in lacZ were quantified. Three mutations in the YVDD motif (V223M, V223S, and V223A) and two mutations in the RNase H domain (S526A and R657S) exhibited frequencies of lacZ inactivation 1.2- to 2.3-fold higher than that for the wild-type MLV RT (P < 0.005). Two mutations (V223I and Y598V) did not affect the frequency of lacZ inactivation. These results establish a sensitive in vivo assay for identification of structural determinants important for accuracy of DNA synthesis and indicate that several structural determinants may have an effect on the in vivo fidelity of MLV RT.
High mutation rates exhibited by retroviruses generate extensive genetic variation and confer high evolutionary potential in retrovirus populations (14, 42, 43, 52–54, 62). Variation in human immunodeficiency virus type 1 (HIV-1) populations may in turn affect the fitness of the species. For example, genetic variation in HIV-1 populations has resulted in the selection of mutations that confer resistance to at least 11 antiretrovirus drugs approved for clinical use and selection of escape mutants that evade the host immune response (15, 46). Genetic variation has also thwarted efforts to develop effective vaccines against HIV-1 (64).
An important mechanism of generating variation in retrovirus populations is the low fidelity of reverse transcriptase (RT) (42, 43, 52–54). Mutation rates of RTs can be measured in vitro or in vivo by various assays (5, 42, 43, 53–55). The measurements of fidelity are often altered by the in vitro assay conditions (18, 39, 44). Previously, the in vitro mutation rate of HIV-1 was estimated to be approximately 20-fold higher than the in vivo mutation rate (4, 42). In vivo forward mutation assays for various RTs have been determined and range from 3.4 × 10−5 to 4.8 × 10−6 mutations/bp/replication cycle (42, 43, 52–54).
Amino acid positions and motifs in RT that may be important for the in vivo fidelity of RT are unknown. However, several studies have identified potential structural determinants that are important for the in vitro fidelity of RT (1–3, 19, 25–27, 35, 37, 45, 50, 51, 56, 57, 60, 66). The YXDD motif is highly conserved in RTs, and mutations in the motif are associated with decreases in enzymatic activity and viral infectivity, changes in the positioning of the primer in the template-primer complex, and resistance to nucleoside analogs (10, 12, 15, 59, 67). In addition, mutations at the methionine of the HIV-1 YMDD motif have been associated with alterations in the in vitro fidelity of RT (1, 2, 19, 25–27, 50, 51, 66). Mutation of the HIV-1 YMDD motif to YVDD (M184V) is associated with high-level resistance to the antiretrovirus drug (−)-2′,3′-dideoxy-3′-thiacytidine (3TC) (9, 20, 63). Several studies have observed large (up to 45-fold) increases in the in vitro fidelity of HIV-1 RT containing the M184V mutation (50, 51, 66). However, another in vitro study using a forward mutation assay has indicated that the YVDD mutant of HIV-1 RT exhibits less than a twofold decrease in the mutation rate (19).
The amino acids involved in binding to the substrate deoxynucleoside triphosphate (dNTP) may also constitute a fidelity determinant of RT (26, 29). Recently, the crystal structure of HIV-1 RT complexed with the template-primer and the dTTP substrate shows six amino acids that contact the substrate dTTP (29). Mutations at three of the positions were associated with alterations in the in vitro fidelity of RT (26, 45). Another motif, the αH helix of the HIV-1 RT thumb domain, binds to the minor groove of the template-primer complex and is associated with alterations in the processivity and fidelity of RT (3, 6, 30). Mutational analysis of the αH helix of HIV-1 RT was associated with lower fidelity in vitro, either through an increase in the rate of RT dissociation from the template DNA or through an increase in strand slippage resulting in lower frameshift fidelity (6). Finally, the RNase H domain may influence the rate of RT processivity and template switching, which may result in an increase in the rate of deletions, deletions with insertions, and duplications (21, 24, 33, 41).
The differences in the fidelities of HIV-1 RT mutants in different in vitro assays demonstrate a need for analyzing the fidelity of RTs during the course of in vivo retrovirus replication. Structural similarities between murine leukemia virus (MLV) and HIV-1 RTs suggests that understanding the structural determinants of MLV RT important for fidelity may provide insights into the fidelity of HIV-1 RT (22, 38). In addition, a greater understanding of the mechanisms of MLV RT fidelity may lead to the development of improved MLV-based vectors and packaging cell lines for gene therapy. In this study, we have developed an in vivo assay for rapid measurement of the fidelity of wild-type or mutated MLV RTs. This assay provides a useful tool to identify structural determinants of MLV RT important for accuracy of DNA synthesis and to determine the extent to which they influence fidelity in vivo.
MATERIALS AND METHODS
Plasmids and retrovirus vector.
The construction of MLV-based retrovirus vector GA-1 was previously described (34). Plasmids pLGPS and pAMS, which expressed the MLV gag and pol genes from a truncated MLV long terminal repeat (LTR) promoter and expressed a replication-competent MLV, respectively, were a kind gift from A. D. Miller (47, 48). Plasmid pSV-A-MLV-env, which expressed the amphotropic MLV envelope gene from the LTR promoter and simian virus 40 enhancer, was obtained from the AIDS Research and Reference Reagents Program (40). Plasmid pBSpac encodes the puromycin N-acetyltransferase gene and confers resistance to puromycin (65). Plasmid pSVα3.6 encodes the α subunit of murine Na+,K+-ATPase gene and confers resistance to ouabain (36).
Generation of MLV RT mutants.
Site-directed mutagenesis of pLGPS was performed by using a Morph kit (5′-3′, Inc.) or Chameleon kit (Stratagene). A detailed description of the mutagenic oligonucleotides and the strategies used to identify mutants is available upon request. Briefly, most of the mutagenic oligonucleotides were designed to introduce additional silent mutations and generate new restriction sites. Restriction digestion analysis was performed to identify plasmids containing mutations. DNA fragments ranging in size from 479 to 1,230 bp from the plasmids containing mutations were subcloned into pLGPS. The inserted fragments were analyzed by DNA sequencing to verify the presence of the desired mutation and the absence of any undesired mutations (ALF automated sequencer; Pharmacia).
Cells, transfections, and infections.
D17 dog osteosarcoma cells were transfected, infected with MLV, and selected for resistance to ouabain or G418 (a neomycin analog) as previously described (33, 34). The D17 cells were selected for resistance to puromycin (final concentration, 3.2 × 10−6 M). The D17-derived ANGIE P cells were maintained, transfected, and selected for drug resistance in a similar manner.
Protocol for determination of in vivo fidelity.
The ANGIE P cells were plated at a density of 2 × 105 cells per 60-mm-diameter dish and 24 h later cotransfected with wild-type or mutated pLGPS and pSVα3.6. The transfected cells were selected for resistance to ouabain, and the resistant colonies were pooled and expanded. The ouabain-resistant cells were plated at a density of 5 × 106 cells per 100-mm-diameter dish; 48 h later, the culture medium containing GA-1 virus was harvested and used to infect D17 target cells. The infected D17 cells that were resistant to G418 were stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), and the frequency of lacZ inactivation was determined by quantification of lacZ-expressing blue colonies and nonexpressing white colonies.
Virus preparation and RT assay.
To isolate and concentrate virus, helper cells containing different constructs were plated at 5 × 106 cells per 100-mm-diameter dish. Viruses were collected 2 days later and centrifuged at 25,000 rpm for 90 min in an SW41 rotor (Beckman) at 4°C. Viral pellets were resuspended in culture medium or phosphate-buffered saline, and virus was stored at −80°C. Concentrated virus was thawed on ice, and the RT activities were determined by modification of a previously described method (23). Briefly, 20-mer oligo(T) (50 μg/ml; Integrated DNA Technologies), poly(A) (100 μg/ml; Pharmacia), and 10 μCi of [3H]dTTP (specific activity, 72 Ci/mmol; ICN) were used in the reactions. The reaction mixtures were precipitated with 10% trichloroacetic acid (Sigma) and filtered through 0.45-μm-pore-size Metricel membranes (Gelman Sciences, Inc.), and the incorporated [3H]dTTP was determined with a scintillation counter.
Western blot analysis.
Western blot analyses to quantitate the amount of protein for the RT assay were performed by standard procedures (58). Briefly, viral proteins were resolved on a sodium dodecyl sulfate–15% polyacrylamide gel, transferred to membranes (Gelman Sciences), and incubated with a 1:10 dilution of a rat anti-MLV capsid monoclonal antibody (13). The membranes were further incubated with anti-rat immunoglobulin G antibody conjugated with horseradish peroxidase (Southern Biotechnology Associates) at a 1:10,000 dilution. The MLV capsid was detected with an ECL (enhanced chemiluminescence) kit (Amersham Pharmacia Biotech) and quantitative autoradiography using a densitometric program (Optimas).
PCR analysis and characterization of proviral DNA.
A total of 161 individual G418-resistant clones infected with GA-1 were isolated and grown in duplicate 24-well tissue culture dishes. Mutant colonies were identified by staining one 24-well dish with X-Gal. Eleven mutant cell clones and six wild-type cell clones were further expanded and lysed, and proviral DNAs were amplified by two rounds of PCR (28, 58).
Three pairs of forward and reverse primers that annealed to six different regions in GA-1 were used to amplify three overlapping fragments of lacZ. DNAs obtained from the first round of PCR were further amplified by a second round of PCR using nested primer sets (58). PCR products (1,216 to 1,326 bp) were further analyzed for smaller deletions or insertions by digestions with various restriction enzymes (BstUI, HaeIII, HhaI, DpnII, HpaII, and NlaIII) and electrophoresis using 7% polyacrylamide gels or 3% MetaPhor gels (MetaPhor agarose; FMC).
Southern analysis of proviral DNA.
Genomic DNA was isolated from the ANGIE P encapsidating cell line, and the proviral structure was analyzed by Southern blot hybridization using standard procedures (16, 58). A 1.2-kb fragment of WH390 containing an internal ribosome entry site (IRES) for expression of the neomycin phosphotransferase gene (neo) was used to generate a probe (specific activity, >109 cpm/μg) as previously described (16). Quantitation of bands was accomplished with the ImageQuant program (Molecular Dynamics).
RESULTS
Construction of ANGIE P encapsidating cell line.
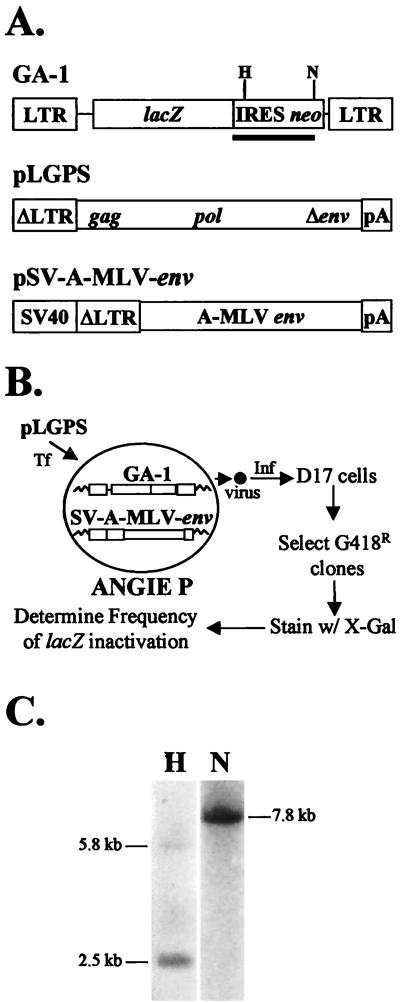
The MLV-based retrovirus vector pGA-1 and the MLV envelope expression construct pSV-A-MLV-env (Fig. 1A) were introduced into D17 cells to generate the ANGIE P encapsidating cell line (Fig. 1B). The vector GA-1 possesses all cis-acting elements needed for viral replication, which include the LTRs, MLV encapsidation signal (Ψ), primer binding site, and polypurine tract. In addition, the vector expresses the Escherichia coli β-galactosidase gene (lacZ) and neo from a single RNA transcript initiating in the 5′ LTR. An IRES of encephalomyocarditis virus is used to express neo (31). The construct pSV-A-MLV-env expresses the amphotropic MLV envelope.
FIG. 1.
Structures of MLV-based constructs and rapid in vivo assay to identify structural determinants in MLV RT important for fidelity. (A) Structures of MLV-based vector pGA-1, pLGPS, and pSV-A-MLV-env. The pGA-1 vector contains LTRs and all cis-acting elements of MLV. pGA-1 transcribes E. coli lacZ and neo from the LTR promoter. The IRES of encephalomyocarditis virus is used to express neo. The construct pLGPS expresses MLV gag and pol from a truncated viral LTR. Finally, pSV-A-MLV-env expresses the amphotropic MLV envelope from a truncated MLV LTR and the simian virus 40 (SV 40) promoter enhancer. (B) Experimental protocol. ANGIE P, a D17-based cell line expressing pGA-1 and amphotropic MLV envelope, was constructed. The wild-type and mutated forms of pLGPS were separately cotransfected (Tf) with pSVα3.6 into the ANGIE P cell line, and the virus produced was used to infect (Inf) D17 cells. The infected cell clones resistant to G418 were stained with X-Gal, and the numbers of blue (wild-type) and white (mutant) colonies were determined. (C) Southern analysis of the ANGIE P encapsidating cell line. Genomic DNA isolated from ANGIE P cells was digested with either HindIII (lane H) or NcoI (lane N). The blots were probed with IRES-neo (black bar in panel A) and quantitated by PhosphorImager analysis.
To construct the ANGIE P encapsidating cell line, pGA-1 was first transfected into the MLV-based packaging cell line PG13, and G418-resistant cells were selected. Virus harvested from the pool of transfected PG13 cells was used to infect D17 cells. After selection for G418 resistance, individual D17 cell clones were isolated and expanded. To ensure that the provirus in the D17 cell clones expressed a functional lacZ, six cell clones were stained with X-Gal and clones that stained blue were selected for further manipulation (data not shown). To verify that the GA-1 provirus was capable of completing one cycle of retrovirus replication, the D17 cell clones were transfected with pAMS, a plasmid that encodes a replication-competent MLV. After transfection, the replication-competent virus was allowed to spread through the culture for 5 to 7 days. Virus was harvested and used to infect fresh D17 cells. The presence of cells that stained blue with X-Gal indicated that the GA-1 provirus was mobilized from the D17 cell clones to the target cells in the presence of the MLV helper virus (data not shown).
Next, the pSV-A-MLV env was cotransfected with pBSpac (a plasmid that confers resistance to puromycin) into a D17 cell clone containing a GA-1 provirus. The transfected cells were selected for resistance to puromycin, and 12 cell clones were isolated and expanded. To verify that the cell clones expressed a functional amphotropic MLV envelope, the cell clones were cotransfected with pLGPS, which expresses the MLV Gag and Pol proteins (Fig. 1A) and pSVα3.6 (a plasmid that confers resistance to ouabain). Ouabain-resistant cells were selected, and virus produced from them was used to infect fresh D17 cells. The infected D17 cells were selected for resistance to G418 and stained with X-Gal (11). The presence of cells that stained blue with X-Gal indicated that the D17 cell clones expressed the amphotropic MLV envelope and mobilized the GA-1 provirus to target cells in the presence of MLV Gag-Pol proteins. All 12 of the D17 cell clones tested for expression of the amphotropic MLV envelope generated virus titers of at least 104 CFU/ml after transfection with pLGPS (data not shown). One of the 12 clones was chosen and named the ANGIE P encapsidating cell line (Fig. 1B).
It was also important to show that the ANGIE P encapsidating cell line did not express a replication-competent MLV, the presence of which could potentially result in multiple rounds of GA-1 replication. Culture supernatant from the ANGIE P encapsidating cell line was used to infect fresh D17 cells, which were selected for resistance to G418. No G418-resistant colonies were observed, indicating that the ANGIE P encapsidating cell line did not harbor a replication-competent virus (data not shown). Finally, the ANGIE P cells were plated at a low density (<100 cells/60-mm-diameter dish) and stained with X-Gal. The frequency of white cells was approximately 1 per 1,500 cells, indicating that the vast majority of the virus was produced from cells containing a functional lacZ (data not shown).
To ensure that the ANGIE P encapsidating cell line contained only one GA-1 provirus, genomic DNA was extracted from the ANGIE P cells and analyzed by Southern blot hybridization (Fig. 1C). The DNA was digested with either HindIII or NcoI and hybridized to a probe containing IRES and neo sequences (Fig. 1A). The HindIII enzyme cuts once in the GA-1 provirus, and two fragments of various lengths, representing 5′ and 3′ portions of the provirus and flanking regions, should be generated from each provirus. The presence of only two detectable fragments after HindIII digestion indicated that only one provirus was present in the ANGIE P cells. Similarly, the NcoI enzyme cuts once in the GA-1 provirus, and one fragment representing the 5′ portion of the provirus and flanking region should be generated from each provirus. The presence of one fragment after NcoI digestion verified that only one provirus was present in the ANGIE P cells (Fig. 1C). Finally, the sequence of the lacZ in ANGIE P cells was determined by PCR amplification and DNA sequencing of both strands to ensure that no mutations were present (data not shown).
Experimental protocol.
The MLV RT expression construct pLGPS was subjected to site-directed mutagenesis, and the ANGIE P cells were used to identify protein determinants important to the accuracy of DNA synthesis during reverse transcription (Fig. 1A and B). First, pLGPS-derived constructs that contained mutations in the MLV RT were separately introduced into the ANGIE P cells by cotransfection with pSVα3.6. Ouabain-resistant colonies were pooled and expanded. Virus was harvested from the pools of transfected cells; serially diluted viruses were used to infect D17 target cells. The infected D17 cells were selected for resistance to G418, and the resulting colonies were stained with X-Gal. The frequency of lacZ inactivation was determined by dividing the number of white colonies by the total number of colonies (blue plus white colonies). In general, the multiplicity of infection was <0.0005, and the probability of double infection was very low (<1/2,000 colonies). The frequency of lacZ inactivation provided a measure of the inactivating mutations introduced into the lacZ gene during one cycle of retroviral replication, which constituted the transfer of genetic information from the GA-1 provirus in the ANGIE P cells to the GA-1 provirus in the D17 target cells. Using this assay, we determined the effect of introducing mutations in the MLV RT on the fidelity of DNA synthesis. In addition, the virus titers obtained after transfections of wild-type or mutated pLGPS constructs were used to determine the effects of mutations on the efficiency of virus replication.
Effects of mutations at the V223 position of the MLV YVDD motif on fidelity.
The V223 of MLV RT was mutated to an alanine (V223A), isoleucine (V223I), methionine (V223M), or serine (V223S), and the effects of these mutations on the frequency of lacZ inactivation were determined. The frequencies of lacZ inactivation with each mutant RT were compared to the frequency of inactivation obtained with wild-type RT in parallel experiments (Table 1).
TABLE 1.
Effects of V223 mutations in the YVDD motif of MLV RT on the frequency of lacZ inactivation
| MLV-RT | No. of expts | No. of Mutant colonies/total coloniesa | Frequency of lacZ inactivation (mean % ± SE)b | Relative change in inactivation of lacZc | Relative viral titerd |
|---|---|---|---|---|---|
| Wild type | 10 | 154/2,860 | 5.2 ± 0.48 | 1.0 | 1.0 |
| Mutant | |||||
| V223A | 6 | 207/2,289 | 8.8 ± 0.45 | 1.7 | 0.03 |
| V223I | 2 | 95/1,764 | 5.3 ± 0.30 | 1.0 | 0.50 |
| V223M | 4 | 182/1,974 | 9.2 ± 0.36 | 1.8 | 0.12 |
| V223S | 4 | 200/1,632 | 12.1 ± 0.43 | 2.3 | 0.01 |
Number of mutant colonies that displayed a white colony phenotype and total number of colonies that were observed in 2 to 10 independent experiments.
Calculated as follows: (number of mutant colonies in each experiment/total number of colonies) × 100. The standard errors were determined by using the two-sample t test.
Calculated as follows: frequency of lacZ inactivation observed with mutant MLV RT/frequency of lacZ inactivation observed with wild-type MLV RT. Statistical analysis using a two-sample t test showed that the V223M, V223S, and V223A mutants of MLV RT displayed mutant frequencies significantly higher than that for wild-type MLV RT (P < 0.001). The mutant frequency obtained with the V223I mutant of MLV RT was not significantly different from that for wild-type MLV RT (P = 0.71).
Virus titer for each experimental group was determined by serial dilutions and infections. The average virus titer obtained with wild-type MLV RT was 4.4 × 104 CFU/ml. The relative virus titer represents a ratio of the virus titer of mutant MLV RT obtained in each experiment divided by the virus titer obtained with wild-type MLV RT in parallel experiments.
The wild-type pLGPS construct inactivated lacZ at an average frequency of 5.2% in one cycle of retrovirus replication. The frequencies of lacZ inactivation obtained in 10 independent experiments were highly reproducible (standard error, ±0.48%). The V223A, V223M, and V223S mutants exhibited 1.7-, 1.8-, and 2.3-fold-higher frequencies of lacZ inactivation, respectively, compared to the wild type. The observed frequencies of lacZ inactivation with the mutants were shown to be statistically different from the wild-type frequency (P < 0.001). Conversely, the V223I mutant showed no change in the frequency of lacZ inactivation (5.3% ± 0.30%). All mutants displayed decreases in viral titers ranging from 2- to 100-fold relative to the wild type (4.4 × 104 CFU/ml) (Table 1 and Fig. 2C). In addition, there was no correlation between reduction in viral titer and fidelity.
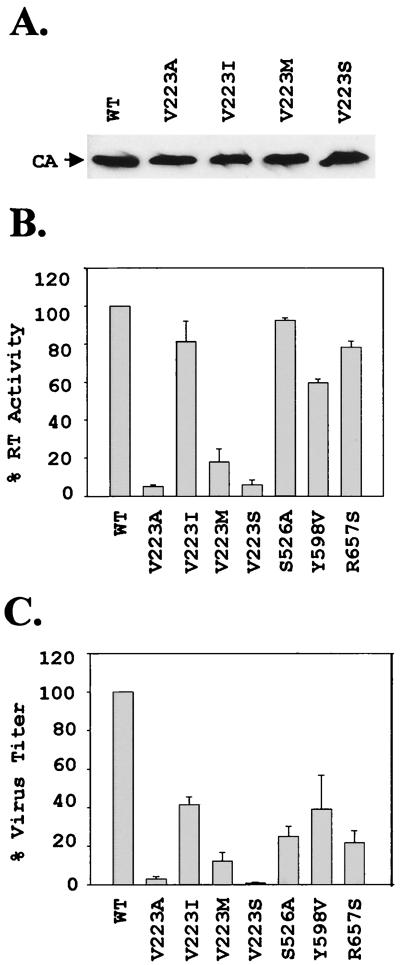
FIG. 2.
Comparison of RT activities and viral titers. (A) Representative Western blot analysis of the wild-type (WT) and V223 mutants used to normalize the amount of viral proteins used in the RT assays. CA, capsid protein. (B) RT activity for virion-associated RT. The activities shown are relative to the RT activity determined for the wild-type MLV RT (set to 100%). The results represent an average of two to three experiments; the error bars represent the standard error of the mean. (C) Summary of virus titers obtained relative to the wild-type viral titer (set to 100%). The results represent an average of 2 to 10 experiments, and the error bars represent the standard error of the mean.
Mutational analysis of the MLV RT RNase H domain.
Mutations S526A, Y598V, and R657S were introduced into the MLV RNase H domain, and the effects of these mutations on the frequency of lacZ inactivation were determined (Table 2). These mutations were selected because it was previously shown that they did not affect the polymerase activity and permitted viral replication to occur (7, 8). Again, the wild-type RT displayed a 5.2% ± 0.30% frequency of lacZ inactivation. Mutants R657S and S526A exhibited frequencies of lacZ inactivation that were increased 1.2- and 1.4-fold, respectively (P < 0.005). In contrast, the Y598V mutant did not exhibit a change in the frequency of lacZ inactivation (5.4% ± 0.39%; P = 0.81). Finally, all three RNase H domain mutants exhibited a 2.5- or 5-fold decrease in the viral titer relative to the wild type (1.2 × 105 CFU/ml) (Table 2 and Fig. 2C). Once again, there was no correlation between reduction in viral titer and fidelity.
TABLE 2.
Effects of mutations in the RNase H domain of MLV RT on the frequency of lacZ inactivation
| MLV RT | No. of expts | No. of mutant colonies/total coloniesa | Frequency of lacZ inactivation (mean % ± SE)b | Relative change in inactivation of lacZc | Relative viral titerd |
|---|---|---|---|---|---|
| Wild type | 9 | 473/8,023 | 5.2 ± 0.30 | 1.0 | 1.0 |
| Mutant | |||||
| S526A | 5 | 429/5,039 | 8.4 ± 0.33 | 1.6 | 0.25 |
| Y598V | 4 | 181/3,534 | 5.4 ± 0.39 | 1.0 | 0.40 |
| R657S | 5 | 344/4,754 | 7.1 ± 0.14 | 1.4 | 0.21 |
Number of mutant colonies that displayed a white colony phenotype and total number of colonies that were observed in four to nine independent experiments.
Calculated as for Table 1.
Calculated as for Table 1. Statistical analysis using a two-sample t test showed that the S526A and R657S mutants of MLV RT displayed mutant frequencies significantly higher than that for wild-type MLV RT (P < 0.005). The mutant frequency obtained with the Y598V mutant of MLV RT was not significantly different from that for the wild-type MLV RT (P = 0.81).
Virus titer for each experimental group was determined by serial dilutions and infections. The average virus titer obtained with wild-type MLV RT was 1.2 × 105 CFU/ml. The relative virus titer represents a ratio of the virus titer of mutant MLV RT obtained in each experiment divided by the virus titer obtained with wild-type MLV RT in parallel experiments.
RT activities of mutant RTs.
Viruses produced from the wild-type and mutant RTs were harvested by ultracentrifugation and characterized by Western blotting analysis using an anti-MLV capsid antibody (13). The Western blots were quantified to estimate the amounts of MLV capsid protein present in the viral preparations. A representative blot is shown in Fig. 2A. The estimated amounts of capsid protein were used to ensure that equivalent amounts of viral proteins were used for determination of RT activities (Fig. 2B). The RT activities ranged from 5% ± 1% for the V223A mutant to 93% ± 1% for the S526A mutant relative to the wild-type enzyme. As expected, all of the RNase H domain mutants exhibited RT activities that were comparable to the wild-type RT activity. With the exception of the V223I mutant, all reductions in RT activities were significantly different from the wild-type level (P < 0.005).
The RT activities were compared to the relative reductions in viral titers observed (Fig. 2C). In general, the reductions in RT activities were proportional to the reductions in viral titers observed for the V223 mutants. As expected, the RNase H mutants exhibited higher RT activities than most of the V223 mutants at the polymerase active site. In general, the RNase H mutants displayed a greater reduction in viral titer relative to RT activities, suggesting that other steps in the reverse transcription process were affected by mutations introduced in the RNase H domain.
Analysis of lacZ-inactivating mutations.
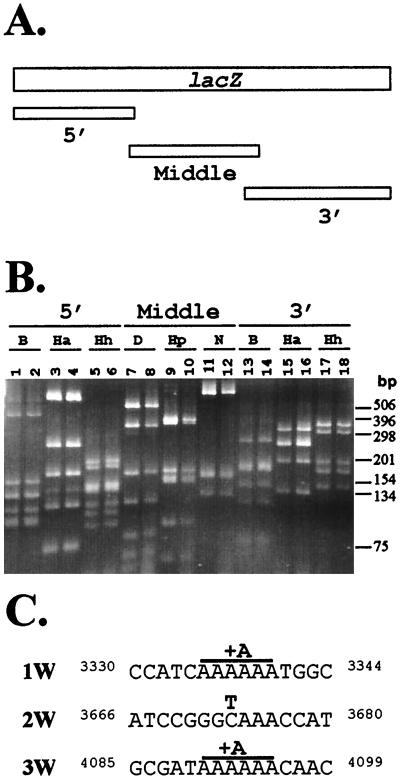
To determine the nature of mutations introduced into the lacZ gene by the wild-type or mutant RTs, 161 infected G418-resistant cell clones were isolated from infections performed with the wild-type RT as well as the V223A, V223M, and Y598V mutants. X-Gal staining of the clones indicated that 11 of 161 cell clones exhibited a white colony phenotype. One mutant clone was derived from infection with the wild-type RT, three were derived from infection with the V223A mutant, three were derived from infection with the Y598V mutant, and four were derived from infection with the V223M mutant. The lacZ sequences from these 11 white colonies was amplified by two rounds of PCR using six different sets of primers. As a result, three overlapping fragments designated the 5′, middle, and 3′ portions were generated (Fig. 3A). The PCR products were approximately 1.2 to 1.3 kb in length. Each of the PCR fragments derived from the 11 cell clones was analyzed by gel electrophoresis and compared to the fragments obtained from a cell clone containing a functional lacZ. This analysis indicated that none of the lacZ genes present in the 11 cell clones were inactivated by large deletions or insertions (>100 bp; data not shown). The amplified lacZ fragments were digested with various restriction enzymes to generate smaller fragments in an effort to identify shorter deletions or insertions (Fig. 3B). The 5′ and 3′ fragments of the lacZ from infected cell clones were digested with BstUI, HaeIII, and HhaI; the middle portion was digested with DpnII, HpaII, and NlaIII. DNA fragments that ranged in size from 75 to 863 bp were analyzed by gel electrophoresis. No differences were observed in any of the restriction digestions, indicating that the lacZ-inactivating mutations did not involve deletions or insertions that were larger than 20 bp (based on the estimated resolution of the restriction analysis).
FIG. 3.
PCR analysis of lacZ mutations in proviral DNAs of infected cell clones. (A) Structure of lacZ and approximate locations of the overlapping PCR-amplified products. The approximate locations of 5′, middle, and 3′ fragments generated during PCR amplification of lacZ are shown. (B) Representative restriction digestion analysis of PCR products obtained from infected cell clone 6W. The 5′ and 3′ fragments were digested with BstUI (B), HaeIII (Ha), and HhaI (Hh); the middle fragment was digested with DpnII (D), HpaII (Hp), and NlaIII (N). Odd-numbered lanes represent PCR products from a cell clone containing wild-type lacZ; even-numbered lanes represent PCR products derived from cell clone 6W containing an inactivated lacZ. (C) lacZ-inactivating mutations in infected cell clones 1W (nucleotides [nt] 3330 to 3343), 2W (nt 3666 to 3680), and 3W (nt 4085 to 4099). Inactivating mutations are shown in bold type. A +1 frameshift mutation in a run of six A nucleotides (+A) and a substitution mutation (C to T; labeled T) are shown.
Since no small deletions or insertions could be observed, both strands of the PCR products derived from three cell clones were sequenced to ensure the accuracy of the sequencing analysis. These three cell clones that displayed a white colony phenotype were derived from an infection with the V223A mutant of MLV RT. The sequencing analysis indicated that two mutants contained a +1 frameshift in runs of six adenosine nucleotides present at different locations in the lacZ gene. One mutant contained a C-to-T transition, which resulted in the generation of a termination codon. These mutations were not the result of errors occurring during the PCRs, since they were identified in independent PCRs (data not shown). These results verified that the white colony phenotype was indeed generated by introduction of mutations in lacZ. In addition, different types of mutations could be identified in this assay (Fig. 3C).
DISCUSSION
This study provides a rapid, accurate, and highly reproducible in vivo assay for identification of MLV RT structural determinants important for fidelity. The in vivo assay described here is rapid compared to the previously described in vivo assays involving shuttle vectors because the mutant frequencies can be determined immediately after the infected cell clones are obtained (42, 43, 53, 54). This assay is also more sensitive and accurate than the previously described in vivo assay utilizing the herpesvirus thymidine kinase reporter gene because it is not necessary to compare viral titers after two different drug selection procedures (52). Due to the large size of the lacZ reporter gene, a high mutant frequency is obtained and a large number of mutants can be identified. The high mutant frequency increases the utility of the assay because the accuracy of RT mutants with low viral titers can be easily determined. In addition, mutations that increase the accuracy of DNA synthesis can also be identified. One limitation of this in vivo assay is that RT mutants with extremely low fidelities may generate highly mutated viruses that cannot confer resistance to G418 and therefore cannot be analyzed.
The results of these studies indicate that mutations at the V223 position of the MLV RT have a statistically significant effect on the in vivo fidelity of RT. These results are consistent with the in vitro forward mutation assays performed with the YVDD mutant of HIV-1 RT (19). For both MLV and HIV-1 RTs, the YVDD motif appears to be approximately twofold more accurate than the YMDD motif. It is possible that even though the overall mutation rate is altered only twofold, the rate of a specific type of mutation is altered to a greater extent. For example, a 10-fold increase in the rate of 1 of the 12 possible substitutions may have a less than twofold effect on the overall mutation rate.
The effect of a twofold alteration in the retrovirus mutation rate on variation in viral populations is undetermined. A model was previously proposed to ascertain the relative impact of mutation rates and selective forces on viral populations (14). Based on the model, it was hypothesized that in a large viral population with a high replicative capacity, small changes in the selective forces will have a greater impact on the viral variants present than large changes in mutation rates. Whether small changes in the mutation rate affect viral evolution will be dependent on the size of the viral population and its growth potential. In this regard, it is significant that the rate of development of drug-resistant variants in cell culture was the same for the YMDD and YVDD variants of HIV-1 (32, 49). These observations, along with our observation that the YVDD variant is only twofold more accurate than the YMDD variant, suggests that twofold alterations in RT fidelity are unlikely to have an effect on the rate of viral evolution.
It is of interest to consider the structural features of the YMDD and YVDD variants and how these features may be related to the statistically significant alterations in fidelity. It has been postulated that the X residue of the YXDD motif may contact the primer and is important for affinity to the template-primer as well as its positioning relative to the catalytic site (10, 17, 59). Repositioning of the template-primer by the YMDD variant may alter flexibility of the dNTP binding pocket, ultimately leading to changes in fidelity (27). In addition, molecular modeling studies have predicted an increased fidelity of YVDD and YIDD variants due to new contacts with the incoming dNTP substrate that are absent from the YMDD variants (29, 51). Therefore, the hydrophobicity and the size of the amino acid side chain at the V223 position may be important for the catalytic function and fidelity of RT. It is interesting that two of the three mutations generated with the V223A mutant RT were +1 frameshifts. Frameshifts account for approximately 10% of all mutations (42, 54), and this limited analysis suggests that the V223A mutation may affect the template-primer affinity and frameshift fidelity, which is consistent with previous observations (10).
The observation that the V223A mutant exhibits a severe reduction in the polymerase activity whereas the V223I mutant has little effect on the polymerase activity is consistent with previously published results obtained with HIV-1 RT (10). Also, the viral titers were consistently reduced to a greater extent than the observed decreases in polymerase activities, suggesting that additional steps during reverse transcription may also be affected by the mutations.
The RNase H mutants S526A and R657S exhibited small but statistically significant alterations in fidelity. As expected, the mutations had little effect on the polymerase activity (8). The changes in fidelity of these mutants may be due to altered processivity of RT, interactions of the RT with the template-primer complex, or the overall structure of the RT (24, 61). The reductions in viral titers associated with these mutants may be due to a reduced efficiency of strand transfer events during reverse transcription (21, 41).
The assay described here may be used to determine the roles of various RT domains, including the dNTP binding site and the thumb domain, in the fidelity of reverse transcription. The importance of other viral proteins to the fidelity of reverse transcription may also be easily determined. Finally, a similar strategy may be used to analyze the effects of mutations in RT on the frequencies of specific types of mutations.
ACKNOWLEDGMENTS
We thank Jeffrey Anderson, Benjamin Beasley, Sara Cheslock, Que Dang, Krista Delviks, Wei-Shau Hu, Carey Hwang, Timor Kubdalov, Yegor Voronin, and Wei-hui Zhang for critical reading of the manuscript and discussion of results. We especially thank Wei-Shau Hu for valuable intellectual input and discussions throughout this project.
This work was supported by the Public Health Service grant CA58875 from the National Institutes of Health.
REFERENCES
- 1.Avidan O, Hizi A. The processivity of DNA synthesis exhibited by drug-resistant variants of human immunodeficiency virus type-1 reverse transcriptase. Nucleic Acids Res. 1998;26:1713–1717. doi: 10.1093/nar/26.7.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakhanashvili M, Avidan O, Hizi A. Mutational studies of human immunodeficiency virus type 1 reverse transcriptase: the involvement of residues 183 and 184 in the fidelity of DNA synthesis. FEBS Lett. 1996;391:257–262. doi: 10.1016/0014-5793(96)00747-8. [DOI] [PubMed] [Google Scholar]
- 3.Beard W A, Bebenek K, Darden T A, Li L, Prasad R, Kunkel T A, Wilson S H. Vertical-scanning mutagenesis of a critical tryptophan in the minor groove binding track of HIV-1 reverse transcriptase. Molecular nature of polymerase-nucleic acid interactions. J Biol Chem. 1998;273:30435–30442. doi: 10.1074/jbc.273.46.30435. [DOI] [PubMed] [Google Scholar]
- 4.Bebenek K, Abbotts J, Roberts J D, Wilson S H, Kunkel T A. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J Biol Chem. 1989;264:16948–16956. [PubMed] [Google Scholar]
- 5.Bebenek K, Abbotts J, Wilson S H, Kunkel T A. Error-prone polymerization by HIV-1 reverse transcriptase. Contribution of template-primer misalignment, miscoding, and termination probability to mutational hot spots. J Biol Chem. 1993;268:10324–10334. [PubMed] [Google Scholar]
- 6.Bebenek K, Beard W A, Casas-Finet J R, Kim H R, Darden T A, Wilson S H, Kunkel T A. Reduced frameshift fidelity and processivity of HIV-1 reverse transcriptase mutants containing alanine substitutions in helix H of the thumb subdomain. J Biol Chem. 1995;270:19516–19523. doi: 10.1074/jbc.270.33.19516. [DOI] [PubMed] [Google Scholar]
- 7.Blain S W, Goff S P. Effects on DNA synthesis and translocation caused by mutations in the RNase H domain of Moloney murine leukemia virus reverse transcriptase. J Virol. 1995;69:4440–4452. doi: 10.1128/jvi.69.7.4440-4452.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blain S W, Goff S P. Nuclease activities of Moloney murine leukemia virus reverse transcriptase. Mutants with altered substrate specificities. J Biol Chem. 1993;268:23585–23592. [PubMed] [Google Scholar]
- 9.Boucher C A, Cammack N, Schipper P, Schuurman R, Rouse P, Wainberg M A, Cameron J M. High-level resistance to (−) enantiomeric 2′-deoxy-3′-thiacytidine in vitro is due to one amino acid substitution in the catalytic site of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 1993;37:2231–2234. doi: 10.1128/aac.37.10.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyer P L, Hughes S H. Analysis of mutations at position 184 in reverse transcriptase of human immunodeficiency virus type 1. Antimicrob Agents Chemother. 1995;39:1624–1628. doi: 10.1128/aac.39.7.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cepko C. XGAL staining of cultured cells. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley Interscience; 1992. pp. 9.11.9–9.11.12. [Google Scholar]
- 12.Chao S F, Chan V L, Juranka P, Kaplan A H, Swanstrom R, Hutchison C A., III Mutational sensitivity patterns define critical residues in the palm subdomain of the reverse transcriptase of human immunodeficiency virus type 1. Nucleic Acids Res. 1995;23:803–810. doi: 10.1093/nar/23.5.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chesebro B, Britt W, Evans L, Wehrly K, Nishio J, Cloyd M. Characterization of monoclonal antibodies reactive with murine leukemia viruses: use in analysis of strains of friend MCF and Friend ecotropic murine leukemia virus. Virology. 1983;127:134–148. doi: 10.1016/0042-6822(83)90378-1. [DOI] [PubMed] [Google Scholar]
- 14.Coffin J M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 15.Cohen J. The daunting challenge of keeping HIV suppressed. Science. 1997;277:32–33. doi: 10.1126/science.277.5322.32. [DOI] [PubMed] [Google Scholar]
- 16.Delviks K A, Hu W S, Pathak V K. Ψ− vectors: murine leukemia virus-based self-inactivating and self-activating retroviral vectors. J Virol. 1997;71:6218–6224. doi: 10.1128/jvi.71.8.6218-6224.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding J, Hughes S H, Arnold E. Protein-nucleic acid interactions and DNA conformation in a complex of human immunodeficiency virus type 1 reverse transcriptase with a double-stranded DNA template-primer. Biopolymers. 1997;44:125–138. doi: 10.1002/(SICI)1097-0282(1997)44:2<125::AID-BIP2>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 18.Dong Q, Copeland W C, Wang T S. Mutational studies of human DNA polymerase alpha. Identification of residues critical for deoxynucleotide binding and misinsertion fidelity of DNA synthesis. J Biol Chem. 1993;268:24163–24174. [PubMed] [Google Scholar]
- 19.Drosopoulos W C, Prasad V R. Increased misincorporation fidelity observed for nucleoside analog resistance mutations M184V and E89G in human immunodeficiency virus type 1 reverse transcriptase does not correlate with the overall error rate measured in vitro. J Virol. 1998;72:4224–4230. doi: 10.1128/jvi.72.5.4224-4230.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao Q, Gu Z, Parniak M A, Cameron J, Cammack N, Boucher C, Wainberg M A. The same mutation that encodes low-level human immunodeficiency virus type 1 resistance to 2′,3′-dideoxyinosine and 2′,3′-dideoxycytidine confers high-level resistance to the (−) enantiomer of 2′,3′-dideoxy-3′-thiacytidine. Antimicrob Agents Chemother. 1993;37:1390–1392. doi: 10.1128/aac.37.6.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garces J, Wittek R. Reverse-transcriptase-associated RNaseH activity mediates template switching during reverse transcription in vitro. Proc R Soc Lond Sev B. 1991;243:235–239. doi: 10.1098/rspb.1991.0037. [DOI] [PubMed] [Google Scholar]
- 22.Georgiadis M M, Jessen S M, Ogata C M, Telesnitsky A, Goff S P, Hendrickson W A. Mechanistic implications from the structure of a catalytic fragment of Moloney murine leukemia virus reverse transcriptase. Structure. 1995;3:879–892. doi: 10.1016/S0969-2126(01)00223-4. [DOI] [PubMed] [Google Scholar]
- 23.Grandgenett D P, Gerard G F, Green M. A single subunit from avian myeloblastosis virus with both RNA-directed DNA polymerase and ribonuclease H activity. Proc Natl Acad Sci USA. 1973;70:230–234. doi: 10.1073/pnas.70.1.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo J, Wu W, Yuan Z Y, Post K, Crouch R J, Levin J G. Defects in primer-template binding, processive DNA synthesis, and RNase H activity associated with chimeric reverse transcriptases having the murine leukemia virus polymerase domain joined to Escherichia coli RNase H. Biochemistry. 1995;34:5018–5029. doi: 10.1021/bi00015a013. [DOI] [PubMed] [Google Scholar]
- 25.Hamburgh M E, Drosopoulos W C, Prasad V R. The influence of 3TC-resistance mutations E89G and M184V in the human immunodeficiency virus reverse transcriptase on mispair extension efficiency. Nucleic Acids Res. 1998;26:4389–4394. doi: 10.1093/nar/26.19.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris D, Kaushik N, Pandey P K, Yadav P N, Pandey V N. Functional analysis of amino acid residues constituting the dNTP binding pocket of HIV-1 reverse transcriptase. J Biol Chem. 1998;273:33624–3334. doi: 10.1074/jbc.273.50.33624. [DOI] [PubMed] [Google Scholar]
- 27.Harris D, Yadav P N, Pandey V N. Loss of polymerase activity due to Tyr to Phe substitution in the YMDD motif of human immunodeficiency virus type-1 reverse transcriptase is compensated by Met to Val substitution within the same motif. Biochemistry. 1998;37:9630–9640. doi: 10.1021/bi980549z. [DOI] [PubMed] [Google Scholar]
- 28.Higuchi R. Simple and rapid preparation of samples for PCR. New York, N.Y: Stockton Press; 1989. [Google Scholar]
- 29.Huang H, Chopra R, Verdine G L, Harrison S C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 30.Jacobo-Molina A, Ding J, Nanni R G, Clark A D, Jr, Lu X, Tantillo C, Williams R L, Kamer G, Ferris A L, Clark P, et al. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jang S K, Krausslich H G, Nicklin M J, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jonckheere H, Witvrouw M, De Clercq E, Anne J. Lamivudine resistance of HIV type 1 does not delay development of resistance to nonnucleoside HIV type 1-specific reverse transcriptase inhibitors as compared with wild-type HIV type 1. AIDS Res Hum Retroviruses. 1998;14:249–253. doi: 10.1089/aid.1998.14.249. [DOI] [PubMed] [Google Scholar]
- 33.Julias J G, Hash D, Pathak V K. E− vectors: development of novel self-inactivating and self-activating retroviral vectors for safer gene therapy. J Virol. 1995;69:6839–6846. doi: 10.1128/jvi.69.11.6839-6846.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Julias J G, Kim T, Arnold G, Pathak V K. The antiretrovirus drug 3′-azido-3′-deoxythymidine increases the retrovirus mutation rate. J Virol. 1997;71:4254–4263. doi: 10.1128/jvi.71.6.4254-4263.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaushik N, Singh K, Alluru I, Modak M J. Tyrosine 222, a member of the YXDD motif of MuLV RT, is catalytically essential and is a major component of the fidelity center. Biochemistry. 1999;38:2617–2627. doi: 10.1021/bi9824285. [DOI] [PubMed] [Google Scholar]
- 36.Kent R B, Emanuel J R, Ben Neriah Y, Levenson R, Housman D E. Ouabain resistance conferred by expression of the cDNA for a murine Na+, K+-ATPase alpha subunit. Science. 1987;237:901–903. doi: 10.1126/science.3039660. [DOI] [PubMed] [Google Scholar]
- 37.Kim B, Hathaway T R, Loeb L A. Fidelity of mutant HIV-1 reverse transcriptases: interaction with the single-stranded template influences the accuracy of DNA synthesis. Biochemistry. 1998;37:5831–5839. doi: 10.1021/bi972672g. [DOI] [PubMed] [Google Scholar]
- 38.Kohlstaedt L A, Wang J, Friedman J M, Rice P A, Steitz T A. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 39.Kunkel T A, Hamatake R K, Motto-Fox J, Fitzgerald M P, Sugino A. Fidelity of DNA polymerase I and the DNA polymerase I-DNA primase complex from Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:4447–4458. doi: 10.1128/mcb.9.10.4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Landau N R, Page K A, Littman D R. Pseudotyping with human T-cell leukemia virus type I broadens the human immunodeficiency virus host range. J Virol. 1991;65:162–169. doi: 10.1128/jvi.65.1.162-169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo G X, Taylor J. Template switching by reverse transcriptase during DNA synthesis. J Virol. 1990;64:4321–4328. doi: 10.1128/jvi.64.9.4321-4328.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mansky L M, Temin H M. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol. 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mansky L M, Temin H M. Lower mutation rate of bovine leukemia virus relative to that of spleen necrosis virus. J Virol. 1994;68:494–499. doi: 10.1128/jvi.68.1.494-499.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez M A, Vartanian J P, Wain-Hobson S. Hypermutagenesis of RNA using human immunodeficiency virus type 1 reverse transcriptase and biased dNTP concentrations. Proc Natl Acad Sci USA. 1994;91:11787–11791. doi: 10.1073/pnas.91.25.11787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin-Hernandez A M, Gutierrez-Rivas M, Domingo E, Menendez-Arias L. Mispair extension fidelity of human immunodeficiency virus type 1 reverse transcriptases with amino acid substitutions affecting Tyr115. Nucleic Acids Res. 1997;25:1383–1389. doi: 10.1093/nar/25.7.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McMichael A J, Phillips R E. Escape of human immunodeficiency virus from immune control. Annu Rev Immunol. 1997;15:271–296. doi: 10.1146/annurev.immunol.15.1.271. [DOI] [PubMed] [Google Scholar]
- 47.Miller A D, Buttimore C. Redesign of retrovirus packaging cell lines to avoid recombination leading to helper virus production. Mol Cell Biol. 1986;6:2895–2902. doi: 10.1128/mcb.6.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller A D, Garcia J V, von Suhr N, Lynch C M, Wilson C, Eiden M V. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J Virol. 1991;65:2220–2224. doi: 10.1128/jvi.65.5.2220-2224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nijhuis M, Schuurman R, de Jong D, van Leeuwen R, Lange J, Danner S, Keulen W, de Groot T, Boucher C A. Lamivudine-resistant human immunodeficiency virus type 1 variants (184V) require multiple amino acid changes to become co-resistant to zidovudine in vivo. J Infect Dis. 1997;176:398–405. doi: 10.1086/514056. [DOI] [PubMed] [Google Scholar]
- 50.Oude Essink B B, Back N K, Berkhout B. Increased polymerase fidelity of the 3TC-resistant variants of HIV-1 reverse transcriptase. Nucleic Acids Res. 1997;25:3212–3217. doi: 10.1093/nar/25.16.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pandey V N, Kaushik N, Rege N, Sarafianos S G, Yadav P N, Modak M J. Role of methionine 184 of human immunodeficiency virus type-1 reverse transcriptase in the polymerase function and fidelity of DNA synthesis. Biochemistry. 1996;35:2168–2179. doi: 10.1021/bi9516642. [DOI] [PubMed] [Google Scholar]
- 52.Parthasarathi S, Varela-Echavarria A, Ron Y, Preston B D, Dougherty J P. Genetic rearrangements occurring during a single cycle of murine leukemia virus vector replication: characterization and implications. J Virol. 1995;69:7991–8000. doi: 10.1128/jvi.69.12.7991-8000.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pathak V K, Temin H M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: deletions and deletions with insertions. Proc Natl Acad Sci USA. 1990;87:6024–6028. doi: 10.1073/pnas.87.16.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pathak V K, Temin H M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proc Natl Acad Sci USA. 1990;87:6019–6023. doi: 10.1073/pnas.87.16.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Preston B D, Poiesz B J, Loeb L A. Fidelity of HIV-1 reverse transcriptase. Science. 1988;242:1168–1171. doi: 10.1126/science.2460924. [DOI] [PubMed] [Google Scholar]
- 56.Rezende L F, Curr K, Ueno T, Mitsuya H, Prasad V R. The impact of multidideoxynucleoside resistance-conferring mutations in human immunodeficiency virus type 1 reverse transcriptase on polymerase fidelity and error specificity. J Virol. 1998;72:2890–2895. doi: 10.1128/jvi.72.4.2890-2895.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rubinek T, Bakhanashvili M, Taube R, Avidan O, Hizi A. The fidelity of 3′ misinsertion and mispair extension during DNA synthesis exhibited by two drug-resistant mutants of the reverse transcriptase of human immunodeficiency virus type 1 with Leu74→Val and Glu89→Gly. Eur J Biochem. 1997;247:238–247. doi: 10.1111/j.1432-1033.1997.00238.x. [DOI] [PubMed] [Google Scholar]
- 58.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 59.Tantillo C, Ding J, Jacobo-Molina A, Nanni R G, Boyer P L, Hughes S H, Pauwels R, Andries K, Janssen P A, Arnold E. Locations of anti-AIDS drug binding sites and resistance mutations in the three-dimensional structure of HIV-1 reverse transcriptase. Implications for mechanisms of drug inhibition and resistance. J Mol Biol. 1994;243:369–387. doi: 10.1006/jmbi.1994.1665. [DOI] [PubMed] [Google Scholar]
- 60.Taube R, Avidan O, Hizi A. The fidelity of misinsertion and mispair extension throughout DNA synthesis exhibited by mutants of the reverse transcriptase of human immunodeficiency virus type 2 resistant to nucleoside analogs. Eur J Biochem. 1997;250:106–114. doi: 10.1111/j.1432-1033.1997.00106.x. [DOI] [PubMed] [Google Scholar]
- 61.Telesnitsky A, Goff S P. RNase H domain mutations affect the interaction between Moloney murine leukemia virus reverse transcriptase and its primer-template. Proc Natl Acad Sci USA. 1993;90:1276–1280. doi: 10.1073/pnas.90.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Temin H M. Retrovirus variation and reverse transcription: abnormal strand transfers result in retrovirus genetic variation. Proc Natl Acad Sci USA. 1993;90:6900–6903. doi: 10.1073/pnas.90.15.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tisdale M, Kemp S D, Parry N R, Larder B A. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc Natl Acad Sci USA. 1993;90:5653–5656. doi: 10.1073/pnas.90.12.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van der Groen G, Nyambi P N, Beirnaert E, Davis D, Fransen K, Heyndrickx L, Ondoa P, Van der Auwera G, Janssens W. Genetic variation of HIV type 1: relevance of interclade variation to vaccine development. AIDS Res Hum Retroviruses. 1998;14(Suppl. 3):S211–S221. [PubMed] [Google Scholar]
- 65.Vara J A, Portela A, Ortin J, Jimenez A. Expression in mammalian cells of a gene from Streptomyces alboniger conferring puromycin resistance. Nucleic Acids Res. 1986;14:4617–4624. doi: 10.1093/nar/14.11.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wainberg M A, Drosopoulos W C, Salomon H, Hsu M, Borkow G, Parniak M, Gu Z, Song Q, Manne J, Islam S, Castriota G, Prasad V R. Enhanced fidelity of 3TC-selected mutant HIV-1 reverse transcriptase. Science. 1996;271:1282–1285. doi: 10.1126/science.271.5253.1282. [DOI] [PubMed] [Google Scholar]
- 67.Wakefield J K, Jablonski S A, Morrow C D. In vitro enzymatic activity of human immunodeficiency virus type 1 reverse transcriptase mutants in the highly conserved YMDD amino acid motif correlates with the infectious potential of the proviral genome. J Virol. 1992;66:6806–6812. doi: 10.1128/jvi.66.11.6806-6812.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]