Abstract
Circular RNAs (circRNAs) are a class of endogenous non-coding RNAs that have been implicated in mediating granulosa cell (GC) proliferation and apoptosis. CircRAB11A was found to have a significantly higher expression in normal follicles compared to atrophic follicles. In this study, we determined that the knockdown of circRAB11A resulted in the inhibition of proliferation and promotion of apoptosis in GCs of chicken. Moreover, circRAB11A was found to act as a sponge for miR-24-5p, both member RAS oncogene family (RAB11A) and epidermal growth factor receptor (EGFR) were revealed to be targets of miR-24-5p through a dual-luciferase reporter assay. RAB11A or EGFR promoted proliferation and suppressed apoptosis in GCs through the phosphatidylinositol-kinase (PI3K)/AKT or extracellular signal-regulated kinase (ERK)1/2 pathway. These findings suggest that circRAB11A may function as a competing endogenous RNA (ceRNA) by targeting the miR-24-5p/RAB11A and miR-24-5p/EGFR axes and activating the ERK1/2 and PI3K/AKT pathways, offering a potential avenue for exploring the mechanism of follicle development.
Key words: apoptosis, circRAB11A, granulosa cells, ERK1/2, PI3K/AKT, proliferation
INTRODUCTION
As a primary livestock, poultry not only provides delicious meat, but also offers high-quality egg products. However, it has been observed that local chickens in China exhibit lower reproductive performance when compared to foreign breeds. The ovarian follicle serves as the fundamental functional unit of the chicken ovary and holds the crucial responsibility of producing mature oocytes and reproducing offspring (Hao et al., 2020). Unlike mammals, which exhibit periodic ovulation after sexual maturity, poultry have evolved a mode of continuous ovulation to ensure daily egg production. The laying hens have pregrade follicles, including small and large white follicles (SWF and LWF), small and large yellow follicles (SYF and LYF), 5 to 6 growing grade follicles (demarcated by volume sequentially as F5, F4, F3, F2, and F1) in their ovaries. When the hens lay eggs, the pre-grade follicles transform into grade follicles, and the dominant follicles in the grade follicles are selected and expelled (Li et al., 2022). After the follicles in the ovaries develop and mature, the egg is discharged (at this time, the egg is called yolk). The yolk gradually forms protein, membrane, and eggshell through the infundibulum, magnum, isthmus, uterus, and vagina, and finally is discharged from the body (Yoshimura and Barua, 2017). During this process, only one dominant follicle is selected to progress to ovulation, while the rest of the follicles undergo atresia after the hen's laying period. Follicular GCs are epithelial cells derived from chicken follicles and can synthesize various types growth factors and hormones regulate the growth, differentiation, and maturation of follicular membrane cells and oocytes, playing an important supporting role in follicular formation and development (Li et al., 2022). The GCs are the earliest developing somatic cells in the ovary and apoptosis of these cells is among other indicator of follicular atresia. To maintain the normal development of poultry follicles, it is essential to reduce GC apoptosis and enhance GC proliferation.
Circular RNAs (circRNAs) are a distinctive class of RNA molecules that differ from the traditional linear RNA molecules. They are characterized by their closed, circular structure that is held together by covalent bonds, lacking the typical 3′ and 5′ ends found in linear RNA molecules (Ling et al., 2022). The discovery and identification of circRNAs dates back to the 1970s (Kolakofsky, 1976), but recent advancements in next-generation sequencing and bioinformatics tools have allowed for a deeper understanding of their functions, which include serving as a sponge for mircoRNA (miRNA) (Panda, 2018), regulating transcription (Zheng, et al., 2020) and encoding protein (Yang, et al., 2018). CircRNAs play a crucial role in regulating a wide range of biological functions, including cell proliferation, apoptosis, and steroid hormone synthesis in GCs. CircRNAs can function as miRNA sponges, which is called competing endogenous RNA (ceRNA) mechanism. The ceRNA mechanism of circRNAs is a hot topic of research in various species. In human research, circRNA-101237 was found to act as a sponge for miR-490-3p, thereby promoting the expression of MAPK1 (Zhang et al., 2020a). circRNA-010383 has been demonstrated to act as a ceRNA, reducing the expression of miR-1335a and contributing to renal fibrosis in diabetic nephropathy (Peng et al., 2021). Another study by Li et al. revealed that circ-ANKHD1, which targets the miR-27a-3p/SFRP1 axis, serves as a sponge for miRNAs and modulates the apoptosis of GCs (Li et al., 2021b). Additionally, circINHA and circSLC41A1 were found to separately inhibit porcine GC apoptosis through the miR-10a-5p/CTGF axis and the miR-9820-5p/SRSF1 axis, respectively (Guo et al., 2019; Wang et al., 2022a).
In our previous sequencing analysis, it was found that circRAB11A was differentially expressed between normal follicles and atrophic follicles in chickens and had significantly higher mRNA abundance in normal developmental grade follicles (He et al., 2022). Therefore, the current study aimed to investigate the specific function and underlying molecular mechanisms of circRAB11A in regulating follicle development in chickens.
MATERIALS AND METHODS
Sample Collection and Cell Culture
In this experiment, we selected 27 breeding hens of Tianfu broilers that were reared under the same conditions, had the same weight and laying time, and were at the peak of egg production. Among them, 3 normal laying hens were used to collect 11 tissue samples, pre-grade follicles and grade follicles. The remaining 24 individuals were used to isolate and culture grade GCs. After collecting 11 tissue samples, including breast muscle, liver, ovary, uterus etc., these tissues were quickly placed in liquid nitrogen. We selected graded follicles to collect granulosa cells for isolation and culture. The graded follicles were categorized as F1, F2, F3, F4, and F5 in descending order of volume (He et al., 2022). DF-1 cells (Fuheng Cell Center, Shanghai, China) were used to verify the targeting relationship between circRNA, miRNA and mRNA, which are chicken fibroblast cell line and cultured in 89% F12 medium (Gibco, Grand Island, NY), 10% fetal bovine serum (Gibco), 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA). For the cell culture, all the cells were incubated with 95% CO2, and in a humid environment at 37°C. GCs were seeded into 6-well, 12-well, 24-well or 96-well plates and cultured in 89% M199 medium (Gibco), 10% fetal bovine serum (Gibco) 1% penicillin/streptomycin (Invitrogen). The animal experiment was conducted under the approval of the Animal Care Committee of Sichuan Agricultural University (Approval No. 2020202028).
Cell Transfection
The sequence of negative controls (NC), circRAB11A siRNA, miR-24-5p mimic, miR-24-5p inhibitor, RAB11A siRNA and EGFR siRNA are shown in n Table S1, and all of these sequences were provided by GenePharma (GenePharma, Shanghai, China). The plasmid sequence of circRAB11A, RAB11A was provided by Tsingke Biotechnology Co., Ltd. Cells were transfected with Lipofectamine 3,000 (Invitrogen) according to the manufacture's protocol. The culture medium was changed 8 h after transfection.
RNA Extraction, Reverse Transcription and Quantitative Real-Time PCR (qPCR) Validation
RNAiso Plus (Takara, Dalian, China) was used to isolate total RNA from tissues or cells according to the manufacturer's instructions. The quantity and quality of RNA was measured using a NanoDrop D2000 spectrophotometer (NanoDrop, DE). RNA that did not pass the quality check was not included in this study. Equal amounts of RNA for each sample were used to synthesize miRNA or cDNA using One-Step miRNA cDNA Synthesis Kit (HaiGene, Haerbin, China) or TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen, Beijing, China). The qPCR assay was performed using TB Green PCR Master Mix (Takara) according to the manufacturer's instructions. Primer 6 was used to design primer in this study. All of primers were synthesized by Tsingke Biotechnology Co., Ltd (Supplementary Table S2). β-actin and U6 as endogenous control were used to normalize gene expression. Relative changes in gene expression were calculated using the 2−ΔΔCt method.
Validation of circRAB11A
Sanger sequencing, RNase R (Epicentre Technologies, Madison, WI) and Oligo d (T) were used to identify whether circRAB11A was looped. For Sanger sequencing assay, divergent primer PCR results were gel purified and submitted to Sanger sequencing at Sangon Biotech Co. Ltd. to verify the junction sequence of circRAB11A. For the RNase R assay, 2 mixed RNA samples with a combined volume of 20 μL were prepared, one with RNase R and the other without RNase R. Then, the samples treat at 37°C for 10 min and inactivate at 90°C for 10 min, the combined RNA samples were reverse transcribed into cDNA for qPCR to prove the presence of circRNA. As for Oligo d (T) assay, because oligo d (T) primers cannot effectively reverse transcribe circRNAs without poly (A) tail, we choose Oligo d (T) primer and random primer to identify circRNAs.
Cell Proliferation Assay
For the cell counting kit-8 (CCK-8) assay, GCs were seeded into 96-well plates. After treatment, 4 time points were set at 12 h intervals, and cell proliferation was checked at each time point using CCK-8 reagent (Multi Sciences, Hangzhou, China) according to the manufacturer's instructions. Absorbance at 450 nm was measured using an enzyme marker (Thermofisher, Varioskan LUX).
For the 5-ethynyl-2′-deoxyuridine (EdU) assay, after transfection for 12 h, GCs were first incubated with EdU reagent A (Ribobio, Guangzhou, China) for 3 h. Then, after fixation with 4% formaldehyde solution for 30 min, the cell permeability was changed with 0.5% Triton X-100. The cell proliferation region and the nucleus were then stained separately. The number of EdU-stained cells was counted by Image-Pro Plus software.
Western Blot Analysis
A total protein extraction kit (BestBio, Shanghai, China) was used to extract total protein from GCs. Protein concentrations were homogenized using the BCA protein assay kit (BestBio). Protein samples were then separated by SDS-PAGE after degeneration. After semi-dry transfer, the target protein was retained on the PVDF membrane. After the PVDF membrane was sealed with sealing liquid for 1 h, the corresponding primary antibody was selected to incubate it overnight at 4°C according to the needs of this study. These PVDF membranes were then incubated with the appropriate secondary antibodies for 1 h at 4°C. ECL is used to detect protein signals. The antibodies used in this study are showed in Supplementary Table S3. Results were quantified using ImageJ software.
Analysis of Cell Apoptosis
The cell apoptosis rate of GCs was measured using a terminal deoxynucleotidyl transferase (TdT) d UTP nick-end labeling (TUNEL) assay kit (Beyotime, Shanghai, China). After transfection for 48 h, cells were fixed with 4% paraformaldehyde for 30 min at room temperature. After washing, 500 µL of 0.3% Triton X-100 was added to each well. Cells were then added with 50 µL of TUNEL detection liquid and incubated in the dark at 37°C for 60 min. Finally, the cells were stained with Hoechst33258. The number of TUNEL-stained cells was counted by Image-Pro Plus software.
Dual-Luciferase Reporter Assay
According to RNAhybrid (Krüger and Rehmsmeier, 2006) and sequencing data, we predicted the potential miRNAs of circRAB11A (Supplementary Figures S2A and S2B). DIANA, TargetScan and miRDB software (Matoušková et al., 2018; Li et al., 2021a) was used to predicted the potential target genes of miR-24-5p, the qPCR was used to identified the potential target genes (Supplementary Figures S2C and S2D). Then, we constructed wild type (WT) and mutant type (MT) by pmirGLO vector, such as pmirGLO-circRAB11A-WT, pmirGLO-circRAB11A-MT, pmirGLO-EGFR-WT-1, pmirGLO-EGFR-MT-1, pmirGLO-EGFR-WT-2 and pmirGLO-EGFR-MT-2. These vectors were synthesized by Sangon Biotech (Shanghai, China). The WT and MT plasmids were co-transfected into DF-1 cells with miR-24-5p mimic or mimic NC. Briefly, after 48 h of transfection, DF-1 cells were lysed and the relative luciferase activities were detected using the Dual-Luciferase Reporter Assay Kit (Beyotime) according to the manufacturer's instructions.
Statistical Analysis
We used independent samples t-test to measure the data from 2 groups, and one-way ANOVA for multiple comparisons. All data in this study were presented as mean ± standard error (SEM). The SPSS 22 (SPSS, Inc., Chicago, IL) was used to analyze the data and GraphPad Prism 8 (GraphPad Software, San Diego, CA) was used to present the figures. Each experiment was performed in triplicate. ∗ P < 0.05, ∗∗ P < 0.01, and a-e P < 0.05 were set as statistical significance.
RESULTS
Identification and Validation of circRAB11A
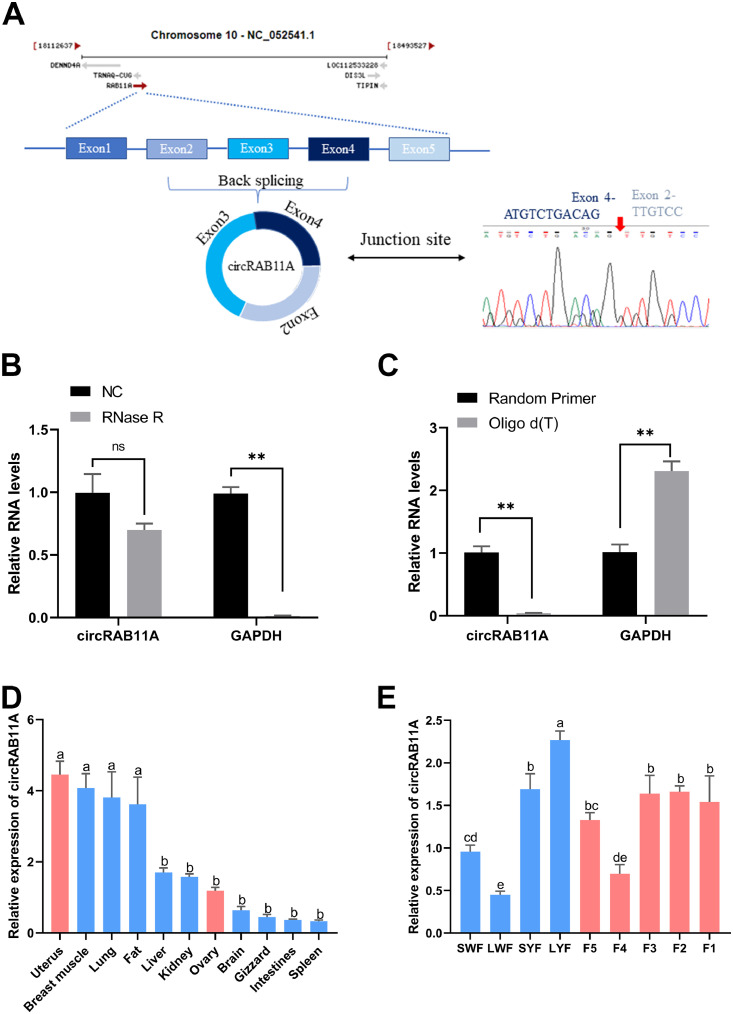
Through a comprehensive circRNA sequencing and bioinformatic analysis (Supplementary Figure S1), we uncovered that circRAB11A is a 471 bp circular RNA generated through the reverse splicing of the RAB11A gene between exons 2 and 4, and is an RNA macromolecule. To validate this finding, we used the Sanger sequencing technology to confirm that circRAB11A is indeed formed through the reverse splicing of exons 2 and 4 of the RAB11A gene (Figure 1A). To assess the stability of circRAB11A, we performed qPCR amplification with and without RNase R digestion. The results indicated that the linear gene GAPDH was significantly reduced (P < 0.01, Figure 1B), but circRAB11A was not significantly altered compared to the control group. Additionally, our reverse transcription reaction revealed that random primers promoted circRAB11A cDNA synthesis more efficiently than Oligo d(T) (P < 0.01, Figure 1C). After confirming the circular structure of circRAB11A, we conducted a qPCR study to assess its expression levels in various tissues and different grades of follicles. The results indicated that circRAB11A was highly expressed in the uterus (a, b P < 0.05, Figure 1D), and its expression was found to be relatively stable in the graded follicles (a-e P < 0.05, Figure 1E).
Figure 1.
Identification and validation of circRAB11A. (A) Schematic diagram of circRAB11A. (B) The expression of circRAB11A and GAPDH in GC after treatment with or without RNase R. n = 9. (C) The expression of circRAB11A and GAPDH in the cDNA of GC generated after treatment with random primer or Oligo d (T). n = 9. (D) The expression of circRAB11A in different chicken tissues. n=9. (E) The expression of circRAB11A in pregrade follicle and grade follicle. n = 9. Values represent mean ± SEM.∗P < 0.05, ∗∗P < 0.01, and a-eP < 0.05.
CircRAB11A Promotes Proliferation and Inhibits Apoptosis of GCs
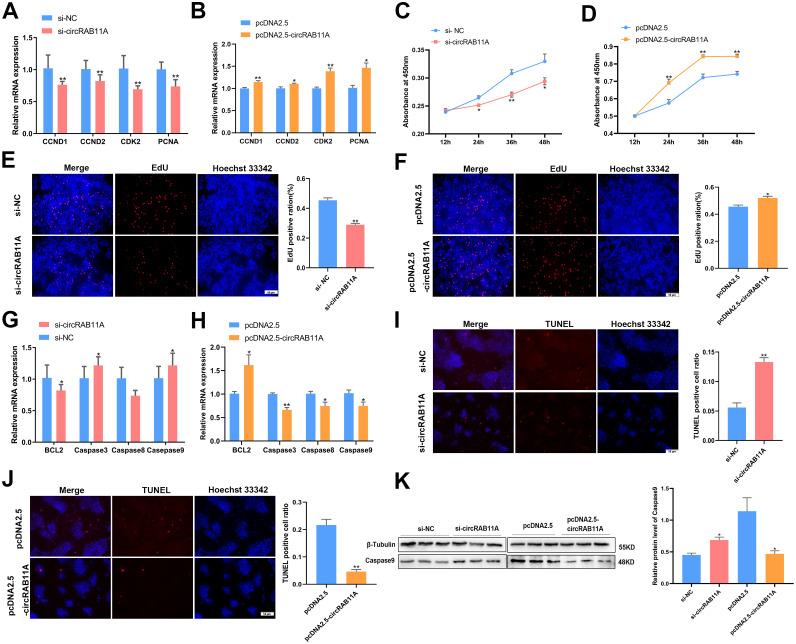
To examine the potential impact of circRAB11A on the proliferation and apoptosis of GCs, we introduced siRNA and overexpression vectors into the GCs (Supplementary Figures S3A and S3B). The qPCR assay revealed that the mRNA levels of CCND1, CCND1, CDK2, and PCNA, which was proliferation-related genes decreased following transfection with si-circRAB11A (P < 0.01, Figure 2A), while the opposite expression of CCND1, CCND1, CDK2, and PCNA was observed with circRAB11A overexpression (P < 0.05, Figure 2B). The CCK-8 assay showed that the proliferation ability of cells transfected with si-circRAB11A was consistently lower than the control group and significant decrease at 24, 36, and 48h time points (P < 0.05, Figure 2C). Conversely, after circRAB11A overexpression, the cell proliferation ability significantly increased compared to the control group at 24, 36, and 48h and continued to rise with time (P < 0.01, Figure 2D). Our EdU assay results indicated that DNA replication activity decreased upon circRAB11A knockdown, but increased after transfection with the circRAB11A vector (P < 0.05, Figures 2E and 2F). We also investigated circRAB11A's effect on GC apoptosis. Results from our qPCR assay indicated that mRNA expression of Caspase3 and Caspase9 decreased with circRAB11A overexpression, but increased in cells transfected with circRAB11A siRNA (P < 0.05, Figures 2G and 2H). The mRNA expression of BCL2 (P < 0.05, Figures 2G and 2H) was opposite to that of the Caspase3 and Caspase9, which corresponds to their respective functions. The TUNEL experiment showed that circRAB11A knockdown or overexpression significantly increased or decreased the number of TUNEL-positive cells compared to the control cells (P < 0.01, Figure 2I and 2J). The western blot assay showed that circRAB11A knockdown increased Caspase9 protein levels, while circRAB11A overexpression reduced them (P < 0.05, Figure 2K). Together, these results suggest that circRAB11A promotes GC proliferation and inhibits GC apoptosis.
Figure 2.
CircRAB11A promoted proliferation and inhibited apoptosis of GCs. (A, B) After transfection with circRAB11A interference or overexpression vector, the mRNA levels of CCND1, CCND2, CDK2, and PCNA were assessed by qPCR. n = 9. (C, D) After transfection with circRAB11A interference or overexpression vector, the growth curve of GCs was evaluated by CCK-8 reagent. n = 8. (E, F) The proliferation status of GCs determined by EdU assay after transfection of circRAB11A. n=3. (G, H) After transfection with circRAB11A interference or overexpression vector, the mRNA levels of BCL2, Caspase3, Caspase8 and Caspase9 were assessed by qPCR. n = 9. (I, J) The apoptosis state of GCs determined by TUNEL assay after transfection of circRAB11A. n = 3. (K) After transfection with circRAB11A interference or overexpression vector, the protein level of β-tubulin, Caspase9 was assessed by western blot. n = 3. Values represent mean ± SEM. * P < 0.05 and ⁎⁎P < 0.01.
CircRAB11A is a Possible Sponge for miR-24-5p
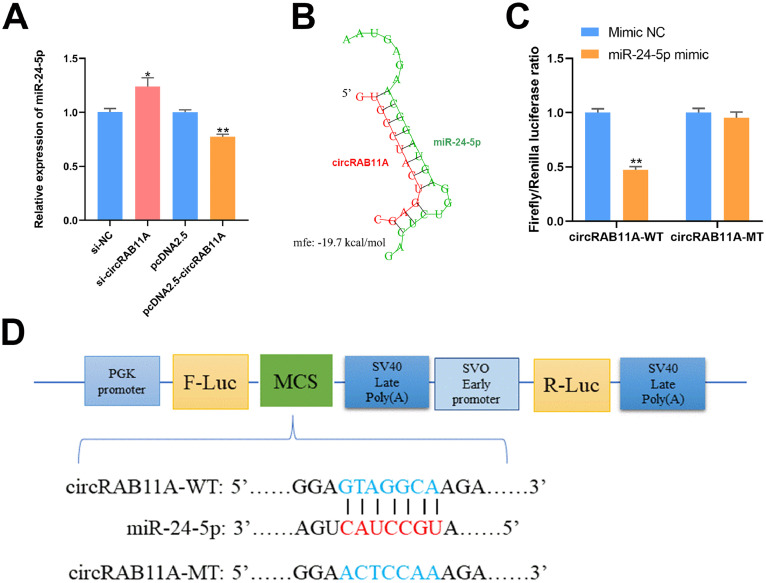
To understand circRAB11A's ability to bind miRNAs, we used RNAhybrid software to identify miRNAs targeted by circRAB11A. After modifying the level of circRAB11A expression through knockdown or overexpression, we analyzed the expression of these miRNAs in GCs. Sequencing data showed that circRAB11A was highly expressed in normal follicles, while miR-24-5p was highly expressed in atresia follicles (Supplementary Figure S2). Our research results indicated that knockdown or overexpression of circRAB11A corresponds to upregulation or downregulation of miR-24-5p mRNA expression (P < 0.05, Figure 3A). The molecular structure revealed a stable binding site between circRAB11A and miR-24-5p (Figure 3B). To further test this interaction, we conducted a luciferase activity experiment using a luciferase expression vector containing circRAB11A that directly binds and regulates miR-24-5p (Figure 3D). The luciferase reporter vector containing the circRAB11A wild-type sequence showed a significant reduction in activity when treated with the miR-24-5p mimic, compared to the control groups. However, the inhibitory effects were completely abolished in DF-1 cells when the putative miR-24-5p binding sites were mutated (P < 0.01, Figure 3C). Finally, the ability of miR-24-5p to interfere with or enhance expression was determined through qPCR (Supplementary Figures S3C and S3D).
Figure 3.
CircRAB11A could bind to miR-24-5p. (A) After transfection with circRAB11A interference or overexpression vector, the mRNA level of the predicted target miRNA of circRAB11A in GCs. n = 9. (B) RNAhybrid software shows the binding structure of miR-24-5p to circRAB11A. (C) Examination of the targeting relationship between circRAB11A and miR-24-5p using a dual-luciferase reporter after transfection of DF-1 cells with circRAB11A-WT/circRAB11A-MT or miR-24-5p mimic/mimic NC. n = 3. (D) Schematic of the circRAB11A-WT or circRAB11A-MT miR-24-5p target site designed for luciferase reporter assays. Values represent mean ± SEM. * P < 0.05 and ⁎⁎P < 0.01.
miR-24-5p resists GC Proliferation and Promotes Apoptosis
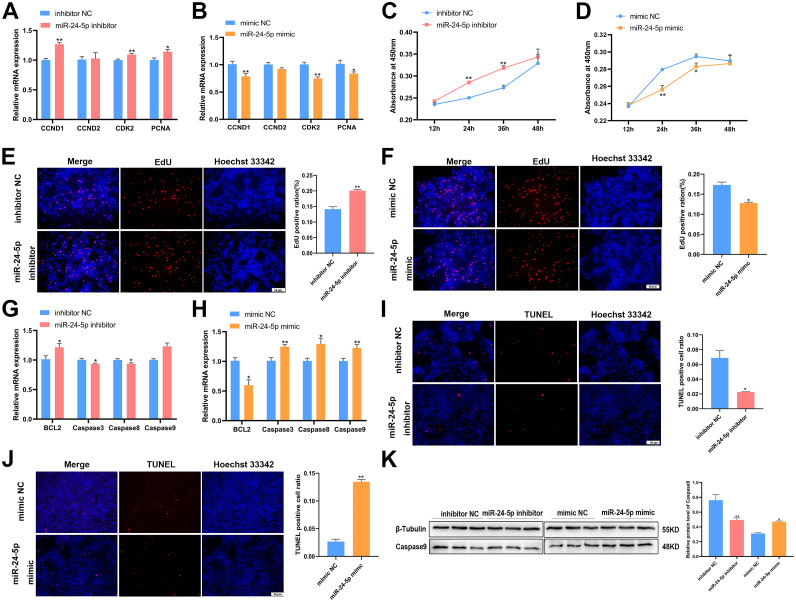
To understand the impact of miR-24-5p on GC proliferation and apoptosis, we manipulated its expression in GCs using miR-24-5p inhibitors or mimics. The results of qPCR showed that the mRNA levels of CCND1, CDK2, and PCNA were increased by the miR-24-5p inhibitor, while they decreased with the miR-24-5p mimic (P < 0.05, Figures 4A and 4B). Neither miR-24-5p inhibitor treatment nor miR-24-5p mimic showed significant expression of CCND2 (P > 0.05, Figures 4A and 4B). This was further confirmed by the CCK-8 and EdU assays, which showed that the miR-24-5p inhibitor promoted GC proliferation and the miR-24-5p mimic inhibited it (P < 0.05, Figures 4C and 4D; P < 0.05, Figures 4E and 4F). However, after miR-24-5p mimic treatment of GCs, the result of CCK-8 was not significant at 36h and 48h, which is a confusing experimental result and requires further experimental exploration. To evaluate the effects of miR-24-5p on GC apoptosis, we first found that the mRNA levels of apoptosis-related genes Caspase3, Caspase8 were decreased with the miR-24-5p inhibitor and increased with miR-24-5p overexpression (P < 0.05, Figures 4G and 4H). Conversely, the mRNA expression of BCL2 was upregulated after miR-24-5p inhibitor treatment, while was downregulated with miR-24-5p mimic. The TUNEL assay revealed that miR-24-5p mimic increased the number of necrotic cells, while the miR-24-5p inhibitor reduced it (P < 0.05, Figures 4I and 4J). Western blot analysis showed that miR-24-5p knockdown decreased the protein level of Caspase9, while miR-24-5p overexpression increased it (P < 0.05, Figure 4K). These findings demonstrate that miR-24-5p can inhibit proliferation and promote apoptosis in GCs.
Figure 4.
MiR-24-5p resists proliferation and promotes apoptosis of GCs. (A, B) After transfection with miR-24-5p inhibitor or mimic, the mRNA levels of CCND1, CCND2, CDK2 and PCNA were assessed by qPCR. n = 9. (C, D) After transfection with miR-24-5p inhibitor or mimic, the growth curve of GCs was evaluated by CCK-8 reagent. n = 8. (E, F) The proliferation status of GCs was determined by EdU assay after transfection of miR-24-5p. n = 3. (G, H) After transfection with miR-24-5p inhibitor or mimic, the mRNA levels of BCL2, Caspase3, Caspase8 and Caspase9 were assessed by qPCR. n = 9. (I, J) The apoptosis state of GCs determined by TUNEL assay after transfection of miR-24-5p 48h. n = 3. (K) After transfection with miR-24-5p inhibitor or mimic, the protein level of β-tubulin, Caspase9 was measured by western blot. n = 3. Values represent mean ± SEM. * P < 0.05 and ⁎⁎P < 0.01.
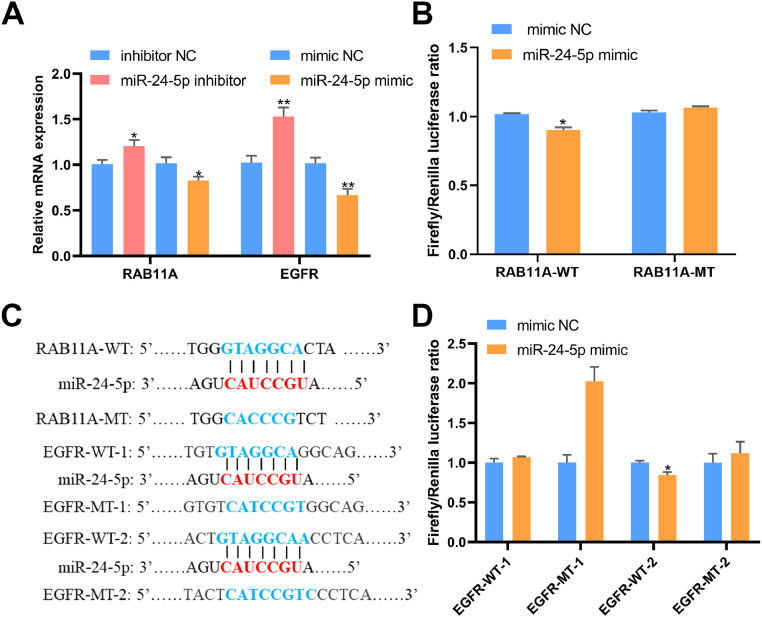
RAB11A and EGFR are Direct Target Genes of miR-24-5p in GCs
We employed DIANA, TargetScan and miRDB to identify potential targets of miR-24-5p and found that RAB11A and EGFR may be regulated by it. Our qPCR results showed that the mRNA levels of RAB11A and EGFR were increased or decreased following treatment with miR-24-5p inhibitor or mimic, respectively (P < 0.05, Figure 5A). Further analysis showed that RAB11A might be a strong target of miR-24-5p as its seed region matches the 3′UTR of RAB11A. To confirm this, we introduced wild-type and mutated RAB11A 3′UTR sequences into a vector and co-transfected it with miR-24-5p mimic or NC. The results showed that the luciferase activity of RAB11A 3′UTR wild-type was significantly decreased following treatment with miR-24-5p mimic, while there was no change with the mutated vector (P < 0.05, Figure 5B). Additionally, the miR-24-5p seed region matches 2 regions of the 3′UTR of EGFR. To verify this, we constructed dual luciferase reporter vectors with the full length of EGFR 3′UTR wild-type or a mutated version without miR-24-5p binding sites (Figure 5C). The results showed a significant reduction in luciferase reporter activity in cells co-transfected with miR-24-5p mimic and the wild-type vector, but not with the mutated vector (P < 0.05, Figure 5D). These findings provide evidence that RAB11A and EGFR are directly regulated by miR-24-5p.
Figure 5.
MiR-24-5p could be combined with RAB11A or EGFR. (A) After transfection with RAB11A interference or overexpression vector, mRNA levels of potential miR-24-5p target genes in GCs. n = 9. (B) Examination of the targeting relationship between miR-24-5p and RAB11A using a dual-luciferase reporter after transfection with RAB11A-WT/RAB11A-MT or miR-24-5p mimic/ mimic NC. n = 3. (C) Illustration of base pairing between miR-24-5p and RAB11A 3′UTR or miR-24-5p and EGFR 3′UTR. (D) Examination of the targeting relationship between miR-24-5p and EGFR using a dual-luciferase reporter after transfection of DF-1 cells with EGFR-WT-1/EGFR-MT-1 (EGFR-WT-2/EGFR-MT-2) or miR-24-5p mimic/ mimic NC. n = 3. Values represent mean ± SEM. * P < 0.05 and ** P < 0.01.
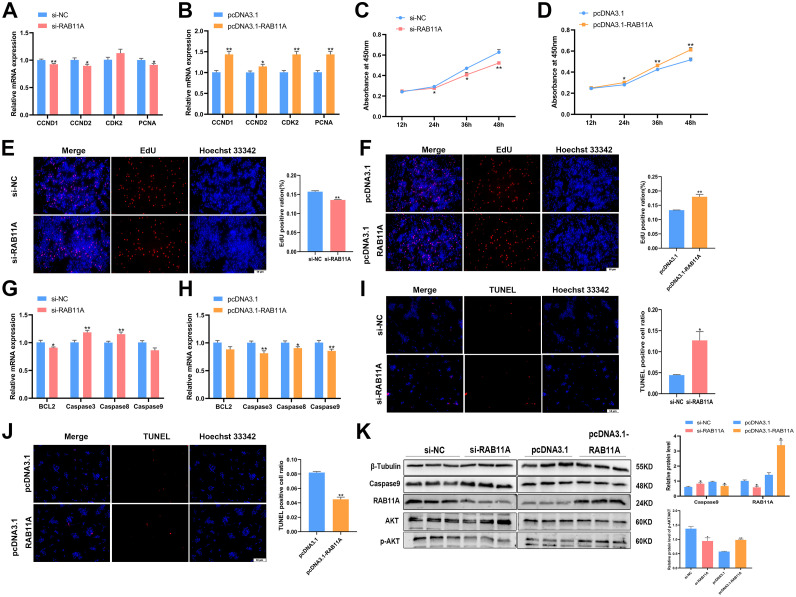
RAB11A Advances GC Proliferation and Suppresses Apoptosis
To understand the impact of RAB11A on GCs, we generated RAB11A overexpression and knockdown constructs (Supplementary Figures S3E and S3F). After transfecting with the pcDNA3.1-RAB11A vector or si-RAB11A, we evaluated the changes in GC proliferation and apoptosis. The qPCR and CCK-8 assay indicated that RAB11A promotes GC proliferation (P < 0.05, Figures 6A–6D). The EdU assay gave similar results with the RAB11A transfected cells (P < 0.01, Figures 6E and 6F). In contrast, qPCR analysis revealed that RAB11A reduces GC apoptosis (P < 0.05, Figures 6G and 6H). Similarly, the TUNEL and western blot assays also confirmed that RAB11A knockdown increases the rate of apoptosis, while RAB11A overexpression suppresses it (P < 0.05, Figures 6I, 6J and 6K). RAB11A has been reported as an activator of the PI3K/AKT signaling pathway (Zhang et al., 2020b). To determine if RAB11A activates this pathway in chicken GCs, we examined its effect on the expression of phosphorylated AKT. Our results from western blot analysis showed that overexpression of RAB11A led to an increase in phosphorylated AKT expression, suggesting that RAB11A activates the PI3K/AKT signaling pathway (Fig. 6K).
Figure 6.
RAB11A advances proliferation and resists apoptosis of GCs. (A, B) After transfection with RAB11A interference or overexpression vector, the mRNA levels of CCND1, CCND2, CDK2 and PCNA were assessed by qPCR. n = 9. (C, D) After transfection with RAB11A interference or overexpression vector, the growth curve of GCs was evaluated by CCK-8 reagent. n = 8. (E, F) The proliferation status of GCs determined by EdU assay after transfection of RAB11A. n = 3. (G, H) After transfection with RAB11A interference or overexpression vector, the mRNA levels of BCL2, Caspase3, Caspase8 and Caspase9 were assessed by qPCR. n = 9. (I, J) The apoptosis state of GCs determined by TUNEL assay after transfection of RAB11A. n = 3. (K) After transfection with RAB11A interference or overexpression vector, the protein levels of β-tubulin, Caspase9, RAB11A, AKT and p-AKT were measured by western bolt. n = 3. Values represent mean ± SEM. * P < 0.05 and ** P < 0.01.
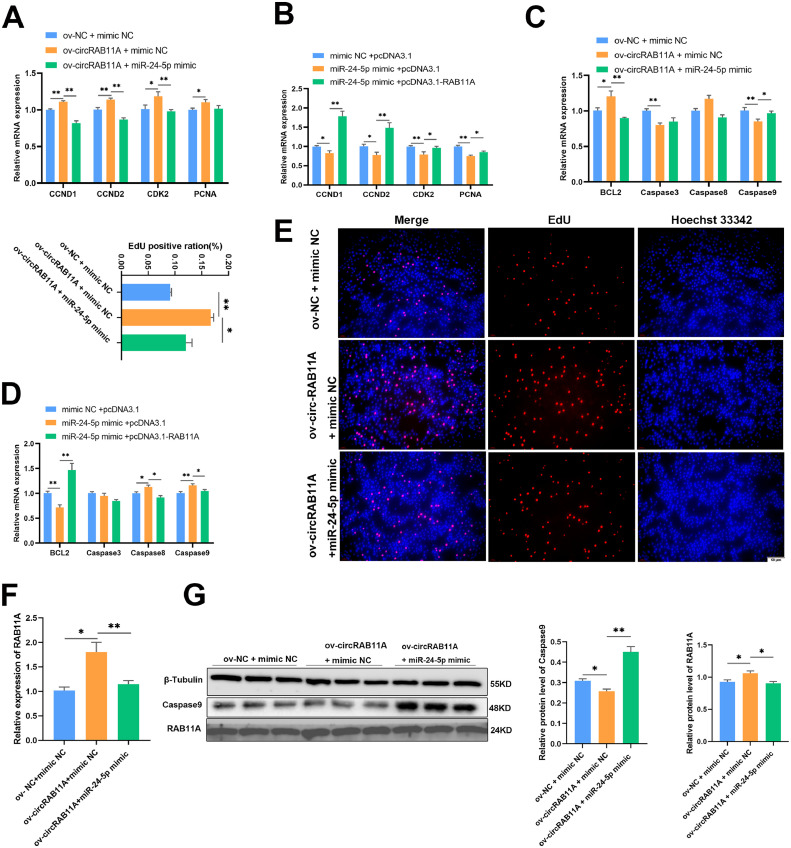
CircRAB11A Regulates GC Proliferation and Apoptosis via miR-24-5p/RAB11A Axis
To determine if circRAB11A influences chicken GC proliferation and apoptosis through the miR-24-5p/RAB11A pathway, we performed qPCR and EdU assays. Our results showed that circRAB11A overexpression significantly increased GC proliferation, which was reversed by the addition of miR-24-5p mimic (P < 0.05, Figures 7A and 7E). Cotransfection with miR-24-5p mimic and RAB11A vector reduced GC proliferation, and this effect was partially reversed by the addition of pcDNA3.1-RAB11A (P < 0.05, Figure 7B). We also evaluated the effect of circRAB11A on GC apoptosis using qPCR and western blot assays. The results showed that circRAB11A reduced GC apoptosis, while co-transfection with circRAB11A vector and miR-24-5p mimic increased the expression of Caspase9 (P < 0.05, Figure 7C). The mRNA level of Caspase9 was highly increased after co-transfection with miR-24-5p mimic and pcDNA3.1 compared to co-transfection with miR-24-5p mimic and RAB11A vector (P < 0.01, Figure 7D). Furthermore, our data showed that circRAB11A increased the expression of RAB11A at both the mRNA and protein levels after co-transfection with mimic NC (P < 0.05, Figures 7F and 7G). These results demonstrate that circRAB11A regulates chicken GC proliferation and apoptosis through the miR-24-5p/RAB11A pathway.
Figure 7.
CircRAB11A regulates GC proliferation and apoptosis through the miR-24-5p/RAB11A axis. (A, B) The mRNA levels of CCND1, CCND2, CDK2 and PCNA were measured by qPCR after co-overexpression. n = 9. (C, D) The mRNA levels of BCL2, Caspase3, Caspase8 and Caspase9 were measured by qPCR after co-overexpression. n = 9. (E) The proliferation status of GCs was determined by EdU assay after co-overexpression. n = 3. (F) The mRNA level of RAB11A was measured by qPCR after co-overexpression. n = 9. (G) The protein levels of β-tubulin, Caspase9 and RAB11A were measured by western bolt after co-overexpression. n = 3. Values represent mean ± SEM. * P < 0.05 and ⁎⁎P < 0.01.
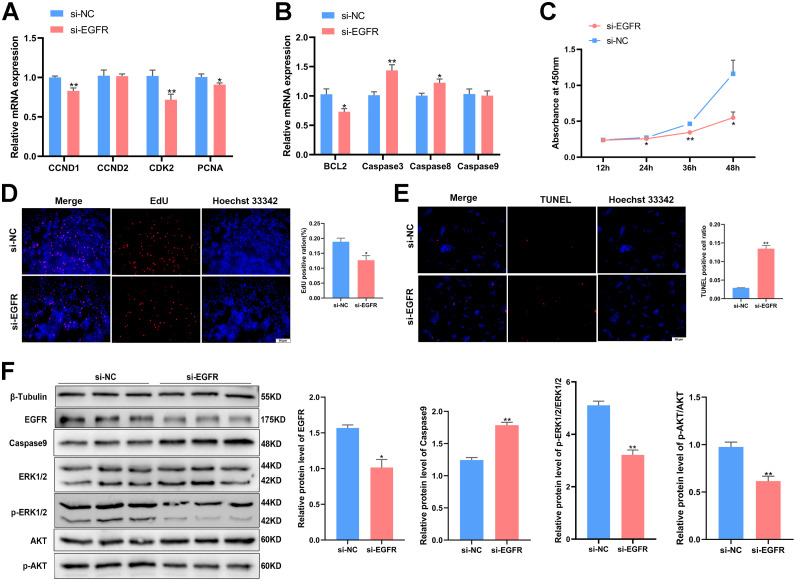
EGFR Regulates GC Proliferation and Apoptosis by Activating ERK1/2 and PI3K/AKT Signaling Pathways
To uncover the role of EGFR in GCs, we utilized siRNA to knock down EGFR expression (Supplementary Figure S3G). After transfection with si-EGFR, we measured the expression of genes related to proliferation and apoptosis. Interestingly, we observed a decrease in the mRNA levels of proliferation-related genes and an increase in the mRNA levels of apoptosis-related genes (P < 0.05, Figures 8A and 8B). Our CCK-8 and EdU assays showed a decrease of GC proliferation after transfection si-EGFR (P < 0.05, Figures 8C and 8D), while the TUNEL assay revealed that GC apoptosis was promoted with si-EGFR (P < 0.05, Figure 8E). The protein level of Caspase9 increased after EGFR knockdown (P < 0.01, Figure 8F). We also investigated the downstream signaling pathways of EGFR, including ERK1/2 and PI3K/AKT. Western blot analysis revealed that when EGFR was knocked down in GCs, there was also a decrease in the expression of EGFR, p-ERK1/2 and p-AKT, while the expression of ERK1/2, AKT remained unchanged (P < 0.05, Figure 8F). These results support the conclusion that EGFR specifically activates the ERK1/2 and PI3K/AKT signaling pathways to regulate GC proliferation and apoptosis.
Figure 8.
CircRAB11A regulates GC proliferation and apoptosis through ERK1/2 and PI3K/AKT signaling pathways by targeting EGFR. (A) After transfection with EGFR interference vector, the mRNA levels of CCND1, CCND2, CDK2 and PCNA were assessed by qPCR. n = 9. (B) After transfection with EGFR interference vector, the mRNA levels of BCL2, Caspase3, Caspase8 and Caspase9 were measured by qPCR. n = 9 (C) After transfection with EGFR interference vector, the growth curves of GC were assessed by CCK-8 reagent. n = 8. (D) The proliferation state of GCs was determined by EdU assay after transfection of si-EGFR. n = 3. (E) The apoptosis state of GCs was determined by TUNEL assay after transfection of si-EGFR. n = 3. (F) After transfection with EGFR interference vector, the protein levels of β-tubulin, EGFR, Caspase9, ERK1/2, p-ERK1/2, AKT and p-AKT were assessed by western blot. n=3. Values represent mean ± SEM. * P < 0.05 and ⁎⁎P < 0.01.
CircRAB11A Regulates Proliferation and Apoptosis of GCs Through miR-24-5p/EGFR Axis
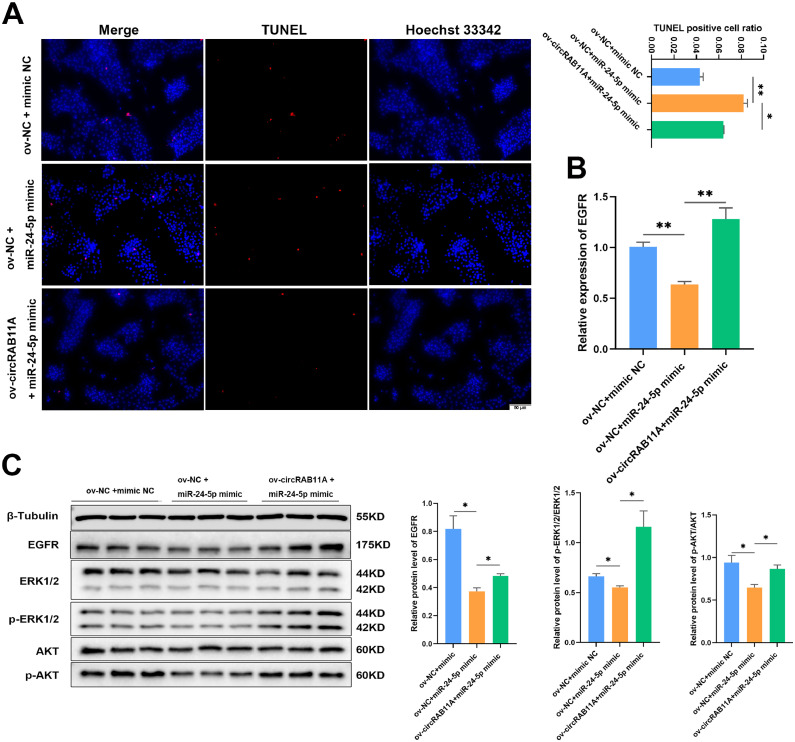
To examine the impact of circRAB11A on the regulation of the miR-24-5p/EGFR axis and ovarian function. The TUNEL assay showed that the miR-24-5p mimic increased DNA death, but the effect was countered by the overexpression of circRAB11A in GC apoptosis (P < 0.05, Figure 9A). The qPCR results showed that miR-24-5p suppressed the expression of EGFR after co-transfection with ov-NC, while ov-circRAB11A increased EGFR expression after co-transfection with miR-24-5p mimic (P < 0.01, Figure 9B). In GCs, we observed that the miR-24-5p mimic decreased the phosphorylation of ERK1/2, and AKT, while not affecting the protein levels of ERK1/2 and AKT. However, after co-transfection with ov-circRAB11A and miR-24-5p mimic, the protein levels of EGFR, p-ERK1/2 and p-AKT increased (P < 0.05, Figure 9C). These results demonstrate that circRAB11A promotes GC proliferation and reduces GC apoptosis through the ERK1/2 and PI3K/AKT pathways by modulating EGFR expression.
Figure 9.
CircRAB11A regulates GC proliferation and apoptosis through the miR-24-5p/EGFR axis. (A) The apoptosis state of GCs determined by TUNEL assay after co-overexpression. n = 3. (B) The mRNA level of EGFR was measured by qPCR after co-overexpression. n = 9. (C) The protein levels of β-tubulin, EGFR, ERK1/2, p-ERK1/2, AKT and p-AKT were measured by western bolt after co-overexpression. n = 3. Values represent mean ± SEM. * P < 0.05 and ⁎⁎P < 0.01.
DISCUSSION
The growth and development of ovarian follicles is closely linked to the growth status of GCs, which are the earliest-developing somatic cells in the follicles (Havelock, et al., 2004). The proliferation and apoptosis of GCs are regulated by many micro factors, such as genes, non-coding RNAs (including mirRNAs, long non-coding RNAs, circRNAs), signaling pathways and so on. It is becoming increasingly evident that numerous circRNAs have been identified as participating in the regulation of the physiological process of GCs proliferation and apoptosis.(Guo, et al., 2019; Xu, et al., 2021; Li, et al., 2023). Despite these findings, the function of circRNAs in chicken GCs is not well understood. Based on the previous circRNA sequencing data available in the group, it can be seen that the expression of circRAB11A was significantly higher in normal follicles than in atretic follicles. In this study, we identified circRAB11A as being generated from RAB11A and specifically enriched in uterus and ovary tissue of chicken, suggesting that circRAB11A may play roles in follicle development. Through a series of molecular biological technologies, we have ascertained that the circRAB11A overexpression increased the proliferation of chicken GCs, while inhibiting their apoptosis.
CircRNAs have been shown to play a crucial role in controlling gene expression through their ability to act as a sponge for miRNAs (Fu et al., 2018; Jia et al., 2018; Huang et al., 2020; Wu et al., 2022). In our study, several lines of evidence indicated that circRAB11A acts as a sponge for miR-24-5p to regulate RAB11A and EGFR expression. In addition, an increasing number of miRNAs are now known to be essential for follicle development. Our previous results showed that miR-23b-3p can inhibit chicken GC proliferation and steroid hormone synthesis by targeting GDF9 (Wei et al., 2022a). Gga-miR-146b-3p promotes apoptosis and attenuates autophagy by targeting AKT1 in chicken GCs (Wei et al., 2022b). Furthermore, miR-30a-5p inhibits GC death by targeting Beclin1 (He, et al., 2022). Here, our results further confirmed that miR-24-5p could inhibit GC proliferation and promote GC apoptosis by directly targeting RAB11A or EGFR in chicken GCs.
The role of miRNA in controlling gene expression and activating signaling pathways has been widely recognized (Li et al., 2023). RAB11A is a GTPase that regulates intracellular trafficking and is important in endometrial function (Kakar-Bhanot et al., 2019). A pervious study found that RAB11A could regulate ovarian cancer cells (Wang et al., 2022b; Wang et al., 2022c). On the other hand, EGFR (also known as ErbB1 or HER1), a single transmembrane protein and receptor tyrosine kinase (RTK), has been shown to regulate various functions of the ovary such as follicle development, oocyte maturation, GC proliferation, differentiation, and steroid production through its interaction with EGF (Zheng, et al., 2017; Zhang, et al., 2022). Additionally, EGFR often regulates the function of GCs through the ERK1/2 and PI3K/AKT pathways in chickens (Wang, et al., 2007; Lin et al., 2011; Wu, et al., 2019). In this study, we observed a reduction in protein expression of p-ERK1/2 after knockdown of EGFR, and a decrease in p-AKT expression after knockdown of both RAB11A and EGFR, suggesting that both RAB11A and EGFR can regulate GC proliferation and apoptosis through the PI3K/AKT pathway.
Furthermore, in this study the ERK1/2 and PI3K/AKT pathway was verified to be involved in GC proliferation and apoptosis by the circRAB11A/miR-24-5p/RAB11A axis and circRAB11A/miR-24-5p/EGFR axis. The ERK1/2 and PI3K/AKT cascade control the intracellular processes that are often involved in the regulation of circRNAs. For instance, circ-0008594 affects non-small-cell lung cancer (NSCLC) growth and lymph node metastasis and accelerates NSCLC progression by controlling the miR-760-mediated PI3K/AKT and MEK/ERK1/2 pathways (Wang et al., 2021). Mechanically, circCDYL2 preserved the activity of the downstream AKT and ERK1/2 by stopping the ubiquitination degradation of GRB7 and improving its interaction with FAK (Ling et al., 2022).
One further implication of this experiment is that RAB11A and EGFR may have a synergistic effect on GC proliferation and apoptosis. Palmieri et al. provide the first evidence that transient transfection of DN-Rab11a alters the output of signaling pathways and differentially modulates cellular proliferative and motility phenotypes in response to EGFR ligands in vitro. In terms of signaling, they reported that transfection of DN-RAB11A augmented ERK1/2 activation in response to EGF (Palmieri, et al., 2006). Additionally, Watanuki et al. demonstrated that afatinib-induced redistribution of EGFR to the cell surface, facilitated by Rab11a-dependent recycling, increased cetuximab binding to the receptor, thus enhancing its effect (Watanuki, et al., 2014). We conjecture that RAB11A may increase the effect of EGFR expression, yet additional experimental data is needed to prove it.
CONCLUSIONS
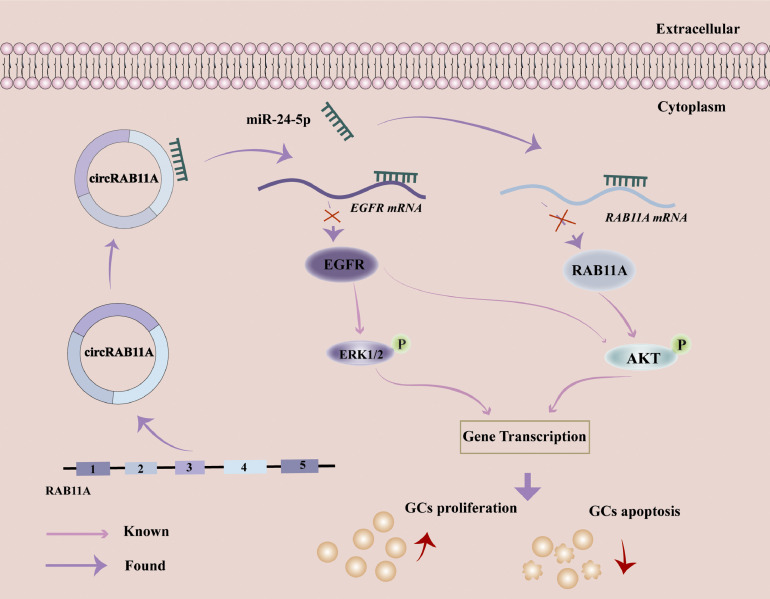
In this study, circRAB11A is a newly discovered transcriptional variant of RAB11A that is expressed in chicken GCs. Its key function is to act as a miR-24-5p sponge, which promotes GC proliferation and represses GC apoptosis by regulating the ERK1/2 and PI3K/AKT pathways. (Fig 10).
Figure 10.
Diagrammatic model of the role of circRAB11A in chicken GC development. circRAB11A promotes proliferation but suppresses apoptosis of chicken GCs by downregulating the expression of EGFR or RAB11A through PI3K/AKT and ERK1/2 signaling pathway.
DISCLOSURES
The authors declare no conflicts of interest.
ACKNOWLEDGMENTS
This research was funded by The National Key Research and Development Program of China, grant number 2021YFD1300600; China Agriculture Research System of MOF and MARA, grant number CARS-40; Sichuan Science and Technology Program, grant number 2021YFYZ0007, 2021YFYZ0031 and 2022YFYZ0005; and and Chengdu Science and Technology Program, grant number 2022-YF05-01014-SN and 2022-YF05-00703-SN.
Author's contributions; Qinyao Wei, Juan Li, Xinyan Li and Huadong Yin conceived and designed the experiments. Qinyao Wei, Juan Li, Xinyan Li, Jialing Xiang and Can Cui performed the experiments. Qinyao Wei, Yao Zhang and Cui Can analyzed the data. Qinyao Wei, Juan Li, Xinyan Li and Can Cui wrote this manuscript. Qinyao Wei, Huadong Yin reviewed and edited this manuscript. Huadong Yin secured the funding.
Consent for Publication: Not applicable.
Ethics Statement: All the animals used for this experiment are approved by the Animal Welfare Committee of Sichuan Agriculture University, and the approval number is 2020202028.
Availability of Data and Material: The datasets used and analyzed during the current study are available form the corresponding author on request.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2024.103841.
Appendix. Supplementary materials
REFERENCES
- Fu Y., Jiang H., Liu J.B., Sun X.L., Zhang Z., Li S., Gao Y., Yuan B., Zhang J.B. Genome-wide analysis of circular RNAs in bovine cumulus cells treated with BMP15 and GDF9. Sci. Rep. 2018;8:7944. doi: 10.1038/s41598-018-26157-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T., Zhang J., Yao W., Du X., Li Q., Huang L., Ma M., Li Q., Liu H., Pan Z. CircINHA resists granulosa cell apoptosis by upregulating CTGF as a ceRNA of miR-10a-5p in pig ovarian follicles. Biochimica et biophysica acta. Gene Regulatory Mechanisms. 2019;1862 doi: 10.1016/j.bbagrm.2019.194420. [DOI] [PubMed] [Google Scholar]
- Hao E.Y., Wang D.H., Chang L.Y., Huang C.X., Chen H., Yue Q.X., Zhou R.Y., Huang R.L. Melatonin regulates chicken granulosa cell proliferation and apoptosis by activating the mTOR signaling pathway via its receptors. Poult. Sci. 2020;99:6147–6162. doi: 10.1016/j.psj.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havelock J.C., Rainey W.E., Carr B.R. Ovarian granulosa cell lines. Mol. Cell. Endocrinol. 2004;228:67–78. doi: 10.1016/j.mce.2004.04.018. [DOI] [PubMed] [Google Scholar]
- He H., Li D., Tian Y., Wei Q., Amevor F.K., Sun C., Yu C., Yang C., Du H., Jiang X., Ma M., Cui C., Zhang Z., Tian K., Zhang Y., Zhu Q., Yin H. miRNA sequencing analysis of healthy and atretic follicles of chickens revealed that miR-30a-5p inhibits granulosa cell death via targeting Beclin1. J. Anim. Sci. Biotechnol. 2022;13:55. doi: 10.1186/s40104-022-00697-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Wu B., Chen M., Hong L., Kong P., Wei Z., Teng X. Depletion of exosomal circLDLR in follicle fluid derepresses miR-1294 function and inhibits estradiol production via CYP19A1 in polycystic ovary syndrome. Aging. 2020;12:15414–15435. doi: 10.18632/aging.103602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W., Xu B., Wu J. Circular RNA expression profiles of mouse ovaries during postnatal development and the function of circular RNA epidermal growth factor receptor in granulosa cells. Metabolism. 2018;85:192–204. doi: 10.1016/j.metabol.2018.04.002. [DOI] [PubMed] [Google Scholar]
- Kakar-Bhanot R., Brahmbhatt K., Chauhan B., Katkam R.R., Bashir T., Gawde H., Mayadeo N., Chaudhari U.K., Sachdeva G. Rab11a drives adhesion molecules to the surface of endometrial epithelial cells. Human Reprod. (Oxford, England) 2019;34:519–529. doi: 10.1093/humrep/dey365. [DOI] [PubMed] [Google Scholar]
- Kolakofsky D. Isolation and characterization of Sendai virus DI-RNAs. Cell. 1976;8:547–555. doi: 10.1016/0092-8674(76)90223-3. [DOI] [PubMed] [Google Scholar]
- Krüger J., Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Ning C., Zhang J., Wang Y., Tang Q., Kui H., Wang T., He M., Jin L., Li J., Lin Y., Zeng B., Yin H., Zhao X., Zhang Y., Xu H., Zhu Q., Li M. Dynamic transcriptome and chromatin architecture in granulosa cells during chicken folliculogenesis. Nature Commun. 2022;13:131. doi: 10.1038/s41467-021-27800-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Liang J., Wang J., Han J., Li S., Huang K., Liu C. Mex3a promotes oncogenesis through the RAP1/MAPK signaling pathway in colorectal cancer and is inhibited by hsa-miR-6887-3p. Cancer Commun. (London, England) 2021;41:472–491. doi: 10.1002/cac2.12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Si S.J., Wu X., Zhang Z.H., Li C., Tao Y.Q., Yang P.K., Li D.H., Li Z.J., Li G.X., Liu X.J., Tian Y.D., Kang X.T. CircEML1 facilitates the steroid synthesis in follicular granulosa cells of chicken through sponging gga-miR-449a to release IGF2BP3 expression. Genomics. 2023;115 doi: 10.1016/j.ygeno.2022.110540. [DOI] [PubMed] [Google Scholar]
- Li X., Gao F., Fan Y., Xie S., Li C., Meng L., Li L., Zhang S., Wei H. A novel identified circ-ANKHD1 targets the miR-27a-3p/SFRP1 signaling pathway and modulates the apoptosis of granulosa cells. Environ. Sci. Pollut. Res. Int. 2021;28:57459–57469. doi: 10.1007/s11356-021-14699-4. [DOI] [PubMed] [Google Scholar]
- Lin J.X., Jia Y.D., Zhang C.Q. Effect of epidermal growth factor on follicle-stimulating hormone-induced proliferation of granulosa cells from chicken prehierarchical follicles. J. Zhejiang Univ. Sci. B. 2011;12:875–883. doi: 10.1631/jzus.B1100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., Gong Y., Sun H., Yang C., Tang Y., Yin L., Zhang D., Wang Y., Yu C., Liu Y. Lipid deposition and progesterone synthesis are increased by miR-181b-5p through RAP1B/ERK1/2 pathway in chicken granulosa cells. J. Agric. Food Chem. 2023;71:12910–12924. doi: 10.1021/acs.jafc.3c03178. [DOI] [PubMed] [Google Scholar]
- Ling Y., Liang G., Lin Q., Fang X., Luo Q., Cen Y., Mehrpour M., Hamai A., Liu Z., Shi Y., Li J., Lin W., Jia S., Yang W., Liu Q., Song E., Li J., Gong C. circCDYL2 promotes trastuzumab resistance via sustaining HER2 downstream signaling in breast cancer. Mol. Cancer. 2022;21:8. doi: 10.1186/s12943-021-01476-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoušková P., Hanousková B., Skálová L. MicroRNAs as potential regulators of glutathione peroxidases expression and their role in obesity and related pathologies. Int. J. Mol. Sci. 2018;19:1199. doi: 10.3390/ijms19041199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri D., Bouadis A., Ronchetti R., Merino M.J., Steeg P.S. Rab11a differentially modulates epidermal growth factor-induced proliferation and motility in immortal breast cells. Breast Cancer Res. Treat. 2006;100:127–137. doi: 10.1007/s10549-006-9244-6. [DOI] [PubMed] [Google Scholar]
- Panda A.C. Circular RNAs Act as miRNA Sponges. Adv. Exp. Med. Biol. 2018;1087:67–79. doi: 10.1007/978-981-13-1426-1_6. [DOI] [PubMed] [Google Scholar]
- Peng F., Gong W., Li S., Yin B., Zhao C., Liu W., Chen X., Luo C., Huang Q., Chen T., Sun L., Fang S., Zhou W., Li Z., Long H. circRNA_010383 acts as a sponge for miR-135a, and its downregulated expression contributes to renal fibrosis in diabetic nephropathy. Diabetes. 2021;70:603–615. doi: 10.2337/db20-0203. [DOI] [PubMed] [Google Scholar]
- Wang H., Zhang Y., Zhang J., Du X., Li Q., Pan Z. circSLC41A1 Resists Porcine Granulosa Cell Apoptosis and Follicular Atresia by Promoting SRSF1 through miR-9820-5p Sponging. Int. J. Mol. Sci. 2022;23:1509. doi: 10.3390/ijms23031509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Yan C., Zhang P., Li G., Zhu R., Wang H., Wu L., Xu G. Microarray identifies a key carcinogenic circular RNA 0008594 that is related to non-small-cell lung cancer development and lymph node metastasis and promotes NSCLC progression by regulating the miR-760-mediated PI3K/AKT and MEK/ERK pathways. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.757541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li J., Ying Wang C., Yan Kwok A.H., Leung F.C. Epidermal growth factor (EGF) receptor ligands in the chicken ovary: I. Evidence for heparin-binding EGF-like growth factor (HB-EGF) as a potential oocyte-derived signal to control granulosa cell proliferation and HB-EGF and kit ligand expression. Endocrinology. 2007;148:3426–3440. doi: 10.1210/en.2006-1383. [DOI] [PubMed] [Google Scholar]
- Wang Y., Li N., Zhao J., Dai C. MiR-193a-5p serves as an inhibitor in ovarian cancer cells through RAB11A. Reprod. Toxicol. (Elmsford, N.Y.) 2022;110:105–112. doi: 10.1016/j.reprotox.2022.04.003. [DOI] [PubMed] [Google Scholar]
- Wang Y., Ren Y., Li N., Zhao J., Zhao S. Rab11a promotes the malignant progression of ovarian cancer by inducing autophagy. Genes Genomics. 2022;44:1375–1384. doi: 10.1007/s13258-022-01314-0. [DOI] [PubMed] [Google Scholar]
- Watanuki Z., Kosai H., Osanai N., Ogama N., Mochizuki M., Tamai K., Yamaguchi K., Satoh K., Fukuhara T., Maemondo M., Ichinose M., Nukiwa T., Tanaka N. Synergistic cytotoxicity of afatinib and cetuximab against EGFR T790M involves Rab11-dependent EGFR recycling. Biochem. Biophys. Res. Commun. 2014;455:269–276. doi: 10.1016/j.bbrc.2014.11.003. [DOI] [PubMed] [Google Scholar]
- Wei Q., Li J., He H., Cao Y., Li D., Amevor F.K., Zhang Y., Wang J., Yu C., Yang C., Du H., Jiang X., Zhu Q., Yin H. miR-23b-3p inhibits chicken granulosa cell proliferation and steroid hormone synthesis via targeting GDF9. Theriogenology. 2022;177:84–93. doi: 10.1016/j.theriogenology.2021.10.011. [DOI] [PubMed] [Google Scholar]
- Wei Q.Y., Xue H.Q., Sun C., Li J., He H., Amevor F.K., Tan B., Ma M., Tian K., Zhang Z., Zhang Y., He H., Xia L., Zhu Q., Yin H., Cui C. Gga-miR-146b-3p promotes apoptosis and attenuate autophagy by targeting AKT1 in chicken granulosa cells. Theriogenology. 2022;190:52–64. doi: 10.1016/j.theriogenology.2022.07.019. [DOI] [PubMed] [Google Scholar]
- Wu G., Xia J., Yang Z., Chen Y., Jiang W., Yin T., Yang J. CircASPH promotes KGN cells proliferation through miR-375/MAP2K6 axis in Polycystic Ovary Syndrome. J. Cell. Mol. Med. 2022;26:1817–1825. doi: 10.1111/jcmm.16231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Xiao H., Pi J., Zhang H., Pan A., Pu Y., Liang Z., Shen J., Du J. EGFR promotes the proliferation of quail follicular granulosa cells through the MAPK/extracellular signal-regulated kinase (ERK) signaling pathway. Cell Cycle (Georgetown, Tex.) 2019;18:2742–2756. doi: 10.1080/15384101.2019.1656952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Xiong F., Bai Y., Xiao J., Zhang Y., Chen J., Li Q. Circ_0043532 regulates miR-182/SGK3 axis to promote granulosa cell progression in polycystic ovary syndrome. Reprod. Biol. Endocrinol. 2021;19:167. doi: 10.1186/s12958-021-00839-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Gao X., Zhang M., Yan S., Sun C., Xiao F., Huang N., Yang X., Zhao K., Zhou H., Huang S., Xie B., Zhang N. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Institute. 2018;110:304–315. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura Y., Barua A. Female reproductive system and immunology. Adv. Exp. Med. Biol. 2017;1001:33–57. doi: 10.1007/978-981-10-3975-1_3. [DOI] [PubMed] [Google Scholar]
- Zhang J.H., Zhan L., Zhao M.Y., Wang J.J., Xie F.F., Xu Z.Y., Xu Q., Cao Y.X., Liu Q.W. Role of EGFR expressed on the granulosa cells in the pathogenesis of polycystic ovarian syndrome. Front. Endocrinol. 2022;13 doi: 10.3389/fendo.2022.971564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.Y., Gao X.H., Ma M.Y., Zhao C.L., Zhang Y.L., Guo S.S. CircRNA_101237 promotes NSCLC progression via the miRNA-490-3p/MAPK1 axis. Sci. Rep. 2020;10:9024. doi: 10.1038/s41598-020-65920-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.Y., Lu M., Liu Z.K., Li H., Yong Y.L., Zhang R.Y., Chen Z.N., Bian H. Rab11a regulates MMP2 expression by activating the PI3K/AKT pathway in human hepatocellular carcinoma cells. Pathol. Res. Pract. 2020;216 doi: 10.1016/j.prp.2020.153046. [DOI] [PubMed] [Google Scholar]
- Zheng Q., Li Y., Zhang D., Cui X., Dai K., Yang Y., Liu S., Tan J., Yan Q. ANP promotes proliferation and inhibits apoptosis of ovarian granulosa cells by NPRA/PGRMC1/EGFR complex and improves ovary functions of PCOS rats. Cell Death Dis. 2017;8:e3145. doi: 10.1038/cddis.2017.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng T., Gan M.L., Shen L.Y., Niu L.L., Guo Z.Y., Wang J.Y., Zhang S.H., Zhu L. circRNA on animal skeletal muscle development regulation. Yi chuan = Hereditas. 2020;42:1178–1191. doi: 10.16288/j.yczz.20-207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.