Abstract
CDH1 encodes for E-cadherin, and its loss of function is the hallmark of invasive lobular carcinoma (ILC). Albeit vanishingly rare, bi-allelic CDH1 alterations may be found in non-lobular breast carcinomas (NL-BCs). We sought to determine the clinicopathologic characteristics and repertoire of genetic alterations of NL-BCs harboring CDH1 bi-allelic genetic alterations. Analysis of 5,842 breast cancers (BCs) subjected to clinical tumor-normal sequencing with an FDA-cleared multi-gene panel was conducted to identify BCs with bi-allelic CDH1 pathogenic/likely pathogenic somatic mutations lacking lobular features. The genomic profiles of NL-BCs with CDH1 bi-allelic genetic alterations were compared to those of ILCs and invasive ductal carcinomas (IDCs), matched by clinicopathologic characteristics. Out of 896 CDH1-altered BCs, 889 samples were excluded based on diagnosis of invasive mixed ductal/lobular carcinoma or ILC or detection of monoallelic CDH1 alterations. Only 7 of 5,842 (0.11%) BCs harbored bi-allelic CDH1 alterations and lacked lobular features. Of these, 4/7 (57%) cases were ER-positive/HER2-negative, 1/7 (14%) was ER-positive/HER2-positive and 2/7 (29%) were ER-negative/HER2-negative. 5/7 (71%) were of Nottingham grade 2 and 2/7 (29%) were of grade 3. The NL-BCs with CDH1 bi-allelic genetic alterations included a mucinous carcinoma (n=1), IDCs with focal nested growth (n=2), IDC with solid papillary (n=1) or apocrine (n=2) features and an IDC of no special type (NST; n=1). E-cadherin expression as detected by immunohistochemistry was absent (3/5) or aberrant (discontinuous membranous/cytoplasmic/granular; 2/5). NL-BCs with CDH1 bi-allelic genetic alterations displayed recurrent genetic alterations including TP53, PIK3CA (57%, 4/7; each), FGFR1 and NCOR1 (28%, 2/7, each) alterations. As compared to CDH1-wildtype IDC-NSTs, NL-BCs less frequently harbored GATA3 mutations (0% vs 47%, p=0.03) but no significant differences were detected when compared to matched ILCs. NL-BCs with CDH1 bi-allelic genetic alterations are vanishingly rare, predominantly comprise IDCs with special histologic features, and have genomic features akin to luminal B ER-positive BCs.
Keywords: Breast Cancer, Lobular, Ductal, CDH1, E-cadherin
INTRODUCTION
Invasive lobular carcinoma (ILC), the most common special subtype of breast cancer, is characterized by discohesiveness of tumor cells due to inactivation of the cell adhesion protein E-cadherin, which is encoded by the CDH1 gene that maps to chromosome 16q22.1 1,2. In ILCs, E-cadherin loss of function (LOF) is caused by bi-allelic alterations affecting the CDH1 gene, including somatic or germline pathogenic mutations coupled with loss of heterozygosity (LOH) of the wild-type allele, CDH1 promoter hypermethylation, or CDH1 homozygous deletions 3.
Approximately 90% of CDH1-altered invasive breast carcinomas can be histologically classified as ILC 4. The remaining (~10%) CDH1-altered breast cancers have been reported to correspond primarily to invasive mammary carcinoma with mixed ductal and lobular features 4–6. Notwithstanding the strong association between CDH1 pathogenic alterations and the lobular phenotype, a small subset of non-lobular invasive breast cancers (NL-BCs) may harbour inactivating bi-allelic CDH1 genetic alterations and loss of E-cadherin protein expression 6; their histologic and genetic characteristics, however, have yet to be characterized. Here, we sought to determine the clinicopathological characteristics and repertoire of genetic alterations of NL-BCs harboring CDH1 pathogenic/likely pathogenic bi-allelic genetic alterations (NL-BCs with CDH1 bi-allelic genetic alterations).
MATERIALS AND METHODS
Case selection and histologic assessment
This study was approved by the Institutional Review Board of Memorial Sloan Kettering Cancer Center (MSK). We retrospectively investigated the presence of LOF genetic alterations affecting CDH1 in targeted sequencing data from 5,842 breast cancers previously subjected to the FDA-cleared MSK Integrated Mutation Profiling of Actionable Targets (MSK-IMPACT) 7 assay in the clinical setting. Five pathologists with an interest in breast pathology (F.D., A.G., S.J., H.Z. and J.S.R.-F.) conducted a detailed histopathologic review of all cases harboring CDH1 pathogenic/likely pathogenic genetic alterations following the criteria put forward by the World Health Organization (WHO) 8. Breast cancers displaying any of the known histologic patterns of classic ILC as well as ‘variant’ ILCs, including dissociated cells arranged in dispersed single-cells, ‘linear streak’, ‘trabecular’, ‘loose alveolar’, ‘targetoid’ or admixed tubular with lobular patterns were excluded from the “NL-BCs with CDH1 bi-allelic genetic alterations” group 9. Pleomorphic ILCs and invasive mixed ductal/lobular carcinoma were also excluded. All breast cancers were graded according to the Nottingham grading system 8,10. Clinical characteristics including diagnostic imaging findings, estrogen receptor (ER), progesterone receptor (PR) and HER2 status were retrieved from the medical records.
Immunohistochemical analysis
The expression of E-cadherin, p120 and beta-catenin was assessed by immunohistochemistry in NL-BCs with CDH1 bi-allelic genetic alterations with available formalin fixed paraffin embedded (FFPE) material (E-cadherin, n=5; p120 and beta-catenin, n=4, each). Immunohistochemical analysis of E-cadherin (clone 36, Ventana, Tucson, AZ), p120 (clone 98, Ventana, Tucson, AZ), and beta-catenin (clone 18, Ventana, Tucson, AZ) were conducted in the Department of Pathology Immunohistochemistry Core at MSK. Appropriate positive and negative controls were included in each slide run. E-cadherin membranous and cytoplasmic expression was evaluated by four pathologists (F.D., A.G., S.J. and H.Z.) and classified as i) negative (i.e., complete loss of expression); ii) retained (i.e., strong, circumferential membranous expression of E-cadherin of the same intensity as observed in non-neoplastic mammary epithelium and/or external control) or iii) aberrant (i.e., discontinuous membranous, cytoplasmic and/or granular expression), as described by Da Silva et al 11 and Choi et al 12. Cytoplasmic and/or membranous expression of p120 was documented, as previously reported 4. Membranous circumferential beta-catenin expression comparable to that seen in normal epithelial cells was considered retained, weak membranous expression was considered reduced, and no expression was considered lost, as previously reported 13.
To investigate phenotypic features of neuroendocrine differentiation, we conducted the assessment of synaptophysin (clone Snp88; BioGenex, Fremont, CA) and chromogranin (clone LK2H10; Ventana, Tucson, AZ) expression by immunohistochemistry in four cases with available material (NL8, NL12, NL3 and NL1). Androgen receptor (AR) stain (clone SP107, Ventana, Tucson, AZ) was performed on available FFPE samples (n=5). Appropriate positive and negative controls were included in each slide run.
Targeted sequencing analysis
Tumor and normal sample sequencing FASTQ files from 5,842 breast cancer patients subjected to targeted sequencing using the Food and Drug Administration (FDA)-authorized MSK-IMPACT assay were retrieved 7. Data were processed utilizing benchmarked and validated state-of-the-art pipeline to detect pathogenic/likely pathogenic somatic mutations affecting CDH1. Somatic CDH1 mutations classified as pathogenic/likely pathogenic or predicted to be likely pathogenic as per OncoKB 14, and homozygous deletions of CDH1 were included in the analysis.
Single nucleotide variants (SNVs) and small insertions and deletions (indels) were detected utilizing the clinically validated pipeline15. In addition, sequencing data were reprocessed using our validated bioinformatics pipeline 16,17 to infer LOH with FACETS 18, as previously described 15,19, and to determine the cancer cell fraction (CCF) using ABSOLUTE 20. Mutations were considered “clonal” if their probability of being clonal was >50% or if the 95% confidence interval lower bound of its CCF was >90% 21. The fraction of genome altered (FGA), defined as the proportion of genome with copy number changes, and the tumor mutation burden (TMB; i.e. the number of non-synonymous mutations per Mb interrogated) were calculated as described previously 22. Cases with high TMB (>50 mutations/Mb) were excluded. Mutational signatures were inferred using SigMA 23, considering all synonymous and non-synonymous somatic mutations in cases with at least 5 single nucleotide variants (SNVs), as previously described 19. Microsatellite instability (MSI) was inferred using MSIsensor 24,25.
Comparison of NL-BCs with CDH1 bi-allelic genetic alterations with CDH1-wild type IDC-NSTs and CDH1-altered ILCs
We compared the repertoire of genetic alterations of NL-BCs with CDH1 bi-allelic genetic alterations (n=7) to that of invasive ductal carcinomas of no special type (IDC-NSTs) lacking CDH1 genetic alterations (n=21) and to CDH1-altered ILCs (n=14), from the study by Razavi et al 26 matched by menopausal status, sample type (primary vs metastatic), Nottingham grade and ER/HER2 status to the cases in our cohort, at a 3:1 and 2:1 ratio, respectively.
RESULTS
Following a retrospective query of 5,842 breast cancers previously subjected to MSK-IMPACT targeted sequencing 7, 15% (896/ 5,842) were found to harbor CDH1 LOF pathogenic/likely pathogenic alterations. Upon histologic review, only 7 out of the 5,842 (0.11%) BCs harbored CDH1 bi-allelic genetic alterations and lacked any histologic features of ILC or mixed ILC.
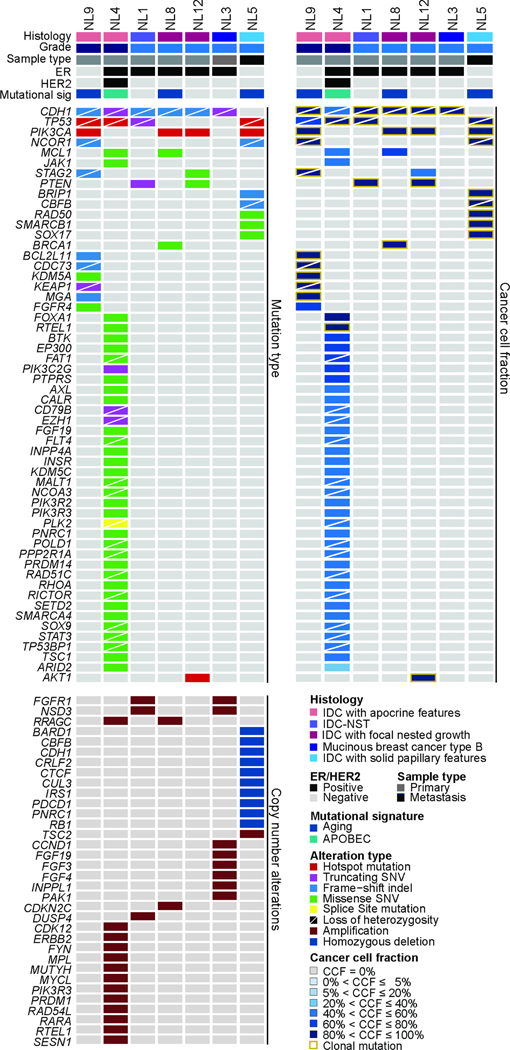
The CDH1 bi-allelic somatic genetic alterations detected in the seven NL-BCs included LOF mutations coupled with LOH of the wild-type allele (n=6) or homozygous deletions (n=1). In five of the six cases harboring CDH1 bi-allelic inactivating mutations, the somatic mutations were clonal (Fig. 1). We next sought to characterize the clinicopathological features of NL-BCs harboring CDH1 bi-allelic genetic alterations. The median age at the time of NL-BC diagnosis was 56 years (range, 39–78). All patients presented with a mass identified on imaging studies (Supplementary Table 1), with a median size of 1.1 cm (range, 0.8–2.1 cm). Three (42%; 3/7) patients presented with axillary nodal metastasis at the time of diagnosis. Six cases were primary cancer samples, whereas one case was a metastatic cancer sample, which was histologically similar to its corresponding primary tumor. The material, however, of the corresponding primary tumor was not available for genetic testing. 4/7 (57%) cases were ER-positive/HER2-negative, 2/7 (29%) cases were ER-negative/HER2-negative and 1/7 (14%) was ER-positive (ER-low [1–10%])/ HER2-positive (Tables 1 and 2). All cases were microsatellite stable (MSS) as per MSIsensor.
Figure 1. Landscape of somatic genetic alterations and clonal compositions in non-lobular breast cancers with CDH1 bi-allelic genetic alterations.
Heatmap showing non-synonymous mutations (top left), matching cancer cell fraction (top right) and copy number alterations (bottom left) in CDH1-altered non-lobular breast cancers. Cases are shown in columns and genes in rows. Histologic type, grade, sample type, estrogen receptor (ER) and HER2 status and dominant mutational signature as inferred by SigMA are shown in phenobars.
Table 1:
Clinicopathologic features of non-lobular breast cancers with CDH1 bi-allelic genetic alterations.
| Clinicopathologic feature | N (%) | |
|---|---|---|
| Median Age (years) | 56 (range 39–78) | |
| Sample Type | Primary | 6 (85%) |
| Metastatic | 1 (15%) | |
| Histologic Grade | 1 | 0 (0%) |
| 2 | 5 (71%) | |
| 3 | 2 (29%) | |
| Hormonal Receptor Status | ER+/HER2- | 4 (57%) |
| ER+/HER2+ | 1 (14%) | |
| ER-/HER2- | 2 (29%) | |
ER, estrogen receptor
Table 2.
Histologic and immunophenotypic features of non-lobular breast cancers with CDH1 bi-allelic genetic alterations
| Case ID | Sample type | Histologic type | Grade/differentiation | ER | HER2 | AR | E-cadherin | p120 | Beta-catenin |
|---|---|---|---|---|---|---|---|---|---|
| NL5 | Metastasis | IDC with solid papillary features | Moderately differentiated | Negative | Negative | NP | NP | NP | NP |
| NL3 | Primary | Mucinous breast cancer type B | 2 | Positive | Negative | Positive | Lost | Cytoplasmic | Lost |
| NL8 | Primary | IDC with focal nested growth | 2 | Positive | Negative | Positive | Lost | Cytoplasmic and membranous | Reduced |
| N12 | Primary | IDC with focal nested growth | 2 | Positive (low) | Negative | Positive | Lost | Cytoplasmic | Lost |
| NL9 | Primary | IDC with apocrine features | 3 | Negative | Negative | Positive | Aberrant (discontinuous membranous and cytoplasmic) | NP | NP |
| NL4 | Primary | IDC with apocrine features | 3 | Positive | Positive | NP | NP | NP | NP |
| NL1 | Primary | IDC-NST | 2 | Positive | Negative | Positive | Aberrant (discontinuous membranous, cytoplasmic and granular) | Cytoplasmic | Reduced |
AR, androgen receptor; ER, estrogen receptor; IDC, invasive ductal carcinoma; IDC-NST, invasive ductal carcinoma of no special type; NP, not performed.
NL-BCs harboring CDH1 bi-allelic genetic alterations comprised a heterogeneous group of cancers (Fig. 2A–2P), including a mucinous carcinoma type B (Fig. 2A–2B), IDC-NSTs with focal nested growth (n=2; Fig. 2C–2F), an IDC with solid papillary features (n=1; Fig. 2G–2J), an IDC-NST (Fig. 2K–2L) and IDCs with apocrine features (n=2; Fig. 2M–2P). We observed focal to extensive histologic features suggestive of neuroendocrine differentiation, including cohesive cells with striking eosinophilic cytoplasm showing varying degrees of granularity arranged in small solid nests or broad trabeculae which were often separated by a limited amount of vascular stroma in four cases (NL3, NL5, NL8 and NL12; Fig. 2A–2J). NL3 was a grade 2 mucinous carcinoma type B composed of large clusters of densely packed and fairly uniform cells with small, dark hyperchromatic nuclei suspended in extracellular mucin, partitioned by delicate fibrous septa in >90% of tumor area. No foci of ILC or lobular features were identified; no lobular carcinoma in situ (LCIS) was present (Fig. 2A–2B and Supplementary Fig. 1). NL8 was and NL12 corresponded to grade 2 IDCs with focal nested architecture (Fig. 2C–2F). NL5 was a moderately differentiated metastatic IDC to brain (NL5-M), with solid papillary features and distinct foamy and granular cytoplasm, which was histologically similar to its corresponding primary breast IDC (NL5-P) with solid papillary architecture, cytoplasmic granules, and nuclear palisading around delicate fibrovascular cores, with an overall tumor grade 2 (Fig. 2G–2J). Despite the focal to extensive histological neuroendocrine features found in four cases (NL3, NL5, NL8 and NL12), none of the cases with available FFPE material to be tested (n=3) were immunoreactive for synaptophysin or chromogranin. One of the NL-BCs with CDH1 bi-allelic genetic alterations (NL1) was a grade 2 IDC-NST forming small glands and nests with occasional histiocytoid features lacking major distinctive histological findings (Fig. 2K–L). Two of the NL-BC with bi-allelic CDH1 alterations (NL4 and NL9) were IDCs with overt apocrine features (Fig. 2M–P). NL4 was a grade 3 IDC with apocrine differentiation and focal insular growth pattern (Fig. 2M–N). NL9 was a grade 3 IDC with apocrine features, growing in sheets and nests (Fig. 2O–P). Androgen receptor immunohistochemical analysis was performed in cases with available FFPE material (n=5) and demonstrated nuclear expression in all 5/5 cases, one of which (NL9) showed overt apocrine features.
Figure 2. Histologic features of non-lobular breast cancers with CDH1 bi-allelic genetic alterations.
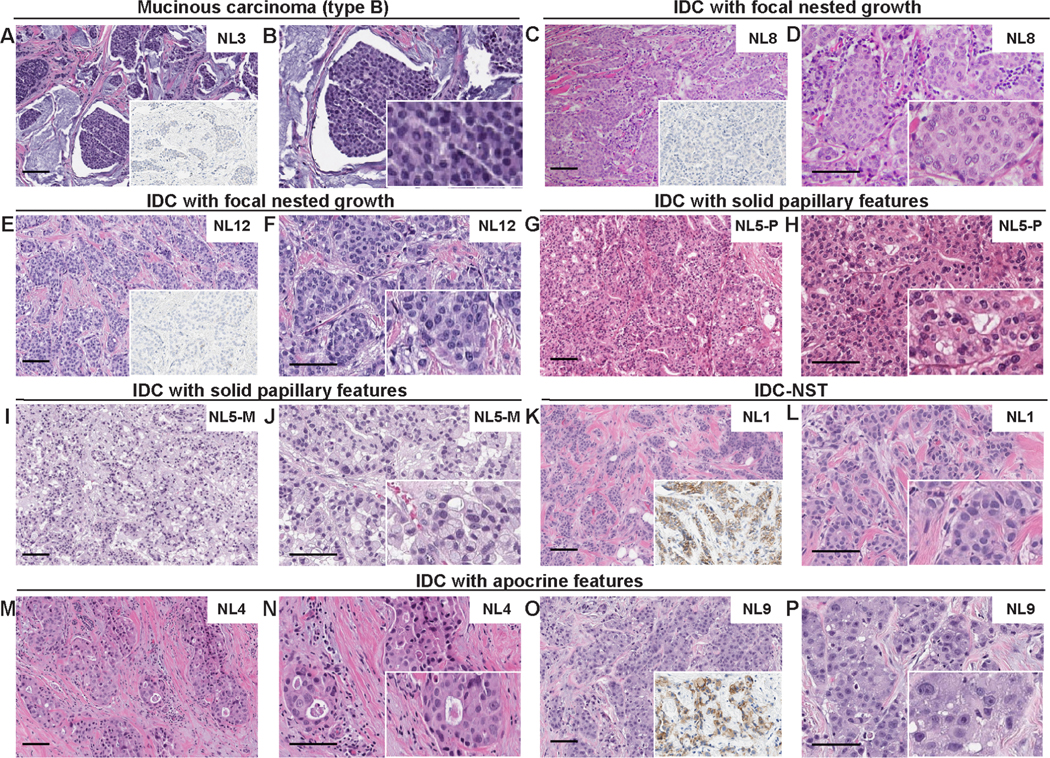
Representative hematoxylin and eosin (H&E) micrographs and E-cadherin expression micrographs (insets) of CDH1-altered non-lobular breast cancers. (A-B) Invasive mucinous breast cancer type B (NL3), (C-F) Invasive ductal carcinomas (IDC) with focal nested growth, (G-J) IDC with solid papillary features (NL5-P and NL5-M), (K-L) Invasive ductal carcinoma of no special type (IDC-NST; NL1), (M-P) IDCs with apocrine features. NL5-M, NL5, metastatic tumor; NL5-P, NL5 primary tumor. Scale bar, 100 microns.
E-Cadherin, p120 and beta-catenin immunohistochemical expression
Given that none of the cases displayed histologic features corresponding to ILCs, no initial E-cadherin, p120 or beta-catenin immunohistochemical analysis was performed by diagnostic pathologists. We conducted immunohistochemical analyses for E-cadherin on 5/7 cases and for p120 and beta-catenin in 4/7 cases with available FFPE material. 3/5 cases (60%) displayed complete absence of E-cadherin staining (E-cadherin-negative) including a mucinous carcinoma type B (NL3), and two IDCs with focal solid nested architecture (NL8 and NL12), whereas 2/5 cases (40%), including an IDC-NST (NL1) and an IDC with apocrine features (NL9), displayed aberrant E-cadherin expression (Fig. 2 and Table 2; see Materials and Methods). 3/4 cases (NL3, NL12 and NL1) showed cytoplasmic p120 expression, whereas one case (1/4; NL8) displayed cytoplasmic and membranous p120 expression. Beta-catenin membranous expression was found to be either lost (2/4; NL3 and NL12) or reduced (2/4; NL8 and NL1), as assessed by immunohistochemistry (Table 2 and Supplementary Fig. 1; see Materials and Methods).
Comparison of CDH1-altered non-lobular breast cancers to CDH1-altered lobular carcinomas (ILC) and CDH1-wild type invasive ductal carcinomas of no special type (IDC-NST)
The most frequently altered genes in NL-BCs with CDH1 bi-allelic genetic alterations were TP53 (4/7, 57%), including 3 hotspot mutations and associated to LOH of the wild-type allele in all (n=4) altered cases, and PIK3CA (4/7, 57%), targeted by mutations affecting the H1047 (n=3) or the Q546 (n=1) hotspot loci. Other recurrently altered genes included FGFR1 (2/7, 28%) affected by gene amplification and NCOR1, affected by frameshift mutations with LOH of the wild-type allele (2/7,28%; Fig. 1 and Fig. 3A)
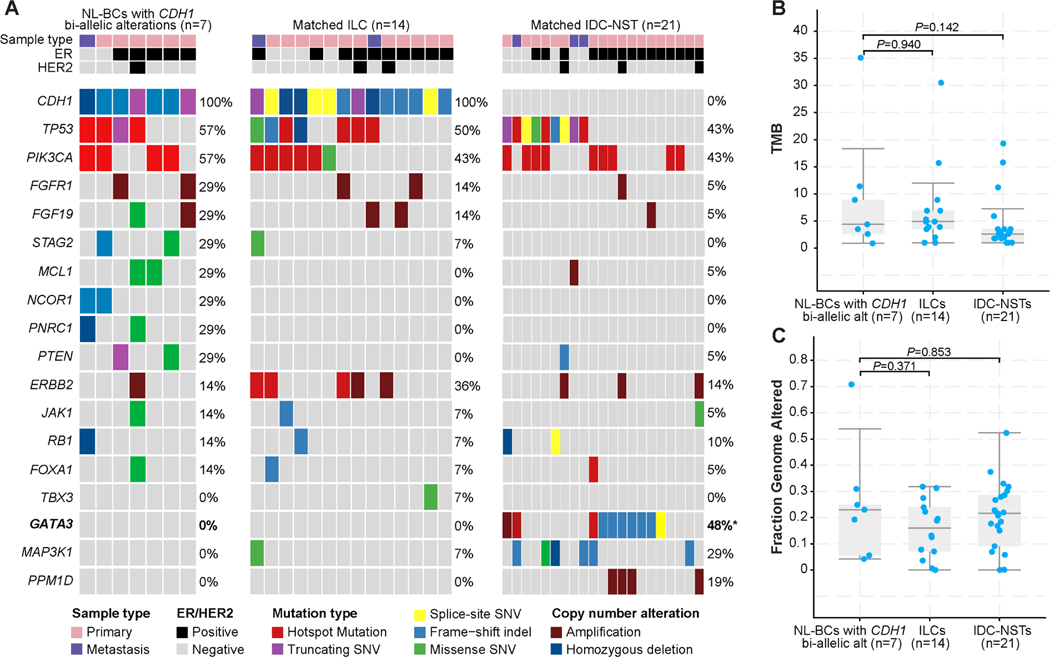
Figure 3. Repertoire of genetic alterations in NL-BCs with CDH1 bi-allelic genetic alterations in comparison with matched CDH1-altered lobular carcinomas and matched invasive ductal carcinoma of no special type.
A. Comparison of the cancer genes most frequently affected by non-synonymous somatic mutations, amplifications, or homozygous deletions in NL-BCs with CDH1 bi-allelic genetic alterations (n=7) and age, menopausal status, sample type, grade, and ER/HER2 receptor status-matched ILCs (n=14) and matched IDC-NSTs (n=21). Cases are shown in columns and genes in rows. Genetic alterations are color-coded according to the legend. ER/HER2 status and sample type are shown on phenobars (down). *P < 0.05, Fisher’s exact test, two-tailed. (B)Tumor mutation burden (TMB) and (C) fraction of genome altered (FGA) in all groups. P values (Mann-Whitney U test) are shown.
We next performed an exploratory, hypothesis-generating analysis to determine whether NL-BCs with CDH1 bi-allelic genetic alterations would differ from CDH1-altered ILCs matched for menopausal status, histologic type, grade and ER/HER2 status at a 1:2 ratio, and from CDH1 wild-type IDC-NSTs matched by clinicopathologic characteristics at a 1:3 ratio. We compared the repertoire of non-synonymous somatic genetic alterations of the primary (n=6) and metastatic (n=1) CDH1-altered NL-BC cases to that of primary (n=12) and metastatic (n=2) ILCs harboring CDH1 genetic alterations, respectively; and to that of primary (n=18) and metastatic (n=3) IDC-NSTs lacking CDH1 genetic alterations, respectively. No differences in the non-synonymous tumor mutation burden (TMB) and fraction of genome altered (FGA) were observed between NL-BCs with CDH1 bi-allelic genetic alterations and the other two groups (p>0.05, Mann-Whitney U test; Fig. 3B–3C). Notably, one of the NL-BCs (NL4) displayed a high TMB (35 mutations/Mb) and a dominant APOBEC mutational signature (Fig. 3B and Fig. 1)
The most frequently altered genes in NL-BCs with CDH1 bi-allelic genetic alterations and CDH1-altered ILCs matched by clinicopathologic characteristics were TP53 (57% and 50%, respectively) and PIK3CA (57% and 43%, respectively); whereas GATA3 (48%) and TP53 and PIK3CA (43%) were the most frequently altered genes in IDC-NSTs lacking CDH1 alterations (Fig. 3A). Notably, GATA3 was found to be mutated at a statistically significant frequency in CDH1-wild type IDC-NSTs compared to NL-BCs with CDH1 bi-allelic genetic alterations (0% vs 48%, p=0.03, Fisher’s exact test). We investigated the frequency of genetic alterations in TBX3 and FOXA1, genes previously reported to be more frequently altered in ILCs vs IDC-NSTs 6,27. We identified FOXA1 mutations in 1/7 (14%) NL-BCs with CDH1 bi-allelic genetic alterations and in 1/14 (7%) matched ILCs and 1/21(5%) matched IDC-NSTs. TBX3 mutation found in 1/14 (7%) matched CDH1-mutant ILC, while none of the NL-BCs with CDH1 bi-allelic genetic alterations (0%) or matched IDC-NSTs (0%) harbored TBX3 mutations (Fig. 3A).
Next, we inferred the dominant mutational signatures using SigMA in cases that had at least five SNVs (n=4). Most (3/4; 75%) NL-BCs with CDH1 bi-allelic genetic alterations displayed a dominant aging/clock-like mutational signature, whereas NL4 (1/4; 25%) had a dominant APOBEC mutational signature (Fig. 1). Although no statistical difference in terms of mutational signatures was observed between the three cohorts, the frequency of cases with dominant APOBEC mutational was numerically higher in CDH1-altered ILCs (5/10, 3 primary, 2 metastatic, 50%) than in NL-BCs with CDH1 bi-allelic genetic alterations (1/6, primary, 11%; p=0.17; Fisher’s exact test).
DISCUSSION
Here we demonstrate that although bi-allelic LOF genetic alterations affecting the CDH1 gene are the hallmark features of breast ILC 6,28,29, CDH1 bi-allelically altered NL-BCs do exist but are vanishingly rare (0.11% of invasive breast cancers). In contrast to ILCs that most frequently present as poorly delimited tumors posing challenges for their detection by imaging 30, all NL-BCs with CDH1 bi-allelic genetic alterations reported here were mass forming lesions identified by ultrasound and/or mammography.
The NL-BCs with CDH1 bi-allelic genetic alterations we identified constituted a histologically heterogenous group, including invasive mucinous carcinoma, IDC with solid papillary features, IDC with focal nested growth, IDC with apocrine features and IDC-NST. The genomic landscape of these tumors was similar to that of matched IDC-NSTs lacking CDH1 genetic alterations, the sole difference being a higher frequency of GATA3 mutations, which have been previously reported to be more frequent in IDC-NSTs than in ILCs 6. Also, the NL-BCs with CDH1 bi-allelic genetic alterations in our study harbored recurrent alterations in TP53, PIK3CA (57%, 4/7, each), FGFR1 and NCOR1 (28%, 2/7, each), resembling the previously described genetic alterations in luminal B ER-positive breast cancer 31.
APOBEC mutagenesis has been implicated in genetic instability, intratumor genetic heterogeneity and hypermutation phenotype in ILCs and appears to play role in progression and development of ILCs 32,33. In line with previous reports32,33, we observed that half of the of CDH1-altered ILCs (50%) had a dominant APOBEC mutational signature, whereas most NL-BCs with CDH1 bi-allelic genetic alterations displayed a dominant aging (75%) signature. These findings suggest that the mutagenesis processes underpinning NL-BCs with CDH1 bi-allelic genetic alterations differ from those of ILCs34.
We and others 27,35 have reported on genetic similarities between breast carcinomas with neuroendocrine differentiation and ILCs such as enrichment of mutations affecting the transcription factors FOXA1 and TBX3, compared to IDC-NSTs. Here, we provide a novel observation that a rare subset of breast carcinomas with histologic features indicative of neuroendocrine differentiation harbor bi-allelic CDH1 genetic alterations, the hallmark of ILC.
Previous studies showed that pure mucinous carcinoma of the breast is not a single homogeneous entity, including two main subtypes mucinous A (or paucicellular), and mucinous B (or hypercellular) tumors, with distinct histological and transcriptional features 36,37. Similarly, different histological subtypes of neuroendocrine carcinoma have been described, including the mucinous type B and solid papillary carcinomas of the breast 38, which share similar transcriptomic profiles 36. In the NL-BCs with CDH1 bi-allelic genetic alterations described in our study, histological features suggestive of neuroendocrine differentiation were present in 4/7 cases, including mucinous type B carcinomas, IDCs with focal nested growth and IDC with solid papillary features, highlighting the overlapping histological features in mucinous carcinomas and carcinomas with histologic neuroendocrine features and signifying the role of bi-allelic CDH1 alterations in at least a rare subset of these entities.
Invasive lobular carcinoma with extracellular mucin (ILCEM) is a rare ILC variant that shows the presence of lobular morphology, classic or variant, in the mucinous and non-mucinous components of a tumor, with no evidence of tubule formation, cohesive nests or micropapillae 39–41. Case NL3 of the cohort we present here was composed of cohesive nests of tumor cells floating in mucin, displayed occasional duct formation, and showed no lobular features, being classified as mucinous carcinoma of the breast. ILCEM, despite its rarity, however, should be considered in the differential diagnosis of mucinous carcinoma of the breast and of classic of ILC 39–41.
It is unknown why despite the CDH1 LOF and E-cadherin loss, p120 cytoplasmic localization and beta catenin loss/reduction, the hallmark histological findings of classic ILC are absent in NL-BCs with CDH1 bi-allelic genetic alterations. It is possible that other epithelial adhesion components or cadherins 42 might compensate the effect of CDH1 LOF in these rare tumors. Various partners, including α-catenin, N-Cadherin 42,43, and vinculin 44 play roles in cadherin-based cell to cell adhesion, and their compensatory upregulation could possibly strengthen the weakened link between E-cadherin and the underlying actin cytoskeleton 44 in this rare cohort.
Our study has several limitations, including the small sample size of the NL-BCs with CDH1 bi-allelic genetic alterations, owing to their rarity, which may limit the identification of statistically significant differences in the comparisons performed between NL-BCs with CDH1 bi-allelic genetic alterations and ILCs and IDC-NSTs. As we could not rule out the type II or β errors, the negative results need to be interpreted with caution. Despite the limitations, our findings demonstrate that NL-BCs with CDH1 bi-allelic genetic alterations, although rare, do exist, are predominantly invasive ductal carcinoma with special histologic features and a genomic landscape resembling that of luminal B ER-positive breast cancers.
Supplementary Material
ACKNOWLEDGEMENTS
FUNDING
S.C., B.W. and F.P. are funded in part by an NIH/NCI P50 CA247749 01 grant. JRSF was partially funded in part by an NIH/NCI P50 CA247749 01 grant, the Breast Cancer Research Foundation and Susan G Komen Leadership grant. BW is partially funded by Breast Cancer Research Foundation and Cycle for Survival grants. S.C. is funded in part by the Breast Cancer Research Foundation. Research reported in this publication was partly funded by a Cancer Center Support Grant of the National Institutes of Health (NIH)/National Cancer Institute (grant No P30CA008748).
Footnotes
ETHICS APPROVAL
The study was approved by the Institutional Review Board of MSK. Written informed consent was obtained as per the approved protocols.
CONFLICT OF INTEREST
J.S.R.-F. reports receiving personal/consultancy fees from Goldman Sachs, Bain Capital, REPARE Therapeutics, Saga Diagnostics and Paige.AI, membership of the scientific advisory boards of VolitionRx, REPARE Therapeutics and Paige.AI, membership of the Board of Directors of Grupo Oncoclinicas, and ad hoc membership of the scientific advisory boards of AstraZeneca, Merck, Daiichi Sankyo, Roche Tissue Diagnostics and Personalis, outside the scope of this study. B.W. reports research funding from Repare Therapeutics, outside the scope of the submitted work. F.P. reports membership of the scientific advisory board of MultiplexDx.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DATA AVAILABILITY
The datasets used and/or analyzed during this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1.Gatza ML, Carey LA. Another Breast Cancer Entity Confirmed: Genomics of Invasive Lobular Breast Cancer. J Clin Oncol. Jun 1 2016;34(16):1838–9. [DOI] [PubMed] [Google Scholar]
- 2.Gall TM, Frampton AE. Gene of the month: E-cadherin (CDH1). J Clin Pathol. Nov 2013;66(11):928–32. [DOI] [PubMed] [Google Scholar]
- 3.Droufakou S, Deshmane V, Roylance R, Hanby A, Tomlinson I, Hart IR. Multiple ways of silencing E-cadherin gene expression in lobular carcinoma of the breast. Int J Cancer. May 1 2001;92(3):404–8. [DOI] [PubMed] [Google Scholar]
- 4.Grabenstetter A, Mohanty AS, Rana S, et al. E-cadherin immunohistochemical expression in invasive lobular carcinoma of the breast: correlation with morphology and CDH1 somatic alterations. Human Pathology. 2020/08/01/ 2020;102:44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu JB, Feng CY, Deng M, et al. E-cadherin expression phenotypes associated with molecular subtypes in invasive non-lobular breast cancer: evidence from a retrospective study and meta-analysis. World J Surg Oncol. Aug 1 2017;15(1):139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciriello G, Gatza ML, Beck AH, et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell. Oct 8 2015;163(2):506–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn. May 2015;17(3):251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.WHO Classification of Tumors Editorial Board. Breast tumours. WHO Classification of Tumors. 5th Edition. 5th Edition ed. Lyon; 2019. [Google Scholar]
- 9.Martinez V, Azzopardi JG. Invasive lobular carcinoma of the breast: incidence and variants. Histopathology. Nov 1979;3(6):467–88. [DOI] [PubMed] [Google Scholar]
- 10.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. Sep 2002;41(3A):154–61. [PubMed] [Google Scholar]
- 11.Da Silva L, Parry S, Reid L, et al. Aberrant expression of E-cadherin in lobular carcinomas of the breast. Am J Surg Pathol. May 2008;32(5):773–83. [DOI] [PubMed] [Google Scholar]
- 12.Choi YJ, Pinto MM, Hao L, Riba AK. Interobserver variability and aberrant E-cadherin immunostaining of lobular neoplasia and infiltrating lobular carcinoma. Mod Pathol. Oct 2008;21(10):1224–37. [DOI] [PubMed] [Google Scholar]
- 13.Gomes DS, Porto SS, Rocha RM, Gobbi H. Usefulness and limitations of E-cadherin and beta-catenin in the classification of breast carcinomas in situ with mixed pattern. Diagn Pathol. Jul 9 2013;8:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chakravarty D, Gao J, Phillips SM, et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol. Jul 2017;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pareja F, Ptashkin RN, Brown DN, et al. Cancer-Causative Mutations Occurring in Early Embryogenesis. Cancer Discov. Apr 1 2022;12(4):949–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pareja F, Brown DN, Lee JY, et al. Whole-Exome Sequencing Analysis of the Progression from Non-Low-Grade Ductal Carcinoma In Situ to Invasive Ductal Carcinoma. Clin Cancer Res. Jul 15 2020;26(14):3682–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pareja F, Lee JY, Brown DN, et al. The Genomic Landscape of Mucinous Breast Cancer. J Natl Cancer Inst. Jul 1 2019;111(7):737–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. Sep 19 2016;44(16):e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pareja F, Ferrando L, Lee SSK, et al. The genomic landscape of metastatic histologic special types of invasive breast cancer. NPJ Breast Cancer. 2020;6:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carter SL, Cibulskis K, Helman E, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. May 2012;30(5):413–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ng CKY, Piscuoglio S, Geyer FC, et al. The Landscape of Somatic Genetic Alterations in Metaplastic Breast Carcinomas. Clin Cancer Res. Jul 15 2017;23(14):3859–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. Jun 2017;23(6):703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gulhan DC, Lee JJ, Melloni GEM, Cortes-Ciriano I, Park PJ. Detecting the mutational signature of homologous recombination deficiency in clinical samples. Nat Genet. May 2019;51(5):912–919. [DOI] [PubMed] [Google Scholar]
- 24.Niu B, Ye K, Zhang Q, et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics. Apr 1 2014;30(7):1015–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwartz CJ, da Silva EM, Marra A, et al. Morphologic and Genomic Characteristics of Breast Cancers Occurring in Individuals with Lynch Syndrome. Clin Cancer Res. Jan 15 2022;28(2):404–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Razavi P, Chang MT, Xu G, et al. The Genomic Landscape of Endocrine-Resistant Advanced Breast Cancers. Cancer Cell. Sep 10 2018;34(3):427–438 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchio C, Geyer FC, Ng CK, et al. The genetic landscape of breast carcinomas with neuroendocrine differentiation. J Pathol. Feb 2017;241(3):405–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Desmedt C, Zoppoli G, Gundem G, et al. Genomic Characterization of Primary Invasive Lobular Breast Cancer. J Clin Oncol. Jun 1 2016;34(16):1872–81. [DOI] [PubMed] [Google Scholar]
- 29.Weigelt B, Geyer FC, Natrajan R, et al. The molecular underpinning of lobular histological growth pattern: a genome-wide transcriptomic analysis of invasive lobular carcinomas and grade- and molecular subtype-matched invasive ductal carcinomas of no special type. J Pathol. Jan 2010;220(1):45–57. [DOI] [PubMed] [Google Scholar]
- 30.Silverstein MJ, Lewinsky BS, Waisman JR, et al. Infiltrating lobular carcinoma. Is it different from infiltrating duct carcinoma? Cancer. Mar 15 1994;73(6):1673–7. [DOI] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. Oct 4 2012;490(7418):61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCart Reed AE, Kalinowski L, Simpson PT, Lakhani SR. Invasive lobular carcinoma of the breast: the increasing importance of this special subtype. Breast Cancer Res. Jan 7 2021;23(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee JY, Schizas M, Geyer FC, et al. Lobular Carcinomas In Situ Display Intralesion Genetic Heterogeneity and Clonal Evolution in the Progression to Invasive Lobular Carcinoma. Clin Cancer Res. Jan 15 2019;25(2):674–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nik-Zainal S, Morganella S. Mutational Signatures in Breast Cancer: The Problem at the DNA Level. Clin Cancer Res. Jun 1 2017;23(11):2617–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ang D, Ballard M, Beadling C, et al. Novel mutations in neuroendocrine carcinoma of the breast: possible therapeutic targets. Appl Immunohistochem Mol Morphol. Feb 2015;23(2):97–103. [DOI] [PubMed] [Google Scholar]
- 36.Weigelt B, Geyer FC, Horlings HM, Kreike B, Halfwerk H, Reis-Filho JS. Mucinous and neuroendocrine breast carcinomas are transcriptionally distinct from invasive ductal carcinomas of no special type. Mod Pathol. Nov 2009;22(11):1401–14. [DOI] [PubMed] [Google Scholar]
- 37.Capella C, Eusebi V, Mann B, Azzopardi JG. Endocrine differentiation in mucoid carcinoma of the breast. Histopathology. Nov 1980;4(6):613–30. [DOI] [PubMed] [Google Scholar]
- 38.Lopez-Bonet E, Alonso-Ruano M, Barraza G, Vazquez-Martin A, Bernado L, Menendez JA. Solid neuroendocrine breast carcinomas: incidence, clinico-pathological features and immunohistochemical profiling. Oncol Rep. Dec 2008;20(6):1369–74. [PubMed] [Google Scholar]
- 39.Cserni G, Floris G, Koufopoulos N, et al. Invasive lobular carcinoma with extracellular mucin production-a novel pattern of lobular carcinomas of the breast. Clinicopathological description of eight cases. Virchows Arch. Jul 2017;471(1):3–12. [DOI] [PubMed] [Google Scholar]
- 40.Soong TR, Dillon DA, Rice-Stitt TL, et al. Invasive lobular carcinoma with extracellular mucin (ILCEM): clinicopathologic and molecular characterization of a rare entity. Mod Pathol. Oct 2022;35(10):1370–1382. [DOI] [PubMed] [Google Scholar]
- 41.Singh K, DiazGomez B, Wang Y, Ou J, Hansen K. Invasive Lobular Carcinoma With Extracellular Mucin: Not All Mucinous Mammary Carcinomas Are Ductal! Int J Surg Pathol. Feb 2019;27(1):55–58. [DOI] [PubMed] [Google Scholar]
- 42.Loh CY, Chai JY, Tang TF, et al. The E-Cadherin and N-Cadherin Switch in Epithelial-to-Mesenchymal Transition: Signaling, Therapeutic Implications, and Challenges. Cells. Sep 20 2019;8(10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaszak I, Witkowska-Pilaszewicz O, Niewiadomska Z, Dworecka-Kaszak B, Ngosa Toka F, Jurka P. Role of Cadherins in Cancer-A Review. Int J Mol Sci. Oct 15 2020;21(20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas WA, Boscher C, Chu YS, et al. alpha-Catenin and vinculin cooperate to promote high E-cadherin-based adhesion strength. J Biol Chem. Feb 15 2013;288(7):4957–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during this study are available from the corresponding author upon reasonable request.