Abstract
We devised an experimental system to examine sequential events by which the human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein (Env) interacts with CD4 and coreceptor to induce membrane fusion. Recombinant soluble CD4 (sCD4) activated fusion between effector cells expressing Env and target cells expressing coreceptor (CCR5 or CXCR4) but lacking CD4. sCD4-activated fusion was dose dependent, occurred comparably with two- and four-domain proteins, and demonstrated Env-coreceptor specificities parallel to those reported in conventional fusion and infectivity systems. Fusion activation occurred upon sCD4 preincubation and washing of the Env-expressing effector cells but not the coreceptor-bearing target cells, thereby demonstrating that sCD4 exerts its effects by acting on Env. These findings provide direct functional evidence for a sequential two-step model of Env-receptor interactions, whereby gp120 binds first to CD4 and becomes activated for subsequent functional interaction with coreceptor, leading to membrane fusion. We used the sCD4-activated system to explore neutralization by the anti-gp120 human monoclonal antibodies 17b and 48d. These antibodies reportedly bind conserved CD4-induced epitopes involved in coreceptor interactions but neutralize HIV-1 infection only weakly. We found that 17b and 48d had minimal effects in the standard cell fusion system using target cells expressing both CD4 and coreceptor but potently blocked sCD4-activated fusion with target cells expressing coreceptor alone. Both antibodies strongly inhibited sCD4-activated fusion by Envs from genetically diverse HIV-1 isolates. Thus, the sCD4-activated system reveals conserved Env-blocking epitopes that are masked in native Env and hence not readily detected by conventional systems.
Human immunodeficiency virus (HIV) enters cells by direct fusion between the surface membranes of the virion and target cell. The fusion process requires a single HIV component, the envelope glycoprotein (Env), as well as two distinct receptor molecules on the surface of the target cell: CD4 (the primary receptor) plus a specific chemokine receptor (the coreceptor, e.g., CCR5 or CXCR4) (reviewed in references 8, 39, and 60). The binding determinants for both CD4 and coreceptor are contained within gp120, the external Env subunit. A plausible concept is that the fusogenic potential of Env is activated only when it interacts with these target cell molecules, thereby conferring target cell specificity for HIV entry. These notions have led to a model in which gp120 binding to CD4 and coreceptor induces conformational changes that culminate in the activation of the fusogenic gp41 subunit of Env (8, 60).
Several lines of evidence suggest that the sequence of gp120 interaction with the target cell receptors is not random but instead involves initial binding to CD4 followed by interaction with coreceptor. First, it has long been known that gp120 can bind with high affinity to CD4 even in the absence of coreceptor, as shown by assays with both soluble and membrane-associated forms of these proteins (39, 60). Second, diverse types of analysis have demonstrated that CD4 binding induces conformational changes in gp120, again in assay systems where coreceptor is not present (39, 60). Third, high-resolution X-ray crystallographic analysis of gp120 (31) coupled with site-directed mutagenesis studies (46) have suggested that CD4 binding induces marked conformational changes in gp120 at both the CD4-interacting and coreceptor-interacting regions. Finally, CD4 has been shown to greatly enhance soluble gp120 binding to coreceptor-bearing cells for HIV type 1 (HIV-1) (6, 21, 28, 29, 33, 35, 47, 53, 57), HIV-2 (28), and simian immunodeficiency virus (SIV) (21, 28, 35, 47, 53), although examples of CD4-independent interaction between Env and coreceptor have been reported for HIV-1 (6, 27, 29, 30, 36), HIV-2 (22, 45), and SIV (21, 35, 47).
These findings with soluble gp120 raise the critical question of whether gp120 in the native membrane-associated Env complex can be activated by CD4 binding to promote functional interaction with coreceptor, leading to membrane fusion. We devised an experimental approach to test this question directly, using an adaptation of our previously described system for quantitating HIV Env-mediated cell fusion (43). This extensively described cell fusion system has been validated to recapitulate essential characteristics of HIV-1 infectivity, including CD4 dependence (43), target cell tropism (9), specific coreceptor requirements (2, 23), and inhibition by antibodies or ligands directed against Env or specific target cell receptors (2, 23, 43). We analyzed the ability of soluble CD4 (sCD4) to induce fusion between effector cells expressing Env and target cells expressing coreceptor but no CD4. Our results provide a direct demonstration of a two-step mechanism in which Env sequentially interacts with CD4 and then coreceptor to induce fusion. Moreover, they reveal important distinctions between these functional fusion studies with membrane-associated Env and previously reported binding studies with soluble gp120.
The sequential nature of the gp120-receptor interactions, coupled with the associated conformational changes in Env, have important implications for antibody blocking of Env function (58, 60). In the present study, the fusion-blocking activities of previously described monoclonal antibodies (MAbs) directed against CD4-induced epitopes on gp120 were compared in the standard versus the sCD4-activated fusion systems. The results reveal the potential of these MAbs to block Env function from genetically diverse primary HIV-1 isolates. The intricate modes by which conserved neutralization determinants on Env are shielded from the humoral immune system have important implications for the design of antibody-based strategies to prevent HIV infection.
MATERIALS AND METHODS
Cells.
HeLa and NIH 3T3 cells were obtained from the American Type Culture Collection and maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum.
Expression of Envs and receptors.
Vaccinia virus expression technology was used (20); except where indicated otherwise, all viral recombinants were derived from the WR parental strain. The following HIV-1 Env-encoding viruses were used, with the indicated Env genes linked to a strong synthetic vaccinia virus promoter (10): vCB-32, SF162 Env (9); vCB-28, JR-FL Env (9); vCB-43, Ba-L Env (9); vCB-41, LAV (Lai) Env (9); and vCB-52, CM235 Env (C. C. Broder and E. A. Berger, unpublished data). For the SIVmac316 Env, vCB-75 (Broder and Berger, unpublished data) was used. In the case of the 89.6 Env, a recombinant derived from the modified vaccinia virus Ankara strain (7) was employed. In some experiments, HIV-1 Env expression was achieved by using the following plasmids containing the strong synthetic vaccinia virus promoter: pCB-41, LAV (Lai) Env (9); pCB-32, SF162 Env (9); pCB-43, Ba-L Env (9); pGA13-89.6, 89.6 Env (G. Alkhatib and E. A. Berger, unpublished data); and pCB-52, CM235 Env (Broder and Berger, unpublished data). Alternatively, plasmids containing the T7 promoter (24, 25) were used for the following primary HIV-1 Envs: pCRII-92HT593.1, pCRII-92UG024.2, pCRII-93BR029.2, pCRII-93BR019.10, pCRII-92UG037.8, and pCRII-93MW965.26 (all obtained from the National Institutes of Health AIDS Research and Reference Reagent Program, Rockville, Md.). Full-length CD4 was expressed by vaccinia virus recombinant vCB-3 (9), and coreceptors were expressed with plasmids pYF1-fusin for CXCR4 (23) and pGA9-CKR5 for CCR5 (2); in these cases, a strong vaccinia virus promoter was used. Alternatively, for enhanced coreceptor expression, a vaccinia virus recombinant expressing either CXCR4 (vCBYF1-fusin) (23) or CCR5 (vvCCR5-1107) (61) was used.
Proteins and antibodies.
The following sCD4 recombinant proteins were donated by S. Johnson, Pharmacia Upjohn, Kalamazoo, Mich.: two-domain (2D) sCD4 (amino acids 1 to 183; 0.47 mM in phosphate-buffered saline [PBS]–0.015% Tween 80) or four-domain (4D) sCD4 (amino acids 1 to 369; 13 μM in PBS–0.015% Tween 80). The human MAbs 17b and 48d were provided by James Robinson, Tulane University, New Orleans, La.; these were derived from HIV-1-infected individuals in the United States (52). Murine MAb D47 (19) was provided by Patricia Earl (National Institute of Allergy and Infectious Diseases, National Institutes of Health). All MAbs were affinity purified from serum-free hybridoma supernatants by using protein G-Sepharose (Pharmacia Upjohn) and were resuspended in PBS.
Cell fusion assays.
Env-mediated cell fusion was quantitated with a previously described vaccinia virus-based reporter gene assay (43). Vaccinia virus expression technology was used to express Env on the designated effector cells and the appropriate receptors on the designated target cells. In addition, one cell population expressed vaccinia virus-encoded bacteriophage T7 RNA polymerase encoded by vP11T7gene 1 (1), and the other contained the lacZ reporter gene under control of the T7 promoter (plasmid pG1NT7β-gal [R. A. Morgan, National Human Genome Research Institute, personal communication]). Specific details are provided for each experiment. In the standard fusion system, target cells expressed both membrane-associated CD4 and the indicated coreceptor; in the sCD4-activated system, the target cells expressed coreceptor only. The cells were maintained overnight at 32°C to allow vaccinia virus-mediated expression of the recombinant proteins. The following day, cells were washed, suspended in medium, and used for fusion assays. Effector and target cells were mixed in duplicate wells of 96-well plates (2 × 105 cells of each type per well). For the sCD4-activated assay, purified sCD4 (2D or 4D) in the buffer indicated above was added at the indicated final concentrations to the appropriate wells; for controls, the equivalent amounts of buffer were added. Plates were incubated for 2.5 h at 37°C, and fusion was quantified by measurement of β-galactosidase activity in nonionic detergent cell lysates, using a 96-well spectrophotometer (Molecular Devices).
For preincubation experiments, sCD4 was preincubated with the indicated cell population for 30 min at 37°C; as negative controls, cells were incubated with buffer alone. The cells were then washed twice in medium to remove excess sCD4, and the two cell populations were mixed. Fusion was quantitated as described above.
Antibody effects on Env function.
Env-expressing effector cells were preincubated with the indicated concentration of purified MAb for 30 min at 37°C prior to addition of sCD4 and target cells. The antibodies were maintained throughout the fusion reaction. Fusion was quantitated as described above.
RESULTS
sCD4 activation of Env-mediated fusion.
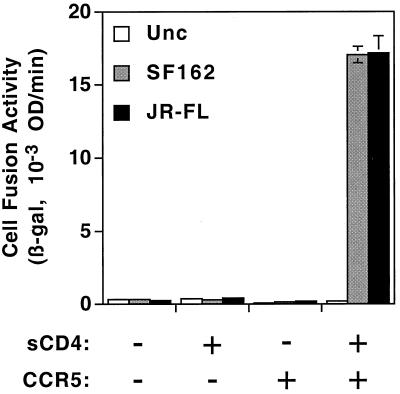
A critical prediction of the two-step model for HIV Env function is that CD4 binding activates Env for functional interaction with coreceptor on target cells; membrane fusion then ensues. To test this question directly, we examined whether sCD4 could induce Env-mediated cell fusion with target cells expressing coreceptor but lacking CD4. Figure 1 shows the results of experiments in which effector cells expressing a prototypic wild-type R5 (macrophage-tropic, non-syncytium-inducing) Env (SF162 or JR-FL) were mixed with CD4-negative NIH 3T3 target cells transfected with either a CCR5-encoding plasmid or a negative control plasmid. Effector cells expressing the nonfusogenic uncleaved Env mutant (Unc) served as an additional negative control. The fusion reactions were performed in the presence or absence of 300 nM sCD4 for 2.5 h; the cells were then lysed, and β-galactosidase was quantitated. No fusion was detected in the absence of sCD4, consistent with many previous studies demonstrating that coreceptors generally do not support infection or fusion when expressed on CD4-negative target cells (2, 12, 14, 16, 17, 23). For both wild-type Envs, but not for the Unc control, sCD4 induced significant fusion with the CCR5-expressing target cells. Fusion was not detected with target cells lacking CCR5, thereby demonstrating strict dependence on the presence of coreceptor.
FIG. 1.
sCD4 activation of Env-mediated, coreceptor-dependent cell fusion. Effector HeLa cells were transfected with indicator plasmid pG1NT7β-gal and infected with recombinant vaccinia viruses encoding the designated Envs (Unc, uncleaved control). Target NIH 3T3 cells were transfected with either pGA9-CKR5 (CCR5: +) or pSC59 (CCR5: −) and infected with vP11T7gene1 encoding T7 RNA polymerase. Four-domain sCD4 (final concentration = 300 nM; sCD4: +) or an equivalent volume of buffer (sCD4: −) was added upon mixing of effector and target cells. Cell fusion was quantitated as described in Materials and Methods. β-gal, β-galactosidase; OD, optical density.
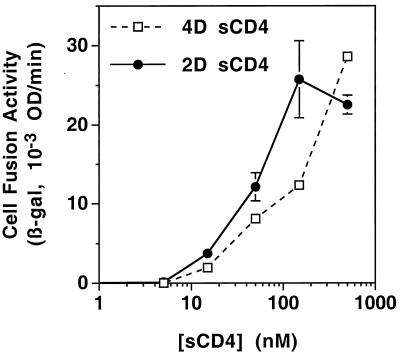
We examined the dose-response relationship between sCD4 concentration and fusion activity. Two sCD4 recombinant proteins, each capable of high-affinity binding to gp120, were compared: 2D sCD4, containing the first two N-terminal extracellular domains; and 4D sCD4, containing all four extracellular domains. As demonstrated in Fig. 2, both sCD4 proteins activated fusion by JR-FL Env with target cells expressing CCR5 but not CD4. The responses were dose dependent for both sCD4 proteins, with increasing effects up to 500 nM. Similar results were observed for the SF162 and 89.6 Envs (data not shown).
FIG. 2.
Dose-response comparison of 4D (sCD4-369) and 2D (sCD4-183) sCD4 for activation of cell fusion. Effector cells were transfected with pG1NT7β-gal and infected with vCB-28 (JR-FL Env). Target cells were transfected with pGA9-CKR5 (encoding CCR5) and infected with vP11T7gene1. At the time of cell mixing, the indicated concentrations were added for each sCD4 construct. The low background value (0.83) obtained in assays using effector cells infected with vCB-16 (Unc Env) was subtracted to give the values shown. Abbreviations are given in the legend to Fig. 1.
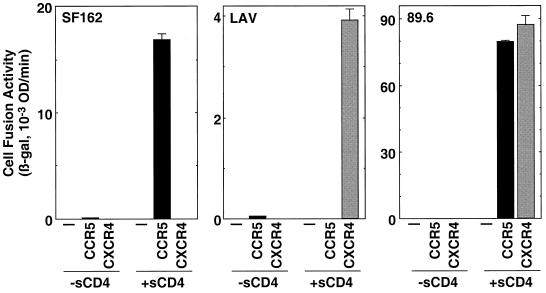
Envs from R5, X4 (T-cell line-tropic, syncytium-inducing), and R5X4 (dualtropic) HIV-1 isolates were analyzed for specificity of coreceptor usage in the sCD4-activated fusion system. As shown in Fig. 3, sCD4-activated fusion was observed for the R5 SF162 Env with target cells expressing CCR5 but not CXCR4, for the X4 LAV Env with CXCR4 but not CCR5, and for the R5X4 89.6 Env with both CCR5 and CXCR4. Thus, for these different classes of Envs, the coreceptor specificities in the sCD4-activated system closely paralleled those reported earlier with various experimental approaches using CD4-positive target cells (2, 12, 14, 16, 17, 23). Fusion induced by sCD4 is not dependent on the high levels of coreceptor achieved with vaccinia virus expression technology; potent sCD4-activated fusion has been obtained for Envs from several primary or laboratory-adapted HIV-1 strains by using target cells expressing endogenous CXCR4 but not CD4 (H. L. Greenstone, B. Dey, and E. A. Berger, unpublished data).
FIG. 3.
Coreceptor specificities of sCD4-activated cell fusion. Effector cells were transfected with pG1NT7β-gal and infected with recombinant vaccinia virus encoding the designated Env. Target cells were transfected with either pSC59 (−), pGA9-CKR5 (CCR5), or pYF1-fusin (CXCR4) and then infected with vP11T7gene1. Four-domain sCD4 (final concentration = 300 nM; +sCD4) or an equivalent volume of buffer (−sCD4) was added upon mixing of effector and target cells. The low background values obtained with the Unc Env, which ranged from 0.31 to 0.55, were subtracted to give the data shown. Abbreviations are given in the legend to Fig. 1.
Env as the target for sCD4.
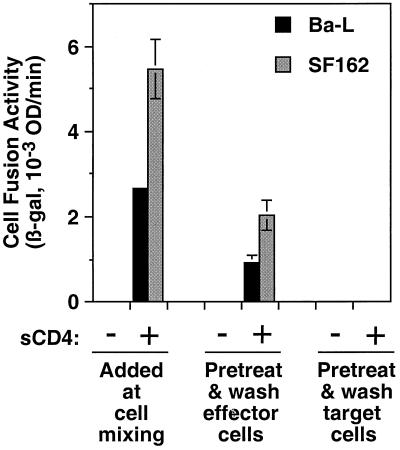
To distinguish whether sCD4 induces fusion by acting on the Env-expressing effector cell and/or the coreceptor-expressing target cell, we performed experiments in which either cell population was preincubated with sCD4 and then washed to remove unbound sCD4 prior to cell mixing. The results shown in Fig. 4 demonstrate that pretreatment of the Env-expressing cells was sufficient to induce fusion; the activity was somewhat less than that observed when sCD4 was continuously present throughout the fusion reaction. By contrast, pretreatment and washing of the target cells did not result in detectable fusion. We conclude that sCD4 promotes fusion by activating Env for functional interaction with coreceptor on the target cell.
FIG. 4.
Comparison of sCD4 pretreatment of effector cells versus target cells. Effector cells were transfected with pG1NT7β-gal and infected with recombinant vaccinia virus encoding the designated Envs. Target cells were transfected with pGA9-CKR5 (CCR5) and infected with vP11T7gene1. The effector cells or target cells were preincubated with either 200 nM 4D sCD4 (sCD4: +) or an equivalent volume of buffer (sCD4: −). The cells were then washed twice prior to cell mixing. As a positive control, sCD4 was added at the time of cell mixing. The background values obtained with Unc, which ranged from 0.45 to 0.75, were subtracted from the values for the indicated Envs. Abbreviations are given in the legend to Fig. 1.
Taken together, the results described above provide direct evidence for a two-step mechanism in which CD4 binding activates Env for functional interaction with coreceptor, leading to membrane fusion.
Epitopes for Env-blocking MAbs revealed by sCD4-activated fusion.
The human MAbs 17b and 48d bind to distinct gp120 epitopes whose exposures are considerably enhanced by CD4 binding (50–52, 55, 58, 59). These MAbs have been shown to block interaction of CD4-bound gp120 to coreceptor (26, 53, 57). High-resolution X-ray crystallographic analysis has revealed that 17b binds to discontinuous determinants on gp120; the conformational epitope is contained within the conserved “bridging sheet” formed by determinants in the C4 region and the stem of the V1/V2 loop of gp120 (31). The C4 region has also been implicated in the 48d epitope (42). Mutational analysis has revealed the critical importance of the C4 region of gp120 for binding of the gp120-sCD4 complex to coreceptor (46). Consistent with the relationships of the 17b and 48d epitopes for coreceptor interaction, these MAbs have been shown to neutralize HIV-1 infection (18, 44, 49–52, 55); however, the neutralizing activities are generally weak (18, 49–52). The proposed interpretation is that the 17b and 48d epitopes are largely masked prior to CD4 binding and are exposed only transiently after CD4 binding and before coreceptor interaction; kinetic and steric constraints might thus be expected to limit neutralization by these MAbs during the normal HIV-1 entry process (31, 58).
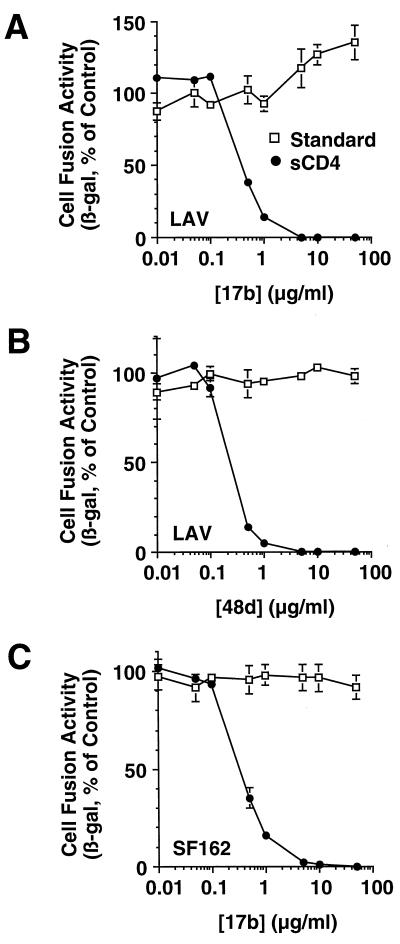
The sCD4-activated fusion system is potentially well suited for detecting the Env-blocking activities of antibodies directed against CD4-induced epitopes, since this system should enable the antibodies to bind to gp120 prior to its interaction with coreceptor on the target cell. In Fig. 5A and B, the dose responses of 17b and 48d, respectively, are shown for the LAV Env, comparing the standard fusion system (target cells expressing both CD4 and CXCR4) with the sCD4-activated fusion system (targets expressing CXCR4 alone). Neither MAb had detectable effects in the standard fusion system at concentrations up to 50 μg/ml. By contrast, both MAbs potently inhibited in the sCD4-activated fusion system; the 50% inhibitory concentrations (IC50s) were 0.4 and 0.1 μg/ml, and fusion was inhibited >95% at 5 and 1 μg/ml for 17b and 48d, respectively.
FIG. 5.
Inhibition of cell fusion by MAbs against CD4-induced epitopes. Effector cells were coinfected with vP11T7gene1 and either vCB-41 (LAV) (A and B) or vCB-32 (SF162) (C). Target cells were first cotransfected with pG1NT7β-gal and either pYF1-fusin (CXCR4) (A and B) or pGA9-CKR5 (CCR5) (C); the targets were then infected with either vCB-3 (standard fusion system) or wild-type WR (sCD4-activated fusion system). Background control targets were cotransfected with pG1NT7β-gal and pSC59 (no coreceptor) and infected in parallel fashion. Env-expressing effector cells were preincubated with twice the indicated final concentration of the designated MAb prior to cell mixing. For sCD4-activated fusion, 2D sCD4 was added just prior to cell mixing to a final concentration of 200 nM. For each Env, the low background values obtained with target cells lacking coreceptor (0.2 for LAV, 0.3 for SF162 in the sCD4-activated system, 0.7 for LAV, and 1.4 for SF162 in the standard system) were subtracted from the signals obtained with target cells expressing coreceptor (14.1 for LAV, 150 for SF162 in the sCD4-activated system, 43.1 for LAV, and 183 for SF162 in the standard system). For each Env, data are expressed as the percentage of the control values obtained in the absence of MAb. β-gal, β-galactosidase.
Presence of 17b and 48d epitopes on genetically diverse HIV-1 strains.
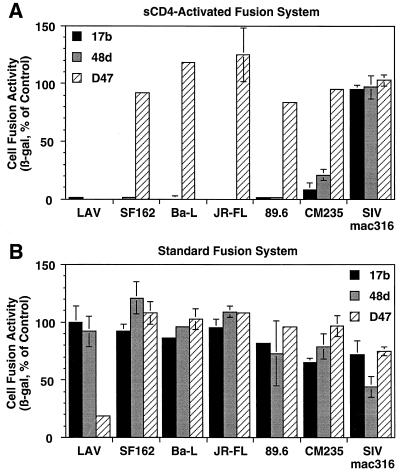
MAbs 17b and 48d were derived from HIV-1-infected individuals in the United States, where clade B is the predominant HIV-1 genetic subtype. The epitopes for these MAbs are considered to be broadly conserved on gp120 from diverse HIV-1 isolates (31, 46, 52, 58, 60). However, while 17b and 48d have been shown to react with Envs from several laboratory-adapted strains of clade B (18, 42, 44, 49–52, 55, 59), these MAbs have been reported to display poor neutralization or binding activities against numerous primary isolates, from clade B as well as other genetic subtypes (18, 40). It is therefore unclear whether the epitopes are present on genetically diverse HIV-1 Envs and whether MAb binding will impair Env function. We used the sCD4-activated fusion system to address this issue. Figure 5C shows a dose-response analysis of the effect of 17b on fusion mediated by Env from SF162 Env (a clade B R5 primary isolate), using target cells expressing CCR5; sCD4-activated fusion was inhibited with potency equivalent to that seen with the LAV Env (IC50 = 0.4 μg/ml, >95% inhibition at 5 μg/ml). We extended these analyses to diverse Envs using different coreceptors. Figure 6A shows that in addition to the LAV and SF162 Envs, both the 17b and 48d MAbs at a final antibody concentration of 50 μg/ml completely inhibited sCD4-activated fusion for Envs from the clade B primary isolates Ba-L (R5), JR-FL (R5), and 89.6 (R5X4); Env from the clade E primary isolate CM235 (R5) was somewhat less sensitive, showing ∼90% inhibition by 17b and ∼80% inhibition by 48d under the same conditions. As negative controls, neither MAb inhibited sCD4-activated fusion mediated by the R5 Env from SIVmac316. Further selectivity was demonstrated with murine MAb D47, directed against the hypervariable V3 loop of LAI (IIIB-BH8 isolate) (19); this MAb selectively inhibited the closely related LAV Env in the sCD4-activated fusion system but had minimal effects on the other Envs. Figure 6B demonstrates that MAbs 17b and 48d were ineffective against any of the Envs in the standard fusion system; only MAb D47 against the LAV Env showed significant inhibitory activity. In Table 1, we further expanded these analyses to several primary Envs from a genetically diverse panel directly cloned by PCR from infected individuals (24, 25). MAb 17b inhibited, in dose-dependent fashion, sCD4-activated fusion for primary Envs from clades A, B, C, D, E, F, and F/B. For Envs from clades A, B, C, D, F, and F/B, the potencies were at least as strong as for the clade B Envs. Consistent with the results described above, the Env from the CM235 clade E primary isolate was somewhat less sensitive, displaying ∼5-fold-weaker IC50 compared to the other Envs (Table 1, experiment 2, and additional dose-response data not shown). From these findings, we conclude that the sCD4-activated fusion system reveals the presence of CD4-induced epitopes that are indeed conserved among genetically distinct HIV-1 subtypes; antibody binding to these epitopes strongly impairs Env function.
FIG. 6.
Breadth and specificity of inhibition of sCD4-activated fusion by MAbs to CD4-induced epitopes versus a V3 loop epitope. Effector and target cells were prepared as for Fig. 5 and mixed in the following Env-coreceptor combinations: LAV-CXCR4, SF162-CCR5, Ba-L–CCR5, JR-FL–CCR5, 89.6-CXCR4, CM235-CCR5, and SIVmac316-CCR5. MAbs 17b and 48d, against CD4-induced epitopes, or MAb D47, against the V3 loop of LAI, were preincubated with Env-expressing effector cells at a concentration of 100 μg/ml; after cell mixing, the final concentrations were 50 μg/ml. For sCD4-activated fusion (A), 2D sCD4 was added just prior to cell mixing to a final concentration of 200 nM. The low background values obtained with target cells transfected with pSC59 instead of coreceptor plasmid were subtracted for each Env. The signal-to-noise ratios ranged from 4 to 167 for the various Envs. For each Env, data are expressed as the percentage of the values obtained in the absence of antibody.
TABLE 1.
Cross-clade inhibition of sCD4-activated fusion by MAb 17b
| Expt | Env | Clade | Coreceptor | % Inhibition at MAb 17b concn of:
|
||
|---|---|---|---|---|---|---|
| 0.1 μg/ml | 0.5 μg/ml | 2.5 μg/ml | ||||
| 1a | LAV | B | CXCR4 | 16 | 65 | |
| SF162 | B | CCR5 | 27 | 62 | ||
| 92HT593.1 | B | CXCR4 | 58 | 100 | ||
| 92UG024.2 | D | CXCR4 | 42 | 80 | ||
| 93BR029.2 | F | CCR5 | 41 | 100 | ||
| 93BR019.10 | F/B | CCR5 | 33 | 80 | ||
| 2b | Ba-L | B | CCR5 | 43 | 90 | |
| 89.6 | B | CXCR4 | 57 | 97 | ||
| 92UG037.8 | A | CCR5 | 47 | 94 | ||
| 93MW965.26 | C | CCR5 | 79 | 98 | ||
| CM235 | E | CCR5 | 0 | 45 | ||
HeLa effector cells were transfected with a plasmid expressing the designated Env and then infected with vP11T7gene1. NIH 3T3 target cells were cotransfected with pG1NT7β-gal and either pYF1-fusin (CXCR4) or pGA9-CKR5 (CCR5), as indicated, and then infected with wild-type WR.
Identical to experiment 1 except that target cells were transfected with pG1NT7β-gal and then infected with either vCBYF1-fusin (CXCR4) or vvCCR5-1107 (CCR5).
DISCUSSION
The awareness that HIV-1 entry requires both CD4 and coreceptor has engendered models for the molecular interactions involved in Env-mediated membrane fusion. In this study, we developed a cell fusion system to examine the distinct steps in the activation pathway for Env-mediated fusion. Our results provide direct demonstration of a sequential two-step model for Env-receptor interactions in the HIV-1 entry mechanism, previously proposed by others (reviewed in references 3, 8, and 60). In the first step, CD4 binds to the gp120 subunit of Env and induces a conformational change(s) which then exposes, creates, or stabilizes the coreceptor binding determinants on gp120. In the second step, the CD4-activated gp120 binds to coreceptor. The gp120-coreceptor interaction presumably triggers newly revealed conformational changes in the gp41 subunit of Env (11, 56), ultimately leading to membrane fusion and virus entry. According to this model, CD4 plays two distinct roles in HIV infection: bringing Env in close apposition to coreceptor on the target cell membrane, and inducing conformational changes in gp120 to enable its binding to coreceptor. Our sCD4-activated system distinguishes these two roles and illustrates that the second can still occur in the absence of the first. The system also enables analysis of the activating properties of sCD4 in the absence of the inhibitory competitive effects seen with target cells expressing both cell-associated CD4 and coreceptor (47).
Our findings represent an important extension of earlier binding experiments using soluble gp120, in which CD4 was shown to significantly enhance the gp120-coreceptor binding interaction (6, 21, 28, 29, 33, 47, 53, 57). The functional demonstration of sCD4-activated fusion is critical, particularly in view of several reports indicating that coreceptor interaction findings obtained with soluble gp120 do not necessarily predict results with the intact cell-associated Env oligomer (5, 15, 34, 38). For example, others have reported that soluble gp120 from T-cell line-adapted strains can interact with CXCR4 in the absence of sCD4 (6, 36, 37). By contrast, we found that sCD4 was absolutely essential to trigger fusion between effector cells expressing Env from the T-cell line-adapted LAV strain and targets expressing CXCR4 (as well as other Env-coreceptor interactions) (Fig. 1, 3, and 4); this is consistent with the strict dependence on CD4 observed in other coreceptor studies using standard infectivity and cell fusion systems (2, 12, 14, 16, 17, 23). Presumably, structural differences between the soluble gp120 monomer and the cell-associated Env oligomer contribute to these differing experimental results.
Our demonstrations of sCD4-activated fusion with both laboratory-adapted and primary HIV-1 isolates extend earlier reports indicating that sCD4 can enhance or enable fusion or infectivity by some HIV and SIV isolates (4, 13, 47–49); these include reports of sCD4-induced entry and infection of CD4-negative, coreceptor-positive cells by both HIV-1 (48) and HIV-2 (45). Moreover, the potency with which 17b inhibited sCD4-activated fusion by diverse Envs in our fusion system parallels closely the demonstrated ability of sCD4 to enhance 17b neutralization of a T-cell line-adapted strain infecting target cells expressing both CD4 and CXCR4 (50). The present results are also consistent with previous evidence that HIV and SIV strains capable of infecting via coreceptor-dependent, CD4-independent mechanisms (6, 21, 22, 27, 30, 35, 36, 45, 47) contain gp120 variants that are permissive for coreceptor binding in the absence of CD4.
In direct comparisons, we found that fusion activity in the sCD4-activated system is typically lower than that in the standard system; the relative activities were variable between different experiments, ranging from ∼10 to ∼100% (data not shown). Reduced activity in the sCD4 system is not surprising, since the function of cell-associated CD4 in bringing Env in close proximity to coreceptor on the target cell is not enabled. Additional experimental efforts are required to delineate the complex experimental variables that might contribute to the relative efficiencies of the two systems (cell type, Env type, levels of relevant molecules, cell density, etc.). A related issue that is well suited for the sCD4-activated system is the functional stability of Env after interaction with CD4. In the experiment shown in Fig. 4, fusion activity of an R5 Env after pretreatment with sCD4 was somewhat less than that observed when sCD4 was added at the time of cell mixing. In preliminary extensions of this finding, we have found that Envs from primary R5 strains retain considerable fusion activity after prolonged (1 to 2 h) preincubation with sCD4, whereas Envs from T-cell line-adapted X4 strains are much more prone to loss of activity (data not shown). The former results are consistent with a recent report demonstrating the long-lived activity of sCD4-activated SIV Env for interaction with CCR5 (47). We are seeking to unravel various factors that may contribute to Env inactivation after interaction with sCD4 (stripping of gp120, instability of gp41, etc.) (39). Another focus of future efforts will be to determine whether sCD4 remains associated with Env following the wash step, or whether Env is capable of maintaining the activated conformation following sCD4 dissociation during washing. Interesting in this regard is the report that Env interaction with CD4 and coreceptor can occur in a trans fashion, with CD4 on one cell activating Env for functional interaction with coreceptor on a different cell (48); perhaps these findings reflect the ability of Env to maintain the CD4-activated conformation after dissociation.
The sCD4-activated system also reflects on recent studies demonstrating that CXCR4 (54) and, to a greater extent, CCR5 (61) form constitutive cell surface associations with CD4 in the absence of gp120. It has been proposed that the native CD4-CCR5 interaction may be important for HIV entry and infection and may represent a new target for anti-HIV drug development (61). The data presented herein indicate that Env-mediated fusion can occur when CD4 is not anchored to the cell surface in normal fashion. This may argue against an obligate role for the constitutive CD4-CCR5 complex in Env function. A reasonable alternative is that the CD4-CCR5 surface interaction is not absolutely essential for Env-mediated fusion but facilitates this process perhaps by increasing the local concentrations of both critical receptors. However, it is also possible that the soluble form of CD4 can engage in the necessary interactions with CCR5 reported for cell-associated CD4; relevant in this regard is the conclusion the CD4-CCR5 interaction occurs via determinants within the first two domains of CD4 (61), coupled with our finding that the truncated 2D sCD4 construct is fully competent to activate Env (Fig. 2).
The broad Env-blocking activities of 17b and 48d revealed by the sCD4-activated fusion system also have important implications for the design of HIV vaccines based on humoral immunity. Perhaps approaches can be devised to elicit antibodies that mimic sCD4 in inducing the epitopes detected by 17b and 48d; combining such approaches with strategies to generate antibodies against these epitopes might prove efficacious in inducing protective humoral immunity. Indeed, human MAbs that enhance exposure of the 17b epitope have been described (41, 59). We are presently examining whether such MAbs can substitute for sCD4 in promoting fusion with target cells bearing coreceptor but not CD4 and whether they can mimic sCD4 in enhancing the fusion-blocking activities of 17b and/or 48d. This concept is also interesting in view of the recent success at generating broadly cross-reactive antibody activity by using complexes of effector cells expressing Env and target cells expressing CD4 plus coreceptor, captured at various stages in the fusion process (32). Perhaps some (all?) of the observed neutralizing activity derives from synergistic effects of multiple antibodies on Env function. These concepts add to the growing optimism that the rapidly expanding knowledge of the Env-CD4-coreceptor interaction may ultimately lead to novel practical strategies for intervention in the AIDS pandemic.
ACKNOWLEDGMENTS
K. Salzwedel and E. D. Smith contributed equally to this work.
We thank the following individuals for supplying essential reagents: J. Robinson for purified MAbs 17b and 48d and the corresponding hybridomas, P. L. Earl for purified MAb D47 and the corresponding hybridoma, L. S. Wyatt and B. Moss for the vaccinia virus recombinant expressing the 89.6 Env (MVA/HIV 89.6 env), and S. Johnson for sCD4 proteins.
K. Salzwedel was supported in part by a National Research Council-National Institutes of Health research associateship. This study was funded in part by the NIH Intramural AIDS Targeted Antiviral Program.
REFERENCES
- 1.Alexander W A, Moss B, Fuerst T R. Regulated expression of foreign genes in vaccinia virus under the control of bacteriophage T7 RNA polymerase and the Escherichia coli lac repressor. J Virol. 1992;66:2934–2942. doi: 10.1128/jvi.66.5.2934-2942.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 3.Allan J S. Receptor-mediated activation of the viral envelope and viral entry. AIDS. 1993;7:S43–S50. [PubMed] [Google Scholar]
- 4.Allan J S, Strauss J, Buck D W. Enhancement of SIV infection with soluble receptor molecules. Science. 1990;247:1084–1088. doi: 10.1126/science.2309120. [DOI] [PubMed] [Google Scholar]
- 5.Baik S S W, Doms R W, Doranz B J. HIV and SIV gp120 binding does not predict coreceptor function. Virology. 1999;259:267–273. doi: 10.1006/viro.1999.9779. [DOI] [PubMed] [Google Scholar]
- 6.Bandres J C, Wang Q F, O'Leary J, Baleaux F, Amara A, Hoxie J A, Zolla-Pazner S, Gorny M K. Human immunodeficiency virus (HIV) envelope binds to CXCR4 independently of CD4, and binding can be enhanced by interaction with soluble CD4 or by HIV envelope deglycosylation. J Virol. 1998;72:2500–2504. doi: 10.1128/jvi.72.3.2500-2504.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belyakov I M, Wyatt L S, Ahlers J D, Earl P, Pendleton C D, Kelsall B L, Strober W, Moss B, Berzofsky J A. Induction of a mucosal cytotoxic T-lymphocyte response by intrarectal immunization with a replication-deficient recombinant vaccinia virus expressing human immunodeficiency virus 89.6 envelope protein. J Virol. 1998;72:8264–8272. doi: 10.1128/jvi.72.10.8264-8272.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger E A, Murphy P M, Farber J M. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 9.Broder C C, Berger E A. Fusogenic selectivity of the envelope glycoprotein is a major determinant of human immunodeficiency virus type 1 tropism for CD4+ T-cell lines vs. primary macrophages. Proc Natl Acad Sci USA. 1995;92:9004–9008. doi: 10.1073/pnas.92.19.9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakrabarti S, Sisler J R, Moss B. Compact, synthetic, vaccinia virus early/late promoter for protein expression. BioTechniques. 1997;23:1094–1097. doi: 10.2144/97236st07. [DOI] [PubMed] [Google Scholar]
- 11.Chan D C, Kim P S. HIV entry and its inhibition. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 12.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, Larosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 13.Clapham P R, McKnight A, Weiss R A. Human immunodeficiency virus type 2 infection and fusion of CD4-negative human cell lines: induction and enhancement by soluble CD4. J Virol. 1992;66:3531–3537. doi: 10.1128/jvi.66.6.3531-3537.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng H K, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 15.Doranz B J, Orsini M J, Turner J D, Hoffman T L, Berson J F, Hoxie J A, Peiper S C, Brass L F, Doms R W. Identification of CXCR4 domains that support coreceptor and chemokine receptor functions. J Virol. 1999;73:2752–2761. doi: 10.1128/jvi.73.4.2752-2761.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doranz B J, Rucker J, Yi Y J, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 17.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y X, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 18.D'Souza M P, Milman G, Bradac J A, McPhee D, Hanson C V, Hendry R M, Corcoran T, Stott J, Fung M, Hanson C, Laman J, Mascola J, Rasheed S, Richman D, Schuitemaker H, Thiriart C, Wainberg M, Weber J, Beddows S, Tilley S, Robinson J, Zolla-Pazner S, Katinger H, Cummins L. Neutralization of primary HIV-1 isolates by anti-envelope monoclonal antibodies. AIDS. 1995;9:867–874. doi: 10.1097/00002030-199508000-00006. [DOI] [PubMed] [Google Scholar]
- 19.Earl P L, Broder C C, Long D, Lee S A, Peterson J, Chakrabarti S, Doms R W, Moss B. Native oligomeric human immunodeficiency virus type 1 envelope glycoprotein elicits diverse monoclonal antibody reactivities. J Virol. 1994;68:3015–3026. doi: 10.1128/jvi.68.5.3015-3026.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Earl P L, Cooper N, Moss B. Expression of proteins in mammalian cells using vaccinia viral vectors. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. 2, suppl. 15. New York, N.Y: John Wiley & Sons; 1991. pp. 16.15.1–16.18.10. [Google Scholar]
- 21.Edinger A L, Blanpain C, Kunstman K J, Wolinsky S M, Parmentier M, Doms R W. Functional dissection of CCR5 coreceptor function through the use of CD4-independent simian immunodeficiency virus strains. J Virol. 1999;73:4062–4073. doi: 10.1128/jvi.73.5.4062-4073.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N C, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by Fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 23.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 24.Gao, F., S. G. Morrison, D. L. Robertson, C. L. Thornton, S. Craig, G. Karlsson, J. Sodroski, M. Morgado, B. Galvao-Castro, H. Vonbriesen, S. Beddows, J. Weber, P. M. Sharp, G. M. Shaw, B. H. Hahn, and WHO and NIAID Networks for HIV Isolation and Characterization. 1996. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. J. Virol. 70:1651–1667. [DOI] [PMC free article] [PubMed]
- 25.Gao F, Yue L, Craig S, Thornton C L, Robertson D L, Mccutchan F E, Bradac J A, Sharp P M, Hahn B H, Osmanov S, Belsey E M, Heyward W, Esparza J, Galvaocastro B, Vandeperre P, Karita E, Wasi C, Sempala S, Tugume B, Biryahwaho B, Rubsamenwaigmann H, Vonbriesen H, Esser R, Grez M, Holmes H, Newberry A, Ranjbar S, Tomlinson P, Bradac J, McCutchan F, Louwagie J, Hegerich P, Lopezgalindez C, Olivares I, Dopazo J, Mullins J I, Delwart E L, Bachmann H M, Goudsmit J, DeWolf F, Saragosti S, Schochetman G, Kalish M, Luo C C, George R, Pau C P, Weber J, Cheingsongpopov R, Kaleebu P, et al. Genetic variation of HIV type 1 in four World Health Organization-sponsored vaccine evaluation sites: generation of functional envelope (glycoprotein 160) clones representative of sequence subtypes A, B, C, and E. AIDS Res Hum Retroviruses. 1994;10:1359–1368. doi: 10.1089/aid.1994.10.1359. [DOI] [PubMed] [Google Scholar]
- 26.Golding H, Dimitrov D S, Manischewitz J, Broder C C, Robinson J, Fabian S, Littman D R, Lapham C K. Phorbol ester-induced down modulation of tailless CD4 receptors requires prior binding of gp120 and suggests a role for accessory molecules. J Virol. 1995;69:6140–6148. doi: 10.1128/jvi.69.10.6140-6148.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hesselgesser J, Halks-Miller M, DelVecchio V, Peiper S C, Hoxie J, Kolson D L, Taub D, Horuk R. CD4-independent association between HIV-1 gp120 and CXCR4: functional chemokine receptors are expressed in human neurons. Curr Biol. 1997;7:112–121. doi: 10.1016/s0960-9822(06)00055-8. [DOI] [PubMed] [Google Scholar]
- 28.Hill C M, Deng H, Unutmaz D, Kewalramani V N, Bastiani L, Gorny M K, Zolla-Pazner S, Littman D R. Envelope glycoproteins from human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus can use human CCR5 as a coreceptor for viral entry and make direct CD4-dependent interactions with this chemokine receptor. J Virol. 1997;71:6296–6304. doi: 10.1128/jvi.71.9.6296-6304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffman T L, Labranche C C, Zhang W T, Canziani G, Robinson J, Chaiken I, Hoxie J A, Doms R W. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proc Natl Acad Sci USA. 1999;96:6359–6364. doi: 10.1073/pnas.96.11.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iyengar S, Schwartz D H, Hildreth J E K. T cell-tropic HIV gp120 mediates CD4 and CD8 cell chemotaxis through CXCR4 independent of CD4: implications for HIV pathogenesis. J Immunol. 1999;162:6263–6267. [PubMed] [Google Scholar]
- 31.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LaCasse R A, Follis K E, Trahey M, Scarborough J D, Littman D R, Nunberg J H. Fusion-competent vaccines broad neutralization of primary isolates of HIV. Science. 1999;283:357–362. doi: 10.1126/science.283.5400.357. [DOI] [PubMed] [Google Scholar]
- 33.Lapham C K, Ouyang J, Chandrasekhar B, Nguyen N Y, Dimitrov D S, Golding H. Evidence for cell-surface association between fusin and the CD4-gp120 complex in human cell lines. Science. 1996;274:602–605. doi: 10.1126/science.274.5287.602. [DOI] [PubMed] [Google Scholar]
- 34.Lee B, Sharron M, Blanpain C, Doranz B J, Vakili J, Setoh P, Berg E, Liu G, Guy H R, Durell S R, Parmentier M, Chang C N, Price K, Tsang M, Doms R W. Epitope mapping of CCR5 reveals multiple conformational states and distinct but overlapping structures involved in chemokine and coreceptor function. J Biol Chem. 1999;274:9617–9626. doi: 10.1074/jbc.274.14.9617. [DOI] [PubMed] [Google Scholar]
- 35.Martin K A, Wyatt R, Farzan M, Choe H, Marcon L, Desjardin E, Robinson J, Sodroski J, Gerard C, Gerard N P. CD4-independent binding of SIV gp120 to rhesus CCR5. Science. 1997;278:1470–1473. doi: 10.1126/science.278.5342.1470. [DOI] [PubMed] [Google Scholar]
- 36.Misse D, Cerutti M, Noraz N, Jourdan P, Favero J, Devauchelle G, Yssel H, Taylor N, Veas F. A CD4-independent interaction of human immunodeficiency virus-1 gp120 with CXCR4 induces their cointernalization, cell signaling, and T-cell chemotaxis. Blood. 1999;93:2454–2462. [PubMed] [Google Scholar]
- 37.Misse D, Cerutti M, Schmidt I, Jansen A, Devauchelle G, Jansen F, Veas F. Dissociation of the CD4 and CXCR4 binding properties of human immunodeficiency virus type 1 gp120 by deletion of the first putative alpha-helical conserved structure. J Virol. 1998;72:7280–7288. doi: 10.1128/jvi.72.9.7280-7288.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mondor I, Moulard M, Ugolini S, Klasse P J, Hoxie J, Amara A, Delaunay T, Wyatt R, Sodroski J, Sattentau Q J. Interactions among HIV gp120, CD4, and CXCR4: dependence on CD4 expression level, gp120 viral origin, conservation of the gp120 COOH- and NH2-termini and V1/V2 and V3 loops, and sensitivity to neutralizing antibodies. Virology. 1998;248:394–405. doi: 10.1006/viro.1998.9282. [DOI] [PubMed] [Google Scholar]
- 39.Moore J P, Jameson B A, Weiss R A, Sattentau Q J. The HIV-cell fusion reaction. In: Bentz J, editor. Viral fusion mechanisms. Boca Raton, Fla: CRC Press; 1993. pp. 233–289. [Google Scholar]
- 40.Moore J P, McCutchan F E, Poon S W, Mascola J, Liu J, Cao Y Z, Ho D D. Exploration of antigenic variation in gp120 from clades A through F of human immunodeficiency virus type 1 by using monoclonal antibodies. J Virol. 1994;68:8350–8364. doi: 10.1128/jvi.68.12.8350-8364.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore J P, Sattentau Q J, Yoshiyama H, Thali M, Charles M, Sullivan N, Poon S W, Fung M S, Traincard F, Pinkus M, Robey G, Robinson J E, Ho D D, Sodroski J. Probing the structure of the V2 domain of human immunodeficiency virus type 1 surface glycoprotein gp120 with a panel of eight monoclonal antibodies: human immune response to the V1 and V2 domains. J Virol. 1993;67:6136–6151. doi: 10.1128/jvi.67.10.6136-6151.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moore J P, Yoshiyama H, Ho D D, Robinson J E, Sodroski J. Antigenic variation in gp120s from molecular clones of HIV-1 LAI. AIDS Res Hum Retroviruses. 1993;9:1185–1193. doi: 10.1089/aid.1993.9.1185. [DOI] [PubMed] [Google Scholar]
- 43.Nussbaum O, Broder C C, Berger E A. Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J Virol. 1994;68:5411–5422. doi: 10.1128/jvi.68.9.5411-5422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parren P W H I, Mondor I, Naniche D, Ditzel H J, Klasse P J, Burton D R, Sattentau Q J. Neutralization of human immunodeficiency virus type 1 by antibody to gp120 is determined primarily by occupancy of sites on the virion irrespective of epitope specificity. J Virol. 1998;72:3512–3519. doi: 10.1128/jvi.72.5.3512-3519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reeves J D, McKnight A, Potempa S, Simmons G, Gray P W, Power C A, Wells T, Weiss R A, Talbot S J. CD4-independent infection by HIV-2 (ROD/B): use of the 7-transmembrane receptors CXCR-4, CCR-3, and V28 for entry. Virology. 1997;231:130–134. doi: 10.1006/viro.1997.8508. [DOI] [PubMed] [Google Scholar]
- 46.Rizzuto C D, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 47.Schenten D, Marcon L, Karlsson G B, Parolin C, Kodama T, Gerard N, Sodroski J. Effects of soluble CD4 on simian immunodeficiency virus infection of CD4-positive and CD4-negative cells. J Virol. 1999;73:5373–5380. doi: 10.1128/jvi.73.7.5373-5380.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Speck R F, Esser U, Penn M L, Eckstein D A, Pulliam L, Chan S Y, Goldsmith M A. A trans-receptor mechanism for infection of CD4-negative cells by human immunodeficiency virus type 1. Curr Biol. 1999;9:547–550. doi: 10.1016/s0960-9822(99)80241-3. [DOI] [PubMed] [Google Scholar]
- 49.Sullivan N, Sun Y, Binley J, Lee J, Barbas C F, Parren P W H I, Burton D R, Sodroski J. Determinants of human immunodeficiency virus type 1 envelope glycoprotein activation by soluble CD4 and monoclonal antibodies. J Virol. 1998;72:6332–6338. doi: 10.1128/jvi.72.8.6332-6338.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sullivan N, Sun Y, Sattentau Q, Thali M, Wu D, Denisova G, Gershoni J, Robinson J, Moore J, Sodroski J. CD4-induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J Virol. 1998;72:4694–4703. doi: 10.1128/jvi.72.6.4694-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thali M, Charles M, Furman C, Cavacini L, Posner M, Robinson J, Sodroski J. Resistance to neutralization by broadly reactive antibodies to the human immunodeficiency virus type 1 gp120 glycoprotein conferred by a gp41 amino acid change. J Virol. 1994;68:674–680. doi: 10.1128/jvi.68.2.674-680.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thali M, Moore J P, Furman C, Charles M, Ho D D, Robinson J, Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 54.Ugolini S, Moulard M, Mondor I, Barois N, Demandolx D, Hoxie J, Brelot A, Alizon M, Davoust J, Sattentau Q J. HIV-1 gp120 induces an association between CD4 and the chemokine receptor CXCR4. J Immunol. 1997;159:3000–3008. [PubMed] [Google Scholar]
- 55.Watkins B A, Buge S, Aldrich K, Davis A E, Robinson J, Reitz M S, Robertguroff M. Resistance of human immunodeficiency virus type 1 to neutralization by natural antisera occurs through single amino acid substitutions that cause changes in antibody binding at multiple sites. J Virol. 1996;70:8431–8437. doi: 10.1128/jvi.70.12.8431-8437.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weissenhorn W, Dessen A, Calder L J, Harrison S C, Skehel J J, Wiley D C. Structural basis for membrane fusion by enveloped viruses. Mol Membr Biol. 1999;16:3–9. doi: 10.1080/096876899294706. [DOI] [PubMed] [Google Scholar]
- 57.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 58.Wyatt R, Kwong P D, Desjardins E, Sweet R W, Robinson J, Hendrickson W A, Sodroski J G. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 59.Wyatt R, Moore J, Accola M, Desjardin E, Robinson J, Sodroski J. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J Virol. 1995;69:5723–5733. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 61.Xiao X D, Wu L, Stantchev T S, Feng Y-R, Ugolini S, Chen H, Shen Z, Riley J L, Broder C C, Sattentau Q J, Dimitrov D S. Constitutive cell surface association between CD4 and CCR5. Proc Natl Acad Sci USA. 1999;96:7496–7501. doi: 10.1073/pnas.96.13.7496. [DOI] [PMC free article] [PubMed] [Google Scholar]